ABSTRACT
Acute kidney injury (AKI) is the most common complication of sepsis. MicroRNAs (miRNAs) play important roles in the sepsis-induced AKI. This paper aimed to explore the role of miRNA 181a-2-3p (miR-181a-2-3p) in the sepsis-induced AKI and the underlying mechanism. Our results revealed that miR-181a-2-3p showed low expression levels in patients with sepsis and mouse models undergoing cecal ligation and puncture (CLP). The addition of miR-181a-2-3p antagonists aggravated the sepsis-induced kidney injuries and inflammatory response in CLP mouse models, as suggested by hematoxylin and eosin (H&E) staining and quantitative real-time PCR (qRT-PCR). In addition, miR-181a-2-3p mimic alleviated the lipopolysaccharide (LPS)-induced inflammatory response, along with apoptosis of TCMK-1. Moreover, results from the GSE46955 data set indicated that GJB2 was highly expressed in septic patients but lowly expressed after recovery. Further, the dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay were carried out, which confirmed that GJB2 was a target of miR-181a-2-3p, and overexpression of GJB2 reversed the anti-inflammatory and antiapoptotic effects of miR-181a-2-3p mimic on the LPS-induced sepsis cell models. In conclusion, miR-181a-2-3p alleviates the inflammatory response and cell apoptosis of septic patients and animal models by upregulating GJB2 expression, which may provide a new therapeutic strategy for sepsis.
KEYWORDS: miR-181a-2-3p, sepsis, inflammatory response, GJB2, apoptosis
INTRODUCTION
Sepsis is a clinical syndrome, infection of which causes host reaction disorder and life-threatening organ dysfunction. Sepsis is associated with a high incidence rate, which accounts for the main cause of death among patients at the intensive care unit (ICU) (1). It has been suggested that sepsis is associated with about 50% of acute kidney injuries (AKI), and up to 60% septic patients have AKI (2). Notably, the mortality rate of septic patients with AKI is significantly higher than that of patients without AKI; in addition, the mortality rate of patients with sepsis-related AKI is higher than that of patients with AKI due to other causes. However, no effective therapeutic strategy is available for septic AKI at present, so it is particularly important to identify the effective therapeutic targets. Overexpression of inflammatory factors (such as tumor necrosis factor alpha [TNF-α], interleukin-1 beta [IL-1β], and IL-6) in sepsis can induce dysfunction of endothelial cells (ECs) and renal tubular epithelial cells (RTECs), leading to cell apoptosis in some severe cases (3). Apoptosis of RTECs induces the collapse of renal tubular epithelium, eventually causing injury of renal function (4). Therefore, RTECs contribute to maintaining the stability of renal function.
MicroRNAs (miRNAs) are a class of single-stranded RNA molecules with length of about 19 to 25 nucleotides which are highly conserved in evolution and extensively distributed in animals and plants. miRNAs can specifically bind to target mRNA to inhibit its expression and regulate gene expression at the posttranscriptional level (5). miRNAs play important roles in the pathophysiological processes of sepsis and the regulation of sepsis-related organ dysfunction. Some miRNAs are involved in the sepsis-related inflammatory response and apoptosis, affecting the occurrence and development of sepsis. For instance, miRNA 146a (miR-146a) is reported to inhibit the sepsis-induced secretion of inflammatory cytokine (TNF-α), thus alleviating the sepsis-related AKI (6). In addition, miR-146a improves the sepsis-induced inflammatory response in cardiomyopathy and cardiac dysfunction through downregulating the expression of TNF-α and IL-1α and suppressing cell apoptosis (7). miR-96-3p promotes the apoptosis of RTECs by targeting SIKIAT1, which aggravates the sepsis-induced AKI (8). In addition, the miR-181 family play critical roles in kidney diseases. For example, the high expression of miR-181d-5p results in increased renal blood flow (9). Moreover, animal