Abstract
American Tegumentary Leishmaniasis (ATL) is an endemic disease in Latin America, mainly caused in Brazil by Leishmania (Viannia) braziliensis. Clinical manifestations vary from mild, localized cutaneous leishmaniasis (CL) to aggressive mucosal disease. The host immune response strongly determines the outcome of infection and pattern of disease. However, the pathogenesis of ATL is not well understood, and host microRNAs (miRNAs) may have a role in this context. In the present study, miRNAs were quantified using qPCR arrays in human monocytic THP-1 cells infected in vitro with L. (V.) braziliensis promastigotes and in plasma from patients with ATL, focusing on inflammatory response-specific miRNAs. Patients with active or self-healed cutaneous leishmaniasis patients, with confirmed parasitological or immunological diagnosis, were compared with healthy controls. Computational target prediction of significantly-altered miRNAs from in vitro L. (V.) braziliensis-infected THP-1 cells revealed predicted targets involved in diverse pathways, including chemokine signaling, inflammatory, cellular proliferation, and tissue repair processes. In plasma, we observed distinct miRNA expression in patients with self-healed and active lesions compared with healthy controls. Some miRNAs dysregulated during THP-1 in vitro infection were also found in plasma from self-healed patients, including miR-548d-3p, which was upregulated in infected THP-1 cells and in plasma from self-healed patients. As miR-548d-3p was predicted to target the chemokine pathway and inflammation is a central to the pathogenesis of ATL, we evaluated the effect of transient transfection of a miR-548d-3p inhibitor on L. (V.) braziliensis infected-THP-1 cells. Inhibition of miR-548d-3p reduced parasite growth early after infection and increased production of MCP1/CCL2, RANTES/CCL5, and IP10/CXCL10. In plasma of self-healed patients, MCP1/CCL2, RANTES/CCL5, and IL-8/CXCL8 concentrations were significantly decreased and MIG/CXCL9 and IP-10/CXCL10 increased compared to patients with active disease. These data suggest that by modulating miRNAs, L. (V.) braziliensis may interfere with chemokine production and hence the inflammatory processes underpinning lesion resolution. Our data suggest miR-548d-3p could be further evaluated as a prognostic marker for ATL and/or as a host-directed therapeutic target.
Keywords: Leishmania braziliensis, microRNA, pathogenesis, active cutaneous leishmaniasis, self-healed cutaneous leishmaniasis, THP-1 cells
Introduction
The leishmaniases are vector-borne diseases caused by protozoan parasites of the genus Leishmania. Transmitted by Phlebotomine sandflies, the leishmaniases are endemic in tropical and subtropical areas, with one million cases/year in 98 countries (Burza et al., 2018). During its life cycle, Leishmania exists as promastigotes (elongated forms with an external flagellum) in the sandfly gut and as amastigotes (round or ovoid forms without an external flagellum) within mononuclear phagocytes of the vertebrate host. After promastigote inoculation in the skin by the vector, the parasites interact primarily with tissue humoral and cellular elements and the infection may progress to overt disease. Depending on the Leishmania species and host characteristics, the disease may manifest as visceral leishmaniasis, affecting organs, such as the liver and spleen, or tegumentary form, causing lesions in the skin and mucosa. More than 15 species may cause cutaneous leishmaniasis, with Leishmania (Viannia) braziliensis the most prevalent species in Brazil, where disease presents as either localized cutaneous leishmaniasis (CL), disseminated cutaneous leishmaniasis, or disfiguring mucosal leishmaniases (Turetz et al., 2002; Machado et al., 2011; Goto and Lauletta Lindoso, 2012). Once diagnosed, most patients are treated with anti-Leishmania drugs but rarely the patients heal without any specific treatment. Comparing active cutaneous and self-healed leishmaniasis patients constitutes a unique opportunity to explore pathogenic mechanisms of lesion development and control that are not fully elucidated.
In human CL, lesion development is not directly related to parasite growth, and few parasites are seen in the skin (Sotto et al., 1989). Instead, Th-1-type immune responses essential for infection control also drive inflammation and lesion development and cause tissue damage if uncontrolled (Vieira et al., 2002). In CL lesions characterized by chronic inflammation, activated CD69+ T cells (Diaz et al., 2002) and regulatory CD4+CD25+FOXP3+ IL-10–producing T cells, granzyme A CD8+ cytotoxic T cells, or even pro-inflammatory CD4+ IFN-γ–producing T cells (Bourreau et al., 2009; Faria et al., 2009) have all been observed. In a recent transcriptomic study of skin samples of cutaneous leishmaniasis patients, delayed or absence of cure was correlated with higher expression of gene sets related to the cytolytic pathway, including mRNAs for granzyme (GZMB), perforin (PRF1), and granulysin (GNLY) (Amorim et al., 2019).
microRNAs (miRNAs), endogenous small non-coding RNAs of ~22 nucleotides, have a fundamental role in shaping the host transcriptome (Baltimore et al., 2008) and act as key regulators in gene expression networks, including those regulating cell cycle, mitosis, apoptosis, differentiation, and immune functions. MicroRNAs mediate gene silencing post-transcriptionally by base-pairing to the 3′-untranslated regions (3’UTR) of their respective target genes. Up/down-regulation of miRNA expression impacts various cellular processes during homeostasis but may also result in dysfunction of cellular activities (Bartel, 2004; Bartel, 2009) and participate in pathological processes including infection and inflammation (O’Connell et al., 2012). In the human immune system, miRNA-clusters have been shown to exert essential roles in the regulation of related gene expression, impacting innate and adaptive immune responses (Hirschberger et al., 2018). Furthermore, as most miRNAs are considered stable in biological fluids and resistant to environmental conditions (Sohel, 2016), miRNAs are suitable for evaluation in plasma samples and represent attractive candidates as biomarkers of disease or therapeutic response.
MicroRNAs can be modulated by different pathogens, such as viruses, bacteria, and protozoan parasites (Chandan et al., 2019; Acuna et al., 2020). Differential expression of diverse miRNA has been identified in Leishmania-host interaction in vitro and experimental in vivo systems with visceral and cutaneous strains of Leishmania (Acuna et al., 2020) as well as human leishmaniasis (Paul et al., 2020). Specifically, in cutaneous leishmaniasis caused by L. braziliensis miR-361-3p, a regulator of GZMB and tumor necrosis factor (TNF) was down-regulated and related to treatment failure (Lago et al., 2018). In contrast, expression of miR-193b and miR-67, involved in regulating expression of triggering receptor expressed on myeloid cells-1 (TREM-1) was positively related to good treatment outcome (Nunes et al., 2018).
In the present study, we searched for differentially expressed microRNA in plasma of patients with active L. (V.) braziliensis infection and self-healed CL patients. In addition, we studied in vitro L. (V.) braziliensis infected-human monocyte-derived THP-1 cells to provide more direct insights into miRNA function. We focused on miRNA related to immune-inflammatory processes given the role of such processes in CL lesion development and resolution. miRNA expression was found to be markedly different between patients with self-healed leishmaniasis compared to healthy controls and cases with active CL. Among various differentially expressed miRNAs in patient plasma and L. braziliensis-infected THP-1 cells, we selected miR-548d-3p that was upregulated in both settings for further validation.
MiR-548d-3p inhibition in THP-1 cells reduced early parasite growth and increased MCP1/CCL2, RANTES/CCL5, and IP-10/CXCL10, whereas in self-healed patients, MCP1/CCL2, RANTES/CXCL5, and IL-8/CXCL8 were decreased, and MIG/CXCL9 and IP-10/CXCL10 were increased compared with active cases. Collectively, these data suggest that L. (V.) braziliensis exploits miRNAs to modulate the production of discrete sets of pro-inflammatory cytokines that are involved in lesion resolution.
Materials and Methods
Ethics Statement
The experimental protocols were approved by the ethics committee of the Faculdade de Medicina, Universidade de São Paulo (CAAE 35670314.0.1001.0065) and are in the accordance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research lnvolving Human Subjects of 1964 with latest amendment of 2013. All individuals agreed to participate by signing the Informed Consent Form.
Patients
Individuals of both gender and age from 15 to 60 years old were selected from endemic areas in Pernambuco state, Northeastern Brazil where L. (V.) braziliensis is the predominant species causing CL. Five patients with active disease were chosen based on the presence of up to five cutaneous lesions, confirmed diagnosis of leishmaniasis and absence of local concomitant bacterial infections, comorbidities such as HIV/aids, diabetes mellitus, dermatitis, peripheral vascular diseases, and previous chemotherapy. The diagnosis of active cases was confirmed by submitting sample to direct microscopic parasitological exam of lesion scrapings, by culture, by inoculation into hamsters for parasitological recovery or by polymerase chain reaction specific for Viannia subgenus (Brito et al., 2009).
Five self-healed patients with a history of previous cutaneous leishmaniasis were also recruited, showing characteristic scars, confirmed diagnosis, and absence of abovementioned comorbidities, co-infections, and previous chemotherapy. Another five healthy individuals represented the control group, being recruited from non-endemic areas and without previous leishmaniasis, abovementioned comorbidities, or co-infections.
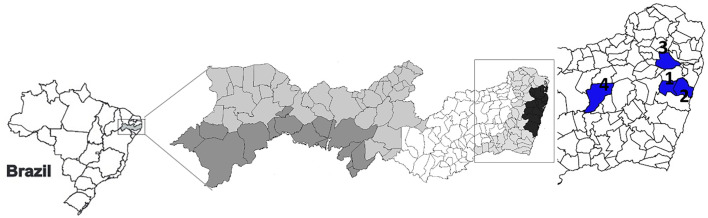
The patients with active or self-healed leishmaniases were from the municipalities of Paudalho, Moreno, Jaboatão, and Bezerros, localities close by preserved remnants of Atlantic forest, in the State of Pernambuco, Northeast Brazil, where intertwine rural and urban environments where they live and work ( Figure 1 ).
Figure 1.
Cartographical representation of the State of Pernambuco, Northeastern Brazil. Regions in blue indicate Moreno (1), Jaboatão (2), Paudalho (3), and Bezerros (4) municipalities.
After confirmed diagnosis, four milliliters of whole blood were collected in EDTA from each individual, and plasma stored at −80°C until use.
