Abstract
Over the last decade, interest in the role of the microbiome in health and disease has increased. The use of germ-free animals and depletion of the microbial flora using antimicrobials are 2 methods commonly used to study the microbiome in laboratory mice. Germ-free mice are born, raised, and studied in isolators in the absence of any known microbes; however, the equipment, supplies, and training required for the use of these mice can be costly and time-consuming. The use of antibiotics to decrease the microbial flora does not require special equipment, can be used for any mouse strain, and is relatively inexpensive; however, mice treated in this manner still retain microbes and they do not live in a germ-free environment. One commonly used antibiotic cocktail regimen uses ampicillin, neomycin, metronidazole, and vancomycin in the drinking water for 2 to 4 wk. We found that the palatability of this mixture is low, resulting in weight loss and leading to removal of mice from the study. The addition of sucralose to the medicated water and making wet food (mash) with the medicated water improved intake; however, the low palatability still resulted in a high number of mice requiring removal. The current study evaluated a new combination of antibiotics designed to reduce the gut microbiota while maintaining body weights. C57BL/6NCrl mice were placed on one of the following drinking water regimens: ampicillin/neomycin/metronidazole/vancomycin water (n = 16), enrofloxacin/ampicillin water (n = 12), or standard reverse osmosis deionized water (RODI) (n = 11). During an 8 day regimen, mice were weighed and water consumption was measured. Feces were collected before and after 8 d of treatment. Quantitative real-time PCR (real-time qPCR) for 16S bacterial ribosome was performed on each sample, and values were compared among groups. The combination of enrofloxacin and ampicillin improved water intake, together with a greater reduction in gut flora.
Interest in the intestinal microbiome and its role in human health has increased dramatically over the last decade. The microbiota has been implicated in metabolic, infectious, and inflammatory disease, and its role has been investigated not only in the gut,2,8,18,38 but also in vasculature,5,6,19,39 kidney,13 liver,28 lung,9,34,37 and brain.12,15 Animal models have been important in furthering our understanding of the microbiota. Two approaches to studying microbiota in mice are the use of germ-free mice22,35,42 and depletion of the flora with oral administration of antibiotics.12,17,18 Both approaches have advantages and disadvantages. Germ-free mice are bred in isolators and are free of microorganisms from birth, allowing studies in mice with no microbes present; mice can then be used to generate gnotobiotic mice in which only known microbes are present. However, to remain germ-free, mice must be maintained in isolators under aseptic housing conditions, which is both costly and labor intensive. In addition, alterations of microbiota in early life may cause sustained effects on body composition10 and lasting negative consequences on the host immune system.31 A more economic approach has been to deplete mouse gut microflora using a combination of broad-spectrum antibiotics given either by oral gavage or in the drinking water. The primary limitation with antibiotic treatment of mice is that not all microbes are eliminated; which can potentially make reproducibility in certain types of studies such as those involving microbial transplantation29 very difficult. However, antibiotic-induced gut dysbiosis can be used on conventionally raised mice without the limitations imposed by maintaining a sterile living environment. Direct handling of the mice is possible, allowing behavioral and imaging assessments, which are not be feasible for mice housed in isolators. Several broad-spectrum antibiotic treatment regimens in the drinking water have been used for gut microbe depletion.7,16,20,25-27,41 One of the more commonly used combinations is comprised of 4 antibiotics (ampicillin, neomycin, metronidazole, and vancomycin) added to the drinking water for periods ranging from 1 to 4 wk.5,9,13,19,23,30,32,34 This cocktail is effective at depleting gut microbes; however, a previous study in our laboratory found it to be highly unpalatable. Dehydration and weight loss can occur in mice receiving antibiotics in the drinking water, and the magnitude of the effect can be significant, depending on the mouse strain.21,30,33 The weight loss can result in a substantial number of mice being removed from studies due to animal welfare concerns as reported in a previous study in which 5 of 5 mice given ampicillin, neomycin, metronidazole, and vancomycin reached eighty percent of baseline body weight and were subsequently removed.33 A reduction in water consumption is also likely to interfere with effective antibiotic treatment and may prolong the time necessary to achieve adequate microbial depletion. Palatability enhancers such as glucose,5,13 sucrose,41 and flavored water23 are sometimes combined with the antibiotics in drinking water. The aim of the current study was to determine whether 8 days of treatment with an alternative mixture comprised of 2 antibiotics (enrofloxacin and ampicillin) was sufficient to deplete the gut flora as compared with the widely used combination of ampicillin, neomycin, metronidazole, and vancomycin. We hypothesized that the combination of 2 antibiotics would be at least equivalent to the combination of 4 antibiotics in reducing the gut flora while causing less weight loss.
Materials and Methods
Animals and Housing.
