Abstract
Although replication repair deficiency, either by mismatch repair deficiency (MMRD) and/or loss of DNA polymerase proofreading, can cause hypermutation in cancer, microsatellite instability (MSI) is considered a hallmark of MMRD alone. By genome-wide analysis of tumors with germline and somatic deficiencies in replication repair, we reveal a novel association between loss of polymerase proofreading and MSI, especially when both components are lost. Analysis of indels in microsatellites (MS-indels) identified five distinct signatures (MS-sigs). MMRD MS-sigs are dominated by multi-base losses, while mutant-polymerase MS-sigs contain primarily single-base gains. MS-deletions in MMRD tumors depend on the original size of the microsatellite and converge to a preferred length, providing mechanistic insight. Finally, we demonstrate that MS-sigs can be a powerful clinical tool for managing individuals with germline MMRD and replication repair deficient cancers, as they can detect the replication repair deficiency in normal cells and predict their response to immunotherapy.
Keywords: Replication repair deficiency, Constitutional Mismatch Repair Deficiency, Microsatellite Instability, Mutational signatures, DNA Polymerase, Immunotherapy response prediction
Introduction
The two major mechanisms that safeguard the fidelity of genome replication are the DNA mismatch repair machinery and DNA polymerase proofreading (ε or δ, encoded by POLE and POLD1)(1–4). Deficiencies of mismatch repair (MMRD) or polymerase proofreading can occur independently or simultaneously, leading to combined DNA replication repair deficiency (RRD). Both mechanisms maintain genome stability by correcting misincorporated non-complementary nucleotides. Thus, when either or both pathways are inactivated, there is an increased rate of replicative mutagenesis that leads to a wide array of hypermutant cancers(1,2,4). The MMR machinery repairs also insertions and deletions in stretches of DNA with small repeated motifs (called microsatellites), which are created as a result of DNA polymerase slippage(4,5). Nevertheless, the impact of the DNA polymerase on the accumulation of these microsatellite insertions and deletions (MS-indels) in human cancer is currently not well-described(6–9).
The increased number of MS-indels in MMRD tumors is marked by microsatellite instability (MSI). MSI is clinically used for tumor stratification, referral to germline testing for cancer predisposition, and has recently been approved as a biomarker for immune checkpoint inhibitors (ICI)(10–13). MSI is commonly detected by comparing microsatellite lengths of a limited panel of microsatellites (5 loci) between a patient’s tumor and germline DNA. The Cancer Genome Atlas (TCGA) used this approach to stratify tumors as microsatellite unstable (MSI-H) or microsatellite stable (MSS) for colorectal, gastric, and endometrial tumors. However, this stratification approach has recently been challenged as being limited in scope, due to the misclassification of some MMRD cancers(6,14). Importantly, using this assay, polymerase mutant cancers do not exhibit high MSI(2,15,16) and therefore, in humans, the role of polymerase mutations in the accumulation of MS-indels in cancer is currently considered largely unknown.
The most common cause of MMRD in human cancers is somatic hypermethylation of the MLH1 promoter, while others are due to loss of heterozygosity from the germline allele in any one of the MMR genes, MSH2, MSH6, MLH1 and PMS2(17),(18). Insight regarding the pathogenesis and clinical behavior of RRD can be gained by studying human syndromes with germline mutations in these genes. Lynch syndrome is caused by germline heterozygous inactivating mutations in one of the MMR genes, and is followed by somatic loss of the remaining allele, leading to MMR deficiency. Since it only requires loss of one allele, Lynch syndrome is associated with earlier cancer onset than somatic MMRD(19). However, although less common, the most aggressive form of MMRD stems from germline biallelic inactivating mutations in one of the MMR genes. This condition, termed constitutional mismatch repair deficiency (CMMRD), is one of the most aggressive cancer syndromes in humans, where virtually all individuals will be affected by a variety of cancers during childhood, and all share hypermutation and specific mutational signatures(1,2,4,20). Similarly, heterogenous germline polymerase mutations in the exonuclease (proofreading) domain of POLE or POLD1(1) result in a cancer syndrome termed polymerase proofreading deficiency (PPD), which is less well-described but also leads to hypermutant cancers(1,2,7,20–22). Importantly, MS-indels in either of these cancer predisposition syndromes have not been previously investigated.
In patients with CMMRD, somatic mutations in the polymerase genes result in combined dysfunction of the replication repair machinery, leading to a dramatic accumulation of single nucleotide variations (SNVs)(1,2,7,20–22). Analysis of SNV-based mutational signatures and their timing also enabled the prediction of early germline events and late treatment-related processes in each cancer that correlate with the type of RRD(2). Moreover, cancers with both MMRD and PPD have unique SNV signatures that are characteristic of the interaction between the two DNA repair mechanisms(7). Nevertheless, SNV-based analyses fail to provide information on several biological and clinical questions related to RRD carcinogenesis. These include insights regarding the mechanisms of mutagenesis during RRD tumor initiation and progression, explaining genotype-phenotype correlations between different mutations in MMR and polymerase genes, and the ability to detect RRD in normal cells. These are highly important for the management of these patients, their tumors, and implementation of surveillance to family members.
While indel-based signatures were recently reported(23), the accumulation of MS-indels and their potential signatures in MMRD during tumorigenesis are not well-described(24–26). Furthermore, the impact of polymerase mutations on the accumulation of MS-indels and their corresponding signatures in cancer is not known(6–9).
To answer the above biological and clinical questions, we used our large cohort of carefully annotated cancers and normal tissues from patients with germline mutations in the MMR and polymerase genes. We analyzed genomic MS-indels from this cohort using whole exome and whole genome sequencing, thoroughly investigating the roles of both replication repair machineries in correcting microsatellite alterations, and their impact on cancer progression and response to therapy. Comparison of these tumors to adult RRD cancers and pediatric non-RRD tumors uncovered: (i) important insights into the accumulation of MS-indels in RRD cancers; (ii) a novel and distinct association between polymerase mutations and MS-indel accumulation; and (iii) a potential mechanism of MMRD that produces large deletions in microsatellites by single events that converge to a microsatellite locus length of ~15bp. We then considered using the activity levels of different MS-indel signatures for tumor stratification, genetic diagnosis, and as biomarkers of response to immunotherapy.
Results
A distinct pattern of microsatellite instability in CMMRD cancers
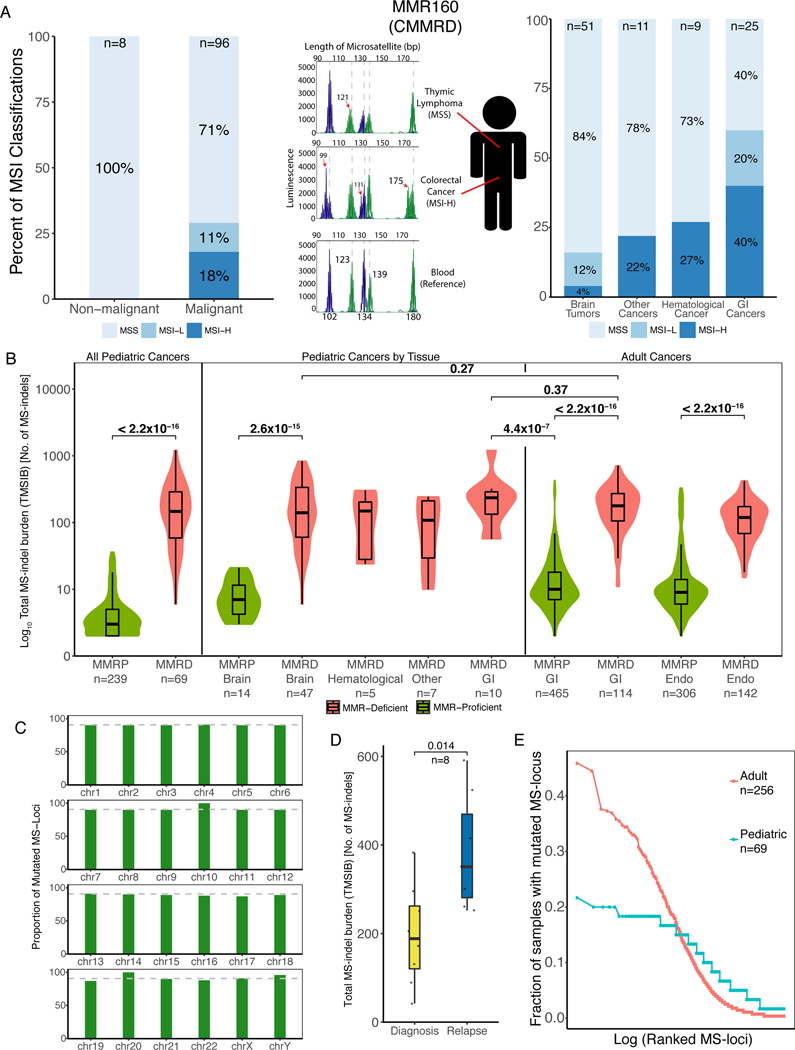
Since all cells from patients with CMMRD have impaired abilities to repair mismatches and MS-indels(1,2), MSI should be prevalent in all normal and cancerous tissues. To test this hypothesis, we analyzed the MSI status of a cohort of 96 cancers and 8 normal tissues using the current gold standard method, the Microsatellite Instability Analysis System (MIAS, Promega)(27,28). Strikingly, only 17 (18%) of the 96 CMMRD cancers and none of the normal tissues were classified as MSI-H (≥2 loci with ≥3bp indels) (Figure 1A). This observation was not related to the specific MMR mutated gene (P=0.95, Kruskal-Wallis, Supplementary Figure 1A) and similarly, different tumors from the same patient resulted in different MSI classifications (Figure 1A, Supplementary Figure 1B, Supplementary Table S1). Interestingly, we observed tissue specificity, with GI cancers having the highest proportion of MSI-H (10/25, 40%) compared to brain tumors (2/51, 4%, P=2×10−4, Chi-squared test).
