ABSTRACT
Endogenous retroviruses (ERVs) are sequences in animal genomes that originated from ancient retrovirus infections; they provide genetic novelty in hosts by being coopted as functional genes or elements during evolution. Recently, we demonstrated that endogenous elements from not only from retroviruses but also nonretroviral RNA viruses are a possible source of functional genes in host animals. The remnants of ancient bornavirus infections, called endogenous bornavirus-like elements (EBLs), are present in the genomes of a wide variety of vertebrate species, and some express functional products in host cells. Previous studies have predicted that the human EBL locus derived from bornavirus nucleoprotein, termed hsEBLN-2, expresses mRNA encoding a protein, suggesting that hsEBLN-2 has acquired a cellular function during evolution. However, the detailed function of the hsEBLN-2-derived product remains to be elucidated. In this study, we show that the hsEBLN-2-derived protein E2 acts as a mitochondrial protein that interacts with mitochondrial host factors associated with apoptosis, such as HAX-1. We also demonstrate that knockdown of hsEBLN-2-derived RNA increased the levels of PARP and caspase-3 cleavage and markedly decreased cell viability. In contrast, overexpression of E2 enhanced cell viability, as well as the intracellular stability of HAX-1, under stress conditions. Our results suggest that hsEBLN-2 has been coopted as a host gene, the product of which is involved in cell viability by interacting with mitochondrial proteins.
IMPORTANCE Our genomes contain molecular fossils of ancient viruses, called endogenous virus elements (EVEs). Mounting evidence suggests that EVEs derived from nonretroviral RNA viruses have acquired functions in host cells during evolution. Previous studies have revealed that a locus encoding a bornavirus-derived EVE, hsEBLN-2, which was generated approximately 43 million years ago in a human ancestor, may be linked to the development of some tumors. However, the function of hsEBLN-2 has not been determined. In this study, we found that the E2 protein, an expression product of hsEBLN-2, interacts with apoptosis-related host proteins as a mitochondrial protein and affects cell viability. This study suggests that nonretroviral RNA viral EVEs have been coopted by hosts with more diverse functions than previously thought, showing a pivotal role for RNA virus infection in evolution.
KEYWORDS: bornavirus, endogenous viral element, paleovirology
INTRODUCTION
Endogenous viral elements are remnants of ancient viral infections in animal genomes. Most of these elements originate from retroviruses called endogenous retroviruses (ERVs), which comprise approximately 8% of the human genome (1, 2). During long-term evolution as host DNAs, the vast majority of ERVs have accumulated mutations and indels in the viral sequence, resulting in inactivation of viral proliferative properties. On the other hand, it has been shown that some ERVs have evolved as functional genes or regulatory elements to play pivotal roles in cellular processes (3). An envelope-like protein derived from human ERV called syncytin is the best-known example of cooption of a viral gene by a host. Syncytin is expressed in placental trophoblasts and functions in human embryo placentation (4). Another example is the neuronal protein Arc, which may have originated from the Gag protein of ancestral retroviruses. Arc acts as a vehicle of mRNA transport into dendrites, which is essential for long-lasting memory in the mammalian brain (5, 6). Antiviral immunity is an acquired function of ERVs. Protein products of certain ERVs can protect host cells from infection or replication of newly invaded exogenous retroviruses (7, 8). Furthermore, regulatory sequences in the long terminal repeats of ERVs control expression of host genes near their integration sites (9). Although ERVs provide beneficial impacts to hosts, it has also been suggested that human ERVs are involved in the pathogenesis of inflammatory diseases or cancers, including lymphoma, leukemia, and carcinomas (10–13). These findings indicate that ERVs, as genomic invaders, have had significant effects on host evolution through assimilation with host genomic DNA.
Recent studies have revealed that integrated nonretroviral RNA virus DNA copies are present in eukaryotic genomes. Endogenous bornavirus-like elements (EBLs) were the first nonretroviral RNA virus elements discovered in vertebrate species (14, 15). The family Bornaviridae includes three genera, Orthobornavirus in all vertebrates except for amphibians, Carbovirus in reptiles, and Cultervirus in fishes. At present, EBLs closely related to orthobornaviruses and carboviruses have been found in many mammals (15). The genome of the primate lineage harbors at least eight independently integrated EBLs. Orthology analyses have revealed that Simiiformes share EBL loci, suggesting that ancient bornaviruses were integrated into our genomes before the divergence of Platyrrhini and Catarrhini approximately 43 million years ago (16). Members of the superorder Afrotheria, which includes elephants, aardvarks, golden moles, and tenrecs, also have identifiable orthologs of EBLs, which were established in the genome of a common ancestor of afrotherians at least 83.3 million years ago, indicating long-term coevolution between bornaviruses and mammals (17).
Recent evidence has shown that, similarly to ERVs, endogenous nonretroviral RNA virus elements might have been coopted by hosts during evolution. Some EBLs still retain relatively long open reading frames (ORFs) and are transcribed into RNAs in host cells. The transcript of an EBL derived from orthobornavirus nucleoprotein (N) in the human genome, named hsEBLN-1, affects the expression of a neighboring gene (18). It has also been demonstrated that hsEBLN-1 is involved in cell cycle transition, cell growth, apoptosis induction, and microtube organization (19, 20). Interestingly, antiviral function against exogenous Borna disease virus 1 (BoDV-1) was found in an EBL in the thirteen-lined ground squirrel genome, itEBLN, and its expression was detected in some tissues of squirrels. The recombinant itEBLN protein strongly suppressed BoDV-1 replication in cultured cells as a dominant negative competitor of the BoDV-1 N protein (21). Furthermore, we demonstrated that both rodent and primate EBLs are present in PIWI-interacting RNA (piRNA)-generating regions of host genomes at a rate that is much more frequent than expected by chance and that they express piRNAs with the potential to protect against bornavirus infection by acting as antisense nucleotides to viral N mRNAs (22). Moreover, bats of the genus Eptesicus have carried a preserved EBL element derived from the ancient bornaviral RNA polymerase gene, with an intact open reading frame (ORF) of 1,718 amino acids, for more than 11.8 million years (23). All of these observations indicate that not only retroviruses but also nonretroviral RNA viruses have served as a source of genetic novelty and have resulted in significant genome evolution in hosts.
