ABSTRACT
Prions are self-perpetuating, alternative protein conformations associated with neurological diseases and normal cellular functions. Saccharomyces cerevisiae contains many endogenous prions, providing a powerful system to study prionization. Previously, we demonstrated that Swi1, a component of the SWI/SNF chromatin-remodeling complex, can form the prion [SWI+]. A small region, Swi11–38, with a unique amino acid composition of low complexity, acts as a prion domain and supports [SWI+] propagation. Here, we further examine Swi11–38 through site-directed mutagenesis. We found that mutations of the two phenylalanine residues or the threonine tract inhibit Swi11–38 aggregation. In addition, mutating both phenylalanines can abolish de novo prion formation by Swi11–38, whereas mutating only one phenylalanine does not. Replacement of half of or the entire eight-threonine tract with alanines has the same effect, possibly disrupting a core region of Swi11–38 aggregates. We also show that Swi11–38 and its prion-fold-maintaining mutants form high-molecular-weight, SDS-resistant aggregates, whereas the double-phenylalanine mutants eliminate these protein species. These results indicate the necessity of the large hydrophobic residues and threonine tract in Swi11–38 in prionogenesis, possibly acting as important aggregable regions. Our findings thus highlight the importance of specific amino acid residues in the Swi1 prion domain in prion formation and maintenance.
KEYWORDS: protein aggregation, prionogenesis, Swi1, prion domain, [SWI+], SWI/SNF, yeast, Saccharomyces cerevisiae
INTRODUCTION
Prions were initially identified as infectious abnormal protein conformations that underpin incurable neurological diseases (1). While the prion concept originally applied to the namesake protein, the idea has grown to encompass additional proteins in a multitude of organisms (2–6). The budding yeast Saccharomyces cerevisiae harbors a number of endogenous proteins that can adopt alternative, heritable protein conformations (7–15). These proteins, termed yeast prions, have greatly contributed to our understanding of the prion phenomena.
One such yeast prion, [SWI+], was identified by our laboratory (11). The protein determinant of [SWI+], Swi1, normally functions as part of the SWI/SNF chromatin-remodeling complex, which modulates the expression of more than 15% of yeast genes (16, 17). Due in part to this role, the prionization of Swi1 leads to multiple phenotypes in yeast, including poor growth on nonglucose carbon sources (e.g., raffinose and glycerol), aggregation of the Swi1 protein, and loss of multicellular features (e.g., flocculation and invasive growth) (11, 18). Swi1 can be divided into three domains (19). The N-terminal, asparagine-rich domain (Swi1N) contains the Swi1 prion domain (PrD), the region necessary and sufficient for prionization. The N region has previously been shown to capably form amyloid fibrils in vitro (19). A middle glutamine-rich domain follows, and a C-terminal, functional domain completes the protein. The expression of this functional domain rescues the poor growth on raffinose phenotype and restores multicellular features (19).
Further research into the Swi1 PrD revealed that the protein’s first 38 amino acids (Swi11–38) could act to maintain and propagate the [SWI+] prion fold (20). Also, deletion of a similarly sized region (residues 2 to 55) from Swi1N prevented coaggregation in yeast containing [SWI+], indicating the critical nature of this region. This extreme N-terminal region is uniquely rich in asparagine residues and devoid of any glutamine residues. Moreover, Swi11–38 could be further truncated, down to Swi11–32, and retain the ability to aggregate and propagate (21). Swi11–38 was also shown to act as a transferable PrD. When fused with Sup35MC, the Sup35 protein without its N-terminal prion domain, for assay purposes, Swi11–38 can de novo form a prion that has been termed [SPS+] (Swi1-conferred [PSI+]). This prion, formed by Swi11–38-MC, exhibits aggregation when visualized by Swi11–38-yellow fluorescent protein (YFP), displays impaired translation termination due to the primary function of Sup35MC, and is curable by treatment with guanidine hydrochloride. Once again, a shorter truncation, Swi11–31, was found to also be capable of prionization. In all, this small N-terminal region of Swi1 stands as the smallest currently identified PrD.
Swi11–38 is highly enriched in polar residues, particularly asparagine. Of the 38 residues, 22 are asparagine and 10 are threonine residues. The inclusion of asparagine and/or glutamine residues is common among currently characterized yeast prions (22, 23). Meanwhile, the six remaining residues comprise a methionine necessary as a start codon, an adjacent aspartate likely playing a role in the protein half-life, an ending proline with probable unimportance for prion capabilities, and three hydrophobic residues: a leucine and two phenylalanines. This largely uncomplicated primary sequence of Swi11–38 gives rise to a protein domain capable of aggregating, maintaining, and propagating an alternative fold and initializing a prion (20, 21). Thus, Swi11–38 exists as a small prion domain and acts as a critical region for supporting [SWI+]. As such, investigation of Swi11–38 may allow the clarification of the prion capabilities of the larger Swi1 protein, which plays an important role in the global regulation of yeast genes and the resulting environmental adaptation (16). To better understand the prionogenicity of Swi11–38, we performed a series of mutagenesis experiments to characterize the contributions of residues to the prionogenic characteristics of this small PrD.
RESULTS
Multiple Swi11–38 mutants cannot coaggregate with Swi1FL in [SWI+] cells.
To dissect the contributions of various residues to the prionogenicity of Swi11–38, we targeted the minority of residues that are nonasparagine amino acids for mutagenesis (Fig. 1A). The first two amino acid residues, methionine and aspartate, were not mutated due to the need for the start codon and the N-end rule, respectively (24). The last amino acid residue, proline, was also not mutated due to our laboratory’s previous work displaying that this residue is not necessary for aggregation, the maintenance of [SWI+], or prionogenesis (21). Additionally, proline is not known to be particularly aggregation or prion promoting.
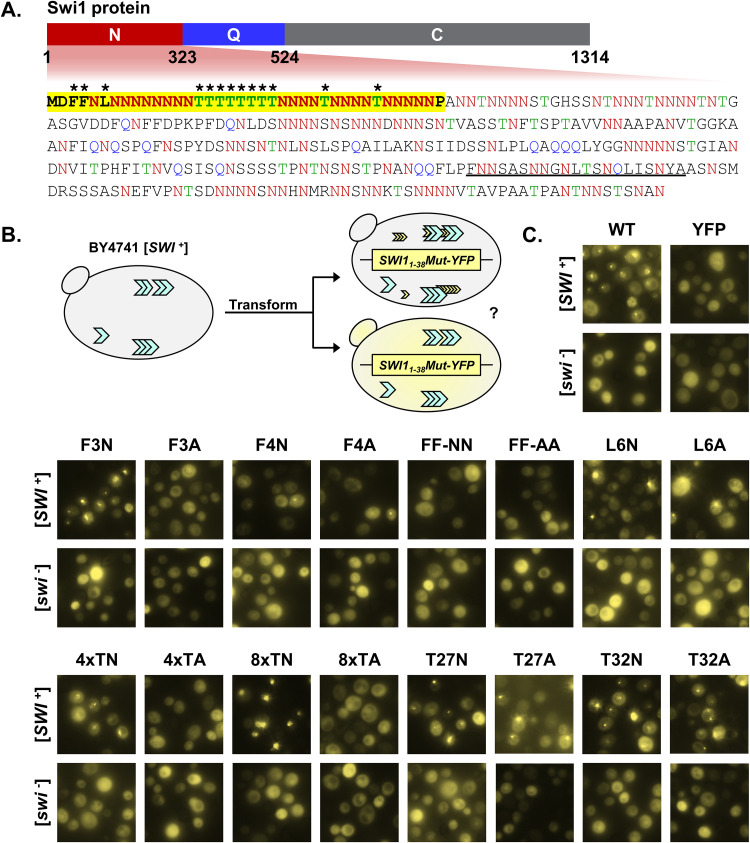
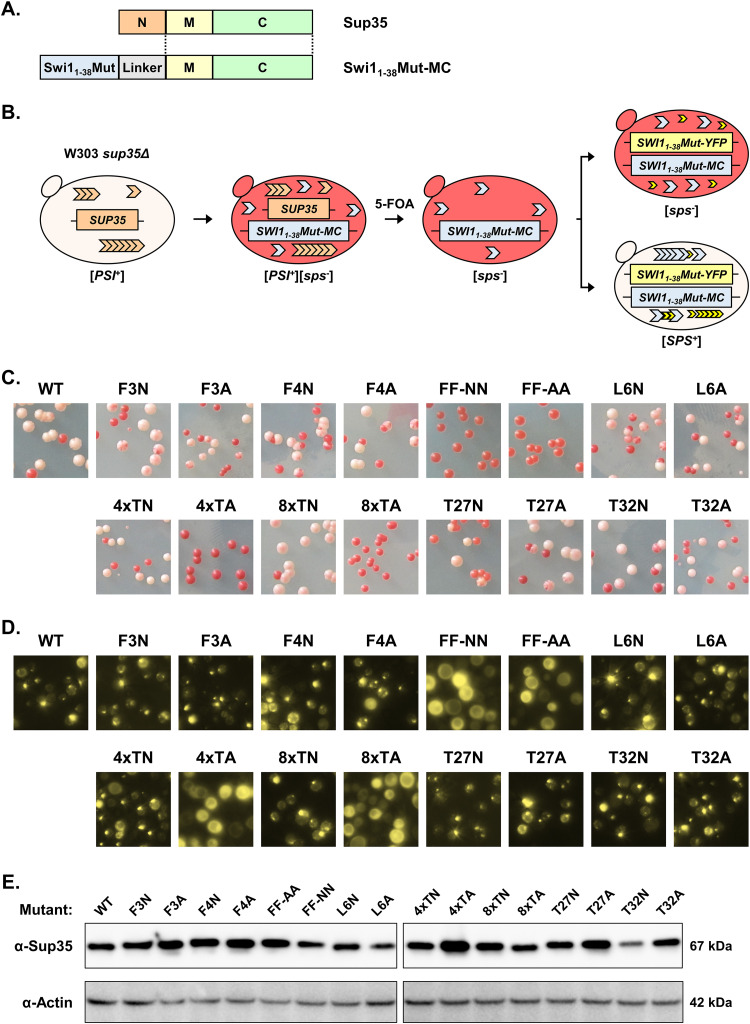
FIG 1.