Figure 1. Microsatellites Accumulate MS-indels in germline MMRD Cancers.
(A) Microsatellite Instability (MSI) status as measured by the Microsatellite Instability Analysis System (MIAS, Promega) of CMMRD samples. Left and right panels: The proportion of MSI high (MSI-H), MSI low (MSI-L) and microsatellite stable (MSS) samples in different tissue types with the number of samples tested (top). The middle section describes a single patient with constitutional mismatch repair deficiency (CMMRD), MMR160, who had a microsatellite stable (MSS) lymphoma and a microsatellite unstable (MSI-H) colorectal cancer. Histograms represent the results of the MIAS, with germline microsatellite lengths of the 5 tested loci (bottom plot) marked as dashed lines in all panels. GI = gastrointestinal.
(B) Comparison of exome-wide total microsatellite indel (MS-indel) burden (TMSIB) between pediatric mismatch repair proficient (MMRP) and MMRD cancers, pediatric cancers by tissue type, and between pediatric and adult MMRD cancers (TMSIB in logarithmic scale). Median TMSIB for each group is written beside each box, and sample numbers are written below each group. Green represents MMRP, and red represents MMRD in children and adults. Statistical significance was calculated by Mann-Whitney U-test. Endo = Endometrial cancers.
(C) Proportion of mutated MS-loci in each chromosome in the entire cohort. The average proportion (90.7%) is labelled and shown as a grey dotted line. The height of the bars in the indicate proportion, and similar heights indicate strong correlation of MS-indels to the number of MS-loci in that region of the chromosome. Supplementary Table S2 shows the similar proportions of MS-loci mutated in each chromosome.
(D) Comparison of the number of MS-indels in diagnosis and relapsed pediatric RRD tumors (n=8). Statistical significance was calculated by the Paired-samples Wilcoxon test.
(E) Analysis of recurrent indels in microsatellite loci (MS-loci), separated by adult MMRD cancers (red, n=256), and pediatric MMRD tumors (blue, n=69). Each point represents the fraction of tumors mutated for a given MS-locus (y-axis) sorted from most- to least-frequently altered (left to right on the x-axis).
It is unclear whether the lack of indels in the five MIAS microsatellite loci (MS-loci) is a unique feature to these loci, or whether MS-loci in CMMRD tumors universally have a low rate of indel accumulation. To test this, we applied MSMuTect to all the MS-loci in the 69 tumor/normal whole-exome pairs of pediatric CMMRD cancers(6). We found that pediatric CMMRD cancers had a significantly higher tumor MS-indel burden (TMSIB) than pediatric MMR-proficient (MMRP, n=239) cancers (P < 2.0×10−15, Mann-Whitney U-test, Figure 1B). Importantly, pediatric CMMRD cases had a similar TMSIB to the adult MMRD cases (n=114, P=0.48), and no significant difference was found in the TMSIB across pediatric cases from different tissues (Figure 1B). On the other hand, the number of SNVs in the pediatric tumors were higher due to the somatic POLE mutations commonly found in CMMRD tumors(2). Overall, our method demonstrates that, as predicted by the lack of MMR, pediatric CMMRD cancers indeed have an increased rate of MS-indels accumulation, similar to adult MMRD cancers.
MS-indels are spread equally on different chromosomes in both adult and pediatric cohorts (Figure 1C, Supplementary Table S2), and increase between initial and relapsed tumors(29) (n=8) (P = 0.014, Paired-samples Wilcoxon Test, Figure 1D). Furthermore, in contrast to adult MMRD tumors which exhibit highly mutated loci(6,14) (hotspots) like those used in the MIAS assay, pediatric MMRD tumors lacked this characteristic. For example, the microsatellite in the ACVR2A gene (chr2:148683686–148683693) is mutated in ~45% of adult MMRD cases, but only 11% in pediatric CMMRD cases (Supplementary Table S3). In addition, while adult tumors have 19 hotspots that are mutated in more than 30% of the cases, the strongest hotspot in pediatric tumors is mutated in ~20% (Figure 1E, Supplementary Table S4). Together, these observations suggest that MS-indels do accumulate in childhood CMMRD cancers, and continue to accumulate as tumors progress. Furthermore, the key differences between adult and childhood MMRD cancers, i.e. the different mutated loci and the lack of MS-loci that are mutated very frequently, may explain why the MIAS assay was unable to detect MSI in pediatric tumors.
MMRD and polymerase mutant tumors have distinct microsatellite indel signatures
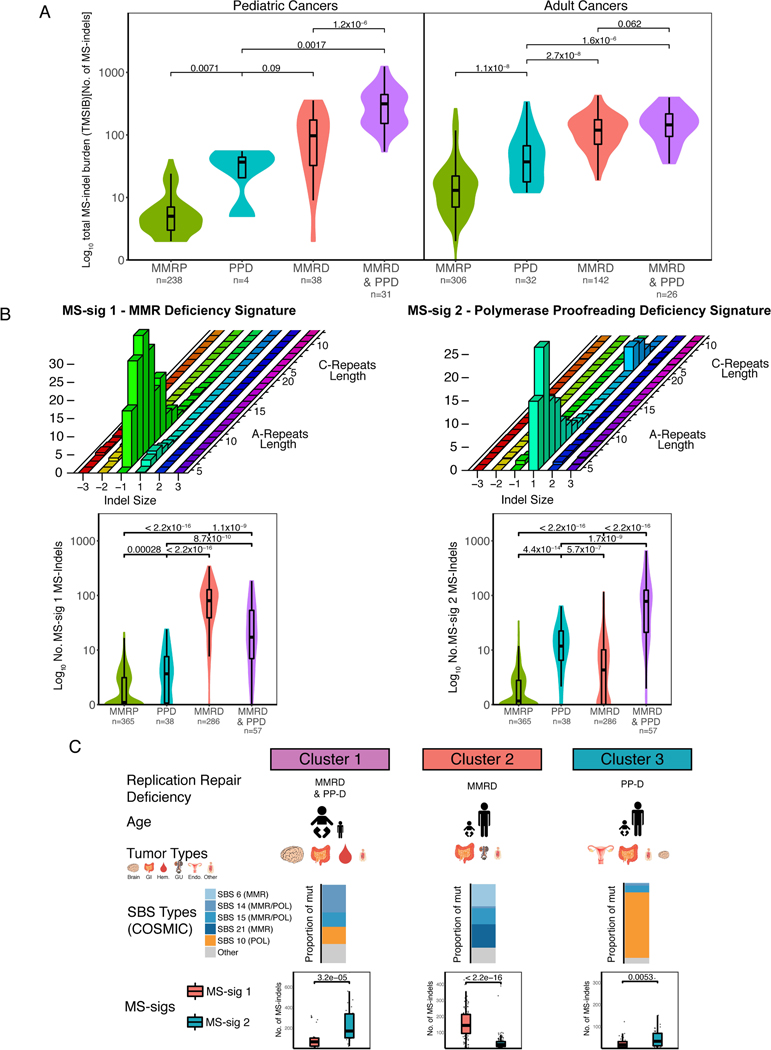
Another striking observation was that childhood tumors had a similar number of MS-indels as adult MMRD cancers, despite being largely MSS in the MIAS analysis (Figure 1A). This was most clearly observed between childhood CMMRD brain tumors and adult GI cancers (P=0.27, Mann-Whitney U-test, Figure 1B), as well as between pediatric and adult GI cancers (P=0.37, Mann-Whitney U-test, Figure 1B). Since CMMRD tumors acquire somatic polymerase mutations, which are known to dramatically increase SNV mutations in cancer and MS-indels in yeast, we hypothesized that mutant polymerases actively contribute to MS-indel accumulation in pediatric CMMRD tumors(3,30,31). We therefore compared MS-indels found in WES of 239 pediatric MMRP, 38 MMRD, 4 polymerase mutant and 31 combined RRD (MMRD& polymerase mutant) tumors to 505 adult endometrial cancers spanning the same four subtypes. Despite proficiency in MMR, polymerase mutant cancers exhibited increased MS-indel burden in both pediatric and adult cancers (P = 0.0071, P=1.1×10−8, respectively; Figure 2A). Moreover, combined RRD cancers have an increased MS-indel burden compared to MMRD cancers in the pediatric cohort (P=1.2×10−6, Figure 2A, left panel), further highlighting the contribution of polymerase mutations to the accumulation of MS-indels.
Figure 2. MMRD and PPD tumors have distinct microsatellite indel signatures.
(A) Comparison of TMSIB between RRD variants (MMRD, PPD, MMRD&PPD) to MMRP in pediatric brain and GI tumors (left) and adult endometrial tumors (right). Sample numbers are written below each group. P values calculated using Mann-Whitney U-test.
(B) Graphical presentation of MS-sig1 (top left) and MS-sig2 (top right) based on indel size (x-axis) and microsatellite locus length, separated by A- or C- repeats (z-axis). The total number of MS-sig1 indels and MS-sig2 indels are compared between the RRD variants in boxplots (bottom left and bottom right). P values calculated using Mann-Whitney U-test.