We have previously shown that some EBLs in the human genome are ubiquitously expressed in several tissues (18). Among them, hsEBLN-2, which encodes a 273-amino acid ORF with high homology to the orthobornavirus N protein, has been reported to interact with other cellular proteins, suggesting that it encodes a functional protein (24). However, the detailed functions of hsEBLN-2 and its product remain to be elucidated. In this study, we show that the hsEBLN-2-derived protein E2 acts as a mitochondrial protein that interacts with mitochondrial host factors associated with apoptosis. Moreover, we demonstrate that knockdown of hsEBLN-2 RNA induces PARP and caspase-3 cleavages and then reduces cell viability. These results suggest that hsEBLN-2 has been exogenously adapted as a host gene and has acquired functions related to cell viability through interactions with other mitochondrial proteins.
RESULTS
Expression profile of hsEBLN-2.
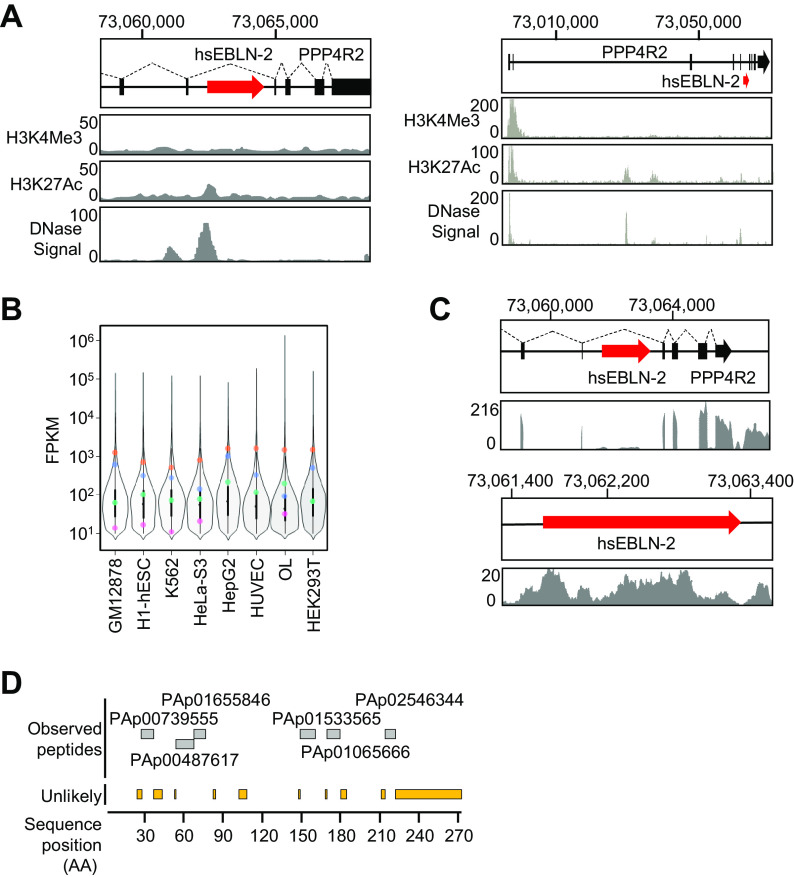
hsEBLN-2, which is located on chromosome 3 at locus 3p13, is inserted within an intron region of the PPP4R2 gene and is flanked by an AluSx element at the 3′ end of the hsEBLN-2 sequence (16). In previous studies, we detected the transcript containing the hsEBLN-2 sequence in various tissues and cell lines (16, 18); however, its detailed profiles have not yet been determined. Based on the UCSC genome browser, H3K4me3/H3K27Ac chromatin immunoprecipitation sequencing (ChIP-seq) around the hsEBLN-2 locus revealed weak H3K27Ac and DNase-sensitive regions upstream of the hsEBLN-2 gene (Fig. 1A). ChIP-Atlas data predict that some enhancer or transcription factors bind upstream of the hsEBLN-2 locus (see Table S1 in the supplemental material), predicting the transcription potential of this locus.
FIG 1.
Expression profile of hsEBLN-2 in the human genome. (A) A schematic diagram showing the position of hsEBLN-2 on human chromosome 3 (chr 3) at locus 3p13 and a UCSC genome browser view (UCSC Genome Browser on Human Assembly; December 2013) (53) showing coverage of H3K4me3 and H3K17Ac chromatin immunoprecipitation sequencing (ChIP-seq) reads and DNase hypersensitive sites of GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, and NHLF cells. (B) Expression level of hsEBLN-2 (pink), PPP4R2 (green), TOMM20 (orange), and HPRT1 (blue) RNAs in human-derived cell lines. Each violin plot and box plot show the distribution of the expression levels of genes in each cell line. (C) IGV genome browser view showing the depth of RNA-seq reads mapped to the genomic region of the hsEBLN-2 locus. (D) Sequences of the hsEBLN-2 region registered in the Peptide Atlas (http://www.peptideatlas.org/) are shown. Displayed peptides are observed from several tissues, including ovary, breast, and blood plasma.
Using publicly available RNA sequencing (RNA-seq) data, we analyzed the expression levels of hsEBLN-2, TOMM20 (which encodes a mitochondrial protein), HPRT1 (a housekeeping gene), and PPP4R2 across eight human cell lines of different origins (GM12878, H1-hESC, K562, HeLa-S3, HepG2, HUVEC, OL, and HEK293T cells). The expression levels were divided into three groups based on FPKM values (Fig. 1B), as follows: the lower quartile was defined as low expression, the interquartile range as moderate expression, and the upper quartile as high expression. hsEBLN-2 RNA was found to be expressed in all cell lines analyzed, and fragments per kilobase per million (FPKM) values for hsEBLN-2 RNA ranged from approximately 0.2 to 3. These data showed that hsEBLN-2 RNA expression is relatively low in most cell lines but moderate in OL cells derived from human oligodendrocytes. In addition, no correlation in the expression levels was found between PPP4R2 and hsEBLN-2 RNAs, suggesting that the hsEBLN-2 RNA is transcribed independently of the PPP4R2 promoter. We therefore performed next-generation sequencing analysis using OL cells and found that hsEBLN-2 RNA is expressed in these cells, albeit at a lower level than that of PPP4R2 exons (Fig. 1C). To further confirm translation of hsEBLN-2 RNA, we searched Peptide Atlas for peptides from hsEBLN-2 and detected expression of multiple peptides within the sequence region in several tissues, such as the ovary, breast, and blood plasma (Fig. 1D). Furthermore, examination of publicly available data sets in GTEx (25) and the Protein Atlas (26) revealed high expression of hsEBLN-2 RNA in the tibial nerve, ovary, and bone marrow. These data suggest that hsEBLN-2 RNA is expressed and translated into protein.
