Abstract
Lipopolysaccharide (LPS) is a bacterial endotoxin present in cigarette smoke. LPS is known to induce inflammation and to increase the size and the multiplicity of lung tumors induced by tobacco-specific nitrosamines. However, the means by which LPS contributes to pulmonary carcinogenesis are not known. One possible mechanism includes LPS-mediated epigenetic deregulation, which leads to aberrant expression of genes involved in DNA repair, tumor suppression, cell cycle progression, and cell growth. In the present work, epigenetic effects of LPS were examined in alveolar type II lung cells of A/J mice. Type II cells were selected because they serve as progenitors of lung adenocarcinomas in smoking induced lung cancer. A/J mice were intranasally treated with LPS, followed by isolation of alveolar type II cells from the lung using cell panning. Global levels of DNA methylation and histone acetylation were quantified by mass spectrometry, while genome-wide transcriptomic changes were characterized by RNA-Seq. LPS treatment was associated with epigenetic changes including decreased cytosine formylation and reduced histone H3K14 and H3K23 acetylation, as well as altered expression levels of genes involved in cell adhesion, inflammation, immune response, and epigenetic regulation. These results suggest that exposure to inflammatory agents in cigarette smoke leads to early epigenetic changes in the lung, which may collaborate with genetic changes to drive the development of lung cancer.
Graphical Abstract
Introduction
Lung cancer is a leading cause of cancer deaths worldwide and is expected to kill 142,670 Americans this year, with over 80% of cases directly attributable to smoking.1 Cigarette smoke contains over 60 known carcinogens2 and many co-carcinogens, including bacterial endotoxin lipopolysaccharide (LPS) (2,400 ng/pack).3 LPS induces immune response in mammalian cells by interacting with toll-like receptor 4 (TLR4).4 This interaction is mediated by MD-2 and CD14, which are necessary for TLR4 recognition and shuttling LPS toward TLR4, respectively.4 Through downstream signaling pathways, TLR4 activates NF-κB, which induces the expression of various pro-inflammatory genes, including those encoding cytokines and chemokines, and also participates in inflammasome regulation.4
Inflammation of the lung, such as that observed in chronic obstructive pulmonary disease (COPD), elevates the risk for lung cancer development in smokers.5, 6 In the A/J mouse model, LPS increases the size and the multiplicity of lung tumors induced by tobacco specific nitrosamine NNK.7 The majority of smoking-induced lung cancers are defined as adenocarcinomas,8 which can arise from the alveolar type II cells in the lung.9 Type II cells have the ability to rapidly proliferate and differentiate following lung injury,9, 10 and can increase in number following exposure to LPS.11 The proliferative ability and the stem-like nature of type II cells make them a key target for investigating the effects of LPS mediated inflammation in connection with lung carcinogenesis.
5-Methylcytosine (MeC) is a key epigenetic modification of DNA with an important role in gene regulation.12 Methylated CpG sites are recognized by methyl-CpG binding proteins, leading to recruitment of histone deacetylases, chromatin remodeling, and in most cases, reduced levels of gene expression.13 5-Hydroxymethylcytosine (hmC) and 5-formyl-dC (fC) are generated from MeC by ten eleven translocation family of proteins (TET 1–3).14, 15 hmC is thought to play a role in DNA demethylation and may possess a separate epigenetic function by directly participating in epigenetic signaling.16 Interestingly, lung adenocarcinomas and other tumors are characterized by reduced global levels of hmC17 and promoter hypermethylation of tumor suppressor genes.18 The biological roles of fC are poorly understood, although it was recently shown to be recognized by a distinct set of protein readers and to form reversible DNA-protein cross-links with histone proteins.19
Smoking-induced lung tumors are characterized by both genetic and epigenetic changes, or so called “epimutations”.20 Genetic changes contributing to lung cancer development include mutations in the p53 tumor suppressor gene and the Kras protooncogene.20 Smoking induces epigenetic changes in DNA methylation an hydroxymethylation, as well as histone acetylation, can lead to aberrant gene expression, which can contribute to carcinogenesis by upregulating protooncogenes and downregulating tumor suppressor genes.21–23 Evidence is accumulating that both genetic abnormalities and epigenetic changes contribute to pulmonary tumorigenesis through induction of abnormal regulation of key signal transduction pathways.24, 25 However, the exact role of inflammation in smoking-induced epigenetic deregulation remains unclear.