(C) Graphical representation of RRD subgroups(2) separated by age of tumor onset, most common tumor types, proportion of RRD-associated COSMIC SBS signatures, and MS-sigs 1 and 2. The size of each graphic corresponds to the proportion of the cancer observed clinically. MS-sigs correlate to the proportion of the COSMIC SBS signatures in each cluster. Statistical significance was calculated using the Mann-Whitney U-test.
We then wondered whether the two components of the replication repair machinery exert distinct alterations in microsatellites. To investigate this, we characterized each MS-indel based on three features, its motif (A or C), the microsatellite length (5–20bp) and the indel size (−3-+3bp) (Online Methods). We then used SignatureAnalyzer(32,33) to infer the different mutational signatures based on these 192 (=2×16×6) configurations, and found five distinct MS-indel signatures (MS-sigs; Figure 2B and Supplementary Figure 2A). When compared to MMR proficient cancers (Figure 2B), MMRD cancers were enriched for two signatures dominated by 1bp deletions in both A- and C-repeats (MS-sigs 1 and 3, Mann-Whitney U-test P<2.2×10−16), whereas polymerase mutations resulted in two signatures dominated by 1bp insertions (MS-sigs 2 and 4, P=4.4×10−14). Additionally, we analyzed RRD cancers with driver mutations in POLD1 (encoding the DNA polymerase that synthesizes the lagging strand(3)) (Supplementary Figures 2B, 2C). POLD1mut cancers were similarly dominated by insertions in MS-indels as POLEmut cancers (Supplementary Figure 2B). To our knowledge, this is the first time that the unique mechanism of MS-indel accumulation by DNA polymerase is described in human cancers. Interestingly, our results are in agreement with several yeast studies, which showed a bias towards 1bp insertions in microsatellites in mutant POLE strains as the leading strand polymerase(9,34,35), but contrast other yeast reports which suggest that mutant POLD1 creates single base deletions in microsatellite sequences(8,36). Together, these findings support that POLE and POLD1 with defective proofreading are associated with increased insertion events (as opposed to the deletion events associated with MMRD) and can contribute to the overall TMSIB in human cancers.
Previously(2), we classified hypermutant cancers into three clinically relevant subtypes of RRD (MMRD, PPD, or both MMRD & PPD) based on their single-base substitution signatures (COSMIC SBS signatures, Supplementary Figure 3). We therefore examined the correlation of the MS-sigs to these clusters. MMRD&PPD (SBS-Cluster 1) are mostly seen in children with germline MMRD that later acquire somatic POLE mutations, and fit a combination of MS-sig1 and MS-sig2. MS-sig1 is specific for MMRD-only cancers (SBS-cluster 2) in both children and adults, and PPD (SBS-Cluster 3) cancers are highly driven by the mutant polymerase and have more MS-sig2 MS-indels than MS-sig1 (P=3.2×10−5, P=0.0053, and P<2.2×10−16 respectively, Figure 2C). These data suggest that both SNV-based signatures and MS-sigs provide additional complementary information to standard mutational calls, and can be used to clinically screen and stratify tumors in the future.
Accumulation of large microsatellite deletions in MMRD cancers with preferred final length
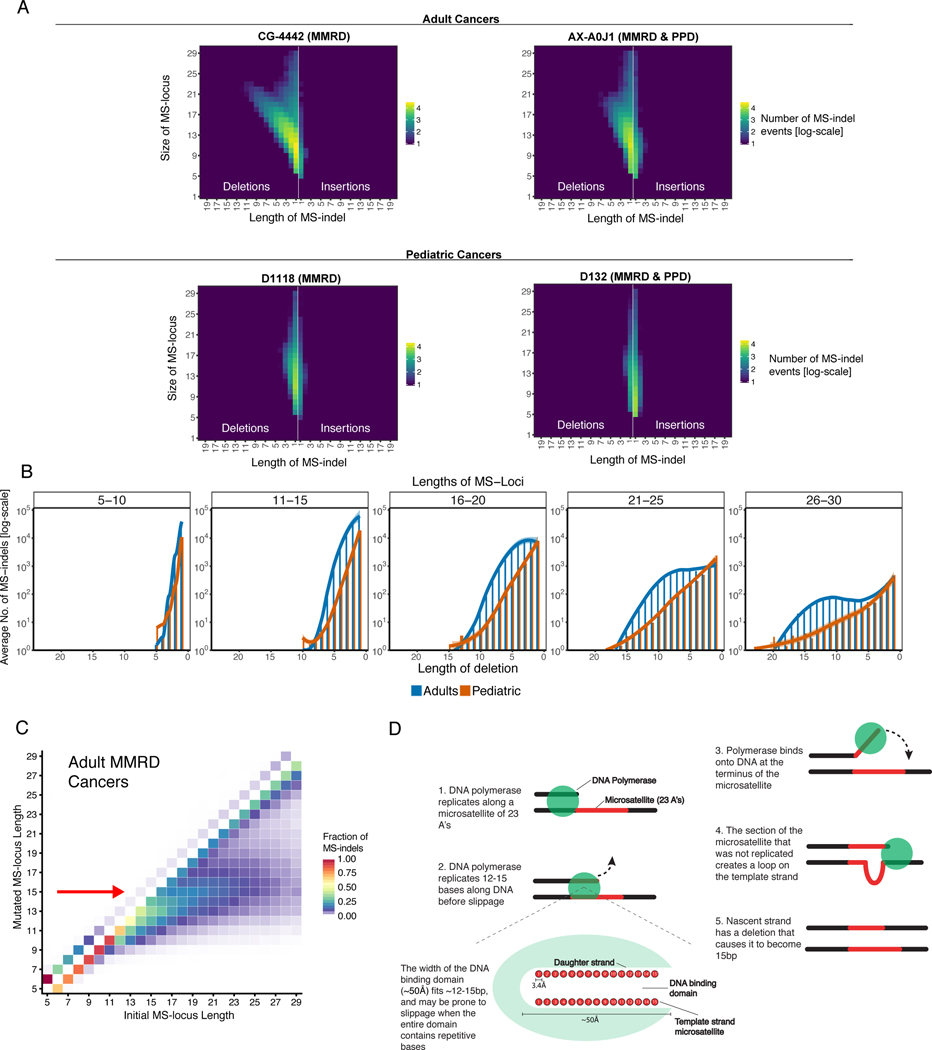
To gain insight into the mechanism of MS-indels accumulation during RRD tumorigenesis, we analyzed the patterns of MS-indels in whole genomes of 46 pediatric RRD cancers (39 brain, 4 colorectal, 1 osteochondroma, 1 Wilm’s tumor, and 1 T-cell lymphoma) and 28 MMRD adult cancers (12 colorectal, 5 gastric, 11 endometrial). The genome provides ~50 times more MS-loci compared to the exome, and has disproportionally higher numbers of longer (>20bp) MS-loci. These two features enable a more refined analysis of the MS-indel distribution. Adult MMRD cancers present (Figure 3A and Supplementary Figures 3, 4A, 4B, Supplementary Table S5) bias towards deletions similar to their behavior in WES (Figure 2B). They also present a new phenomenon of accumulating long deletions (>3bases), which increase in size with increasing MS-loci length. In contrast, pediatric CMMRD cases have mostly 1bp deletions and MMRD&PPD cancers are enriched with 1bp insertions (Figure 3A and Supplementary Figures 3, 4A, 4B, 4C, Supplementary Table S5) with a lack of long deletions. The lack of long MS-indels in pediatric cancers is likely the reason for the inability of the conventional electrophoresis methodologies (e.g. MIAS, Promega, Figure 1A) to detect MSI in these cancers, as the standard assay cannot robustly identify short indels and only considers indels of ≥3 bases as robust events.
Figure 3. Accumulation of large microsatellite deletions in MMRD cancers towards a preferred final length.
(A) Examples of genomic MS-indel heatmaps of germline MMRD and MMRD & PPD cancers in adults (TCGA) and children. Pediatric cancers have an enrichment of single base MS-indels, while adult cancers accumulate insertions of one base and large (>3bp) deletions that increase in size (x-axis) with increasing locus length (y-axis). Each color represents the log10 count of MS-indel events.
(B) The total number of MS-deletions in both pediatric and adults by locus length (each panel presents a range of 5 bases) and mutation size (x-axis). The number of deletions were averaged for each deletion length.
(C) The fraction of MS-indels by initial (x-axis) and final/post-mutation (y-axis) lengths in adult tumors revealing the convergence of MS-loci to 15bp (indicated by red arrow). Fraction of indels was calculated by dividing the number of indels corresponding to each initial and mutated locus length by total number of MS-indels in each initial MS-locus length.
(D) Schematic representation of the emergence of 15bp-long microsatellites as a targeted locus length in adult MMRD cancers. The polymerase is prone to slippage after replicating a tract of 15bp-repeats, resulting in a loop in the template strand upon rebinding. If unrepaired, the loop can result in a deletion that causes the nascent strand to become 15bp, irrespective of the original locus length.
To better understand the kinetics of the accumulation of large deletions in MS-loci, we compared two known models. (i) A two-phase model where each mutational event can alter either a single repeat motif or multiple motifs; or a simple (ii) stepwise-mutation model, which assumes only one repeat motif is altered per mutational event, making large deletions the amalgamation of multiple deletions(37,38). The stepwise model predicts that the relationship between the MS-indel size and the frequency of the mutational events of that size will be unimodal (i.e. Poisson). On the other hand, the two-phase model enables a bimodal relationship between the MS-indel size and frequency (Methods).
The pediatric tumors follow the predictions of the stepwise model(39) and present a sharp decrease in the frequency of MS-indels as the deletion size increases. In contrast, the adult tumors follow the stepwise model only for short loci (<12bp), and from 12bp and onwards, they deviate from the stepwise model and resemble the two-phase model(37) (Figure 3B, Supplementary Figures 4D and 4E).
