Abstract
Langhans multinucleated giant cells (LGCs) are a specific type of multinucleated giant cell (MGC) containing a characteristic horseshoe-shaped ring of nuclei that are present within granulomas of infectious etiology. Although cytokines that trigger macrophage activation such as IFN-γ induce LGC formation, it is not clear whether cytokines that trigger macrophage differentiation contribute to LGC formation. Here, we found that IL-15, a cytokine that induces M1 macrophage differentiation, programs human peripheral blood adherent cells to form LGCs. Analysis of the IL-15 treated adherent cell transcriptome identified gene networks for “T cells”, “DNA damage and replication” as well as “interferon (IFN)-inducible genes” that correlated with IL-15 treatment and LGC-type MGC formation. Gene networks enriched for myeloid cells were anti-correlated with IL-15 treatment and LGC formation. Functional studies revealed that T cells were required for IL-15 induced LGC formation, involving direct contact with myeloid cells via CD40L-CD40 interaction as well as IFN-γ release. These data indicate that IL-15 induces LGC formation via the direct interaction of activated T cells and myeloid cells.
Introduction
Multinucleated giant cells (MGCs) are a classic hallmark of granulomas, organized collections of activated macrophages and lymphocytes. MGCs were first described by Langhans early in 1868 and were regarded as a sign of tuberculosis (Langhans, 1868). Subsequently, MGCs were found in various granulomatous conditions including infectious diseases such as leprosy (Helming and Gordon, 2007). MGCs are classified as two main types: Langhans-type giant cells (LGCs) and foreign body-type giant cells (FBGCs).
LGCs are found within granulomas of infectious and non-infectious etiology. In leprosy, caused by the intracellular bacterium M. leprae, LGCs are present in the tuberculoid but not lepromatous lesions, therefore associated with the ability of the host to restrict the pathogen (Ridley and Jopling, 1966). Morphologically, these LGCs are circular or ovoid in shape with a limited number of nuclei that are arranged in a circular or “horse-shoe” pattern. These MGCs seldom exceed 50 microns and typically contain no more than 10–20 nuclei per cell (McNally and Anderson, 2011). FBGCs result from the macrophage response to indigestible substances. They exhibit an irregularly-shaped cytoplasm, may contain hundreds of nuclei per cell and may exceed 1 mm in diameter (McNally and Anderson, 2011).
It has been widely thought that MGCs originate from fusion of macrophages, similar to the formation of osteoclasts (Chambers, 1978, Helming and Gordon, 2007). Several cytokines and molecules have been reported to trigger the induction of different types of MGCs. The Th1 cytokine IFN-γ, differentially expressed in tuberculoid leprosy lesions (Cooper et al., 1989, Yamamura et al., 1991), induce cells that are morphologically similar to LGCs (Enelow et al., 1992, Fais et al., 1994, Takashima et al., 1993, Weinberg et al., 1984, 1985). Formation of LGCs can also be induced by granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage-CSF (M-CSF), and IL-3. Inducers of FBGCs include IL-4, IL-13, DAP12, and α-tocopherol (McNally and Anderson, 1995, Mizuno et al., 2001). IL-15 is produced largely by innate immune cells including monocytes in response to IFN-γ and microorganisms (Carson et al.,1995). In particular, IFN-γ up-regulates the expression of the transcription factor interferon-regulatory factor 1 (IRF1) by activating STAT1, and IRF1 subsequently induces IL-15 production by binding the promoter of the IL15 gene (Honda et al., 2006). And IL-15 induced greater expression of the co-stimulatory molecule CD40, while both CD209+ cell populations expressed macrophage specific markers CD14, CD16 (FcγRIII), and CD64 (FcγRI) (Montoya et al., 2009). Here, we examined the role of IL-15 in LGC formation given that IL-15 is a potent inducer of macrophage differentiation (Krutzik et al., 2005, Montoya et al., 2009).
Results
IL-15 treatment of adherent monocytes induces LGCs
Given that we previously found that IL-15 differentiated adherent monocytes from PBMC into M1-like macrophages which have a similar phenotype to macrophages in T-lep lesions (Krutzik et al., 2005) where LGCs are found, we investigated whether IL-15 induced MGC formation by treating adherent monocytes in vitro. At the same time, we compared the ability of GM-CSF to induce MGC formation, given that this cytokine has also been shown to induce an M1-like macrophage (Fleetwood et al., 2007). Although GM-CSF did not induce MGC formation, IL-15 induced LGC formation as early as day 3, with increased numbers on day 6, such that on day 10 approximately 40% of the total cells were LGCs more than control and GM-CSF group (Figure 1a–c, 1m). The combination of IL-15 and GM-CSF had little additional effect on LGC formation as compared to IL-15 alone but led to aggregates of LGCs and small lymphoid appearing cells resembling granulomas (Figure 1a–c and 1m).
Figure 1.
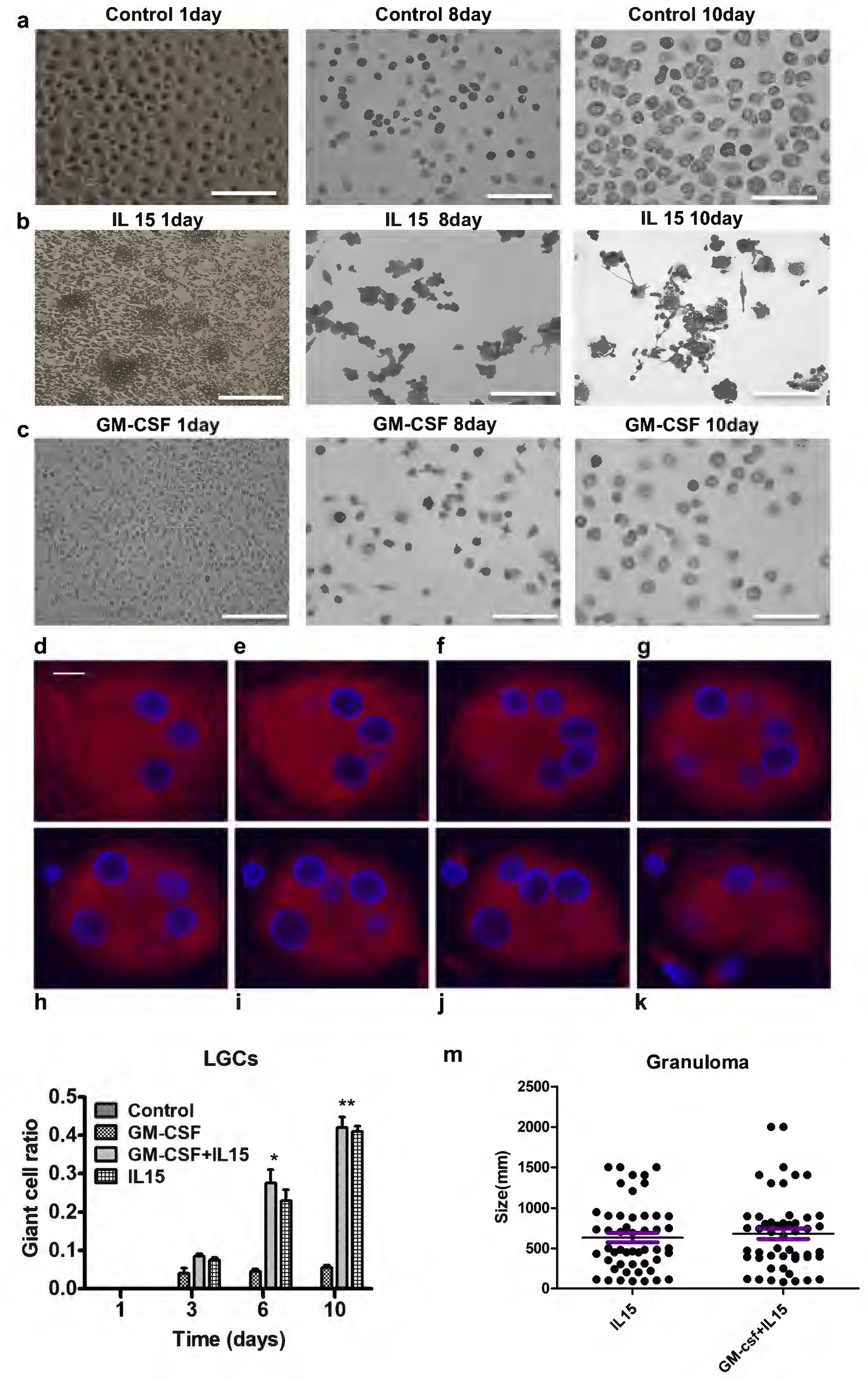
rhIL-15 treatment the adherent monocytes for LGC formation. Adherent monocytes were cultured with 10 days. (a-c) Microscopic imaging of adherent cells cultured with rhIL15 and medium control at day 1 (a,b) and Giemsa stain of rhIL-15 treated adherent monocytes at day 8 and 10 (c). (d-k) Z-stack confocal analysis revealed that nuclei of LGC induced by rhIL15 are organized in different levels inside of the cytosol. (l, m) Co-treatment of adherent monocytes with rhIL-15 or rhIL-15 plus GM-CSF detected the ration of LGCs and granulomas. Scale bars: a-c (150μm); d-k (10μm)
Microscopic imaging of the IL-15 treated adherent monocytes at days 8 and 10 indicated LGCs were ovoid and contained approximately 3–10 nuclei per cell. Visual cues suggested to us a three-dimensional structure to the LGCs (Figure 1d–k, 2i). We imaged series of longitudinally cross-sectioned planes through the LGCs from tuberculoid leprosy lesions and IL-15 treated monocytes, finding that the nuclei are located at the cell periphery as is typical of LGCs (Figure 1d–k, 2 a–h, 2i). Hence, we refer to IL-15 induced MGCs as LGCs.