E2 protein localizes to mitochondria.
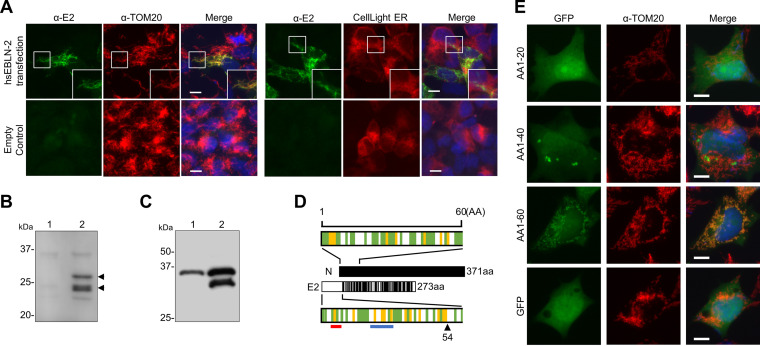
To understand the possible function of the hsEBLN-2 product, we first generated an E2 protein expression plasmid and transfected it into OL cells. Intracellular localization of E2 was detected using the anti-E2 polyclonal antibody we generated. As shown in Fig. 2A, the recombinant E2 protein clearly colocalized with the mitochondrial protein TOM20 but not with the endoplasmic reticulum (ER) marker CellLight ER-red fluorescent protein (RFP), indicating that E2 is a mitochondrial protein. Western blot analysis revealed that two bands at approximately 24 and 30 kDa were induced in transfected cells (Fig. 2B, arrowheads). It has been shown that the vast majority of mitochondrial proteins have a mitochondrial targeting signal (MTS) at the N terminus of their precursor, which is cleaved upon/after mitochondrial import (27); this suggests that the two bands detected by Western blotting represent the precursor and cleaved forms of E2, respectively. To confirm this, we generated an E2 protein that was Flag tagged at either the N or C terminus. As shown in Fig. 2C, a 24-kDa band was detected only in cells transfected with C-terminally tagged E2. This result indicates that the N terminus of the E2 protein was cleaved. We next predicted the cleavage region at the E2 N terminus by using Mitoprot and MitoFates (27–29), which can predict a mitochondrial targeting sequence and cleavage site in the N-terminal region of input proteins. As illustrated in Fig. 2D, a cleavage site was predicted at amino acid 54. The recombinant protein in which green fluorescent protein (GFP) is fused to amino acids 1 to 60 of E2 (AA1-60) at the N terminus colocalized with TOM20 in transfected cells (Fig. 2E), confirming that the N-terminal region of E2 functions as an MTS. The fact that the N-terminal region of the E2 protein lacks detectable homology with the BoDV-1 N protein (Fig. 2D) indicates that the MTS was acquired after hsEBLN-2 integration into the genome.
FIG 2.
Mitochondrial targeting of the E2 protein. (A) Subcellular localization of E2. OL cells were transfected with the E2 expression plasmid or empty plasmid and stained with an anti-E2 antibody. An anti-TOM20 antibody and CellLight endoplasmic reticulum (ER)-red fluorescent protein (RFP) were used to visualize mitochondria and the ER, respectively. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and observed by fluorescence microscopy. Bars, 10 μm. (B and C) The E2 protein is cleaved in transfected cells. (B) 293T cells were transfected with the control (lane 1) or E2 expression plasmid (lane 2), and the E2 protein was detected by Western blotting with an anti-E2 antibody. (C) 293T cells were transfected with the E2 expression plasmid tagged with Flag at the C (lane 1) or N terminus (lane 2) of the protein, and the E2 protein was detected by Western blotting with an anti-Flag antibody. (D) The E2 protein contains a mitochondrial targeting signal. The full-length and N-terminal 60 amino residues of the Borna disease virus 1 (BoDV-1) N and E2 proteins are shown in the schematic diagram. Sites in the E2 protein homologous or similar to those in the N protein are indicated by black lines. The green line indicates hydrophobic amino acids, and the yellow line indicates basic amino acids. The arrowheads indicate the predicted cleavage positions by mitochondrial processing peptidase predicted by both Mitoprot and MitoFates. The red and blue lines indicate the binding site of TOM20 and the positively charged amphiphilicity high score region predicted by MitoFates, respectively. N, BoDV-1 N; E2, hsEBLN-2. (E) The N terminus of BoDV-1 N contains a putative mitochondrial targeting signal (MTS). OL cells were transfected with plasmids expressing green fluorescent protein (GFP) fused with the N-terminal 20 amino acids (aa) (AA1-20), 40 aa (AA1-40), and 60 aa (AA1-60), and their localization was detected by fluorescence microscopy after staining with an anti-TOM20 antibody. Bars, 10 μm.
E2 interacts with mitochondrial proteins.
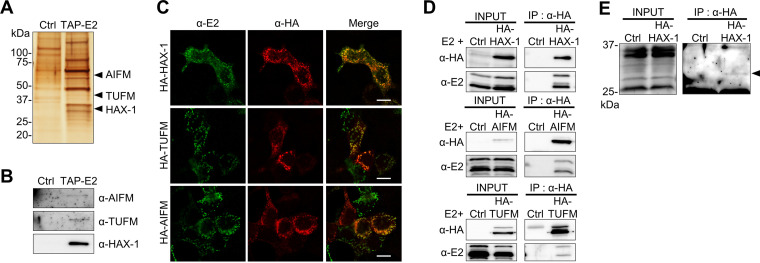
In a previous study, several proteins were reported to interact with the E2 protein using a large-scale human protein-protein interaction mapping assay (Table 1) (24); however, the certainty and details of the interaction between each protein were not analyzed. To better understand the function of E2, we sought to determine binding partners by an immunoprecipitation assay with tandem affinity purification-tagged E2 (TAP-E2). Here, we used 293T cells, which have a higher transfection efficiency than that of OL cells. As shown in Fig. 3A and B, SDS-PAGE followed by mass spectrometry revealed that the E2 protein interacts with TUFM (Tu translation elongation factor, mitochondrial), AIFM (apoptosis-inducing factor mitochondria-associated 1), and HAX-1 (HCLS1-associated protein X-1), all of which are known as mitochondrially localized proteins. Furthermore, we observed colocalization of E2 with these interacting proteins at mitochondria in 293T cells transfected with hemagglutinin (HA)-tagged interacting proteins (Fig. 3C). A coimmunoprecipitation assay using cells transfected with the E2 expression plasmid and each HA-tagged host protein confirmed their interactions (Fig. 3D). Moreover, we detected the endogenous E2 protein as a faint band by immunoprecipitation with the HA-HAX protein (Fig. 3E). These observations suggest that the E2 protein plays a role with known interacting partners as a mitochondrial protein.
