Abstract
Mitochondria are increasingly being recognized as information hubs that sense cellular changes and transmit messages to other cellular components, such as the nucleus, the endoplasmic reticulum (ER), the Golgi apparatus, and lysosomes. Nonetheless, the interaction between mitochondria and the nucleus is of special interest because they both host part of the cellular genome. Thus, the communication between genome-bearing organelles would likely include gene expression regulation. Multiple nuclear-encoded proteins have been known to regulate mitochondrial gene expression. On the contrary, no mitochondrial-encoded factors are known to actively regulate nuclear gene expression. MOTS-c (mitochondrial open reading frame of the 12S ribosomal RNA type-c) is a recently identified peptide encoded within the mitochondrial 12S ribosomal RNA gene that has metabolic functions. Notably, MOTS-c can translocate to the nucleus upon metabolic stress (e.g., glucose restriction and oxidative stress) and directly regulate adaptive nuclear gene expression to promote cellular homeostasis. It is hypothesized that cellular fitness requires the coevolved mitonuclear genomes to coordinate adaptive responses using gene-encoded factors that cross-regulate the opposite genome. This suggests that cellular gene expression requires the bipartite split genomes to operate as a unified system, rather than the nucleus being the sole master regulator.
Keywords: gene regulation, metabolism, mitochondria, mitochondrial-derived peptides, mitochondrial open reading frame of the 12S ribosomal RNA type-c (MOTS-c), mitonuclear communication, stress responses
1. Introduction
Eukaryotic cells are multigenomic by virtue of the unique union forged one to two billion years ago between our ancestral cell (thought to have derived from the archaea Lokiarchaeota[1]) and free-living bacteria (thought to originate from a proteobacterial lineage[2]).[3,4] Multiple prokaryotic signatures are still present in the mitochondria, including a circular genome and the usage of a distinct genetic code. The mitochondrial and nuclear (mitonuclear) genomes have coevolved, continuously adapting to each other to establish a unified dual-genomic cellular system. The survival advantage of a dual genome over a single genome, in which the genetic information is consolidated in the nucleus, is unclear. From an economic and administrative perspective, a single centralized genome seems more sensible, especially considering that lateral transfer of mitochondrial DNA (mtDNA) to the nuclear genome occurred multiple times, as evidenced by the degenerate copies of mtDNA sequences scattered throughout the nuclear genome.[5] Also, only 13 protein-coding genes remain hosted in the mtDNA, entailing that >98% of mitochondrial proteins are encoded in the nuclear genome. However, the recent discovery of small peptides or microproteins that are encoded in the mtDNA and have regulatory roles[6] indicates a more complex mitochondrial transcriptome.[7] Such mitochondrial-derived peptides (MDPs) are encoded in the mtDNA as short open reading frames (sORFs) and have various biological actions.[6,8] We have recently reported that one particular MDP, MOTS-c (mitochondrial open reading frame of the 12S ribosomal RNA type-c), can translocate to the nucleus to regulate adaptive gene expression in response to cellular stress.[9–12] Here, we discuss the implications of MOTS-c, a mitochondrial-encoded regulator of the nuclear genome, and propose that the dual mitonuclear genomes should be considered a singular genetic system that is coordinated through dynamic intergenomic communication using reciprocal gene-encoded regulators. We suggest a more inclusive approach to gene network analyses for basic research and therapeutic development.
2. How Do Mitochondria Communicate?
A cell must constantly adapt to the ever-changing environment to maintain homeostasis. Such dynamic responses require a highly coordinated network of processes and signals that are largely compartmentalized to specific organelles and subcellular complexes. To orchestrate the vastly complex cellular system, communication between organelles is paramount.[13] Mitochondria act as a cellular hub that senses and integrates environmental changes, and as a coordinator of adaptive responses through constant communication with other cellular compartments,[14] including the nucleus, regulate key cellular processes that have a significant impact on health span and life span.[4,15–33] Many of the known mitochondrial communication mechanisms include those that respond to cellular stress and help to maintain cellular homeostasis.[4,15,33] Some prominent pathways include the mitochondrial unfolded response (UPRmt), whereby mitochondrial perturbations activate stress-responsive transcriptional responses in the nucleus via factors, such as activating transcription factor associated with stress-1 (ATFS-1) in Caenorhabditis elegans (C. elegans) and ATFS-5 in mammals,[34,35] reactive oxygen species (ROS) signaling that has a broad cellular impact including nuclear gene regulation,[23] and mitochondrial damage-associated molecular patterns that consist of molecules released from injured mitochondria.[36,37] Well-described mediators of mitochondrial retrograde communication can be largely categorized as i) primary metabolites (e.g., acetyl-CoA),[38,39] secondary metabolites (e.g., ROS),[40,41] transient molecules (e.g., Ca2+),[42] damaged mitochondrial components (e.g., fragmented mtDNA),[43,44] or nuclear-encoded proteins (e.g., cytochrome c).[45] However, none of these signals, despite their importance, are inherently encoded in the mtDNA. We hypothesize that cellular homeostasis is maintained through a balancing act between regulators independently encoded in each genome, unifying the dual genomes to operate as a single genetic system. MDPs such as MOTS-c may represent archaic communication peptides that are transmitted to deliver specific messages to the nucleus[9] and perhaps other subcellular compartments.
3. The Instructions for Life Are Split and Encoded in Two Separate Genomes
3.1. The Coevolution of Two Genomes in Eukaryotic Cells
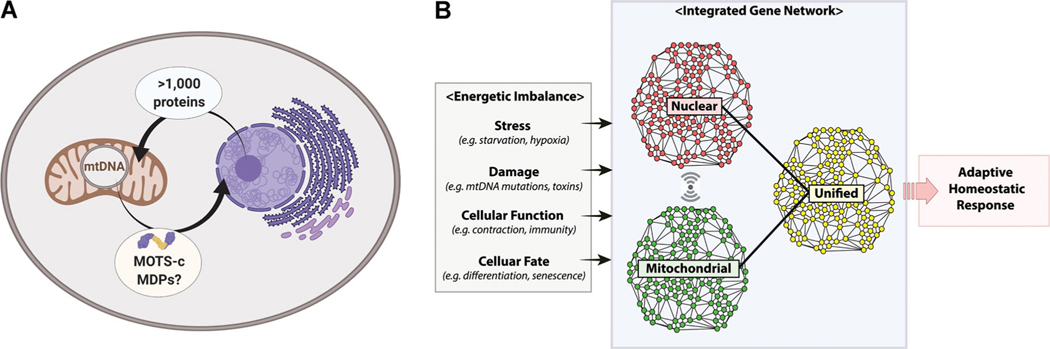
Eukaryotic cells are composed of various compartmentalized organelles that must closely function together to maintain homeostasis and survive.[13] Organellar coordination is possible through a tight network of communication mechanisms. Of the various organelles, the communication between mitochondria and the nucleus is special in that they both possess independent genomes. Owing to their endosymbiotic bacterial ancestry, mitochondria presumably retained part of their prokaryote-derived genome in coevolution, first with the ancestral archaeal genome and later the nuclear genome. This suggests that an ancient bidirectional communication system encoded in both genomes may have been the basis of eukaryotic cellular gene regulation, in addition to other possibilities, such as redox regulation of gene expression.[46] However, whereas the nuclear genome is known to encode over 1000 proteins that regulate the mitochondria, there were no known factors encoded in the mtDNA with active roles in regulating the nuclear genome. In fact, all 13 proteins encoded in the mtDNA are structural components of the electron transport chain (ETC) with no evident regulatory roles.
For the past one to two billion years, mitochondria have retained their prokaryotic circular genome that is equipped with a unique genetic system.[3,4] Although the exact period when a fully developed nucleus emerged is still unclear,[47,48] the mitochondrial and nuclear genomes have been coevolving to coordinate complex biological networks at the cellular and organismal levels. The evolutionary benefits of operating cells based on a bigenomic system, which would be energetically unfavorable compared to a unigenomic system, are unclear. Further, coordinating gene expression between two genomes adds regulatory complexity compared to an operation based on a single consolidated genome. This is especially puzzling as lateral gene transfer from mitochondrial genomes to the nuclear genome has evidently taken place over time[49] and is observed even today in cancer cells.[50] Such highly homologous, yet degenerate, mitochondrial sequences within the nuclear genome, collectively referred to as nuclear mitochondrial transfer,[5] provide strong evidence that genetic consolidation in the nucleus was, and is, possible. The question then remains on the survival advantage that a dual-genome system confers and the regulatory mechanisms underlying bigenomic coupling.