Parasites
Leishmania parasites were previously isolated from a patient with mucosal leishmaniasis at Corte de Pedra, Bahia, Brazil, and characterized as L. (V.) braziliensis by the Leishmania Collection at Fundação Oswaldo Cruz—CLIOC/FIOCRUZ. To preserve the infectivity, the parasites were inoculated via intraperitoneal route and maintained through regular passages in hamster (Mesocricetus auratus). Amastigotes were then purified from the spleen of hamster and expanded in axenic culture with Schneider’s insect medium (Sigma-Aldrich, USA) containing 100 UI/ml penicilin and 100 μg/ml streptomycin and supplemented with 10% heat-inactivated fetal calf serum (FCS) (Cultilab, Brazil) at 26°C. The amastigote-derived promastigotes were cryopreserved in aliquots and thawed for use in specific experiments. Promastigotes were cultured in Schneider’s insect medium (Sigma-Aldrich, USA) containing 100 UI/ml penicilin and 100 μg/ml streptomycin and supplemented with 10% heat-inactivated fetal calf serum (FCS) (Cultilab, Brazil) at 26°C. The parasites used in the experiments were at the stationary phase of growth and with no more than four passages in culture.
Infection of Macrophages With L. (V.) braziliensis
THP-1 monocytic cell line (ATCC) was maintained in RPMI 1640 medium (Sigma-Aldrich, USA) supplemented with 2 mM l-glutamine, 1 mM sodium piruvate, 0.2% sodium bicarbonate, and 5% FCS (complete medium). Then 106 cells in 1 ml of RPMI 1640 medium were plated onto 24-well plates (Costar, USA) and incubated in the presence of 20 ng/ml phorbol myristate acetate (PMA; Sigma-Aldrich, USA) for 48 h at 37°C in a humid atmosphere with 5% CO2 to allow differentiation into macrophages (Tsuchiya et al., 1982). In experiments for parasite load analysis, round coverslips were placed in the well. Non-adherent cells were then removed and L. (V.) braziliensis promastigotes were added to the wells in triplicates (parasite:cell ratio = 5:1) and incubated for 4 h at 33°C in humid atmosphere with 5% CO2 to allow infection of macrophages. Non-infected cells were maintained only with culture medium, being the negative control of the experiment. After washing out the non-internalized parasites, complete RPMI medium was added to the wells, beginning the experimental period (0 h). The plates were then maintained for 6 or 24 h at 37°C in a humid atmosphere with 5% CO2.
Evaluation of Parasite Load in Macrophages
Glass coverslips were removed from the wells and stained with panoptic dyes (Instant Newprov, Brazil) and mounted on glass slides for evaluation of parasitism. A total of 900 cells were counted for each experimental condition, 300 cells/coverslip, under light microscope (Carl Zeiss, Germany), and the number of parasites per 100 cells calculated as [(number of parasites/number of infected cells) × (number of infected cells/total number of cells) × 100].
RNA Extraction, Reverse Transcription, and Pre-Amplification
Total RNA extraction from adherent THP-1 cells was performed using the miRVana PARIS isolation kit (Thermo Fisher, USA), according to the manufacturer’s instructions, and RNA integrity was determined in spectrophotometer as an OD260/280 absorption ratio between 1.8 and 2.1. The total RNA purification in plasma samples was performed using the miRNeasy Serum/Plasma kit (Qiagen, USA), with the addition of a spike-in control (Caenorhabditis elegans cel-miR-39) to ensure the quality of the procedure and to allow qPCR normalization, according to the manufacturer’s instructions. Complementary DNA (cDNA) to template RNA purified from THP-1 cells and plasma samples was synthesized with miScript II RT kit (Qiagen, USA). Briefly, 250 ng of total RNA from THP-1 cells were added to 2 μl of 5× miScript HiSpec Buffer, 1 μl of 10× Nucleics Mix, and 1 μl of miScript Reverse Transcriptase Mix. RNase-free water was added to a final volume of 10 μl. The RNA was incubated for 60 min at 37°C to insert poly-A tail downstream of the miRNA sequence and anneal a T-tail tag for the cDNA elongation. The enzyme was inactivated at 95°C for 5 min. The reaction was performed in the Mastercycler Gradient thermal cycler (Eppendorf, Germany), and the product was stored at −20°C until use. The reverse transcription reaction for plasma samples followed the same protocol, with the manufacturer’s instructions to add 4.5 µl of the purified total RNA. Then, 40 µl of DEPC water was added into each 10 µl RT-PCR product and submitted to a pre-amplification reaction (preAmp), using the miScript PreAmp PCR Kit (Qiagen, USA) according to the manufacturer’s instructions. Then the samples were diluted 10× and stored at −20°C.
Quantitative Real-Time PCR for miRNA
miRNA expression was evaluated with the miScript microRNA PCR array (Qiagen, USA), focusing on inflammation, and auto-immunity pathway-related molecules (MIHS-105Z). Ready-to-use qPCR plates containing a set of 84 specific primers for miRNAs and 12 internal controls were filled in with the previously prepared master mix containing PCR Buffer, SYBR Green, and the 10-fold diluted cDNA for in vitro infected THP-1 macrophages or preAmp samples of plasma samples. Quantitative PCR conditions were 40 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. Normalization of miRNA expression in THP-1-derived macrophages was performed using SNORD95 and RNU6-6p as reference genes amplified in the qPCR plate. The relative expression levels were calculated using the Comparative Ct method, with non-infected cells being considered as the calibrator group.
For plasma samples, miRNA expression was also evaluated by relative quantification after previous normalization described by Marabita et al. (2016). The cel-miR-39 spike-in control was considered as a technical reference. Simultaneously, a geometric mean of all expressed miRNAs was used as a normalization factor to calculate relative expression to a calibrator group, which varied depending on the analysis.
In Silico miRNA Target Prediction
Target prediction strategy was performed in two different platforms, considering the miRNAs differentially expressed in the in vitro experiment. For an initial screening in human leishmaniasis pathway, we used DIANA-miRpath 3.0 server in the reverse search module (Vlachos et al., 2015), with Targetscan (Agarwal et al., 2015) as the chosen algorithm. To discover potential interactions with other biological pathways related to human leishmaniasis pathogenesis, we performed a second analysis using MiEAA (MiRNA Enrichment Analysis and Annotation), which integrates data from different databases such as miRBase, miRWalk, and miRTarBase (Backes et al., 2016).
In Vitro miRNA Inhibition
The inhibition of miR-548d-3p in THP-1-derived macrophages was performed in an in vitro infection experiment through a transient transfection protocol. Assays with three different concentrations (3, 10, and 30 nM) of the mirVana® miR-548d3p inhibitor (Ambion, USA) or mirVana® miRNA Mimic, scramble Negative Control (Ambion, USA) were performed, and 10 nM concentration was chosen for further use ( Figure S1 ). At the end of the experiment, the cell viability was evaluated by Trypan blue exclusion test when viability higher than 95% was seen in all conditions. Before the addition of L. (V.) braziliensis promastigotes, a solution containing the miR-548d-3p inhibitor or the negative control together with 3 μl of FUGENE transfection reagent (Promega, USA) diluted in 500 μl of RPMI medium previously incubated for 20 min at room temperature was added into each well, and maintained for 24 h. Simultaneously, non-transfected cells received only complete RPMI medium. The experiment continued with promastigote infection for evaluation of parasitism, and chemokine levels in supernatants collected and stored at −80°C until use.
Evaluation of Chemokine Production
Chemokine quantification in culture supernatants was performed using CBA – Human Chemokine Kit (BD Biosciences, USA) in accordance with manufacturer’s instructions. Briefly, 50 µl of capture beads for MCP1/CCL2, RANTES/CCL5, IL-8/CXCL8, MIG/CXCL9, and IP10/CXCL10, 50 µl of Detection Reagent, and 50 µl of the studied sample or standard were added consecutively to each sample tube and incubated for 3 h at room temperature, in the dark. Next, the samples were washed with 1 ml of Wash buffer, and centrifuged. After discarding the supernatant, the pellet was resuspended in 300 µl buffer and analyzed in a FACS LSR Fortessa flow cytometer (BD Biosciences, USA). Raw data was then analyzed using FCAP Array software (BD Biosciences, USA). The detection limits of each chemokine were as follows: 2.7 pg/ml for MCP1/CCL2, 1.0 pg/ml for RANTES/CCL5, 0.2 pg/ml for IL-8/CXCL8, 2.5 pg/ml for MIG/CXCL9, and 2.8 pg/ml for IP10/CXCL10.
Statistical Analysis
Regarding in vitro miRNA expression, statistical analyses were performed with Qiagen miScript miRNA PCR Array Data Analysis online software, where data were submitted to an integrated Student’s t test under the manufacturer’s recommendation that was applied in the previous similar work (Muxel et al., 2017). Ex vivo data were also submitted to Student’s t test, with Bonferroni’s correction, using Microsoft Excel 365. Parasite load data were analyzed by ANOVA with Tukey’s post-test, and data from chemokine quantification by Kruskal-Wallis test with Bonferroni’s correction. The differences were considered significant when P < 0.05.
Results
miRNA Expression in L. (V.) braziliensis Promastigote-Infected THP-1 Cells
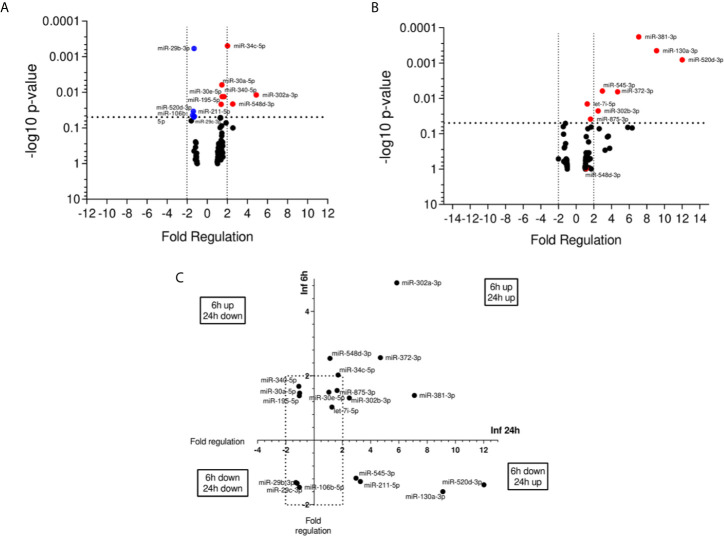
We evaluated miRNA expression at 6 and 24 h post infection with L. (V.) braziliensis promastigotes, considering non-infected THP-1 cells as the calibrator group. 19 out of 84 miRNAs presented significant alteration in expression (P < 0.05). At 6 h p.i., seven miRNAs were upregulated while five were down-regulated ( Figure 2A ). In contrast, eight miRNAs were up regulated in 24 h ( Figure 2B ). From these results, we observed that these miRNAs expression was modulated through time and classified them into four groups. MiR-106b-5p, miR-29b-3p, and miR-29c-3p were the down-regulated molecules at both 6 and 24 h, while miR-195-5p, miR-30a-5p, and miR-340-5p were up-regulated at 6 h and down-regulated at 24 h. There were also down-regulated miRNAs at 6 h and upregulated at 24 h, such as miR-130a-3p, miR-211-5p, miR-520d-3p, and miR-545-3p, whereas let7i-5p, miR-30e-5p, miR-302a-3p, miR-302b-3p, miR-34c-5p, miR-372-3p, miR-381-3p, miR-548d-3p, and miR-875-5p were upregulated at both time-points ( Figure 2C ).