Mutation of the phenylalanine residues or threonine tract disrupts Swi11–38 coaggregation with Swi1FL. (A) Diagram of Swi1 protein domains. The N region (Swi11–323) includes the Swi1 prion domain, and the sequence of this region is presented. The amino acid residues asparagine (N), glutamine (Q), and threonine (T) are in red, blue, and green, respectively. The first 38 amino acid residues (Swi11–38) are in boldface type and highlighted in yellow. A previously predicted amyloid core region (Swi1239–259) is underlined. Asterisks indicate those residues that were targeted for mutagenesis. (B) Diagram of the experiment. BY4741 [SWI+] or [swi−] cells were transformed with p415TEF-SWI11–38-YFP (WT), p415TEF-SWI11–38Mut-YFP, or p415TEF-YFP. Transformants were observed using fluorescence microscopy for aggregate foci or diffuse signals. (C) Fluorescence images of BY4741 [SWI+] or [swi−] cells transformed with p415TEF-SWI11–38-YFP (WT), p415TEF-SWI11–38Mut-YFP, or p415TEF-YFP. See Table 1 for amino acid sequences of the mutants. For each construct, 3 independent transformations of both [SWI+] and [swi−] yeast cells were conducted. On average, approximately 900 cells (from across 3 colonies) were observed per transformation. Shown are representative views, and quantitative results are shown in Fig. 2C.
The remaining nonasparagine residues, including phenylalanine (F), threonine (T), and leucine (L), in Swi11–38 were mutagenized. Noticeably, these amino acids are not overrepresented in the asparagine/glutamine-rich PrDs of identified yeast prions, although some of them have been reported as amyloidogenic (23, 25). Codons for individual amino acids were swapped via PCR mutagenesis to codons for either asparagine (N) or alanine (A) (Table 1). Asparagine was selected due to its importance in prionogenicity and the fact that Swi11–38 is already very N rich: small changes to the number of N residues are unlikely to have a sizeable effect (23). The replacement of the phenylalanine(s) in Swi11–38 with the polar, uncharged asparagine allows us to determine the value of the singular and/or duplicated phenylalanine(s) and the prionogenic hydrophobicity provided by it. Furthermore, replacing threonine residues allows us to evaluate whether Swi11–38 requires unique contributions of threonine or its tandem tract. On the other hand, alanine was selected due to its lack of prionogenicity and its simple and small structure, particularly compared to amino acids such as phenylalanine with a large aromatic side chain. The phenylalanine residues at positions 3 and 4 were mutated singularly or in tandem, producing the mutants F3N, F3A, F4N, F4A, FF-NN, and FF-AA. The leucine residue at position 6 was mutated singularly to construct the mutants L6N and L6A. For the threonine tract in the center of Swi11–38, the last 4 threonine residues (positions 19 to 22) were replaced with either all asparagine or all alanine residues to produce 4×TN or 4×TA, respectively. The entire threonine tract (positions 15 to 22) was mutated to be either entirely asparagine residues (8×TN) or entirely alanine residues (8×TA). The interspersed threonine residues in the back portion of Swi11–38 were singularly mutated to create T27N, T27A, T32N, and T32A. Together with the wild-type (WT) Swi11–38 construct, these mutants were initially assayed for their ability to coaggregate with full-length Swi1 (Swi1FL).
TABLE 1.
Swi11–38 mutants
| Name | DNA mutation | Amino acid sequencea |
|---|---|---|
| WT | MDFFNLNNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP | |
| F3N | TTC → AAC | MDNFNLNNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP |
| F3A | TTC → GCC | MDAFNLNNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP |
| F4N | TTT → AAC | MDFNNLNNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP |
| F4A | TTT → GCC | MDFANLNNNN NNNNTTTTTT TTNNNNTNNNNTNNNNNP |
| FF-NN | TTCTTT → AACAAC | MDNNNLNNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP |
| FF-AA | TTCTTT → GCCGCC | MDAANLNNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP |
| L6N | TTG → AAC | MDFFNNNNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP |
| L6A | TTG → GCG | MDFFNANNNN NNNNTTTTTT TTNNNNTNNN NTNNNNNP |
| 4×TN | ACTACTACTACC → AACAACAACAAC | MDFFNLNNNN NNNNTTTTNN NNNNNNTNNN NTNNNNNP |
| 4×TA | ACTACTACTACC → GCAGCAGCAGCA | MDFFNLNNNN NNNNTTTTAA AANNNNTNNN NTNNNNNP |
| 8×TN | ACTACTACTACTACTACTACTAAC → AACAACAACAACAACAACAACAAC | MDFFNLNNNN NNNNNNNNNN NNNNNNTNNN NTNNNNNP |
| 8×TA | ACTACTACTACTACTACTACTAAC → GCAGCAGCAGCAGCAGCAGCAGCA | MDFFNLNNNN NNNNAAAAAA AANNNNTNNN NTNNNNNP |
| T27N | ACT → AAT | MDFFNLNNNN NNNNTTTTTT TTNNNNNNNN NTNNNNNP |
| T27A | ACT → GCT | MDFFNLNNNN NNNNTTTTTT TTNNNNANNN NTNNNNNP |
| T32N | ACT → AAC | MDFFNLNNNN NNNNTTTTTT TTNNNNTNNN NNNNNNNP |
| T32A | ACT → GCT | MDFFNLNNNN NNNNTTTTTT TTNNNNTNNN NANNNNNP |
Boldface type indicates mutated amino acid residues that differ from the wild-type sequence.
Each mutant was tagged with yellow fluorescent protein (YFP) and individually transformed into BY4741 [SWI+] and [swi−] yeast (Fig. 1B). This process was repeated for three biological replicates. Wild-type Swi11–38-YFP served as a positive control, specifically aggregating in [SWI+] cells, while YFP alone served as a negative control. While F3N displayed aggregation similar to that of the WT, the other phenylalanine mutants displayed greatly hampered aggregation formation (Fig. 1C). Indeed, the FF-NN and FF-AA constructs were not observed to have any puncta visible. The other mutant constructs that resulted in deficient aggregation were 4×TA and 8×TA; however, 4×TN and 8×TN displayed aggregation akin to that of the WT. Thus, the replacement of these threonine residues with alanine removed the polar side groups that are aggregation prone and greatly disrupted aggregation. On the other hand, maintaining that polarity via mutation to the similarly polar asparagine allowed aggregation. The remaining mutations (L6N, L6A, T27N, T27A, T32N, and T32A) had aggregation similar to that of the WT. All constructs did not produce observable aggregates in [swi−] cells, indicating that the observed aggregation was specific to mutant Swi11–38-YFP (Swi11–38Mut-YFP) adopting the prion fold of the existing [SWI+] and not amorphous aggregates forming solely due to overexpression.
Overexpression of Swi1FL allows coaggregation of additional Swi11–38 mutants.
To further examine the aggregation capabilities of the Swi11–38 mutant constructs, we performed the same coaggregation assay in the presence of higher Swi1FL expression levels (Fig. 2A). We started with BY4741 swi1Δ/p416TEF-SWI1FL [SWI+] and [swi−] yeast for this experiment. In these cells, instead of expressing SWI1FL from its chromosomal locus under the control of its endogenous promoter, SWI1FL was expressed from a plasmid under the control of the significantly stronger TEF promoter. These conditions should provide an overexpression context for Swi1FL and an additional opportunity for the Swi11–38 mutants to decorate the existing [SWI+] aggregates.
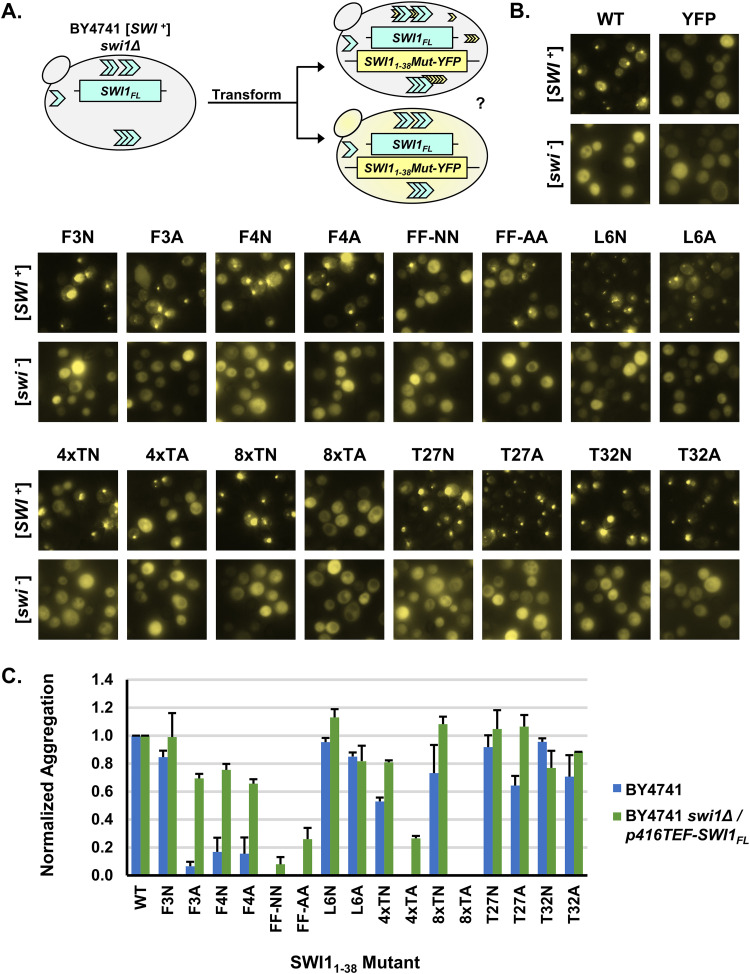
FIG 2.