The human mtDNA encodes for 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), 13 proteins (all structural components of the respiratory chain complexes), and multiple small peptides (see Section 4). Over 1000 proteins that are required for diverse mitochondrial functions are encoded in the nuclear genome and imported into the mitochondria. With 13 mitochondrial-encoded proteins involved in oxidative phosphorylation (OXPHOS), nuclear-encoded proteins constitute more than 98% of the currently known mitochondrial proteome. Some nuclear-encoded mitochondrial proteins synthesized by mitochondria-associated cytoplasmic ribosomes are cotranslated and translocated into the mitochondria.[51,52] Interestingly, switching yeast to a different carbon source triggers a synchronous mitochondrial and cytosolic translation program, especially for complex III and IV subunits.[53] The orchestration of mitonuclear translation is likely layered and context-specific. Also, because of insufficient tRNAs encoded within mtDNA, a range of cytosolic (nuclear-encoded) tRNAs must be imported in many species,[54] including tRNAGln in humans.[55]
Although smaller in size compared to its nuclear counterpart, mutations in mtDNA are linked to several diseases, including MELAS (mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes), cancer, and drug-induced deafness.[56,57] mtDNA exists in high copy numbers, often reaching hundreds to thousands of copies.[58] Thus, a single mitochondrion can frequently harbor a mix of wild-type and mutant mtDNA, a phenomenon known as heteroplasmy.[59] mtDNA point mutations and deletions increase with age and drive heteroplasmy,[60–68] which is thought to underlie various pathophysiological aspects of aging. The mitochondrial free radical theory of aging posits that ROS, formed by free radicals generated during mitochondrial respiration, promote mtDNA mutations due to their proximity and relatively primitive mtDNA repair mechanism.[69] However, recent reports have shown that the major source of mtDNA point mutations in aged tissues arises from replication infidelity (i.e., DNA polymerase errors) rather than ROS-induced damage.[66–68] Thus, ROS-induced mtDNA damage does not seem to increase with age. In fact, mtDNA are i) protected by tightly bound protein clusters (i.e., nucleoids) and ii) positioned more distal to ROS-producing ETC complexes than previously thought.[70–72] So, if not mtDNA damage per se, how do mitochondria contribute to the regulation of life span and/or health span? ROS are pleiotropic. They cause oxidative stress at higher concentrations (pathological), but act as signaling molecules at lower levels (physiological).[73] In fact, physiological ROS response has been suggested as eustress[74] that increases adaptive responses to promote health and extend life span, referred to as mitohormesis.[75] Increasing interest in ROS as signaling molecules that regulate normal physiological processes has provided insight into its role in regulating life span and/or health span.[19,23,74] In fact, ROS is likely a component of a broader integrated mitochondrial signaling program elicited in response to mtDNA damage accumulation. Yet, how the level of age-dependent mtDNA mutations and heteroplasmy is sensed and communicated by mitochondria is an area of ongoing research.
3.2. Mitonuclear Epistasis: Transgenomic Interactions Are Important for Cellular Fitness
The importance of mitonuclear genomic compatibility in cellular fitness has been demonstrated in various organisms, even in closely related species. In yeast, artificially coupling incompatible mitonuclear genomes of two species (Saccharomyces bayanus and Saccharomyces cerevisiae) caused the hybrids to be sterile.[76] In flies, mtDNA introgression (i.e., Drosophila simulans mtDNA on a Drosophila melanogaster nuclear genome background) showed variable life spans, in which some combinations were long-lived while others were short-lived, indicating the importance of mitonuclear genomic compatibility on gene expression.[77] Also, mitonuclear mismatch in flies affected the expression of nuclear genes in a sex-dependent manner, plausibly due to the maternal inheritance of mtDNA.[78] Further, polymorphisms in a mitochondrial-encoded tRNA and a nuclear-encoded tRNA synthetase, which are independently silent, cause significant developmental and reproductive defects in flies when combined.[79] In worms, association mapping in a panel of hybrids, generated by a cross between C. elegans strains (N2 and HW), indicated that the quantitative trait of aging may depend on genome-wide quantitative variations in mitonuclear genomic compatibility.[80] Interspecies cytoplasmic–nuclear hybrids (cybrids) within Caenorhabditis briggsae showed that mitonuclear discord can reduce fecundity, increase adiposity, and elevate ROS levels.[81] Chimeric cybrids generated from mouse cells that harbor rat mitochondria showed greatly reduced respiration even though rat mtDNA could replicate and translate mitochondrial-encoded proteins, indicating subtle but nontrivial mitonuclear incompatibility.[82] Mitochondrial nuclear exchange (MNX) mice, in which mitonuclear genomes derived from different strains of mice are cross-paired, demonstrate the significance of intergenomic compatibility in organismal fitness.[83,84] MNX mice exhibit altered cellular metabolism and oxidative stress levels and resistance to cardiac damage and an atherogenic diet.[84,85] In addition, mtDNA replacement between conplastic mice (i.e., Mus musculus domesticus and Mus musculus musculus) can cause varying degrees of sex-dependent infertility to embryonic loss and stillbirths.[86] These observations support functional mitonuclear genome incompatibility as a key factor to reproductive fitness and a contributor to reproductive isolation. In humans, the impact of mitonuclear incompatibility on health and life span is an ongoing field of research, but it has been reported to affect certain disease phenotypes, including cardiomyopathy and insulin resistance.[87] Notably, specific nuclear gene expression can also be affected by specific mitochondrial variants. A mutation in the adenine nucleotide translocase (ANT1) gene, which transports adenosine diphosphate (ADP)/ adenosine triphosphate (ATP) in mitochondria, can cause autosomal-recessive cardiomyopathy. However, the ANT1 mutation can differentially manifest depending on the mtDNA it resides with.[88] Further, discordant mitochondrial and nuclear protein subunits of the respiration complexes may underwrite mitonuclear incompatibility;[89,90] the subunits of ETC complexes are encoded in both mitochondrial and nuclear genomes, and a suboptimal match will compromise energy production and other mitochondrial functions and even impact speciation.
4. Regulatory Peptides Encoded in the Mitochondrial Genome: New Ancient Cellular Alphabets?
The discovery of humanin (HN), an sORF encoded within the mitochondrial 16S rRNA locus,[91] suggested a more complex bacterial-like polycistronic mitochondrial genome that encodes for short gene-within-genes, adding an entirely novel layer to mitochondrial signaling and gene expression.[27] The need for intracellular and endocrine messages to be encoded in the mitochondrial genome is just beginning to be unveiled. Nonetheless, considering that i) hundreds to thousands of mitochondria can exist in a given cell, ii) mitochondria are dynamic (fusion and fission), and iii) are mobile, it would be challenging for the nucleus to keep track of each individual mitochondrion without inherent mitochondrial retrograde signals to initiate the communication. Peptides, or microproteins, encoded in the nuclear and mitochondrial genomes are increasingly being identified, owing to technological and computational advances[92–94] that aid in the discovery of sORFs that fell short of the minimum 100-amino-acid (a.a.) cutoff for a bona fide protein-coding gene during earlier genome annotation predictions.[95,96] Together with the fact that the mitochondrial transcriptome is far more complex and richer in RNA species than previously thought,[97] the identification of additional MDPs is very likely and will provide a higher resolution of the mitochondrial proteome. To date, eight different MDPs have been reported, including HN,[91,98,99] SHLP1–6 (small HN-like peptides 1–6),[100] and MOTS-c.[101] Although the genetics of MDP expression is currently being investigated by several groups, it is clear that each MDP has distinct and redundant functions.[102]
4.1. Humanin
HN was the first MDP to be identified. It was discovered in 2001 from an intact occipital lobe of an Alzheimer’s disease (AD) patient as a 75 bp ORF sequence within the mitochondrial 16S rRNA gene yielding a 24-a.a. peptide.[91,99,103] Functional studies have shown HN to be protective against AD-related toxins.[91,99] HN was then cloned by a second group as a binding partner of the insulin-like growth factor-binding protein-3, which regulates the bioavailability of IGF-1 via its heparin-binding domain.[98] We subsequently showed that circulating HN levels are under the regulation of IGF-1, hence making a connection between MDPs and aging.[104] HN also binds to Bax, which triggers apoptosis once it moves to the mitochondria. HN keeps Bax from translocating to the mitochondria, thereby acting as an antiapoptotic factor.[99] The cytoprotective effects of HN have been demonstrated in various pathophysiological conditions, including the brain,[105–110] the cardiovascular system,[111–118] eyes,[114,119–121] bone,[122,123] metabolism,[124,125] and cellular senescence.[126]