Figure 2.
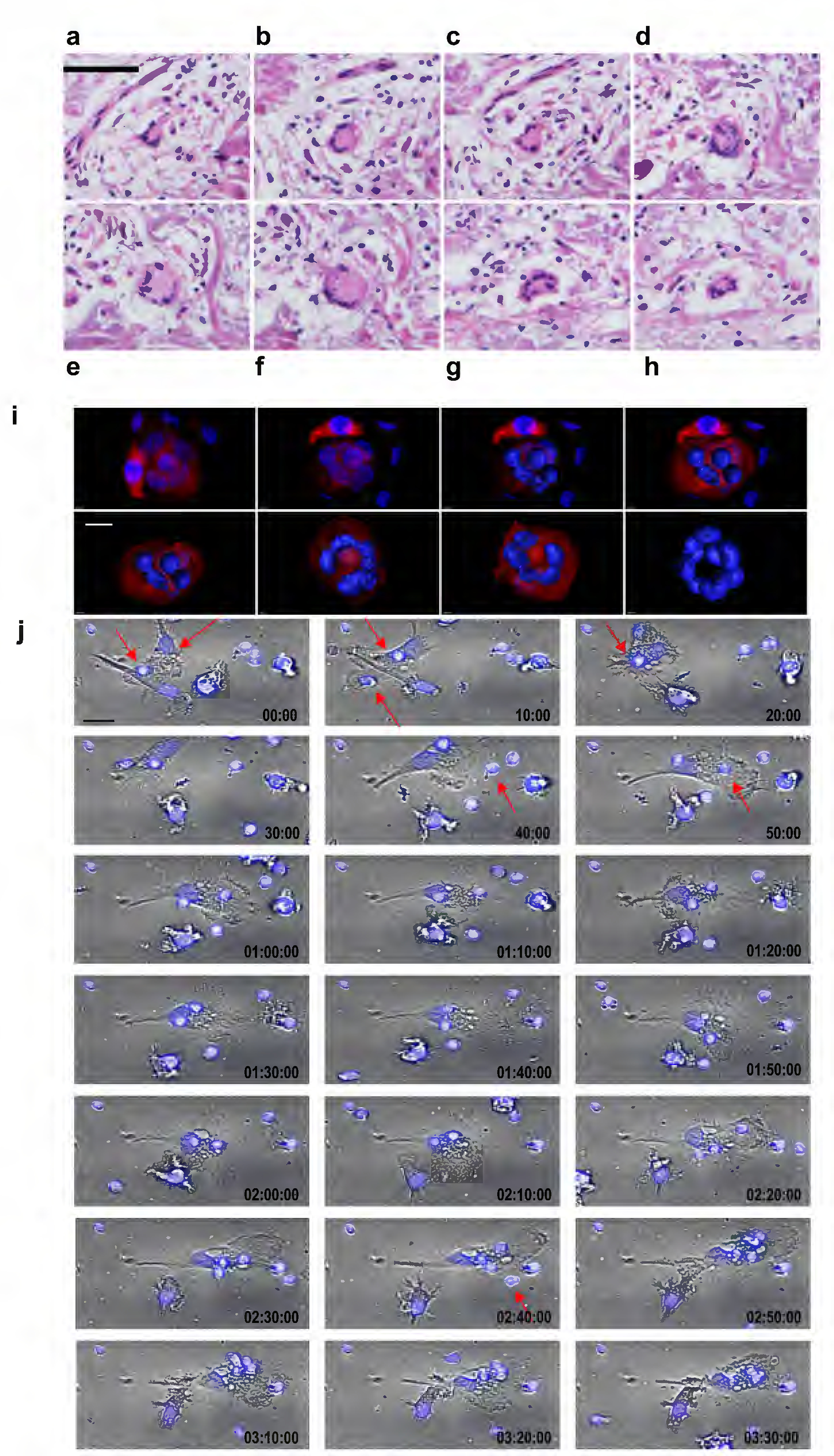
Structure and formation of the LGCs. (a-h) The three-dimensional structure of MGCs from T-lep lesions. Series of cross-sectioned planes through an LGC from a T-lep lesions confirming that the LGC indeed is a three-dimensional sphere. (i) Each image represents one step of the 3D modeling of IL-15 derived LGCs. The first image shows the original confocal image, second to seventh images show different angles during the modeling and last image shows the 3-dimensional model of the nuclei ring. 3D modeling was performed using Imaris 8.0 software. (j) The formation of LGCs by IL-15 was measured by time lapse microscopy. Cell imaging capture was assessed each 10min in total of 3h by PerkinElmer Operetta (EPA, USA). Scale bars: a-h (100μm); i (10μm); j (50μm).
Z-stack confocal analysis of LGCs revealed that nuclei are organized at the peripheral of the cytosol (Figure 1d–k, 2i). A 3D reconstruction based upon 12 z slices (Figure 2i), revealed that the spherical nuclei are in close proximity to each other, forming a torus in section studied, with proximity to the cell membrane. To evaluate the formation of LGCs by IL-15, we used time lapse microscopy. At day 5, we observed that large ovoid cells resembling monocytes/macrophages were in proximity to smaller round cells (Figure 2j and S3). Subsequently, within 10 minutes, smaller cells appeared organized around the monocytes/macrophages, after 50 minutes the nuclei of smaller cells aggregated within monocytes/macrophage. Over the next three hours, four nuclei were detected in the newly formed LGC, in addition to the original cell nucleus (Movie S1).
Gene expression analysis of LGC formation.
To investigate the mechanism of IL-15 induced LGC formation, adherent monocytes from three donors were treated with IL-15. Control cultures consisted of adherent monocytes in media alone and treatment with IL-4, known to induce FBGCs (McNally and Anderson, 1995). The percentage of LGCs was quantified at days 1, 3, 6 and 10 (Figure 1i) and gene expression profiles determined by RNA-seq. After filtering out background expression, a dataset 15,368 genes were obtained (GSE132270). Principal component analysis (PCA) of the DESeq2 normalized counts was used to first identify samples displaying similar trends in gene expression (Figure 3a) (Langfelder and Horvath, 2008, Lopez et al., 2017). PCA indicated that gene expression data formed four major groups: media (day 0), media (days 1, 3, 6), IL-4 treatment (days 1, 3, 6, 10) and IL-15 treatment (days 1, 3, 6, 10). The data for media day 10 grouped with the IL-4 group.