Substantially higher Swi1FL expression levels promote aggregation of Swi11–38 mutants in [SWI+] cells. (A) Diagram of the experiment. BY4741 swi1Δ/p416TEF-SWI1FL [SWI+] or [swi−] cells were transformed with p415TEF-SWI11–38Mut-YFP. Transformants were observed using fluorescence microscopy for aggregate foci or diffuse signals. (B) Fluorescence images of BY4741 swi1Δ/p416TEF-SWI1FL [SWI+] or [swi−] cells transformed with p415TEF-SWI11–38-YFP (WT), p415TEF-SWI11–38Mut-YFP, or p415TEF-YFP. See Table 1 for amino acid sequences of the mutants. Similar to that described in the legend of Fig. 1C, for each construct, 3 independent transformations of both [SWI+] and [swi−] yeast cells were conducted. On average, approximately 900 cells (from across 3 colonies) were observed per transformation. Shown are representative views, and quantitative results are shown in panel C. (C) Quantification of Swi11–38Mut-YFP aggregation observed in panel B and Fig. 1C. Cells were manually counted using Fiji software. Normalized aggregation is defined as the number of aggregate-containing cells divided by the total number of cells with YFP fluorescence and then normalized to the WT (which had a raw aggregation percentage of ∼50 to 80%) per biological replicate. The mean number of cells with YFP fluorescence observed per mutant per replicate was approximately 900. Error bars represent standard errors of the means.
Once again, based on three biological replicates, several mutants exhibited aggregation akin to that of the WT (Fig. 2B). These mutants included F3N, L6N, L6A, 4×TN 8×TN, T27N, T27A, T32N, and T32A. These results once again highlighted that most of the singular mutants were capable of coaggregating and adopting the prion conformation of Swi1FL. Interestingly, the other singular phenylalanine mutants (F3A, F4N, and F4A) that had low (<20%) aggregation rates in the initial assay displayed an increased aggregation frequency (∼60%) under Swi1FL overexpression conditions (Fig. 2C). On the other hand, the double-phenylalanine mutants (FF-NN and FF-AA) displayed a greatly impaired ability to decorate Swi1FL aggregates; however, there were low levels of observable puncta. Another mutant, 4×TA, also displayed a low aggregation frequency (∼20%) in the Swi1FL overexpression context, whereas aggregates were not seen under non-Swi1FL-overexpression conditions (∼0%). The 8×TA mutant was unable to form notable aggregates even under Swi1FL overexpression conditions, suggesting that replacing the polar threonine tract with small, hydrophobic, nonpolar alanine residues in the center of Swi11–38 likely was disruptive to the aggregation core of the protein. No construct resulted in consistent aggregation in [swi−] cells, although single cells with puncta were observed in a minimal (<5%) number of colonies. These rare instances in the originally [swi−] cells likely reflect randomly generated Swi11–38 or Swi1FL aggregates from the very favorable overexpression conditions.
Although there was an increase in Swi11–38 mutant coaggregation when Swi1FL was overexpressed, the most deleterious mutants still had significant effects. The removal of aromatic groups via the replacement of the phenylalanine residues displayed a stepwise effect on the aggregation frequency, with the removal of both leading to a greater decrease than the removal of just one. Additionally, a peculiar site-specific effect was observed as the F4N mutation readily decreased observed aggregation whereas the F3N mutation did not. Moreover, whether the aggregates of these and other Swi11–38 mutants were stable without the presence of Swi1FL aggregates remained an open question.
Swi11–38 requires a phenylalanine to maintain [SWI+].
We proceeded to use the BY4741 swi1Δ/p416TEF-SWI1FL/p415TEF-SWI11–38Mut-YFP [SWI+] transformants to investigate the maintenance of the prion fold by the various Swi11–38 mutants in the absence of Swi1FL (Fig. 3A). To do so, isolates containing aggregates were transferred to medium containing 5-fluoroorotic acid (5-FOA). Cells containing the URA3 marker, in this case on the plasmid p416TEF-SWI1FL, would process 5-FOA into a toxic chemical, killing the cells. Thus, using this selection system, we generated cells with the swi1Δ/p415TEF-SWI11–38Mut-YFP genotype that have no full-length Swi1 present.
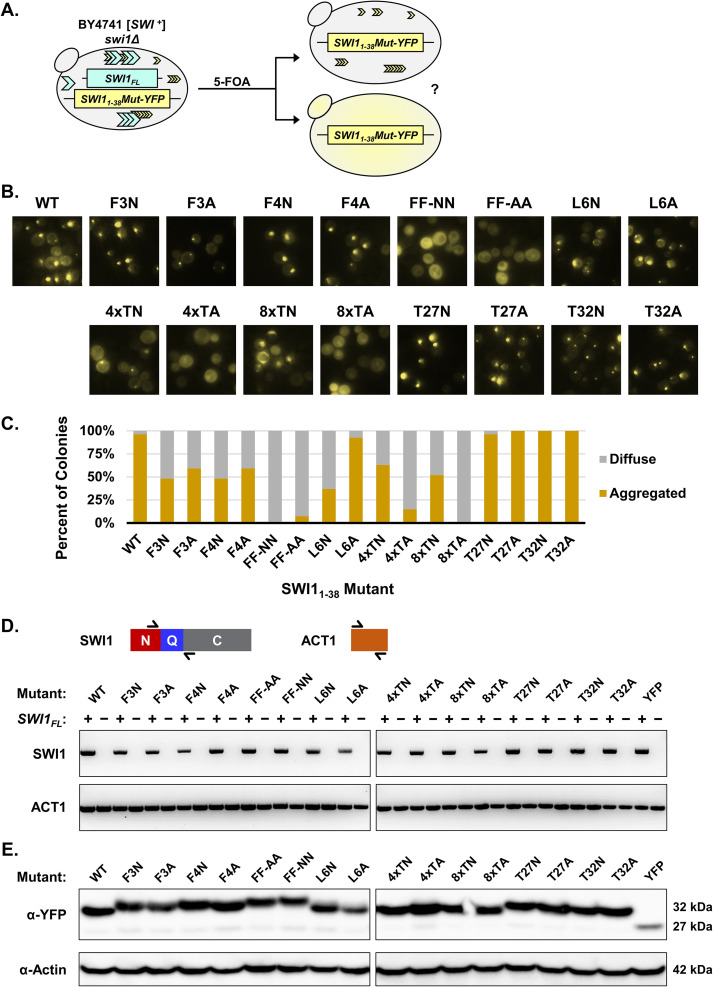
FIG 3.
Mutation of both phenylalanine residues leads to Swi11–38 being unable to maintain the prion fold in the absence of Swi1FL. (A) Diagram of the experiment. BY4741 swi1Δ/p416TEF-SWI1FL/p415TEF-SWI11–38Mut-YFP [SWI+] cells containing aggregates were treated with 5-FOA to select against cells containing the p416TEF-SWI1FL plasmid. The resulting BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells were observed using fluorescence microscopy for aggregate foci or diffuse signals. (B) Representative fluorescence images of the resulting BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. See Table 1 for amino acid sequences of the mutants. For each construct, 3 aggregate-containing BY4741 swi1Δ/p416TEF-SWI1FL/p415TEF-SWI11–38Mut-YFP isolates were treated with 5-FOA to drop out the full-length Swi1 expression plasmid. From there, 9 colonies for each isolate (for a total of 27 colonies) were examined for each construct. Shown are representative views, and quantitative results are shown in panel C. (C) Quantification of yeast colonies retaining Swi11–38Mut-YFP aggregates after the removal of Swi1FL via 5-FOA treatment. For experiments quantified in this graph, aggregated colonies had >25% of cells containing aggregates, and diffuse colonies showed aggregation in <5% of cells. The percentage of colonies was calculated as the number of each of the two types of colonies (aggregated and diffuse) divided by the total number of colonies examined for each construct. (D) Diagram of RT-PCR primer targets and the resulting RT-PCR amplification visualized by agarose gel. Primers flanking the Q region of SWI1 were used to confirm the loss of SWI1FL in the BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. Primers covering ACT1 were used as a positive control. Samples labeled as + correspond to the pre-5-FOA BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. Samples labeled as − correspond to the post-5-FOA BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. (E) Western blot of BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. The membrane was probed with either anti-GFP or antiactin. Estimated molecular weights based on the sequence are listed at the right.
Among these newly generated yeast isolates, we examined whether individual colonies contained aggregates, indicating maintenance of an adopted prion fold (Fig. 3B and C). The single-phenylalanine mutants (F3N, F3A, F4N, and F4A) displayed observable puncta in cells in ∼50% of colonies. However, once both phenylalanine residues were replaced with either asparagine or alanine as in FF-NN and FF-AA, aggregation was almost completely abolished: only one colony was found to contain any aggregation. This result indicates that the phenylalanine residues play a pivotal role in maintaining Swi11–38 aggregation.
Other mutants also displayed deficiencies in maintaining aggregation once Swi1FL was removed. The 4×TA mutant had aggregates in ∼10% of colonies, while no observed 8×TA colonies contained aggregates (Fig. 3C). The substitution of the polar threonine residues with the nonpolar alanine residues likely disrupted the stability of any prion fold adopted by Swi11–38. Meanwhile, the 4×TN and 8×TN mutants both presented a reduced maintenance ability (∼60% and ∼50% of colonies, respectively), suggesting that the presence of the threonine tract remains important but is not required for Swi11–38 aggregation. Once again, the T27N, T27A, T32N, and T32A mutants did not present any meaningful deviation from the WT control. In addition, the L6A mutant did not demonstrate impairment in maintaining aggregation. The L6N mutant displayed a substantial decrease in the number of colonies with cells containing aggregates. This difference between the L6A and L6N mutants may be due to the similarities between alanine and leucine, both nonpolar, hydrophobic amino acids, as opposed to the polarity introduced by an asparagine residue.