4.2. Small Humanin-Like Peptides
In addition to HN, the 16S rRNA hosts six additional peptides (20–38 a.a.), named SHLP1–6.[100] SHLP2 and SHLP3 were cytoprotective, whereas SHLP6 promoted cellular apoptosis. Akin to HN, SHLP2 protected primary mouse cortical neurons from β-amyloid (Aβ1–42) toxicity. SHLP2 and SHLP3 accelerated insulin-dependent adipocyte differentiation. However, the intracerebroventricular treatment of SHLP2, but not SHLP3, enhanced insulin responsiveness as indicated by increased glucose clearance and suppressed hepatic glucose production. This indicates that SHLP2, but not SHLP3, may affect hypothalamic response to peripheral insulin. However, in cells, both SHLP2 and SHLP3 enhanced mitochondrial respiration and ATP production, indicating distinct metabolic roles for these peptides in vivo. Notably, SHLP2 and HN can bind to and prevent the misfolding of islet amyloid polypeptide, which is implicated in type 2 diabetes (T2D) mellitus.[127] Further, circulating SHLP2 levels were negatively correlated with prostate cancer risk in white men, but not in black men; circulating SHLP2 levels above 350 pg mL–1 ruled out prostate cancer in both races with >95% accuracy, indicating a key role in the development and racial disparity of prostate cancer.[128]
4.3. Mitochondrial Open Reading Frame of the 12S rRNA Type-c
MOTS-c is a 16-a.a. peptide that is encoded within the mitochondrial 12S rRNA gene as a 51 bp ORF. MOTS-c translation obligatorily occurs in the cytoplasm using the standard genetic code because mitochondrial translation, using the mitochondria-specific genetic code, will face tandem start and stop codons.[101] Because selective depletion of mitochondrial RNA causes a time-dependent loss of MOTS-c,[101] it is plausible that the MOTS-c transcript is generated in the mitochondria, which is then exported to the cytosol by an unknown mechanism. Interestingly, MOTS-c localizes to the mitochondria under resting conditions.[9,101] As described earlier, some mitochondrial proteins synthesized by mitochondrial-associated cytoplasmic ribosomes are cotranslated and translocated into the mitochondria.[51,52] It is possible that the exported MOTS-c transcript may be cotranslated and translocated to the mitochondria using such mitochondrial-associated cytoplasmic ribosomes. Work is also in progress to understand cellular cues that induce MOTS-c expression, and metabolic alterations[129–133] and inflammation[134,135] may be key. Similar to HN, MOTS-c has both intracellular and endocrine roles.[25,101] Its role as a regulator of adaptive nuclear gene expression will be discussed in detail in Section 5. Initially, MOTS-c was identified while screening, both genetically and pharmacologically, for metabolic regulators in human cells.[101] Indeed, multiple studies have confirmed its metabolic actions in various pathophysiological conditions (Table 1). In cells, MOTS-c increased glucose uptake and glycolysis in a 5′adenosine monophosphate-activated protein kinase (AMPK)dependent and sirtuin-1-dependent manner.[101] In mice, MOTS-c targeted skeletal muscle to improve insulin sensitivity and i) prevented diet-induced obesity and insulin resistance and ii) reversed age-dependent muscle insulin resistance.[101] In humans, circulating MOTS-c levels were significantly lower (20.3%) only in obese male children/adolescents (5–14 years) of Chinese descent but not in their female counterparts.[131] Further, the male cohort exhibited a negative correlation between MOTS-c and body mass index, waist circumference, waist-to-hip ratio, fasting insulin level, homeostasis model assessment of insulin resistance (HOMA-IR), and glycated hemoglobin (HbA1c). Another study in adults (31–38 years) showed that plasma MOTS-c levels were comparable between lean and obese subjects, but only the lean cohort showed a significant (anti)correlation between circulating MOTS-c and blood lactate (r = 0.65), HOMA-IR (r = 0.75), as well as the Matsuda index (r = –0.65).[130] MOTS-c also plays a role in endothelial health. Coronary endothelial function was significantly correlated with circulating MOTS-c levels in humans.[134] Notably, MOTS-c improved mouse and rat endothelial function in vitro.[134] Further, in a mouse ovariectomy model, MOTS-c improved i) metabolic function by reducing adipose accumulation[144] and ii) bone density by inhibiting osteoclast formation in an AMPK-dependent manner.[137] However, MOTS-c can also improve bone density by promoting the differentiation of bone mesenchymal stem cells (BMSCs) to osteoblasts via the transforming growth factor-β (TGF-β)/Smad pathway.[140]
Table 1.
Summary of Literature on MOTS-c
| Author | Year | Ref | Main Findings | Model |
|---|---|---|---|---|
| Lee, C. et. al. | 2015 | [101] | Identification of MOTS-c as a mitochondrial-encoded regulator of metabolic homeostasis. Reverses age-dependent muscle insulin resistance and diet-induced obesity/insulin resistance in mice. Regulates cellular glucose, nucleotide, and fatty acid metabolism | Mammalian cells, Mice |
| Fuku, N. et. al. | 2015 | [180] | A biological link between a MOTS-c polymorphism (m.1382A>C; K14Q variant) and exceptional longevity in a Japanese population | Humans |
| Ming, W. et. al. | 2016 | [137] | Inhibition of osteoporosis via MOTS-c-regulated osteoclastogenesis in an AMPK-dependent manner. | Mammalian cells, Mice |
| Zempo, H. et. al. | 2016 | [129] | Correlation between MOTS-c variant (m.1382A>C; K14Q variant) and diabetes in Japanese men. | Humans |
| Thevis, M. et. al. | 2016 | [181] | Development of qualitative identification of MOTS-c using mass spectrometry. | Mass Spectrometry |
| Zhai, D. et. al. | 2017 | [139] | Therapeutic efficacy of MOTS-c against MRSA | Mammalian cells, Mice |
| Cataldo, L.R. et. al. |
2018 | [130] | Positive association between circulating MOTS-c and insulin sensitivity in lean, but not obese, (adult) individuals | Humans |
| Du, C. et. al. | 2018 | [131] | Reduced circulating MOTS-c levels in obese male children/adolescents and negative correlation to insulin resistance and obesity. | Humans |
| Hu, B.T. et. al. | 2018 | [138] | Bone mesenchymal stem cell (BMSC) differentiation to osteoblasts by MOTS-c via the TGF-β/Smad pathway. | Mammalian cells |
| Kim, K.H. et. al. | 2018 | [9] | Stress-responsive nuclear translocation of MOTS-c and direct regulation of adaptive gene expression in the nucleus. | Mammalian cells |
| Qin, Q. et. al. | 2018 | [134] | Positive correlation between circulating MOTS-c levels and coronary endothelial function. MOTS-c improves endothelial function in vitro. | Rats, Humans |
| Che, N. et. al. | 2019 | [182] | Collagen (type 1) synthesis promotion by MOTS-c in osteoblasts via the TGF-β/Smad pathway. | Mammalian cells |
| Knoop, A. et. al. | 2019 | [183] | Development of a mass spectrometry-based method to detect MOTS-c in plasma samples for doping control purposes. | Mass Spectrometry |
| Li, Q. et. al. | 2019 | [184] | Prevention of fat accumulation in D-galactose-treated mice by MOTS-c. | Mice |
| Lu, H. et. al. | 2019 | [136] | Prevention of ovariectomy-induced fat accumulation and insulin resistance and regulation of adipose tissue homeostasis by MOTS-c. | Mammalian cells, Mice |
| Raijmakers, R.P. et. al. |
2019 | [135] | Differential expression of MOTS-c in circulating monocytes of Q fever fatigue syndrome (GFS) patients. | Humans |
| Ramanjaneya, M. et. al. |
2019 | [132] | Significantly reduced serum MOTS-c levels in type 2 diabetes (T2D) patients. Serum MOTS-c negatively correlates with HbA1c and age. | Humans |
| Ramanjaneya, M. et. al. |
2019 | [133] | Lipid infusion significantly increases circulating MOTS-c levels, which is negated by insulin infusion. | Humans |
Interestingly, MOTS-c has a significant impact on inflammatory pathways. DNA microarray analysis on HEK293 cells treated with MOTS-c showed substantial anti-inflammatory gene signatures[101] and mice systemically treated with MOTS-c showed reduced levels of circulating interleukin-6 and tumor necrosis factor-α.[101,139] Notably, in a 1983 study, the majority of complementary DNA clones (>75%) from interferoninduced human monoblast cells (KG-1) were mapped to the mitochondrial 12S and 16S rDNA loci, although no specific genes were identified at that time.[145] We referenced this work during the early stages of MOTS-c identification.[101] Consistent with this work, we recently reported that MOTS-c significantly downregulated interferon-related genes under metabolic stress.[9] The role of MOTS-c may fall in the interface between immune and metabolic, or immunometabolic signaling.