Figure 3.
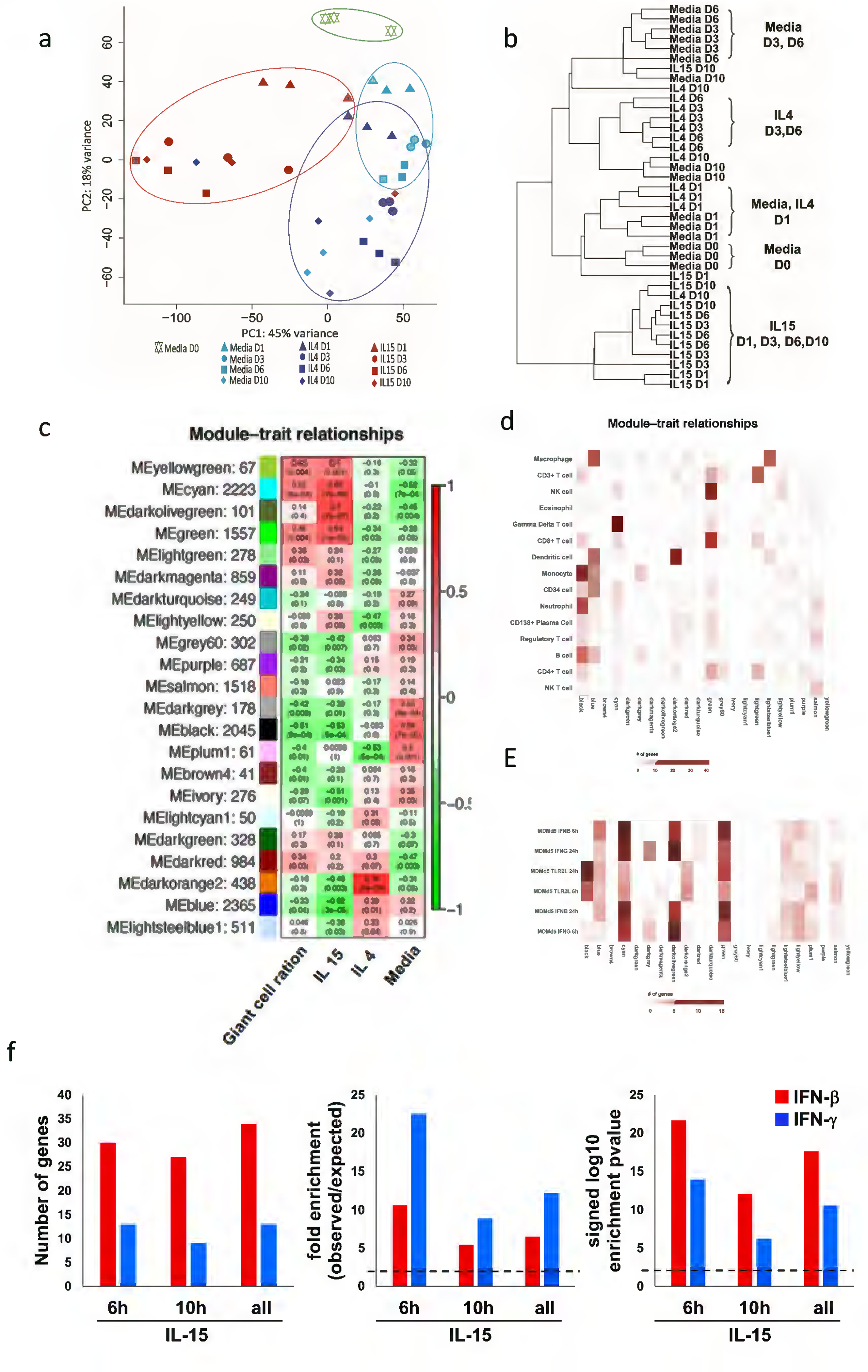
Gene expression analysis of LGC formation. (a) Principal component analysis (PCA) of the DESeq2 normalized counts was used to displaying similar trends in gene expression. (b) Hierarchical clustering for the characteristics of the relationships amongst samples. (c) Identification of LCG gene modules. WGCNA eigengene modules correlated to at least one condition (p ≤ 0.05). Red indicates positive correlation, and green indicates inverse correlation. Module eigengenes, as well as the corresponding number of genes in each module, are labeled on the y axis, and conditions are labeled on the x axis. (d) Integration of WGCNA gene modules with cell-type–specific gene signatures. For each of the modules of related genes derived from WGCNA analysis, enrichment for cell-type–specific gene signatures for cell types with immune or structural functions were calculated and displayed in a heatmap of Z scores. Cell-type names are provided in rows, and WGCNA module are provided in the column. (e) Integration of WGCNA for interferon signature. For the significant modules derived from WGCNA, enrichment for MDM IFN-γ and IFNβ specific downstream genes (2h, 6h and 24h) were calculated and displayed in a heatmap of Z scores. (f) Enrichment analysis of overlap between IFN-γ and IFN-β- specific upregulated genes identified in IL-15 treated- human monocytes time points transcripts (fold change ≥ 2 and P ≤ 0.05). Dotted lines indicate either the expected fold enrichment of one (left) or the hypergeometric enrichment P value of 0.05 (log P = 1.3, right). Hypergeometric analyses were performed to determine fold enrichment (observed/expected) and signed log enrichment P value (negative for deenriched). The Bonferroni multiple hypothesis test correction was applied for each group.
A second unsupervised analysis, hierarchical clustering, was also performed to characterize the relationships between samples (Figure 3b). Consistent with the PCA, 10 of the 12 samples for IL-15 treatment of adherent monocytes clustered together. The media samples for day 3 and 6 clustered together, as did the IL-4 samples for day 3 and 6. The media samples from day 0 and 1 clustered together but separately from the other media samples. There was not a clear clustering pattern for the day 10 samples.
To further define the potential interaction between genes associated with LGCs formation, we used weighted gene co-expression network analysis (WGCNA), an unbiased approach that defines modules of highly interconnected genes based on pairwise correlations (Langfelder and Horvath, 2008). We focused on the modules that were specifically induced by IL-15 and correlated with LGC formation. This was accomplished by first encoding the cytokines as a binary vector that was one for one cytokine at all time-points and zero for the other conditions. A vector was also created for the percentage of LGCs in the cultures at each time-point. The module expression levels were then correlated with these binary vectors to identify specific module/ligand associations. In total, 22 modules were identified, of which four were correlated with IL-15 treatment (Figure 3c), yellowgreen, cyan, darkolivegreen and green. Of these modules, three were associated with LGC formation, yellowgreen, cyan, and green. Two modules, lightgreen and darkred correlated with LGC formation but not with IL-15 treatment.
The WGCNA modules were associated with particular cell types using SAVANT (Lopez et al., 2017), which compares each module to cell type signature genes. Several modules were associated with T cell genes, consistent with the presence of T cells in the adherent monocytes. The cyan module was strongly enriched for gamma delta T cell genes (Figure 3d). The green module was linked to NK cells, CD3+ T cells and CD8+ T cells. The yellowgreen, darkolivegreen and darkred modules were not associated with a particular cell type. The lightgreen module correlated with CD3+ T cells, consistent with the functional analysis showing T cell activation. The black and blue modules, which were anti-correlated with both LGCs formation and IL-15 treatment, were enriched for monocytes and macrophages, respectively.
