ABSTRACT
Measles virus (MeV), an enveloped RNA virus in the family Paramyxoviridae, is still an important cause of childhood morbidity and mortality worldwide. MeV usually causes acute febrile illness with skin rash, but in rare cases persists in the brain, causing a progressive neurological disorder, subacute sclerosing panencephalitis (SSPE). The disease is fatal, and no effective therapy is currently available. Although transsynaptic cell-to-cell transmission is thought to account for MeV propagation in the brain, neurons do not express the known receptors for MeV. Recent studies have shown that hyperfusogenic changes in the MeV fusion (F) protein play a key role in MeV propagation in the brain. However, how such mutant viruses spread in neurons remains unexplained. Here, we show that cell adhesion molecule 1 (CADM1; also known as IGSF4A, Necl-2, and SynCAM1) and CADM2 (also known as IGSF4D, Necl-3, SynCAM2) are host factors that enable MeV to cause membrane fusion in cells lacking the known receptors and to spread between neurons. During enveloped virus entry, a cellular receptor generally interacts in trans with the attachment protein on the envelope. However, CADM1 and CADM2 interact in cis with the MeV attachment protein on the same cell membrane, causing the fusion protein triggering and membrane fusion. Knockdown of CADM1 and CADM2 inhibits syncytium formation and virus transmission between neurons that are both mediated by hyperfusogenic F proteins. Thus, our results unravel the molecular mechanism (receptor-mimicking cis-acting fusion triggering) by which MeV spreads transsynaptically between neurons, thereby causing SSPE.
IMPORTANCE Measles virus (MeV), an enveloped RNA virus, is the causative agent of measles, which is still an important cause of childhood morbidity and mortality worldwide. Persistent MeV infection in the brain causes a fatal progressive neurological disorder, subacute sclerosing panencephalitis (SSPE), several years after acute infection. However, how MeV spreads in neurons, which are mainly affected in SSPE, remains largely unknown. In this study, we demonstrate that cell adhesion molecule 1 (CADM1) and CADM2 are host factors enabling MeV spread between neurons. During enveloped virus entry, a cellular receptor generally interacts in trans with the attachment protein on the viral membrane (envelope). Remarkably, CADM1 and CADM2 interact in cis with the MeV attachment protein on the same membrane, triggering the fusion protein and causing membrane fusion, as viral receptors usually do in trans. Careful screening may lead to more examples of such “receptor-mimicking cis-acting fusion triggering” in other viruses.
KEYWORDS: SSPE, cis-acting, evolution, fusion triggering, host factor, measles virus, receptor, subacute sclerosing panencephalitis
INTRODUCTION
Measles, an acute febrile illness with skin rash, is still an important cause of childhood morbidity and mortality worldwide (1, 2). The causative agent of the disease is measles virus (MeV), an enveloped RNA virus in the family Paramyxoviridae. Persistent MeV infection in the brain causes subacute sclerosing panencephalitis (SSPE), a progressive fatal neurological disorder (2, 3). The first symptoms of SSPE, behavioral and intellectual disabilities, insidiously emerge 4 to 10 years after acute measles, followed by paroxysmal movements, myoclonic jerks, and/or negative myoclonus (2). Progressive neurological deterioration and eventual death generally occur within 4 years of diagnosis (2). The incidence of SSPE is 6.5 to 11 per 100,000 cases of measles (2), and the disease remains incurable due to the lack of effective therapies (4).
MeV bears two envelope glycoproteins, the hemagglutinin (H) and fusion (F) proteins (3, 5). Upon binding a receptor on the cell surface, the H protein triggers conformational changes of the F protein, causing the fusion of the envelope with the cell membrane and subsequent delivery of the viral genome into the cell. The H and F proteins are also expressed on the surface of infected cells, inducing syncytia via cell-to-cell fusion with neighboring receptor-positive cells.
The signaling lymphocytic activation molecule family member 1 (SLAMF1; also known as SLAM and CD150) on immune cells and nectin-4 on epithelial cells act as receptors for MeV (6–8). MeV initially infects SLAMF1+ immune cells in the respiratory tract and subsequently spreads to the whole lymphatic organs (9). At the later stage of infection, MeV is transmitted to nectin-4+ epithelial cells, resulting in shedding into the airway lumen and transmission to the next host (7, 8). Thus, SLAMF1 and nectin-4 nicely explain how MeV causes measles. On the other hand, MeV mainly infects neurons lacking SLAMF1 and nectin-4 in SSPE (3, 10, 11), suggesting the presence of unidentified neuronal receptors.
Importantly, genomic changes are required for persistent MeV brain infection, because wild-type (WT) MeV isolates from acute measles are not neurotropic (12). The genomes of MeV isolates from SSPE patients are indeed hypermutated (13). Since the mutations generally preclude the production of free MeV particles (13), transsynaptic cell-to-cell transmission is thought to account for MeV propagation in neurons (14–17). The mutations also cause specific substitutions in the ectodomain of the F protein (e.g., T461I), enabling the virus to spread in primary human neurons in vitro as well as in the brains of experimentally infected mice and hamsters (4, 12, 17–22). These substitutions destabilize the prefusion form of the F protein, rendering it hyperfusogenic (12, 23). F proteins containing such substitutions, but not the WT F protein, can induce membrane fusion in SLAMF1- and nectin-4-negative cells, when expressed together with the WT H protein (19).
Recently, by employing so-called “receptor-blind” MeV H proteins that have substitutions within the receptor binding sites, we demonstrated that the weak interaction between the SLAMF1-blind H protein and SLAMF1 or between the nectin-4-blind H protein and nectin-4 could support membrane fusion mediated by hyperfusogenic and unstable mutant F proteins, but not the WT F protein (24). Unexpectedly, the cis, but not trans, interaction of the SLAMF1-blind H protein with SLAMF1 could trigger the hyperfusogenic F(T461I) protein for membrane fusion where cell-to-cell contact is maintained (24).
We reasoned that neuronal MeV receptors could also weakly interact in cis with the H protein. In this study, we examined both cis- and trans-acting molecules and identified cell adhesion molecule 1 (CADM1; also known as IGSF4A, Necl-2, and SynCAM1) and CADM2 (also known as IGSF4D, Necl-3, and SynCAM2) as host factors that allow neuropathogenic MeVs to cause membrane fusion through cis-acting fusion triggering.
RESULTS
CADM1 and CADM2 induce membrane fusion with hyperfusogenic F proteins.
We first looked for MeV neuronal receptors using recombinant MeVs bearing the hyperfusogenic mutant F proteins, but failed to identify them despite years of effort. This made us adopt a different approach. SLAMF1 is a member of the SLAM family of receptors (25), whereas nectin-4 is a member of the nectin family of cell adhesion molecules (26). These families belong to the immunoglobulin superfamily, and their members have closely related structures and sequences, exhibiting homophilic and/or heterophilic interactions within respective families (25, 26). Thus, we hypothesized that other molecules expressed in human brains from the SLAM and nectin families act as neuronal receptors in combination with hyperfusogenic F proteins, because they may weakly interact with the WT H protein.
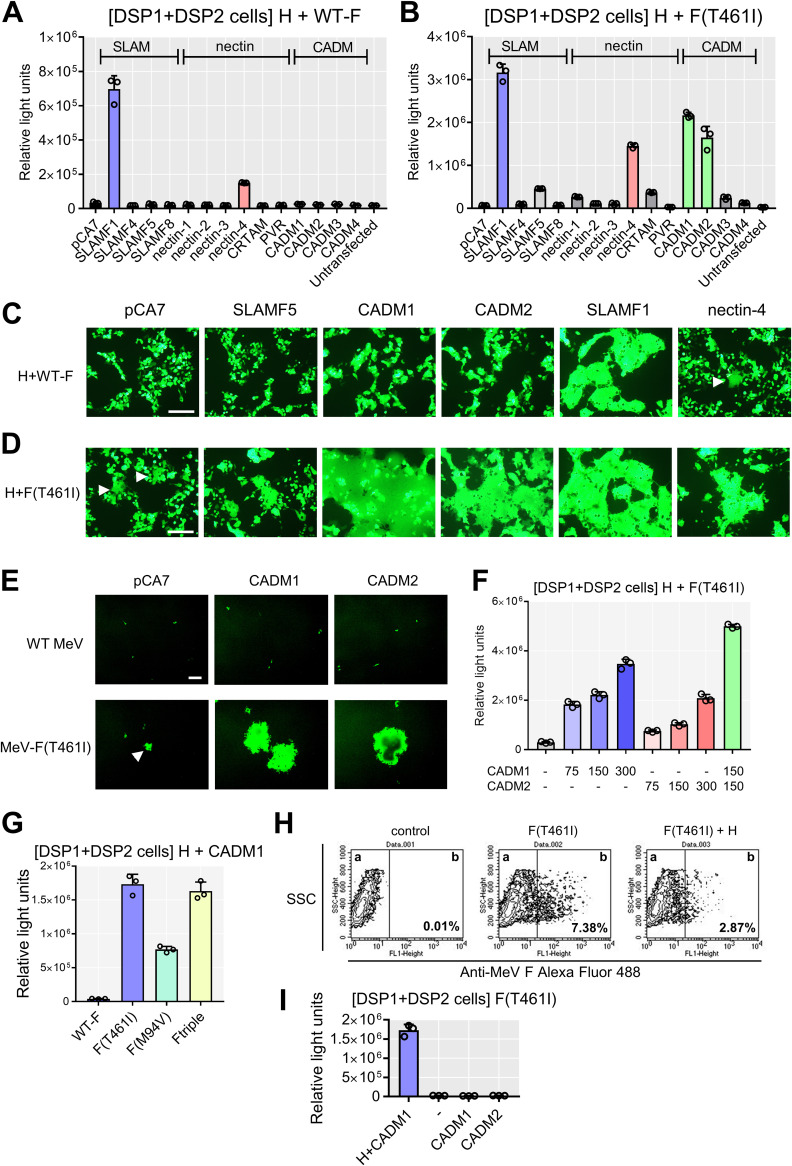
The dual split protein (DSP) assay using 293FT cells was employed to quantitate levels of cell-to-cell fusion (24, 27–29). In this system, a pair of chimeric reporter proteins, DSP1 and DSP2, each comprised of the split Renilla luciferase (RLUC) and split green fluorescent protein (GFP), are stably expressed in 293FT cells, respectively (293FT/DSP1 and 293FT/DSP2 cells). When cell-to-cell fusion is induced between 293FT/DSP1 and 293FT/DSP2 cells, Renilla luciferase and GFP activities are restored by the association of DSP1 and DSP2. We examined SLAMF4, SLAMF5, and SLAMF8 in the SLAM family and nectin-1, nectin-2, nectin-3, major histocompatibility complex class I (MHC-I)-restricted T-cell associated molecule (CRTAM), and poliovirus receptor (PVR) in the nectin family by the DSP assay. These molecules are expressed in the brain. The CADM family is structurally and evolutionally related to the nectin family, and CADM family molecules are expressed in various tissues, including the brain (30). We therefore additionally examined all CADM family molecules. None of the SLAM and nectin family molecules were able to induce the levels of luciferase activities comparable to those with SLAMF1 and nectin-4, whether in combination with the WT F protein (Fig. 1A) or in combination with the F(T461I) protein (Fig. 1B). However, CADM1 and CADM2 induced increased levels of luciferase activity when expressed with the F(T461I) protein (Fig. 1B). The results indicate that CADM1 and CADM2, like SLAMF1 and nectin-4, have the unique activity to trigger membrane fusion, whereas other molecules examined do not.