5. MOTS-c: a Mediator in a Two-Way Genomic Dialogue?
5.1. Mitochondria Are Regulators of Nuclear Gene Expression
Mitochondria dynamically sense the constantly changing intra- and extracellular environmental milieu, including substrate/nutrient availability and oxygen levels, and relay messages to other subcellular compartments, including the nucleus. The perturbation of cellular homeostasis, either because of increased cellular functions that change the energetic demand and balance (e.g., skeletal muscle during exercise) or environmental challenges (e.g., starvation), can trigger mitochondria to propagate distress calls. In that sense, mitonuclear communication is uniquely important in that it occurs between organelles that possess independent genomes, indicating that gene regulation is a likely endpoint for both. Mild mitochondrial perturbation in C. elegans can extend the life span by activating UPRmt that remodels chromatin through histone modifications, including H3K9 methylation by the histone methyltransferase MET-2 and the nuclear cofactor LIN-65[146] and H3K27 demethylation by histone demethylases (jmjd-1.2 and jmjd-3.1).[147] Multiple proteins that are nuclear-encoded reside in the mitochondria, which then translocate to the nucleus in response to various stimulations. This is reminiscent of a reconnaissance factor of sorts that reports back information to the nucleus that mitochondria sense. ATFS-1 is a duallocalizing basic leucine zipper transcription factor, which contains both a mitochondrial targeting sequence and a nuclear localization sequence in C. elegans.[148] ATFS-1 is imported into mitochondria under resting conditions, where it gets degraded by the Lon protease. However, under stress conditions mitochondrial import of ATFS-1 is reduced, causing it to translocate to the nucleus.[148] In the nucleus, ATFS-1 promotes the expression of mitochondrial stress-adaptive genes that are involved in protein folding, glycolysis, and antioxidant pathways.[148] In mammals, ATF5, a functional ortholog of ATFS-1, mediates UPRmt.[149] There are multiple other nuclear-encoded mitochondrial-targeted proteins that can also reside in the nucleus as a means to transmit mitochondrial signals.[150] ROS can also lead to epigenetic alternations.[151,152] ROS can regulate the chromatin-binding capacity of histone demethylase Rph1p, thereby extending the chronological life span in yeast.[153] In addition, mitochondrial metabolites, such as tricarboxylic acid (TCA) cycle intermediates, provide various substrates required for epigenetic modification of chromatin (e.g., acetyl-CoA for histone acetylation) and consequently regulate the gene expression.[154] Notably, some TCA cycle enzymes can translocate to the nucleus and provide an onsite synthesis of metabolite substrates for epigenetic remodeling.[155]
5.2. MOTS-c: Mitochondrial-Encoded Regulators of Nuclear Gene Expression?
As mentioned above, mitochondria originate from endosymbiotic bacteria. It is not unusual to find endosymbiotic bacteria even today, but receiving tenure as an organelle is undoubtedly an exceptional event. Communication between endosymbiotic bacteria and the archaeal ancestral cell is likely to have played a crucial role in coordinating the extraordinary merger. It is plausible that each entity attempted to communicate using an already existing mechanism encoded within their genome. Further, it is likely that such communication mechanisms reciprocally targeted gene expression. Even today, bacteria communicate to other bacteria using gene-encoded peptides or small molecules that are synthesized by gene-encoded processes (e.g., quorum sensing and niche protection).[156–158] Notably, many bacterial communicators target key cellular processes, such as ribosome synthesis (translation inhibition), cell membrane synthesis, and DNA synthesis,[159] setting up a foundation for complex eukaryotic cellular regulators. If such factors were crucial for effective communication not only during the endosymbiotic period and the establishment of mitochondria, but also throughout eukaryotic evolution, the selection pressure would have been strong. Also, sORFs can evolve faster[160,161] and are less conserved[161,162] than larger canonical genes. This suggests that sORFs support species-specific adaptive evolution and fine-tuning of gene regulation,[163] especially considering that they yield peptides with specific functions.[161] On this line, MDPs presumably derived from archaic communication mechanisms originally encoded within the protomitochondrial bacterial genome and have been selected or evolved over time for functional adaptation. One such function of MDPs could be nuclear gene regulation, especially because the communication between genome-containing organelles likely involves cross-genomic regulation. However, whereas “nucleus-to-mitochondria” gene regulation is known, active and direct “mitochondria-to-nucleus” gene regulation has been largely unknown.
In resting conditions, MOTS-c is largely extranuclear and colocalizes mainly to the mitochondria.[9,101] However, MOTS-c can rapidly translocate to the nucleus within 30 min upon metabolic or oxidative stress in an AMPK-dependent manner.[9] AMPK was also required for the physiological effects of MOTS-c,[101,137,144] suggesting that these effects may have been exerted, in part, through nuclear gene regulation. Further, the hydrophobic core, but not the cationic tail, was required for MOTS-c to enter the nucleus,[9] indicating potential interaction with other proteins.[164] In the nucleus, MOTS-c was able to bind to i) chromatin using its hydrophobic and cationic regions and ii) adaptive stress response transcription factors, including nuclear factor erythroid 2-related factor 2 (NFE2L2/NRF2) and activating transcription factor-1 (ATF1).[9] Using RNA sequencing, we found that MOTS-c significantly regulated 802 genes in response to glucose restriction.[9] Further, motif analysis in the promoters of regulated genes revealed a number of putative coregulators of MOTS-c nuclear actions, including motifs related to the interferon signaling pathway.[9] Ultimately, MOTS-c protected cells from glucose and serum restriction, an in vitro model of energetic stress driven by a disrupted supply and demand balance.[9] In summary, metabolic stress triggered MOTS-c to dynamically translocate to the nucleus, bind to chromatin, and regulate nuclear gene expression to regain homeostasis (Figure 1A). Although MOTS-c may be the first mitochondrial-encoded message to be identified that is sent to the nucleus to mount an adaptive stress response, considering the redundant nature of various cellular signaling pathways, it may be a token of multiple MDPs with diverse nuclear roles.
Figure 1.
Mitonuclear genome cross-regulation and the composite gene network. A) The mitochondrial proteome consists of over 1000 proteins that are encoded in the nuclear genome. On the contrary, the nuclear proteome was devoid of regulatory factors encoded in the mitochondrial genome. MOTS-c is the first MDP that has been shown to enter the nucleus and directly engage in adaptive nuclear gene expression. MOTS-c may be a harbinger of many other MDPs with nuclear roles. This notion would be consistent with the observation that the coevolved mitonuclear genomes independently encode for reciprocal gene-regulatory factors to coordinate gene regulation. B) In line with (A), eukaryotic gene networks are dynamically built based on a unified, yet split, bipartite genome compartmentalized in the nucleus and mitochondria. The unified mitonuclear gene network may provide a richer cellular context during adaptive gene responses to diverse pathophysiological cellular stimuli (stress, damage, function, fate) that are often accompanied by energetic imbalance. Thus, integrated gene analyses that account for the interaction of the two genomes would provide a more comprehensive biological perspective.
Notably, in yeast, the nuclear 25S rRNA gene encodes a peptide, Tar1p, that acts in the mitochondria.[165,166] Tar1p has been suggested to suppress the formation of extrachromosomal ribosomal circles,[167] which accumulate in yeast with mitochondrial deficiency via the retrograde response protein[168] and replicative aging.[169] Although no homolog of Tar1p has been identified in humans, the fact that both mitonuclear rDNAs encode for peptides that function in opposing subcellular compartments suggests a general paradigm where these types of peptides may sense and cross-regulate ribosomal functions.[10] This is especially interesting considering that mitonuclear translation is tightly coordinated, as mentioned above.[53] Further, translation is a highly energy-dense process that is subject to regulation under metabolic stress,[170] a condition that triggers MOTS-c to migrate to the nucleus. Thus, MOTS-c may have multiple roles in the nucleus, including ribosome metabolism and assembly in response to stress.
6. What Does a Dual-Genome System Mean for Health?
6.1. Our Dual Genomes Are Programmed for a Singular Unified Genetic Network
Traditionally, the nucleus was thought to host all gene-encoded regulators for both genomes, providing an image of a nuclear-centric unidirectional communication between the mitonuclear genomes. However, the recent discovery of the nuclear actions of MOTS-c suggests that gene expression can be coordinated by factors encoded in each genome that regulate the other genome (Figure 1A). A possible role for MDPs would be to fine-tune the adaptation of mitonuclear gene expression (Figure 1B). Therefore, the ability of a cell to differentiate between subtly distinct and complex cellular cues (e.g., stress response, differentiation, and proliferation) and to precisely mount the appropriate response is dependent on the detailed coordination of the dual genomes, and in effect, MDPs, such as MOTS-c, may present a mitochondrial context to the nucleus.