We analyzed whether the gene modules could be further differentiated by immunologic characteristics using Savant to detect correlations with immune perturbation signatures (Lopez et al., 2017). The most striking associations for the WGCNA modules were found for the signatures of IFN-activated monocyte-derived macrophages (Figure 3e). The cyan module, which was associated with T cells, was strongly associated with IFN treatment of MDMs, greater for IFN-β than IFN-γ. The green module, associated with NK and T cells, was more strongly associated with IFN-β vs. IFN-γ treated MDMs. The darkolivegreen module, although not associated with a specific cell type was associated with IFN-β and IFN-γ treated MDM, although the association with IFN-γ was stronger.
To examine whether IFN-induced genes were part of the IL-15 treated adherent cell gene expression profile, we overlapped the IL-15 upregulated genes at 6 h, 10 h and both 6 and 10h (FC>1.5, p<0.05) with an IFN-specific transcriptomes from the RNA-seq data of IFN-β and IFN-γ-stimulated MDMs (Figure 3f) (Teles et al., 2013). Of the total 177 upregulated genes by IL-15 at 6h of stimulation, 30 genes overlapped with the 245 genes in the IFN-β signature and 13 genes overlapped with the 50 genes in the IFN-γ signature, significantly enriched by 11-fold (p= 2.08e-22) and 23-fold (p= 1.01e-14), respectively. For the 311 upregulated genes by IL-15 at 24h; 27 genes overlapped with IFN-β and nine genes overlapped with IFN-γ, enriched by 5-fold (p= 8.03e-13) and 9-fold (p=6.24e-07); respectively. The same trend was observed when we analyzed both time points together (Figure 3f).
We performed functional analysis of the WGCNA modules using ClueGo to further characterize the gene networks (Figure 4). The green and lightgreen modules were both associated with T cell functional pathways. The green module which was associated with IL-15 treatment and LGC formation, and well as IFN-inducible genes, was linked to the “T cell receptor complex” pathway including CD8A, and the T cell activation marker CD40L. The lightgreen module was associated with LGC formation was enriched for “α/β T cell activation”, including CD28, CD3E, GATA3, RORA and RORC. The darkolivegreen module was enriched for genes in the functional pathways involving IFN signaling including “IFN-γ-stimulated genes” and “IFN-a/b signaling”. The darkred module was linked to oxidative pathways in mitochondria. The cyan module, which was associated with IL-15 treatment and LGC formation, contained IFNG and was enriched for IFN-inducible genes was associated with DNA replication and damage pathways. The yellowgreen module was associated with pathways related to cell division.
Figure 4.
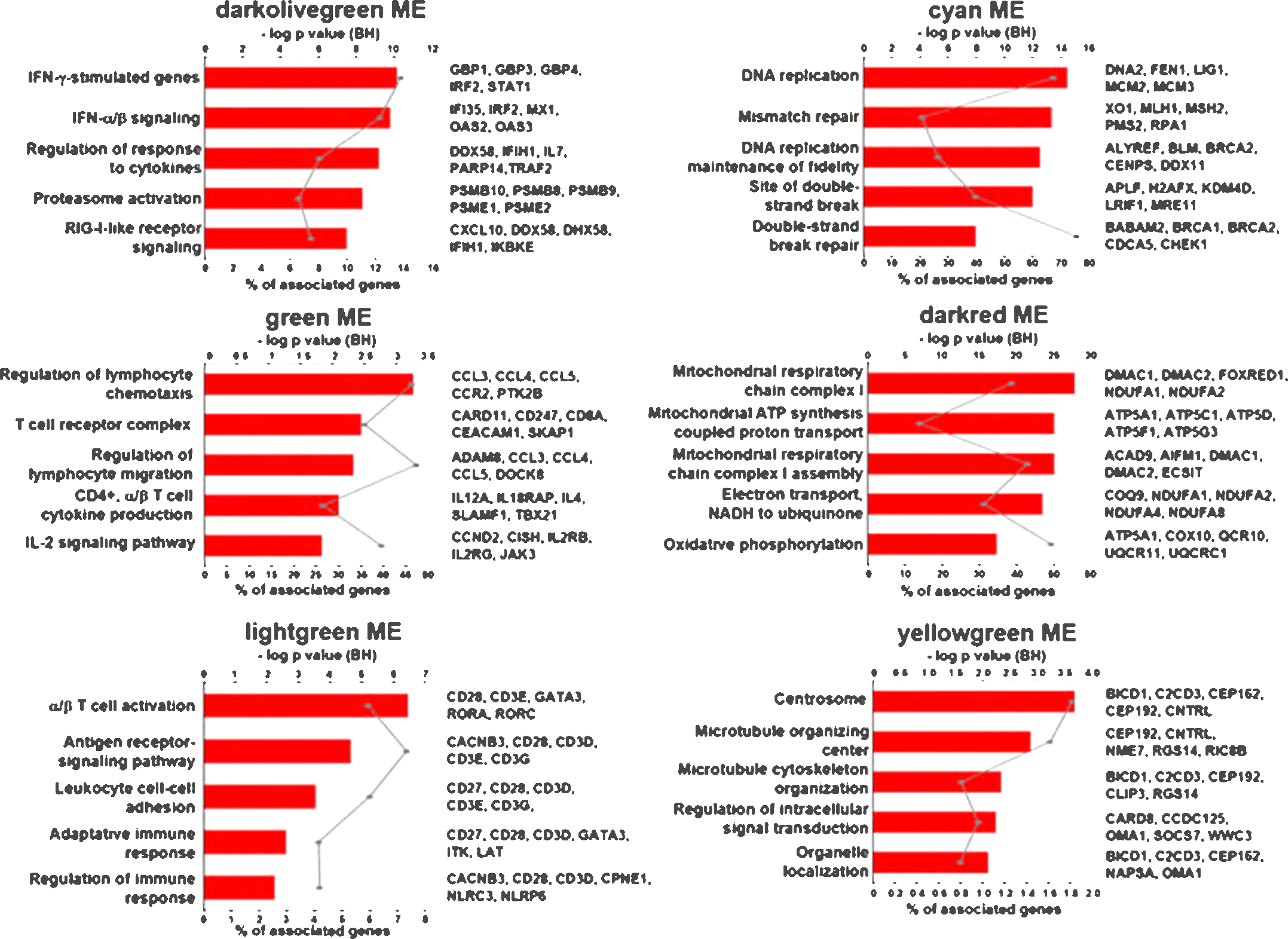
Functional analysis of the IL-15 positively and negatively correlated modules. Top 5 functional GO terms for the specific modules. Graphs show the number of associated genes, −log p value and 5 hits for each GO term. Padj was calculated with B-H multiple testing for the association of the functional term with the gene-expression data.
Role of T cells in LGC formation
Given that we had observed small lymphoid cells in close proximity to LGCs and the RNA-seq analysis demonstrating a module cyan that was associated with T cells containing genes typical of DNA replication, we determined the role of T cells in LGCs formation. Initially, we measured T cell viability in RPMI 1640 with 20% serum, both with or without cytokine supplementation. We cultured peripheral adhered monocytes which contain T cells from three healthy donors in RPMI 1640 with 20% FBS, with 50ng/mL rhIL15, in the presence or absence of 50ng/mL rhIL-2 and rhIL-7 for ten days.
CD4+ and CD8+ T cells were quantified by flow cytometry. We measured cell viability using the Zombie green fixable viability kit at day0, day3, day6 and day10. We found that the percentage of CD8+ T cells were increased in two groups of cytokine treatment: rhIL-15 treatment, as well as rhIL-15, rhIL-2 and rhIL-7 co-treatment, at both day6 and day10 (Figure S1 a–d). While in the mock group we found the percentage of CD8+ T cells decreased with time. The percentage of CD4+ T cells was maintained in the different groups and time points (Figure S1 a–d). We also found there were <6% dead cells in the mock group for both CD4+ and CD8+ T cells by the Zombie green fixable viability kit, but a higher percentage in the rhIL-15 treatment, as well as the rhIL-15, rhIL-2 and rhIL-7 co-treatment groups and time points (Figure S1 d).