All animal procedures were reviewed and approved by National Institute of Environmental Health Sciences Animal Care and Use Committee. In this study, 6 to 8-wk old female C57BL/6 mice (n = 40) (Charles River Laboratories, Raleigh, NC) were identified by ear punch and housed 4 per cage in individually ventilated cages on a double-sided rack (Techniplast, Exton, PA). Females were used due to availability, as they were purchased from an investigator who had an excess number of mice and could readily be cohoused. Prior to use, each cage was autoclaved with hardwood bedding (Sani-chips, PJ Murphy, Montville, NJ), NIH-31 rodent diet (Harlan Laboratories, Madison, WI), Enviro-dri nesting material (Shepherd Specialty Papers, Watertown, TN) and cotton squares (Ancare, Bellmore, NY). Autoclaved caging was used throughout the study, with cages changed on day 0, day 4, and day 7. Cages were changed on day 4 and day 7 to minimize bacteria in the environment. Experiments were conducted in an AAALAC, International accredited facility under constant environmental conditions (70 to 73 °F, relative humidity 40% to 60%, 12:12 h light:dark cycle (0700 to 1900 EST). All mice were negative on receipt from the vendor for rodent murine pathogens that include EDIM, MHV, MNV, MPV, MVM, MS-1, M. pulmonis, PVM, Reovirus, Sendai, TMEV, CARbacillus, Ectromelia, E. cuniculi, Hantavirus, K virus, LCMV, LDEV, Adenovirus 1 and 2, MCMV, MTV, POL, Helicobacter spp, and endo and ectoparasites. Sentinel mice remained negative during the experiment.
Antibiotics.
Antibiotics were dissolved in autoclaved reverse osmosed, deionized (RODI) water. All antibiotics used were pharmaceutical grade except for neomycin, which we could not find in a USP grade that would readily dissolve in water.
Three cages (n = 12 mice) received drinking water containing 0.575 mg/mL of enrofloxacin (Enroflox, Norbook Laboratories Limited, Newry, Northern Ireland, UK) and 1 mg/mL ampicillin (Putney, Portland, ME) (2-AB group).
Four cages (n = 16 mice) received RODI containing 1mg/mL each of ampicillin (Putney, Portland, ME) neomycin (Millipore, Burlington, MA), metronidazole (Letco Medical, LLC, Decatur, AL), and 0.5 mg/mL vancomycin (Mylan Pharmaceuticals, Morgantown, WV) sweetened with the addition of 1:50 MediDrop Sucralose (Clear H2O, Westbrook, ME) (4-AB group). The 4-AB group also received supplemental mash prepared daily using ground feed and 4-AB water to counter poor palatability and water consumption.
Three cages (n = 11 mice) were the control group and received autoclaved RODI water (RODI group). One mouse in the RODI water control group died prior to the start of the study (necropsy did not reveal cause of death); therefore, 2 cages contained 4 mice and one cage contained 3 mice.
Body weight, water consumption, and feces collection.
Body weights were recorded on days 0, 1, 4, 5, 6, 7 and 8, and water consumption was measured on days 1, 5, 6, 7 and 8. The volume of water consumed was determined by taking the weight of the bottle and subtracting it from the measurement obtained at the previous measurement. Fresh feces were collected aseptically from each mouse, flash frozen with liquid nitrogen and stored at -80 °C on day 0 and day 8. On day 0, mice were manually restrained, and feces were collected directly from the anus using forceps. Forceps were soaked in 70% EtOH and allowed to dry between animals. On day 8, mice were euthanized by CO2 asphyxiation, cervically dislocated, and feces were collected from the rectum and distal colon to ensure adequate volume for DNA extraction and quantification. Sterile 2 mL screw cap microtubes (Sarstedt, Nümbrecht, Germany) were used for flash freezing and storage.
Macroscopic assessment of cecum.
A photograph of the gastrointestinal tract was taken of one animal from each of the 3 groups after euthanasia and before feces collection on day 8 to record gross changes after antibiotic treatment.
DNA Extraction and real-time qPCR.
DNA extraction and quantitative, real-time PCR (real-time qPCR) targeting the hypervariable region 3 of the 16S ribosomal RNA (HV3-16S) gene was done on collected feces. Total genomic DNA was extracted from feces using the Qiagen QIAmp PowerFecal DNA Kit (Hilden, Germany). DNA quantity was determined fluorometrically using the DeNovix – dsDNA Broad Range – Fluorescence Quantification Assay and the DeNovix DS-11+ Fluorometer (Wilmington, DE). DNA samples were diluted to a final concentration of 1 ng/µL using UltraPure DNase/RNase-Free Distilled Water (Invitrogen, Thermo Fisher Scientific, Waltham, MA). Real-time qPCR was performed on each sample in triplicate using 8 µL (8 ng of total DNA template) in combination with SsoAdvanced Universal Inhibitor-Tolerant SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) in accordance with the manufacture's recommended guidelines. The forward HV3-16S primer 5′CCAGACTCCTACGGGAGGCAG-3′ and the reverse HV3-16S primer 5′-CGTATTACCGCGGCTGCTG-3′ (10µM) were added to the supermix. Real-time qPCR was carried out using a Bio-Rad CFX96 thermal cycler (Bio-Rad, Hercules, CA). Reaction mixtures (20 µL total volume) were held at 98 °C for 3 min, followed by 40 cycles at 98 °C for 15 s and 60 °C for 30 s followed by a melting curve to verify nonspecific amplification. Plasmids verified to carry a HV3-16S insert for standard curve generation were constructed using a TOPO TA Cloning Kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA) and gene inserts were obtained using genomic DNA from E. coli strain ATCC 10536.