In bioinformatic analyses, it is currently a common practice to analyze datasets (e.g., chromatin immunoprecipitation sequencing and assay for transposase accessible chromatin with high-throughput sequencing) only after filtering out repetitive sequences, including key mitochondrial sequences of interest (e.g., those encoding MDPs). Indeed, any ChIP- or ATAC-enriched regions identified on the mitochondrial chromosome are usually discarded as spurious findings or even used to estimate the background noise of the method.[171] However, growing evidence suggests that some nuclear factors may be imported into the mitochondria and bind mtDNA in a sequence-specific manner (e.g., JunD, MafK, and Rfx5)[172] (Figure 1A). In addition, sequence motifs seemingly bound by nuclear transcription factors in the mitochondria show evidence of negative selection, thus supporting their functionality.[173] Together, these observations support the existence of a complex dialogue between the nuclear and mitochondrial genomes, far more involved and dynamic than previously estimated. Because of the sheer amount of rRNA sequences in cellular transcriptomes (usually 95% of total RNAs), rRNA depletion of both nuclear and mitochondria-encoded sequences has been adopted as a cost-effective measure to analyze gene expression regulation.[174,175] In addition, any repetitive sequences remaining in RNA-sequencing datasets are usually eliminated from downstream analyses as well. Because of this experimental analytical pragmatism, very little is known about the differential regulation of mitochondrial rRNA sequences (encoding many known MDPs), and more largely of mitochondrial genomes, as well as their impact on cellular physiology. Together, this suggests that appropriate tools and pipelines are still missing to fully grasp the importance and prevalence of bidirectional genomic communication between the nuclear and mitochondrial compartments.
As mentioned above, mitonuclear epistasis can have profound effects on cellular and organismal fitness. Artificially placing entire mitochondrial and nuclear genomes together, which have a wide range of nucleotide variants, can cause significant cellular dysfunction and even death.[89] However, specific nuclear gene expression can also be affected by specific mitochondrial variants. For instance, a mutation in the ANT1 gene can manifest as a cardiomyopathy of differing degrees depending on the specific mtDNA variant with which it is coupled.[88] Thus, mitonuclear linkage disequilibrium (LD) may provide a more detailed and comprehensive intergenomic interaction map. Although mitonuclear LD in humans is currently largely unexplored, a recent analysis indicated weak, albeit significant, correlation using SNPs associated with annotated genes; interestingly, mitonuclear LD were substantially higher in X-linked genes.[176] With the increasing appreciation for previously nonannotated sORFs, the (re) discovery of new species of noncoding RNA genes (i.e., long noncoding RNA, circular RNA), and the potential roles of transposable elements (previously labeled as “junk DNA”) in the nuclear and mitochondrial genomes, it will be interesting to examine mitonuclear LD in this new light.
Hence, mitonuclear compatibility and communication has direct and timely translational implications. Mitochondrial replacement therapy (MRT) has been developed to help women who are carriers of pathogenic mtDNA mutations to conceive healthy offspring to prevent transmission of diseases. The procedure involves the transfer of the nuclear DNA from an oocyte with mtDNA mutations into an enucleated donor oocyte bearing healthy mitochondria.[177] Children born from MRT will have three genetic parents; hence, they are also referred to as “three-parent babies.” MRT is no simple over-the-counter battery pack exchange, but an extensive form of germline gene therapy that requires much consideration at the biological and ethical levels.[89,178–180] Human cybrids have altered mtDNA copy number, ATP turnover rates, ROS production, and OXPHOS gene expression.[181] Incompatible mitonuclear combinations are thought to be selected against the oocyte stage, leading to readsorption of failed development.[179] The results from mice and invertebrates suggest that many deleterious effects of MRT may not be revealed until adulthood[178] and may further be exacerbated with aging.
7. Conclusions and Outlook
Nuclear gene regulation by mitochondrial-encoded MOTS-c adds another gene-centric layer to mitonuclear communication. However, MOTS-c may just represent the proverbial tip of the iceberg in this case, the first in a long list of many other MDPs that may regulate the nuclear genome. It is plausible that various MDPs may have distinct, or even multiple, tasks in the nucleus, being targeted at various compartments such as nucleoli, RNAprocessing sites (a.k.a. “transcription factories”), DNA repair foci, nuclear bodies (e.g., promyelocytic leukemias or speckles), and nuclear envelope. The biology of nuclear-residing MOTS-c, and perhaps other MDPs, is just starting to unravel, and several outstanding questions remain. A fundamental question lies on the chromatin state (i.e., eu- or heterochromatin) and the precise locations that MOTS-c binds to. MOTS-c is likely to function as an adaptive cofactor involved in providing context to larger transcriptional programs and may dynamically interact with chromatin depending on the type of cell and stimuli. This hypothesis is consistent with the fact that genes regulated upon MOTS-c overexpression were enriched for various promoter motifs, likely underlying distinct partners. We reported MOTS-c as an adaptive transcriptional cofactor upon metabolic stress, but the mechanism that we have described could apply to any cellular condition where the energetic balance is shifted, including differentiation, proliferation, or even immune responses.
Another key question is the mechanism by which MOTS-c crosses the nuclear envelope. Based on the requirement of the hydrophobic core, MOTS-c may require other partners, which could be regulated by AMPK, to facilitate the shuttling. The connection to AMPK is also clear, but its role as a nuclear gatekeeper for MOTS-c is an intriguing phenomenon. Note that because AMPK is mainly known as a serine/threonine kinase, residues devoid of MOTS-c, it is likely that intermediates are involved. The identification of MOTS-c-specific binding partners may provide further insight into the coordinated network of mitochondrial- and nuclear-encoded signals.
Technological advances will also accelerate our understanding of MOTS-c. mtDNA editing technology is currently possible for targeting heteroplasmy using mitochondrial nucleases, including mitoTALEN[182] and mitochondrially targeted zinc finger-nuclease.[183] However, widely accepted methods for targeted mtDNA mutagenesis, akin to clustered regularly interspaced short palindromic repeats for nuclear genes, have yet to be developed.[184] In addition, an easily accessible and highly sensitive method to confirm homoplastic targeted mtDNA alteration in a cell population will be required. Nonetheless, targeted mtDNA mutagenesis will prove to be an invaluable genetic tool to further explore not only the role of MDPs, but also a wide range of mitochondrial pathophysiology. Further, peptidomic advances will greatly aid in the analysis of known MDPs and the identification of additional peptides.[93,94]
The role of MDPs in regulating diverse cellular and organismal functions has just begun to unravel and the field is in its infancy. Until recently, mitochondrial-encoded gene products (i.e., proteins, tRNA, and rRNA) have not been thought to play regulatory roles. Consequently, mtDNA was not considered a source of active regulators of aging and age-related chronic diseases, which have a genetic basis. This is reflected, in part, by the absence of FDA-approved drugs that are based on mtDNA-encoded factors, even though mitochondria are strongly implicated in multiple diseases. Further studies on MDPs at multiple levels, including peptide chemistry, regulation of expression, mechanisms of interfacing with subcellular compartments and plasma membrane, and nuclear genome regulation, will add an enriching layer to basic research and translational development.
Acknowledgements
This work was funded by grants from NIH (R01AG052558), the Ellison Medical Foundation, the American Federation for Aging Research (AFAR), and the Hanson-Thorell family (C.L.), and NIH grant R00AG049934, the Hanson-Thorell family, and the NAVIGAGE foundation (B.A.B.). Figure 1 was replaced on August 26, 2019 after initial publication online.
Footnotes
Conflict of Interest
C.L. is a consultant for and a shareholder of CohBar Inc. The remaining authors declare no competing interests.
Contributor Information
Bérénice A. Benayoun, Leonard Davis School of Gerontology, University of Southern California, Los Angeles, CA 90089, USA USC Norris Comprehensive Cancer Center, Epigenetics and Gene Regulation Program, Los Angeles, CA 90089, USA; USC Stem Cell Initiative, Los Angeles, CA 90089, USA.
Changhan Lee, Leonard Davis School of Gerontology, University of Southern California, Los Angeles, CA 90089, USA USC Norris Comprehensive Cancer Center, Epigenetics and Gene Regulation Program, Los Angeles, CA 90089, USA; Biomedical Sciences, Graduate School, Ajou University, Suwon 16499, Republic of Korea.