Figure 2.
miRNA profiles of L. braziliensis infected THP-1-macrophages. Volcano plot of differential expression of miRNA in L. (V.) braziliensis promastigote-infected THP-1 macrophages in relation to non-infected cells at 6 h (A) and 24 h (B) post-infection compared to uninfected-macrophages. Each dot represents one miRNA. Red dots indicate the up-regulated miRNAs, and the blue dots represent the down-regulated miRNAs (P < 0.05). Black dotted line corresponds to p = 0.05, log 10. The relative up- and down-regulation of miRNAs, expressed as boundaries of 2 or -2 of Fold Regulation, respectively. P-value was determined based on two-tailed Student’s t test. Significantly expressed miRNAs in different times distributed in four groups (C). Experiments were repeated three times.
miRNA Profiling of Serum From ATL Patients
The participants of this study were predominantly male and the active disease patients were younger (mean 20 and median 20 years old, minimum-maximum:18-22 ya) than self-healed patients (mean 26.2 and median 27 years old, minimum-maximum: 21-30 ya). The number of present or past lesions, and their localization were similar in both groups. The patients with active disease presented lesions characteristic of the localized form of CL (rounded, ulcerated, with well-defined and elevated edges and granulomatous bottom) on uncovered parts of the body, with evolution ranging from 15 to 28 days. The self-healed individuals presented typical scars without previous anti-Leishmania chemotherapy, with healing time ranging from three to nine months ( Table 1 ).
Table 1.
Demographic and clinical data of active disease and self-healed patients.
| Code | Gender/Age (years old) | Occupation | Locality | Clinical form | Lesions (n) | Size (cm) | Evolution | Lesion site | Parasite search | PCR | Isolation Parasite | IDRM | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Active disease patients | 0001 | M/19 | Military | Paudalho/PE | Ulcerated | 01 | 1.0 × 2.0 | 15 days | Left leg | + | + | ND | + |
| 0002 | M/21 | Military | Paudalho/PE | Ulcerated | 01 | 0.5 × 0.5 | 15 days | Right flank | + | + | – | + | |
| 0003 | M/22 | Military | Paudalho/PE | Ulcerated | 01 | 0.5 × 0.5 | 15 days | Right hand | + | – | + | + | |
| 0004 | M/20 | Military | Paudalho/PE | Ulcerated | 01 | 0.6 × 0.6 | 1 month | Right hand | + | – | – | + | |
| 0005 | M/18 | Student | Moreno/PE | Ulcerated | 01 | 1.5 × 1.5 | 15 days | Left Leg | + | + | + | + | |
| Code | Gender/Age | Occupation | Provenance | Reported clinical form | Scars (n) | Size (cm) | Healing time | Scar sites | Parasite search | PCR | Parasite Isolation | IDRM | |
| Self-healed patients | 0006 | M/27 | Farmer | Moreno/PE | Ulcerated | 02 | 2.0 × 3.0; 1.0 × 1.0 | 4 months | Right foot | + | – | – | ND |
| 0007 | M/29 | Machine operator | Jaboatão/PE | Ulcerated | 01 | 4.0 × 3.0 | 9 months | Left leg | − | + | – | ND | |
| 0008 | M/30 | Engineer | Bezerros/PE | Ulcerated | 02 | 2.0 × 2.0 | 6 months | Left leg | − | + | – | ND | |
| 0009 | F/24 | Housewife | Moreno/PE | Ulcerated | 01 | 1.5 × 1.5 | 4 months | Right leg | + | + | – | ND | |
| 0010 | M/21 | Farmer | Moreno/PE | Ulcerated | 01 | 2.0 × 2.0 | 3 months | Right thigh | − | + | + | ND |
M, male; F, female; L, left; R, right; PCR, polymerase chain reaction; IDRM, Montenegro skin test; ND, not done.
The miRNA expression in plasma samples of patients with active disease was not significantly different compared to the control group ( Figure 3A ). In contrast, self-healed individuals presented 14 differentially expressed miRNAs, eight of them up-regulated (miR-15b-3p, miR-29b-3p, miR-181-5p, miR-202-3p, miR-211-5p, miR-302c-5p, miR-373-3p, miR-449-5p), and six down-regulated (miR-19b-3p, miR-21-5p, miR-29c-3p, miR-30e-5p, miR-656-3p, miR-93-5p) in relation to the control group ( Figure 3B ). When comparing self-healed patients with the active disease group, we found a total of 23 significantly altered miRNAs, with 14 of these up-regulated (miR-15b-5p, miR-20b-5p, miR-29b-3p, miR-181a-5p, miR-181d-3p, miR-202-3p, miR-211-5p, miR-300, 302c-5p, miR-410-3p, miR-548d-3p, miR-875-3p, miR-655-3p, miR-1324), and nine down-regulated (miR-16-5p, miR-17-5p, miR-19b-3p, miR-21-5p, miR-29c-3p, miR-30e-5p, miR-93-5p, miR-454-3p, miR-656-3p) ( Figure 3C ). Our data suggest distinct miRNA profiles in plasma samples of active and self-healed ATL patients.
Figure 3.
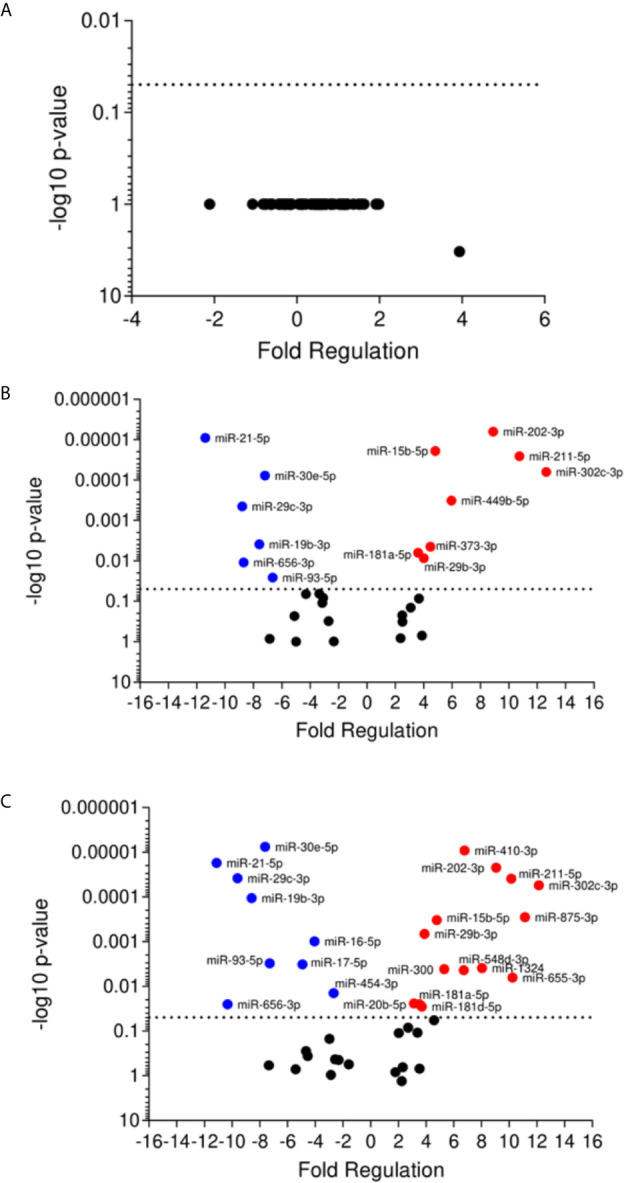
Volcano plot of differential expression of miRNA in plasma of cutaneous leishmaniasis patients. Volcano plot of differential expression of miRNA in plasma samples of active disease patients (A) and self-healed (B) compared to healthy individuals, and self-healed (C) compared to active disease patients. Each dot represents one miRNA. Red dots indicate the up-regulated miRNAs, and the blue dots represent the down-regulated miRNAs (P < 0.05). Black dotted line corresponds to p=0.05, log 10. The relative up- and down-regulation of miRNAs, expressed as boundaries of 2 or -2 of Fold Regulation, respectively. P-value was determined based on two-tailed Student’s t test. P < 0.05 (Student t test and Bonferroni correction).
Concomitant Altered Expression of miRNAs in In Vitro and Ex Vivo Experiments
To focus on understanding the miRNA modulation and function during infection, we searched for correlations between up- and down-regulated miRNAs. We found some miRNAs differentially expressed in infected THP-1 cells and plasma samples of ATL patients ( Table 2 ). The miR-548d-3p and miR-875 were upregulated in self-healed patients and in vitro at 6 and 24 h of incubation post-infection. Despite that, miR-211-5p and miR-29b-3p were upregulated in self-healed patients, but down-regulated at 6 h of incubation post-infection in vitro. Down-regulated expression was observed for miR-29c-3p ex vivo, showing similar modulation in the in vitro experiment at both time points. Finally, miR-30e-5p was upregulated in vitro at 6 and 24 h of incubation post-infection and down-regulated in plasma samples ( Figures 2C and 3C ).
Table 2.