FIG 1.
CADM1 and CADM2 induce membrane fusion with hyperfusogenic F proteins. (A and B) The DSP assay with host molecules using the WT F (A) or F(T461I) protein (B). pCA7 expression plasmids respectively encoding MeV H and MeV F proteins with pCA7 encoding a host protein or with pCA7 alone as a control were transfected into cocultured 293FT/DSP1 and 293FT/DSP2 cells [DSP1 + DSP2 cells]. (C and D) The fusion assays with host molecules using the WT F (C) or F(T461I) protein (D). pCA7 expression plasmids respectively encoding MeV H, MeV F, and EGFP with pCA7 encoding a host protein or pCA7 alone were transfected into 293FT cells. White arrowheads indicate small syncytia. (E) 293FT cells expressing CADM1 or CADM2 infected with WT MeV or MeV-F(T461I). pCA7, control plasmid. A white arrowhead indicates a small syncytium. (F) The DSP assay with the H and F(T461I) proteins using different amounts of plasmids encoding CADM1/2 (0 to ∼300 ng). (G) The DSP assay using various F proteins with the H and CADM1 proteins. (H) The F(T461I) protein was transiently expressed with or without the H protein in 293FT cells. Surface expression of the F protein was analyzed by flow cytometry. Percentages of the F protein-positive population in compartment b are shown. (I) CADM1 or CADM2 was expressed with the F(T461I) protein with or without the H protein in the DSP assay. n = 3; mean ± standard deviation (SD) (A, B, F, G, and I). Scale bar = 200 μm (C, D, and E).
Cell-to-cell fusion was also evaluated by syncytium formation. Expression plasmids respectively encoding the WT H protein, the WT F or F (T461I) protein, enhanced GFP (EGFP), and one of SLAMF5, CADM1, CADM2, SLAMF1, and nectin-4 proteins were transfected into 293FT cells, and the cells were observed 36 h after transfection. With the WT F protein, syncytia were observed only when SLAMF1 or nectin-4 was expressed (Fig. 1C). In contrast, with the F(T461I) protein, syncytia were detected not only after expression of SLAMF1 or nectin-4 but also after expression of CADM1 or CADM2 (Fig. 1D). Syncytia were also induced in 293FT cells transiently expressing CADM1 or CADM2 after infection with the recombinant MeV possessing the F(T461I) protein [MeV-F(T461I)], but not with the WT MeV (Fig. 1E). It should be noted that small syncytia were observed in cells expressing the WT H and F(T461I) proteins even without exogenous expression of receptors (indicated by white arrows in Fig. 1D and E). They may be caused by endogenously expressed CADM1 (see Fig. 3A below). These findings are consistent with the results of the DSP assay (Fig. 1A and B). With different amounts of plasmids encoding CADM1 and CADM2 in the DSP assay, luciferase activities were found to increase in a dose-dependent manner, but coexpression of CADM1 and CADM2 showed little synergistic effect (Fig. 1F).
We then examined whether CADM1 can induce cell-to-cell fusion in combination with two other hyperfusogenic F proteins, F(M94V) and F-triple (S103I N462S N465S). These substitutions in the F protein have been detected in multiple MeV isolates from SSPE patients (12, 19). They also induced increased luciferase activity, like the F(T461I) protein (Fig. 1G). Expression of the F(T461I) protein without the H protein did not induce cell-to-cell fusion with CADM1 or CADM2, although its surface expression level was rather increased compared with that in the presence of the H protein (Fig. 1H and I). The result indicates that the H protein is necessary for CADM1 and CADM2 to induce membrane fusion.
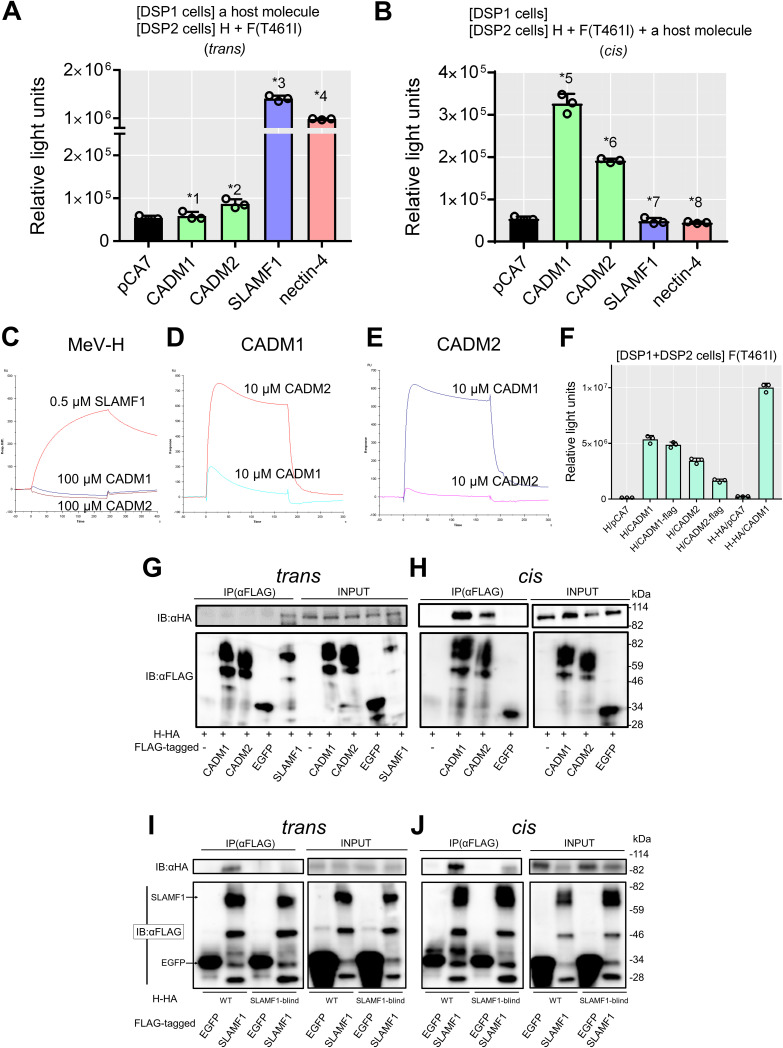
CADM1 and CADM2 act in cis with respect to MeV envelope proteins.
We then examined whether CADM1 and CADM2 act in trans (in cells different from those expressing MeV envelope proteins, like the usual receptors) or in cis (in cells also expressing MeV envelope proteins). CADM1 and CADM2 were expressed in trans or in cis with respect to H and F proteins in the DSP assay. When expressed in trans, CADM1 and CADM2 did not induce increased levels of luciferase activity, unlike SLAMF1 and nectin-4 (Fig. 2A). However, when expressed in cis, they did induce increased levels of luciferase activity (Fig. 2B). These data indicate that CADM1 and CADM2 act in cis, but not in trans, to trigger membrane fusion. In these experiments, the interaction of overexpressed CADM1 or CADM2 with endogenously expressed CADM1 likely mediated cell-to-cell contact (see Fig. 3A below).
FIG 2.