References
- [1].Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG, Nature 2015, 521, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG, Nature 2018, 557, 101. [DOI] [PubMed] [Google Scholar]
- [3].Sunnucks P, Morales HE, Lamb AM, Pavlova A, Greening C, Front. Genet 2017, 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Quiros PM, Mottis A, Auwerx J, Nat. Rev. Mol. Cell Biol 2016, 17, 213. [DOI] [PubMed] [Google Scholar]
- [5].Bensasson D, Zhang D-X, Hartl DL, Hewitt GM, Trends Ecol. Evol 2001, 16, 314. [DOI] [PubMed] [Google Scholar]
- [6].Kim S-J, Xiao J, Wan J, Cohen P, Yen K, Physiol J. 2017, 595, 6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood A-MJ, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS, Cell 2011, 146, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Son JM, Lee C, BMB Rep. 2019, 52, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim KH, Son JM, Benayoun BA, Lee C, Cell Metab. 2018, 28, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mangalhara KC, Shadel GS, Cell Metab. 2018, 28, 330. [DOI] [PubMed] [Google Scholar]
- [11].Wong W, Sci. Signaling 2018, 11, eaav4285. [Google Scholar]
- [12].Yong CQY, Tang BL, Cells 2018, 7, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gottschling DE, Nyström T, Cell 2017, 169, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vyas S, Zaganjor E, Haigis MC, Cell 2016, 166, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chandel NS, Cell Metab. 2015, 22, 204. [DOI] [PubMed] [Google Scholar]
- [16].Bratic A, Larsson NG, J. Clin. Invest 2013, 123, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lopez-Otin C, Galluzzi L, Freije JMP, Madeo F, Kroemer G, Cell 2016, 166, 802. [DOI] [PubMed] [Google Scholar]
- [18].Durieux J, Wolff S, Dillin A, Cell 2011, 144, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sun N, Youle RJ, Finkel T, Mol. Cell 2016, 61, 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Melber A, Haynes CM, Cell Res. 2018, 28, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Karpac J, Jasper H, Cell 2013, 154, 271. [DOI] [PubMed] [Google Scholar]
- [22].Hill S, Sataranatarajan K, Remmen HV, Front. Genet 2018, 9, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shadel GS, Horvath TL, Cell 2015, 163, 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].da Cunha FM, Torelli NQ, Kowaltowski AJ, Oxid. Med. Cell. Longevity 2015, 2015, 482582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zarse K, Ristow M, Cell Metab. 2015, 21, 355. [DOI] [PubMed] [Google Scholar]
- [26].Woo DK, Shadel GS, Cell 2011, 144, 11. [DOI] [PubMed] [Google Scholar]
- [27].Lee C, Yen K, Cohen P, Trends Endocrinol. Metab 2013, 24, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yin F, Cadenas E, Antioxid. Redox Signaling 2015, 22, 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chandel NS, BMC Biol. 2014, 12, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G, Circ. Res 2012, 111, 1198. [DOI] [PubMed] [Google Scholar]
- [31].Antico Arciuch VG, Elguero ME, Poderoso JJ, Carreras MC, Antioxid. Redox Signaling 2012, 16, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xu X, Duan S, Yi F, Ocampo A, Liu G-H, Belmonte JCI, Cell Metab. 2013, 18, 325. [DOI] [PubMed] [Google Scholar]
- [33].Topf U, Wrobel L, Chacinska A, Trends Cell Biol. 2016, 26, 577. [DOI] [PubMed] [Google Scholar]
- [34].Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM, Science 2012, 337, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Qureshi MA, Haynes CM, Pellegrino MW, J. Biol. Chem 2017, 292, 13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Galluzzi L, Kepp O, Kroemer G, Nat. Rev. Mol. Cell Biol 2012, 13, 780. [DOI] [PubMed] [Google Scholar]
- [37].Wilkins HM, Weidling IW, Ji Y, Swerdlow RH, Front. Immunol 2017, 8, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Menzies KJ, Zhang H, Katsyuba E, Auwerx J, Nat. Rev. Endocrinol 2016, 12, 43. [DOI] [PubMed] [Google Scholar]
- [39].Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED, Cell 2014, 158, 84. [DOI] [PubMed] [Google Scholar]
- [40].Ma Q, Annu. Rev. Pharmacol. Toxicol 2013, 53, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Merry TL, Ristow M, Physiol J. 2016, 594, 5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].De Stefani D, Rizzuto R, Pozzan T, Annu. Rev. Biochem 2016, 85, 161. [DOI] [PubMed] [Google Scholar]
- [43].West AP, Shadel GS, Ghosh S, Nat. Rev. Immunol 2011, 11, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ, Nature 2010, 464, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu X, Kim CN, Yang J, Jemmerson R, Wang X, Cell 1996, 86, 147. [DOI] [PubMed] [Google Scholar]
- [46].Allen JF, Proc. Natl. Acad. Sci. U. S. A 2015, 112, 10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pittis AA, Gabaldón T, Nature 2016, 531, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ettema TJG, Nature 2016, 531, 39. [DOI] [PubMed] [Google Scholar]
- [49].Roger AJ, Muñoz-Gómez SA, Kamikawa R, Curr. Biol 2017, 27, R1177. [DOI] [PubMed] [Google Scholar]
- [50].Ju YS, Curr. Opin. Genet. Dev 2016, 38, 23. [DOI] [PubMed] [Google Scholar]
- [51].Lesnik C, Cohen Y, Atir-Lande A, Schuldiner M, Yoav A, Nat. Commun 2014, 5, 5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Williams CC, Jan CH, Weissman JS, Science 2014, 346, 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Couvillion MT, Soto IC, Shipkovenska G, Churchman LS, Nature 2016, 533, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Schneider A, Annu. Rev. Biochem 2011, 80, 1033. [DOI] [PubMed] [Google Scholar]
- [55].Rubio MAT, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, Alfonzo JD, Proc. Natl. Acad. Sci. U. S. A 2008, 105, 9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wallace DC, Science 1999, 283, 1482. [DOI] [PubMed] [Google Scholar]
- [57].Lightowlers RN, Taylor RW, Turnbull DM, Science 2015, 349, 1494. [DOI] [PubMed] [Google Scholar]
- [58].Lewis SC, Uchiyama LF, Nunnari J, Science 2016, 353, aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Stewart JB, Chinnery PF, Nat. Rev. Genet 2015, 16, 530. [DOI] [PubMed] [Google Scholar]
- [60].Pikó L, Hougham AJ, Bulpitt KJ, Mech. Ageing Dev 1988, 43, 279. [DOI] [PubMed] [Google Scholar]
- [61].Cortopassi GA, Arnheim N, Nucleic Acids Res. 1990, 18, 6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM, Am. J. Hum. Genet 2006, 79, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Larsson N-G, Annu. Rev. Biochem 2010, 79, 683. [DOI] [PubMed] [Google Scholar]
- [64].Greaves LC, Nooteboom M, Elson JL, Tuppen HA, Taylor GA, Commane DM, Arasaradnam RP, Khrapko K, Taylor RW, Kirkwood TBL, Mathers JC, Turnbull DM, PLoS Genet. 2014, 10, e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gardner K, Payne BA, Horvath R, Chinnery PF, Eur. J. Hum. Genet 2015, 23, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA, Nat. Genet 2007, 39, 540. [DOI] [PubMed] [Google Scholar]
- [67].Ameur A, Stewart JB, Freyer C, Hagström E, Ingman M, Larsson N-G, Gyllensten U, PLoS Genet. 2011, 7, e1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kennedy SR, Salk JJ, Schmitt MW, Loeb LA, PLoS Genet. 2013, 9, e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yakes FM, Van Houten B, Proc. Natl. Acad. Sci. U. S. A 1997, 94, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, Cell 2013, 155, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cogliati S, Enriquez JA, Scorrano L, Trends Biochem. Sci 2016, 41, 261. [DOI] [PubMed] [Google Scholar]