Set of miRNAs significantly expressed both in in vitro and ex vivo contexts.
| Up regulated | Down regulated |
|---|---|
| miR-410-3p | miR-30e-5p |
| miR-202-3p | miR-21-5p |
| miR-211-5p | miR-29c-3p |
| miR-302c-3p | miR-19b-3p |
| miR-15b-5p | miR-16-5p |
| miR-875-3p | miR-17-5p |
| miR-29b-3p | miR-93-5p |
| miR-548d-3p | miR-454-3p |
| miR-300 | miR-656-3p |
| miR-1324 | |
| miR-655-3p | |
| miR-181a-5p | |
| miR-181d-5p | |
| miR-20b-5p |
miRNAs in bold, molecules modulated in similar ways in both experiments.
miRNA Predicted Targets and Interactions With Biological Pathways Related to ATL Pathogenesis
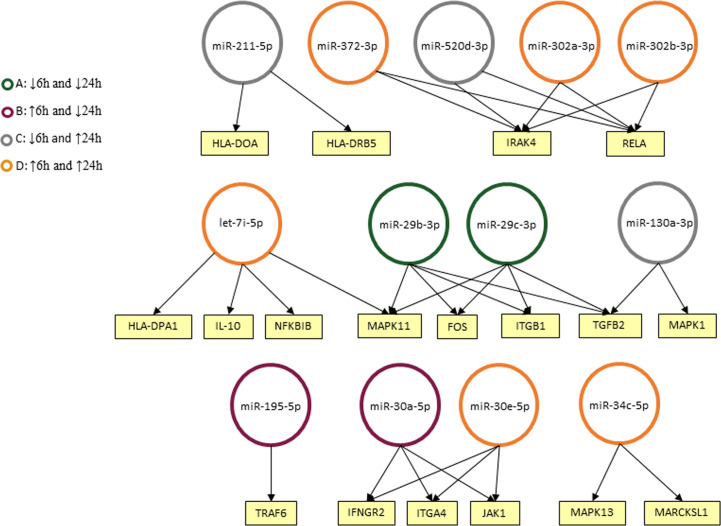
We used Diana MiRPath 3.0 with TargetScan2 as the chosen algorithm to predict miRNA/mRNA interactions, focusing on miRNAs modulated in infected THP-1 cells. Among the interactions predicted in the initial analysis using the Diana platform, there were cytokines encoded by the TGFB2 and IL10 genes, MHC class II proteins (HLA-DPA1, HLA-DRB5, and HLA-DOA) and genes related to signaling pathways (e.g. MAPK1, MAP3K7, IRAK4) ( Figure 4 ).
Figure 4.
Predicted interactions between the set of differentially expressed microRNAs in THP-1 cells after 6 and 24 h post infection. The genes on which they are suggested to act in the Leishmaniasis pathway in humans are shown as seen in KEGG. miRNAs were classified into four groups according to their modulation through time: down-regulated in 6 and 24 h (A), up-regulated on 6 h and down-regulated in 24 h (B), down-regulated in 6 h and up-regulated in 24 h (C) and up-regulated in both timepoints (D).
Further predictions made in MiEAA platform showed, in more than one classification system (PANTHERDB, WikiPathways, and KEGG), some pathways known to be important in the parasite-host interaction that can be regulated by the expressed microRNAs in L. braziliensis-infected THP-1 cells ( Figure 5A ) and in self-cured patients plasma compared with active patient sample ( Figure 5B ). Cytokine signaling pathways, such as IFN-γ, TNF-α, and TGF-β, are known to be involved in the immune response against Leishmania. Also, signal transduction pathways such as JAK-STAT and PI3K were putative targets of differentially expressed miRNAs, as well as the VEGF, Wnt, and HIF-1 pathways. There is also a potential interference of miRNAs expressed in the oxidative stress pathway. Finally, the signaling cascade activated by IGF receptors may be influenced by the differentially expressed microRNAs. Searching predicted pathways targeted by the circulating microRNAs present in plasma, we observed the inflammation mediated by chemokines and cytokines and the chemokine signaling pathways. Besides, important pathways involved in B cell development like B cell activation and mTOR signaling pathways were predicted. Pathways involved in Th17 and Th2 differentiation and T cell proliferation such as T cell activation, IL-4, IL-6, and IL-2 signaling were evidenced.
Figure 5.
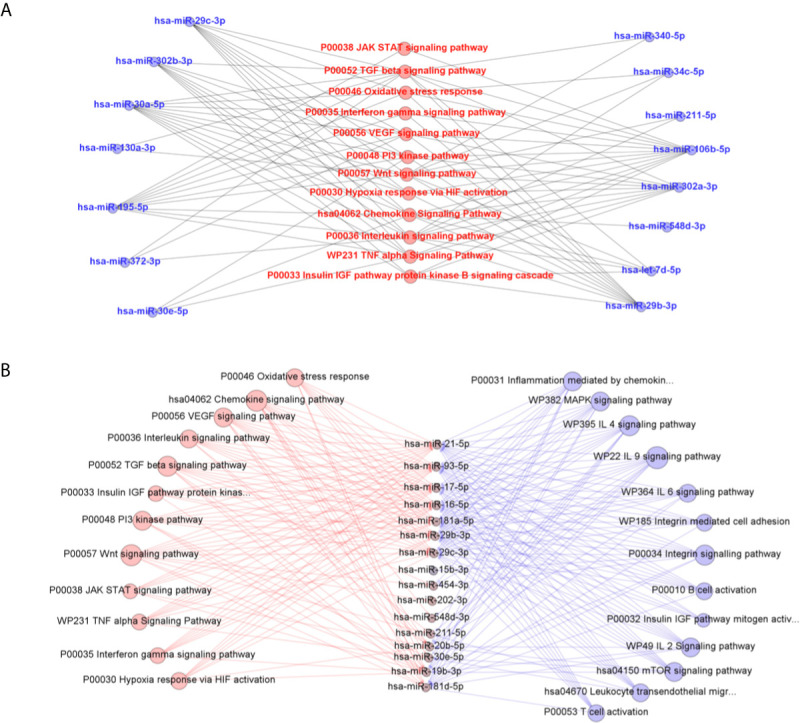
Predicted interactions between the set of differentially expressed microRNAs in THP-1 cells after 6 and 24 h post infection with L. braziliensis (A) and in plasma samples from self-healed patients compared to active disease individuals (B) and the biological pathways related to inflammatory response on which they are suggested to act according to MiEAA algorithms. In (B), pathways in red were predictably targeted by in vitro and ex vivo miRNA sets, while the others in blue were evidenced only in miRNAs significantly quantified in plasma.
Among various differentially expressed miRNAs, we selected miR-548d-3p that was upregulated in patient plasma and L. braziliensis-infected THP-1 cells and targets only two pathways for further validation.
Effect of miR-548d-3p Inhibition on Parasite Load in THP-1 Infection With L. (V.) braziliensis
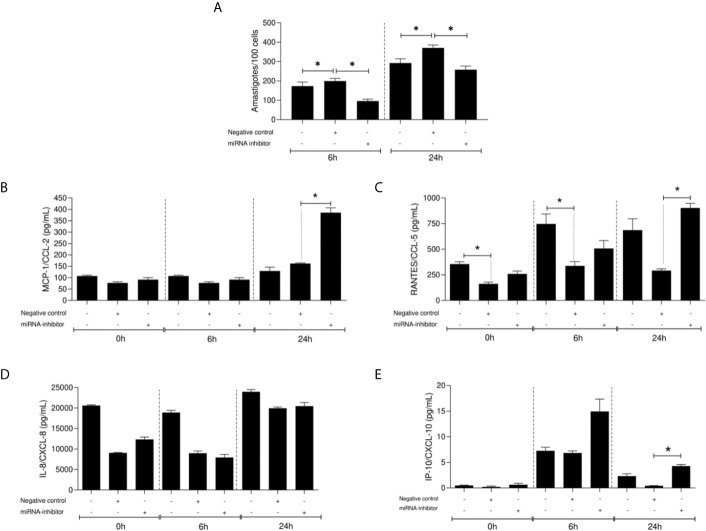
The function of miR548d-3p during L. braziliensis infection was evaluated using 10 nM specific inhibitor or scrambled miRNA. At both 6 and 24 h post-infection, a significant decrease was observed in parasite load when miR-548d-3p was inhibited (P < 0.05), compared to transfection with scrambled RNA, negative control ( Figure 6A ).
Figure 6.
Parasite load (number of amastigotes/100 cells) (A) and chemokine levels (pg/ml) (B–E) in L. (V.) braziliensis promastigote-infected THP-1 cells transiently transfected with miR-548d-3p inhibitor at 6 and 24 h post-infection. One experiment was carried out by adding the miR-548d-3p inhibitor (10 nM) or negative control (scrambled miRNA; 10 nM) with the transfection reagent diluted in RPMI medium or only RPMI medium (non-transfected cells) to wells containing 106 THP-1 adherent cells and maintained for 24 h at 37°C (5% CO2) then infected with L. (V.) braziliensis promastigotes. (A) * = P < 0.05 (one way ANOVA and Student t test). MCP1/CCL2 (B), RANTES/CXCL5 (C), IL-8/CXCL8 (D), and IP-10/CXCL10 (E) concentrations were measured by flow-cytometry using the CBA kit. (B–E) * = p <0.05 (Kruskal-Wallis and Bonferroni tests).
miR-548d-3p Inhibition on Chemokine Production in L. (V.) braziliensis–Infected THP-1 Cells
Inhibition of miR-548d-3p did not affect the production of CCL2 by infected THP-1 cells at 6 h p.i. but led to a >2-fold increase in secretion of CCL2 at 24 h p/i/compared to both untreated infected cells and cells treated with the scrambled RNA-negative control ( Figure 6B ). In contrast, CCL5 production appeared more susceptible to modulation by transfection of the scrambled control RNA and use of the inhibitor tended to normalize the production to that seen in untransfected cells ( Figure 6C ). The production of CXCL8 and CXCL10 were not significantly affected by the miR-548d-3p inhibitor in comparison to untransfected cells but a small but significant increase in CXCL10 was observed compared to the scrambled inhibitor at 24 h p.i. ( Figures 6D, E ).
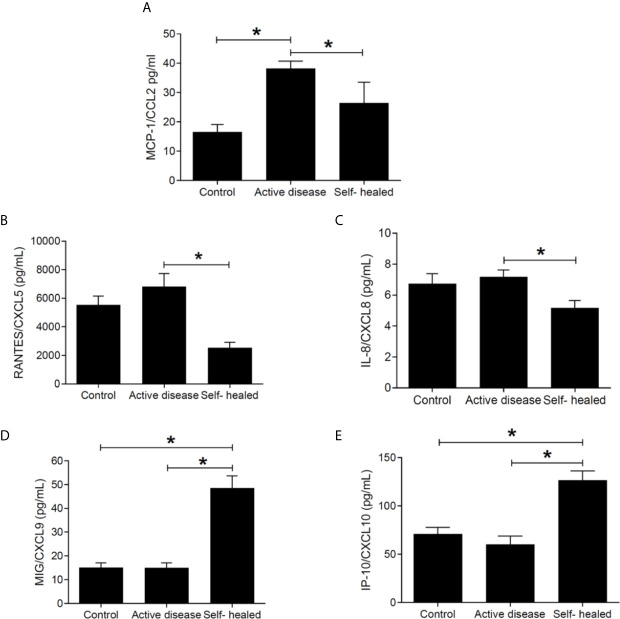
Chemokine Levels in Plasma of ATL Patients
CCL2 was found at significantly higher concentration in plasma samples of patients with active disease compared to self-healed and control groups ( Figure 7A ), whereas CXCL5 and CXCL8 were decreased in self-healed cases compared to active cases ( Figures 7B, C ). Significantly higher concentrations of CXCL9 ( Figure 7D ) and IP-10 ( Figure 7E ) were seen in self-healed patients in relation to healthy individuals and patients with active disease (P < 0.05).