experiments have revealed that treatment with miR-181 agonist significantly rescues kidney injury induced by unilateral ureteral obstruction (10). The above research results show that the miR-181 family have a protective effect on kidney injury. miR-181a-2-3p belongs to the miR-181 family. As discovered by Kim et al., the expression of miR-181a-2-3p decreased significantly in chronic obstructive pulmonary disease (COPD), and silencing miR-181a-2-3p enhanced the COPD-induced inflammatory response (11). In addition, the authors found that downregulation of miR-181a-2-3p led to a decreased number of cancer stem cells (CSCs). Therefore, we speculate that miR-181a-2-3p is an miRNA related to inflammation, cell proliferation, and cell apoptosis, but its underlying mechanism in sepsis should be further investigated.
RESULTS
miR-181a-2-3p expression was downregulated in LPS-induced cell model and CLP mouse models.
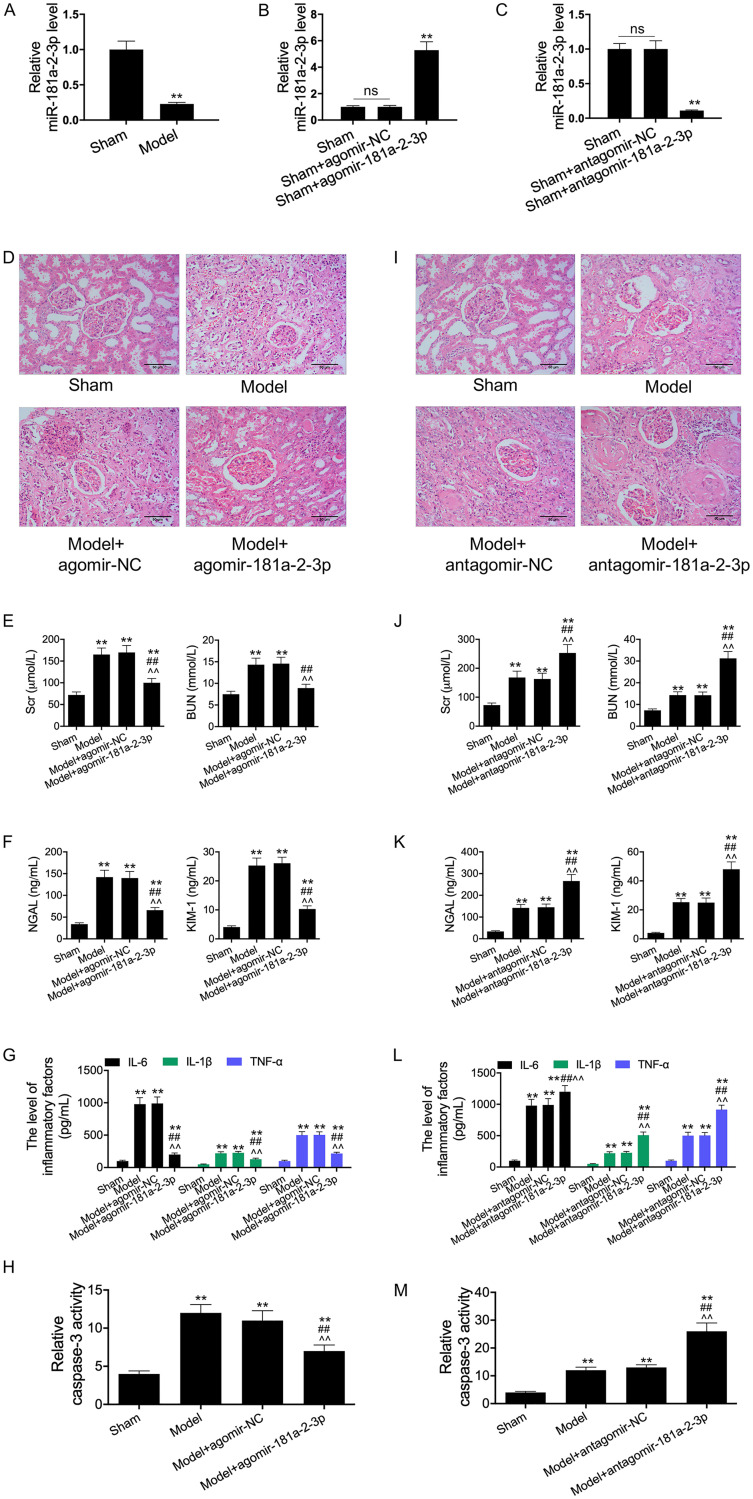
To test the potential role of miR-181a-2-3p in sepsis, a lipopolysaccharide (LPS)-induced sepsis cell model and the standardized sepsis cecal ligation and puncture (CLP) mouse models were constructed. Then, we examined whether miR-181a-2-3p participated in the inflammatory response and apoptosis in vivo. According to our results, miR-181a-2-3p was also downregulated in kidney tissues of mice with sepsis (Fig. 1A). Thereafter, CLP mouse models were transfected with the agomir-181a-2-3p and antagomir-181a-2-3p. As a result, transfection with agomir-181a-2-3p significantly increased the miR-181a-2-3p expression, while transfection with antagomir-181a-2-3p remarkably inhibited the miR-181a-2-3p expression in kidney tissues of CLP mice (Fig. 1B and C). Subsequently, we performed pathological analysis on kidney tissues of mice. As revealed by H&E staining of the kidney, the cell morphology of CLP mouse models was severely damaged and inflammatory response was observed, indicating the successful construction of the sepsis-AKI mouse models. However, relative to transfection with agomir-NC, transfection with agomir-181a-2-3p reduced inflammatory cell infiltration and recovered the tissue structure (Fig. 1D). In addition, results of enzyme-linked immunosorbent assay (ELISA) revealed that the levels of creatinine (Cr), blood urea nitrogen (BUN), neutrophil gelatinase-associated lipocalin (NGAL), and kidney injury molecule 1 (KIM-1) in CLP mouse models significantly increased, suggesting the disordered kidney function in CLP mouse models; meanwhile, the addition of agomir-181a-2-3p reversed this phenomenon, demonstrating that agomir-181a-2-3p had a certain protective effect on the sepsis-induced kidney dysfunction (Fig. 1E and F). Similarly, the addition of agomir-181a-2-3p reversed the sepsis-induced upregulated inflammatory cytokines (TNF-α, IL-1β, and IL-6) and enhanced caspase-3 activity. In addition, the addition of antagomir-181a-2-3p aggravated the sepsis-induced AKI (Fig. 1I to M). Thus, our results suggested that 181a-2-3p might participate in the sepsis-induced injury to RTECs.
FIG 1.
miR-181a-2-3p expression was decreased in LPS-induced cell model and CLP mice models. (A) The expression of miR-181a-2-3p in sepsis mice and sham mice was measured by qRT-PCR. RT-PCR assay was used to detect miR-181a-2-3p expression in agomir-181a-2-3p transfection group (B) and antagomir-181a-2-3p transfection group (C). (D and I) The photomicrographs of kidney tissues stained by H&E (200×). (E, F, G, J, K, and L) ELISA was performed to examine the levels of Cr, BUN, NGAL, KIM-1, IL-1β, IL-6, and TNF-α in the serum of each group. (H and M) ELISA was performed to examine the caspase-3 activity. **, P < 0.01; ##, P < 0.01; ^^, P < 0.01.
Effects of miR-181a-2-3p on the LPS-induced secretion of inflammatory factors and apoptosis of TCMK-1 cells.
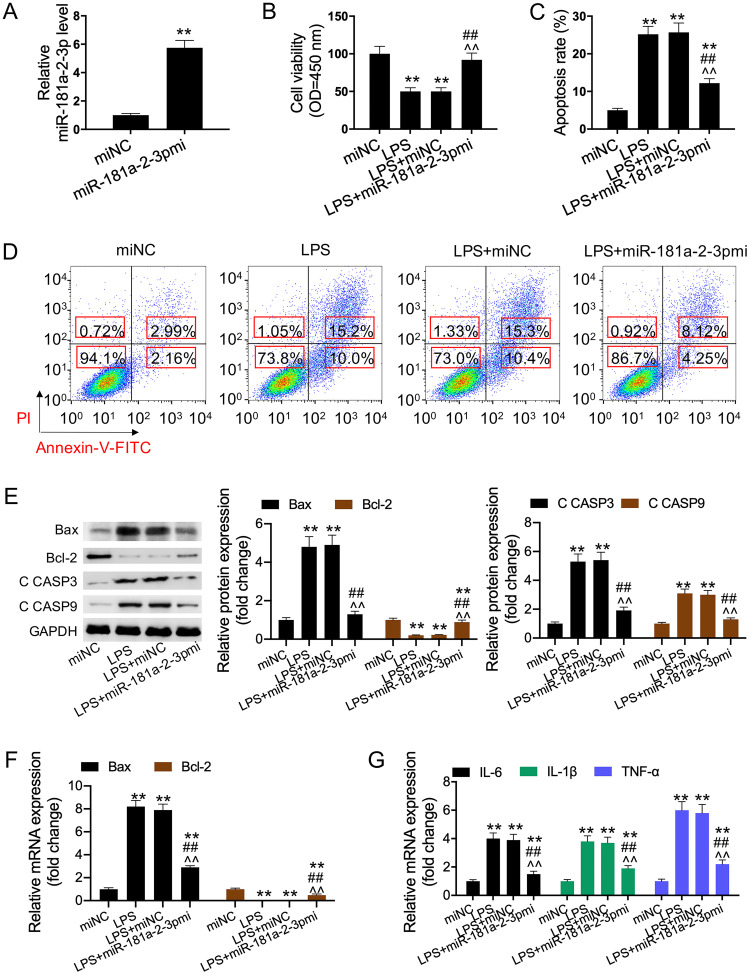
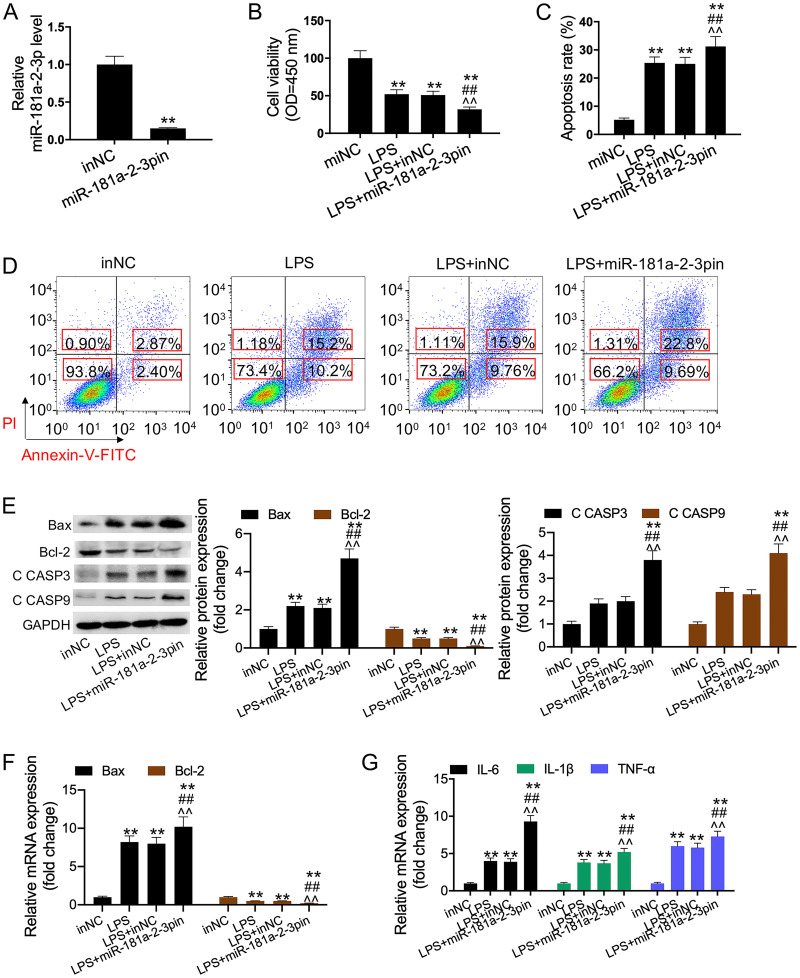