- [72].Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF, Proc. Natl. Acad. Sci. U. S. A 2012, 109, 6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shadel GS, Horvath TL, Cell 2015, 163, 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sarsour EH, Kalen AL, Goswami PC, Antioxid. Redox Signaling 2014, 20, 1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ristow M, Nat. Med 2014, 20, 709. [DOI] [PubMed] [Google Scholar]
- [76].Lee H-Y, Chou J-Y, Cheong L, Chang N-H, Yang S-Y, Leu J-Y, Cell 2008, 135, 1065. [DOI] [PubMed] [Google Scholar]
- [77].Rand DM, Fry A, Sheldahl L, Genetics 2006, 172, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mossman JA, Tross JG, Li N, Wu Z, Rand DM, Genetics 2016, 204, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL, PLoS Genet. 2013, 9, e1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhu Z, Lu Q, Zeng F, Wang J, Shi H, Sci. Rep 2015, 5, 17303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chang CC, Rodriguez J, Ross J, G3: Genes, Genomes, Genet. 2015, 6, 209. [Google Scholar]
- [82].Yamaoka M, Isobe K, Shitara H, Yonekawa H, Miyabayashi S, Hayashi JI, Genetics 2000, 155, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Dunham-Snary KJ, Ballinger SW, Science 2015, 349, 1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, Sammy MJ, Johnson M, Dunham-Snary KJ, Cao X, Bradley WE, Zhang J, Wei CC, Chacko B, Schurr TG, Kesterson RA, Dell’italia LJ, Darley-Usmar VM, Welch DR, Ballinger SW, Biochem. J 2013, 455, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Betancourt AM, King AL, Fetterman JL, Millender-Swain T, Finley RD, liva CR, Crowe DR, Ballinger SW, Bailey SM, Biochem. J 2014, 461, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ma H, Marti Gutierrez N, Morey R, Van Dyken C, Kang E, Hayama T, Lee Y, Li Y, Tippner-Hedges R, Wolf DP, Laurent LC, Mitalipov S, Cell Metab. 2016, 24, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zaidi AA, Makova KD, Nat. Ecol. Evol 2019, 3, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].McManus MJ, Picard M, Chen H-W, De Haas HJ, Potluri P, Leipzig J, Towheed A, Angelin A, Sengupta P, Morrow RM, Kauffman BA, Vermulst M, Narula J, Wallace DC, Cell Metab. 2019, 29, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lane N, BioEssays 2011, 33, 860. [DOI] [PubMed] [Google Scholar]
- [90].Hill GE, Mol. Biol. Evol 2015, 32, 1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I, Proc. Natl. Acad. Sci. U. S. A 2001, 98, 6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rathore A, Martinez TF, Chu Q, Saghatelian A, Expert Rev. Proteomics 2018, 15, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Makarewich CA, Olson EN, Trends Cell Biol. 2017, 27, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Saghatelian A, Couso JP, Nat. Chem. Biol 2015, 11, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Rothnagel J, Menschaert G, Proteomics 2018, 18, 1700035. [DOI] [PubMed] [Google Scholar]
- [96].Couso J-P, Patraquim P, Nat. Rev. Mol. Cell Biol 2017, 18, 575. [DOI] [PubMed] [Google Scholar]
- [97].Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS, Cell 2011, 146, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T, Nishimoto I, Cohen P, Proc. Natl. Acad. Sci. U. S. A 2003, 100, 13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC, Nature 2003, 423, 456. [DOI] [PubMed] [Google Scholar]
- [100].Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J, Muzumdar R, Barzilai N, Cohen P, Aging 2016, 8, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P, Cell Metab. 2015, 21, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hill S, Sataranatarajan K, Remmen HV, Front. Genet 2018, 9, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hashimoto Y, Ito Y, Niikura T, Shao Z, Hata M, Oyama F, Nishimoto I, Biochem. Biophys. Res. Commun 2001, 283, 460. [DOI] [PubMed] [Google Scholar]
- [104].Lee C, Wan J, Miyazaki B, Fang Y, Guevara-Aguirre J, Yen K, Longo V, Bartke A, Cohen P, Aging Cell 2014, 13, 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Yen K, Wan J, Mehta HH, Miller B, Christensen A, Levine ME, Salomon MP, Brandhorst S, Xiao J, Kim S-J, Navarrete G, Campo D, Harry GJ, Longo V, Pike CJ, Mack WJ, Hodis HN, Crimmins EM, Cohen P, Sci. Rep 2018, 8, 14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lue Y, Gao C, Swerdloff R, Hoang J, Avetisyan R, Jia Y, Rao M, Ren S, Atienza V, Yu J, Zhang Y, Chen M, Song Y, Wang Y, Wang C, Am. J. Physiol.: Heart Circ. Physiol 2018, 315, H634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kumfu S, Charununtakorn ST, Jaiwongkam T, Chattipakorn N, Chattipakorn SC, Alzheimer’s Dis J. 2018, 61, 1343. [DOI] [PubMed] [Google Scholar]
- [108].Han K, Jia N, Zhong Y, Shang X, J. Cell. Biochem 2017, 119, 3111. [DOI] [PubMed] [Google Scholar]
- [109].Kim S-J, Guerrero N, Wassef G, Xiao J, Mehta HH, Cohen P, Yen K, Oncotarget 2016, 7, 46899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T, Nishimoto I, Cohen P, Proc. Natl. Acad. Sci. U. S. A 2003, 100, 13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Qin Q, Mehta H, Yen K, Navarrete G, Brandhorst S, Wan J, Delrio S, Zhang X, Lerman LO, Cohen P, Lerman A, Am. J. Physiol.: Heart Circ. Physiol 2018, 315, H1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Blasco N, Cámara Y, Núñez E, Beà A, Gisel B, Carles F, Marisol R-M, Cristina G, Ignasi B, Elena G-A, David G-D, Jesús V, Ramon M, Marta L, Daniel S, Redox Biol. 2018, 16, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Thummasorn S, Shinlapawittayatorn K, Khamseekaew J, Jaiwongkam T, Chattipakorn SC, Chattipakorn N, Mitochondrion 2018, 38, 31. [DOI] [PubMed] [Google Scholar]
- [114].Nashine S, Cohen P, Chwa M, Lu S, Nesburn AB, Kuppermann BD, Kenney MC, Cell Death Dis. 2017, 8, e2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zhang X, Urbieta-Caceres VH, Eirin A, Bell CC, Crane JA, Tang H, Jordan KL, Oh Y-K, Zhu X-Y, Korsmo MJ, Bachar AR, Cohen P, Lerman A, Lerman LO, Life Sci. 2012, 91, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zacharias DG, Kim SG, Massat AE, Bachar AR, Oh YK, Herrmann I, Rodriguez-Porcel M, Cohen P, Lerman LO, Lerman A, PLoS One 2012, 7, e31065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Oh YK, Bachar AR, Zacharias DG, Kim SG, Wan J, Cobb LJ, Lerman LO, Cohen P, Lerman A, Atherosclerosis 2011, 219, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Bachar AR, Scheffer L, Schroeder AS, Nakamura HK, Cobb LJ, Oh YK, Lerman LO, Pagano RE, Cohen P, Lerman A, Cardiovasc. Res 2010, 88, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Minasyan L, Sreekumar PG, Hinton DR, Kannan R, Oxid. Med. Cell. Longevity 2017, 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Matsunaga D, Sreekumar PG, Ishikawa K, Terasaki H, Barron E, Cohen P, Kannan R, Hinton DR, PLoS One 2016, 11, e0165150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Sreekumar PG, Ishikawa K, Spee C, Mehta HH, Wan J, Yen I, Cohen P, Kannan R, Hinton DR, Invest. Ophthalmol. Visual Sci 2016, 57, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Eriksson E, Wickström M, Perup LS, Johnsen JI, Eksborg S, Kogner P, Savendahl L, Natl J. Cancer Inst. 2014, 106, 459. [DOI] [PubMed] [Google Scholar]
- [123].Zaman F, Zhao Y, Celvin B, Mehta HH, Wan J, Chrysis D, Ohlsson C, Fadeel B, Cohen P, Sävendahl L, FASEB J. 2019, 33, 4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Qin Q, Jin J, He F, Zheng Y, Li T, Zhang Y, He J, Biochem. Biophys. Res. Commun 2018, 497, 292. [DOI] [PubMed] [Google Scholar]
- [125].Muzumdar RH, Huffman DM, Atzmon G, Buettner C, Cobb IJ, Fishman S, Budagov T, Cui L, Einstein FH,Poduval A, Hwang D, Barzilai N, Cohen P, PLoS One 2009, 4, e6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kim S-J, Mehta HH, Wan J, Kuehnemann C, Chen J, Hu J-F, Hoffman AR, Cohen P, Aging 2018, 10, 1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Okada AK, Teranishi K, Lobo F, Isas JM, Xiao J, Yen K, Cohen P, Langen R, Sci. Rep 2017, 7, 7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Xiao J, Howard L, Wan J, Wiggins E, Vidal A, Cohen P, Freedland SJ, Oncotarget 2017, 8, 94900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Zempo H, Fuku N, Nishida Y, Higaki Y, Naito H, Hara M, Tanaka K, FASEB. J 2016, 30, 956. [Google Scholar]