Mice/ Samples Removed from Study.
One mouse from the 2-AB group and 3 mice from the 4-AB group exceeded the weight loss limit of 20% of starting weight and were removed from the study, therefore, fecal data for these 4 mice was not included in the analysis. The mouse from the 2-AB group was removed from study on day 5 and the 3 mice from the 4-AB group were removed on day 7. DNA extraction was unsuccessful for two 2-AB samples and one 4-AB sample on day 0, so these samples were excluded from the analysis.
Statistics.
Statistical analyses were performed with Prism version 8.2.1 (GraphPad, La Jolla, CA). A 2-way repeated-measures analysis of variance (ANOVA) was used to compare differences in water consumption, and a mixed-effects model was used to compare differences in body weights with group and day as the main factors. Post hoc comparisons using Tukey test were done to compare water consumption and body weights between the groups. The number of copies of bacteria present in the feces were log-transformed before statistical analysis due to the skewed distribution. An ordinary one-way ANOVA was used to compare differences in the log-transformed numbers; post hoc comparisons using Tukey test were done to compare differences between and among the groups before and after antibiotic treatment. Differences were considered significant at the 0.05 level (P < 0.05). Results are expressed as mean ± SE.
Results
Water Consumption.
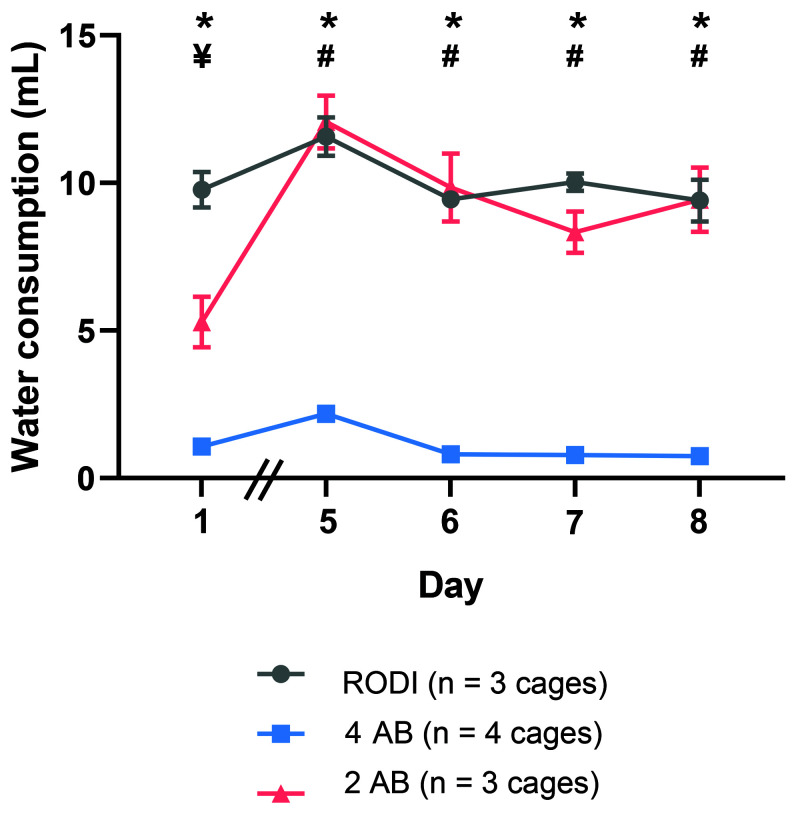
Water consumption was monitored throughout the study. Water consumption was used as an indirect measure of the palatability of the water. Mice were group housed; therefore, water consumption was monitored from the whole cage rather than for individual mice. A significant difference in water consumption (F (2,7) = 93.96; P < 0.0001) was detected among the groups (Figure 1). The 4-AB group consumed significantly less water than the RODI group at each timepoint (P < 0.05) and less water than the 2-AB group on days 5 to 8 (P < 0.05). The 2-AB group consumed less water than the RODI group on day 1 (P < 0.05) but this decrease was not observed on subsequent days. These data show that water consumption is higher in the 2-AB group than the 4-AB group, suggestive of differences in palatability.
Figure 1.