To further validate the previous results, cells were transfected with miR-181a-2-3p mimic. Quantitative real-time PCR (qRT-PCR) assay was performed to measure the cell transfection efficiency of miR-181a-2-3p mimic. The results indicated that the miR-181a-2-3p expression significantly increased in cells transfected with miR-181a-2-3p mimic compared to that of cells transfected with the mimic control (Fig. 2A). Furthermore, our results showed that overexpression of miR-181a-2-3p partially reversed the LPS-induced inhibition of cell viability and promotion of HK2 cell apoptosis (Fig. 2B to D). We also detected the expression levels of apoptosis-related proteins (Bax, Bcl-2, and C CASP-3/9) and the mRNA expression of inflammatory cytokines (IL-1β, IL-6, and TNF-α). As a result, LPS was found to induce inflammatory response and cell apoptosis, while the addition of miR-181a-2-3p reversed this effect (Fig. 2E to G). In the subsequent study, the addition of miR-181a-2-3p inhibitor was discovered to aggravate the LPS-induced cell apoptosis and inflammatory response (Fig. 3A to G).
FIG 2.
miR-181a-2-3p overexpression relieved LPS-induced apoptosis and inflammatory response. (A) qRT-PCR was used to detect miR-181a-2-3p expression in miR-181a-2-3p mimic-transfected TCMK-1 cells. (B) Cell viability was detected using CCK-8 assay in each group. (C and D) Cell apoptosis was detected using flow cytometry analysis in each group. (E) Bax, Bcl-2, and C CASP3/-9 expression was determined using Western blotting. (F and G) qRT-PCR was performed to examine the levels of Bax, Bcl-2, IL-1β, IL-6, and TNF-α in each group. **, P < 0.01; ##, P < 0.01; ^^, P < 0.01. OD, optical density.
FIG 3.
miR-181a-2-3p inhibition aggravated LPS-induced apoptosis and inflammatory response. (A) qRT-PCR was used to detect miR-181a-2-3p expression in miR-181a-2-3p inhibitor-transfected TCMK-1 cells. (B) Cell viability was detected using CCK-8 assay in each group. (C and D) Cell apoptosis was detected using flow cytometry analysis in each group. (E) Bax, Bcl-2, and C CASP3/-9 expression were determined using Western blotting. (F and G) qRT-PCR was performed to examine the levels of Bax, Bcl-2, IL-1β, IL-6, and TNF-α in each group. **, P < 0.01; ##, P < 0.01; ^^, P < 0.01.
GJB2 was a target of miR-181a-2-3p.
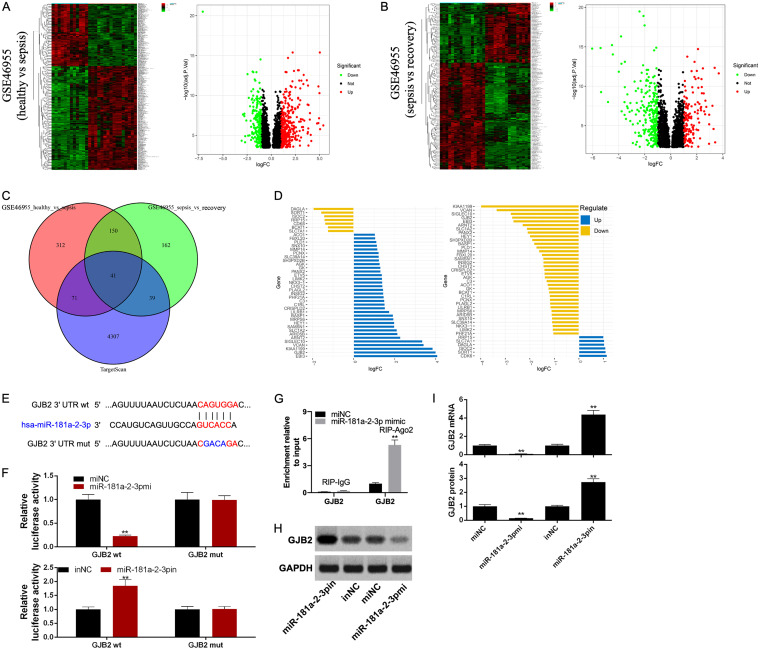
GSE46955 was a sepsis-related data set. There were 574 differentially expressed genes (DEGs) identified between the healthy group and the sepsis group, including 215 downregulated genes and 359 upregulated genes (Fig. 4A); in addition, 392 DEGs were identified between the sepsis group and the recovery group, including 252 downregulated genes and 140 upregulated genes (Fig. 4B). Altogether, 4,458 target genes of miR-181a-2-3p were found in the TargetScan database. Next, the Venn plot showed 41 intersected target genes (Fig. 4C). However, it was interesting to observe that GJB2 was highly expressed in septic patients but lowly expressed in recovering patients (Fig. 4D). Figure 3E shows the binding sites between the 3′ untranslated region (UTR) of GJB2 and miR-181a-2-3p detected by the TargetScan database. Typically, the direct interaction between miR-181a-2-3p and GJB2 was further verified by the dual-luciferase reporter system. As a result, the luciferase activity of GJB2-WT was significantly reduced by miR-181a-2-3p mimic and remarkably enhanced by miR-181a-2-3p inhibitor. However, the luciferase activity of GJB2-MUT remained unchanged (Fig. 4F). Moreover, RNA immunoprecipitation (RIP) assay was performed to test whether GJB2 interacted with miR-181a-2-3p. It was discovered that GJB2 was substantially enriched in RIPAgo2 upon the addition of miR-181a-2-3p mimic relative to the RIP-IgG control group (Fig. 4G), further indicating the interaction of GJB2 with miR-181a-2-3p. Thereafter, HK2 cells were transfected with miR-181a-2-3p mimic or miR-181a-2-3p inhibitor to examine the effect of miR-181a-2-3p expression on the GJB2 level. Results of Western blotting implied that the GJB2 protein expression was downregulated by the overexpression of miR-181a-2-3p but upregulated by the inhibition of miR-181a-2-3p (Fig. 4H and I).