TABLE 1.
Predicted and confirmed interaction partners of E2 protein
| Gene symbol | Full gene name | Main locationa | Related biological processb |
|---|---|---|---|
| Predicted interaction partners | |||
| STK16 | Serine/threonine kinase 16 | Cytosol | Protein autophosphorylation |
| TUSC2 | Tumor suppressor candidate 2 | Cytosol | Cell cycle |
| CAB39 | Calcium binding protein 39 | Nucleoli fibrillar center | Cell cycle arrest |
| FANCC | Fanconi anemia complementation group C | Nucleoplasm | DNA damage, DNA repair |
| AP1S1 | Adaptor-related protein complex 1 sigma 1 subunit | Golgi apparatus and vesicles | Protein transport |
| WRAP73 | WD repeat containing, antisense to TP73 | Not available | Cilium biogenesis/degradation |
| C20orf24 | Chromosome 20 open reading frame 24 | Cytosol | Mitochondrial respirasome assembly |
| ATG12 | Autophagy related 12 | Not available | Autophagosome assembly |
| PRKAB1 | Protein kinase AMP-activated noncatalytic subunit beta 1 | Cytosol | Fatty acid biosynthesis |
| Confirmed interaction partners | |||
| HAX-1 | HCLS1-associated protein X-1 | Mitochondria | Negative regulation of apoptotic process |
| AIFM | Apoptosis-inducing factor mitochondria-associated 1 | Mitochondria | Positive regulation of apoptotic process |
| TUFM | Tu translation elongation factor, mitochondrial | Mitochondria | Mitochondrial translational elongation |
Information on main location and related biological process were obtained from The Human Protein Atlas (https://www.proteinatlas.org/) (26).
Information on main location and related biological process were obtained from UniProt (https://www.uniprot.org/) (52).
FIG 3.
The E2 protein interacts with mitochondrial proteins. (A) Tandem affinity purification (TAP) assay of the E2 protein. 293T cells were transfected with pCTAP-E2 (TAP-E2) or the control empty plasmid (Ctrl) and processed for TAP assays. After SDS-PAGE, the gel was stained by silver staining, and the specific bands were evaluated by mass spectrometry. (B) TAP detection of proteins of interest was confirmed by Western blotting with specific antibodies against each protein. (C) Plasmids expressing E2 and hemagglutinin (HA)-tagged interacting proteins were cotransfected into 293T cells, and localizations were detected using anti-E2 and anti-HA antibodies. Bars, 10 μm. (D) Pulldown assay of E2-interacting proteins. 293T cells were cotransfected with plasmids expressing E2 and HA-tagged E2-interacting proteins. Immunoprecipitation was conducted using an anti-HA antibody and detected with anti-HA and E2 antibodies. (E) Exogenous expression of the E2 protein. 293T cells were transfected with the HA-tagged HAX-1 plasmid, and 24 h after transfection, immunoprecipitation was performed with an anti-HA antibody. An arrowhead indicates full-length endogenous E2 protein detected with the anti-E2 antibody.
A possible link of E2 protein to the cellular stress response.
Among the interacting proteins identified, AIFM and HAX-1 have been reported to be associated with apoptosis; AIFM binds to the mitochondrial intima and is thought to act as an NADH oxidase (30). In addition, AIFM is cleaved by proteases activated downstream of apoptosis-inducing signals, such as PARP, and migrates to the nucleus, causing DNA fragmentation and caspase-independent apoptosis (31). In contrast, HAX-1, located in both the ER and the mitochondria, is associated with the maintenance of mitochondrial function and protection from apoptosis (32). HAX-1 is known to promote cell survival by attenuating signals from mitochondria under stress conditions (33).
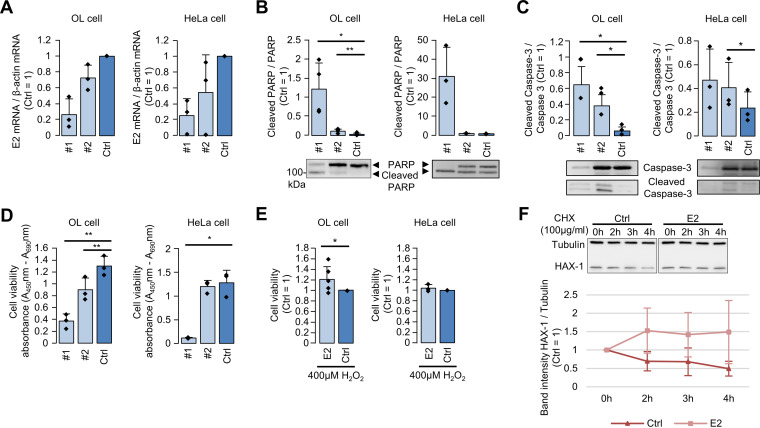
As both AIFM and HAX-1 are involved in apoptosis, we focused on whether the function of E2 is also related to cell viability and apoptosis. To examine this, we knocked down endogenous hsEBLN-2 RNA by using antisense oligonucleotides (ASOs) in OL and HeLa cells and investigated PARP and caspase-3 cleavages, hallmarks of apoptosis. As depicted in Fig. 4A to C, ASOs reduced hsEBLN-2 RNA and significantly increased the levels of PARP and caspase-3 cleavages in both cell lines. Furthermore, the viability of cells treated with ASOs was markedly decreased (Fig. 4D). In contrast, OL cells overexpressing E2 showed slightly but significantly increased viability compared with that of control cells after stress induction by H2O2 treatment (Fig. 4E). Note that overexpression of E2 appeared not to increase the cell viability in HeLa cells, since the cells are relatively sensitive to cell damage induced by plasmid transfection. These results indicate that expression of E2 may confer durability to cellular stress by suppressing apoptosis.
FIG 4.