Figure 7.
Chemokine concentration (pg/ml) in plasma of cutaneous leishmaniasis patients and controls. MCP1/CCL2 (A), RANTES/CXCL5 (B), IL-8/CXCL8 (C), MIG/CXCL9 (D), and IP-10/CXCL10 (E) concentrations were measured by flow-cytometry using the CBA kit. * = p < 0.05 (Kruskal-Wallis and Bonferroni tests). N =5 patients and controls per group.
Discussion
Macrophages exert a dual role in the pathogenesis of CL, being both host cell and also the main effector cell for parasite clearance (Tomiotto-Pellissier et al., 2018). The disease outcome depends on the interplay between Leishmania and the host immune responses which govern these opposing macrophage functions. Leishmania employ strategies to evade the host immune response, including altering the miRNAs expression (Lemaire et al., 2013; Muxel et al., 2017; Muxel et al., 2018; Nunes et al., 2018; Fernandes et al., 2019; Paul et al., 2020). In this context, having access to L. braziliensis-infected active and self-healed CL patients, we searched for differentially expressed miRNAs in plasma and in parallel conducted an in vitro study using L. (V.) braziliensis infected-human monocyte-derived THP-1 cells. With this approach, we sought to attribute altered miRNA profiles to mechanisms of disease pathogenesis.
Alterations of miRNA expression were seen in self-healed patient samples compared with active cutaneous leishmaniasis cases and healthy controls, the latter being indistinguishable by miRNA profile. Thus, these data suggest that host cells from individuals that cure without treatment are more active in altering miRNA expression upon L. (V.) braziliensis infection, although we cannot rule out at this preliminary stage in our investigation whether this is confounded by other factors unrelated to infection e.g., environmental exposure or host genetics. Concerning environment, the patients were from endemic areas, from nearby cities with similar climate and environmental characteristics. Differentially expressed miRNAs have been related to inflammatory chemokine levels, and this may contribute to the self-healing nature of these patients. An additional weakness of the study is that parasites from these patients were not genotyped or functionally evaluated. This may be important given that some strains of L. braziliensis are more susceptible to oxidative stress than others, and induce lesions with a higher tendency to spontaneous healing (Souza et al., 2010; Sarkar et al., 2012). In addition, we have to consider a proper balance between regulatory and pro-inflammatory mediators, especially IFN-γ and IL-10, that may differ depending on the hosts background, important for the lesion to heal (Gomes-Silva et al., 2007; de Assis Souza et al., 2013).
Evaluation of altered miRNA expression in vitro L. (V.) braziliensis infected-human monocyte-derived THP-1 cells showed a set of miRNAs also found altered in plasma of leishmaniasis patients. In silico prediction of THP-1 expressed miRNA targets and interactions with biological pathways suggested a link between the differentially expressed miRNAs and altered expression targeted cytokine, chemokine, and signaling pathways, signal transduction pathways, and others.
A total of 19 out of 84 miRNAs exhibited an altered in vitro expression compared with non-infected THP-1 cells either at 6 or 24 h of incubation after infection, showing that Leishmania can modulate these molecules in a temporally distinct manner during the early stages of in vitro infection as seen by others (Guerfali et al., 2008; Bazzoni et al., 2009; Lemaire et al., 2013). In the in-silico predictions using DIANA miRPath 3.0, considering Leishmania infection pathway, upregulated miR-195-5p at 6 h may target tumor-necrosis factor receptor-associated factor 6 (TRAF6), an important player in signal transduction of both the TNF receptor (TNFR) superfamily and the interleukin-1 receptor (IL-1R). These are crucial to ultimately activate transcription factors, such as nuclear factor kappa B (NF-κB) and interferon-regulatory factor (IRF), to induce immune and inflammatory responses (Ye et al., 2002; Wang et al., 2010). In addition, two isoforms of miR-30 family, miR-30a-5p, and miR-30e-5p, were suggested to target Interferon gamma receptor 2 (IFNGR2), Janus kinase 1 (JAK1), Integrin subunit alpha 4 (ITGA4) genes throughout time. These predicted interactions suggest participation in parasite control mechanism and inflammatory process. Other important events of the immune response such as Toll-like receptor signaling and antigen presentation were also predicted to be compromised by the influence of let-7i-5p, miR-130a-3p, miR-520d-3p, and two isoforms of miR-302.
Other relevant pathways that are known to play a role in the adaptive immune response in cutaneous leishmaniasis were targeted by miRNAs identified exclusively in plasma samples from self-healed patients compared with active disease subjects. Pathways related to T and B cell activation including mTOR pathway that can modulate B cell development (Limon and Fruman, 2012; Iwata et al., 2017) were predicted. Cytokine related pathways such as IL-2, IL-4, IL-6, and IL-9 were also evidenced. These different pathways potentially targeted by circulating microRNAs might reflect the diversity of cells participating in the immune response in humans, in contrast to exclusively monocyte/macrophage in vitro experiment.
Among altered miRNAs in the present study, other miRNAs belonging to the same family were previously analyzed in Leishmania infection. The modulation of miR-29b, miR-29c, and miR-30e were observed in L. amazonensis-infected murine macrophages (Muxel et al., 2017; Muxel et al., 2018; Fernandes et al., 2019). Upregulation of miR-29b was observed in L. major-infected human monocyte-derived macrophages, whereas this miRNA was down-regulated in L. donovani infection (Geraci et al., 2015). Stimulation of the nucleotide-binding oligomerization domain containing 2 (NOD2) induces miR-29 family upregulation, resulting in downregulation of IL-12p40 without alteration to IL-6, TGF-β or IL-10 production (Yu et al., 2002; Guo et al., 2012; Reveneau et al., 2012). However, NOD2-stimulation increases the levels of IL-1β, IL-6, and IL-23 cytokines, Nitric Oxide Synthase 2 (NOS2) expression and nitric oxide (NO) production during Leishmania infection (Lima-Junior et al., 2013).
We also searched for other biological pathways that could be affected during Leishmania infection, and our predictions using the MiEAA platform pointed to some involved in inflammation and wound healing as follows. Also, our predictions highlighted TNF, IFN-γ, and TGF-β signaling pathways, cytokines with respective proinflammatory and regulatory roles in Leishmania infection (Souza et al., 2012; de Assis Souza et al., 2013; Souza et al., 2016). The oxidative stress response pathway was also revealed once reactive oxygen and nitrogen species (ROS and RNS) produced during an inflammatory response are an important part of host-defense strategies of organisms to kill the parasite (Kocyigit et al., 2005).
Many characteristics of leishmanial lesions such as microcirculation impairment, metabolic demand for leukocytes, parasite proliferation, and secondary bacterial infection are indicators of a hypoxic event in those lesions (Fraga et al., 2012). Related to this condition, changes in miRNAs that regulate Hypoxia-inducible factor 1 (HIF-1) activation in response to hypoxia were also identified in silico. Other possible consequence of a hypoxic, inflammatory microenvironment is the induction of vascular remodeling via Vascular endothelial growth factor A/Vascular endothelial growth factor receptor (VEGF-A/VEGFR) expression by HIF-1 influence, which are elevated in the skin of humans and mice infected with Leishmania parasites (Fraga et al., 2012; Araujo and Giorgio, 2015). Differentially expressed miRNA affecting VEGF were also observed in our data. Our in-silico predictions also showed that some of the altered miRNAs targets the IGF-I signaling pathway. The role of this hormone in Leishmania infection has been long studied with pleiotropic effect in innate and adaptive immune response and pathogenesis in leishmaniases (Reis et al., 2021).
Cutaneous lesions are characterized by chronic inflammation where concur activated CD69+ T cells (Diaz et al., 2002), regulatory CD4+CD25+FOXP3+ IL-10–producing T cells, granzyme A CD8+ cytotoxic T cells, CD4+ IFN-γ–producing T cells (Bourreau et al., 2009; Faria et al., 2009) and where higher expression of gene sets related to the cytolytic pathway is observed (Amorim et al., 2019). The influx of cells into the lesion reflects the role of chemokines and one of miRNA seen altered in vitro and patients’ plasma in the present study was miR-548d-3p. miR-548d-3p and others from the same family were reported related to wound healing and inflammation in rheumatoid arthritis and Leishmania donovani infection (Wang et al., 2018; Huang et al., 2020) thus we proceeded with functional validation of the miR-548d-3p in L. braziliensis infected-THP-1 cells. The miR-548 family is a larger and poorly conserved, encompassing 69 human miR-548 genes located in almost all human chromosomes (Liang et al., 2012). Previous studies showed that miR-548d are processed from the same encoded hairpin cluster of miR-548aa1 (GenBank ID 100500863) and that miR-548d-3p belongs to the cluster family of hsa-miR-548-d1 (miRbase ID MI0003668) (Cummins et al., 2006; Landgraf et al., 2007) transcribed from negative strand of intronic region of ATPase family AAA domain containing 2 (ATAD2, gene ID NM_014109.4) gene located into chromosome 8 (search in miRIAD toll) (Cummins et al., 2006). The transcription of miR-548d1 is related to transcription of the ATAD2 gene, as observed upon glucocorticoid stimulation (Rainer et al., 2009). This information showed the complex changes in miRNA/miRtron expression regulation upon distinct stimuli. ATAD2 has a ATP-binding site and ATPase activity, regulating the assembly of protein complexes (Morozumi et al., 2016), as CREB-binding promoter region or regulating histone hyperacetylation (Koo et al., 2016; Lazarchuk et al., 2020), suggesting the ATAD2/miR-548d can alter gene transcription during infection. ATAD2 inhibits the expression of vascular endothelial growth factor A (VEGFA) by altering miR-520a levels (Hong et al., 2018), linking miR-548d expression to modulation of other miRNAs. Also, ATAD2 can be a target of miRNAs, including miRNAs modulated during Leishmania infection including molecules described in our study such as miR-302, miR-373, and miR-93 (Bragato et al., 2018; Fernandes et al., 2019; Kumar et al., 2020).
miR-548d-3p was shown to enhance cell proliferation and inhibit apoptosis in breast cancer cells (Song et al., 2016), suggesting a possible role in inhibition of apoptosis seen in L. donovani-infected macrophages (Moore and Matlashewski, 1994). The miR-548 family can regulate expression of High mobility group box1 (HMBG1) a non-histone nuclear protein, a potent stimulator of tissue damage and inflammation through expression of pro-inflammatory cytokines (Martinotti et al., 2015; Son et al., 2019). The miR-548d-3p was seen previously in healing and inflammatory processes. In post-burn wound healing, the vascular endothelial growth factor-A (VEGFA) a key factor involved in the wound healing process was shown to likely be targeted by miR-548d-3p (Huang et al., 2020). In rheumatoid arthritis, an autoimmune inflammatory disease, another member of the miR-548 family, miR-548a-3p, was significantly down-regulated in serum samples targeting Toll-like receptor 4/nuclear factor kappa B (TLR4/NF-kappaB) signaling pathway (Wang et al., 2018). In THP-1 cells infected with promastigotes isolates from post-kala-azar dermal leishmaniasis, other members of the 548-miRNA family, miR-548at-5p, miR-548t-3p, were upregulated when compared to THP-1 cells infected with promastigotes isolated from visceral leishmaniasis patients (Kumar et al., 2020).