Recently, Vaz et al. reported that early cigarette smoke-induced epigenetic alterations can sensitize human bronchial epithelial cells for transformation by a single oncogene.26 Therefore, it is of interest to characterize early epigenetic changes in the lung. In previous study, we examined epigenetic effects of LPS in lung tumors of A/J mice mice co-treated with LPS and tobacco specific nitrosamine NNK for 22 weeks.27 In this study, 2,957 genes were identified to change expression including cytokines, chemokines, putative oncogenes, and tumor suppressor genes.27 However, that earlier study did not focus on specific cell types28, 29 and did not examine epigenetic changes in alveolar type II cells, which serve as precursors to lung adenocarcinoma.30
In the present study, we investigated early epigenetic responses of alveolar type II cells towards LPS-induced inflammation in the A/J mouse model. Mice were intranasally treated with LPS, and global changes in DNA methylation and histone acetylation were quantified by mass spectrometry, while genome wide transcriptome changes were characterized by RNA-Seq. Our results reveal that acute LPS exposure induces early epigenetic changes in type II cells as manifested in decreased cytosine hydroxymethylation, reduced histone H3 acetylation, and altered expression levels of a number of genes involved in cell adhesion, inflammation, immune response, and epigenetic regulation.
Materials and Methods
Chemicals and Enzymes
PDE I, PDE II, and DNase I were purchased from Worthington Biochemical Corp. (Lakewood, NJ), while calf intestinal alkaline phosphatase was from Sigma-Aldrich, (Madison, WI). Nanosep10K filters were acquired from Pall corp. (Port Washington, NY). O-(Biotinyl-carbazoylmethyl) hydroxylamine was obtained from Cayman Chemical (Ann Arbor, MI). Isotopically labeled D3-hmC was purchased from Cambridge Isotope Labs (Cambridge, MA). 13C10,15N2-5-methyl-2ʹ-deoxycytidine was synthesized as described elsewhere.31 All other chemicals used were bought from Sigma-Aldrich (Milwaukee, WI) or Fisher Scientific (Fairlawn, NJ).
Animal Treatments
Female A/J mice were obtained from Jackson Laboratory (Bar Harbor, ME) and housed in specific-pathogen-free animal quarters at Research Animal Resources, University of Minnesota Academic Health Center. All animal experiments were performed according to the U.S. National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee, University of Minnesota.
LPS treatment:
Female A/J mice (8 weeks of age) were divided into 2 groups (10 per group) and intranasally treated with LPS or vehicle control under isoflurane anesthesia. Mice in Group 1 were treated with physiological saline (50 μL) every day for 4 days. Mice in group 2 were treated with lipopolysaccharide (LPS, P. aeruginosa 10), 2 μg in 50 μL of physiological saline on day 1 and 5 μg in 50 μL saline on days 2–4. Mice were euthanized in a CO2 chamber on day 5. Lung tissues were harvested and immediately used for cell panning.
Isolation of Type II alveolar cells from lung tissue
Type II alveolar cells were isolated according to published procedures.11 In brief, murine lung tissues were perfused with 30 mL of PBS. The whole lung tissue was then injected with 3 mL dispase (5 U/mL, Stem Cell Technology, Vancouver Canada) and placed on ice. Tissues were finely minced, and additional 60 μL dispase (5 U/mL) and 7.5 μL DNase I (2 U/μL, Thermo Fisher Scientific, Fairlawn, NJ) was added prior to incubation at 37 °C for 45 min, with inverting every 5 min. Dissociated cells were pushed through a 100 micron cell strainer, followed by a 40 micron cell strainer (Corning, Corning, NY) to remove connective tissue. To the resulting cells, RBC lysis buffer (e-Biosciences, San Diego, CA) was added and mixed every 5 min at room temperature for a total of 30 min. The suspension was centrifuged, the supernatant was removed, and the cells were resuspended in 50:50 DMEM/F-12 media. The cells were plated on dual antibody coated plates prepared 24 h in advance with anti-mouse CD16/CD32 and anti-mouse CD45.1 (e-Biosciences). After incubation for 2 hours at 37 °C to facilitate negative selection for type II alveolar cells, the media containing the suspended type II alveolar cells was removed and pelleted by centrifugation. Cells were lysed and stored at −80 °C in 750 μL of Trizol Reagent (Thermo Fisher, Fairlawn, NJ).