Water consumption in a cage housing 4 mice day 1 and days 5–8. Mice were placed on antibiotic water on day 0. Mice given the 4 AB cocktail consumed significantly less water than mice given either the 2 AB cocktail or RODI water (P < 0.0001). A significant decrease in water consumption was observed in mice given the 2 AB cocktail compared with mice given RODI water on day 1. No difference was observed between these 2 groups on days 5–8. Mice given the 4AB cocktail consumed significantly less water than mice given the RODI water on day 1 and days 5–8. The 4AB group also consumed less water than the 2 AB group on days 5–8. *P < 0.05, comparison between RODI water control group and 4 AB group. ¥P < 0.05, comparison between RODI water control group and 2 AB group. #P < 0.05, comparison between 2 AB group and 4 AB group. Data represents mean ± SEM.
Body Weight.
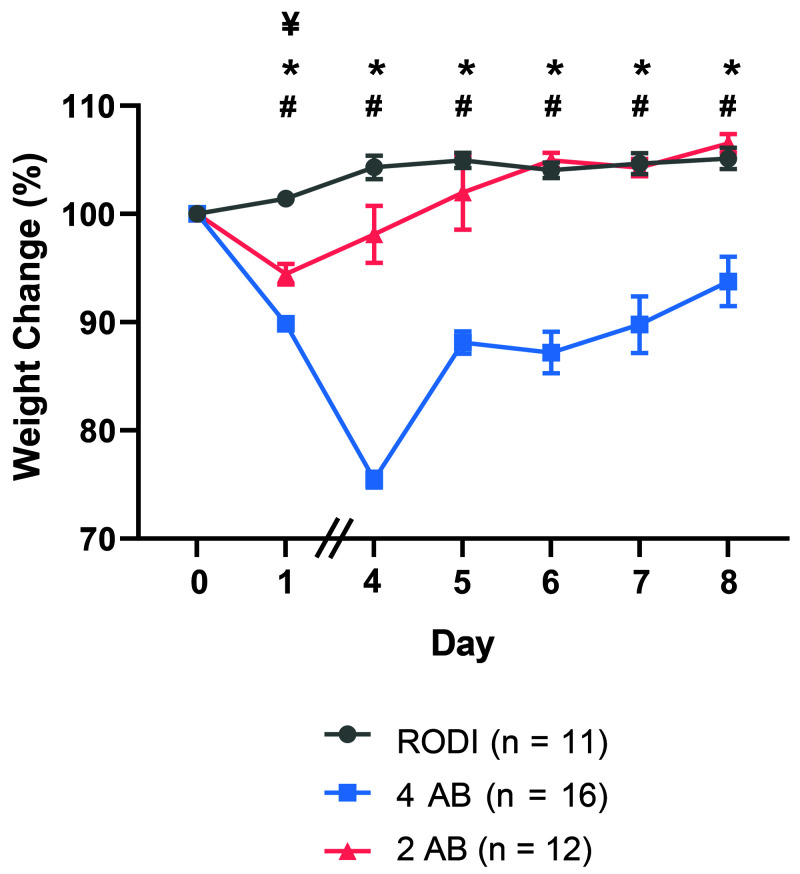
The percent weight loss was monitored to compare the effects of the 4-AB and 2-AB antibiotic water combinations on changes in body weight. On day 0, mice in the RODI group had a mean body weight of 17.8 g, mice in the 4 AB group weighed 18.4 g, and the 2 AB group weighed 17.9 g. On day 8, mice in these groups had mean weights of 18.7 g, 17.1 g, and 19.1 g, respectively. A significant difference in body weight (F (2,36) = 47.09; P < 0.0001) was detected among the groups (Figure 2). The 4-AB group lost significantly more weight than did the RODI group on day 1 (P < 0.0001) and on days 4 to 8 (P < 0.05). The 4-AB also lost significantly more body weight than the 2-AB group on day 1 (P < 0.01) and days 4 to 8 (P < 0.05). The 2-AB group lost significantly more weight than did the RODI group on day 1 (P < 0.0001). No differences were detected between the RODI and 2-AB groups on days 4 to 8. Weight loss associated with the 4-AB water was not observed with the 2-AB water after day 1.
Figure 2.
Percent weight change on day 1 and days 4–8 compared with starting weight (day 0). Mice were placed on antibiotic water on day 0. Mice given the 4 AB cocktail lost significantly more weight than mice given either the 2 AB cocktail or the RODI water (P < 0.0001). Mice given the 4AB cocktail lost significantly more weight than mice given RODI water and the 2 AB cocktail on day 1 and days 4–8. Mice given the 2 AB cocktail lost a significant amount of weight compared with mice given RODI water on day 1. *P < 0.05, comparison between RODI water control group and 4 AB group. #P < 0.05, comparison between 2 AB group and 4 AB group. ¥P < 0.05, comparison between RODI water control group and 2 AB group. Data represents mean ± SEM.
Real-time qPCR for Microbial Flora.