Both the rhIL-15 group and the rhIL-15, rhIL-2, rhIL-7 co-treatment group showed a significant induction of IFN-γ expression at day3, day6 and day10 (Figure S2 a). There was no IFN-γ in the mock treated cells, indicating that the amount of cytokine in the serum was minimal. Similarly, IL-15, IL-2 and IL-7 were not detected in the mock treated cells (Figure 2S b–d). IL-2 and IL-7 were also not detected in the rhIL-15 treated cells. The detection of IL-15, IL-2 and IL-7 in the cultures in which these cytokines were added served as a positive control. IFN-β was not significantly increased in any group (Figure 2S e).
These data indicate that T cells survive in different groups and time points, even in mock groups, although the survival of CD8+ T cells was lower than CD4+ T cells in the mock group. The number of CD8+ T cells were increased in two groups of rhIL-15 treatment, rhIL-15, rhIL-2 and rhIL-7 co-treatment at day6 and day10 (Figure 1S a, b, c, f), so that the decreased ability of CD8+ T cells vs. CD4+ T cells to promote multinucleated cell formation as shown in Figure 5 is not related to cell death and capacity to make IFN-γ.
Figure 5.
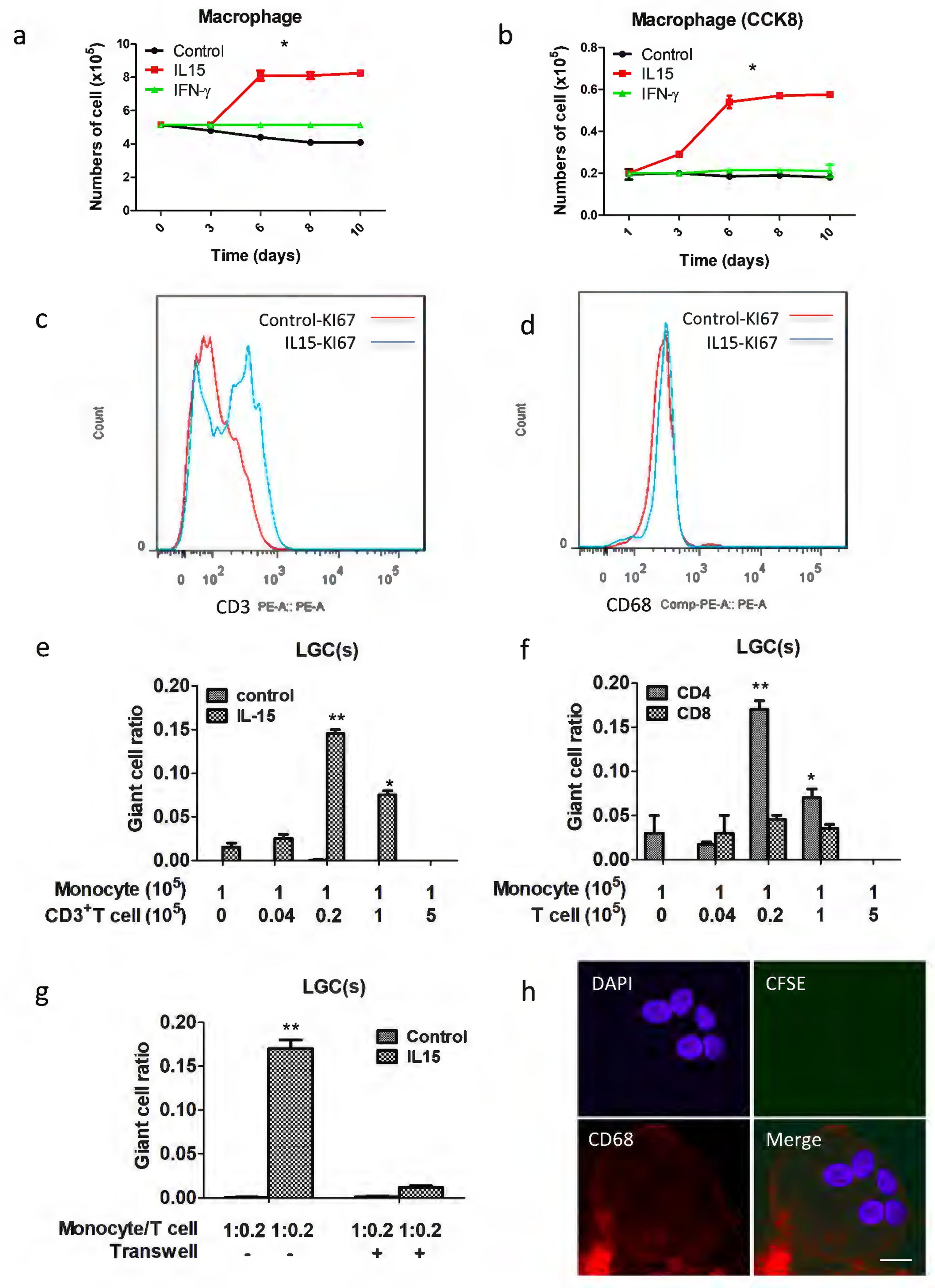
T cell proliferation analysis and human monocytes (CD14+ cells isolated from PBMCs after depletion of T cells using anti-CD14 microbeads) cultured with autologous T cells induces LGC formation. Cells were counted (a), CCK8 added (b) and stained by KI67 with CD68 (c) and CD3 (d) to detect proliferation from 1 day to 10 days. (e) the mean total number of LGCs by the different ratio between monocytes and T cells. (f) CD4+ T cells induced LGCs more efficiently than CD8+ T cells. ‘Monocyte’ refers to the number of monocytes x 105, and “T cell” refers to the number of T cells x 105. (g) Transwell analyses the relationship between monocyte with autologous T cells for LGC formation. (h) Pre-stain T cells with CFSE, and then cultured with monocytes, laser scanning confocal microscopy analysis the LGCs formation at 10 days with CD86-APC (red), CFSE (green) and DAPI (blue), scale bar: 10μm. Values represent the mean giant cell ratio calculated without distinguishing between monocytes and T cells, and error bars indicate the standard mean of the error of the indicated ratio of monocytes to T cells.
We noted that the cell count in the IL-15 treated adherent monocytes, but not the media control cultures, increased from day 3 to day 6 and then plateaued (Figure 5a). At the same time, we observed that CCK8 expression increased indicating cell proliferation (Figure 5b). We further found that IL-15 induced a significant increase in KI67 expression of CD3+ T cells (Figure 5c) but had no effect on CD68+ cells (Figure 5d), further indicating T cell proliferation.
We next purified CD3+ T cells and CD14+ monocytes and performed co-cultures in the presence of IL-15 using different ratios of T cells to monocytes (Figure 5e). Even in the absence of T cells, IL-15 did induce some monocytes to form LGCs at day 10. At a T cell: monocyte ratio of 0.04, LGC formation was low, while it was greatest at a T cell: monocyte ratio of 0.2. Notably, compared to LGCs induced from adherent monocytes and T cells, those induced from purified CD3+ T cells and CD14+ monocytes exhibited significantly lower rates of LGC formation. We believe this may be related to a decrease in cell viability caused by the positive selection process used to purify those cells.
The addition of CD4+ T cells resulted in an increase of LGC formation which was much greater than that found using CD8+ T cells (Figure 5f). In addition, using transwell cultures, we found that direct interaction between CD14+ cells and CD3+ T cells was required for LGC formation (Figure 5g). Finally, using CFSE staining, we determined that there are no proliferating monocytes present within the LGCs at day 8 (Figure 5h).
Role of IFN-γ in LGC formation
Given that LGCs are only present in the form of leprosy with predominant local expression of IFN-γ, we focused on IFN-γ in this study, and tested the role of IFNG in the IL-15 induced LGC pathway. Both anti-IL-15 and anti-IFN-γ neutralizing monoclonal antibodies blocked IL-15 induced LGC formation by approximately 90%. Anti-IFN-γ antibodies blocked the ability of GM-CSF plus IFN-γ to induce LGC formation; however anti-IL-15 neutralizing antibodies had no effect (Figures 6a and 6b). Further, given that CD40L was expressed in the green module associated with LGC formation and T cell activation, we determine the role of CD40L-CD40 interaction in LGC formation. We found that anti-CD40L antibodies also greatly reduced the ability of IL-15 to induce LGC formation (Figure 6c), indicating a role for CD40L in LGC formation.