Interestingly, the large deletions in adult tumors reduced the length of long (>15bp) MS-loci to become ~15bp (Figure 3C and Supplementary Figure 4F). These findings suggest that there is an underlying mechanism of MS deletions that gives larger loci the tendency to converge to 15bp in length. A potential model for this phenomenon is that after the DNA polymerase replicates 12–15bp on a microsatellite locus, it may become more susceptible to slip off from the template strand. The width of the DNA-binding domain of the polymerase is known to be ~50Å, which is approximately the combined length of 15 nucleotides (51Å) in a DNA double helix(40). We predict that because 15 nucleotides span the entire DNA-binding domain, the weak association of the polymerase to repeated nucleotides makes the polymerase especially prone to detach from the template strand. As the new strand then rebinds to the template strand, the template strand may generate a loop to become fully complementary to the ~15bp of the nascent strand(41,42) (Figure 3D).
This process is restricted to deletions, hinting that it may be specific to MMR deficient cells, whereas insertions in polymerase mutant cancers did not reveal the same large events (Supplementary Figure 4A).
Clinical applications of MS-sigs
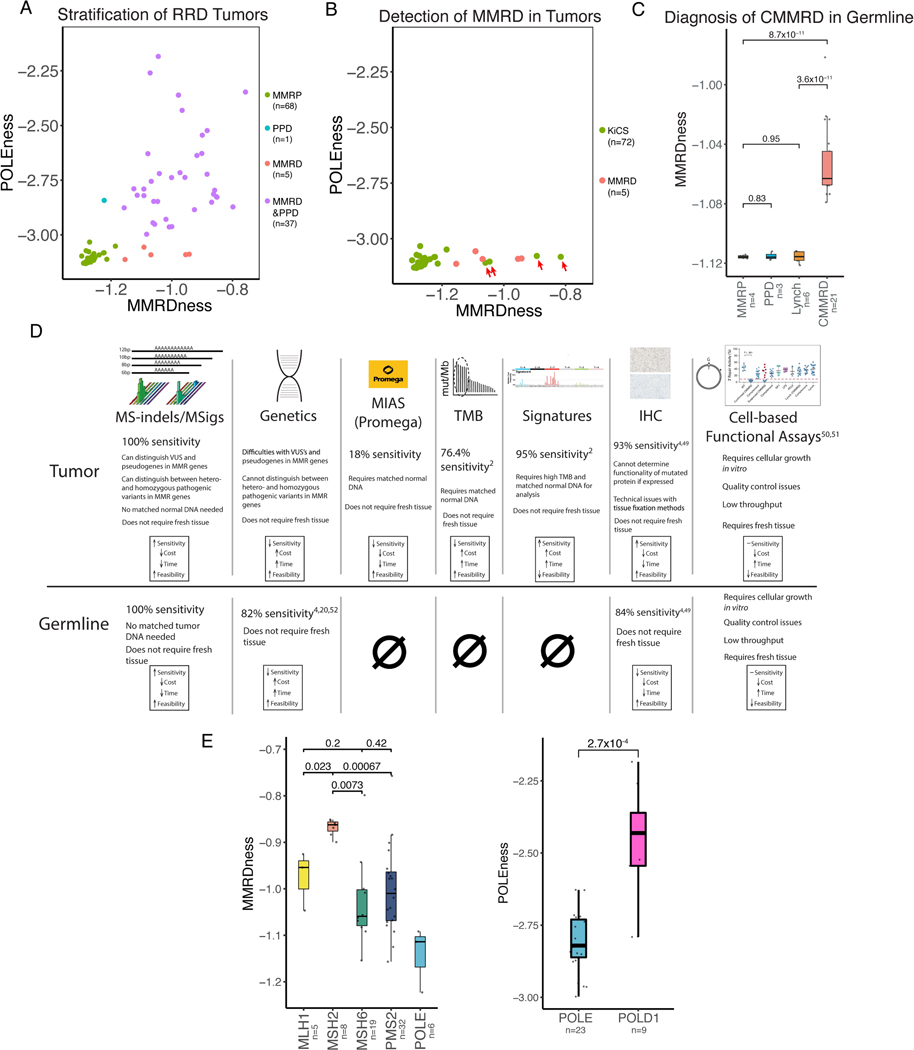
To begin answering the potential diagnostic and predictive role of MS-indels in patients with RRD cancers, we first compared the ability of MS-sigs to diagnose RRD cancers with currently used SNV-based clinically approved tools(2). The MS-indel signatures presented above (MS-sig1 and MS-sig2) are based on detecting MS-indels by comparing pairs of tumor and normal samples. However, the characteristic patterns of MS-indels and the large number of microsatellites in the genome may enable analyzing tumors without the need for matched normal samples. To test this, we designed scores that reflect the prevalence of MS-sig1 (MMRDness score) and MS-sig2 (POLEness score, Methods) in any given sample. Both scores were calculated by taking the logarithm (base 10) of the ratio of the number of −1 deletions (MMRDness) or +1 insertions (POLEness) divided by the total number of MS-loci within the specified lengths (10–15bp for MMRDness and 5–6bp for POLEness, Methods). Higher scores indicate higher prevalence of the MMRDness and POLEness signatures in each sample. Only MS-sig1 and MS-sig2 were used due to the overwhelming dominance of A-repeat MS-loci in the genome compared to C-repeats, which are represented by MS-sigs 3 and 4. We calculated the two scores for a well-characterized set of tumors and were able to separate the four subtypes that we analyzed (MMRP, MMRD, PPD and MMRD&PPD) (Figure 4A).
Figure 4. Clinical applications of MS-sigs.
(A) Two-dimensional plot separating all types of replication repair deficient (RRD) from wildtype (MMRP) cancers using whole genome sequencing. Green represents MMRP cancers, red represents mismatch repair deficient (MMRD) cancers, blue is polymerase proofreading deficient (PPD) cancers, and purple is combined MMRD and PPD cancers. Each dot represents an individual tumor.
(B) Discovery of unexpected MMRD tumors using MS-sigs. Two-dimensional plot of presumed MMRP tumors from the Sickkids Cancer Sequencing (KiCS) program. KiCS samples shown in green, and MMRD tumors in red. Four tumors from the KiCS cohort that had increased MMRDness are highlighted by red arrows. These were later confirmed to be MMRD (Supplementary Table S6).
(C) Detection of germline constitutional mismatch repair deficiency (CMMRD) using ultra low-passage WGS MMRDness scores. Blood samples from CMMRD and relevant control patients were used. Statistical significance was calculated using the Mann-Whitney U-test.
(D) Comparison of MS-sigs to other clinically approved (CLIA) methods to diagnose MMRD in normal and malignant cells. Sensitivity was calculated by dividing the number of positive MMRD cases in each assay by the number of samples that were tested(2,4,20,62–65).
(E) Genotype phenotype differences between different replication repair genes. Comparison of MMRDness between MMR gene mutations, and POLEness in POLEmut vs. POLD1mut cancers. P values were calculated using the Mann-Whitney U-test.
We then validated whether the MMRDness and POLEness scores can be used as a more cost-effective clinical tool for detecting MMRD and PPD. We sequenced 52 MMRP and RRD tumors with a small fraction of the standard genomic coverage (0.5X instead of the typical 30X), and calculated their MMRDness and POLEness. We observed similar clustering of the RRD subtypes (Supplementary Figure 5A), validating their ability to accurately and cost-effectively classify RRD tumors. Subsequently, we tested whether the MS-sigs can improve the current SNV-based methods used to screen and diagnose RRD tumors(4). We assessed the MMRDness and POLEness scores (Methods) of 72 tumors for which WGS was available through our clinical KiCS sequencing program(2). The MMRDness score uncovered four tumors (Figure 4B, Supplementary Figure 5B) that were not known to be MMRD by the sequencing center, as they had relatively low tumor mutational burdens and no SNV-based MMRD signatures (3–17 Mut/Mb, Supplementary Table S6). Genetic testing revealed that three of them have canonical Lynch syndrome germline mutations and the remaining one carries a novel germline MMR mutation (Supplementary Table S6). This suggests that MMRDness and POLEness can be both a more accurate and less expensive test compared to current methods for detecting RRD in tumors.
Analysis of Whole Genome Microsatellite Instability Signatures can Diagnose CMMRD in normal cells
Having established the ability of MS-sigs and the corresponding MMRDness score to identify MMRD tumors, we tested the ability of the tool to identify germline (CMMRD) mutations non-malignant (blood) samples. All non-malignant samples from CMMRD patients (n=34) had an elevated MMRDness score compared to MMR proficient, polymerase mutant, and Lynch syndrome patients (P=8.7×10−11, Mann-Whitney U-test, Figure 4C, Supplementary Figure 5C). Notably, the Promega panel assay was completely unable to detect CMMRD in non-malignant tissues (Figure 1A) and, similarly, SNV-based signatures are not able to detect hypermutation in normal cells. The MMRDness score neither requires a patient-matched normal control nor high sequencing depth to detect the MS-sig patterns throughout the genome that correspond to CMMRD. The high specificity and sensitivity of the MMRDness score (Figure 4D) enables it to be an inexpensive and robust assay for screening and diagnosing CMMRD prior to cancer, which can be used to implement early surveillance protocols to improve the survival of CMMRD patients. On the other hand, the diagnostic role of POLEness is limited to RRD tumors (Figure 4A), and further investigation is required to assess its ability to detect PPD in the germline.