CADM1/2 act in cis. (A) Examination of trans-acting fusion-triggering functions of CADM1, CADM2, SLAMF1, and nectin-4 by the DSP assay (n = 3; mean ± SD). 293FT/DSP1 cells [DSP1 cells] were transfected with pCA7 encoding CADM1, CADM2, SLAMF1, or nectin-4 or with pCA7 alone as a control. The cells were mixed with 293FT/DSP2 cells [DSP2 cells] transfected with pCA7 plasmids respectively encoding the H and the F(T461I) proteins. (B) Examination of cis-acting fusion-triggering functions of CADM1, CADM2, SLAMF1, and nectin-4 by the DSP assay (n = 3; mean ± SD). 293FT/DSP1 cells were transfected with the empty pCA7 plasmid. The cells were mixed with 293FT/DSP2 cells transfected with pCA7 plasmids respectively encoding the H and F(T461I) proteins with pCA7 encoding CADM1, CADM2, SLAMF1, or nectin-4 or with pCA7 alone. Significance by unpaired multiple t test: *1, P = 0.52; *2, P = 0.0080; *3, P = 0.0000023; *4, P = 0.000000028; *5, P = 0.000036; *6, P = 0.0000034; *7, P = 0.32; *8, P = 0.038. (C to E) SPR analysis does not detect interactions of the MeV H protein with CADM1 and CADM2. The ectodomain of the WT H protein (C) was immobilized on research-grade CM5 chips. Then, 100 μM ectodomain of CADM1 or CADM2 or 0.5 μM ectodomain of SLAMF1 in HBS-P buffer was injected over the immobilized WT H protein. The ectodomain of CADM1 (D) or CADM2 (E) was immobilized on research-grade CM5 chips. Then, 10 μM ectodomain of CADM1 or CADM2 in HBS-P buffer was injected over the immobilized protein. (F) The WT H or H-HA protein was expressed together with the F(T461I) protein, with or without CADM1, CADM2, CADM1-FLAG or CADM2-FLAG in the DSP assay (n = 3; mean ± SD). (G and H) Interactions of the H-HA protein with CADM1-FLAG, CADM2-FLAG, SLAMF1-FLAG, and EGFP-FLAG in trans (G) or in cis (H) examined by coimmunoprecipitation assay using anti-FLAG antibody (Ab). Total lysates (INPUT) and immunoprecipitates (IP) were detected by immunoblotting (IB) using anti-HA (upper) Ab or anti-FLAG Ab (lower). (I and J) Interactions of the WT or SLAMF1-blind (R533A) H-HA protein with SLAMF1-FLAG and EGFP-FLAG in trans (I) or in cis (J) were examined by coimmunoprecipitation assay using anti-FLAG antibody (Ab). Total lysates (INPUT) and immunoprecipitates (IP) were detected by immunoblotting (IB) using anti-HA (upper) or anti-FLAG (lower) Ab.
We next examined the interaction of the WT H protein with CADM1 or CADM2 using surface plasmon resonance (SPR) analysis. The ectodomains of the WT H protein (residues 149 to 617), CADM1, CADM2, and SLAMF1 were prepared and analyzed. The WT H protein was found to bind SLAMF1, but not CADM1 or CADM2 at all, while CADM1 and CADM2 bound each other as reported previously (31) (Fig. 2C,D and E). The interaction of the WT H protein with CADM1 or CADM2 was also examined by coimmunoprecipitation analysis. For this analysis, the hemagglutinin (HA) tag-fused H protein (H-HA) and FLAG tag-fused proteins (CADM1-FLAG, CADM2-FLAG, SLAMF1-FLAG, and EGFP-FLAG) were used. These HA and FLAG peptide tags did not influence the functions of the proteins in membrane fusion (Fig. 2F). When the H-HA protein and FLAG tag-fused proteins were expressed in trans, the interaction of the H protein with SLAMF1, but not with CADM1, CADM2, or EGFP, was observed (Fig. 2G). In contrast, when the H-HA protein and the FLAG tag-fused proteins were expressed in cis (in the same cells), the H protein was found to coimmunoprecipitate with CADM1 and CADM2, but not with EGFP (Fig. 2H). Thus, the H protein interacts in cis, but not in trans, with CADM1 and CADM2. The WT H protein also interacted in cis with SLAMF1 (Fig. 2I and J), although SLAMF1 did not induce membrane fusion in cis because of the downregulation of the H protein from the cell surface (24). The SLAMF1-blind H protein interacted in cis, but not in trans, with SLAMF1.
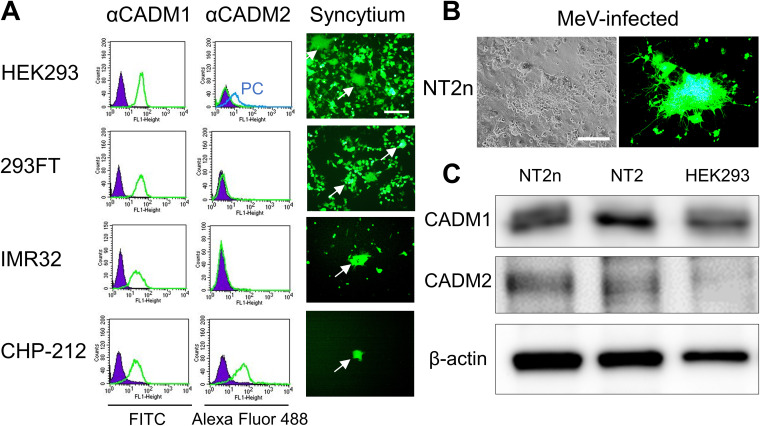
Expression of CADM1 and CADM2 in cells.
Expression of CADM1 and CADM2 and syncytium formation induced by the WT H and F(T461I) proteins were examined in several SLAMF1- and nectin-4-negative human cell lines (Fig. 3A). HEK293 is derived from human embryonic kidney cells, but its most likely origin is an embryonic adrenal precursor cell (32). 293FT is a derivative of HEK293 and was used for the DSP assay. IMR32 and CHP-212 are derived from human neuroblastoma cells. CADM1 was expressed in all the cell lines, while CADM2 was detected only in CHP-212 cells. When the WT H and F(T461I) proteins were transfected, all these cell lines developed syncytia. The human embryonal carcinoma cell line NTERA-2cl.D1 (NT2) can be differentiated into neurons (NT2n) following treatment with retinoic acid (17, 33). Our previous study has shown that while WT MeV and MeV-F(T461I) infect NT2n cells at the comparable low efficiencies, MeV-F(T461I), but not WT MeV, spreads between NT2n cells (17). We confirmed the efficient spread of MeV-F(T461I) between NT2n cells (Fig. 3B) and detected expression of CADM1 and CADM2 in them (Fig. 3C). Thus, the expression patterns of CADM1 and CADM2 support the hypothesis that these molecules are individually or jointly involved in syncytium formation and virus spread mediated by hyperfusogenic F proteins in SLAMF1- and nectin-4-negative cells.
FIG 3.
Expression of CADM1/2 in cell lines. (A) Expression of CADM1 and CADM2 (left) and syncytia induced by the WT H and F(T461I) proteins (right) in different cell lines. Green and purple lines indicate cells immunostained with anti-CADM1 or -CADM2 Ab and with control Ab, respectively. A blue line indicates positive-control (PC) cells in which CADM2 was expressed transiently. For the induction of syncytia, pCA7 expression plasmids respectively encoding H, F(T461I), and EGFP were transfected into each cell line. White arrows indicate syncytia. (B) NT2n cells observed 3 days after infection with MeV-F(T461I) under a phase-contrast or fluorescence microscope. Scale bar = 200 μm (A and B). (C) Western blotting for CADM1, CADM2, and β-actin in different cells.
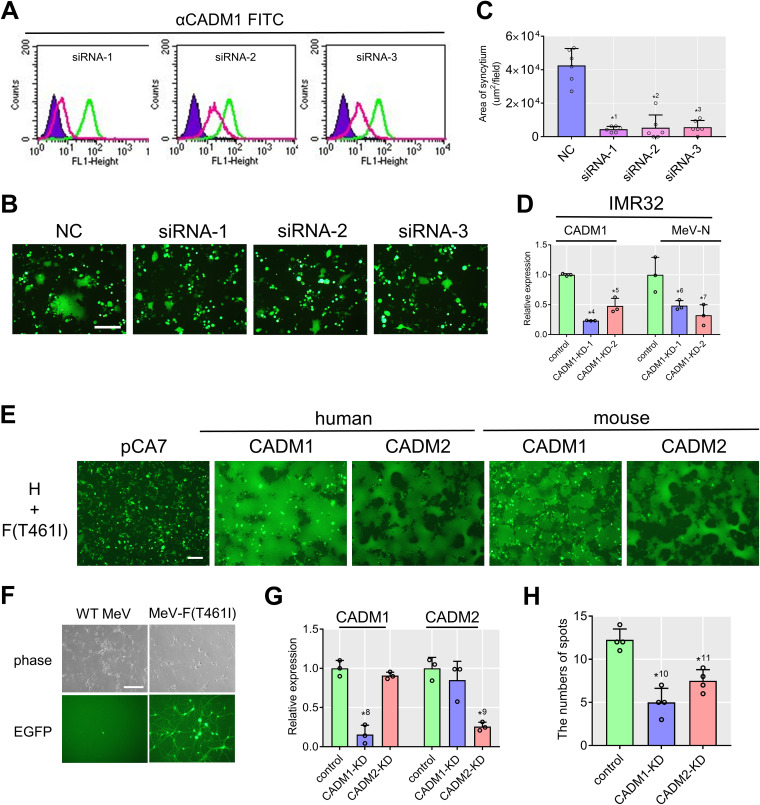
Knockdown of CADM1 and CADM2 inhibits syncytium formation and virus spread mediated by the hyperfusogenic F protein.