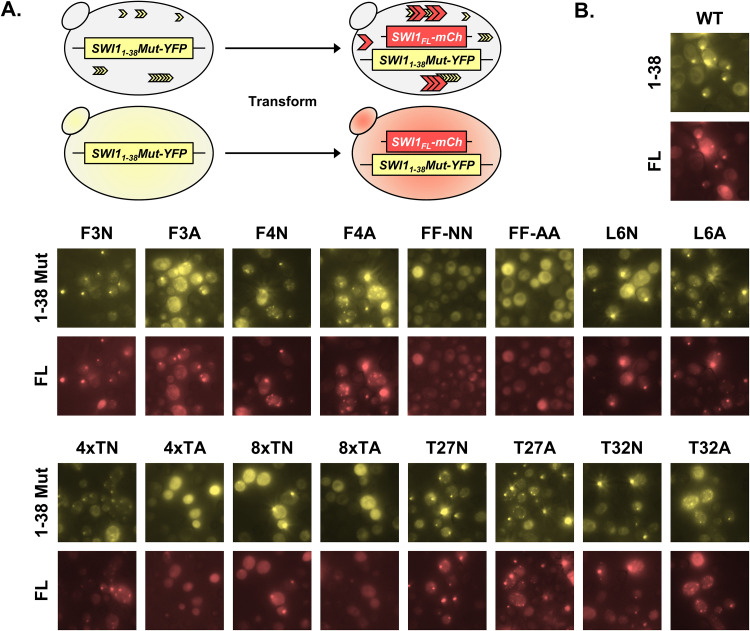
Cells containing Swi11–38Mut-YFP aggregates were examined for the curability of this aggregation. For the Swi11–38 WT and the various mutants, multiple isolates were streaked onto selective medium containing 5 mM guanidine hydrochloride (GdnHCl). GdnHCl cures or rids cells of many endogenous yeast prions, including [SWI+], through the inactivation of the molecular chaperone Hsp104 (26). Treatment with GdnHCl resulted in the loss of Swi11–38Mut-YFP aggregation for the WT and all aggregate-maintaining mutants (data not shown). This result indicates that the aggregation was of a prion form. Additionally, we tested whether the aggregated Swi11–38Mut-YFP could transmit its prion fold back to Swi1FL by transforming the BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells with p416TEF-SWI1FL-mCherry, which allows the expression of a fluorescently tagged version of Swi1FL (Fig. 4A). Colonies containing cells with Swi11–38Mut-YFP aggregates also displayed Swi1FL-mCherry aggregates when visualized via fluorescence microscopy, and the puncta of Swi11–38Mut-YFP and Swi1FL-mCherry were largely colocalized (Fig. 4B). Mutants unable to maintain aggregation without Swi1FL, and thus not having aggregates to support the transmission of a prion fold back to Swi1FL-mCherry, did not display any mCherry foci. Thus, these aggregates present in the BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells could transmit a prion fold back to Swi1FL, indicating that the observed Swi11–38Mut-YFP aggregates are of a prion form.
FIG 4.
Swi11–38 aggregates can transmit the prion fold back to Swi1FL. (A) Diagram of the experiment. BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells were transformed with p416TEF-SWI1FL-mCherry. Transformants were observed using fluorescence microscopy for aggregate foci or diffuse signals. (B) Fluorescence images of the resulting BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP/p16TEF-SWI1FL-mCherry cells. See Table 1 for amino acid sequences of the mutants. Samples were imaged with the appropriate filters for Swi11–38-YFP/Swi11–38Mut-YFP (1–38/1–38 Mut) and Swi1FL-mCherry (FL). For each construct, 3 different transformants were examined. Shown are representative views.
To confirm the validity of these results, the yeast isolates were checked for SWI1FL mRNA via reverse transcription-PCR (RT-PCR) using a pair of primers in the SWI1 coding region but downstream of SWI11–38 to verify the absence of SWI1FL expression (Fig. 3D). We confirmed that none of the examined isolates after 5-FOA treatment contained SWI1FL (Fig. 3D). The expression of the mutant Swi11–38 constructs was also examined at the protein level to address the possibility that the results could be influenced by variations in expression levels rather than the mutations. No notable differences were observed when assessed via Western blotting (Fig. 3E). Slight variations in band locations were seen on the blot, but these differences were likely due to the changes in the molecular weight (MW) and electrophoretic mobility due to the mutations combined with a high acrylamide percentage on a gradient gel.
Loss of both phenylalanine residues disrupts de novo prion formation by Swi11–38.
We next examined if these Swi11–38 mutants were able to de novo form a prion. Our laboratory’s previous research established that Swi11–38 can act as a bona fide prion domain and de novo form a prion termed [SPS+] (21). To examine the ability of the Swi11–38 mutants to do so, we employed the widely used Sup35 assay, in which a prion or prion-like domain of interest is attached to the MC regions of Sup35 in place of its own prion-domain-containing N region (10, 22).
Sup35 functions normally as a translation terminator in yeast, and this function combined with a genetic alteration to the ADE1 gene (ade1-14) provides a useful tool for evaluating prionogenesis (27). Under nonprion conditions, Sup35 acts as an efficient translation terminator, recognizes the premature stop codon introduced with ade1-14, and prevents the creation of a necessary enzyme in the adenine synthesis pathway. This prevention results in the buildup of an adenine precursor that provides the yeast cells with a red hue. When prionized, Sup35 can no longer efficiently function as a translation terminator, and readthrough of the premature stop codon results in the production of the requisite enzyme. This situation results in the synthesis of adenine and little buildup of the red adenine precursor, leading to the yeast colonies being white or light pink.
To initialize the assay, Swi11–38 mutants were linked to Sup35MC and transformed into W303 sup35Δ/p316SUP35 [PSI+] yeast provided by the Weissman laboratory (Fig. 5A and B). After confirmation of a white-to-red color change indicating that the Swi11–38Mut-MC fusions were functional, the SUP35 plasmid was removed via treatment with 5-FOA. From there, three red isolates for each Swi11–38Mut-MC fusion were transformed with p415TEF-SWI11–38Mut-YFP in order to provide an overexpression of Swi11–38Mut to induce de novo prion formation at a high rate. Additionally, we confirmed via Western blotting that the expression of Swi11–38Mut-MC was consistent among the different mutants (Fig. 5E).
FIG 5.
Swi11–38 requires at least one of its phenylalanine residues for de novo prion formation. (A) Diagram of the fusion proteins created. Swi11–38Mut was linked to Sup35MC via a DPGGPGGG linker to allow the use of the Sup35 assay for de novo prion formation. (B) Diagram of the experiment. W303 sup35Δ/p316SUP35FL [PSI+] cells were transformed with p415TEF-SWI11–38-MC (WT) or p415TEF-SWI11–38Mut-MC. The transformants were treated with 5-FOA to select against cells containing the p316SUP35FL plasmid. The resulting W303 sup35Δ/p415TEF-SWI11–38Mut-MC cells were then transformed with p416TEF-SWI11–38-YFP (WT) or the corresponding p416TEF-SWI11–38Mut-YFP plasmids. Transformants were grown, spread onto −LU plates, and checked for a color change corresponding to the prionization of the Swi11–38Mut-MC protein. (C) Representative images of W303 sup35Δ/p415TEF-SWI11–38Mut-MC/p416TEF-SWI11–38Mut-YFP colonies on −LU plates. Images are representative of full-plate images captured from 3 biological replicates. (D) Representative fluorescence images of W303 sup35Δ/p415TEF-SWI11–38Mut-MC/p416TEF-SWI11–38Mut-YFP yeast cells. Prion-forming constructs display aggregation visualized from cells from white or light-pink colonies. Constructs unable to form [SPS+] (FF-NN, FF-AA, 4×TA, and 8×TA) display diffuse signals as seen in cells from red colonies. Images are representative of multiple examined colonies for each mutant. (E) Western blot of W303 sup35Δ/p415TEF-SWI11–38Mut-MC/p416TEF-SWI11–38Mut-YFP cell lysates. The membrane was probed with either anti-Sup35 or antiactin. Estimated molecular weights based on the sequence are listed at the right.
The majority of the Swi11–38 constructs produced colonies with colors indicative of prionization, e.g., white, light-pink, and sectored with multiple hues (Fig. 5C). In addition to the WT, these constructs included the single-phenylalanine mutants (F3N, F3A, F4N, and F4A), the other single-residue mutants (L6N, L6A, T27N, T27A, T32N, and T32A), and the threonine tract asparagine mutants (4×TN and 8×TN). When treated with GdnHCl, the majority of nonred colonies could be reverted to red, indicating curability (data not shown). While replacing one phenylalanine with an asparagine or alanine did not disrupt prion formation, replacing both phenylalanine residues (FF-NN and FF-AA) completely abolished de novo prion formation by Swi11–38. The aromaticity of particular amino acid side chains may play a crucial role in nucleating the prion fold, explaining the lack of prion formation of the FF-NN and FF-AA mutants. The 4×TA and 8×TA mutants also led to Swi11–38 losing its prion-forming ability. In this case, the addition of multiple alanine residues with their small methyl side chains in what otherwise would be a long stretch of polar residues proved deleterious, as swapping one polar amino acid for another polar amino acid (as in 4×TN and 8×TN) did not result in a similar prionization impairment.
We examined the generated sup35Δ/p415TEF-SWI11–38Mut-MC/p416TEF-SWI11–38Mut-YFP colonies for aggregation using fluorescence microscopy. There was a small number of wholly white colonies that presented on the FF-NN, FF-AA, 4×TA, and 8×TA plates. All such colonies were checked for aggregate formation, and none contained puncta of any sort, indicating that they were nonprion cells containing mutations in the adenine synthetic pathway (Fig. 5D and data not shown). On the other hand, randomly selected white, light-pink, or sectored colonies from among all other mutants and the WT displayed aggregates (Fig. 5D).