FIG 4.
GJB2 was a target of miR-181a-2-3p. (A) Heat map and volcano plot of DEGs between healthy group and sepsis group in GSE46955 data set. (B) Heat map and volcano plot of DEGs between sepsis group and recovery group in GSE46955 data set. (C) Venn plot of intersecting genes between GSE46955 data set and TargetScan database. (D) The expression of 41 intersected target genes in GSE46955 data set. (E) The binding sites and mutant binding sites between miR-181a-2-3p and GJB2. (F) After transfection with miR-181a-2-3p mimic or inhibitor, the luciferase activity of GJB2-WT/MUT was tested by dual-luciferase reporter assay. (G) RIP assay was performed to detect the enrichment of GJB2 in RIP-Ago2 or RIP-IgG. (H and I) The GJB2 protein level and mRNA level were tested via Western blotting and qRT-PCR assay to assess the effect of miR-181a-2-3p mimic and inhibitor on GJB2 expression. **, P < 0.01.
GJB2 was involved in the sepsis-induced AKI and inflammatory response in vivo.
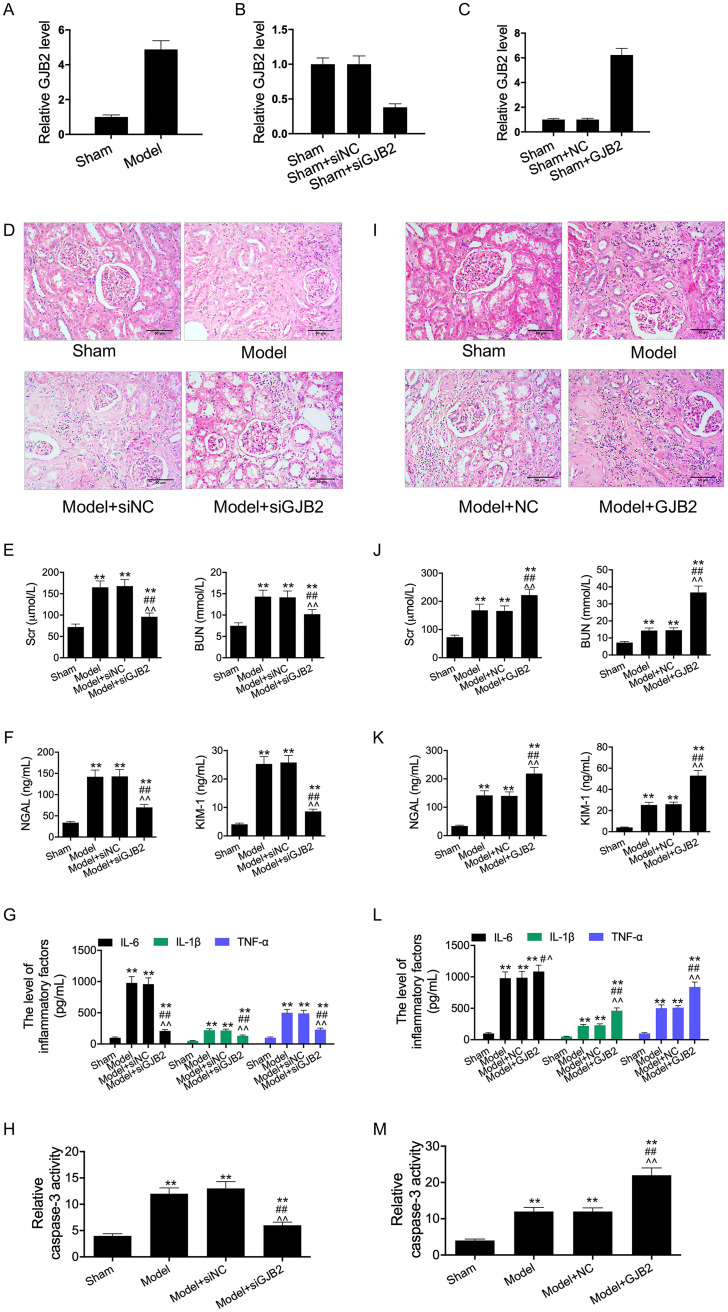
Our results suggested that the expression of GJB2 was significantly upregulated in mice with sepsis (Fig. 5A). CLP mouse models were transfected with siRNA-GJB2 and GJB2 expression vector. As a result, GJB2 silencing significantly decreased GJB2 expression, whereas GJB2 overexpression remarkably increased GJB2 expression in the kidney tissues of CLP mice (Fig. 5B and C). Subsequently, H&E staining of the kidney demonstrated that GJB2 silencing reduced inflammatory cell infiltration and recovered tissue structure compared with transfection with agomir-NC (Fig. 5D). In addition, results of ELISA revealed that GJB2 silencing downregulated the levels of Cr, BUN, NGAL, and KIM-1 and decreased the activity of caspase-3 in CLP mouse models (Fig. 5E to H). Meanwhile, GJB2 overexpression aggravated the sepsis-induced AKI (Fig. 5I to M). Taken together, our results suggested that GJB2 might participate in the sepsis-induced injury of RTECs.
FIG 5.
GJB2 expression was increased in LPS-induced cell model and CLP mice models. (A) The expression of GJB2 in sepsis mice and sham mice was measured by qRT-PCR. RT-PCR assay was used to detect GJB2 expression in siRNA-GJB2 transfection group (B) and vector expressing GJB2 transfection group (C). (D and I) The photomicrographs of kidney tissues stained by H&E (200×). (E, F, G, J, K, and L) ELISA was performed to examine the levels of Cr, BUN, NGAL, KIM-1, IL-1β, IL-6, and TNF-α in the serum of each group. (H and M) ELISA was performed to examine the caspase-3 activity. **, P < 0.01; ##, P < 0.01; ^^, P < 0.01.
Overexpression of GJB2 reversed the LPS-induced anti-inflammatory and antiapoptotic effects of miR-181a-2-3p mimic on HK2 cells.