To strengthen the above hypothesis, HEK293 cells were transfected with three kinds of small interfering RNAs (siRNA-1, -2, -3) targeting different sequences of the CADM1 gene. All the siRNAs knocked down the expression of CADM1 to various degrees (Fig. 4A), and upon expression of the WT H and F(T461I) proteins, CADM1 knocked-down (KD) cells formed fewer syncytia than those transfected with the control siRNA (Fig. 4B and C). CADM1 was also knocked down in IMR32 cells by using lentivirus vectors (CADM1-KD-1 and CADM1-KD-2) carrying small hairpin RNAs (shRNAs) against CADM1. At 72 h after infection with MeV-F(T461I), expression of the MeV nucleocapsid (N) gene, as determined by quantitative PCR (qPCR), was decreased in CADM1 KD IMR32 cells (Fig. 4D).
FIG 4.
Knockdown of CADM1 inhibits syncytium formation and virus spread. (A) CADM1 knockdown efficiencies in HEK293 cells analyzed by flow cytometry. Green and red lines indicate cells transfected with nontargeting control (NC) and CADM1-targeting siRNAs, respectively. Unstained controls are also shown (purple). (B) Syncytia in CADM1 KD cells induced by the WT H and F(T461I) proteins. pCA7 expression plasmids respectively encoding H, F(T461I), and EGFP were transfected into the cells. (C) Syncytium formation was quantitated by measuring the total area of syncytia (>2,000 μm2) per field using a BZ-X710 microscope with the BZ-X analyzer (Keyence) (n = 6; mean ± SD). (D) Relative expression of CADM1 and MeV N mRNAs in CADM1 KD IMR32 cells after infection with MeV-F(T461I) (n = 3; mean ± SD). (E) Syncytia in 293FT cells induced by human/mouse CADM1 or CADM2. 293FT cells were transfected with expression plasmids respectively encoding H, F(T461I), EGFP, and human/mouse CADM1 or CADM2. (F) Mouse primary neurons infected with WT MeV or MeV-F(T461I) as examined by a phase-contrast or fluorescence microscope. While a singly infected cell is found in the left lower panel, an infected spot (a local area with multiple infected cells) is detected in the right lower panel. (G) Relative expression of CADM1 and CADM2 mRNAs in KD mouse primary neurons infected with MeV-F(T461I) (n = 3; mean ± SD). (H) Numbers of infected spots in CADM1 or CADM2 KD mouse primary neurons (n = 4; mean ± SD). Scale bar = 200 μm (B, E, and F). Significance by unpaired multiple t test: *1, P = 0.0000039; *2, P = 0.000031; *3, P = 0.0000087; *4, P = 0.00000029; *5, P = 0.0022; *6, P = 0.042; *7, P = 0.026; *8, P = 0.00072; *9, P = 0.0010; *10, P = 0.00041; *11, P = 0.0019.
To determine the effect of CADM1 and CADM2 knockdown on MeV spread between neurons, we examined mouse primary neurons, as human NT2n cells were severely damaged by transfection or lentivirus infection. The use of mouse primary neurons is warranted by the finding that recombinant MeVs possessing hyperfusogenic F proteins, but not the WT MeV, spread in mouse brains (12). Indeed, when expressed together with the WT H and F(T461I) proteins, mouse CADM1 and CADM2 induced syncytia in 293FT cells as efficiently as human CADM1 and CADM2 (Fig. 4E). The results suggest that hyperfusogenic MeVs spread in human and mouse brains in the same manner. MeV-F(T461I), but not the WT MeV, efficiently spread in mouse primary neurons isolated from the cerebral cortex of fetal C57BL/6 mice (E17) (Fig. 4F). While singly infected cells were found upon infection with the WT MeV, local areas with multiple infected cells (here referred to as infected spots) were detected only after infection with MeV-F(T461I) (Fig. 4F). Mouse primary neurons were infected with the lentivirus vector carrying shRNAs against CADM1 or CADM2 and then 3 days later infected with MeV-F(T461I). After an additional 4 days, the cells were observed with a fluorescence microscope and then collected for RNA isolation to evaluate knockdown efficiency by qPCR. The targeted molecule (CADM1 or CADM2) was significantly knocked down (Fig. 4G), and the number of infected spots was reduced both in CADM1 KD neurons and in CADM2 KD neurons (Fig. 4H).
DISCUSSION
SSPE is a fatal complication of MeV infection, and no effective therapy is currently available. Our results show that CADM1 and CADM2 are host molecules required for MeV spread in neurons lacking SLAMF1 and nectin-4. How neurons are initially infected by MeV is currently unknown. It has been shown that the WT viruses can infect SLAMF1- and nectin-4-negative cells at low efficiencies (2 to 3 logs lower than those for SLAMF1-positive cells) (34). Thus, during the viremic phase, MeV-infected immune cells (lymphocytes, dendritic cells, and macrophages) may transfer the virus to neurons by occasionally invading the brain through the blood-brain barrier. A recent study also suggested that transendocytosis elicited by nectins may account for transmission of MeV from infected epithelial cells to neurons (35). However, without any mechanism allowing MeV spread within the brain, the infection would be limited to the first infected neurons. Thus, MeV adaptation, including hyperfusogenic changes in the F protein and use of CADM1 and CADM2, may be required for persistence.
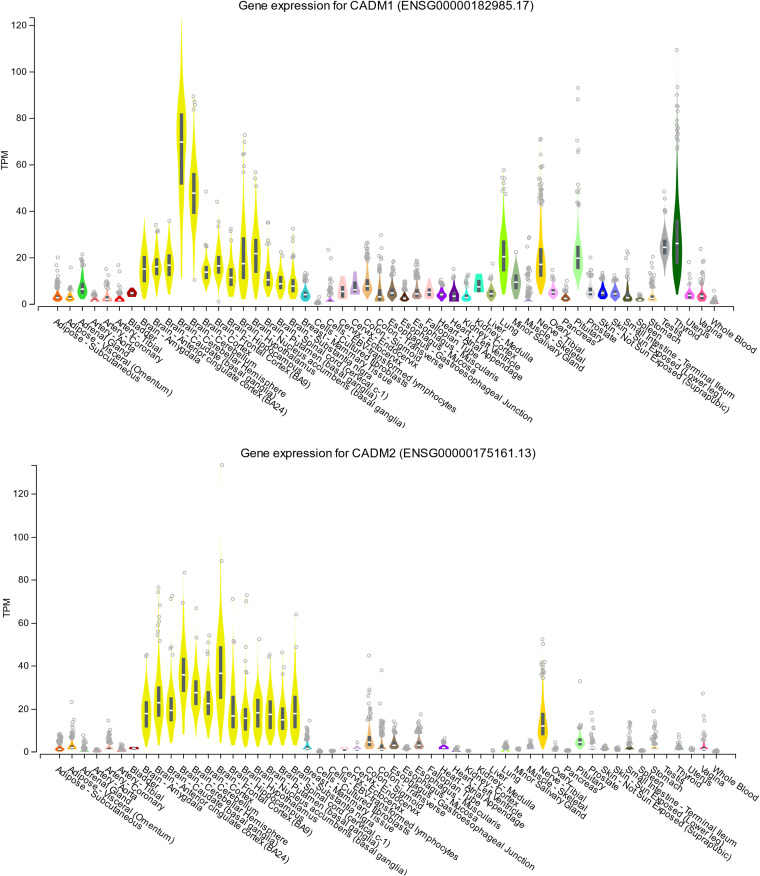
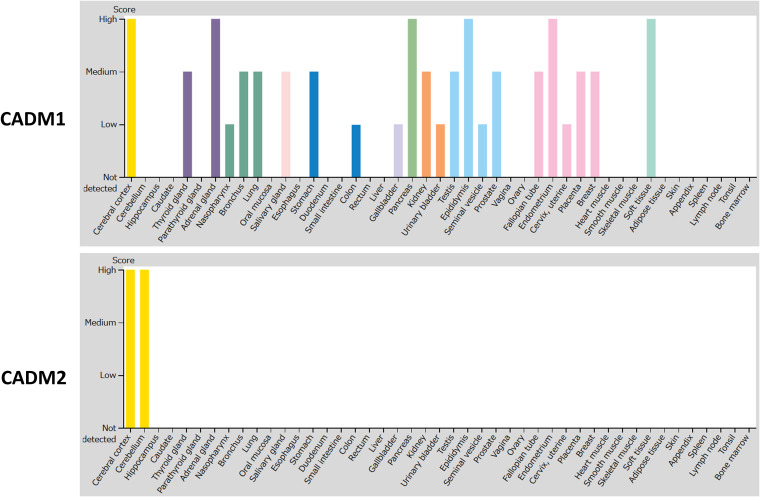
CADM 1- and CADM 2-mediated MeV spread may be a promising target for therapeutic antiviral drugs. Tissue distributions of CADM1 and CADM2 at the mRNA (Fig. 5) and protein (Fig. 6) levels indicate that they are abundantly expressed in human brains. The members of the CADM family have been implicated in diverse physiological processes, including cell adhesion (30), spermatogenesis (36, 37), tumor suppression (38), and development of the nervous system (30). They are located on both sides of the synaptic cleft, participating in the formation of synapses (39). Interestingly, CADM1 is a marker of human T cell lymphotropic virus 1-infected cells (40).
FIG 5.
Tissue expression patterns of CADM1 and CADM2 at the mRNA level. Tissue expression patterns of CADM1 (upper) and CADM2 (lower) at the mRNA level were obtained from the Genotype-Tissue Expression (GTEx) consortium using the GTEx Portal (https://www.gtexportal.org/home/gene/CADM1 and https://www.gtexportal.org/home/gene/CADM2). TPM, transcripts per million.