The E2 protein is involved in cell viability. (A) OL and HeLa cells were transfected with antisense oligonucleotides (ASOs) against hsEBLN-2 RNA (ASO 1 and 2) and control ASO (Ctrl), and relative levels of hsEBLN-2 RNA were quantified by real-time reverse transcription-PCR (RT-PCR). Values are presented as the mean ± standard error (SE) of three independent experiments. (B and C) OL and HeLa cells were transfected with each ASO, and PARP and caspase-3 cleavages were detected with an anti-PARP antibody and an anti-caspase-3 antibody. Relative expression levels of cleaved proteins were calculated by the band intensity based on Western blotting. Values are presented as the mean ± SE of three independent experiments. Statistical significance was analyzed by two-tailed t test. *, P < 0.05’ **, P < 0.01. (D) Knockdown of hsEBLN-2 RNA reduces cell viability. OL and HeLa cells were transfected with ASOs, and cell viability was measured by the WST-1 assay. Values are presented as the mean ± SE of three independent experiments. Statistical significance was analyzed by two-tailed t test. *, P < 0.05; **, P < 0.01. (E) E2 overexpression increases stress resistance. OL and HeLa cells were transfected with E2 expression (E2) and control empty (Ctrl) plasmids, and 24 h after transfection, the cells were treated with 400 μM H2O2 for 24 h. Cell viability was measured by the WST-1 assay. Values are presented as the mean ± SE of six or three independent experiments. Statistical significance was analyzed by two-tailed t test. *, P < 0.05. (F) OL cells were transfected with E2 or control empty plasmid (Ctrl) and treated with 100 μg/ml cycloheximide (CHX) for 2, 3, and 4 h. Expression levels of HAX-1 and tubulin were detected by Western blotting, and band intensities were measured by ImageJ. Values are presented as the mean ± SE of three independent experiments.
Finally, to investigate the mechanism underlying the function of the E2 protein, we further focused on HAX-1 because it has been reported to interact with several virus-derived proteins, including human immunodeficiency virus 1 (HIV-1) Vpr and hepatitis C virus (HCV) core protein (34, 35). Previous reports have revealed that interaction among viral proteins affects the stability or cellular localization of HAX-1, leading to disruption of HAX-1 function (35). As shown in Fig. 4F, overexpression of E2 markedly enhanced the intracellular stability of HAX-1 when cells were treated with cycloheximide, which is an inhibitor of protein biosynthesis. This result suggests that interaction of the E2 protein with HAX-1 may enhance the stability of the latter in mitochondria, enhancing cell viability.
DISCUSSION
In this study, we demonstrated that the E2 protein may be involved in cell viability as a mitochondrial protein. It has been shown that hsEBLN-2 integrated into an intron of the PPP4R2 gene and then evolved as a host genomic sequence for at least 43 million years (16). Nevertheless, hsEBLN-2 remains a long ORF and has the potential to express transcripts. Consistent with previous studies showing hsEBLN-2 transcription in many tissues and cultured cells (18, 23), we observed expression of hsEBLN-2 by RNA-seq analysis using OL cells, albeit at a low level (Fig. 1). Although the promoter sequence for hsEBLN-2 transcription has not yet been identified, it will be of interest to elucidate whether transcription of hsEBLN-2 is regulated by epigenetics or is coregulated by the host genes. Further studies are needed to understand the host gene transcription associated with the expression of hsEBLN-2.
We found that E2 localizes to mitochondria and carries a mitochondrial localization signal, suggesting the possibility that E2 is cleaved after entering the mitochondria. The parent gene of hsEBLN-2 is the bornaviral N gene, which encodes nucleoprotein and binds to viral genomic RNA to form the nucleocapsid. The N proteins of current bornaviruses have a nuclear localization signal (NLS) at the N terminus and localize at the nucleus, where the replication of bornaviruses takes place (36, 37). E2 does not encode the NLS in the corresponding region of bornaviral N proteins and indeed does not exhibit nuclear localization under the conditions examined. The mitochondrial localization signal of E2 is located in the approximately 60-amino-acid extension region of the N terminus, which has no sequence homology to bornaviral N (Fig. 2). This suggests that hsEBLN-2 is integrated immediately downstream of the mitochondrial localization signal. Alternatively, the MTS sequence might have been acquired during integration via template switching by LINE-1. In any case, the acquisition of the MTS may have changed its subcellular localization and conferred a new function to E2, and the NLS may have become inactive by accumulation of mutations. The fact that the MTSs are conserved across the primate species, including chimpanzees (GenBank accession number XP_009444139.2), gorillas (GenBank accession number XP_004035951.1), and macaque monkeys (GenBank accession number NP_001252880.1), indicates that EBLN-2 may have acquired an MTS at the time of integration more than 43 million years ago, before the divergence of Platyrrhini and Catarrhini (15). Note that we cannot exclude the possibility that the sequence has subsequently evolved into the MTS during evolution.
We identified mitochondrial proteins, namely, TUFM, AIFM, and HAX-1, as interacting partners of the E2 protein (Fig. 3). AIFM, a known apoptosis-associated protein, migrates to the nucleus after apoptosis induction and promotes chromosome condensation and fragmentation (31). Although TUFM functions as a translation elongation factor in mitochondria, it has been reported that expression of TUFM is involved in the transition from adenoma to carcinoma in colorectal cancer (38). On the other hand, HAX-1 is a protein known to regulate mitochondrial membrane potential and inhibit apoptosis, and it has been reported to interact with several proteins, including some viral proteins (34, 35, 39–43). In addition, only a faint band of endogenous E2 protein was observed by the immunoprecipitation assay with HAX-1. Because E2 protein has a predicted PEST sequence at amino acid residues 53 to 67, it may be relatively unstable. This may make it difficult to detect the endogenous E2 protein. It is interesting to note that all of the host factors found to interact with E2 are localized to mitochondria and play important roles in cell proliferation and survival. In our analysis, we observed that knockdown of hsEBLN-2-derived RNA reduced cell viability and increased cell resistance to stress by E2 protein expression (Fig. 4). As hsEBLN-2 RNA knockdown also promoted PARP cleavage, these results suggest that the E2 protein has the ability to negatively regulate apoptosis. In fact, we showed that E2 protein expression decreases HAX-1 degradation (Fig. 4), suggesting that the E2 protein may function in concert with other mitochondrial proteins involved in cell proliferation and survival. On the other hand, considering that hsEBLN-2 is located in the intron of PPP4R2, we cannot deny the possibility that ASO directly affected the pre-mRNA expression of PPP4R2. PPP4R2 has been reported to suppress DNA damage; PPP4R2 knockdown increased DNA damage and apoptosis under etoposide treatment in NSC-34 cells, a mouse nervous system cell line (44). However, PPP4R2 knockdown treatment alone did not induce apoptosis (44). Another report showed that PPP4R2 knockdown increased apoptosis in murine leukemic bone marrow cells (45), but there was no significant difference by clonogenic assay (45), suggesting that knockdown treatment alone is unlikely to cause strong cytotoxicity. Therefore, it is more likely that the induction of apoptosis is caused by knockdown of hsEBLN-2. Further experiments using hsEBLN-2 knockout cells would reveal the function of E2 protein and its evolution in detail.