HPLC-ESI+-MS/MS quantitation of MeC, hmC, and fC
Genomic DNA was extracted using Trizol extraction protocol (Invitrogen, Carlsbad CA) according to the manufacturer’s protocol. DNA purity and concentration were determined by UV spectroscopy. Genomic DNA (1–2 μg) was subjected to hydrolysis with PDE I (3.6 U, 5 μg), PDE II (3.2 U), DNase I (50 U), and alkaline phosphatase (10 U) in 10 mM Tris HCl/15 mM MgCl2 buffer (pH 7) at 37 °C overnight. The hydrolysates were spiked with 13C1015N2-5-methyl-2′-deoxycytidine (1 pmol), 5-hydroxymethyl-d2-2′-deoxycytidine-6-d1 (900 fmol), and 13C1015N2-5-formyl-2′-deoxycytidine (500 fmol) (internal standards for mass spectrometry, synthesized in our laboratory)31 and filtered through Nanosep 10K Omega filters (Pall Corporation, Port Washington, NY).
DNA hydrolysates were dissolved in 100 mM ammonium acetate buffer (pH 4.5) containing 100 mM aniline and 400 μM O-(biotinylcarbazoylmethyl) hydroxylamine (Cayman Chemical. Ann Arbor, MI) and allowed to react for 24 h to convert fC to biotinyl-fC.32 This was necessary due to the relatively low abundance of fC in lung tissues and its co-elution with other nucleosides during HPLC separation (results not shown). The resulting mixture was dried, followed by offline HPLC to enrich for MeC, hmC, biotinyl-fC, and caC. Atlantis T3 column (Waters, 4.6 × 150 mm, 3 μm) was eluted at a flow rate of 0.9 mL/min with a linear gradient of 5 mM ammonium formate buffer, pH 4.0 (A) and methanol (B). Solvent composition was changed linearly from 3 to 30% B over 15 min, increased to 80% over the next 3 min, maintained at 80% B for the next 2 min, and brought back to 3 % B. The column was equilibrated for 7 min prior to the next injection. dC was quantified by HPLC-UV using calibration curves obtained by analyzing authentic dC standards. HPLC fractions corresponding to MeC, hmC, caC, and biotinyl-fC (7–8.6 min for hmC and caC, 9–10.5 min for MeC, and 18.7–20.2 min for biotinyl-fC) were combined, dried, and analyzed by isotope dilution HPLC-ESI-MS/MS.