We treated the [SPS+] colonies with 5-FOA to select against the p416TEF-SWI11–38Mut-YFP plasmid. This process removes a portion of the overexpression conditions by theoretically halving the overall expression of SWI11–38Mut. After treatment with 5-FOA, some colonies, regardless of mutation, stably maintained the [SPS+] phenotypes, while others did not (data not shown). This result was not surprising as the higher-overexpression conditions with the p416TEF-SWI11–38Mut-YFP plasmid present likely supported the maintenance of weaker variants. Thus, the inability of the double-phenylalanine mutants to de novo form [SPS+] even under the highly favorable two-plasmid overexpression conditions indicates that these mutations indeed abolish the prion-forming capability of Swi11–38-MC.
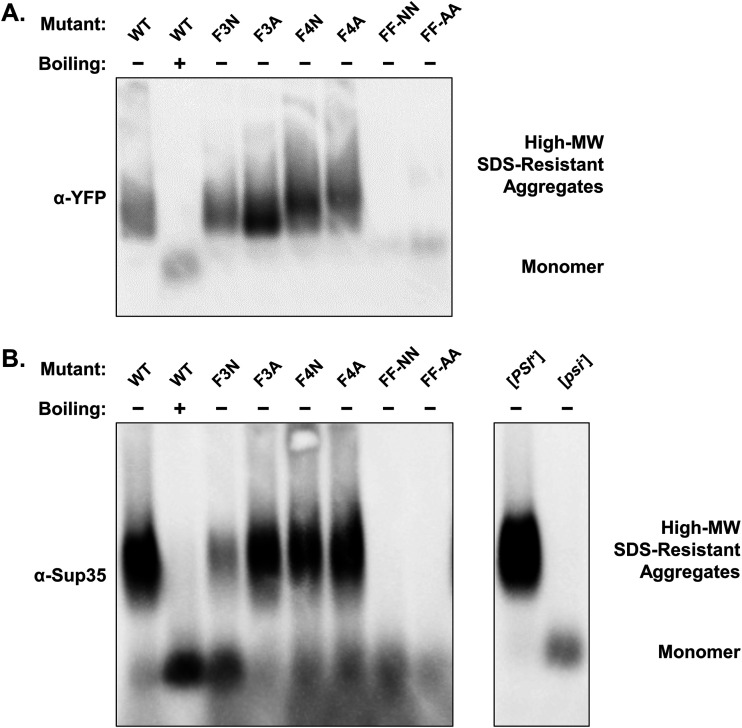
Double-phenylalanine mutants do not form high-MW, SDS-resistant aggregates.
With aggregation, maintenance of the [SWI+] prion fold, and de novo prion formation by Swi11–38 being deleteriously affected by the replacement of its phenylalanine residues, we next examined these mutants via semidenaturing detergent agarose gel electrophoresis (SDD-AGE). This technique allows the identification of high-molecular-weight, detergent-resistant protein aggregates. We cultivated both BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP and W303 sup35Δ/p415TEF-SWI11–38Mut-MC/p416TEF-SWI11–38Mut-YFP yeast cells to check for such aggregates of Swi11–38Mut-YFP and Swi11–38Mut-MC, respectively.
In the presence of 2% SDS, we found that the WT Swi11–38-YFP protein formed a noticeable smear of high-MW, SDS-resistant species when probed with anti-YFP (Fig. 6A). Upon boiling, the high-MW species dissembled to become low-MW monomers (Fig. 6A). The single-phenylalanine mutants (F3N, F3A, F4N, and F4A) all form a similar smear. On the other hand, both FF-NN and FF-AA display only a faint lower banding that corresponds to where monomeric species are found (as seen in the boiled WT sample). These results correlate with the minimal aggregation observed in the FF-NN and FF-AA samples and the inability of these mutants to maintain the [SWI+] fold in the absence of Swi1FL.
FIG 6.
Swi11–38 no longer forms high-molecular-weight, SDS-resistant aggregates when both phenylalanine residues are replaced. (A) Blot of SDD-AGE of BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells with (+) or without (−) boiling. The membrane was probed with anti-GFP to detect Swi11–38Mut-YFP. (B) Blot of SDD-AGE of W303 sup35Δ/p415TEF-SWI11–38Mut-MC/p416TEF-SWI11–38Mut-YFP cells with (+) or without (−) boiling and W303 sup35Δ/p316SUP35FL cells. The membrane was probed with anti-Sup35 to detect Swi11–38Mut-MC or Sup35FL.
Similarly, high-MW, SDS-resistant forms of WT Swi11–38-MC were seen by SDD-AGE (Fig. 6B). The single-phenylalanine mutants displayed much the same pattern as the WT, and the double-phenylalanine mutants showed a signal only in the monomeric region. This loss of high-MW, SDS-resistant species mirrored the loss of de novo prion formation by FF-NN and FF-AA, indicating the inability of these mutants to adopt stable prion aggregates.
DISCUSSION
Our laboratory initially discovered the [SWI+] prion and documented the existence of the Swi1 prion domain within the protein’s N region (11, 19). Further study revealed that Swi11–38 could recapitulate the aggregation phenotype of [SWI+] and function as a bona fide prion domain (20, 21). This extreme N-terminal region is a uniquely asparagine-rich but glutamine-free, small prion domain extensively demonstrated to function in vivo for prion formation and maintenance. Investigating the functioning and characteristics of Swi11–38 is important to understanding the prionization of Swi1, which regulates over 15% of the yeast genome and plays a significant role in modulating yeast multicellularity (16–18). In this study, we further dissected Swi11–38 and its ability to aggregate, maintain the [SWI+] prion fold, and de novo form a prion.
Multiple mutants of Swi11–38 that we created via the replacement of singular nonasparagine residues with either asparagine or alanine had no significant effect. Previous work by our laboratory showed that Swi11–31, a truncation of Swi11–38 that did not include T32, could still transmit the [SWI+] prion fold and form a prion when fused with Sup35MC. The lack of necessity for this end portion of Swi11–38 suggests that mutations at this location would likely be more easily tolerated than those at other locations. Indeed, we observed that the T32N and T32A mutations maintained similarity to the WT throughout our various assays. Interestingly, T27N and T27A also did not demonstrate significant deviations from the WT in aggregation, maintenance, or prionization. Meanwhile, the L6A mutation did not generate meaningful differences in the functioning of Swi11–38 as a prion domain. However, L6N displayed decreases in maintaining the [SWI+] prion fold in the absence of Swi1FL. This difference between the mutation to alanine and the mutation to asparagine may indicate that the decrease in hydrophobicity interrupts a buried region of aggregated Swi11–38.
The threonine tract mutants (4×TN, 4×TA, 8×TN, and 8×TA) demonstrated a dichotomy based on maintaining the polarity of the residues versus losing said polarity. The 4×TN and 8×TN mutants where the tract was partially or wholly replaced with the similarly polar, uncharged asparagine showed little variance from WT Swi11–38. Conversely, the 4×TA and 8×TA mutants exhibited severely reduced aggregation in both genetic backgrounds as well as completely abolished de novo prion formation. Threonine tracts of lengths similar to the one found in Swi11–38 can be found in some adhesins or flocculins in various yeasts (28–30). In those contexts, such polythreonine stretches are thought to be important for the formation of β-sheet structures and the surface-binding properties of the proteins. The threonine tract of Swi11–38 may also play a similar role for its aggregation, although the exact structure of this prion domain has yet to be determined. However, threonine and other uncharged polar residues such as asparagine and serine have a noted role in the promotion of aggregation and amyloidogenesis. Thus, the threonine tract of Swi11–38 may provide a stable core for the formation of the high-MW, SDS-resistant species observed in this study.
Mutating the two phenylalanine residues at the beginning of Swi11–38 resulted in a fairly direct relationship between the number of phenylalanines and maintenance of the [SWI+] prion fold as well as prionization. Replacing one phenylalanine residue (F3N, F3A, F4N, and F4A) led to ∼50% of colonies maintaining aggregates in the BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells (Fig. 3C), and replacing the second phenylalanine residue (FF-NN and FF-AA) led to an almost complete loss of the prion as observed via aggregation. If the ability of Swi11–38 to maintain a prion fold is dependent on the aromaticity present in these phenylalanine residues, then perhaps the replacement of the phenylalanine residues with other aromatic amino acids (i.e., tryptophan or tyrosine) may have no effect versus the WT. De novo prion formation by Swi11–38-MC also relied on the presence of at least one phenylalanine residue, with neither FF-NN nor FF-AA being capable of prionization. This reliance of the prionogenicity of Swi11–38 on a single amino acid residue being present belies the fact that the jump from aggregable to prionogenic can be extremely small. Indeed, previous research found that just a small number of mutations could lead to an existing asparagine/glutamine-rich domain to gain prion capabilities (31). The mutations presented in that research primarily relied on replacing nonprionogenic residues (e.g., charged amino acids) and the introduction of hydrophobic and/or aromatic residues (e.g., phenylalanine), much the opposite of some of the deleterious mutants produced in Swi11–38.
Given the impact of the removal of the aromatic side groups on Swi11–38, we also examined whether either the FF-NN or FF-AA mutation affected aggregation in the context of longer regions, such as Swi1N or Swi1NQ. However, no change in aggregation was observed in BY4741 [SWI+] cells (data not shown). This result indicates that other residues or regions of Swi1N can stand in for the loss of the two phenylalanine residues at positions 3 and 4. Indeed, multiple aromatic amino acids can be found downstream of Swi11–38 (i.e., positions 73, 76, 77, and 82). These other aromatic-containing residues may indeed provide the necessary underpinning of the region’s prion-forming capacity when the first two phenylalanine residues are replaced. Additionally, Sant’Anna et al. showed that a predicted amyloidogenic region (Swi1239–259) can in fact form amyloid in vitro (32). Regions such as Swi1239–259 likely provide any required stabilization needed to offset the destabilization of Swi11–38, allowing the maintenance and propagation of the prion fold. Taken together, the presence of multiple aromatic residues and the amyloidogenic region located downstream of the Swi11–38 PrD suggests that [SWI+] formation is likely a favorable event in S. cerevisiae. In this regard, it has been shown that [SWI+] can confer fungicide resistance and tolerance to certain alcohols and can aid yeast to adapt to environmental changes (16, 22, 33).