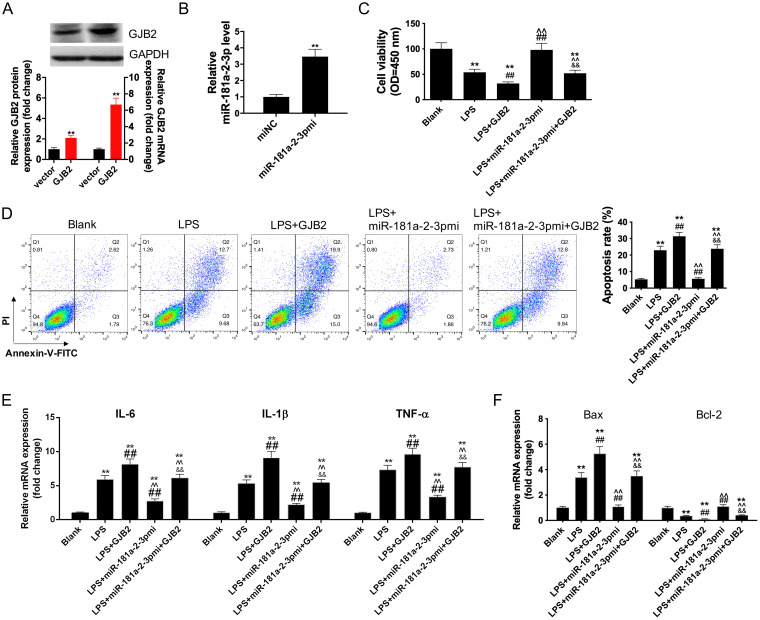
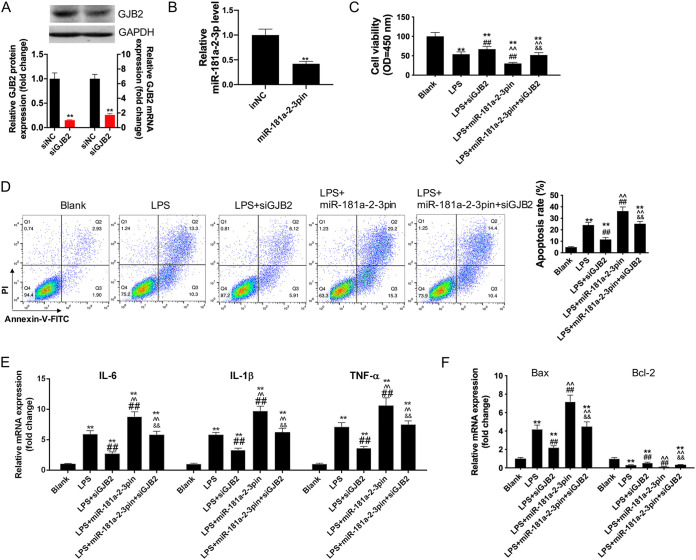
First, siRNA-GJB2 was transfected into HK2 cells to inhibit GJB2 expression (Fig. 6A), while the GJB2 expression pcDNA3.1 vector was transfected into HK2 cells to upregulate GJB2 expression, as verified by Western blotting and qRT-PCR assays (Fig. 7A). Next, the transfection of miR-181a-2-3p mimics upregulated miR-181a-2-3p mRNA level of HK2 cells (Fig. 6B) and the transfection of miR-181a-2-3p inhibitors inhibited miR-181a-2-3p expression of HK2 cells (Fig. 7B). To verify whether miR-181a-2-3p was involved in the inflammatory response and cell apoptosis in sepsis by targeting GJB2, we cotransfected HK2 cells with GJB2 expression pcDNA3.1 vector and miR-181a-2-3p and later treated cells with LPS. The results showed that the addition of GJB2 expression pcDNA3.1 vector promoted the effect of LPS on aggravating cell apoptosis and inflammatory response (Fig. 6C to F). Subsequently, our results indicated that overexpression of GJB2 reversed the suppression effect of miR-181a-2-3p mimic on the inflammatory cytokines (IL-1β, IL-6, and TNF-α) and cell apoptosis of LPS-induced macrophages. In addition, silencing GJB2 abolished the promotion effect of miR-181a-2-3p inhibitor on the inflammatory cytokines (IL-1β, IL-6, and TNF-α) and cell apoptosis of LPS-induced macrophages (Fig. 7C to F).
FIG 6.
Overexpression of GJB2 reversed the anti-inflammatory and antiapoptosis effects of miR-181a-2-3p mimic on LPS-induced HK2. (A) Western blotting and qRT-PCR were used to detect GJB2 expression in vector expressing GJB2-transfected HK2 cells. (B) Western blotting and qRT-PCR were used to detect miR-181a-2-3p expression in miR-181a-2-3p mimic-transfected HK2 cells. Cotransfection of miR-181a-2-3p mimic and GJB2 treatment with LPS-induced HK2, cell viability (C), cell apoptosis (D), the levels of IL-1β, IL-6, and TNF-α (E), and the expression of Bax and Bcl-2 (F) were detected using CCK-8 assay, flow cytometry analysis, and qRT-PCR. **, P < 0.01; ##, P < 0.01; ^^, P < 0.01; &&, P < 0.01.
FIG 7.
Overexpression of GJB2 reversed the anti-inflammatory and antiapoptosis effects of miR-181a-2-3p mimic on LPS-induced HK2. (A) Western blotting and qRT-PCR were used to detect GJB2 expression in siRNA-GJB2-transfected HK2 cells. (B) Western blotting and qRT-PCR were used to detect miR-181a-2-3p expression in miR-181a-2-3p inhibitor-transfected HK2 cells. Cotransfection of miR-181a-2-3p inhibitors and siRNA-GJB2 treatment with LPS-induced HK2, cell viability (C), cell apoptosis (D), the levels of IL-1β, IL-6, and TNF-α (E), and the expression of Bax and Bcl-2 (F) were detected using CCK-8 assay, flow cytometry analysis, and qRT-PCR. **, P < 0.01; ##, P < 0.01; ^^, P < 0.01; &&, P < 0.01.
DISCUSSION
An increasing number of studies have indicated that miRNAs play vital roles in inflammatory disease. It is reported that miR-181a-2-3p silencing aggravates the COPD-induced inflammatory responses and cell apoptosis (11, 12). Similarly, in this study, we tested the miR-181a-2-3p expression in CLP mouse models and found that miR-181a-2-3p expression was reduced. In addition, we carried out H&E staining to evaluate kidney injuries and discovered that there was obvious inflammatory cell infiltration and severe vacuolated degradation of RTECs; while after transfection with agomir-181a-2-3p, the tissue condition was markedly improved. Cr and BUN, the important indicators to judge kidney injury, were highly expressed in sepsis-induced AKI (13). Meanwhile, NGAL and KIM-1, the vital biomarkers for AKI, were also highly expressed in the early stage of AKI. Therefore, they were used to assess the kidney function in CLP mouse models (14). Further, the higher IL-1β, IL-6, and TNF-α levels and greater caspase-3 activity were observed in sepsis mouse models, but their levels were significantly reduced by the addition of agomir-181a-2-3p, suggesting that agomir-181a-2-3p mimic markedly alleviated the sepsis-induced kidney injury and inflammatory response. Afterwards, this study discovered that miR-181a-2-3p mimic reduced the LPS-induced apoptosis and inflammatory response in kidney tissues. The above experimental results proved that miR-181a-2-3p played an important role in the sepsis-induced AKI animal model, but its role in human RTECs should be further verified.