Quantitation of MeC, hmC, and biotinyl-fC was performed using a Dionex Ultimate 3000 UHPLC (Thermo Fisher, Waltham MA) interfaced with a Thermo TSQ Vantage mass spectrometer (Thermo Fisher). Chromatographic separation was achieved on a Zorbax SB-C18 column (0.5 × 150 mm, 3 μm, Agilent) eluted at a flow rate of 15 μL/min with a gradient of 2 mM ammonium formate (A) and methanol (B). Solvent composition was maintained at 5% B for the first 3 min and linearly changed from 5 to 40% B for 7 min. Solvent composition was returned to 5% B, and the column was re-equilibrated for 4 min. Under these conditions, MeC and 13C1015N2-MeC eluted at 4.1 min, hmC and its internal standard (d3-hmC) eluted at 3.5 min, while biotinyl-fC and its internal standard (13C1015N2-biotinyl-fC) eluted at 7.0 min. Quantitation was achieved by monitoring the MS/MS transitions m/z 258.1 [M + H+] → m/z 141.1 [M – deoxyribose + H+] for hmC, m/z 261.2 [M + H+] → m/z 145.1 [M – deoxyribose + H+] for D3-hmC, m/z 242.1 [M + H+] → m/z 126.1 [M + H+] for MeC, m/z 254.2 [M + H+] → m/z 133.1 [M + H+] for 13C1015N2-MeC, m/z 569.1 [M + H+] → m/z 453.3 [M – deoxyribose + H+] for biotinyl-5fC, m/z 581.2 [M + H+] → m/z 460.4 [M – deoxyribose + H+] for Biotinyl-13C1015N2-5fC. Mass spectrometry parameters were optimized upon infusion of authentic standards. Typical settings on the mass spectrometer were: spray voltage of 2700 V, sheath gas of 15 units, declustering voltage at 5 V, and the ion transfer tube temperature at 350 °C. The full-width at half-maximum (FWHM) was maintained at 0.7 for both Q1 and Q3. MS/MS fragmentation was induced using argon as a collision gas at 1.0 mTorr and a collision energy of 10.3 V. Using 1 μg of DNA, the method’s LOD was 3.1 fmol for MeC, 6.2 fmol for hmC, and 1.2 fmol for fC, while the corresponding LOQ values for MeC, hmC, and fC were 6.2 fmol, 9.3 fmol, and 4.9 fmol, respectively.
Histone acetylation analysis
Type II alveolar cells isolated from the lungs of control and treated A/J mice were homogenized with a glass douncer on ice. Total histones were extracted from the lysates as previously described.33, 34 Histone proteins were acetylated with (13C2, d3)-acetyl N-hydroxysuccinimide ester to block all unmodified lysines and subsequently digested by trypsin.34 The peptides were desalted with C18 Stage Tips (3M Corporation, St. Paul, MN) prior to analysis.35
Tryptic peptides were analyzed by nanoflow liquid chromatography electrospray tandem mass spectrometry (nanoLC-ESI-MS/MS) using a Thermo Scientific Orbitrap Fusion mass spectrometer (Thermo Scientific, San Jose, CA) coupled to a Proxeon Easy nLC 1000 UPLC system (Thermo Fisher Scientific, Odense, Denmark). Each sample was dissolved in HPLC buffer A (0.1% formic acid in water) and loaded onto a column (25 cm x 75 μm I.D.) packed with ReproSil-Pur Basic C18 stationary phase (2.5 μm, Dr. Maisch GmbH). Peptides were eluted with a gradient of 5% to 15% B (0.1% formic acid in acetonitrile) over 26 min, then 15% to 35% HPLC buffer B over 16 min at 300 nL/min.
Tryptic peptides were analyzed using an FT survey scan from 300–1600 m/z at a resolution of FWHM 120,000 (at 200 m/z), followed by HCD MS/MS scans using the top speed mode (3 seconds per cycle) at a resolution of FWHM 15,000 (at 200 m/z) and a normalized collision energy at 35%. Targeted MS/MS data acquisition was achieved with an inclusion list for fully labeled histone tryptic peptides that covered known lysine acetylation sites. For each peptide, modification isomers with all possible combinations of light/heavy lysine acetylation (Δ M of 42.010565 and 47.036094 Da, respectively) at detectable charge states were considered in the inclusion list for targeted fragmentations.
MS data were searched against the Uniprot Mus musculus proteome database (http://www.uniprot.org) using MaxQuant search engine (v1.4.1.2) as previously described.36 Heavy and light acetylation on lysine as well as methionine oxidation were included as variable modifications, with 6 ppm specified as the precursor mass error and 0.025 Da as the fragment mass error. All peptide spectra matches were filtered at 1% False Discovery Rate with a minimum Andromeda score cutoff of 40. HPLC elution profiles were manually evaluated to ensure accurate quantifications. Only peptides that were confidently identified were selected for stoichiometry analysis. Acetylation stoichiometries of specific sites were calculated using in-house developed scripts based on the extracted peak areas of each modification isomer and quantification of modification-specific fragment ions.34 Statistical significance analysis of site-specific acetylation stoichiometry dynamics between control and treated samples was conducted with two-sided Student’s t-test using SAS statistical software 9.3 (SAS Institute Inc., Cary, NC).