Intriguingly, many of the phenylalanine residues in the Swi1N region, those that were mutated at positions 3 and 4 as well as those closely downstream at positions 73, 76, 77, and 82, are conserved in other Saccharomyces species (data not shown). For example, the Swi1 genes in S. boulardii, S. paradoxus, and S. pastorianus all contain the above-mentioned phenylalanine residues (34–36). The asparagine contents of the corresponding Swi1N regions across these species are highly similar (∼31 to 34%), although S. cerevisiae Swi1N contains a greater number of asparagine residues by raw count. Moreover, in the case of S. boulardii, the threonine tract can also be found within the corresponding Swi11–38 region. It should be noted that additional charged amino acid residues present in S. pastorianus may prevent the extreme N terminus of Swi1 from acting similarly to Swi11–38 examined in this study. In all, we do not currently know if Swi1 exhibits prionogenicity in these other species; however, the gene appears to retain the components that likely provide the basis for prionogenicity in S. cerevisiae. Further research may elucidate the possibility of [SWI+] existence in other species.
Although the structure of aggregated or prionized Swi11–38 (or its various fusions) is as yet unknown, our laboratory has previously demonstrated that Swi1N can form amyloid. In this study, we have demonstrated that Swi11–38 forms high-molecular-weight, SDS-resistant aggregates in the case of either Swi11–38-YFP initially aggregated alongside Swi1FL in the [SWI+] prion form or the [SPS+] prion de novo formed by Swi11–38-MC. It is likely that these protein species visualized by SDD-AGE are of an amyloid variety as such patterning mirrors that of larger Swi1 constructs, Sup35 (Fig. 6B), and other amyloid-forming proteins.
In all, select amino acids in Swi11–38 are crucial for this prion domain’s ability to aggregate, maintain the [SWI+] prion fold, and de novo form a prion. While the overall asparagine-rich composition of Swi11–38 provides a basis for prionization, this region depends on the presence of its two phenylalanine residues for the ability to prionize, although these two specific residues are not vital in the context of Swi1N or Swi1NQ. The other nonasparagine residues, which are mainly threonine residues, likely maintain the favorable uncharged, polar side chains that favor disorder but also aggregation. As such, it remains likely that like other prionogenic proteins, Swi11–38 and its larger iterations, Swi1N, Swi1NQ, and full-length Swi1, achieve their prionogenicity largely via overall composition.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used in this study are listed in Table 2. The W303 sup35Δ/SUP35::TRP1/p316SUP35FL strain was provided by the Weissman laboratory (University of California, San Francisco).
TABLE 2.
Yeast strains used in this study
Yeast cells were grown according to established protocols at 30°C in either yeast extract-peptone-dextrose (YPD) or synthetic complete (SC) medium minus the appropriate amino acids (e.g., leucine [-L] or leucine-uracil [-LU]) (37). When indicated, medium was supplemented with 1 g/liter 5-FOA for counterselection against a URA3-carrying plasmid or with 5 mM GdnHCl for the inactivation of Hsp104 to disrupt prion propagation.
Plasmid construction.
Plasmids used in this study are listed in Table 3. Briefly, the p415TEF-SWI11–38-YFP plasmid (20) was used as the template to produce the various mutant SWI11–38 plasmids via PCR. See Table 4 for primer information. For mutations in the first portion of SWI11–38, the mutant PCR product was digested with SpeI/XhoI for cloning back into similarly digested plasmid p415TEF-SWI11–38-YFP to produce p415TEF-SWI11–38F3N-YFP, p415TEF-SWI11–38F3A-YFP, p415TEF-SWI11–38F4N-YFP, p415TEF-SWI11–38FF-NN-YFP, p415TEF-SWI11–38FF-AA-YFP, p415TEF-SWI11–38L6N-YFP, and p415TEF-SWI11–38L6A-YFP. For mutations in the middle of SWI11–38, the mutant PCR product was cloned back into p415TEF-SWI11–38-YFP via SacI/XhoI sites to produce p415TEF-SWI11–384×TN-YFP, p415TEF-SWI11–384×TA-YFP, p415TEF-SWI11–388×TN-YFP, and p415TEF-SWI11–388×TA-YFP. For mutations in the back portion of SWI11–38, the mutant PCR product was cloned back into p415TEF-SWI11–38-YFP via the SacI/BamHI sites to produce p415TEF-SWI11–38T27N-YFP, p415TEF-SWI11–38T27A-YFP, p415TEF-SWI11–38T32N-YFP, and p415TEF-SWI11–38T32A-YFP. To produce the collection of mutant p416TEF-SWI11–38-YFP plasmids, the mutant SWI11–38-YFP was cloned from the respective p415TEF-SWI11–38-YFP plasmids and into p416TEF via SpeI/XhoI sites.
TABLE 3.
Plasmids used in this study
| Plasmid | Marker | Replicon | Promoter | Use | Reference or source |
|---|---|---|---|---|---|
| p415TEF-SWI11–38-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38-YFP | 20 |
| p415TEF-SWI11–38F3N-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F3N-YFP | This study |
| p415TEF-SWI11–38F3A-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F3A-YFP | This study |
| p415TEF-SWI11–38F4N-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F4N-YFP | This study |
| p415TEF-SWI11–38F4A-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F4A-YFP | This study |
| p415TEF-SWI11–38FF-NN-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38FF-NN-YFP | This study |
| p415TEF-SWI11–38FF-AA-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38FF-AA-YFP | This study |
| p415TEF-SWI11–38L6N-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38L6N-YFP | This study |
| p415TEF-SWI11–38L6A-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38L6A-YFP | This study |
| p415TEF-SWI11–384×TN-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–384×TN-YFP | This study |
| p415TEF-SWI11–384×TA-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–384×TA-YFP | This study |
| p415TEF-SWI11–388×TN-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–388×TN-YFP | This study |
| p415TEF-SWI11–388×TA-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–388×TA-YFP | This study |
| p415TEF-SWI11–38T27N-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T27N-YFP | This study |
| p415TEF-SWI11–38T27A-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T27A-YFP | This study |
| p415TEF-SWI11–38T32N-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T32N-YFP | This study |
| p415TEF-SWI11–38T32A-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T32A-YFP | This study |
| p415TEF-YFP | LEU2 | CEN6/ARSH4 | TEF1 | Expression of YFP | 18 |
| p416TEF-SWI1FL | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi1 | 11 |
| p416TEF-SWI1FL-mCherry | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi1-mCherry | 20 |
| p416TEF-SWI11–38-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38-YFP | This study |
| p416TEF-SWI11–38F3N-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F3N-YFP | This study |
| p416TEF-SWI11–38F3A-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F3A-YFP | This study |
| p416TEF-SWI11–38F4N-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F4N-YFP | This study |
| p416TEF-SWI11–38F4A-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F4A-YFP | This study |
| p416TEF-SWI11–38FF-NN-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38FF-NN-YFP | This study |
| p416TEF-SWI11–38FF-AA-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38FF-AA-YFP | This study |
| p416TEF-SWI11–38L6N-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38L6N-YFP | This study |
| p416TEF-SWI11–38L6A-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38L6A-YFP | This study |
| p416TEF-SWI11–384×TN-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–384×TN-YFP | This study |
| p416TEF-SWI11–384×TA-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–384×TA-YFP | This study |
| p416TEF-SWI11–388×TN-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–388×TN-YFP | This study |
| p416TEF-SWI11–388×TA-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–388×TA-YFP | This study |
| p416TEF-SWI11–38T27N-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T27N-YFP | This study |
| p416TEF-SWI11–38T27A-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T27A-YFP | This study |
| p416TEF-SWI11–38T32N-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T32N-YFP | This study |
| p416TEF-SWI11–38T32A-YFP | URA3 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T32A-YFP | This study |
| p415TEF-SWI11–38-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38-MC | This study |
| p415TEF-SWI11–38F3N-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F3N-MC | This study |
| p415TEF-SWI11–38F3A-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F3A-MC | This study |
| p415TEF-SWI11–38F4N-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F4N-MC | This study |
| p415TEF-SWI11–38F4A-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38F4A-MC | This study |
| p415TEF-SWI11–38FF-NN-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38FF-NN-MC | This study |
| p415TEF-SWI11–38FF-AA-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38FF-AA-MC | This study |
| p415TEF-SWI11–38L6N-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38L6N-MC | This study |
| p415TEF-SWI11–38L6A-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38L6A-MC | This study |
| p415TEF-SWI11–384×TN-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–384×TN-MC | This study |
| p415TEF-SWI11–384×TA-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–384×TA-MC | This study |
| p415TEF-SWI11–388×TN-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–388×TN-MC | This study |
| p415TEF-SWI11–388×TA-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–388×TA-MC | This study |
| p415TEF-SWI11–38T27N-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T27N-MC | This study |
| p415TEF-SWI11–38T27A-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T27A-MC | This study |
| p415TEF-SWI11–38T32N-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T32N-MC | This study |
| p415TEF-SWI11–38T32A-MC | LEU2 | CEN6/ARSH4 | TEF1 | Expression of Swi11–38T32A-MC | This study |
| p316Sup35FL | URA3 | CEN6/ARSH4 | SUP35 | Expression of Sup35 | Weissman lab |
TABLE 4.