Therefore, the LPS-induced HK2 cells were used for further verification (15), and the effects of miR-181a-2-3p mimic on cell apoptosis and inflammatory response were consistent with those observed in the CLP mouse models. Therefore, it was concluded in this study that overexpression of miR-181a-2-3p had anti-inflammatory and antiapoptotic effects in sepsis. To explore the mechanism of miR-181a-2-3p in mediating inflammatory response and cell apoptosis in sepsis, bioinformatics analysis was conducted, which identified a target gene named GJB2 in the GSE46955 data set. Interestingly, GJB2 was highly expressed in septic patients but lowly expressed in recovering patients, suggesting that GJB2 was an important target gene of miR-181a-2-3p in the development of sepsis. In addition, the high expression of GJB2 predicts dismal prognosis (16). miR-27a regulates the apoptosis of cochlear sensory epithelium via GJB2 (17). On the other hand, caspase cascade and the Bcl-2 families play important roles in the process of apoptosis (18, 19), while the antiapoptotic protein Bcl-2 suppresses apoptosis via the mitochondrion-dependent caspase-9 pathway (20). Furthermore, miR-513a-5p is reported to decrease Bcl-2 expression and promotes the dichlorvos-induced apoptosis of HK2 cells via the Bcl-2/Bax–caspase-3 pathway (21). Therefore, caspase-3/9 and bax/bcl-2 can serve as the most powerful markers to reflect apoptosis. In this study, LPS significantly increased the expression of cleaved caspase-3/9 and bax but inhibited that of bcl-2. Meanwhile, overexpression of miR-181a-2-3p reversed the above effect on the LPS-induced HK2 cells. However, overexpression of GJB2 restored the effect of LPS on HK2 cells by counteracting the functions of miR-181a-2-3p mimic in LPS-induced HK2 cells, thereby upregulating the expression of Bax and inhibiting that of Bcl-2. These results proved that miR-181a-2-3p was involved in the LPS-induced cell apoptosis by targeting GJB2. Consequently, GJB2 was verified as an important regulator of cells whose expression was related to cell apoptosis, inflammatory response, and organ injury degree in sepsis.
Therefore, we investigated whether miR-181a-2-3p played a role in the specific regulation of GJB2 expression, inflammatory cytokine release, and HK2 cell apoptosis. Results of Western blotting showed that miR-181a-2-3p mimic promoted the expression of GJB2 but inhibited that of inflammatory cytokines and cell apoptosis of LPS-induced HK2 cells. Moreover, GJB2 overexpression reversed these effects, indicating that miR-181a-2-3p upregulation might be related to the downregulation of GJB2 and the occurrence of sepsis.
Conclusion.
Collectively, our results concluded that the miR-181a-2-3p expression was downregulated in sepsis, which regulated the sepsis-induced injury of RTECs through targeting GJB2. Our study strongly supported that miR-181a-2-3p functioned as a marker in sepsis, providing a therapeutic target for the early detection of sepsis.
MATERIALS AND METHODS
Construction of cecal ligation and puncture (CLP) mouse models and transfection.
The C57BL/6J male mice weighing 20 to 25 g that were used in this study were provided by the Medical Laboratory Animal Center (Guangdong, China). All mice were randomly divided into 10 groups (n = 5). After mice were anesthetized with pentobarbital, the peritoneum was opened to expose the cecum. Thereafter, the middle part of the cecum was ligated with a 5.0-mm silk thread, and the ipsilateral cecum was perforated twice with a 20-gauge needle. Then, the cecum was returned to the abdominal cavity and the peritoneum was sutured. Mice in sham operation group underwent the same procedure except for ligation and perforation. In addition, subcutaneous injection of Carprofen at a dose of 5 mg/kg (MedChemExpress, New Jersey, USA) was adopted for pain control after the CLP procedure.
After surgery, in vivo transfection was performed using the Entranster-in vivo reagent (Engreen Biosystem, Ltd., Beijing, China). In brief, 100 μl Entranster-in vivo reagent, 50 μg miR-181a-2-3p antagomir, or miR-181a-2-3p agomir and 30 μg siRNA-GJB2 or vector expressing GJB2 were injected into mice via the tail vein. After 24 h, mice were anesthetized with 2% methoxyflurane and sacrificed. Blood samples were collected from the heart, and the kidney tissues were harvested from the mice for follow-up tests. All animal experiments were approved by the Animal Research Committee of Gannan Medical University and performed in accordance with the guidelines of the National Animal Protection and Ethics Institute.
Hematoxylin and eosin staining.
Mice were sacrificed at 24 h post the establishment of CLP mice models. The kidney tissues were extracted and fixed with 10% formalin for 24 h. After dehydration, all tissues were sectioned in paraffin and stained with hematoxylin and eosin. Cell morphology in tissue is observed and photographed under an optical microscope (magnification, ×200).
Enzyme-linked immunosorbent assay.
Blood was collected from the eyeball of the mice and coagulated for 30 min at 37°C. The serum was obtained after centrifugation at 3,000 rpm for 15 min. The levels of IL-1β, IL-6, TNF-α (BioLegend, USA), creatinine (Cr), blood urea nitrogen (BUN) (Jiancheng, Nanjing, China), neutrophil gelatinase-associated lipocalin (NGAL), and kidney injury molecule 1 (KIM-1) (Beyotime, Shanghai, China) were detected by ELISA kit according to the manufacturer’s instruction.
Cell culture and constructing sepsis cell models.
TCMK-1 and HK2 cells were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technology, Wuhan, China) containing 10% fetal bovine serum (FBS; Life Technology, Wuhan, China) and 1% penicillin/streptomycin (Life Technology, Wuhan, China), in 37°C, 5% CO2 incubator. When the cells fused 80% to 90%, the cells were subcultured. The new medium was replaced the next day after subculture, and the medium was replaced every 2 to 3 days after that. Then, LPS (50 or 100 μg/ml) was added into TCMK-1 and HK2 cells for 24 h to induce apoptosis.
Flow cytometry.
The cell apoptosis was determined via fluorescein isothiocyanate (FITC) annexin V and propidium iodide (PI) double staining kit (Invitrogen, Carlsbad, USA). After different treatments, cells were collected and resuspended and then incubated with 5 μl of annexin V and 5 μl of PI. After incubation for 20 min in the dark at 37°C, the cells were stained for 15 min and flow cytometer (BD Biosciences, San Jose, CA, USA) was used to detect apoptosis. The final test data were analyzed using Flowjo software (Flowjo, Ashland, OR, USA).
Cell viability assay.
Cell counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan) was used to detect the cell viability according to its instructions. Briefly, cells (2 × 104 per well) were added in 96-well plate and cultured for 24 h. Following transfection and 24 h of incubation, CCK-8 solution was added into each well and incubated for another 3 h. The colorimetric analysis at 450 nm was measured using a spectrophotometer (Multiskan MK3; Thermo Fisher Scientific).
Cell transfection.
miR-181a-2-3p mimic or miR-181a-2-3p inhibitor and the respective control (miNC or inNC), small interfering RNA (siRNA) against-GJB2 (si-GJB2), or GJB2 expression pcDNA3.1 vector and its control (si-NC/NC) were purchased from GenePharma Co., Ltd. (Shanghai, China). These oligonucleotides were then transfected into cells by using the Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. At 24 h later, cells were harvested for further experiments.
RNA isolation and quantitative real-time PCR.