RNA-Seq
Following isolation, type II alveolar cells were stored in Trizol (Thermo Fisher) at −80 °C to minimize RNA degradation. RNA was isolated using Qiagen miRNeasy kits (Qiagen, Hilden Germany) according to the manufacturer’s instructions. RNA was quantified using a fluorometric RiboGreen assay (Thermo Fisher). RNA integrity was confirmed using Agilent Bioanalyzer (Agilent, Santa Clara CA). Samples with RNA integrity scores higher than 7 were selected for RNA-Seq analysis. Total RNA samples were converted to Illumina sequencing libraries using SMARTer Stranded Total RNA-Seq Kit – Pico Mammalian Input (Takara Bio USA, Mountain View CA). RNA was reverse transcribed into cDNA, and Illumina adapters were added using PCR. rRNA was cleaved with ZapR and R-Probes. Final library size distribution was validated using capillary electrophoresis and quantified using fluorimetry (PicoGreen) and via Q-PCR. Indexed libraries were normalized, pooled, and size selected to 320bp +/− 5% using Caliper’s XT instrument (PerkinElmer, Waltham MA). Truseq libraries were hybridized to a single read flow cell, and individual fragments were clonally amplified by bridge amplification on the Illumina cBot using the HiSeq SR Cluster Kit v4 cBOT (Illumina, San Diego CA). Sequencing was performed on the Illumina HiSeq 2000 sequencing system using Illumina’s SBS chemistry. Primary analysis and de-multiplexing were performed using Illumina’s CASAVA software 1.8.2.
50 bp FastQ paired-end reads (n = 27.8 Million per sample) were trimmed using Trimmomatic (v 0.33) enabled with the optional “-q” option; 3bp sliding-window trimming from 3’ end requiring minimum Q30. Quality control on raw sequence data for each sample was performed with FastQC. Read mapping was performed via Bowtie (v2.2.4.0) using the mouse genome (mm10) as reference. Gene quantification was done via Feature Counts (Subread Package v1.4.6). Differentially expressed genes were identified using the edgeR (negative binomial) feature in CLC Genomics Workbench 9.5.2 (CLC Bio, Qiagen, Boston, MA) using raw read counts. The edgeR parameters used included trimmed mean of M-values (TMM) for normalization of RNA composition, estimate common dispersion (1e-14), estimate Tagwise dispersion (10), aveLogCPM of 2 and finally an exact Test. False discovery rate (FDR) Benjamini-Hochberg corrected values of less than 0.05 and fold change of greater or equal to 2.0 were used as criteria for significantly regulated genes. Principal component analysis was performed using Emperor.37 Significantly dysregulated genes were clustered with Cluster 3.0, and visualized with Java TreeView. Pathway analysis and gene ontology (Ingenuity Pathway Analysis; IPA, Ingenuity Systems Inc., Redwood City, CA) were performed to identify potential diseases and functions associated with differentially regulated genes. Weighted gene co-expression network analysis (WGCNA)38 analysis of data was conducted to correlate the changes of DNA modifications, histone modifications and expressions of genes caused by LPS.
RNA-Seq validation via qRT–PCR
The purity and integrity of total RNA were confirmed using the Agilent Bioanalyzer (Agilent, Santa Clara CA). The first-strand complementary DNA was synthesized by using SuperScript IV VILO Master Mix (Invitrogen, Carlsbad, CA) with 2 micrograms of RNA in 20 μL reaction. The first-strand complementary DNA mixture was further diluted to 200 μL with RNase-free water and stored at −20 °C until use.