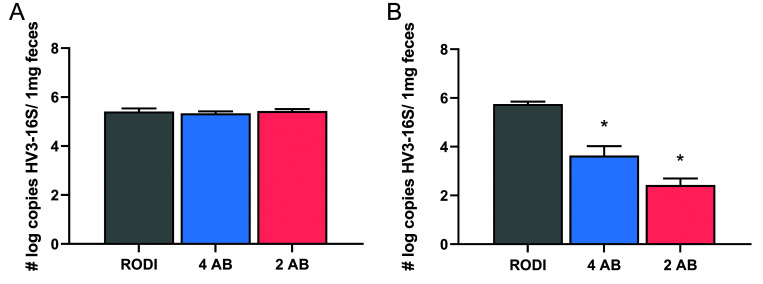
Real-time qPCR was conducted using feces collected on day 0 and day 8 to compare the efficacy of the 2 AB and 4 AB antibiotic combinations in reducing the microbial flora in the gut. Due to the skewed distribution, the copy numbers per mg of feces were log-transformed before comparison. A significant difference (F (5,56) = 34.08; P < 0.0001) was detected in the number of bacteria in the feces between and among groups (Figure 3). Mice in the 2-AB group (P < 0.0001) and 4-AB group (P < 0.0001) both had significant reductions in bacteria as compared with the RODI group on day 8. A significant reduction in bacteria was also found in the 2-AB group on day 8 compared with day 0 and in the 4-AB group on day 8 as compared with day 0 (both P < 0.0001). The 2-AB group had significantly fewer bacteria than the 4-AB group on day 8 (P < 0.01). No changes were detected in microbial flora in the RODI group on day 8 compared with day 0, and no differences observed in bacterial load among the 3 groups on day 0.
Figure 3.
Number log copies of HV3-16S ribosomal RNA gene per 1mg feces in mice before and after 8 d of antibiotic water treatment. Feces were collected on the day antibiotic treatment began (day 0) (RODI n = 11; 4 AB n = 12; 2 AB n = 9) and after 8 d of antibiotic treatment (day 8) (RODI n = 11; 4 AB n = 13; 2 AB n = 11). Microbial numbers were significantly lower in the 4 AB group (P < 0.0001) and 2 AB group (P < 0.0001) compared with the RODI group on day 8. A significant reduction in microbial numbers was also found within the 2 AB and 4 AB groups on day 8 compared with Day 0 (P < 0.0001). On day 8, the 2 AB group had significantly fewer bacteria in their feces than the 4 AB group (P = 0.0033). No differences were observed within the RODI group between day 0 and day 8 and no differences between any groups on day 0. *P < 0.05; Data represents mean ± SEM.
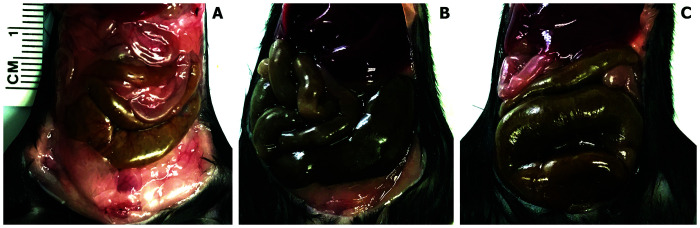
Macroscopic assessment of cecum.
Cecum enlargement was observed at necropsy on day 8 in both the 2-AB and 4-AB groups as compared with the RODI group (Figure 4).
Figure 4.
Gross examination of the cecum in the RODI water control group (A), 2 AB group (B), and 4 AB group (C). Enlargement was observed in both (2 AB and 4 AB) antibiotic treated groups compared with the control group following 8 d of treatment through the drinking water.
Discussion
As the role of the microbiome and its effects on human health and disease continue to be investigated, studies in animals will likely increase in the coming years. Germ-free mouse models are generally considered to be the gold standard in human gut microbiota research, but due to the significant limitations of this approach, including the expense associated with housing and contamination monitoring24 alternate models including oral administration of antibiotics are often used. Two commonly used oral administration routes are delivering the antibiotics via gavage and adding them to the drinking water. Gavage allows for a higher, more precise dose of antibiotics than administration through the drinking water; however, this approach is more labor intensive and the process, especially when repeated, can be stressful for the mice.3 One investigation showed that antibiotic administration via oral gavage effectively depleted intestinal microbiota, producing a similar phenotype to that found in germ-free mice but required twice daily dosing for a 17 d period.33 The germ free-like phenotype was not observed in mice placed on antibiotic water.33 However, discrepancies in research findings have been identified when using germ-free and antibiotic models.6,15 Administration of antibiotics through the drinking water does not produce as robust a response in gut flora depletion as does delivery via gavage, nor will it necessarily produce the same phenotypes seen in germ-free mice. However, it has nonetheless been an acceptable complimentary model to germ-free mice36 and is the most common way of administering gut flora depleting cocktails in mice at our institute.