Importantly, miR-548d-3p was induced in both self-healed leishmaniasis patient samples and in vitro L. braziliensis-infected THP-1 cells. Because the miR-548-3p was found in patients’ plasma, it is likely that it is secreted by L. braziliensis-infected THP-1 cells, an aspect deserving further studies. It is known that THP-1 cell line can actively secrete microvesicles and exosomes that may contain miRNAs, such as miR-150 (Zhang et al., 2010) and miR‐103‐3p (Chen et al., 2020). Further, the content of microvesicles and exosomes may be modified by inflammatory conditions, infections including Leishmania, apoptosis, etc (Silverman et al., 2010; Baxter et al., 2019; Yao et al., 2019).
Inhibiting miR-548d-3p in THP-1 cells we observed a decrease in parasite load, and an increase in the production of MCP1/CCL2, RANTES/CCL5, and IP-10/CXCL10. In parallel, in plasma of self-healed patients, MCP1/CCL2, RANTES/CCL5, and IL-8/CXCL8 were decreased but increased MIG/CXCL9 and IP-10/CXCL10. We should be cautious to relate the in vitro experimental data to the evaluation in plasma. However, we observe a dichotomy impact of miR-548d, when upregulated in the early stage of in vitro infection of THP-1 derived monocytes by L. braziliensis (6–24 h) that is apparently reducing MCP-1 and RANTES at the infection site, contributing to the control of local inflammatory response, but at the same time, it is enabling parasite growth subverting the inflammatory response and lesion wound healing. These findings, considering the possibility of secretion of miR548d-3p by macrophages, are in line with the upregulated miR548d-3p found in the self-healed plasma patients that may reduce the MCP-1 and RANTES at systemic levels, contributing positively to wound healing modulating the inflammation. High IP-10 and MIG secretion in self-healed patients suggests that the miR-548-3p is not able to control the secretion of these cytokines. Previously, we observed higher levels of IP-10 and MIG, IFN-γ, and TNF in active and self-healed cutaneous leishmaniasis regulating parasite growth control (Souza et al., 2012; de Assis Souza et al., 2013).
Other studies have reported the role of these chemokines in cutaneous leishmaniasis. RANTES/CCL5, together with KC/CXCL1 and MIP-2/CXCL2 (Ohmori and Hamilton, 1994; Lebovic et al., 2001) participate in neutrophil, monocyte, and lymphocyte recruitment to inflammatory focus and interfere in the persistence of cutaneous leishmaniasis lesions (Teixeira et al., 2005; Costa-Silva et al., 2014). In experimental cutaneous leishmaniasis, the upregulation of miR-294 regulated Ccl2/Mcp-1 mRNA levels and infectivity in L. amazonensis infected BALB/c bone marrow-derived macrophages (Fernandes et al., 2019). Similarly, the downregulation of chemokines CCL2, CCL5, CXCL10, CXCL11, and CXCL12 was seen with upregulation of let-7a, miR-25, miR-26a, miR-132, miR-140, miR-146a, and miR-155 in L. major-infected human macrophages (Guerfali et al., 2008).
miRNAs are promising tools for diagnosis, treatment, and prognostic markers. Product for diagnosis is a reality mainly for cancers. No miRNA-based therapeutic formulations like miRNA mimics and antagomirs have reached the pharmaceutical breakthrough, but some are currently in clinical trials. In CL caused by L. braziliensis, miR-361-3p was appointed as a prognostic marker related to therapeutic failure. The miR-548d-3p evaluated in the present study was shown to exert tumor-suppressive effects in osteosarcoma cells and proposed as a therapeutic tool for osteosarcoma (Chen et al., 2019). Based on our findings, further studies are warranted to more clearly establish a role for miR-548d-3p as a prognostic marker and therapeutic target in cutaneous leishmaniasis.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Comite de Etica e Pesquisa da Faculdade de Medicina da Universidade de São Paulo. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HG: Conceptualization, study design, project and researcher supervision, manuscript preparation. MS, and ER-S: Conceptualization, study design, experimental work, data analysis, manuscript preparation. LF-W: study design, researcher supervision, manuscript preparation. SM: study design, experimental work, data analysis, manuscript preparation. LR: experimental work, manuscript preparation. RZ: experimental work. VP and MB: coordination of sample and data collection in endemic area, data interpretation. DL and PK: data analysis, manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (grants 2018/23512-0, 2018/14398-0, and 2018/24693-9, fellowship 2014/14756-2 to MS and 2019/25393-1 to LR), Medical Research Council (grants MR/P024661/1 and MR/S019472), the Conselho Nacional de Pesquisa (research fellowship to HG), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; fellowship to MS) and LIM 38 (Hospital das Clínicas, Faculdade de Medicina, Universidade de São Paulo).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Alexis Germán Murillo Carrasco for providing technical assistance with the bioinformatics approach.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.687647/full#supplementary-material
Effect of different concentrations of miR-548d-3p inhibitor or negative control (scrambled miRNA) transiently transfected on parasite load (number of amastigotes/100 cells) in L. (V.) braziliensis promastigote-infected THP-1 cells at 6 h (A) and 24 h (B) post-infection. The experiment was carried out by adding the synthetic molecules at 3nM, 10nM or 30nM with the transfection reagent diluted in RPMI medium or only RPMI medium (non-transfected cells) to wells containing 106 THP-1 adherent cells and maintained for 24 h at 37°C (5% CO2) then infected with L. (V.) braziliensis promastigotes. (A) * = P < 0.05 (one way ANOVA and student t test).
References
- Acuna S. M., Floeter-Winter L. M., Muxel S. M. (2020). MicroRNAs: Biological Regulators in Pathogen-Host Interactions. Cells 9 (1), 113. 10.3390/cells9010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V., Bell G. W., Nam J. W., Bartel D. P. (2015). Predicting Effective microRNA Target Sites in Mammalian mRNAs. Elife 4, e05005. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim C. F., Novais F. O., Nguyen B. T., Misic A. M., Carvalho L. P., Carvalho E. M., et al. (2019). Variable Gene Expression and Parasite Load Predict Treatment Outcome in Cutaneous Leishmaniasis. Sci. Transl. Med. 11 (519), eaax4204. 10.1126/scitranslmed.aax4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo A. P., Giorgio S. (2015). Immunohistochemical Evidence of Stress and Inflammatory Markers in Mouse Models of Cutaneous Leishmaniosis. Arch. Dermatol. Res. 307 (8), 671–682. 10.1007/s00403-015-1564-0 [DOI] [PubMed] [Google Scholar]
- Backes C., Khaleeq Q. T., Meese E., Keller A. (2016). miEAA: microRNA Enrichment Analysis and Annotation. Nucleic Acids Res. 44 (W1), W110–W116. 10.1093/nar/gkw345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D., Boldin M. P., O’Connell R. M., Rao D. S., Taganov K. D. (2008). MicroRNAs: New Regulators of Immune Cell Development and Function. Nat. Immunol. 9 (8), 839–845. 10.1038/ni.f.209 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116 (2), 281–297. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell 136 (2), 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A. A., Phan T. K., Hanssen E., Liem M., Hulett M. D., Mathivanan S., et al. (2019). Analysis of Extracellular Vesicles Generated From Monocytes Under Conditions of Lytic Cell Death. Sci. Rep. 9 (1), 7538. 10.1038/s41598-019-44021-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., et al. (2009). Induction and Regulatory Function of miR-9 in Human Monocytes and Neutrophils Exposed to Proinflammatory Signals. Proc. Natl. Acad. Sci. U. S. A. 106 (13), 5282–5287. 10.1073/pnas.0810909106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourreau E., Ronet C., Darsissac E., Lise M. C., Marie D. S., Clity E., et al. (2009). In Leishmaniasis Due to Leishmania Guyanensis Infection, Distinct Intralesional Interleukin-10 and Foxp3 mRNA Expression Are Associated With Unresponsiveness to Treatment. J. Infect. Dis. 199 (4), 576–579. 10.1086/596508 [DOI] [PubMed] [Google Scholar]
- Bragato J. P., Melo L. M., Venturin G. L., Rebech G. T., Garcia L. E., Lopes F. L., et al. (2018). Relationship of Peripheral Blood Mononuclear Cells miRNA Expression and Parasitic Load in Canine Visceral Leishmaniasis. PloS One 13 (12), e0206876. 10.1371/journal.pone.0206876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito M. E., Andrade M. S., Mendonca M. G., Silva C. J., Almeida E. L., Lima B. S., et al. (2009). Species Diversity of Leishmania (Viannia) Parasites Circulating in an Endemic Area for Cutaneous Leishmaniasis Located in the Atlantic Rainforest Region of Northeastern Brazil. Trop. Med. Int. Health 14 (10), 1278–1286. 10.1111/j.1365-3156.2009.02361.x [DOI] [PubMed] [Google Scholar]
- Burza S., Croft S. L., Boelaert M. (2018). Leishmaniasis. Lancet 392 (10151), 951–970. 10.1016/S0140-6736(18)31204-2 [DOI] [PubMed] [Google Scholar]