It is known that E2 interacts with several host factors (24) (Table 1); however, no interaction with these proteins was identified in our study. Among the previously identified proteins, TUSC2/FUS1 appears to be an interesting candidate for analyzing the function of E2 in mitochondria. TUSC2/FUS1 is a mitochondrially localized protein with the ability to regulate cellular oxidative stress by balancing reactive oxygen species production and mitochondrial homeostasis (46). Further study to investigate the possible interaction between E2 and TUSC2/FUS1 may be important to understand the mitochondrion-related function of the E2 protein.
In summary, this study revealed that a nonretroviral endogenous RNA viral element in the human genome that is at least 43 million years old may produce a mitochondrial protein associated with cell viability. Although the function of the E2 protein in vivo remains to be elucidated, it is noteworthy that the E2 function identified in this study appears to correlate with its possible role as a tumor suppressor gene, which has been reported in different perspective studies. Further analysis of hsEBLN-2 will not only be necessary to understand coevolution between hosts and bornaviruses but will also provide insight into the evolutionary significance of why hosts preserve RNA viral genes as pseudogenes in their genomes.
MATERIALS AND METHODS
Cells and transfection.
293T cells and HeLa cells were cultured in Dulbecco’s modified Eagle's medium (DMEM)-high glucose (4.5%) supplemented with 10% fetal bovine serum (FBS). OL cells, derived from a human oligodendrocyte, were cultured in DMEM-high glucose (4.5%) supplemented with 5% FBS. Plasmid transfections were conducted using Lipofectamine LTX (Thermo Fisher Scientific) and ASO transfections using Oligofectamine (Thermo Fisher Scientific) following the manufacturer’s instructions.
Plasmid construction.
The E2 protein-expressing plasmid was constructed by inserting the hsEBLN-2 ORF, which was amplified by PCR from OL cell cDNA, into the pEF4A vector or pCTAP vector. To construct an E2 protein-expressing plasmid with GFP fused at the N terminus, the hsEBLN-2 ORF was cloned into the pAcGFP1-N1 plasmid (Clontech). HAX-1, AIFM, and TUFM ORFs were amplified from cDNA of 293T cells using N-terminal or C-terminal HA-tagged primers. After amplification, the PCR products were cloned into the pcDNA3 vector. The nucleotide sequences were confirmed by DNA sequencing.
Immunofluorescence assay.
OL or 293T cells were seeded onto 8-well chamber slides. One day after seeding, expression plasmids were transfected into the cells. For ER staining, the cells were treated with CellLight ER-RFP (Thermo Fisher Scientific) together with plasmid transfection. At 24 h after transfection, the cells were fixed for 15 min in 4% paraformaldehyde, permeabilized by incubation for 5 min in phosphate-buffered saline (PBS) containing 0.4% Triton X-100, and treated with 1% bovine serum albumin in PBS for blocking. After incubation with anti-TOM20 (catalog no. sc-17764; Santa Cruz Biotechnology), anti-HA (catalog no. H9658; Sigma-Aldrich), or anti-E2 antibodies for 1 h at 37°C, the cells were washed three times and incubated with appropriate Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 1 h at 37°C. The antibodies were each diluted 1:1,000 in 1% bovine serum albumin in PBS.
Western blotting.
Samples were subjected to 12% SDS-PAGE, and Western blotting was performed. The proteins were transferred onto a polyvinylidene fluoride membrane that was then blocked in 5% low-fat milk in PBS-0.1% Tween 20. Anti-E2, anti-HA, anti-AIFM (catalog no. ab110327; Abcam), anti-TUFM (catalog no. H00007284-B01P; Abnova), anti-caspase-3 (catalog no. GTX110543; GeneTex), and anti-HAX-1 (catalog no. GTX110543; BD Biosciences) antibodies were each diluted 1:1,000 in 5% low-fat milk in PBS-0.1% Tween 20, incubated with the membranes for 1 h at room temperature, and then washed three times. The membrane was incubated with the appropriate horseradish peroxidase-coupled antibody (Bio-Rad), which was diluted 1:5,000 in 5% low-fat milk in PBS-0.1% Tween 20, for 1 h at room temperature. After three washes, the membrane was visualized with ECL Prime Western blot detection reagents (GE Healthcare).
TAP assay.
The TAP assay was performed with the InterPlay C-terminal mammalian TAP system (Agilent Technologies) according to the manufacturer’s instructions. Briefly, the hsEBLN-2 ORF-inserted pCTAP plasmid was transfected into 293T cells. One day after transfection, the cells were collected and lysed with lysis buffer and purified with streptavidin resin and calmodulin resin. The samples that were purified by the TAP assay were subjected to 12% SDS-PAGE, and the proteins were detected by silver staining. To identify binding partners of the hsEBLN-2 protein, mass spectrometry for each band was performed using a gel slice.
Coimmunoprecipitation.
Coimmunoprecipitation was conducted in a manner similar to that described previously (47). Briefly, 293T cells were seeded into 10-cm dishes 1 day before transfection. The cells were transfected with each expression plasmid using Lipofectamine LTX. At 24 h posttransfection, the cells were collected and lysed using lysis buffer, and the cell lysates were sonicated and centrifuged. The supernatants were incubated with 10 μg of anti-HA antibody for 2 h with rotation at 4°C. After incubation, 1 mg SureBeads Protein G (Bio-Rad) was added and incubated for 2 h with rotation at 4°C. The beads were washed three times with 1 ml of lysis buffer and boiled at 98°C for 5 min to elute the proteins.
Real-time PCR.