Primers used in this study
| Primer | Sequence (5′–3′) | Resulting plasmid or descriptiona |
|---|---|---|
| Swi1 F3N For | AGAACTAGTATGGATAACTTTAATTTGAAT | p415TEF-SWI11–38F3N-YFP |
| Swi1 F3A For | AGAACTAGTATGGATGCCTTTAATTTGAAT | p415TEF-SWI11–38F3A-YFP |
| Swi1 F4N For | AGAACTAGTATGGATTTCAACAATTTGAAT | p415TEF-SWI11–38F4N-YFP |
| Swi1 F4A For | AGAACTAGTATGGATTTCGCCAATTTGAAT | p415TEF-SWI11–38F4A-YFP |
| Swi1 FF-NN For | AGAACTAGTATGGATAACAACAATTTGAAT | p415TEF-SWI11–38FF-NN-YFP |
| Swi1 FF-AA For | AGAACTAGTATGGATGCCGCCAATTTGAAT | p415TEF-SWI11–38FF-AA-YFP |
| Swi1 L6N For | ACTAGTATGGATTTCTTTAATAACAATAATAATAATAATAATAATAATACTACTACT | p415TEF-SWI11–38L6N-YFP |
| Swi1 L6A For | ACTAGTATGGATTTCTTTAATGCGAATAATAATAATAATAATAATAATACTACTACT | p415TEF-SWI11–38L6A-YFP |
| Swi1 4×TN For | ACTACTAACAACAACAACAATAACAATAATACTAATAATAATAATACT | p415TEF-SWI11–384×TN-YFP |
| Swi1 4×TN Rev | GTTATTGTTGTTGTTGTTAGTAGTAGTAGTATTATTATTATTATTATTATTATTCAA | p415TEF-SWI11–384×TN-YFP |
| Swi1 4×TA For | ACTACTGCAGCAGCAGCAAATAACAATAATACTAATAATAATAATACT | p415TEF-SWI11–384×TA-YFP |
| Swi1 4×TA Rev | GTTATTTGCTGCTGCTGCAGTAGTAGTAGTATTATTATTATTATTATTATTATTCAA | p415TEF-SWI11–384×TA-YFP |
| Swi1 8×TN For | AATAATAACAACAACAACAACAACAACAACAATAACAATAATACT | p415TEF-SWI11–388×TN-YFP |
| Swi1 8×TN Rev | GTTGTTGTTGTTGTTGTTATTATTATTATTATTATTATTATTCAA | p415TEF-SWI11–388×TN-YFP |
| Swi1 8×TA For | AATAATGCAGCAGCAGCAGCAGCAGCAGCAAATAACAATAATACT | p415TEF-SWI11–388×TA-YFP |
| Swi1 8×TA Rev | TGCTGCTGCTGCTGCTGCATTATTATTATTATTATTATTATTCAA | p415TEF-SWI11–388×TA-YFP |
| Swi1 T27N Rev | GGTGGATCCGGATTATTATTATTATTAGTATTATTATTATTATTATTATTGTTATTGGT | p415TEF-SWI11–38T27N-YFP |
| Swi1 T27A Rev | GGTGGATCCGGATTATTATTATTATTAGTATTATTATTATTAGCATTATTGTTATTGGT | p415TEF-SWI11–38T27A-YFP |
| Swi1 T32N Rev | GGTGGATCCGGATTATTATTATTATTGTTATTATTATTATTAGT | p415TEF-SWI11–38T32N-YFP |
| Swi1 T32A Rev | GGTGGATCCGGATTATTATTATTATTAGCATTATTATTATTAGT | p415TEF-SWI11–38T32A-YFP |
| p415TEF-SWI11–38-YFP For | TTATCTACACGACGGGGAGTCA | Multiple SWI11–38 mutants |
| p415TEF-SWI11–38-YFP Rev | AATGTAAGCGTGACATAACTAATTACATGA | Multiple SWI11–38 mutants |
| p415TEF-SWI11–38-YFP For 4× | CAAGACGATAGTTACCGGATAAGG | Multiple SWI11–38 mutants |
| p415TEF-SWI11–38-YFP Rev 4× | TGGATTTTGATGTAATTGTTGGGATTC | Multiple SWI11–38 mutants |
| p415TEF-SWI11–38-YFP For 4× HT | AAGACGATAGTTACCGGATAAGGCGCA | Multiple SWI11–38 mutants |
| p415TEF-SWI11–38-YFP Rev 4× HT | AGAATAGACCGAGATAGGGTTGAGTGTTGT | Multiple SWI11–38 mutants |
| SpeI-SWI1–38 For | GGTTCAAGCTATGCGTCAGACCCCGTAGAAAAGATCAAAGG | p415TEF-SWI11–38-MC |
| SWI1–38-BamHI-Linker Rev | ACCACCACCAGGACCACCTGGATCCGGATTATTATTATTATTAGTATTATTATTATTAGT | p415TEF-SWI11–38-MC |
| Linker-SUP35MC For | GGTGGTCCTGGTGGTGGTATGTCTTTGAACGACTTTCAAAAGC | p415TEF-SWI11–38-MC |
| SUP35MC-XhoI Rev | CTGCGAGCCCTCGAGTTACTCGGCAATTTTAACAATTTTACCAATTGCT | p415TEF-SWI11–38-MC |
| SpeI-SWI1–38 For Short | TCAGACCCCGTAGAAAAGATCAAAGG | p415TEF-SWI11–38-MC |
| SUP35MC-XhoI Rev Short | CTGCGAGCCCTCGAGTTACTC | p415TEF-SWI11–38-MC |
| SWI1 SRT For | TCTAACTCTACTCCGAATGCAAATC | NA |
| SWI1 SRT Rev | ACGTTGATATTAATATTGCTATTCAAGCT | NA |
| ACT1 RT For | TTGGTTATTGATAACGGTTCTGGTATG | NA |
| ACT1 RT Rev | GGTGAACGATAGATGGACCACTT | NA |
NA, not applicable.
The p415TEF-SWI11–38-MC plasmid was produced by PCR amplifying SWI11–38 from p415TEF-SWI11–38-YFP with the SpeI-SWI11–38 For and SWI11–38-BamHI-Linker Rev primers and PCR amplifying MC from p316SUP35FL with the Linker-SUP35MC For and SUP35MC-XhoI Rev primers. These two PCR products were then linked by using a mixture of both as the template and the SpeI-SWI11–38 For Short and SUP35MC-XhoI Rev Short primers, producing the full-length SWI11–38-Linker-MC product where the DPGGPGGG linker contains a BamHI site. SWI11–38-Linker-MC was subsequently cloned into p415TEF via SpeI/XhoI sites. The collection of mutant p415TEF-SWI11–38-MC plasmids was generated by cloning the mutant SWI11–38 from the respective p415TEF-SWI11–38-YFP plasmids into p415TEF-SWI11–38-MC via SacI/BamHI sites.
Site-directed mutagenesis.
The suite of SWI11–38 mutants was produced via the incorporation of base substitutions in PCR primers (Table 4) and the usage of p415TEF-SWI11–38-YFP as the template. All PCRs were conducted using PrimeSTAR HS DNA polymerase (TaKaRa Bio, Mountain View, CA, USA) according to the manufacturer’s recommended protocols. Custom primers were ordered from Integrated DNA Technologies (Coralville, IA, USA), and annealing temperatures were estimated via the Integrated DNA Technologies OligoAnalyzer tool.
Yeast transformation.
Yeast cells were transformed as previously described (37). In brief, cells were spun down at 2,500 rpm for 3 min, the supernatant was removed, and cells were resuspended in 1 ml H2O. Cells were then spun down again at 2,500 rpm for 3 min, the supernatant was removed, and cells were resuspended in 1 ml of 0.1 M lithium acetate. After 10 min, the cells were pelleted again, the supernatant was removed, and cells were resuspended in 100 μl of Li-PEG (0.1 M lithium acetate, 30% polyethylene glycol 3350 in H2O). From this mixture, 94.5 μl of resuspended cells was combined with 3.5 μl of single-stranded DNA (ssDNA) and 2.0 μl of the appropriate plasmid. The transformation mixture was then incubated at 42°C for 30 min. Thereafter, the transformation mixture was moved to ice for 5 min before spreading onto the appropriate selective medium.
Microscopy.
Images were captured using a Zeiss Axiovert 200 epifluorescence microscope with an attached camera and AxioVision AC software (Zeiss, Oberkochen, Germany). Cell samples were visualized with a 100× objective and the appropriate filters for differential interference contrast (DIC), mCherry, or yellow fluorescent protein (YFP). Images were analyzed using Fiji software (38, 39).
RT-PCR.
Yeast samples for RT-PCR were grown overnight in selective medium (3 ml). The next day, the cultures were spun down at 2,500 rpm for 5 min, and the medium was removed. The cell pellet was resuspended in 1 ml of H2O before spinning down again at 2,500 rpm for 5 min. The supernatant was once again removed, and the pellet was resuspended in 600 μl of RLT buffer from the Qiagen RNeasy minikit (Qiagen, Hilden, Germany). The resuspended cells were transferred to a screw-cap tube with silica beads, and additional RLT buffer was added to fill the tube to maximum. A Mini-Beadbeater 16 instrument (BioSpec Products, Bartlesville, OK, USA) was used to lyse the suspended cells by beating five times in 1-min intervals, with resting on ice for 1 min in between. Tubes were spun down at 8,000 × g for 15 s.
The clarified lysates were transferred to microcentrifuge tubes, and thereafter, the Qiagen RNeasy minikit protocol was followed. The RNA concentration was quantified using a Take3 microvolume plate with a Synergy HT plate reader and Gen5 software (BioTek, Winooski, VT, USA). The corresponding cDNA was synthesized using the SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA, USA). The resulting cDNA was immediately used for PCR using the SWI1 SRT For and Rev primers and the ACT1 RT For and Rev primers.
SDS-PAGE.
Yeast samples for SDS-PAGE were grown overnight in selective medium (3 ml) and prepared via alkaline lysis similarly to a method previously described (40). The following day, the optical density at 600 nm (OD600) of the cultures was measured, and a volume of culture equal to an OD600 of 2.0 was transferred to a microcentrifuge tube. Cells were pelleted at 13,000 rpm for 1 min, the medium was removed, and the cells were washed with 500 μl of ice-cold water before being pelleted again. The washed cell pellet was resuspended in 200 μl of 0.1 M NaOH and incubated at room temperature for 10 min. After another centrifugation step at 13,000 rpm for 1 min, the pellet was resuspended in 50 μl of 2× Laemmli buffer (Bio-Rad, Hercules, CA, USA). Samples were boiled for 10 min prior to loading onto a 4 to 20% Mini-Protean TGX precast protein gel (Bio-Rad, Hercules, CA, USA). After the completion of electrophoresis, samples were transferred to a polyvinylidene difluoride (PVDF) membrane using an iBlot dry blotting system (Invitrogen, Carlsbad, CA, USA).