qRT–PCR was performed by Light Cycler 96 (Roche, Indianapolis, IN) using SYBR Select Master Mix (Applied Biosystems, Foster City, CA) and gene-specific primers (see Supplementary Information, Table S1). Twenty-five nanograms of complementary DNA sample were added to a 10 μL reaction to achieve the final concentration of each primer of 0.5 μM. For the PCR amplification, a program of initial denaturation at 95 °C for 15 min, followed by 45 cycles consisting of denaturation at 94 °C for 15 s, annealing at 50 °C for 30 s, and extension at 72 °C for 34 s was used. All samples were normalized to an internal control gene, β-actin. Comparative Ct method was used to assess the relative gene expression.39 Values were expressed as relative units compared with the vehicle control mouse lung tissue and the respective standard deviation.
Results
Global cytosine methylation, hydroxymethylation, and formylation changes in alveolar type II cells of mice treated with LPS
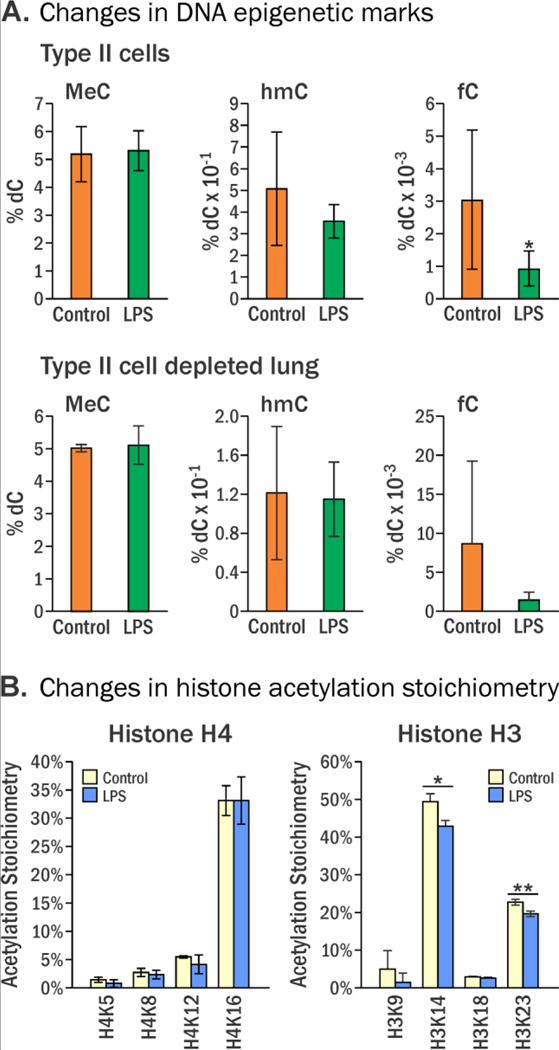
Isotope dilution HPLC-ESI-MS/MS revealed that global cytosine methylation (MeC) levels in DNA isolated from alveolar type II cells of control and LPS treated mice were similar: 5.19 ± 0.99 and 5.32 ± 0.71 % of Cs, respectively (Figure 1A). Global cytosine hydroxymethylation was lower in genomic DNA isolated from type II cells of LPS treated mice (0.36 ± 0.08% of Cs) as compared to controls (0.51 ± 0.26), but these differences did not reach statistical significance (p = 0.13) (Figure 1A). The global levels of fC in DNA isolated from type II cells showed a statistically significant decrease in LPS treated animals relative to controls (0.0030 ± 0.002 vs 0.0009 ± 0.0005 % of Cs, respectively, p = 0.039) (Figure 1A).
Figure 1.
Global levels of epigenetic DNA marks in type II alveolar lung cells of female A/J mice treated intranasally with LPS (4 μg/ day) for 4 days. Data represent mean values ± SD of at least three animals. A, MeC (n = 9), hmC (n = 9), and fC (n = 7), in genomic DNA isolated from type II cells. Statistics were calculated using the Wilcoxon-Mann-Whitney test, p-values: MeC (0.836), hmC (0.366), and fC (0.035). B, LPS-induced changes in histone lysine acetylation stoichiometry presented as percent acetylated. Statistical significance was evaluated between treated and control samples using a two-sided Student’s t-test (* p< 0.05, ** p<0.01). Histone acetylation stoichiometries were assessed to determine global gene activation by deacetylation due to exposure. Measured acetylation sites include lysines 5, 8, 12, and 16 on histone H4 and lysines 9, 14, 18, and 23 on Histone H3.