The blood samples were collected in a centrifuge tube containing the separation gel and clot activator. The samples were centrifuged at 3,000 rpm for 15 min at 37°C, and the supernatant was collected in a new centrifuge tube for another centrifugation at 15,000 rpm for 30 min. The collected supernatant was stored at −80°C. Blood total RNA isolation kit (BioTeke Corporation, Beijing, China) was used to isolate mRNA in order to detect miR-181a-2-3p expression or GJB2 expression in mouse.
Total RNA was extracted from cells and kidney tissues using TRIzol reagent (ThermoFisher, MA, USA). Extracted RNA was used to synthesize complementary DNA (cDNA) using a cDNA reverse transcription kit (Thermo Fisher Scientific). The expression levels of mRNAs were measured using a SYBR green master mix kit (Thermo Fisher Scientific). TaqMan microRNA RT kit (Thermo Fisher Scientific) was used to measure the miR-181a-2-3p level with U6 small nuclear RNA (U6-snRNA) as the internal control. The relative expression levels of GJB2, IL-1β, IL-6, TNF-α, Bax, and Bcl-2 were calculated using the 2−ΔΔCt method with GAPDH as the internal control. The special primers for miR-181a-2-3p or U6 were purchased from GeneCopoeia (Rockville, MD, USA), and primers for miR-181a-2-3p, U6, GJB2, IL-1β, IL-6, TNF-α, Bax, Bcl-2, and GAPDH are listed in Table 1.
TABLE 1.
Primer sequences used for quantitative RT-PCR
| cDNA | Primer directiona | Sequence |
|---|---|---|
| mmu-miR-181a-2-3p | F | 5′-GCTAATGAACGATCGGCCGG-3′ |
| R | 5′-ACAGAGCGATTAAGCCTCAC-3′ | |
| mmu-GJB2 | F | 5′-CAACCGCCATCCTTGAAGT-3′ |
| R | 5′-TGCGTTGATCACCTGATC-3′ | |
| mmu-IL-1β | F | 5′-ATTCTGTTCCGAGGCCCTG-3′ |
| R | 5′-AAGCTTCGGCAGTGTGC-3′ | |
| mmu-IL-6 | F | 5′-CTTAAATGGCTTCTATCGGC-3′ |
| R | 5′-TGGTGAGGGGAAACCTTTATCTT-3′ | |
| mmu-TNF-α | F | 5′-GAAGTGCATCCTTCCGTCTTCCG-3′ |
| R | 5′-ACTTCTTGTGGGCCCGAGCTCT-3′ | |
| mmu-Bax | F | 5′-TGCGAACGTGCCTGAA-3′ |
| R | 5′-AACGCGCTGCTGTGTCCCG-3′ | |
| mmu-Bcl-2 | F | 5′-ACTGCCCTGCACAAACTTTCGCGC-3′ |
| R | 5′-TGCCTGACACAACTTGTGGCTGAA-3′ | |
| mmu-GAPDH | F | 5′-TTTGGAGGGGATTTCTCATCTCG-3′ |
| R | 5′-TCAAGAGTTGGTCGTATCGGATTG-3′ | |
| mmu-U6 | F | 5′-GGCGCTTATATCAGCATTCGCAC-3′ |
| R | 5′-CATCCTGCCTTGATGTTCGGAGAG-3′ | |
| hsa-miR-181a-2-3p | F | 5′-ACTGCTGCTGTGTTGATGATGCTT-3′ |
| R | 5′-CCTGTCGTGTGCCAAAAGT-3′ | |
| hsa-GJB2 | F | 5′-CGTGTGTCACAGTGTCCCACACAA-3′ |
| R | 5′-CTGTAAAACATGACTGCATGCATGAA-3′ | |
| hsa-IL-1β | F | 5′-CTGTCATGTGCATGTCA-3′ |
| R | 5′-TGTCGTGTGTCATGTCATGAAACTCC-3′ | |
| hsa-IL-6 | F | 5′-CTGTGTCGTGCTCTCTCTCGGTCAA-3′ |
| R | 5′-CTGTACAATGAAACATAA-3′ | |
| hsa-TNF-α | F | 5′-AACATGACTGAACTGACA-3′ |
| R | 5′-CTGTAGTGCTACCATGCTGAAC-3′ | |
| hsa-Bax | F | 5′-CTGTAGTGACATGCAAATGA-3′ |
| R | 5′-GTGTGTACCTGCATGTGACAA-3′ | |
| hsa-Bcl-2 | F | 5′-ACGGCTGCCGTGACTGTGA-3′ |
| R | 5′-CCTGTGTAGTCATGTGATGA-3′ | |
| hsa-GAPDH | F | 5′-TCGTGTGATGTGTAGTAGATAAATGA-3′ |
| R | 5′-CTGCCTGTAGTGATGATGATAGTT-3′ | |
| hsa-U6 | F | 5′-GGGTGATCTGATCATGATA-3′ |
| R | 5′-GCTGACATGATGAAAATGATAAAA-3′ |
F, forward; R, reverse.
Dual-luciferase reporter assay.
Luciferase reporter system was performed using dual-luciferase reporter assay kit (Promega, Madison, WI, USA). HEK293 cells were transfected with GJB2-WT and GJB2-MUT reporters (Promega) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Luciferase activities were measured by dual-luciferase reporter assay system protocol (Promega, Madison, USA) at 48 h posttransfection.
Caspase-3 activity assay.
Protein from kidney tissues was used to determine caspase-3 activity. The activity of relative caspase-3 in kidney was conducted by using the caspase-3 activity kit (Bestbio, China) according to the manufacturer’s instruction.
RNA immunoprecipitation assay.
After transfection with miR-133a or miR-NC for 48 h, RIP assay was conducted by adopting the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) following the manufacturer’s protocols. In brief, cells were lysed with the RIP buffer containing magnetic beads and incubated with anti-IgG (Millipore) or anti-argonaute2 (Ago2, Millipore) antibody bound to the magnetic beads. In addition, the immunoprecipitated RNA was isolated by using the TRIzol reagent (Thermo Fisher Scientific), and SIRT1 enrichment in the IgG or Ago2 immunoprecipitated complex was detected by qRT-PCR.
Western blotting.
Cells were lysed using RIPA lysis buffer (Beyotime, Shanghai, China) with 1 mmol/liter phenylmethylsulfonyl fluoride (PMSF). The total protein was quantified using bicinchoninic acid (BCA) protein assay kit (Beyotime) according to the manufacturer’s instructions. Total proteins were separated using SDS-PAGE gel, transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA), and blocked with 5% nonfat milk for 1 h at 37°C. Subsequently, the membranes were incubated with the primary antibodies against Bax (1:1,000; Beyotime), Bcl-2 (1:600; Beyotime), cleaved casapase-3 (1:1,000; Abcam), cleaved caspase-9 (1:800; Abcam), and GAPDH (1:2,000; Beyotime) overnight at 4°C and then incubated with secondary antibody labeled with horseradish peroxidase (HRP; 1:1,000; Abcam) for 1 h at 37°C. The membranes were then washed with Tris-buffered saline with Tween 20 (TBST) buffer 4 times and treated with western-light substrate for 5 min. Protein signals were detected by Image Lab software (Bio-Rad, Hercules, CA, USA).
Data collection and study design.