FIG 6.
Tissue expression patterns of CADM1 and CADM2 at the protein level. Tissue expression patterns of CADM1 and CADM2 at the protein level were obtained from the Human Protein Atlas (HPA) (54–56) using the webpage of HPA (http://www.proteinatlas.org). Images are available from https://www.proteinatlas.org/ENSG00000182985-CADM1/tissue and https://www.proteinatlas.org/ENSG00000175161-CADM2/tissue (v19.3.proteinatlas.org). Protein expression scores are based on a best estimate of the “true” protein expression from a knowledge-based annotation, described in more detail at https://www.proteinatlas.org/about/assays+annotation#ihk.
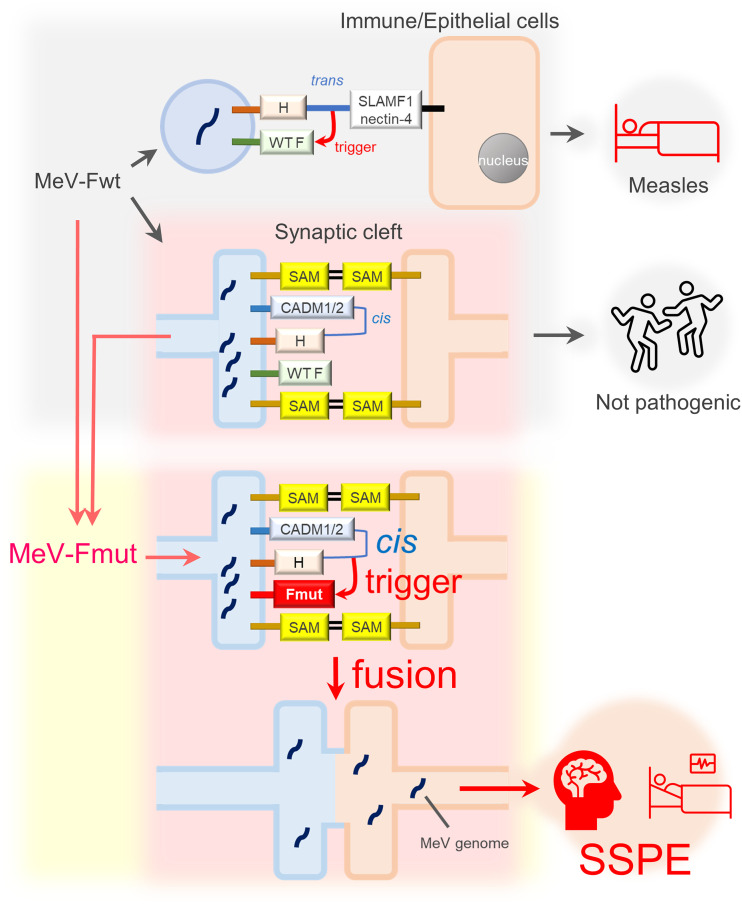
Importantly, CADM1 and CADM2 act only in cis, but not in trans (Fig. 2A and B). The data indicate that CADM1 and CADM2 induce membrane fusion in cis by mimicking part of receptor function. Like SLAMF1 and nectin-4, CADM1 and CADM2 bind the H protein, thereby triggering the conformational changes of the F protein for membrane fusion (although the F protein must be a hyperfusogenic mutant). However, CADM1 and CADM2 act in cis on the H protein present on the same membrane (Fig. 7). Thus, CADM1 and CADM2 are different from usual receptors that act in trans on viral attachment proteins present on different membranes. Virus receptors usually connect two separate membranes by interacting with attachment proteins. Since the contact between pre- and postsynaptic membranes is maintained by various synaptic adhesion molecules (including CADMs themselves) (41), the cis interaction between the H protein and CADM1/CADM2 on the same membrane may suffice to cause virus transmission at synapses (Fig. 7).
FIG 7.
A schematic of MeV transmission between neurons. Fwt and Fmut are WT and hyperfusogenic mutant F proteins, respectively. In addition to other molecules, CADM1 and CADM2 may also act as synaptic adhesion molecules (SAM).
In the experiments in Fig. 1A and B, the cell-to-cell contact was likely mediated by the interaction of overexpressed CADM1 or CADM2 with endogenously expressed CADM1 in 293FT cells. In the literature, heterophilic interactions of CADM1 with CADM2, CADM3, CADM4, nectin-3, CRTAM, and PVR have been reported (26, 42, 43). However, it is still possible that the cis-acting function of the other molecules examined in Fig. 1A and B was masked due to the lack of appropriate binding partners to bring cell membranes into close contact. This possibility will be carefully investigated in our future studies. We have previously shown that SLAMF1 could interact in cis with the SLAMF1-blind H protein on the same membrane, causing fusion triggering and membrane fusion (24). The present study demonstrates the first real example of “receptor-mimicking” cis-acting fusion triggering, which could occur where cell-to-cell contact is naturally maintained.
Coimmunoprecipitation detected the interactions of the H protein with CADM1 and CADM2 only when the molecules were expressed in the same cells (Fig. 2G and H). The cis interactions may be mediated by the specific conformations formed only when these molecules are present in the same cells, the structures at their stalk, transmembrane and cytoplasmic regions, and/or other supporting molecules. In our previous study, the WT H protein was not able to trigger the hyperfusogenic F protein, upon binding to SLAMF1 on the same cell (24). The copresence of SLAMF1 in the same cell downregulated the strongly interacting WT H protein from the cell surface, but not the weakly interacting SLAMF1-blind H protein (24). Even if the interaction between the SLAMF1-blind H protein and SLAMF1 is weak, the local enrichment of these molecules expressed in the same cell might allow for their functional interaction. In fact, coimmunoprecipitation detected the interaction of the SLAMF1-blind H protein with SLAMF1 in cis, but not in trans (Fig. 2I and J). Similarly, the cis interaction of the H protein with CADM1/CADM2 may be relatively weak. Further physical, biochemical, and structural studies are needed to solve this issue.
Besides CADM1 and CADM2 at neurological synapses, other cis-acting molecules may also play a role in cell-to-cell transmission of enveloped viruses where close cell-to-cell contacts exist so that trans interactions are not essential for membrane fusion (e.g., polarized epithelia). cis interactions of receptors with their ligands expressed on the same cell have been described in other biological systems (44, 45). Interactions between these cis-acting molecules and virus attachment proteins may be considered to belong to the same category.
Unlike MeV isolates from acute measles patients that cannot spread in neurons, viruses in the brains of SSPE patients acquire specific substitutions in the F protein, not in the receptor-binding H protein, to utilize receptor-mimicking cis-acting fusion triggering. This is a novel evolutionary mechanism for the expansion of viral tropism and pathogenicity where authentic receptors do not exist.
MATERIALS AND METHODS
Cell lines and culture conditions.
293FT, HEK293, IMR32, CHP-212, and NT2 cells as well as Vero cells stably expressing human SLAMF1 (Vero/hSLAM cells) (46) were maintained in Dulbecco's modified Eagle medium (DMEM; Fujifilm Wako Pure Chemical Corporation) supplemented with 10% fetal bovine serum (FBS). The 293FT line is a derivative of 293T and was obtained from Invitrogen. 293FT cells stably expressing DSP1 (293FT/DSP1) or DSP2 (293FT/DSP2) (27–29) (gifts from Z. Matsuda) were maintained in DMEM supplemented with 10% FBS and 1 μg/ml puromycin (InvivoGen). A protocol of neuronal differentiation of NT2 cells was described previously (17, 33). Mouse primary neurons were isolated from the cortex of the C57BL/6 mouse at embryonic day 17 and cultured according to the protocol described previously (47).
Plasmids.
The eukaryotic expression vector pCA7 is a derivative of pCAGGS (48, 49). The pCA7 plasmids respectively encoding WT H (the IC-B strain), Ed-H (the Edmonston strain), WT F, F(T461I), F(M94V), F-triple (S103I N462S N465S), SLAMF1 (accession no. NM_003037.5), and nectin-4 (NM_030916.3) were described previously (19, 24, 50). Plasmids encoding the SLAM, nectin, and CADM family molecules were purchased from K.K. DNAFORM under the following clone ID numbers: 100071880 (SLAMF4; accession no. NM_016382.4), 100004338 (SLAMF5; accession no. NM_003874.4), 40004601 (SLAMF8; accession no. NM_020125.3), 100072606 (nectin-1; accession no. NM_002855.5), 100009475 (nectin-2; accession no. NM_002856.3), 100070930 (CRTAM; accession no. NM_019604.4), 3902226 (PVR; accession no. NM_006505.5), 40118330 (CADM1; accession no. NM_001098517.2), 5259422 (CADM2; accession no. NM_001167675.2), 100008462 (CADM3; accession no. NM_001127173.3), 100014610 (CADM4; accession no., NM_145296.2), 5372277 (mouse CADM1; accession no. NM_001025600.1), and A830029E02 (mouse CADM2; accession no. NM_178721.4). The synthesized DNA encoding nectin-3 (accession no. NM_015480.3) was purchased from Eurofins Genomics K.K. DNA fragments respectively encoding SLAMF4, SLAMF5, SLAMF8, nectin-1, nectin-2, nectin-3, CRTAM, PVR, CADM1, CADM2, CADM3, CADM4, mouse CADM1, and mouse CADM2 were PCR amplified and inserted into pCA7 predigested with EcoRI and NotI. For coimmunoprecipitation analysis, the HA tag (YPYDVPDYA) and FLAG tag (DYKDDDDK) sequences were fused to the C termini of the WT H protein and other proteins (CADM1, CADM2, SLAMF1, and EGFP), respectively. shRNA lentivirus plasmid vectors (pLVRH1MP) against human CADM1 were purchased from GeneCopoeia (catalog no. CSHCTR001-1-LVRH1MP [negative control], HSH065250-LVRH1MP-a [CADM1-KD-1], and HSH065250-LVRH1MP-b [CADM1-KD-2]). pLLX is a dual-promoter lentivirus vector constructed by inserting the U6 promoter-driven shRNA cassette 5′ to the ubiquitin-C promoter in the FUIGW plasmid (51, 52). The pLLX plasmid and support plasmids for lentivirus vector production, pCMV-VSV-G-RSV-REV and pCAG-HIVgp, were provided by Z. Zhou and M. E. Greenberg. shRNA lentivirus plasmid vectors against Renilla luciferase (control), mouse CADM1, and mouse CADM2 were generated by inserting annealed primer dimers into the HpaI and XhoI restriction sites in the pLLX plasmid. The shRNA sequences introduced were 5′-CGGCCTCTTCTTATTTATGGTTCAAGAGACCATAAATAAGAAGAGGCCGC-3′ for Renilla luciferase (RLUC), 5′-GGGAGGAGATTGAAGTCAACTTTCAAGAGAAGTTGACTTCAATCTCCTCCC-3′ for mouse CADM1 (CADM1-KD), and 5′-GTGCTAGAAATTCACTATACACCTTCAAGAGAGGTGTATAGTGAATTTCTAGCAC-3′ for mouse CADM2 (CADM2-KD).