- Chandan K., Gupta M., Sarwat M. (2019). Role of Host and Pathogen-Derived MicroRNAs in Immune Regulation During Infectious and Inflammatory Diseases. Front. Immunol. 10, 3081. 10.3389/fimmu.2019.03081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yan C., Yu H., Zhen S., Yuan Q. (2019). miR-548d-3p Inhibits Osteosarcoma by Downregulating KRAS. Aging (Albany N. Y.) 11 (14), 5058–5069. 10.18632/aging.102097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Yao X., Yao H., Ji Q., Ding G., Liu X. (2020). Exosomal miR-103-3p From LPS-Activated THP-1 Macrophage Contributes to the Activation of Hepatic Stellate Cells. FASEB J. 34 (4), 5178–5192. 10.1096/fj.201902307RRR [DOI] [PubMed] [Google Scholar]
- Costa-Silva M. F., Gomes L. I., Martins-Filho O. A., Rodrigues-Silva R., Freire Jde M., Quaresma P. F., et al. (2014). Gene Expression Profile of Cytokines and Chemokines in Skin Lesions From Brazilian Indians With Localized Cutaneous Leishmaniasis. Mol. Immunol. 57 (2), 74–85. 10.1016/j.molimm.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Cummins J. M., He Y., Leary R. J., Pagliarini R., Diaz L. A., Jr, Sjoblom T., et al. (2006). The Colorectal microRNAome. Proc. Natl. Acad. Sci. U. S. A. 103 (10), 3687–3692. 10.1073/pnas.0511155103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Assis Souza M., de Castro M. C., de Oliveira A. P., de Almeida A. F., de Almeida T. M., Reis L. C., et al. (2013). Cytokines and NO in American Tegumentary Leishmaniasis Patients: Profiles in Active Disease, After Therapy and in Self-Healed Individuals. Microb. Pathog. 57, 27–32. 10.1016/j.micpath.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Diaz N. L., Zerpa O., Ponce L. V., Convit J., Rondon A. J., Tapia F. J. (2002). Intermediate or Chronic Cutaneous Leishmaniasis: Leukocyte Immunophenotypes and Cytokine Characterisation of the Lesion. Exp. Dermatol. 11 (1), 34–41. 10.1034/j.1600-0625.2002.110104.x [DOI] [PubMed] [Google Scholar]
- Faria D. R., Souza P. E., Duraes F. V., Carvalho E. M., Gollob K. J., Machado P. R., et al. (2009). Recruitment of CD8(+) T Cells Expressing Granzyme A Is Associated With Lesion Progression in Human Cutaneous Leishmaniasis. Parasite Immunol. 31 (8), 432–439. 10.1111/j.1365-3024.2009.01125.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J. C. R., Aoki J. I., Maia Acuna S., Zampieri R. A., Markus R. P., Floeter-Winter L. M., et al. (2019). Melatonin and Leishmania Amazonensis Infection Altered miR-294, miR-30e, and Mir-302d Impacting on Tnf, Mcp-1, and Nos2 Expression. Front. Cell Infect. Microbiol. 9:60. 10.3389/fcimb.2019.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga C. A., Oliveira M. V., Alves L. R., Viana A. G., Sousa A. A., Carvalho S. F., et al. (2012). Immunohistochemical Profile of HIF-1alpha, Vegf-a, VEGFR2 and MMP9 Proteins in Tegumentary Leishmaniasis. Bras. Dermatol. 87 (5), 709–713. 10.1590/s0365-05962012000500006 [DOI] [PubMed] [Google Scholar]
- Geraci N. S., Tan J. C., McDowell M. A. (2015). Characterization of microRNA Expression Profiles in Leishmania-Infected Human Phagocytes. Parasite Immunol. 37 (1), 43–51. 10.1111/pim.12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Silva A., de Cassia Bittar R., Dos Santos Nogueira R., Amato V. S., da Silva Mattos M., Oliveira-Neto M. P., et al. (2007). Can Interferon-Gamma and Interleukin-10 Balance Be Associated With Severity of Human Leishmania (Viannia) Braziliensis Infection? Clin. Exp. Immunol. 149 (3), 440–444. 10.1111/j.1365-2249.2007.03436.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Lauletta Lindoso J. A. (2012). Cutaneous and Mucocutaneous Leishmaniasis. Infect. Dis. Clin. North Am. 26 (2), 293–307. 10.1016/j.idc.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Guerfali F. Z., Laouini D., Guizani-Tabbane L., Ottones F., Ben-Aissa K., Benkahla A., et al. (2008). Simultaneous Gene Expression Profiling in Human Macrophages Infected With Leishmania Major Parasites Using SAGE. BMC Genomics 9, 238. 10.1186/1471-2164-9-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Shao L., Zheng L., Du Q., Li P., John B., et al. (2012). miRNA-939 Regulates Human Inducible Nitric Oxide Synthase Posttranscriptional Gene Expression in Human Hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 109 (15), 5826–5831. 10.1073/pnas.1118118109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberger S., Hinske L. C., Kreth S. (2018). MiRNAs: Dynamic Regulators of Immune Cell Functions in Inflammation and Cancer. Cancer Lett. 431, 11–21. 10.1016/j.canlet.2018.05.020 [DOI] [PubMed] [Google Scholar]
- Hong S., Chen S., Wang X., Sun D., Yan Z., Tai J., et al. (2018). ATAD2 Silencing Decreases VEGFA Secretion Through Targeting has-miR-520a to Inhibit Angiogenesis in Colorectal Cancer. Biochem. Cell Biol. 96 (6), 761–768. 10.1139/bcb-2018-0081 [DOI] [PubMed] [Google Scholar]
- Huang M., Huang X., Jiang B., Zhang P., Guo L., Cui X., et al. (2020). Linc00174-EZH2-ZNF24/Runx1-VEGFA Regulatory Mechanism Modulates Post-Burn Wound Healing. Mol. Ther. Nucleic Acids 21, 824–836. 10.1016/j.omtn.2020.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T. N., Ramirez-Komo J. A., Park H., Iritani B. M. (2017). Control of B Lymphocyte Development and Functions by the mTOR Signaling Pathways. Cytokine Growth Factor Rev. 35, 47–62. 10.1016/j.cytogfr.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocyigit A., Keles H., Selek S., Guzel S., Celik H., Erel O. (2005). Increased DNA Damage and Oxidative Stress in Patients With Cutaneous Leishmaniasis. Mutat. Res. 585 (1-2), 71–78. 10.1016/j.mrgentox.2005.04.012 [DOI] [PubMed] [Google Scholar]
- Koo S. J., Fernandez-Montalvan A. E., Badock V., Ott C. J., Holton S. J., von Ahsen O., et al. (2016). ATAD2 Is an Epigenetic Reader of Newly Synthesized Histone Marks During DNA Replication. Oncotarget 7 (43), 70323–70335. 10.18632/oncotarget.11855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Vijaykumar S., Dikhit M. R., Abhishek K., Mukherjee R., Sen A., et al. (2020). Differential Regulation of Mirna Profiles of Human Cells Experimentally Infected by Leishmania Donovani Isolated From Indian Visceral Leishmaniasis and Post-Kala-Azar Dermal Leishmaniasis. Front. Microbiol. 11, 1716. 10.3389/fmicb.2020.01716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago T. S., Silva J. A., Lago E. L., Carvalho E. M., Zanette D. L., Castellucci L. C. (2018). The miRNA 361-3p, a Regulator of GZMB and TNF Is Associated With Therapeutic Failure and Longer Time Healing of Cutaneous Leishmaniasis Caused by L. (Viannia) Braziliensis. Front. Immunol. 9, 2621. 10.3389/fimmu.2018.02621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., et al. (2007). A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 129 (7), 1401–1414. 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarchuk P., Hernandez-Villanueva J., Pavlova M. N., Federation A., MacCoss M., Sidorova J. M. (2020). Mutual Balance of Histone Deacetylases 1 and 2 and the Acetyl Reader ATAD2 Regulates the Level of Acetylation of Histone H4 on Nascent Chromatin of Human Cells. Mol. Cell Biol. 40 (9), e00421–19. 10.1128/MCB.00421-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovic D. I., Chao V. A., Martini J. F., Taylor R. N. (2001). Il-1beta Induction of RANTES (Regulated Upon Activation, Normal T Cell Expressed and Secreted) Chemokine Gene Expression in Endometriotic Stromal Cells Depends on a Nuclear Factor-Kappab Site in the Proximal Promoter. J. Clin. Endocrinol. Metab. 86 (10), 4759–4764. 10.1210/jcem.86.10.7890 [DOI] [PubMed] [Google Scholar]
- Lemaire J., Mkannez G., Guerfali F. Z., Gustin C., Attia H., Sghaier R. M., et al. (2013). MicroRNA Expression Profile in Human Macrophages in Response to Leishmania Major Infection. PloS Negl. Trop. Dis. 7 (10), e2478. 10.1371/journal.pntd.0002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Guo L., Liu C. (2012). Genome-Wide Analysis of mir-548 Gene Family Reveals Evolutionary and Functional Implications. J. BioMed. Biotechnol. 2012, 679563. 10.1155/2012/679563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Junior D. S., Costa D. L., Carregaro V., Cunha L. D., Silva A. L., Mineo T. W., et al. (2013). Inflammasome-Derived IL-1beta Production Induces Nitric Oxide-Mediated Resistance to Leishmania. Nat. Med. 19 (7), 909–915. 10.1038/nm.3221 [DOI] [PubMed] [Google Scholar]
- Limon J. J., Fruman D. A. (2012). Akt and mTOR in B Cell Activation and Differentiation. Front. Immunol. 3, 228. 10.3389/fimmu.2012.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado P. R., Rosa M. E., Costa D., Mignac M., Silva J. S., Schriefer A., et al. (2011). Reappraisal of the Immunopathogenesis of Disseminated Leishmaniasis: In Situ and Systemic Immune Response. Trans. R. Soc. Trop. Med. Hyg. 105 (8), 438–444. 10.1016/j.trstmh.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabita F., de Candia P., Torri A., Tegner J., Abrignani S., Rossi R. L. (2016). Normalization of Circulating MicroRNA Expression Data Obtained by Quantitative Real-Time RT-PCR. Brief Bioinform. 17 (2), 204–212. 10.1093/bib/bbv056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinotti S., Patrone M., Ranzato E. (2015). Emerging Roles for HMGB1 Protein in Immunity, Inflammation, and Cancer. Immunotarg. Ther. 4, 101–109. 10.2147/ITT.S58064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. J., Matlashewski G. (1994). Intracellular Infection by Leishmania Donovani Inhibits Macrophage Apoptosis. J. Immunol. 152 (6), 2930–2937. [PubMed] [Google Scholar]