Ampicillin, neomycin, metronidazole, and vancomycin are frequently used as a broad-spectrum oral antibiotic treatment and are bactericidal against gram positive (ampicillin and vancomycin), gram negative (ampicillin and neomycin), and anaerobic bacteria (ampicillin and metronidazole). A 4 wk regiment of oral antibiotics is necessary for our investigators to get adequate depletion of the microflora. In our experience, mice can exhibit severe weight loss under these circumstances. The weight loss observed in mice placed on drinking water containing these 4 antibiotics raises welfare concerns that reduced water and food intake necessitates placing enough mice on study to offset those removed due to weight loss limits.
The aim of the current study was to compare the commonly used 4-AB cocktail in the drinking water to a 2-AB cocktail in the drinking water in terms of depleting the number of microbes and determining how many days would be required to reduce the number of microbes to a level acceptable to our investigators. For this study we included an extra cage of mice (n = 4) to the 4-AB group because we anticipated having to remove mice due to weight loss and wanted to ensure that we had at least 12 mice for day 8 feces collection. We also wanted to identify a combination of antibiotics that was more readily consumed by mice. Water consumption has been used as an indicator of palatability.14,15 Preliminary work in our laboratory found that enrofloxacin and ampicillin were well tolerated in the drinking water. The combination of enrofloxacin and ampicillin is effective against gram negative and gram-positive species and some anaerobes. We compared the efficacy of this 2-antibiotic combination to the ampicillin, neomycin, metronidazole, and vancomycin cocktail with regard to decreasing the number of gut microbiota.
In this study we found a significant reduction in water consumption in mice given the 4-AB cocktail compared with the control group at all time points. The 2-AB group consumed less water than did the control group during the first 24 h. This may have been due to neophobia, as the water consumption was not different from the control group at all other time points. The 4-AB group consumed less water than the 2-AB group on days 5 to 8. As water consumption was used as an indirect measure of palatability;14 these results suggest the 4-AB cocktail was less palatable than the 2-AB cocktail. The reduced water consumption by the 4-AB group coincided with a reduction in body weight. The 4-AB group lost a significant amount of weight compared with the RODI group on all days and the 2-AB group on day 1 and days 4 to 8. This weight loss occurred despite access to supplemental mash that was not provided to the other 2 groups. The observed weight loss was likely a result of diminished food consumption and dehydration associated with reduced water intake. Our data show that improving the palatability of the water will not only reduce the need to remove mice from study due to weight loss but also improves overall animal welfare.
Cecum enlargement has been previously reported in mice treated with antibiotics.17,33 We saw similar changes in the cecum in both antibiotic groups as compared with the RODI group. Cecal distention has also been reported in germ-free rats.40 Prolonged gastric emptying has been reported in germ-free mice as compared with conventionally raised mice1 which likely accounts for the gross observations. The mechanisms affecting intestinal motility are speculated to include the release of bacterial substances or end products of bacterial fermentation, intestinal neuroendocrine factors, and mediators released by the gut immune system.4
Real-time qPCR analysis of feces showed a significant reduction in gut microbiota in both the 2-AB and 4-AB groups as compared with the RODI group after 8 d of treatment with antibiotics in the drinking water. This depletion was more robust in the 2-AB group than in the 4-AB group, suggesting that the 2-AB cocktail is more efficient at depleting gut microbiota than is the widely used 4-AB cocktail. Significant reductions were also found in the 2-AB and 4-AB groups on day 8 as compared with day 0. A longer course of treatment using the 4-AB cocktail would likely be necessary to achieve reductions similar to those seen in the 2-AB group. Most of the protocols we reviewed using the 4-AB cocktail in the drinking water ranged from 2 to 4 wk.9,13,19,23,32,34 Based on our findings, we would not recommend using the 4-AB cocktail for 8 d. Although significant reductions in bacteria were achieved in the 4-AB group on day 8 as compared with day 0, levels may not have been sufficiently depleted for certain microbiome studies, depending on the hypothesis being tested. We achieved a significantly greater reduction using the 2-AB cocktail within the 8 day timeframe due to greater antibiotic consumption ass compared with the 4-AB group. However, further reduction of the microbiome might be achieved in the 4-AB. In our experience, mice are removed from study due to extreme weight loss (greater than 20%), resulting in poor animal welfare and requiring the use of more mice. A more extended treatment with 2-AB cocktail could further reduce the microflora and may be examined in the future. Finding a shorter treatment period to allow our investigators to shorten their experimental timeline was one aim of this study. In addition, a shorter treatment regimen may be preferential because prolonged antibiotic administration is a significant risk factor for the emergence and colonization of antibiotic-resistant bacteria.11
We did not examine specific phyla of bacteria that were reduced. 16S does not identify the bacterial species, nor does it differentiate between live and dead bacteria. The species ratio (anaerobes: aerobes) of remaining microflora may be altered between the 2 treatment groups, as metronidazole is included in the 4-AB cocktail to target anaerobic bacteria.