Virus preparation.
Two recombinant MeVs encoding EGFP, IC323-EGFP and IC323-EGFP-F(T461I), were described previously (19, 34). Vero/hSLAM cells cultured in a 15-cm dish were infected with each recombinant MeV at a multiplicity of infection of 0.001. The cells were harvested and then lysed by three freeze-thaw cycles. Lysates were centrifuged at 2,500 × g for 10 min at 4°C. Supernatants were collected and concentrated by centrifugation at 6,000 × g overnight at 4°C. Pellets were suspended in 2 ml each of Neurobasal medium (Thermo Fisher Scientific) and stored at –80°C.
Lentivirus vector preparation.
Each lentivirus vector plasmid carrying shRNA (20 μg) was transfected, together with pCMV-VSV-G-RSV-REV (4 μg) and pCAG-HIVgp (4 μg), into 293FT cells cultured in a 15-cm dish, using PEI-MAX (84 μl; Polysciences). The medium was exchanged for fresh medium 24 h after transfection. Then, supernatants were harvested and filtered through a 0.45-μm-pore filter (Thermo Fisher Scientific) at 2 days after transfection. Collected supernatants were centrifuged at 6,000 × g overnight at 4°C for concentration. Pellets were suspended in 1.5 ml each of Neurobasal medium and stored at –80°C.
DSP assay.
DSP1 and DSP2 are split proteins of Renilla luciferase and green fluorescent protein (GFP), which become functional when reassociated with each other after 293FT/DSP1 and 293FT/DSP2 cells are fused. The protocol of the DSP assay was described previously (24). Briefly, pCA7 expression plasmids respectively encoding MeV H and MeV F proteins with pCA7 encoding a host protein or pCA7 alone as a control was transfected into cocultured 293FT/DSP1and 293FT/DSP2 cells in 24-well plates using Lipofectamine LTX (Thermo Fisher Scientific). The Renilla luciferase activity was analyzed 24 h after transfection using the Renilla luciferase assay system (Promega).
To evaluate trans-acting functions of host molecules in fusion triggering, 293FT/DSP1 cells were transfected with pCA7 encoding CADM1, CADM2, SLAMF1, or nectin-4 or with pCA7 alone as a control. The cells were mixed with 293FT/DSP2 cells transfected with pCA7 plasmids respectively encoding the WT H and the F(T461I) proteins. To evaluate cis-acting functions, 293FT/DSP1 cells were transfected with the empty pCA7 plasmid. The cells were mixed with 293FT/DSP2 cells transfected with pCA7 plasmids respectively encoding the WT H and F(T461I) proteins with pCA7 encoding CADM1, CADM2, SLAMF1, or nectin-4 or with pCA7 alone. The Renilla luciferase activity was analyzed 24 h after the cells were mixed.
A small increase of luciferase activity (compared with the background level) is observed when the MeV H and hyperfusogenic F proteins are used for the assay, and cell-to-cell contact is maintained by the interactions between some adhesion molecules (24). This presumably occurs because 293FT cells endogenously express CADM1 that can interact with the H protein, triggering the hyperfusogenic F protein for membrane fusion (as shown in the present study). When SLAMF1, nectin-4, CADM1, or CADM2 is overexpressed, a much higher level of luciferase activity is observed.
Fusion assay.
Cells cultured in 24-well plates were transfected with different combinations of pCA7 plasmids respectively encoding MeV H (WT H or Ed-H) (0.1 μg), MeV F [WT F, F(T461I), F(M94V) or F-triple] (0.1 μg), EGFP (0.1 μg), and a host molecule (SLAMF1, SLAMF5, nectin-4, CADM1, CADM2, or none) (0.3 μg) using Lipofectamine LTX (1 μl). The cells were observed 30 to 48 h after transfection by fluorescence microscopy.
Infection of 293FT cells transiently expressing CADM1/2 with MeV.
293FT cells cultured in 6-well plates were transfected with pCA7 plasmids respectively encoding CADM1 and CADM2 (2 μg) using Lipofectamine LTX (4 μl) and subsequently infected with WT MeV or MeV-F(T461I) 24 h after transfection. The cells were observed 48 h after infection by fluorescence microscopy.
Flow cytometry.
For evaluation of levels of CADM1 and CADM2 expression on the surfaces of cells, HEK293, 293FT, IMR32, and CHP-212 cells were collected and fixed with phosphate-buffered saline (PBS) containing 4% paraformaldehyde for 15 min at room temperature. After being washed with PBS, cells were then incubated with chicken monoclonal antibody (Ab) against CADM1 (CM005-3; Medical & Biological Laboratories) or rabbit polyclonal Ab against CADM2 (15690-T32; Sino Biological) for 1 h on ice, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated anti-chicken IgY (abcam) or Alexa Fluor 488-conjugated anti-rabbit IgG (Thermo Fisher Scientific), respectively. For evaluation of the surface expression of the MeV F protein, 293FT cells were transfected with pCA7 encoding the F(T461I) protein together with that encoding the WT H protein or pCA7 alone for control. The cells were incubated with mouse polyclonal Ab against the MeV F protein 48 h after transfection, followed by Alexa Fluor 488-conjugated anti-mouse IgG (Thermo Fisher Scientific). Cells were then analyzed on the FACSCalibur HD flow cytometer and the BD CellQuest Pro version 5.2.1 software (BD Biosciences). Target cell populations were distinguished from cell debris by forward scatter (FSC) and side scatter (SSC) gating, followed by detection of the target protein-positive populations.
Coimmunoprecipitation assay.
To evaluate trans-interactions of the WT H protein with host molecules, 293FT cells cultured on 6-well plates were transfected with pCA7 encoding CADM1-FLAG, CADM2-FLAG, EGFP-FLAG, or SLAMF1-FLAG or with empty pCA7 as a control. 293FT cells cultured on 6-well plates were separately transfected with pCA7 encoding the H-HA protein or the SLAMF1-blind H(R533A)-HA protein (24). At 3 h after transfection, these two sets of transfected cells were mixed and cultured for an additional 21 h. To evaluate cis-interactions, 293FT cells cultured on 6-well plates were transfected with pCA7 encoding the H-HA protein or the SLAMF1-blind H(R533A) protein and that encoding CADM1-FLAG, CADM2-FLAG, EGFP-FLAG, or SLAMF1-FLAG or with none as a control. At 24 h after transfection, the cells were washed with PBS, and lysed in 500 μl of the immunoprecipitation (IP) lysis buffer (Thermo Fisher Scientific) containing the protease inhibitor cocktail (Sigma-Aldrich). After incubation for 15 min on ice, the lysates were centrifuged at 12,000 × g for 30 min at 4°C. Then, 50 μl each of supernatant was mixed with an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer (125 mM Tris-HCl [pH 6.8], 10% 2-mercaptoethanol, 4% SDS, 0.1% bromophenol blue, and 20% glycerol), boiled for 5 min, and stocked as the input samples at –20°C. For preclearing of the lysate, the rest of the supernatant was incubated for 30 min at 4°C with protein G-Sepharose (GE Healthcare AB). The precleared cell lysate was collected by centrifugation, and the bead pellet was discarded. The cell lysate was treated with mouse anti-FLAG monoclonal Ab (F1804; Sigma-Aldrich) and protein G-Sepharose for 3 h at 4°C. The samples were centrifuged and washed three times with the IP lysis buffer. Pellets were mixed with 30 μl of the 2× SDS loading buffer, boiled for 5 min, and stocked as the IP samples at –20°C.
Western blotting.