Figure 6.
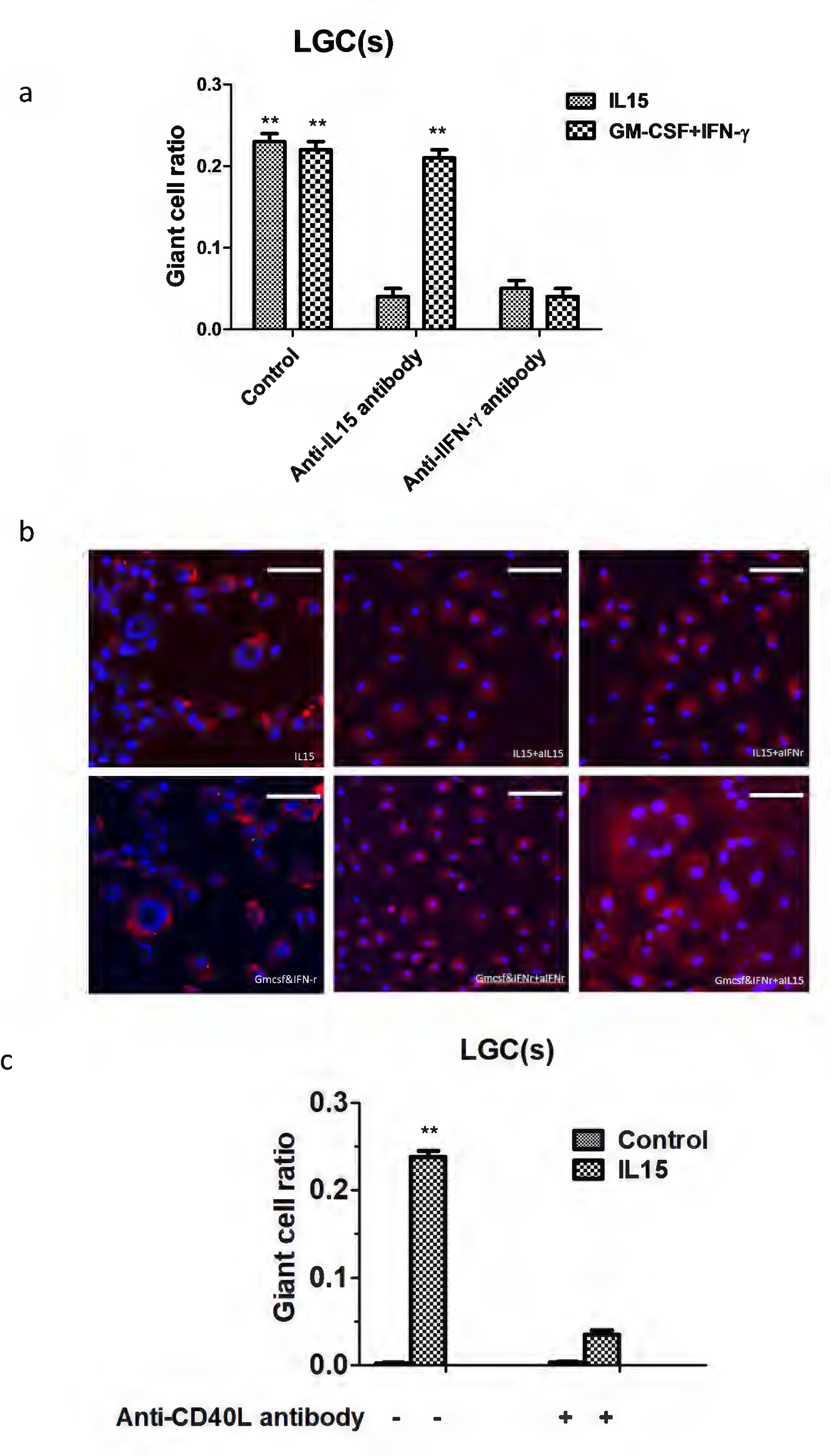
The CD40-CD40L axis and IFN-γ were required for LGC formation. Highly purified monocytes isolated from T cell-depleted PBMC were cultured with the indicated concentration of rhIL15. The indicated antibodies (10 ug/ml) were added to the culture medium (a, b), in addition to exogenous sCD40L (3 ug/ml) (c). Values represent the mean giant cell ratio, and error bars indicate the standard mean of the error (n = 3 independent cultures). Scale bar: 100μm
Phagocytic capacity of LGCs
To further characterize IL-15 induced LGCs, we studied their expression of typical macrophage and dendritic surface markers, i.e. CD68, CD86, CD209, CD163, CD11c and CD36. As expected, we found that they were all expressed on LGCs; however, CD163 was only weakly expressed (Figures S3). Finally, both IL-15 induced LGCs and macrophages exhibited phagocytic capacity for BCG (Figures S4a, b) and M. leprae (Figure S4e). The phagocytic capacity of LGCs and macrophages for both bacteria was similar (Figures S4c, d).
Discussion
The differentiation of monocytes into macrophages at the site of microbial infection is a critical part of the host defense response. Here we examined whether the differentiation of monocytes into M1-like macrophages by IL-15 (Krutzik et al., 2005, Montoya et al., 2009) resulted in the formation of LGCs. We found that the treatment of human peripheral blood adherent cells with IL-15 resulted in the formation of LGCs. Analysis of the IL-15 induced transcriptome in of adherent cells, which contains ~20% lymphocytes, identified gene networks for T cells, DNA damage and replication as well as interferon (IFN)-inducible genes. The ability of IL-15 to induce LGC formation required the presence of T cells in direct contact with myeloid cells and was dependent on CD40L-CD40 interaction as well as IFN-γ release. The ability of IFN-γ and CD40L-CD40 interaction to contribute to the mechanism of IL-15 induced LGC formation is consistent with the ability of IFN-γ alone to induce monocytes to form LGC (Weinberg et al., 1984) and the role of IFN-γ and CD40L-CD40 in ConA-induced LGC formation in cultures of monocytes and T cells (Sakai et al., 2012). Our transcriptome data indicate a role for lymphocytes including CD4+, CD8+ and γδ T cells as well as NK cells in LGC formation. We clearly demonstrate a role for T cells in IL-15 induced LC formation, as IL-15 induced LGC formation was lower in CD14 selected monocytes than adherent cells, but the response was augmented by the addition of CD3+, CD4+ and CD8+ T cells. In addition, depletion of T cells prior to enrichment for monocytes reduced LGC formation. IL-15 also induces IFN-γ production by γδ T cells and NK cells, and these cells express CD40L such that they are capable of contributing to LGC formation in a similar manner as αβ T cells. While further experiments are needed to clarify the functional role of γδ T cells and NK cells in LGC formation. Our data indicate that IL-15 induction of LGC formation involves the direct interaction of activated T cells and myeloid cells.
We studied the role of LGC formation in IL-15 induced cultures not only because IL-15 induces monocytes to differentiate into M1-like macrophages but also IL-15 expression is greater in the self-limiting vs. the progressive forms of leprosy (Jullien et al., 1997). IL-15 is induced via activation of TLR2/1 on monocytes by mycobacterial lipopeptides (Krutzik et al., 2008) and this pathway could possibly contribute to LGC formation at the site of disease. Mycobacterial lipopeptides as well as the cell wall muramyl dipeptide (MDP) have been shown to induce LGC formation (Herrtwich et al., 2016). In addition, the role of mycobacterial lipids to induce LGC formation has been examined. The M. tuberculosis glycolipid lipomannan also induces the formation of LGCs (Yamagami et al., 2001), involving a process that is mediated by TLR2 (Puissegur et al., 2007).