- [130].Cataldo LR, Fernandez-Verdejo R, Santos JL, Galgani JE, J. Investig. Med 2018, 66, 1019. [DOI] [PubMed] [Google Scholar]
- [131].Du C, Zhang C, Wu W, Liang Y, Wang A, Wu S, Zhao Y, Hou L, Ning Q, Luo X, Pediatr. Diabetes 2018, 19, 1058. [DOI] [PubMed] [Google Scholar]
- [132].Ramanjaneya M, Bettahi I, Jerobin J, Chandra P, Abi Khalil C, Skarulis I, Atkin SL, Abou-Samra A-B, Front. Endocrinol 2019, 10, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ramanjaneya M, Jerobin J, Bettahi I, Bensila M, Aye M, Siveen KS, Sathyapalan T, Skarulis M, Abou-Samra A-B, Atkin SL, Clin. Endocrinol 2019. [DOI] [PubMed] [Google Scholar]
- [134].Qin Q, Delrio S, Wan J, Jay Widmer R, Cohen P, Lerman LO, Lerman A, Int. J. Cardiol 2018, 254, 23. [DOI] [PubMed] [Google Scholar]
- [135].Raijmakers RPH, Jansen AFM, Keijmel SP, Ter Horst R, Roerink ME, Novakovic B, Joosten LAB, Van Der Meer JWM, Netea MG, Bleeker-Rovers CP, J. Transl. Med 2019, 17, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Fuku N, Pareja-Galeano H, Zempo H, Alis R, Arai Y, Lucia A, Hirose N, Aging Cell 2015, 14, 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Ming W, Lu G, Xin S, Huanyu L, Yinghao J, Xiaoying L, Chengming X, Banjun R, Li W, Zifan L, Biochem. Biophys. Res. Commun 2016, 476, 412. [DOI] [PubMed] [Google Scholar]
- [138].Thevis M, Schanzer W, Rapid Commun. Mass Spectrom 2016, 30, 635. [DOI] [PubMed] [Google Scholar]
- [139].Zhai D, Ye Z, Jiang Y, Xu C, Ruan B, Yang Y, Lei X, Xiang A, Lu H, Zhu Z, Yan Z, Wei D, Li Q, Wang L, Lu Z, Mol. Immunol 2017, 92, 151. [DOI] [PubMed] [Google Scholar]
- [140].Hu BT, Chen WZ, Eur. Rev. Med. Pharmacol. Sci 2018, 22, 7156. [DOI] [PubMed] [Google Scholar]
- [141].Che N, Qiu W, Wang J, Sun XX, X Xu L, Liu R, Gu L, Eur. Rev. Med. Pharmacol. Sci 2019, 23, 3183. [DOI] [PubMed] [Google Scholar]
- [142].Knoop A, Thomas A, Thevis M, Rapid Commun. Mass Spectrom 2019, 33, 371. [DOI] [PubMed] [Google Scholar]
- [143].Li Q, Lu H, Hu G, Ye Z, Zhai D, Yan Z, Wang L, Xiang A, Lu Z, Biochem. Biophys. Res. Commun 2019, 513, 439. [DOI] [PubMed] [Google Scholar]
- [144].Lu H, Wei M, Zhai Y, Li Q, Ye Z, Wang L, Luo W, Chen J, Lu Z, J. Mol. Med 2019, 97, 473. [DOI] [PubMed] [Google Scholar]
- [145].Tsuzuki T, Nomiyama H, Setoyama C, Maeda S, Shimada K, Pestka S, Biochem. Biophys. Res. Commun 1983, 114, 670. [DOI] [PubMed] [Google Scholar]
- [146].Tian JoeY., Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A, Cell 2016, 165, 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, Murillo V, Wolff SC, Shaw RJ, Auwerx J, Dillin A, Cell 2016, 165, 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM, Science 2012, 337, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, Haynes CM, Curr. Biol 2016, 26, 2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Monaghan RM, Whitmarsh AJ, Trends Biochem. Sci 2015, 40, 728. [DOI] [PubMed] [Google Scholar]
- [151].Munoz-Najar U, Sedivy JM, Antioxid. Redox Signaling 2011, 14, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Rando TA, Chang HY, Cell 2012, 148, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Schroeder EA, Raimundo N, Shadel GS, Cell Metab. 2013, 17, 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lu C, Thompson CB, Cell Metab. 2012, 16, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Nagaraj R, Sharpley MS, Chi F, Braas D, Zhou Y, Kim R, Clark AT, Banerjee U, Cell 2017, 168, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Sturme MHJ, Kleerebezem M, Nakayama J, Akkermans ADL, Vaughan EE, De Vos WM, Antonie van Leeuwenhoek 2002, 81, 233. [DOI] [PubMed] [Google Scholar]
- [157].Bassler BL, Curr. Opin. Microbiol 1999, 2, 582. [DOI] [PubMed] [Google Scholar]
- [158].Miravet-Verde S, Ferrar T, Espadas-García G, Mazzolini R, Gharrab A, Sabido E, Serrano L, Lluch-Senar M, Mol. Syst. Biol 2019, 15, e8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Cotter PD, Ross RP, Hill C, Nat. Rev. Microbiol 2013, 11, 95. [DOI] [PubMed] [Google Scholar]
- [160].Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD, PLoS Genet. 2013, 9, e1003860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Couso J-P, Patraquim P, Nat. Rev. Mol. Cell Biol 2017, 18, 575. [DOI] [PubMed] [Google Scholar]
- [162].Ladoukakis E, Pereira V, Magny EG, Eyre-Walker A, Couso JP, Genome Biol. 2011, 12, R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Smith NGC, Eyre-Walker A, Nature 2002, 415, 1022. [DOI] [PubMed] [Google Scholar]
- [164].Keskin O, Gursoy A, Ma B, Nussinov R, Chem. Rev 2008, 108, 1225. [DOI] [PubMed] [Google Scholar]
- [165].Coelho PSR, Genes Dev. 2002, 16, 2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Bonawitz ND, Chatenay-Lapointe M, Wearn CM, Shadel GS, Curr. Genet 2008, 54, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Poole AM, Kobayashi T, Ganley ARD, BioEssays 2012, 34, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Borghouts C, Genetics 2004, 166, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Sinclair DA, Guarente L, Cell 1997, 91, 1033. [DOI] [PubMed] [Google Scholar]
- [170].Lindqvist LM, Tandoc K, Topisirovic I, Furic L, Curr. Opin. Genet. Dev 2018, 48, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Wu S, Wang J, Zhao W, Pounds S, Cheng C, Theor. Biol. Med. Modell 2010, 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Marinov GK, Wang YE, Chan D, Wold BJ, PLoS One 2014, 9, e84713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Blumberg A, Sri Sailaja B, Kundaje A, Levin L, Dadon S, Shmorak S, Shaulian E, Meshorer E, Mishmar D, Genome Biol. Evol 2014, 6, 2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].O’Neil D, Glowatz H, Schlumpberger M, Curr. Protoc. Mol. Biol 2013, 103, 4.19.1. [DOI] [PubMed] [Google Scholar]
- [175].Herbert ZT, Kershner JP, Butty VL, Thimmapuram J, Choudhari S, Alekseyev YO, Fan J, Podnar JW, Wilcox E,Gipson J, Gillaspy A, Jepsen K, Bondurant SS, Morris K, Berkeley M, Leclerc A, Simpson SD, Sommerville G, Grimmett L, Adams M, Levine SS, BMC Genomics 2018, 19, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Sloan DB, Fields PD, Havird JC, Proc. Biol. Sci 2015, 282, 20151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Couzin-Frankel J, Science 2015, 348, 14. [DOI] [PubMed] [Google Scholar]
- [178].Reinhardt K, Dowling DK, Morrow EH, Science 2013, 341, 1345. [DOI] [PubMed] [Google Scholar]
- [179].Morrow EH, Reinhardt K, Wolff JN, Dowling DK, EMBO Rep. 2015, 16, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Coller BS, Annu. Rev. Med 2019, 70, 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, Tarek M, Del Carpio JC, Nesburn AB,Boyer DS, Kuppermann BD, Vawter MP, Jazwinski SM, Miceli MV, Wallace DC, Udar N, Biochim. Biophys. Acta 2014, 1842, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Bacman SR, Kauppila JHK, Pereira CV, Nissanka N, Miranda M, Pinto M, Williams SL, Larsson N-G, Stewart JB, Moraes CT, Nat. Med 2018, 24, 1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Gammage PA, Viscomi C, Simard M-L, Costa ASH, Gaude E, Powell CA, Van Haute L, McCann BJ, Rebelo-Guiomar P, Cerutti R, Zhang L, Rebar EJ, Zeviani M, Frezza C, Stewart JB, Minczuk M, Nat. Med 2018, 24, 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Gammage PA, Moraes CT, Minczuk M, Trends Genet. 2018, 34, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]