ASOs targeting hsEBLN-2 RNA (ASO 1, GACUGGGCTGGGTGTGGTGGUUCA; ASO 2, AUCACUCATCACCGCACTCCAGCC) or luciferase, which was used as a control, were transfected into OL and HeLa cells. At 48 h after transfection, total RNA was extracted using TriPure isolation reagent (Sigma-Aldrich) and reverse transcribed using a Verso cDNA synthesis kit (Thermo Fisher Scientific) according to the instructions. Real-time PCR was conducted using Thunderbird probe quantitative PCR (qPCR) mix (Toyobo) with hsEBLN-2-specific primers (forward, GAATGTTTCTAGATCAGTGCATGGA; reverse, TCTCCTCAAAGACTGGGATCTCA) and a probe (6-carboxyfluorescein [FAM]-CTCCATTGCTCTACGGGCCATTGTG-black hole quencher 1 [BHQ1]) or β-actin-specific primers (forward, AGCCTCGCCTTTGCCG; reverse, CTGGTGCCTGGGGCG) and a probe (FAM-CCGCCGCCCGTCCACACCCGCC-BHQ1). The PCR cycling parameters were as follows: denaturation at 95°C for 1 min, followed by 45 cycles of 95°C for 15 s and 60°C for 30 s. hsEBLN-2 RNA expression was calculated by the threshold cycle (ΔΔCT) method.
WST-1 assay.
OL and HeLa cells were seeded into 24-well plates and transfected with ASOs 1 day after seeding. At 60 or 48 hours after transfection, cell viability was measured using the Premix WST-1 cell proliferation assay system (TaKaRa) according to the instructions. OL cells were seeded into 24-well plates; 1 day after seeding, the hsEBLN-2 expression plasmid or empty plasmid was transfected into the cells. At 24 hours after transfection, the cells were treated with 400 μM H2O2 for 24 h, and cell viability was measured using the Premix WST-1 cell proliferation assay system.
Next-generation sequencing.
Sequences of an OL cell total RNA sample after depletion of rRNA were analyzed by next-generation sequencing. Publicly available RNA-seq data for human cell lines (GM12878, K562, H1-hESCs, HeLa-S3, and HepG2 cells [GEO series GSE33480]; 293T cells [GEO series GSE60559]) were downloaded from National Center for Biotechnology Information (NCBI) (48). These RNA-seq reads were mapped to the human genome assembly GRCh38 using Gencode v24 annotation (49) and TopHat v2.0.13 (50). Expression level of each gene was calculated by Cufflinks v2.2.1 (51). Expression higher than 1 FPKM was considered positive expression.
CHX assay.
OL cells were seeded into 24-well plates and transfected with the hsEBLN-2 expression plasmid or empty plasmid 1 day later. At 24 h after transfection, the cells were treated with 100 mg/ml cycloheximide (CHX). The cells were collected at 2, 3 and 4 h after CHX treatment with sample buffer.
Statistics.
Statistical significance was analyzed by two-tailed t test.
ACKNOWLEDGMENTS
This work was supported in part by JSPS KAKENHI grants JP18K15172 (K.F.), JP19K22530 (K.T.), and JP20H05682 (K.T.); by MEXT KAKENHI grants JP16H06429, JP16K21723, and JP16H06430 (all to K.T.); by the JSPS Core-to-Core Program (K.T.); and by the Joint Usage/Research Center Program on inFront, Kyoto University.
Footnotes
Supplemental material is available online only.
Contributor Information
Keizo Tomonaga, Email: tomonaga@infront.kyoto-u.ac.jp.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Bock M, Stoye JP. 2000. Endogenous retroviruses and the human germline. Curr Opin Genet Dev 10:651–655. 10.1016/S0959-437X(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths DJ. 2001. Endogenous retroviruses in the human genome sequence. Genome Biol 2:reviews1017.1. 10.1186/gb-2001-2-6-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson WE. 2019. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat Rev Microbiol 17:355–370. 10.1038/s41579-019-0189-2. [DOI] [PubMed] [Google Scholar]
- 4.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 5.Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T. 2018. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172:262–274.e11. 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, Yoder N, Belnap DM, Erlendsson S, Morado DR, Briggs JAG, Feschotte C, Shepherd JD. 2018. The neuronal gene Arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell 172:275–288.e18. 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A 101:11117–11122. 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoye JP. 1998. Fv1, the mouse retrovirus resistance gene. Rev Sci Tech Oie 17:269–277. 10.20506/rst.17.1.1080. [DOI] [PubMed] [Google Scholar]
- 9.Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448:105–114. 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Boese A, Sauter M, Galli U, Best B, Herbst H, Mayer J, Kremmer E, Roemer K, Mueller-Lantzsch N. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328–4336. 10.1038/sj.onc.1203794. [DOI] [PubMed] [Google Scholar]
- 11.Depil S, Roche C, Dussart P, Prin L. 2002. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia 16:254–259. 10.1038/sj.leu.2402355. [DOI] [PubMed] [Google Scholar]
- 12.Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223–3228. 10.1038/sj.onc.1208543. [DOI] [PubMed] [Google Scholar]
- 13.Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E, Stadhouders R, Johrens K, Wurster KD, Callen DF, Harte MF, Giefing M, Barlow R, Stein H, Anagnostopoulos I, Janz M, Cockerill PN, Siebert R, Dorken B, Bonifer C, Mathas S. 2010. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med 16:571–579. 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 14.Belyi VA, Levine AJ, Skalka AM. 2010. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog 6:e1001030. 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, Tomonaga K. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84–87. 10.1038/nature08695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horie M, Tomonaga K. 2019. Paleovirology of bornaviruses: what can be learned from molecular fossils of bornaviruses. Virus Res 262:2–9. 10.1016/j.virusres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi Y, Horie M, Nakano A, Murata K, Itou T, Suzuki Y. 2016. Exaptation of bornavirus-like nucleoprotein elements in afrotherians. PLoS Pathog 12:e1005785. 10.1371/journal.ppat.1005785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sofuku K, Parrish NF, Honda T, Tomonaga K. 2015. Transcription profiling demonstrates epigenetic control of non-retroviral RNA virus-derived elements in the human genome. Cell Rep 12:1548–1554. 10.1016/j.celrep.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 19.He P, Sun L, Zhu D, Zhang H, Zhang L, Guo Y, Liu S, Zhou J, Xu X, Xie P. 2016. Knock-down of endogenous bornavirus-like nucleoprotein 1 inhibits cell growth and induces apoptosis in human oligodendroglia cells. Int J Mol Sci 17:435. 10.3390/ijms17040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers KN, Barone G, Ganesh A, Staples CJ, Howard AE, Beveridge RD, Maslen S, Skehel JM, Collis SJ. 2016. The bornavirus-derived human protein EBLN1 promotes efficient cell cycle transit, microtubule organisation and genome stability. Sci Rep 6:35548. 