This study has shown that a 2-antibiotic combination (enrofloxacin and ampicillin) can be used to reduce the number of microbial flora more effectively than does the commonly used 4-antibiotic cocktail (ampicillin, neomycin, metronidazole, and vancomycin) when administered in the drinking water for 8 d. In addition, we have shown that this 2-antibiotic cocktail is more readily consumed by mice with minimal changes in body weight and fluid consumption. With improved palatability, mice more readily drank the water, increasing their daily dose and decreasing the time required to achieve sufficient microbiota depletion. This 2-AB regimen is recommended over the 4-AB regimen for laboratories using antibiotics to reduce the gut flora, as the 2-AB regimen quickly reduces the microbial flora with minimal animal welfare issues.
Acknowledgments
This research was supported [in part] by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. The authors are grateful for the real-time qPCR training and consultation with Michael Johnston in the Quality Assurance Laboratory, animal husbandry assistance from Jenetta Jackson in the Animal Resource Section, statistical analyses by Dr Min Shi in the Biostatistics & Computational Biology Branch, and the literature review assistance from Sharon Bolger and Stacey Mantooth in the NIEHS Library.
References
- 1.Abrams GD, Bishop JE. 1967. Effect of the normal microbial flora on gastrointestinal motility. Proc Soc Exp Biol Med 126:301–304. 10.3181/00379727-126-32430. [DOI] [PubMed] [Google Scholar]
- 2.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. 2008. ATP drives lamina propria T(H)17 cell differentiation. Nature 455:808–812. 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 3.Balcombe JP, Barnard ND, Sandusky C. 2004. Laboratory routines cause animal stress. Contemp Top Lab Anim Sci 43:42–51. [PubMed] [Google Scholar]
- 4.Barbara G, Stanghellini V, Brandi G, Cremon C, Di Nardo G, De Giorgio R, Corinaldesi R. 2005. Interactions between commensal bacteria and gut sensorimotor function in health and disease. Am J Gastroenterol 100:2560–2568. 10.1111/j.1572-0241.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 5.Battson ML, Lee DM, Jarrell DK, Hou S, Ecton KE, Weir TL, Gentile CL. 2018. Suppression of gut dysbiosis reverses Western diet-induced vascular dysfunction. Am J Physiol Endocrinol Metab 314:E468–E477. 10.1152/ajpendo.00187.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer F, Ascher S, Pontarollo G, Reinhardt C. 2019. Antibiotic treatment protocols and germ-free mouse models in vascular research. Front Immunol 10:1–7. 10.3389/fimmu.2019.02174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrello C, Garavaglia F, Cribiu FM, Ercoli G, Bosari S, Caprioli F, Facciotti F. 2018. Short-term oral antibiotics treatment promotes inflammatory activation of colonic invariant natural killer t and conventional CD4+ T cells. Front Med (Lausanne) 5:1–12. 10.3389/fmed.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho Y, Osgood RS, Bell LN, Karoly ED, Shore SA. 2019. Ozone-induced changes in the serum metabolome: Role of the microbiome. PLoS One 14:1–26. 10.1371/journal.pone.0221633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zarate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. 2014. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158:705–721. 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennesen PJ, van der Ven AJ, Kessels AG, Ramsay G, Bonten MJ. 2001. Resolution of infectious parameters after antimicrobial therapy in patients with ventilator-associated pneumonia. Am J Respir Crit Care Med 163:1371–1375. 10.1164/ajrccm.163.6.2007020. [DOI] [PubMed] [Google Scholar]
- 12.Desbonnet L, Clarke G, Traplin A, O'Sullivan O, Crispie F, Moloney RD, Cotter PD, Dinan TG, Cryan JF. 2015. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav Immun 48:165–173. 10.1016/j.bbi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Emal D, Rampanelli E, Stroo I, Butter LM, Teske GJ, Claessen N, Stokman G, Florquin S, Leemans JC, Dessing MC. 2017. Depletion of gut microbiota protects against renal ischemia-reperfusion injury. J Am Soc Nephrol 28:1450–1461. 10.1681/ASN.2016030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ezell PC, Papa L, Lawson GW. 2012. Palatability and treatment efficacy of various ibuprofen formulations in C57BL/6 mice with ulcerative dermatitis. J Am Assoc Lab Anim Sci 51:609–615. [PMC free article] [PubMed] [Google Scholar]
- 15.Fröhlich EE, Farzi A, Mayerhofer R, Reichmann F, Jacan A, Wagner B, Zinser E, Bordag N, Magnes C, Frohlich E, Kashofer K, Gorkiewicz G, Holzer P. 2016. Cognitive impairment by antibiotic-induced gut dysbiosis: Analysis of gut microbiota-brain communication. Brain Behav Immun 56:140–155. 10.1016/j.bbi.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. 2012. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 37:171–186. 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Ge X, Ding C, Zhao W, Xu L, Tian H, Gong J, Zhu M, Li J, Li N. 2017. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J Transl Med 15:1–9. 10.1186/s12967-016-1105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasa L, Abecia L, Forcen R, Castro M, de Jalon JA, Latorre E, Alcalde AI, Murillo MD. 2015. Antibiotic-induced depletion of murine microbiota induces mild inflammation and changes in toll-like receptor patterns and intestinal motility. Microb Ecol 70:835–848. 10.1007/s00248-015-0613-8. [DOI] [PubMed] [Google Scholar]