Histone acetylation changes in type II cells of mice treated with LPS
As mentioned above, cytosine methylation status can influence the recruitment of histone deacetylases and thus affect chromatin structure. Acetylation of histone proteins is a key regulatory mechanism that maintains epigenetic memory in eukaryotic cells.40 Acetylation of histones H3 and H4 correlates site-specifically with the global gene expression profiles and transcriptional activation,41, 42 while the dysregulation of histone acetylation contributes to tumorigenesis and disease progression in lung cancer.41, 43
To determine whether LPS treatment induced global changes in histone acetylation in type II cells, we employed a chemical proteomics strategy that enabled site-specific identification of lysine acetylation stoichiometry.34 Histone proteins were extracted from type II cells isolated from LPS treated and control A/J mice, isotopically labeled, and analyzed by nanoHPLC-ESI+-MS/MS as described previously.33 The data were analyzed by in-house developed scripts to calculate site-specific acetylation stoichiometries.34
We found that treatment LPS decreased acetylation stoichiometry of H3K14 and H3K23, while H3K9, H3K18, H4K5, H4K8, H4K12, and H4K16 showed no significant change (Figure 1B). The observed changes could be functionally important: in budding yeast, H3K14 acetylation is required for activation of G2-M DNA damage checkpoint by directly regulating the compaction of chromatin and by recruiting chromatin remodeling protein complex RSC.44 LPS-induced loss of H3K14Ac may interfere with G2-M checkpoint activation, potentially contributing to mutagenesis and cell proliferation.
Gene expression changes in type II cells from LPS treated A/J mice
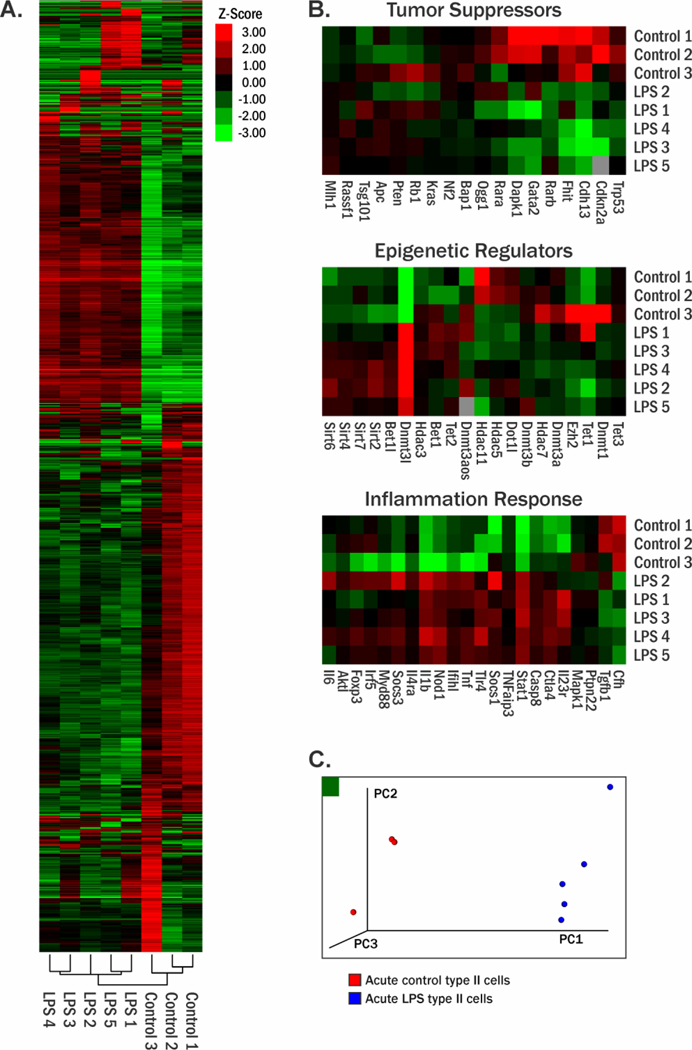
To determine whether LPS treatment led to changes in gene expression, we conducted RNA-Seq analyses using mRNA isolated from type II alveolar cells of LPS-treated A/J mice. Total sequencing reads for each sample were within at least 95% of the targeted 20 million reads for each sample. One LPS treated sample had 68.6 million total number of reads, with the rest ranging from 19.3 – 29.1 million reads. With gene expression profiles, unsupervised principal component analysis was conducted. PCA analysis indicated that control (red) and LPS treated samples (blue) separated into distinct clusters (Figure 2C). This result reveals significant changes in gene expression patterns in type II cells following exposure to LPS.