10.1038/srep35548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujino K, Horie M, Honda T, Merriman DK, Tomonaga K. 2014. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc Natl Acad Sci U S A 111:13175–13180. 10.1073/pnas.1407046111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parrish NF, Fujino K, Shiromoto Y, Iwasaki YW, Ha H, Xing J, Makino A, Kuramochi-Miyagawa S, Nakano T, Siomi H, Honda T, Tomonaga K. 2015. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA (New York, NY) 21:1691–1703. 10.1261/rna.052092.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horie M, Kobayashi Y, Honda T, Fujino K, Akasaka T, Kohl C, Wibbelt G, Muhldorfer K, Kurth A, Muller MA, Corman VM, Gillich N, Suzuki Y, Schwemmle M, Tomonaga K. 2016. An RNA-dependent RNA polymerase gene in bat genomes derived from an ancient negative-strand RNA virus. Sci Rep 6:25873. 10.1038/srep25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. 2007. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3:89. 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–585. 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. 2015. Tissue-based map of the human proteome. Science 347:1260419–1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 27.Schatz G. 1996. The protein import system of mitochondria. J Biol Chem 271:31763–31766. 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 28.Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786. 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 29.Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K. 2015. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14:1113–1126. 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cande C, Cohen I, Daugas E, Ravagnan L, Larochette N, Zamzami N, Kroemer G. 2002. Apoptosis-inducing factor (AIF): a novel caspase-independent death effector released from mitochondria. Biochimie 84:215–222. 10.1016/S0300-9084(02)01374-3. [DOI] [PubMed] [Google Scholar]
- 31.Hong SJ, Dawson TM, Dawson VL. 2004. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci 25:259–264. 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Fadeel B, Grzybowska E. 2009. HAX-1: a multifunctional protein with emerging roles in human disease. Biochim Biophys Acta 1790:1139–1148. 10.1016/j.bbagen.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Vafiadaki E, Arvanitis DA, Pagakis SN, Papalouka V, Sanoudou D, Kontrogianni-Konstantopoulos A, Kranias EG. 2009. The anti-apoptotic protein HAX-1 interacts with SERCA2 and regulates its protein levels to promote cell survival. Mol Biol Cell 20:306–318. 10.1091/mbc.e08-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee A, Saito K, Meyer K, Banerjee S, Ait-Goughoulte M, Ray RB, Ray R. 2009. Hepatitis C virus core protein and cellular protein HAX-1 promote 5-fluorouracil-mediated hepatocyte growth inhibition. J Virol 83:9663–9671. 10.1128/JVI.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yedavalli VS, Shih HM, Chiang YP, Lu CY, Chang LY, Chen MY, Chuang CY, Dayton AI, Jeang KT, Huang LM. 2005. Human immunodeficiency virus type 1 Vpr interacts with antiapoptotic mitochondrial protein HAX-1. J Virol 79:13735–13746. 10.1128/JVI.79.21.13735-13746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda T, Tomonaga K. 2013. Nucleocytoplasmic shuttling of viral proteins in Borna disease virus infection. Viruses 5:1978–1990. 10.3390/v5081978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T, Shoya Y, Koda T, Takashima I, Lai PK, Ikuta K, Kakinuma M, Kishi M. 1998. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology 243:188–197. 10.1006/viro.1998.9049. [DOI] [PubMed] [Google Scholar]
- 38.Xi HQ, Zhang KC, Li JY, Cui JX, Zhao P, Chen L. 2017. Expression and clinicopathologic significance of TUFM and p53 for the normal-adenoma-carcinoma sequence in colorectal epithelia. World J Surg Oncol 15:90. 10.1186/s12957-017-1111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN. 2008. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 452:98–102. 10.1038/nature06604. [DOI] [PubMed] [Google Scholar]
- 40.Han J, Goldstein LA, Hou W, Froelich CJ, Watkins SC, Rabinowich H. 2010. Deregulation of mitochondrial membrane potential by mitochondrial insertion of granzyme B and direct Hax-1 cleavage. J Biol Chem 285:22461–22472. 10.1074/jbc.M109.086587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johns HL, Doceul V, Everett H, Crooke H, Charleston B, Seago J. 2010. The classical swine fever virus N-terminal protease Npro binds to cellular HAX-1. J Gen Virol 91:2677–2686. 10.1099/vir.0.022897-0. [DOI] [PubMed] [Google Scholar]
- 42.Kang YJ, Jang M, Park YK, Kang S, Bae KH, Cho S, Lee CK, Park BC, Chi SW, Park SG. 2010. Molecular interaction between HAX-1 and XIAP inhibits apoptosis. Biochem Biophys Res Commun 393:794–799. 10.1016/j.bbrc.2010.02.084. [DOI] [PubMed] [Google Scholar]
- 43.Kawaguchi Y, Nakajima K, Igarashi M, Morita T, Tanaka M, Suzuki M, Yokoyama A, Matsuda G, Kato K, Kanamori M, Hirai K. 2000. Interaction of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) with HS1-associated protein X-1: implication of cytoplasmic function of EBNA-LP. J Virol 74:10104–10111. 10.1128/JVI.74.21.10104-10111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosio Y, Berto G, Camera P, Bianchi F, Ambrogio C, Claus P, Di Cunto F. 2012. PPP4R2 regulates neuronal cell differentiation and survival, functionally cooperating with SMN. Eur J Cell Biol 91:662–674. 10.1016/j.ejcb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Herzig JK, Bullinger L, Tasdogan A, Zimmermann P, Schlegel M, Teleanu V, Weber D, Rucker FG, Paschka P, Dolnik A, Schneider E, Kuchenbauer F, Heidel FH, Buske C, Dohner H, Dohner K, Gaidzik VI. 2017. Protein phosphatase 4 regulatory subunit 2 (PPP4R2) is recurrently deleted in acute myeloid leukemia and required for efficient DNA double strand break repair. Oncotarget 8:95038–95053. 10.18632/oncotarget.21119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yazlovitskaya EM, Voziyan PA, Manavalan T, Yarbrough WG, Ivanova AV. 2015. Cellular oxidative stress response mediates radiosensitivity in Fus1-deficient mice. Cell Death Dis 6:e1652. 10.1038/cddis.2014.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujino K, Horie M, Honda T, Nakamura S, Matsumoto Y, Francischetti IM, Tomonaga K. 2012. Evolutionarily conserved interaction between the phosphoproteins and X proteins of bornaviruses from different vertebrate species. PLoS One 7:e51161. 10.1371/journal.pone.0051161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 46:D8–D13. 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22:1760–1774. 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.UniProt C. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D et al . 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download JVI.02030-20-s0001.xlsx, XLSX file, 14 KB (14KB, xlsx)