Proteins in samples were separated by SDS-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membranes (Hybond-P; Amersham Biosciences). The membranes were then incubated with primary Abs for 1 h. Rabbit polyclonal Abs against FLAG (F7425; Sigma), HA (Y-11; Santa Cruz Biotechnology), CADM1 (orb48299; Biorbyt), and CADM2 (MBS9604993; MyBioSource) and mouse monoclonal Ab against β-actin (3598R-100; Santa Cruz Biotechnology) were used, respectively. The membranes were washed with Tris-buffered saline containing 0.05% Tween 20 (TBS-T) and incubated with horseradish peroxidase-conjugated goat anti-mouse (Bio-Rad) or anti-rabbit (Zymed) IgG Ab for 1 h at room temperature. After being washed with TBS-T, the membranes were treated with the ECL Plus reagent (Amersham Biosciences), and chemiluminescent signals were detected and imaged using a VersaDoc 5000 imager (Bio-Rad).
SPR.
SPR experiments were performed by using BIAcore3000 (BIAcore) as described previously (53). Briefly, streptavidin was covalently coupled onto research-grade CM5 chips (BIAcore), and the biotinylated WT H protein (residues 149 to 617) was immobilized on the streptavidin-coupled chips. The His-tagged purified ectodomains of CADM1 and CADM2 were also covalently coupled onto research-grade CM5 chips. The His-tagged purified ectodomains of SLAMF1, CADM1, and CADM2 in HBS-P (10 mM HEPES; 150 mM NaCl; 0.005% surfactant P20, pH 7.4) buffer were respectively injected over the immobilized the WT H protein, CADM1, or CADM2.
Knockdown by siRNAs.
siRNAs against human CADM1 were purchased from Dharmacon (ID no. D-001810-10-05 [nontargeting control], J-016565-05-0002 [siRNA-1], J-016565-06-0002 [siRNA-2], and J-016565-07-0002 [siRNA-3]), and 40 pmol each of RNA interference (RNAi) duplex was diluted in 400 μl DMEM without serum in a well of 6-well culture plate. Then, 4 μl of Lipofectamine RNAiMAX (Thermo Fisher Scientific) was added to each well containing the diluted RNAi molecules. After incubation for 20 min at room temperature, 2 ml of the diluted HEK293 cells (1 × 105 cells/ml) was added to each well. After 48 h of culture, the cells were transfected with pCA7 plasmids respectively encoding the WT H protein (0.7 μg), F(T461I) protein (0.7 μg), and EGFP (0.7 μg) using Lipofectamine LTX (4 μl). The cells were observed at 48 h after transfection by fluorescence microscopy. Syncytium formation was quantitated by measuring the total area of syncytia (>2,000 μm2) per field using a BZ-X710 microscope with the BZ-X analyzer (Keyence).
Knockdown by lentivirus vectors carrying shRNAs.
IMR32 cells cultured in 24-well plates (2 × 105 cells per well) were infected with 500 μl of lentivirus vectors carrying shRNAs against human CADM1. At 2 days after lentivirus infection, the culture medium was replaced by 500 μl of medium containing MeV-F(T461I). The cells were collected for RNA isolation at 3 days after MeV infection.
Mouse primary neurons cultured in 24-well plates (1 × 105 cells per well) were infected with 500 μl of lentivirus vectors carrying shRNAs against mouse CADM1 or CADM2 at the day of isolation. Half of the culture medium was replaced by 250 μl of medium containing the concentrated MeV-F(T461I) stock at 3 days after lentivirus infection. At 3 days after MeV infection, half of the culture medium was replaced by 250 μl of culture medium. At 4 days after MeV infection (7 days after shRNA treatment), the number of infected spots (defined in the text) was counted under a fluorescence microscope, and the cells were collected for RNA isolation.
RNA isolation and reverse transcription.
Cells cultured in 24-well plates were washed with PBS and lysed in 500 μl of the TRIzol reagent (Thermo Fisher Scientific), and extracted RNAs were resuspended in 50 μl of nuclease-free water. Then, 1 μl each of RNA samples was mixed with 0.5 μl of 50 μM oligo(dT) primer, 0.5 μl of 10 mM deoxynucleoside triphosphate (dNTP) mixture, and 4.5 μl of nuclease-free water. Samples were incubated at 65°C for 5 min and then placed on ice for 1 min. After addition of 3.5 μl of cDNA synthesis mixture (2 μl of 5× First-Strand buffer, 0.5 μl of 0.1 M dithiothreitol [DTT], 0.5 μl of RNaseOUT [Thermo Fisher Scientific], and 0.5 μl of Superscript III reverse transcriptase [Thermo Fisher Scientific]), samples were incubated at 50°C for 50 min and then at 85°C for 5 min to terminate reactions. Then, 1 U of RNase H (TaKaRa) was added to each sample, followed by incubation for 20 min at 37°C. Forty microliters of nuclease-free water was added to each sample, and cDNAs were stored at –20°C.
qPCR.
qPCR (45 cycles of 95°C for 15 s and 60°C for 30 s) was performed with Thunderbird SYBR qPCR mixture (Toyobo) using a CFX96 real-time system (Bio-Rad). The primers used for this assay were 5′- CCATCAGTTGCCAAGTCAATAAG-3′ and 5′-GGCCTGAAGTCCCTGAAATAA-3′ for human CADM1 mRNA, 5′-GGTGTGAACCATGAGAAGTATGA-3′ and 5′-GAGTCCTTCCACGATACCAAAG-3′ for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, 5′-CCCAACAGGCAGACCATTTA-3′ and 5′-GTGTAGAGCTGGCAGAAGTATC-3′ for mouse CADM1 mRNA, 5′-GACAGTACACCTGCTCCTTATT-3′ and 5′-AATCCACTAATCTGAGGCTTCTC-3′ for mouse CADM2 mRNA, 5′-AACAGCAACTCCCACTCTTC-3′ and 5′-CCTGTTGCTGTAGCCGTATT-3′ for mouse GAPDH mRNA, and 5′-GGAAGATAGGAGGGTCAAACAG-3′ and 5′-ATGCAGTGTCAATGTCTAGGG-3′ for MeV N mRNA, respectively. The amounts of individual mRNAs were normalized to that of GAPDH mRNA, and the levels of relative expression of control samples were set to 1.0. Data were analyzed with the CFX Maestro software (Bio-Rad).
Measurements and statistical tests.
All measurements were taken from distinct samples. All statistical tests were performed using GraphPad Prism 7 software (a two-tailed unpaired multiple t test [the Holm-Šídák test]).
Animal experiments.
All animal experiments were performed in accordance with procedures approved by the Animal Care and Use Committee of Kyushu University (permit no. A19-383-0) and were in compliance with all relevant ethical regulations.
Data availability.
The data sets generated and/or analyzed during the current study are available from the corresponding authors on reasonable request.
ACKNOWLEDGMENTS
We thank Z. Matsuda for providing the DSP assay system, Z. Zhou and M. E. Greenberg for providing the pLLX vector, and N. Kurisaki, H. Harada, S. Watanabe, and R. Andino for discussions. We also appreciate technical assistance from the Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences.
This work was supported by JSPS KAKENHI grant no. JP20K07527 (to Y.S.) and JP20H00507 (to Y.Y.), Qdai-jump Research Program Wakaba Challenge FA79903505 (to Y.S.), and Japan Agency for Medical Research and Development (AMED) grant JP20wm0325002 (to T.H.). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Contributor Information
Yuta Shirogane, Email: yuuta@virology.med.kyushu-u.ac.jp.