The mechanism of LGC formation is thought to involve both cell fusion and abnormal cell division. Lipomannan induced LGC formation is dependent on the β1 integrin /ADAM9 cell fusion machinery (Puissegur et al., 2007), but few papers provide experimental evidence for cell fusion. Herrtwich et al investigated fusion using monocytes differing in CD45 isoforms (Cui et al., 2007). RANKL-stimulated macrophage precursors to undergo cell fusion (Cui et al., 2007); however, chronic exposure to bacterial lipopeptides activate LGC formation via TLR2 but independent of cell fusion (Herrtwich et al., 2016). Instead, bacterial lipoprotein-induction of LGCs involved DNA replication and damage, leading to mitotic defects and faulty cell division (Herrtwich et al., 2016). The authors stated “scRNA-seq at the same time point (day 6) revealed upregulated expression of the mitotic regulators Ccnb1, Ccnd1 and Cdk1 as well as increased expression of the DNA replication licensing factor Mcm6 and the DNA synthesis promoting genes Rrm1 and Rrm2 in F>4c macrophages”. However, in IL-15 treated adherent cells, we found these genes which regulate mitosis and DNA synthesis in the cyan module which was linked to T cells and IFN-γ downstream genes. It remains to be determined whether IL-15 induced LGC formation involves cell fusion and/or dysregulation of cell replication.
Our data indicating the IL-15-derived LGCs were able to phagocytose M. leprae and BCG, similarly to IL-15 differentiated macrophages. The functional consequence of the phagocytosis of mycobacteria by LGC is not clear, and has been difficult to study in the absence of pure populations of these cells. Instead the function of LGC has been inferred by clinical association. As mentioned, in leprosy LGCs are found in self-limiting tuberculoid leprosy but not disseminated lepromatous lesions (Ridley and Jopling, 1966). Tuberculosis-associated giant cells have been associated with restriction of cell-to-cell spread of mycobacteria, which is beneficial to the host (Brodbeck and Anderson, 2009, McNally and Anderson, 2011). LGCs may also be detrimental because their secretion of metalloproteinase or other molecules may promote tissue destruction (Belton et al, 2016). Another possibility proposed was that multinucleation sequesters a non-phagocytosible foreign body to protect host tissues from the adverse consequences of an on-going chronic inflammatory response (Chambers, 1978). Nevertheless, IL-15, in addition to inducing M1-like macrophage differentiation and LGC formation induces an antimicrobial pathway is part of the innate immune response in humans (Krutzik et al., 2008, Montoya et al., 2014). Finally, IL-15 induces the expansion of CD8+ cytolytic T lymphocytes that express granzyme B, perforin and granulysin, so called ‘tri-cytotoxic CTL” that are antimicrobial in function (Balin et al., 2018). Together, IL-15 is a potent driver of both innate and adaptive human immune responses in mycobacterial infection. However, at present there is still a lack of an ideal in vivo LGC model, to investigate the relationship of IL-15, T cells, IFN-γ, and LGC.
Methods
Study approval
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the local Institutional Review Boards. All donors provided written informed consent for the collection of peripheral blood and skin specimens for subsequent scientific analysis. All donors were recruited with approval from the Institutional Review Board of University of California, Los Angeles and Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Nanjing, Jiangsu, China (No. 2016-KY-023).
Reagents and antibodies
Blocking antibodies against human CD40L, IFN-g, IL-15 and rhIL-15 and rhIFN-g were purchased from R&D systems (Minneapolis, MN, USA). DAPI, CFSE, and CellMask deep red plasma membrane stain purchased from Thermo Fisher Scientific (USA). All fluorescent antibodies and isotype control IgG were purchased from BD Biosciences (Franklin Lakes, NJ, USA).
Differentiation of LGCs from peripheral adhered monocytes
Blood was collected from healthy volunteer donors after informed consent was obtained in accordance with the guidelines of the Institute of Dermatology, CAMS. PBMCs were isolated from whole blood using the Lymphoprep system (Axis-Shield PoC, Oslo, Norway). The adhered monocytes were re-suspended at a density of 1×106 cells per well in 24-well plates in RPMI-1640 medium containing 2% fetal bovine serum (FBS), 2 mM L-glutamine, 50 uM b-mercaptoethanol, 100 U ml −1 penicillin and 100 U ml−1 streptomycin. After one hour, cells were washed three times and cultured in the RPMI-1640 medium containing 20% fetal bovine serum (FBS) in the presence of 50ng/mL rhIL15. and then supplemented with medium every 2 days. Nuclei were detected by Giemsa stain.
We measured cell proliferation by flow cytometry using CCK8 and Ki-67. One T-lep tissue sample was fixed in 10% paraformaldehyde, decalcified with Plank–Rychlo’s solution (0.5 M aluminum chloride containing 8.5% hydrochloric acid and 5.4% formic acid), embedded in paraffin, and cut into 5 μm sections. The sections were stained with hematoxylin and eosin (HE) for histopathological examination. Microscopic fields which contained LGCs were examined
The cultured cells were stained by 1ug/mL DAPI and 1ug/mL Cellmask red plasma membrane and examined under an Olympus IX81 microscope with magnification 5um (Olympus, Tokyo, Japan), and the ration of nuclei and LGCs in the representative area was counted. The Giant cell ratio was calculated according to the following formula: Giant cell ratio = (number of LGCs) / (total number of cell counted). More than 300 cells were counted for each experiment.
LGCs from peripheral CD14+ monocytes and T cells
CD14+ cells were isolated from PBMCs after anti-CD3 microbead depletion of T cells using anti-CD14 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). The purity of the isolated cells was demonstrated as >95% by flow cytometry using a FACS Verse system (BD Biosciences). The isolated CD14+ monocytes were re-suspended at a density of 200 000 cells with different ratios of T cells in 24-well plates. Transwell was used to separate CD14+ monocytes and CD3+ T cells, and then detect LGC formation.
RNA sequencing and analysis
RNA was harvested from cell cultures and isolated with RNeasy micro kit (Qiagen). RNA was quantified by Nanodrop and quality assessed by Agilent 2100 Bioanalyzer. Libraries were created from high quality RNA using the TruSeq RNA Library Prep Kit v2 (Illumina). Libraries were quantified by Qubit and sequenced in duplicate on a HiSeq 2500 Sequencing System (Illumina).
Sequenced reads were aligned to the human reference genome as described (Montoya et al., 2014) with raw counts were calculated with HTseq using the hg19 Ensemble annotation. Normalization and differential expression analyses were performed using the DESeq2 package for R. FDR was controlled by applying the Benjamin-Hochberg correction to P-values. Differentially expressed genes were identified using the cutoffs FC > 2 vs Media and adjusted P adjust-value < 0.05. Hierarchical clustering was performed with “hclust”, and principle component analysis via “prcomp” in R (version 3.2.4).
Gene expression profiles were obtained and analyzed for modules of highly interconnected genes using weighted gene correlation network analysis (WGCNA, “wgcna” package in R) as described by (Montoya et al., 2014). P values for overlap of modules with cell-type–specific signatures were calculated using the hypergeometric distribution and were corrected using a Bonferroni adjustment (n = 30). Cell-specific enrichment was calculated using SaVanT (Signature Visualization Tool) (Lopez et al., 2017). Interferon induced genes were identified using the gene expression profile data of IFN-treated MDMs (GSE82227 and GSE125352) as previously described (Teles et al., 2013).
Functional, network and transcriptional regulator analysis were performed using ClueGO plug in (Cytoscape) (Bindea et al., 2009). The significance values for the canonical pathways is calculated by B-H Multiple testing p value. Functionally similar GO terms were grouped into simplified representative terms.
3D modeling
Monocytes were cultured for 10 days in the presence of IL-15 (50ng/ml) and then confocal microscopy was performed. Z slices images of all volume of the cell were scanned and a z stack total volume image of the cell were used to 3D modeling animation using Imaris 8.0 (Bitplane). For the formation of LGCs by IL-15, we started at day 5, stained with DAPI and then used time lapse microscopy. Cell imaging capture was assessed each 10min in total of 3h by PerkinElmer Operetta (EPA, USA).