Different alterations of the MMR and polymerase genes have varying effects on MMRDness and POLEness
The four genes MLH1, MSH2, MSH6 and PMS2, that when mutated lead to MMRD, play different roles in the MMR mechanism(43). However, thus far, no footprint has been found in the mutational landscape for the four different genes. For example, germline mutations in MSH2 and MLH1 have higher penetrance than MSH6 and PMS2, and are found most commonly in adult Lynch Syndrome cases. In contrast, for individuals with CMMRD, the reverse is observed. PMS2 is the most commonly mutated gene, followed by MSH6, and biallelic MSH2 germline mutations are extremely rare. These cancers have similar tumor mutation burdens (TMB) irrespective of the mutated MMR gene(1,2); thus, the different penetrance cannot be simply explained by a higher mutation rate. Other functional assays have also failed to explain this genotype-phenotype relationship despite the different mechanistic roles of the MMR genes. Similarly, PPD is almost strictly observed with POLE but not POLD1 germline mutations - an observation that also cannot be explained by differences in TMB or other functional assays.
We therefore evaluated whether the deficiency in different genes may have distinct effects on the MMRDness score (Figure 4E). We found that MMRD cancers with biallelic inactivating mutations in MSH2 had the strongest MMRDness score, followed by MLH1, and was much lower in PMS2 mutant cancers (P=6.7×10−4, Mann-Whitney U test, Figure 4E). This fits well with the clinical observations of mismatch repair deficiency, as MSH2 and MLH1 mutations have higher cancer penetrance than PMS2 and MSH6. This also correlates with the known mechanism of mismatch repair, as the MSH2 protein initially recognizes mismatches during replication and is crucial for both the mutSα and mutSβ complexes, whereas MLH1 is an important factor in the mutL complex(44–46).
Next, we investigated whether the two polymerase genes (POLE and POLD1), have a distinct effect on the POLEness score. POLD1mut cancers had significantly higher POLEness score than POLEmut cancers (P=2.7×10−4, Mann-Whitney U-test, Figure 4E), supporting our previous finding from the exomes (P=2.3×10−4, Supplementary Figure 2C), where POLD1mut cancers had a higher TMSIB than POLEmut. This can be explained by the replication of the lagging strand inherently having more dissociation and slippage of the polymerase from the DNA(16,47).
POLEness Score Predicts Response to Immunotherapy in RRD Cancers
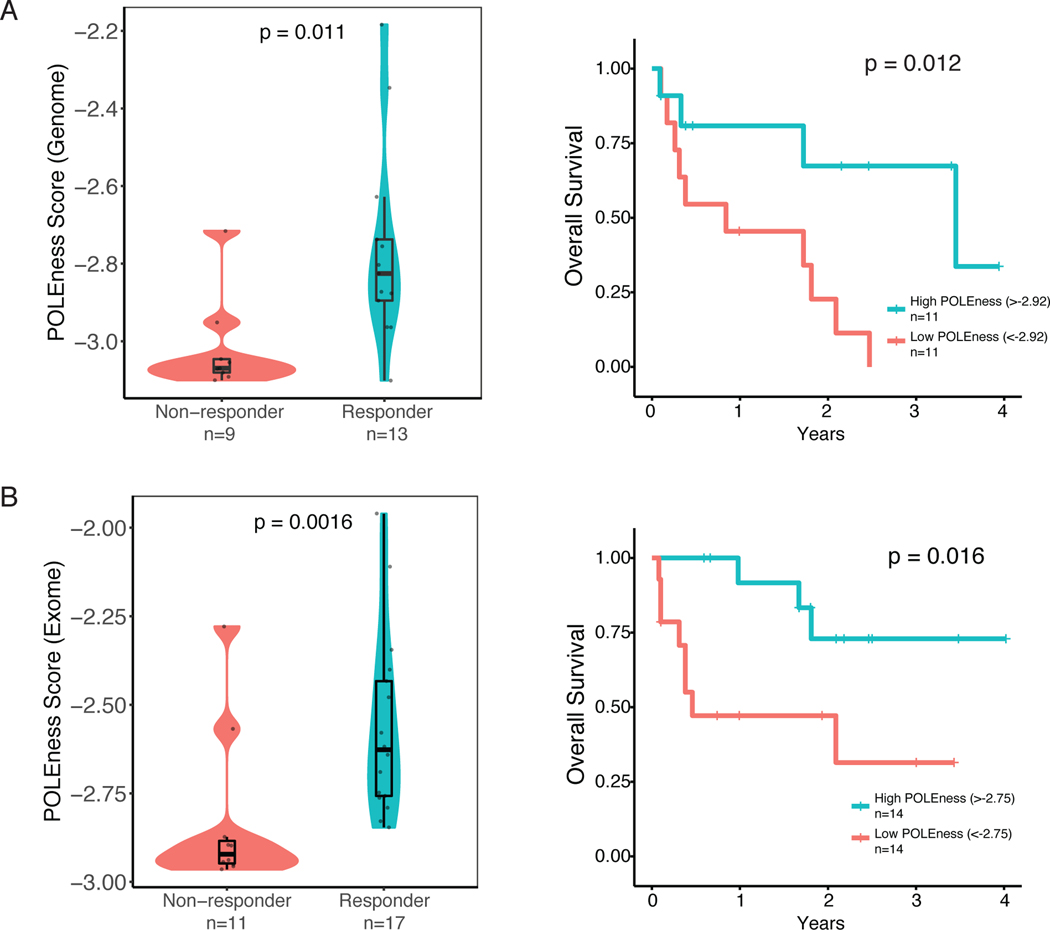
It was recently shown that RRD tumors tend to respond to ICI therapy(48). However, not all patients respond, and currently there is no adequate method to distinguish responders from non-responders (Supplementary Figure 6A). As MS-indels are highly immunogenic(10,49–52), we hypothesized that within the context of RRD cancers, MMRDness and/or the POLEness score could predict response to ICI therapy. To test this, we initially used WGS data of 22 RRD tumors undergoing ICI as a part of our consortium registry study(48). We observed that the POLEness score of responders was higher than non-responders to ICI (P=0.011, Mann-Whitney U-test, Figure 5A) and tumors with a high POLEness score (>−2.92 POLEness, Methods) had a significantly higher survival than the low POLEness group (P=0.012, Log-Rank test, Figure 5A). We therefore tested the ability of the POLEness score to predict response to ICI using exomes from a larger cohort of patients (n=28). POLEness scores were higher in responders to ICI therapy than in non-responders (P=0.0016, Mann-Whitney U-test, Figure 5B). However, exome and genome MMRDness was not shown to have a significant difference between responders and non-responders, potentially because all tumors in the cohort are MMRD (Supplementary Figure 6B). The high-POLEness groups in both genome and exome had significantly longer survival times than the low-POLEness groups (P=0.012 and 0.016, Log-Rank Test, Figures 5A, 5B). To our knowledge this is the first time that a mutational signature was suggested as a biomarker to distinguish between responders and non-responders to ICI in RRD tumors(10,49–51). This result sheds new light on the immunogenicity of MS-indels and the use of their signatures as novel biomarkers for response to immune checkpoint inhibition in the context of RRD.
Figure 5. POLEness Score Predicts Response to Immunotherapy in RRD Cancers.
(A) Genomic POLEness scores of 22 RRD cancers stratified by response to immune checkpoint inhibition. Overall survival of patients on immunotherapy separated by the median Genomic POLEness scores (median POLEness = −2.92, Methods). P values for comparing responders to non-responders were calculated using the Mann-Whitney U-test, and Kaplan-Meier curves were generated for survival with p values calculated using the Log Rank test.
(B)Same as (A) using exomic POLEness scores from 28 RRD patients (median POLEness = −2.75, Methods).
Discussion
This report uncovers roles for both components of the replication repair machinery in MS-indel accumulation in cancers. These observations address important fundamental biological questions regarding accumulation of MS-indels during carcinogenesis, which can be translated to clinical use.
Our data reveal that in addition to defects in MMR, loss of proofreading by replication-specific DNA polymerases is also associated with an increase in MS-indels. MS-indels produced by the deficiencies of the two replicative repair mechanisms have distinct characteristics. While MMRD generates mainly deletions (MS-sig1 and MS-sig3), PPD results predominantly in insertions (MS-sig2 and MS-sig4), suggesting a strand-bias repair process (Supplementary Figure 7). The MS-sigs described were comparable to the ID-1 and ID-2 indel signatures reported in Alexandrov et al., (2020), which were common in tissues that are associated with replication repair deficiency(23). However, Alexandrov et al. did not analyze MS-indels in loci longer than 5 repeated units, and therefore were not able to find the unique MS-sigs present in larger MS-loci.
We hypothesize that the proofreading domain of the polymerase repairs extra bases on the nascent strand by changing the conformation of the DNA to push the substrate into the exonuclease domain of the polymerase. This would activate the exonucleolytic mode of DNA polymerase to excise the loop. Since mutations in the proofreading domain of the polymerase would prevent the DNA substrate from shifting into the exonuclease site, these remain unrepaired as replication continues(34,53), creating a permanent insertion. As larger insertions were not observed in PPD cancers, we postulate that larger indel loops within the nascent strand do not fit into the exonuclease domain of the polymerase (Supplementary Figure 7). In contrast, the MMR system is more effective in detecting loops on the parental strand(3,45,54,55). These loops would result in deletions if unrepaired, which translated into the surge of MS-sig1 and MS-sig3 in MMRD cancers in our cohort.