Yusuke Yanagi, Email: yyanagi@virology.med.kyushu-u.ac.jp.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Coughlin MM, Beck AS, Bankamp B, Rota PA. 2017. Perspective on global measles epidemiology and control and the role of novel vaccination strategies. Viruses 9:11. 10.3390/v9010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mekki M, Eley B, Hardie D, Wilmshurst JM. 2019. Subacute sclerosing panencephalitis: clinical phenotype, epidemiology, and preventive interventions. Dev Med Child Neurol 61:1139–1144. 10.1111/dmcn.14166. [DOI] [PubMed] [Google Scholar]
- 3.Griffin DE. 2013. Measles virus, p 1042–1069. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Watanabe S, Shirogane Y, Sato Y, Hashiguchi T, Yanagi Y. 2019. New insights into measles virus brain infections. Trends Microbiol 27:164–175. 10.1016/j.tim.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Lamb RA, Parks GD. 2013. Paramyxoviridae, p 957–995. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893–897. 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 7.Noyce RS, Bondre DG, Ha MN, Lin L-T, Sisson G, Tsao M-S, Richardson CD. 2011. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog 7:e1002240. 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VHJ, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533. 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laksono BM, de Vries RD, McQuaid S, Duprex WP, de Swart RL. 2016. Measles virus host invasion and pathogenesis. Viruses 8:210. 10.3390/v8080210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reymond N, Fabre S, Lecocq E, Adelaı¨de J, Dubreuil P, Lopez M. 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J Biol Chem 276:43205–43215. 10.1074/jbc.M103810200. [DOI] [PubMed] [Google Scholar]
- 11.McQuaid S, Cosby SL. 2002. An immunohistochemical study of the distribution of the measles virus receptors, CD46 and SLAM, in normal human tissues and subacute sclerosing panencephalitis. Lab Invest 82:403–409. 10.1038/labinvest.3780434. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe S, Ohno S, Shirogane Y, Suzuki SO, Koga R, Yanagi Y. 2015. Measles virus mutants possessing the fusion protein with enhanced fusion activity spread effectively in neuronal cells, but not in other cells, without causing strong cytopathology. J Virol 89:2710–2717. 10.1128/JVI.03346-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter MA. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255–265. 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki Y, Koprowski H. 1974. Cell to cell transmission of virus in the central nervous system. I. Subacute sclerosing panencephalitis. Lab Invest 31:187–196. [PubMed] [Google Scholar]
- 15.Allen IV, McQuaid S, McMahon J, Kirk J, Mcconnell R. 1996. The significance of measles virus antigen and genome distribution in the CNS in SSPE for mechanisms of viral spread and demyelination. J Neuropathol Exp Neurol 55:471–480. 10.1097/00005072-199604000-00010. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence DM, Patterson CE, Gales TL, D'Orazio JL, Vaughn MM, Rall GF. 2000. Measles virus spread between neurons requires cell contact but not CD46 expression, syncytium formation, or extracellular virus production. J Virol 74:1908–1918. 10.1128/jvi.74.4.1908-1918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato Y, Watanabe S, Fukuda Y, Hashiguchi T, Yanagi Y, Ohno S. 2018. Cell-to-cell measles virus spread between human neurons is dependent on hemagglutinin and hyperfusogenic fusion protein. J Virol 92:e02166-17. 10.1128/JVI.02166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirogane Y, Watanabe S, Yanagi Y. 2012. Cooperation between different RNA virus genomes produces a new phenotype. Nat Commun 3:1235. 10.1038/ncomms2252. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe S, Shirogane Y, Suzuki SO, Ikegame S, Koga R, Yanagi Y. 2013. Mutant fusion proteins with enhanced fusion activity promote measles virus spread in human neuronal cells and brains of suckling hamsters. J Virol 87:2648–2659. 10.1128/JVI.02632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayata M, Takeuchi K, Takeda M, Ohgimoto S, Kato S, Sharma LB, Tanaka M, Kuwamura M, Ishida H, Ogura H. 2010. The F gene of the Osaka-2 strain of measles virus derived from a case of subacute sclerosing panencephalitis is a major determinant of neurovirulence. J Virol 84:11189–11199. 10.1128/JVI.01075-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurgens EM, Mathieu C, Palermo LM, Hardie D, Horvat B, Moscona A, Porotto M. 2015. Measles fusion machinery is dysregulated in neuropathogenic variants. mBio 6:e02528-14. 10.1128/mBio.02528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angius F, Smuts H, Rybkina K, Stelitano D, Eley B, Wilmshurst J, Ferren M, Lalande A, Mathieu C, Moscona A, Horvat B, Hashiguchi T, Porotto M, Hardie D. 2018. Analysis of a subacute sclerosing panencephalitis genotype B3 virus from the 2009–2010 South African measles epidemic shows that hyperfusogenic F proteins contribute to measles virus infection in the brain. J Virol 93:e01700-18. 10.1128/JVI.01700-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashiguchi T, Fukuda Y, Matsuoka R, Kuroda D, Kubota M, Shirogane Y, Watanabe S, Tsumoto K, Kohda D, Plemper RK, Yanagi Y. 2018. Structures of the prefusion form of measles virus fusion protein in complex with inhibitors. Proc Natl Acad Sci U S A 115:2496–2501. 10.1073/pnas.1718957115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirogane Y, Hashiguchi T, Yanagi Y. 2019. Weak cis and trans interactions of the hemagglutinin with receptors trigger fusion proteins of neuropathogenic measles virus isolates. J Virol 94:e01727-19. 10.1128/JVI.01727-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannons JL, Tangye SG, Schwartzberg PL. 2011. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol 29:665–705. 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 26.Mandai K, Rikitake Y, Mori M, Takai Y. 2015. Nectins and nectin-like molecules in development and disease. Curr Top Dev Biol 112:197–231. 10.1016/bs.ctdb.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Kondo N, Miyauchi K, Meng F, Iwamoto A, Matsuda Z. 2010. Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J Biol Chem 285:14681–14688. 10.1074/jbc.M109.067090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa H, Meng F, Kondo N, Iwamoto A, Matsuda Z. 2012. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng Des Sel 25:813–820. 10.1093/protein/gzs051. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Li X, Nakane S, Liu S, Ishikawa H, Iwamoto A, Matsuda Z. 2014. Co-expression of foreign proteins tethered to HIV-1 envelope glycoprotein on the cell surface by introducing an intervening second membrane-spanning domain. PLoS One 9:e96790. 10.1371/journal.pone.0096790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori M, Rikitake Y, Mandai K, Takai Y. 2014. Roles of nectins and nectin-like molecules in the nervous system. Adv Neurobiol 8:91–116. 10.1007/978-1-4614-8090-7_5. [DOI] [PubMed] [Google Scholar]
- 31.Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T. 2007. SynCAMs organize synapses through heterophilic adhesion. J Neurosci 27:12516–12530. 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YC, Boone M, Meuris L, Lemmens I, Van Roy N, Soete A, Reumers J, Moisse M, Plaisance S, Drmanac R, Chen J, Speleman F, Lambrechts D, Van De Peer Y, Tavernier J, Callewaert N. 2014. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun 5:4767. 10.1038/ncomms5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paquet-Durand F, Tan S, Bicker G. 2003. Turning teratocarcinoma cells into neurons: rapid differentiation of NT-2 cells in floating spheres. Brain Res Dev Brain Res 142:161–167. 10.1016/s0165-3806(03)00065-8. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto K, Ono N, Tatsuo H, Minagawa H, Takeda M, Takeuchi K, Yanagi Y. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J Virol 76:6743–6749. 10.1128/jvi.76.13.6743-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Generous AR, Harrison OJ, Troyanovsky RB, Mateo M, Navaratnarajah CK, Donohue RC, Pfaller CK, Alekhina O, Sergeeva AP, Indra I, Thornburg T, Kochetkova I, Billadeau DD, Taylor MP, Troyanovsky SM, Honig B, Shapiro L, Cattaneo R. 2019. Trans-endocytosis elicited by nectins transfers cytoplasmic cargo, including infectious material, between cells. J Cell Sci 132:jcs235507. 10.1242/jcs.235507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamada D, Yoshida M, Williams YN, Fukami T, Kikuchi S, Masuda M, Maruyama T, Ohta T, Nakae D, Maekawa A, Kitamura T, Murakami Y. 2006. Disruption of spermatogenic cell adhesion and male infertility in mice lacking TSLC1/IGSF4, an immunoglobulin superfamily cell adhesion molecule. Mol Cell Biol 26:3610–3624. 10.1128/MCB.26.9.3610-3624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Weyden L, Arends MJ, Chausiaux OE, Ellis PJ, Lange UC, Surani MA, Affara N, Murakami Y, Adams DJ, Bradley A. 2006. Loss of TSLC1 causes male infertility due to a defect at the spermatid stage of spermatogenesis. MCB 26:3595–3609. 10.1128/MCB.26.9.3595-3609.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakami Y. 2005. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci 96:543–552. 10.1111/j.1349-7006.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC. 2002. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297:1525–1531. 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 40.Manivannan K, Rowan AG, Tanaka Y, Taylor GP, Bangham CRM. 2016. CADM1/TSLC1 identifies HTLV-1-infected cells and determines their susceptibility to CTL-mediated lysis. PLoS Pathog 12:e1005560. 10.1371/journal.ppat.1005560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalva MB, McClelland AC, Kayser MS. 2007. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci 8:206–220. 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakayama T, Ohashi K, Mizuno K, Iseki S. 2001. Cloning and characterization of a novel mouse immunoglobulin superfamily gene expressed in early spermatogenic cells. Mol Reprod Dev 60:158–164. 10.1002/mrd.1072. [DOI] [PubMed] [Google Scholar]
- 43.Ito T, Kasai Y, Kumagai Y, Suzuki D, Ochiai-Noguchi M, Irikura D, Miyake S, Murakami Y. 2018. Quantitative analysis of interaction between CADM1 and its binding cell-surface proteins using surface plasmon resonance imaging. Front Cell Dev Biol 6:86. 10.3389/fcell.2018.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Held W, Mariuzza RA. 2008. Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat Rev Immunol 8:269–278. 10.1038/nri2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Held W, Mariuzza RA. 2011. Cis-trans interactions of cell surface receptors: biological roles and structural basis. Cell Mol Life Sci 68:3469–3478. 10.1007/s00018-011-0798-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol 75:4399–4401. 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakashima H, Tsujimura K, Irie K, Ishizu M, Pan M, Kameda T, Nakashima K. 2018. Canonical TGF-β signaling negatively regulates neuronal morphogenesis through TGIF/Smad complex-mediated CRMP2 suppression. J Neurosci 38:4791–4810. 10.1523/JNEUROSCI.2423-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 49.Takeda M, Ohno S, Seki F, Nakatsu Y, Tahara M, Yanagi Y. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J Virol 79:14346–14354. 10.1128/JVI.79.22.14346-14354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tahara M, Takeda M, Seki F, Hashiguchi T, Yanagi Y. 2007. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J Virol 81:2564–2572. 10.1128/JVI.02449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868–872. 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Z, Hong EJ, Cohen S, Zhao W, Ho H-YH, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JAJ, Weitz CJ, Greenberg ME. 2006. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52:255–269. 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A 104:19535–19540. 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson IM, Edlund K, Lundberg E, Navani S, Szigyarto CAK, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, Von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, Von Heijne G, Nielsen J, Pontén F. 2015. Proteomics. Tissue-based map of the human proteome. Science 347:1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 55.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, Von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnström H, Glimelius B, Sjöblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. 2017. A pathology atlas of the human cancer transcriptome. Science 357:eaan2507. 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 56.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, Bäckström A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson A, Sjöstedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Pontén F, Von Feilitzen K, Lilley KS, Uhlén M, Lundberg E. 2017. A subcellular map of the human proteome. Science 356:eaal3321. 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and/or analyzed during the current study are available from the corresponding authors on reasonable request.