BCG and M. leprae infection of IL-15 induced LGCs
BCG were infected with pMV261-GFP vector. Live bacilli were grown in Middlebrook 7H9 medium with OADC at 37°C. M. leprae were labelled using the PKH26 green Fluorescent cell linker kit (Sigma, USA). Monocytes treated with rhIL-15 for 10 days were infected with M .leprae or BCG-GFP (10:1) for 6h, after that removed the supernatant and washed one time, then fresh medium was added and incubated overnight at 37°C in humidified atmosphere of 5% CO2. After the incubation period, cells were fixed with 4% paraformaldehyde in the dark for 10min, followed by two washing steps with PBS. The cell membranes were stained using CellMask™ Deep Red Plasma membrane Stain (ThermoFisher, USA). The slides were visualized using a confocal microscope (Olympus, Japan).
Data availability
Datasets related to this article can be found at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132270, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82227; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125352, These datasets are hosted at Gene Expression Omnibus (GEO)
Supplementary Material
Acknowledgements
This work has been supported by Milstein Medical Asian American Partnership Foundation, the CAMS Initiative for Innovative Medicine (grant 2016-I2M-1-005, 2017-I2M-B&R-14), and the National Natural Science Foundation of China (grant 81371751,81972950).
Footnotes
Conflict of interest
The authors have declared that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balin SJ, Pellegrini M, Klechevsky E, Won ST, Weiss DI, Choi AW, et al. Human antimicrobial cytotoxic T lymphocytes, defined by NK receptors and antimicrobial proteins, kill intracellular bacteria. Sci Immunol 2018;3(26). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25(8):1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck WG, Anderson JM. Giant cell formation and function. Curr Opin Hematol 2009;16(1):53–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ. Multinucleate giant cells. J Pathol 1978;126(3):125–48. [DOI] [PubMed] [Google Scholar]
- Cooper CL, Mueller C, Sinchaisri TA, Pirmez C, Chan J, Kaplan G, et al. Analysis of naturally occurring delayed-type hypersensitivity reactions in leprosy by in situ hybridization. J Exp Med 1989;169(5):1565–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Cuartas E, Ke J, Zhang Q, Einarsson HB, Sedgwick JD, et al. CD200 and its receptor, CD200R, modulate bone mass via the differentiation of osteoclasts. Proc Natl Acad Sci U S A 2007;104(36):14436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enelow RI, Sullivan GW, Carper HT, Mandell GL. Induction of multinucleated giant cell formation from in vitro culture of human monocytes with interleukin-3 and interferon-gamma: comparison with other stimulating factors. Am J Respir Cell Mol Biol 1992;6(1):57–62. [DOI] [PubMed] [Google Scholar]
- Fais S, Burgio VL, Silvestri M, Capobianchi MR, Pacchiarotti A, Pallone F. Multinucleated giant cells generation induced by interferon-gamma. Changes in the expression and distribution of the intercellular adhesion molecule-1 during macrophages fusion and multinucleated giant cell formation. Lab Invest 1994;71(5):737–44. [PubMed] [Google Scholar]
- Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol 2007;178(8):5245–52. [DOI] [PubMed] [Google Scholar]
- Helming L, Gordon S. The molecular basis of macrophage fusion. Immunobiology 2007;212(9–10):785–93. [DOI] [PubMed] [Google Scholar]
- Herrtwich L, Nanda I, Evangelou K, Nikolova T, Horn V, Sagar, et al. DNA Damage Signaling Instructs Polyploid Macrophage Fate in Granulomas. Cell 2016;167(5):1264–80.e18. [DOI] [PubMed] [Google Scholar]
- Jullien D, Sieling PA, Uyemura K, Mar ND, Rea TH, Modlin RL. IL-15, an immunomodulator of T cell responses in intracellular infection. J Immunol 1997;158(2):800–6. [PubMed] [Google Scholar]
- Krutzik SR, Hewison M, Liu PT, Robles JA, Stenger S, Adams JS, et al. IL-15 links TLR2/1-induced macrophage differentiation to the vitamin D-dependent antimicrobial pathway. J Immunol 2008;181(10):7115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik SR, Tan B, Li H, Ochoa MT, Liu PT, Sharfstein SE, et al. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat Med 2005;11(6):653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans T Ueber Riesenzellen mit wandständigen Kernen in Tuberkeln und die fibröse Form des Tuberkels. Archiv für pathologische Anatomie und Physiologie und für klinische Medicin 1868;42(3):382–404. [Google Scholar]
- Lopez D, Montoya D, Ambrose M, Lam L, Briscoe L, Adams C, et al. SaVanT: a web-based tool for the sample-level visualization of molecular signatures in gene expression profiles. BMC Genomics 2017;18(1):824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally AK, Anderson JM. Interleukin-4 induces foreign body giant cells from human monocytes/macrophages. Differential lymphokine regulation of macrophage fusion leads to morphological variants of multinucleated giant cells. Am J Pathol 1995;147(5):1487–99. [PMC free article] [PubMed] [Google Scholar]
- McNally AK, Anderson JM. Macrophage fusion and multinucleated giant cells of inflammation. Adv Exp Med Biol 2011;713:97–111. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Okamoto H, Horio T. Muramyl dipeptide and mononuclear cell supernatant induce Langhans-type cells from human monocytes. J Leukoc Biol 2001;70(3):386–94. [PubMed] [Google Scholar]
- Montoya D, Cruz D, Teles RM, Lee DJ, Ochoa MT, Krutzik SR, et al. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe 2009;6(4):343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya D, Inkeles MS, Liu PT, Realegeno S, Teles RM, Vaidya P, et al. IL-32 is a molecular marker of a host defense network in human tuberculosis. Sci Transl Med 2014;6(250):250ra114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissegur MP, Lay G, Gilleron M, Botella L, Nigou J, Marrakchi H, et al. Mycobacterial lipomannan induces granuloma macrophage fusion via a TLR2-dependent, ADAM9- and beta1 integrin-mediated pathway. J Immunol 2007;178(5):3161–9. [DOI] [PubMed] [Google Scholar]
- Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr 1966;34:255–73. [PubMed] [Google Scholar]
- Sakai H, Okafuji I, Nishikomori R, Abe J, Izawa K, Kambe N, et al. The CD40-CD40L axis and IFN-gamma play critical roles in Langhans giant cell formation. Int Immunol 2012;24(1):5–15. [DOI] [PubMed] [Google Scholar]
- Takashima T, Ohnishi K, Tsuyuguchi I, Kishimoto S. Differential regulation of formation of multinucleated giant cells from concanavalin A-stimulated human blood monocytes by IFN-gamma and IL-4. J Immunol 1993;150(7):3002–10. [PubMed] [Google Scholar]
- Teles RM, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 2013;339(6126):1448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JB, Hobbs MM, Misukonis MA. Recombinant human gamma-interferon induces human monocyte polykaryon formation. Proc Natl Acad Sci U S A 1984;81(14):4554–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JB, Hobbs MM, Misukonis MA. Phenotypic characterization of gamma interferon-induced human monocyte polykaryons. Blood 1985;66(6):1241–6. [PubMed] [Google Scholar]
- Yamagami H, Matsumoto T, Fujiwara N, Arakawa T, Kaneda K, Yano I, et al. Trehalose 6,6’-dimycolate (cord factor) of Mycobacterium tuberculosis induces foreign-body- and hypersensitivity-type granulomas in mice. Infect Immun 2001;69(2):810–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura M, Uyemura K, Deans RJ, Weinberg K, Rea TH, Bloom BR, et al. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science 1991;254(5029):277–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets related to this article can be found at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132270, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82227; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125352, These datasets are hosted at Gene Expression Omnibus (GEO)