Another intriguing observation related to the role of MMR on the template strand is that larger deletions in adult MMRD cancers converged to a common length of microsatellites of ~15bp (Figure 3C). We propose that this 15bp convergence is due to the size of the DNA binding domain of the polymerase complex being similar to the length of 15bp, which is about 50Å(40). Indeed, these large events can explain the ease of detection of MSI-H in adult MMRD cancers by the standard electrophoresis-based assay that specifically looks for long (≥3bp) MS-indels. Furthermore, pediatric tumors lacked hotspots or commonly-mutated MS-loci even within specific tissues (Figure 1E, Supplemental Table S3). In adults, MMRD cells can acquire MS-indels in the ACVR2A gene, which may lead to a selection advantage in the GI system, but not in other tissue types. Thus, the contrast between pediatric and adult cases may be due to the selective pressures that are present in different tissue types, although comparative analysis between pediatric and adult GI cancers still showed unique MS-indel profiles (Supplementary Figure 4B). Additionally, the distinct MS-deletions in adult and pediatric cancers might be due to unknown differences in DNA damage sensing or response mechanisms. Further experimental investigation could clarify these phenomena, and can hint towards a better understanding of the kinetics and patterns of MS-indel accumulation.
The biological observations described above can have a major clinical impact on the management of patients with RRD cancers. Firstly, the standard methods for MSI classification (e.g. MIAS Promega) cannot detect MSI in most pediatric MMRD tumors (Figure 1A) and possibly in tumors in which MMRD occurs late in carcinogenesis due to other mutational processes. The latter includes treatment-related MMRD cancers such as leukemias, gliomas and POLEmut-driven tumors(2,56,57), where MMRD occurs later, and are therefore falsely termed MSS(6,15,16). Both SNV- and MS-indel-based methods may be more sensitive and specific to classify such tumors correctly for management and potential genetic testing. Additionally, MS-sigs was able to differentiate between MMRD and PPD tumor genotypes (Figure 4E), which has not been previously shown through SNV or the canonical MSI analysis method. The increased MMRDness signature in MSH2 and MLH1 mutant tumors (Figure 4E) correlates with the more aggressive phenotype seen in Lynch Syndrome cases and the biological importance of the two proteins in the repair mechanism of the mutSα and mutSβ MMR complexes(44–46). The ability to use low-pass whole-genome sequencing from tumors, without their matched normal samples, can make MS-sigs the basis for an inexpensive tool for RRD classification. There has been a recent report that used deep sequencing of a panel of 277 MS-loci to diagnose CMMRD using blood DNA(58). However, our method of ultra low-pass whole-genome sequencing (ULP-WGS) at 0.1–0.5X coverage is less expensive per sample, and covers between 10% to 50% of the ~23 million MS-loci, respectively. The ULP-WGS approach can be more sensitive and specific than deep sequencing of specific loci, depending on the depth of sequencing in each of the approaches. MS-sigs can also provide additive information to the clinical diagnosis, such as the POLEness signature that can predict response to immunotherapy (Figure 5). Ongoing and future investigations will compare the effectiveness of the two methods in detecting CMMRD.
Secondly, we showed that MS-sigs can be used to detect MMRD in non-malignant samples of individuals with CMMRD. The potential to detect MMRD from non-malignant samples, perhaps even before cancer development, has important clinical applications. In many cases, genetic diagnosis of CMMRD is difficult due to the large number of variants of unknown significance (VUS) and pseudogenes in the mismatch repair and DNA polymerase genes. Furthermore, MS-sigs was found to have extremely high sensitivity and specificity (both 100% in our cohort) in detecting MMRD in tumors and CMMRD in normal tissue. MS-sigs can distinguish functionally disruptive mutations from VUS’s and pseudogenes in the MMR and polymerase genes, and can be conducted using low quantities (<150ng) of unmatched DNA from FFPE-embedded or frozen tissue (Figure 4D). We suggest here that MS-sigs is an assay that can assess the MMRD phenotype from accessible, non-malignant tissues, which may become a frontline tool for the clinical diagnosis of RRD tumors and for monitoring patients at high risk. Further validation on clinical cohorts is required.
Finally, our data provide further support to the notion that MS-indels may be strongly immunogenic in RRD hypermutant cancers(59). In our cohort, MS-sig based POLEness score, but not MMRDness, was a predictive biomarker for response to immune checkpoint inhibition. This adds to the recent report which correlates MSI status to anti-PD-1 immunotherapy response(49). Since our cohort includes responders from brain and other tumor sites, which are not considered MSI-H using conventional methods, the robustness of Next-Generation Sequencing based MS-sig analysis may be superior to panel-based methods. Our observations suggest that the single repeat insertions represented by the POLEness signature likely yield highly immunogenic neoantigens, which in turn result in a robust immunogenic response against RRD tumor cells(48,60,61). Future large studies comparing MSI based signatures to other genomic features of RRD cancers, as well as investigating differences in immunotherapy response between MMRD, PPD and MMRD&PPD cancers will be required to validate our findings.
In summary, our data adds a new dimension to the role of RRD in human cancer mutagenesis in the form of microsatellite maintenance and instability. Studies of cancers emerging from rare, germline childhood cancer syndromes provide important insights and may be applied to more common adult cancers. Further mechanistic analysis will potentially explain the different types and sizes of MS-indels that are introduced during tumorigenesis. Future studies will determine the impact of both MMRD and PPD on microsatellites, which can lead to advancements in the classification, diagnosis, and determination of biomarkers in the management of RRD cancers and individuals.
Methods
Sample Collection and DNA Extraction for Whole Exome and Genome Sequencing of Replication Repair Deficient Tumors
As described in previous reports, patients with replication repair deficiency have been routinely consented and registered into the International Replication Repair Consortium with written, informed consent. The study was conducted in accordance with the Canadian Tri-Council Policy Statement II (TCPS II), and was approved by the Institutional Research Ethics Board (REB approval number 1000048813), and all data were centralized in the Division of Haematology/Oncology at The Hospital for Sick Children (SickKids). Tumor and blood samples were collected from the Sickkids tumor bank, and diagnosis of replication repair deficiency was confirmed via sequencing and immunohistochemistry of the four MMR genes and sequencing of the POLE and POLD1 genes by a clinically approved laboratory. DNA was extracted using the Qiagen DNeasy Blood & Tissue kit for frozen tissues, and QIAamp DNA FFPE Tissue Kit for paraffin-embedded tissues.
Whole Exome and Genome Sequencing Data of Pediatric Tumors in the KiCS Program
Whole exome and genome sequencing data of pediatric tumors were obtained through the Sickkids’ Cancer Sequencing (KiCS) program. Detailed information about KiCS can be found at: www.kicsprogram.com.
Whole Exome Sequencing Data of Pediatric Brain Tumors
Written informed consent was provided for patients with brain tumors treated at The Hospital for Sick Children (Toronto, Canada), and was approved by the institutional review board. DNA from the tumors and non-malignant samples were extracted for whole-exome sequencing. Further details on the DNA extraction protocol and exome-sequencing pipeline are available in previous reports.
Whole Exome and Genome Sequencing Data and MSI Status Identification of Adult Tumors in The Cancer Genome Atlas Database
Whole exome sequencing and whole genome sequencing data for CRC (colorectal carcinoma), STAD (stomach adenoma), and UCEC (uterine corpus endometrial carcinoma) were downloaded from TCGA (The Cancer Genome Atlas), which were sequenced on the Illumina platform. As described in a previous report, their MSI status were annotated by TCGA.
Whole Exome Sequencing Data of Pediatric Neuroblastoma from the TARGET Database
Whole exome sequencing data of pediatric neuroblastoma was acquired from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative.
Microsatellite Instability Status of Pediatric Tumors Determined by the Microsatellite Instability Analysis System (Promega)
DNA extracted from tumor and matched normal tissues were quantified with Nanodrop (Thermo Scientific, Wilmington, DE), and amplified with Platinum™ Multiplex PCR Master Mix (ThermoFisher Scientific, Wilmington, DE) and MSI 10X Primer Pair Mix (Promega) in a Veriti™ 96-Well Thermal Cycler, as per the manufacturer’s recommendations for PCR cycling conditions. The primer mix targets a panel of five mononucleotide loci, termed BAT-25, BAT-26, NR-21, NR-24, MONO-27, and following amplification, the products were run in a 3130 Genetic Analyzer for fluorescent capillary electrophoresis. Electrophoretograms were subsequently visualized using the Peak Scanner Software (v1.0, ThermoFisher Scientific, Wilmington, DE), and the highest peaks that were flanked by lower peaks were selected to be the representative alleles for each of the five loci in the panel. Each tumor-normal pair were compared by their allelic lengths. Tumors were considered MSI-High (MSI-H) if two or more loci were unstable (≥3bp shift from the normal allele), MSI-Low (MSI-L) if one locus was unstable, and MSI stable (MSS) if all five loci were stable (<2bp shift from the normal allele)(27,28).
Exome Sequencing of Replication Repair Deficient Pediatric Tumors
All high-throughput sequencing and mutation identification were performed at The Centre for Applied Genomics at the Hospital for Sick Children, as described in previous reports(1,2). Briefly, tumor and matched non-malignant tissue DNA were sequenced on an Illumina HiSeq2500 machine using Agilent’s exome enrichment kit (Sure Select V4/V5; with > 50% baits above 25x coverage). Processing into FASTQ files was done using CASAVA and/or HAS, which were aligned to UCSC’s hg19 GRCh37 with BWA. The realignment and recalibration of aligned reads were done using SRMA and/or GATK and the Genome Analysis Toolkit26 (v1.1–28), respectively. Whole-exome tumor mutation burden was calculated using MuTect (v1.1.4) and filtered using dbSNP (v132), the 1000 Genomes Project (Feb 2012), a 69-sample Complete Genomics dataset, the Exome Sequencing Project (v6500), and the ExAc database for common single-nucleotide polymorphisms.