- Morozumi Y., Boussouar F., Tan M., Chaikuad A., Jamshidikia M., Colak G., et al. (2016). Atad2 is a Generalist Facilitator of Chromatin Dynamics in Embryonic Stem Cells. J. Mol. Cell Biol. 8 (4), 349–362. 10.1093/jmcb/mjv060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muxel S. M., Acuna S. M., Aoki J. I., Zampieri R. A., Floeter-Winter L. M. (2018). Toll-Like Receptor and Mirna-Let-7e Expression Alter the Inflammatory Response in Leishmania amazonensis-Infected Macrophages. Front. Immunol. 9, 2792. 10.3389/fimmu.2018.02792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muxel S. M., Laranjeira-Silva M. F., Zampieri R. A., Floeter-Winter L. M. (2017). Leishmania (Leishmania) Amazonensis Induces Macrophage miR-294 and miR-721 Expression and Modulates Infection by Targeting NOS2 and L-arginine Metabolism. Sci. Rep. 7, 44141. 10.1038/srep44141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes S., Silva I. B., Ampuero M. R., de Noronha A. L. L., de Souza L. C. L., Correia T. C., et al. (2018). Integrated Analysis Reveals That miR-193b, miR-671, and TREM-1 Correlate With a Good Response to Treatment of Human Localized Cutaneous Leishmaniasis Caused by Leishmania Braziliensis. Front. Immunol. 9, 640. 10.3389/fimmu.2018.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R. M., Rao D. S., Baltimore D. (2012). microRNA Regulation of Inflammatory Responses. Annu. Rev. Immunol. 30, 295–312. 10.1146/annurev-immunol-020711-075013 [DOI] [PubMed] [Google Scholar]
- Ohmori Y., Hamilton T. A. (1994). IFN-Gamma Selectively Inhibits Lipopolysaccharide-Inducible JE/monocyte Chemoattractant Protein-1 and KC/GRO/Melanoma Growth-Stimulating Activity Gene Expression in Mouse Peritoneal Macrophages. J. Immunol. 153 (5), 2204–2212. [PubMed] [Google Scholar]
- Paul S., Ruiz-Manriquez L. M., Serrano-Cano F. I., Estrada-Meza C., Solorio-Diaz K. A., Srivastava A. (2020). Human microRNAs in Host-Parasite Interaction: A Review. 3 Biotech. 10 (12), 510. 10.1007/s13205-020-02498-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainer J., Ploner C., Jesacher S., Ploner A., Eduardoff M., Mansha M., et al. (2009). Glucocorticoid-Regulated microRNAs and Mirtrons in Acute Lymphoblastic Leukemia. Leukemia 23 (4), 746–752. 10.1038/leu.2008.370 [DOI] [PubMed] [Google Scholar]
- Reis L. C., Ramos-Sanchez E. M., Araujo F. N., Leal A. F., Ozaki C. Y., Sevillano O. R., et al. (2021). Pleiotropic Effect of Hormone Insulin-Like Growth Factor-I in Immune Response and Pathogenesis in Leishmaniases. J. Immunol. Res. 2021, 6614475. 10.1155/2021/6614475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveneau S., Petrakis T. G., Goldring C. E., Chantome A., Jeannin J. F. (2012). Oct-1 Cooperates With the TATA Binding Initiation Complex to Control Rapid Transcription of Human Inos. Cell Mol. Life Sci. 69 (15), 2609–2619. 10.1007/s00018-012-0939-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Ghosh S., Pakrashi S., Roy D., Sen S., Chatterjee M. (2012). Leishmania Strains Causing Self-Healing Cutaneous Leishmaniasis Have Greater Susceptibility Towards Oxidative Stress. Free Radic. Res. 46 (5), 665–673. 10.3109/10715762.2012.668186 [DOI] [PubMed] [Google Scholar]
- Silverman J. M., Clos J., de’Oliveira C. C., Shirvani O., Fang Y., Wang C., et al. (2010). An Exosome-Based Secretion Pathway Is Responsible for Protein Export From Leishmania and Communication With Macrophages. J. Cell Sci. 123 (Pt 6), 842–852. 10.1242/jcs.056465 [DOI] [PubMed] [Google Scholar]
- Sohel M. A. (2016). Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achievements Life Sci. 10, 175–186. 10.1016/j.als.2016.11.007 [DOI] [Google Scholar]
- Song Q., Song J., Wang Q., Ma Y., Sun N., Ma J., et al. (2016). miR-548d-3p/TP53BP2 Axis Regulates the Proliferation and Apoptosis of Breast Cancer Cells. Cancer Med. 5 (2), 315–324. 10.1002/cam4.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son G. H., Kim Y., Lee J. J., Lee K. Y., Ham H., Song J. E., et al. (2019). MicroRNA-548 Regulates High Mobility Group Box 1 Expression in Patients With Preterm Birth and Chorioamnionitis. Sci. Rep. 9 (1), 19746. 10.1038/s41598-019-56327-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotto M. N., Yamashiro-Kanashiro E. H., da Matta V. L., de Brito T. (1989). Cutaneous Leishmaniasis of the New World: Diagnostic Immunopathology and Antigen Pathways in Skin and Mucosa. Acta Trop. 46 (2), 121–130. 10.1016/0001-706x(89)90006-5 [DOI] [PubMed] [Google Scholar]
- Souza M. A., Almeida T. M., Castro M. C., Oliveira-Mendes A. P., Almeida A. F., Oliveira B. C., et al. (2016). American Tegumentary Leishmaniasis: mRNA Expression for Th1 and Treg Mediators Are Predominant in Patients With Recent Active Disease. Immunobiology 221 (2), 253–259. 10.1016/j.imbio.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Souza M. A., Castro M. C., Oliveira A. P., Almeida A. F., Reis L. C., Silva C. J., et al. (2012). American Tegumentary Leishmaniasis: Cytokines and Nitric Oxide in Active Disease and After Clinical Cure, With or Without Chemotherapy. Scand. J. Immunol. 76 (2), 175–180. 10.1111/j.1365-3083.2012.02717.x [DOI] [PubMed] [Google Scholar]
- Souza A. S., Giudice A., Pereira J. M., Guimaraes L. H., de Jesus A. R., de Moura T. R., et al. (2010). Resistance of Leishmania (Viannia) Braziliensis to Nitric Oxide: Correlation With Antimony Therapy and TNF-Alpha Production. BMC Infect. Dis. 10, 209. 10.1186/1471-2334-10-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. J., Fernandes J. D., Teixeira C. R., Andrade B. B., Pompeu M. L., Santana da Silva J., et al. (2005). Distinct Leishmania Braziliensis Isolates Induce Different Paces of Chemokine Expression Patterns. Infect. Immun. 73 (2), 1191–1195. 10.1128/IAI.73.2.1191-1195.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiotto-Pellissier F., Bortoleti B., Assolini J. P., Goncalves M. D., Carloto A. C. M., Miranda-Sapla M. M., et al. (2018). Macrophage Polarization in Leishmaniasis: Broadening Horizons. Front. Immunol. 9, 2529. 10.3389/fimmu.2018.02529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S., Kobayashi Y., Goto Y., Okumura H., Nakae S., Konno T., et al. (1982). Induction of Maturation in Cultured Human Monocytic Leukemia Cells by a Phorbol Diester. Cancer Res. 42 (4), 1530–1536. [PubMed] [Google Scholar]
- Turetz M. L., Machado P. R., Ko A. I., Alves F., Bittencourt A., Almeida R. P., et al. (2002). Disseminated Leishmaniasis: A New and Emerging Form of Leishmaniasis Observed in Northeastern Brazil. J. Infect. Dis. 186 (12), 1829–1834. 10.1086/345772 [DOI] [PubMed] [Google Scholar]
- Vieira M. G., Oliveira F., Arruda S., Bittencourt A. L., Barbosa A. A., Jr., Barral-Netto M., et al. (2002). B-Cell Infiltration and Frequency of Cytokine Producing Cells Differ Between Localized and Disseminated Human Cutaneous Leishmaniases. Mem Inst Oswaldo Cruz 97 (7), 979–983. 10.1590/s0074-02762002000700009 [DOI] [PubMed] [Google Scholar]
- Vlachos I. S., Zagganas K., Paraskevopoulou M. D., Georgakilas G., Karagkouni D., Vergoulis T., et al. (2015). Diana-miRPath v3.0: Deciphering microRNA Function With Experimental Support. Nucleic Acids Res. 43 (W1), W460–W466. 10.1093/nar/gkv403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang P., Liu Y., Cheng G. (2010). TRAF-Mediated Regulation of Immune and Inflammatory Responses. Sci. China Life Sci. 53 (2), 159–168. 10.1007/s11427-010-0050-3 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zheng F., Gao G., Yan S., Zhang L., Wang L., et al. (2018). MiR-548a-3p Regulates Inflammatory Response Via TLR4/NF-kappaB Signaling Pathway in Rheumatoid Arthritis. J. Cell Biochem. 120 (2), 1133–1140 10.1002/jcb.26659 [DOI] [PubMed] [Google Scholar]
- Yao Z., Jia X., Megger D. A., Chen J., Liu Y., Li J., et al. (2019). Label-Free Proteomic Analysis of Exosomes Secreted From THP-1-Derived Macrophages Treated With IFN-alpha Identifies Antiviral Proteins Enriched in Exosomes. J. Proteome Res. 18 (3), 855–864. 10.1021/acs.jproteome.8b00514 [DOI] [PubMed] [Google Scholar]
- Ye H., Arron J. R., Lamothe B., Cirilli M., Kobayashi T., Shevde N. K., et al. (2002). Distinct Molecular Mechanism for Initiating TRAF6 Signalling. Nature 418 (6896), 443–447. 10.1038/nature00888 [DOI] [PubMed] [Google Scholar]
- Yu Z., Zhang W., Kone B. C. (2002). Signal Transducers and Activators of Transcription 3 (STAT3) Inhibits Transcription of the Inducible Nitric Oxide Synthase Gene by Interacting With Nuclear Factor Kappab. Biochem. J. 367 (Pt 1), 97–105. 10.1042/BJ20020588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., et al. (2010). Secreted Monocytic miR-150 Enhances Targeted Endothelial Cell Migration. Mol. Cell 39 (1), 133–144. 10.1016/j.molcel.2010.06.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of different concentrations of miR-548d-3p inhibitor or negative control (scrambled miRNA) transiently transfected on parasite load (number of amastigotes/100 cells) in L. (V.) braziliensis promastigote-infected THP-1 cells at 6 h (A) and 24 h (B) post-infection. The experiment was carried out by adding the synthetic molecules at 3nM, 10nM or 30nM with the transfection reagent diluted in RPMI medium or only RPMI medium (non-transfected cells) to wells containing 106 THP-1 adherent cells and maintained for 24 h at 37°C (5% CO2) then infected with L. (V.) braziliensis promastigotes. (A) * = P < 0.05 (one way ANOVA and student t test).
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.