SDD-AGE.
Yeast samples for SDD-AGE were grown overnight in selective medium (3 ml) and prepared similarly to a method previously described (41). The next day, the culture was diluted into a larger volume of selective medium (30 ml total) and grown over approximately 4 h at 30°C with shaking at 225 rpm. Yeast was harvested afterward by spinning down at 2,500 rpm for 5 min. The medium was removed, and the resulting cell pellet was washed with 10 ml of H2O. After another spin down, the H2O was removed, and 800 μl of cell lysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2, 5% glycerol, 10 mM phenylmethylsulfonyl fluoride [PMSF], cOmplete Mini protease inhibitor cocktail [Roche, Basel, Switzerland]) was added. The cell suspension was transferred to a 2.0-ml screw-cap tube filled halfway with silica beads, and additional cell lysis buffer was added to fill the tube to maximum. A Mini-Beadbeater 16 instrument was used to lyse the suspended cells by beating five times in 1-min intervals, with resting on ice for 1 min in between. The resulting samples were then used for SDD-AGE.
SDD-AGE was conducted as described previously (42). Briefly, yeast lysates were first mixed with 4× Laemmli sample buffer (2× Tris-acetate-EDTA [TAE], 20% glycerol, 8% SDS, 0.1% bromophenol blue). Samples were either incubated at room temperature for 7 min or boiled for 10 min. Samples were loaded onto 1.5% agarose–0.1% SDS gels. After the completion of electrophoresis, samples were transferred to a PVDF membrane using capillary action and 1× Tris-buffered saline (TBS).
Immunoblotting.
Membranes were blocked via incubation in 5% milk in phosphate-buffered saline (PBS) at either 4°C overnight or room temperature for 2 h. Blots were washed three times for 5 min with PBS plus 0.01% Tween 20 before probing with primary antibody for 2 h at room temperature. The following primary antibodies were used for detection: JL-8 anti-green fluorescent protein (anti-GFP) antibody (Clontech, Mountain View, CA, USA), anti-Sup35 antibody (gift from the Liebman laboratory, University of Nevada, Reno, NV, USA), or antiactin antibody clone C4 (Chemicon, Temecula, CA, USA). All primary antibodies were used at a 1:2,500 dilution. Blots were washed three times for 5 min with PBS plus 0.01% Tween 20 before probing with horseradish peroxidase-conjugated rat anti-mouse secondary antibody (Cell Signaling Technology, Danvers, MA, USA) for 1 h at room temperature. Blots were washed three times for 5 min with PBS plus 0.01% Tween 20 before incubation with the Clarity Western ECL substrate (Bio-Rad, Hercules, CA, USA). Blots were imaged using a ChemiDoc imaging system (Bio-Rad, Hercules, CA, USA).
De novo prion formation assay.
W303 sup35Δ/p316SUP35FL [PSI+] cells were independently transformed with each of the p415TEF-SWI11–38-MC wild-type and mutant constructs. Transformants were grown on −LU medium, and a color change to red was observed, indicating that the fusion proteins were functional in translational termination. Three red colonies were selected for each construct and streaked onto −L medium plus 5-FOA to select against p316SUP35FL. The resulting colonies were selected and restreaked onto both −L and −LU media to confirm the loss of p316SUP35FL. Afterward, three different colonies from each of the three sup35Δ/p415TEF-SWI11–38Mut-MC isolates were transformed with the corresponding p416TEF-SWI11–38Mut-YFP plasmids. The resulting plates of sup35Δ/p415TEF-SWI11–38Mut-MC/p416TEF-SWI11–38Mut-MC colonies were then checked for coloration and aggregation via fluorescence microscopy.
ACKNOWLEDGMENTS
We thank J. S. Weissman (University of California, San Francisco) for the W303 sup35Δ/p316SUP35FL yeast strain.
This work was supported by grants from the National Institutes of Health (R01GM110045) to L.L. and from the National Institutes of Health (R01GM126318) to Z.D.
The funders had no role in the study design, data collection and interpretation, or decision to submit the work for publication.
Contributor Information
Zhiqiang Du, Email: z-du@northwestern.edu.
Liming Li, Email: limingli@northwestern.edu.
REFERENCES
- 1.Prusiner SB. 2013. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet 47:601–623. 10.1146/annurev-genet-110711-155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liebman SW, Chernoff YO. 2012. Prions in yeast. Genetics 191:1041–1072. 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabortee S, Kayatekin C, Newby GA, Mendillo ML, Lancaster A, Lindquist S. 2016. Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc Natl Acad Sci U S A 113:6065–6070. 10.1073/pnas.1604478113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai X, Chen J, Xu H, Liu S, Jiang Q-X, Halfmann R, Chen ZJ. 2014. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156:1207–1222. 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan AH, Hochschild A. 2017. A bacterial global regulator forms a prion. Science 355:198–201. 10.1126/science.aai7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, Miller TM, Grinberg LT, Seeley WW, Diamond MI. 2014. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82:1271–1288. 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox BS. 1965. Ψ, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20:505–521. 10.1038/hdy.1965.65. [DOI] [Google Scholar]
- 8.Lacroute F. 1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol 106:519–522. 10.1128/JB.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edskes HK, Gray VT, Wickner RB. 1999. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci U S A 96:1498–1503. 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sondheimer N, Lindquist S. 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 5:163–172. 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 11.Du Z, Park K-W, Yu H, Fan Q, Li L. 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 40:460–465. 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel BK, Gavin-Smyth J, Liebman SW. 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 11:344–349. 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki G, Shimazu N, Tanaka M. 2012. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336:355–359. 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- 14.Halfmann R, Wright JR, Alberti S, Lindquist S, Rexach M. 2012. Prion formation by a yeast GLFG nucleoporin. Prion 6:391–399. 10.4161/pri.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes DL, Lancaster AK, Lindquist S, Halfmann R. 2013. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 153:153–165. 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Z, Regan J, Bartom E, Wu W-S, Zhang L, Goncharoff DK, Li L. 2020. Elucidating the regulatory mechanism of Swi1 prion in global transcription and stress responses. Sci Rep 10:21838. 10.1038/s41598-020-77993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malovichko YV, Antonets KS, Maslova AR, Andreeva EA, Inge-Vechtomov SG, Nizhnikov AA. 2019. RNA sequencing reveals specific transcriptomic signatures distinguishing effects of the [SWI+] prion and SWI1 deletion in yeast Saccharomyces cerevisiae. Genes (Basel) 10:212. 10.3390/genes10030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Z, Zhang Y, Li L. 2015. The yeast prion [SWI+] abolishes multicellular growth by triggering conformational changes of multiple regulators required for flocculin gene expression. Cell Rep 13:2865–2878. 10.1016/j.celrep.2015.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du Z, Crow ET, Kang HS, Li L. 2010. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol Cell Biol 30:4644–4655. 10.1128/MCB.00225-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crow ET, Du Z, Li L. 2011. A small, glutamine-free domain propagates the [SWI+] prion in budding yeast. Mol Cell Biol 31:3436–3444. 10.1128/MCB.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valtierra S, Du Z, Li L. 2017. Analysis of small critical regions of Swi1 conferring prion formation, maintenance, and transmission. Mol Cell Biol 37:e00206-17. 10.1128/MCB.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158. 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toombs JA, McCarty BR, Ross ED. 2010. Compositional determinants of prion formation in yeast. Mol Cell Biol 30:319–332. 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachmair A, Finley D, Varshavsky A. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179–186. 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 25.Du Z. 2011. The complexity and implications of yeast prion domains. Prion 5:311–316. 10.4161/pri.5.4.18304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 40:1357–1369. 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 27.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268:880–884. 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 28.Teunissen AW, Steensma HY. 1995. Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11:1001–1013. 10.1002/yea.320111102. [DOI] [PubMed] [Google Scholar]
- 29.Willaert RG. 2018. Adhesins of yeasts: protein structure and interactions. J Fungi (Basel) 4:119. 10.3390/jof4040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, Gaur NK, Lipke PN. 2006. Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot Cell 5:1664–1673. 10.1128/EC.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul KR, Hendrich CG, Waechter A, Harman MR, Ross ED. 2015. Generating new prions by targeted mutation or segment duplication. Proc Natl Acad Sci U S A 112:8584–8589. 10.1073/pnas.1501072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sant’Anna R, Fernández MR, Batlle C, Navarro S, de Groot NS, Serpell L, Ventura S. 2016. Characterization of amyloid cores in prion domains. Sci Rep 6:34274. 10.1038/srep34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newby GA, Lindquist S. 2017. Pioneer cells established by the [SWI+] prion can promote dispersal and out-crossing in yeast. PLoS Biol 15:e2003476. 10.1371/journal.pbio.2003476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khatri I, Tomar R, Ganesan K, Prasad GS, Subramanian S. 2017. Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Sci Rep 7:371. 10.1038/s41598-017-00414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue JX, Li J, Aigrain L, Hallin J, Persson K, Oliver K, Bergström A, Coupland P, Warringer J, Lagomarsino MC, Fischer G, Durbin R, Liti G. 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet 49:913–924. 10.1038/ng.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar AN, Gorter de Vries AR, van den Broek M, Brouwers N, de la Torre Cortès P, Kuijpers NGA, Daran J-MG, Abeel T. 2019. Chromosome level assembly and comparative genome analysis confirm lager-brewing yeasts originated from a single hybridization. BMC Genomics 20:916. 10.1186/s12864-019-6263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amberg DC, Burke DJ, Strathern JN. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kushnirov VV. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857–860. . [DOI] [PubMed] [Google Scholar]
- 41.Fan Q, Park KW, Du Z, Morano KA, Li L. 2007. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177:1583–1593. 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halfmann R, Lindquist S. 2008. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J Vis Exp 2008:838. 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]