Genome Sequencing of Pediatric Tumors
Whole genome sequencing was done using the Illumina HiSeq2500 or Illumina HiSeqX at ≥30X coverage, as well as on the Illumina NovaSeq6000 at 0.5X coverage. Read realignment and recalibration were conducted using the same pipeline as exome sequencing.
Exomic and Genomic Microsatellite Definition and Identification and Mutation Calling via MSMuTect
The detailed methods of the MSMuTect algorithm was previously reported(6). Briefly, repeats of five or more nucleotides were considered to be MS loci, and using the PHOBOS algorithm and the lobSTR approach, tumor and normal BAM files were aligned with their 5’ and 3’ flanking sequences. Each MS-locus allele was estimated using the empirical noise model,, which is the probability of observing a read with a microsatellite (MS) length k and motif m, where the true length of the allele is j with the motif m. This was used to call the MS alleles with the highest likelihood of being the true allele at each MS-locus. The MS alleles of each tumor and matched normal pair were called individually, which were compared to identify the mutations on the tumor MS-loci. The Akaike Information Criterion (AIC) score was assigned to both the tumor and normal models, and a threshold score that was determined by using simulated data was applied to make the final call of the MS-indel.
Microsatellite Indel Signatures Discovered by SignatureAnalyzer
For the WES samples noted in Supplementary Table S7, we quantified the number of MS-indels it has using different parameters, defined by the length of the microsatellite locus in the normal sample, the size of the indel (one base insertion/deletion, two base insertion/deletion, etc.), and the MS-locus motif (A- and C-repeats, Figure 2B, Supplementary Figure 2A). We then used the BayesianNMF algorithm (SignatureAnalyzer)(32,33) with its default parameters to infer the different mutational processes operating in our samples. We did not include other repeat motifs, as they do not have sufficient mutational events.
Calculation of MMRDness and POLEness Scores
Exome- Genome-wide microsatellite indels in each sample were quantified and segregated into deletions and insertions of up to three bases, and by locus size from five to 40 bases. The number of MS-indels in each sample was normalized to its own sum of total MS-indels. MMRDness scores were calculated by taking log10 of the sum of the average proportions of one base deletions in loci sized from 10 to 15 bases. The POLEness scores were similarly calculated by taking log10 of the sum of the average proportions of one base insertions in loci sized 5 to 6 bases. Both scores were normalized to the total number of MS-loci with the respective lengths.
Whole Exome Sequencing Microsatellite Indel Quantification and Comparison
Exome-wide microsatellite indels for each tumor/normal pair were identified and quantified using MSMuTect(6). We processed the MSMuTect output using the Rstudio graphical user interface (v1.1.447). The processing included summing the total number of MS-indels and calculating the length change of each microsatellite in each tumor compared to their matched normal samples. The medians number of MS-indels in the exome of each tumor type and in each MS-indel signature cluster were compared using the Mann-Whitney U-test in Rstudio, and statistical significance was considered at p<0.05.
Whole Genome Sequencing Microsatellite Indel Quantification
Genome-wide microsatellite indels were summed for each sample according to the reference length of each locus and the change in size/length of the indel. Genome-wide deletions were identified and summed for both adult and pediatric samples, and were averaged for each deletion length. The mean and standard deviations were calculated using Rstudio (v1.1.447) at each deletion length in adult and pediatric samples independently. Fractions of indels based on initial and mutated loci size in adult cancers were calculated by dividing the number of events pertaining to a specific initial and mutated locus size by the sum of all MS-indels in all adult tumors. The same procedure was followed for pediatric cancers.
MMRDness and POLEness Score Comparison Between Deficient MMR and Polymerase Proofreading
MMRDness medians were calculated for pediatric replication repair deficient tumors with the same mutated MMR gene, and compared between the four genes using the Mann-Whitney U-test. Similarly, POLEness medians were calculated for POLE or POLD1 mutants and compared.
MMRDness and POLEness Score Calculation and Comparison for Ultra Low Coverage Whole Genome Sequencing Samples
MMRDness and POLEness scores were calculated as explained above for the ultra-low pass coverage sequencing samples. The tumors were separated according to their replication repair deficiency type (MMRP, PPD, MMRD, and MMRD&PPD), and medians of their MMRDness were calculated. Comparison of MMRDness between groups was done using the Mann-Whitney U-test. Germline samples were also segregated based on their replication repair deficiency statuses (MMRP, PPD, Lynch, CMMRD), and their MMRDness scores were compared using the same method as the tumors.
Immunotherapy Response Thresholds for POLEess scores
POLEness scores were determined from 28 tumors where WES was available, and from 22 of them that had WGS available. These tumors were collected from patients with RRD cancers who underwent immunotherapy. POLEness scores were calculated as described above. For survival analysis, tumors were separated by their respective median POLEness scores (−2.92 in genome and −2.75 in exome). P values were determined using the Log Rank test.
Supplementary Material
Significance.
Exome- and genome-wide microsatellite instability analysis reveals novel signatures that are uniquely attributed to mismatch repair and DNA polymerase. This provides new mechanistic insight on microsatellite maintenance and can be applied clinically for diagnosis of replication repair deficiency and immunotherapy response prediction.
Acknowledgements
The SickKids Cancer Sequencing (KiCS) program is supported by the Garron Family Cancer Centre with funds from the SickKids Foundation. This research is supported by Meagan’s Walk (MW-2014-10), b.r.a.i.n.child Canada, LivWise, an Enabling Studies Program grant from BioCanRx - Canada’s Immunotherapy Network (a Network Centre of Excellence), the Canadian Institutes for Health Research (CIHR) grant (PJT-156006), the CIHR Joint Canada-Israel Health Research Program (MOP – 137899), and a Stand Up to Cancer (SU2C) – Bristol-Myers Squibb Catalyst Research (SU2C-AACR-CT07-17) research grant. Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. G.G. and Y.E.M were partially funded by G.G. funds at Massachusetts General Hospital. G.G. was partially funded by the Paul C. Zamecnick Chair in Oncology at the Massachusetts General Hospital Cancer Center.
Footnotes
Data Availability
Existing whole exome and/or whole genome sequencing data have previously been deposited in the European Genome Phenome Archive under the study accession numbers EGAS00001000579, EGAS00001001112. New sequencing data from this report have been deposited in the European Genome Phenome Archive, and the accession number is EGAS00001004816. Public datasets used include the TCGA database (https://portal.gdc.cancer.gov/), TARGET database (https://ocg.cancer.gov/programs/target), and the Sickkids KiCS program (https://kicsprogram.com/). Supplementary table 7 includes all of the raw data used in all analyses within this manuscript. Further information and/or requests for data will be fulfilled by the corresponding author, Uri Tabori (uri.tabori@sickkids.ca).
Code Availability
The software and pipelines used for data collection are the following: MSMuTect (Maruvka et al., 2017)(https://github.com/getzlab/MSMuTect/)(6), SignatureAnalyzer (https://software.broadinstitute.org/cancer/cga/msp), MuTect (https://software.broadinstitute.org/cancer/cga/mutect). All data was analyzed using Rstudio (v1.1.447). All R packages and other statistical code used for analysis are: Scales (https://CRAN.R-project.org/package=scales), ggplot2 (https://CRAN.R-project.org/package=ggplot2), dplyr (https://CRAN.R-project.org/package=dplyr), Reshape2 (https://CRAN.R-project.org/package=reshape2), RColorBrewer (https://CRAN.R-project.org/package=RColorBrewer), ggpubr (https://CRAN.R-project.org/package=ggpubr), tibble (https://CRAN.R-project.org/package=tibble), epade (https://CRAN.R-project.org/package=epade), Plot3D (https://CRAN.R-project.org/package=plot3D), forcats (https://CRAN.R-project.org/package=forcats), survival (https://CRAN.R-project.orag/package=survival), survminer (https://CRAN.R-project.org/package=survminer). Scripts used to generate the figures is accessible upon request from the corresponding author.
Conflicts of Interest:
G.G. is a founder, consultant and holds privately held equity in Scorpion Therapeutics. G.G. receives research funds from IBM and Pharmacyclics. G.G. is an inventor on patent applications related to MuTect, ABSOLUTE, MutSig and POLYSOLVER. Y.E.M and G.G. are inventors on patent applications related to MSMuTect and MSMutSig. G.G., Y.E.M., U.T., and J.C. are inventors on patent applications related to MSIdetect. Other authors declare no competing interests.