- 19.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA, Lusis AJ, Hazen SL. 2015. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem 290:5647–5660. 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, Shapiro H, Dori-Bachash M, Pevsner-Fischer M, Lorenzo-Vivas E, Keren-Shaul H, Paul F, Harmelin A, Eberl G, Itzkovitz S, Tanay A, Di Santo JP, Elinav E, Amit I. 2016. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166:1231–1246.13. 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 21.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. 2010. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 3:148–158. 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jäckel S, Kiouptsi K, Lillich M, Hendrikx T, Khandagale A, Kollar B, Hörmann N, Reiss C, Subramaniam S, Wilms E, Ebner K, Bruhl MV, Rausch P, Baines JF, Haberichter S, Lammle B, Binder CJ, Jurk K, Ruggeri ZM, Massberg S, Walter U, Ruf W, Reinhardt C. 2017. Gut microbiota regulate hepatic von Willebrand factor synthesis and arterial thrombus formation via Toll-like receptor-2. Blood 130:542–553. 10.1182/blood-2016-11-754416. [DOI] [PubMed] [Google Scholar]
- 23.Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. 2017. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood 129:729–739. 10.1182/blood-2016-03-708594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy EA, King KY, Baldridge MT. 2018. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol 9:1–16. 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15:374–381. 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SH, Cho BH, Kiyono H, Jang YS. 2017. Microbiota-derived butyrate suppresses group 3 innate lymphoid cells in terminal ileal Peyer's patches. Sci Rep 7:1–12. 10.1038/s41598-017-02729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, Pevsner-Fischer M, Shapiro H, Christ A, Harmelin A, Halpern Z, Latz E, Flavell RA, Amit I, Segal E, Elinav E. 2015. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163:1428–1443. 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Hao X, Chen Y, Bai L, Gao X, Lian Z, Wei H, Sun R, Tian Z. 2017. The microbiota maintain homeostasis of liver-resident γδT-17 cells in a lipid antigen/CD1d-dependent manner. Nat Commun 7:13839. 10.1038/ncomms13839. Erratum: Nat Commun 2017. 8: 15265. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundberg R, Toft MF, August B, Hansen AK, Hansen CHF. 2016. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes 7:68–74. 10.1080/19490976.2015.1127463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. 2009. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol 183:6041–6050. 10.4049/jimmunol.0900747 PubMed [DOI] [PubMed] [Google Scholar]
- 31.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489–493. 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118:229–241. 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen FE. 2011. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 6:1–13. 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuijt TJ, Lankelma JM, Scicluna BP, Sousa e Melo FD, Roelofs JJTH, Daan de Boer J, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, de Vos WM, van der Poll T, Joost Wiersinga W. 2016. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65:575–583. 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skye SM, Zhu W, Romano KA, Guo CJ, Wang Z, Jia X, Kirsop J, Haag B, Lang JM, DiDonato JA, Tang WHW, Lusis AJ, Rey FE, Fischbach MA, Hazen SL. 2018. Microbial transplantation with human gut commensals containing CutC is sufficient to transmit enhanced platelet reactivity and thrombosis potential. Circ Res 123:1164–1176. 10.1161/CIRCRESAHA.118.313142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley C, Kaiser T, Beura LK, Hamilton MJ, Weingarden AR, Bobr A, Kang J, Masopust D, Sadowsky MJ, Khoruts A. 2017. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5:1–13. 10.1186/s40168-017-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ, Boon ACM, Lenschow DJ, Stappenbeck TS. 2017. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357:498–502. 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Tian F, Wang P, Zheng H, Zhang Y, Tian H, Zhang L, Gao X, Wang X. 2018. Gut Microbiota as a modulator of paneth cells during parenteral nutrition in mice. JPEN J Parenter Enteral Nutr 42:1280–1287. 10.1002/jpen.1162. [DOI] [PubMed] [Google Scholar]
- 39.Winek K, Engel O, Koduah P, Heimesaat MM, Fischer A, Bereswill S, Dames C, Kershaw O, Gruber AD, Curato C, Oyama N, Meisel C, Meisel A, Dirnagl U. 2016. Depletion of cultivatable gut microbiota by broad-spectrum antibiotic pretreatment worsens outcome after murine stroke. Stroke 47:1354–1363. 10.1161/STROKEAHA.115.011800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wostmann B, Bruckner-Kardoss E. 1959. Development of cecal distention in germ-free baby rats. Am J Physiol 197:1345–1346. 10.1152/ajplegacy.1959.197.6.1345. [DOI] [PubMed] [Google Scholar]
- 41.Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, Sartor BR, Aliprantis AO, Charles JF. 2016. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci U S A 113:E7554–E7563. 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL. 2016. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165:111–124. 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]