The GSE46955 microarray data set, including 12 healthy control samples, 16 sepsis samples, and 16 recovery samples, was downloaded from the Gene Expression Omnibus (GEO) database of the NCBI database (https://www.ncbi.nlm.nih.gov/). The data sets were performed on the Illumina humanRef-8 v2.0 platform, and the former was used to screen the differentially expressed genes (DEGs). Additionally, we screened DEGs between 12 healthy control samples and 16 sepsis samples, as well as between 16 sepsis samples and 16 recovery samples.
Data preprocessing and DEGs screening.
All the raw expression profiling data were processed by quality control, background correction, normalization, logarithmic conversion, and batch effect removal using the “limma” function in R package (22). The “limma” function in R package was used to screen DEGs from the GSE46955 data set. Typically, DEGs were screened upon the thresholds of false discovery rate (FDR) of <0.05 and absolute value of log2 fold change (FC) of ≥1.
Statistical analysis.
The SPSS 17.0 software (SPSS, Chicago, IL, USA) was employed for statistical analysis. All quantitative data are presented as the mean ± standard deviation (SD). Differences between two groups were compared using Student’s t test when they had a normal distribution. A one-way analysis of variance (ANOVA) was used to compare data among groups when they had a normal distribution and homogeneous variances, followed by Tukey’s post hoc test. A difference of P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We declare that we have no financial conflicts of interest.
This work was supported by National Natural Science Foundation of China (81701933).
REFERENCES
- 1.Rello J, Valenzuela-Sánchez F, Ruiz-Rodriguez M, Moyano S. 2017. Sepsis: a review of advances in management. Adv Ther 34:2393–2411. 10.1007/s12325-017-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peerapornratana S, Manrique-Caballero CL, Gómez H, Kellum JA. 2019. Acute kidney injury from sepsis: current concepts, epidemiology, pathophysiology, prevention and treatment. Kidney Int 96:1083–1099. 10.1016/j.kint.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao Y, Sun F, Lei M. 2018. miR-25 inhibits sepsis-induced cardiomyocyte apoptosis by targetting PTEN. Biosci Rep 38:BSR20171511. 10.1042/BSR20171511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie D, Xu Y, Jing W, Juxiang Z, Hailun L, Yu H, Zheng DH, Lin YT. 2017. Berberine nanoparticles protects tubular epithelial cells from renal ischemia-reperfusion injury. Oncotarget 8:24154–24162. 10.18632/oncotarget.16530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. 2019. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci 20:6249. 10.3390/ijms20246249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funahashi Y, Kato N, Masuda T, Nishio F, Kitai H, Ishimoto T, Kosugi T, Tsuboi N, Matsuda N, Maruyama S, Kadomatsu K. 2019. miR-146a targeted to splenic macrophages prevents sepsis-induced multiple organ injury. Lab Invest 99:1130–1142. 10.1038/s41374-019-0190-4. [DOI] [PubMed] [Google Scholar]
- 7.Zhou YL, Yang SH, Zhang C, Zhang B, Yang XJ. 2018. LncRNA MALAT1 modulates the immunoreaction of rats with lipopolysaccharide-induced sepsis by targeting the miR-146a/NF-κB P65 pathway. Sichuan Da Xue Xue Bao Yi Xue Ban 49:865–870. [PubMed] [Google Scholar]
- 8.Lu S, Wu H, Xu J, He Z, Li H, Ning C. 2020. SIKIAT1/miR-96/FOXA1 axis regulates sepsis-induced kidney injury through induction of apoptosis. Inflamm Res 69:645–656. 10.1007/s00011-020-01350-0. [DOI] [PubMed] [Google Scholar]
- 9.Klein JD, Wang XH. 2018. Electrically stimulated acupuncture increases renal blood flow through exosome-carried miR-181. Am J Physiol Renal Physiol 315:F1542–F1549. 10.1152/ajprenal.00259.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Yang Z, Heng Y, Miao C. 2019. MicroRNA-181 exerts an inhibitory role during renal fibrosis by targeting early growth response factor-1 and attenuating the expression of profibrotic markers. Mol Med Rep 19:3305–3313. 10.3892/mmr.2019.9964. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Kim DY, Heo HR, Choi SS, Hong SH, Kim WJ. 2019. Role of miRNA-181a-2-3p in cadmium-induced inflammatory responses of human bronchial epithelial cells. J Thorac Dis 11:3055–3069. 10.21037/jtd.2019.07.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan S, Ren Y, Zhang W, Zhang H, Wang C. 2021. Long non-coding maternally expressed gene 3 regulates cigarette smoke extract-induced apoptosis, inflammation and cytotoxicity by sponging miR-181a-2-3p in 16HBE cells. Oncol Lett 21:45. 10.3892/ol.2020.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Sun A, Meng T, Zhu Y. 2021. Expression and role of ABIN1 in sepsis: in vitro and in vivo studies. Open Med (Wars) 16:33–40. 10.1515/med-2021-0008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Seibert FS, Sitz M, Passfall J, Haesner M, Laschinski P, Buhl M, Bauer F, Babel N, Pagonas N, Westhoff TH. 2018. Prognostic value of urinary calprotectin, NGAL and KIM-1 in chronic kidney disease. Kidney Blood Press Res 43:1255–1262. 10.1159/000492407. [DOI] [PubMed] [Google Scholar]
- 15.Zheng C, Zhou Y, Huang Y, Chen B, Wu M, Xie Y, Chen X, Sun M, Liu Y, Chen C, Pan J. 2019. Effect of ATM on inflammatory response and autophagy in renal tubular epithelial cells in LPS-induced septic AKI. Exp Ther Med 18:4707–4717. 10.3892/etm.2019.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Zhang YJ, Wu ZH. 2020. High GJB2 mRNA expression and its prognostic significance in lung adenocarcinoma: a study based on the TCGA database. Medicine (Baltimore, MD) 99:e19054. 10.1097/MD.0000000000019054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Lin C, He Y, Li A, Ni W, Sun S, Gu X, Li J, Li H. 2016. Mir-27a promotes apoptosis of cochlear sensory epithelium in Cx26 knockout mice. Front Biosci (Landmark Ed) 21:364–373. 10.2741/4393. [DOI] [PubMed] [Google Scholar]
- 18.Fan TJ, Han LH, Cong RS, Liang J. 2005. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai) 37:719–727. 10.1111/j.1745-7270.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 19.Kvansakul M, Hinds MG. 2015. The Bcl-2 family: structures, interactions and targets for drug discovery. Apoptosis 20:136–150. 10.1007/s10495-014-1051-7. [DOI] [PubMed] [Google Scholar]
- 20.Sharifi S, Barar J, Hejazi MS, Samadi N. 2014. Roles of the Bcl-2/Bax ratio, caspase-8 and 9 in resistance of breast cancer cells to paclitaxel. Asian Pac J Cancer Prev 15:8617–8622. 10.7314/APJCP.2014.15.20.8617. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Xu YN, Niu X, Li Z, Wang JF. 2018. miR-513a-5p targets Bcl-2 to promote dichlorvos induced apoptosis in HK-2 cells. Biomed Pharmacother 108:876–882. 10.1016/j.biopha.2018.09.101. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]