References
- 1.Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet 2015;47(3):257–62 doi 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 2.Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017;171(5):1042–56 e10 doi 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunkel TA, Erie DA. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu Rev Genet 2015;49:291–313 doi 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabori U, Hansford JR, Achatz MI, Kratz CP, Plon SE, Frebourg T, et al. Clinical Management and Tumor Surveillance Recommendations of Inherited Mismatch Repair Deficiency in Childhood. Clin Cancer Res 2017;23(11):e32–e7 doi 10.1158/1078-0432.Ccr-17-0574. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harbor symposia on quantitative biology 2009;74:91–101 doi 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maruvka YE, Mouw KW, Karlic R, Parasuraman P, Kamburov A, Polak P, et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol 2017;35(10):951–9 doi 10.1038/nbt.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haradhvala NJ, Kim J, Maruvka YE, Polak P, Rosebrock D, Livitz D, et al. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun 2018;9(1):1746 doi 10.1038/s41467-018-04002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. The Journal of biological chemistry 2005;280(33):29980–7 doi 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 9.Kirchner JM, Tran H, Resnick MA. A DNA Polymerase ε Mutant That Specifically Causes +1 Frameshift Mutations Within Homonucleotide Runs in Yeast. Genetics 2000;155(4):1623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372(26):2509–20 doi 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med 2000;342(2):69–77 doi 10.1056/nejm200001133420201. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita H, Nakayama K, Ishikawa M, Nakamura K, Ishibashi T, Sanuki K, et al. Microsatellite instability is a biomarker for immune checkpoint inhibitors in endometrial cancer. Oncotarget 2018;9(5):5652–64 doi 10.18632/oncotarget.23790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016;22(4):813–20 doi 10.1158/1078-0432.Ccr-15-1678. [DOI] [PubMed] [Google Scholar]
- 14.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22(11):1342–50 doi 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497(7447):67–73 doi 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prindle MJ, Loeb LA. DNA polymerase delta in DNA replication and genome maintenance. Environ Mol Mutagen 2012;53(9):666–82 doi 10.1002/em.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer 2015;15(3):181–94 doi 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 18.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nature reviews Clinical oncology 2010;7(3):153–62 doi 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umar A, Boland CR, Terdiman JP, Syngal S, Chapelle Adl, Rüschoff J, et al. Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. JNCI: Journal of the National Cancer Institute 2004;96(4):261–8 doi 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wimmer K, Kratz CP, Vasen HF, Caron O, Colas C, Entz-Werle N, et al. Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium ‘care for CMMRD’ (C4CMMRD). J Med Genet 2014;51(6):355–65 doi 10.1136/jmedgenet-2014-102284. [DOI] [PubMed] [Google Scholar]
- 21.Wimmer K, Kratz CP. Constitutional mismatch repair-deficiency syndrome. Haematologica 2010;95(5):699–701 doi 10.3324/haematol.2009.021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abedalthagafi M. Constitutional mismatch repair-deficiency: current problems and emerging therapeutic strategies. Oncotarget 2018;9(83):35458–69 doi 10.18632/oncotarget.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578(7793):94–101 doi 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer research 1997;57(21):4749–56. [PubMed] [Google Scholar]
- 25.Thibodeau SN, French AJ, Roche PC, Cunningham JM, Tester DJ, Lindor NM, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer research 1996;56(21):4836–40. [PubMed] [Google Scholar]
- 26.Gologan A, Sepulveda AR. Microsatellite instability and DNA mismatch repair deficiency testing in hereditary and sporadic gastrointestinal cancers. Clinics in laboratory medicine 2005;25(1):179–96 doi 10.1016/j.cll.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Jarvinen H, et al. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer research 2001;61(11):4545–9. [PubMed] [Google Scholar]
- 28.Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, et al. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn 2006;8(3):305–11 doi 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Loga K, Woolston A, Punta M, Barber LJ, Griffiths B, Semiannikova M, et al. Extreme intratumour heterogeneity and driver evolution in mismatch repair deficient gastro-oesophageal cancer. Nature Communications 2020;11(1):139 doi 10.1038/s41467-019-13915-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pursell ZF, Kunkel TA. DNA polymerase epsilon: a polymerase of unusual size (and complexity). Progress in nucleic acid research and molecular biology 2008;82:101–45 doi 10.1016/s0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran HT, Gordenin DA, Resnick MA. The 3’-->5’ exonucleases of DNA polymerases delta and epsilon and the 5’-->3’ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Molecular and cellular biology 1999;19(3):2000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N, et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nature communications 2015;6:8866- doi 10.1038/ncomms9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nature genetics 2016;48(6):600–6 doi 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing X, Kane DP, Bulock CR, Moore EA, Sharma S, Chabes A, et al. A recurrent cancer-associated substitution in DNA polymerase epsilon produces a hyperactive enzyme. Nat Commun 2019;10(1):374 doi 10.1038/s41467-018-08145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aksenova A, Volkov K, Maceluch J, Pursell ZF, Rogozin IB, Kunkel TA, et al. Mismatch Repair–Independent Increase in Spontaneous Mutagenesis in Yeast Lacking Non-Essential Subunits of DNA Polymerase ε. PLOS Genetics 2010;6(11):e1001209 doi 10.1371/journal.pgen.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdulovic AL, Hile SE, Kunkel TA, Eckert KA. The in vitro fidelity of yeast DNA polymerase delta and polymerase epsilon holoenzymes during dinucleotide microsatellite DNA synthesis. DNA repair 2011;10(5):497–505 doi 10.1016/j.dnarep.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sainudiin R, Durrett RT, Aquadro CF, Nielsen R. Microsatellite mutation models: insights from a comparison of humans and chimpanzees. Genetics 2004;168(1):383–95 doi 10.1534/genetics.103.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995;139(1):457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dieringer D, Schlötterer C. Two distinct modes of microsatellite mutation processes: evidence from the complete genomic sequences of nine species. Genome Res 2003;13(10):2242–51 doi 10.1101/gr.1416703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, Sauer-Eriksson AE, et al. Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat Struct Mol Biol 2014;21(1):49–55 doi 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- 41.Levinson G, Gutman GA. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic acids research 1987;15(13):5323–38 doi 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlotterer C, Tautz D. Slippage synthesis of simple sequence DNA. Nucleic acids research 1992;20(2):211–5 doi 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martín-López JV, Fishel R. The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome. Fam Cancer 2013;12(2):159–68 doi 10.1007/s10689-013-9635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, et al. Reconstitution of 5′-Directed Human Mismatch Repair in a Purified System. Cell 2005;122(5):693–705 doi 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 45.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human Mismatch Repair: RECONSTITUTION OF A NICK-DIRECTED BIDIRECTIONAL REACTION. The Journal of biological chemistry 2005;280(48):39752–61 doi 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A Defined Human System That Supports Bidirectional Mismatch-Provoked Excision. Molecular Cell 2004;15(1):31–41 doi 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 47.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. The Journal of biological chemistry 2009;284(7):4041–5 doi 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouffet E, Larouche V, Campbell BB, Merico D, Borja Rd, Aronson M, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol 2016;34(19):2206–11 doi 10.1200/jco.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 49.Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science (New York, NY) 2019;364(6439):485–91 doi 10.1126/science.aau0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19(3):133–50 doi 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (New York, NY) 2017;357(6349):409–13 doi 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51(2):202–6 doi 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkash V, Kulkarni Y, Ter Beek J, Shcherbakova PV, Kamerlin SCL, Johansson E. Structural consequence of the most frequently recurring cancer-associated substitution in DNA polymerase epsilon. Nat Commun 2019;10(1):373 doi 10.1038/s41467-018-08114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Modrich P. Mechanisms in eukaryotic mismatch repair. The Journal of biological chemistry 2006;281(41):30305–9 doi 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haye JE, Gammie AE. The Eukaryotic Mismatch Recognition Complexes Track with the Replisome during DNA Synthesis. PLOS Genetics 2015;11(12):e1005719 doi 10.1371/journal.pgen.1005719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Thuijl HF, Mazor T, Johnson BE, Fouse SD, Aihara K, Hong C, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol 2015;129(4):597–607 doi 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diouf B, Cheng Q, Krynetskaia NF, Yang W, Cheok M, Pei D, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat Med 2011;17(10):1298–303 doi 10.1038/nm.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González-Acosta M, Marín F, Puliafito B, Bonifaci N, Fernández A, Navarro M, et al. High-sensitivity microsatellite instability assessment for the detection of mismatch repair defects in normal tissue of biallelic germline mismatch repair mutation carriers. J Med Genet 2020;57(4):269–73 doi 10.1136/jmedgenet-2019-106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. The Lancet Oncology 2017;18(8):1009–21 doi 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 60.Maletzki C, Huehns M, Bauer I, Ripperger T, Mork MM, Vilar E, et al. Frameshift mutational target gene analysis identifies similarities and differences in constitutional mismatch repair-deficiency and Lynch syndrome. Molecular carcinogenesis 2017;56(7):1753–64 doi 10.1002/mc.22632. [DOI] [PubMed] [Google Scholar]
- 61.Maby P, Galon J, Latouche JB. Frameshift mutations, neoantigens and tumor-specific CD8(+) T cells in microsatellite unstable colorectal cancers. Oncoimmunology 2016;5(5):e1115943 doi 10.1080/2162402x.2015.1115943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakry D, Aronson M, Durno C, Rimawi H, Farah R, Alharbi QK, et al. Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency consortium. Eur J Cancer 2014;50(5):987–96 doi 10.1016/j.ejca.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Shuen AY, Lanni S, Panigrahi GB, Edwards M, Yu L, Campbell BB, et al. Functional Repair Assay for the Diagnosis of Constitutional Mismatch Repair Deficiency From Non-Neoplastic Tissue. Journal of Clinical Oncology 2019;37(6):461–70 doi 10.1200/jco.18.00474. [DOI] [PubMed] [Google Scholar]
- 64.Bodo S, Colas C, Buhard O, Collura A, Tinat J, Lavoine N, et al. Diagnosis of Constitutional Mismatch Repair-Deficiency Syndrome Based on Microsatellite Instability and Lymphocyte Tolerance to Methylating Agents. Gastroenterology 2015;149(4):1017–29.e3 doi 10.1053/j.gastro.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 65.Durno C, Boland CR, Cohen S, Dominitz JA, Giardiello FM, Johnson DA, et al. Recommendations on Surveillance and Management of Biallelic Mismatch Repair Deficiency (BMMRD) Syndrome: A Consensus Statement by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2017;152(6):1605–14 doi 10.1053/j.gastro.2017.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.