Figure 2.
Effects of LPS induced inflammation in the lungs of A/J mice on gene expression in type II alveolar cells. A, Gene expression heat map represented as row z-score and unsupervised hierarchal clustering. B, Heat maps showing mRNA expression changes in tumor suppressor genes, epigenetic regulators, and inflammation-associated genes. C, Principle component analysis of the gene expression changes observed in type II cells, controls (red) and LPS treated (blue).
RNA-Seq data was interrogated using edgeR. A total of 1,064 genes exhibited significantly altered levels of expression (fold change > 2) in type II cells of LPS treated mice as compared to controls. A subset of dysregulated genes is shown in Figure 2A as an expression heat map with sample clustering. These data show the control samples clustered together, and treated samples clustered together. Data for the subsets of genes associated with tumor suppression, epigenetic regulation, and the inflammation response are shown in more detail in Figure 2B. The most upregulated genes were Stfa3, Gm15056, Stfa2, Saa3, and Ly6i, while the most downregulated genes were Cyp2a5, Esr2, Grin2b, Cpne5, Cd207, Cdh4, Chil4, Nxf7, and (Table 1). The Stfa2 and Stfa3 genes are both protease inhibitors of papain-like cysteine proteases in the cytoplasm, which prevents the degradation of proteins.45 Saa3 is a protumorigenic mediator in fibroblasts associated with pancreatic cancer,46 and the Ly6i protein is associated with inflammation.47 Cdh4 functions as a tumor suppressor gene in lung cancer48 and has been shown to be hypermethylated in other cancers.49 Among the downregulated genes, Chil4 gene is associated with tissue remodeling and wound healing.50 Nxf7 is part of a family of nuclear export factors and function in mRNA processing in the cytoplasm.51
Table 1.
Genes showing the largest changes in gene expression. Ten most upregulated and 10 most downregulated genes were selected, regardless their function.
| Increased Expression | Decreased Expression | ||||
|---|---|---|---|---|---|
| Fold Change | p-value | Fold Change | p-value | ||
| Stfa3 | 524.14 | 1.52E-82 | Cyp2a5 | −20.40 | 1.23E-11 |
| Gm15056 | 444.25 | 3.91E-51 | Esr2 | −21.00 | 0.028924 |
| Stfa2 | 354.61 | 1.62E-37 | Grin2b | −21.36 | 0.047951 |
| Saa3 | 344.77 | 5.27E-19 | Cpne5 | −21.78 | 9.92E-11 |
| Ly6i | 249.96 | 3.41E-43 | Cd207 | −21.87 | 1.56E-13 |
| BC100530 | 232.73 | 3.77E-84 | Pcdhac2 | −22.28 | 0.009135 |
| BC117090 | 207.32 | 9.05E-50 | Cdh4 | −28.78 | 0.032902 |
| 2010005H15Rik | 159.70 | 2.4E-12 | Fabpl | −31.47 | 7.18E-14 |
| Gm5416 | 139.40 | 5.53E-63 | Nxf7 | −86.82 | 0.017719 |
| Gm5483 | 112.38 | 5.01E-62 | Chil4 | −287.30 | 2.08E-07 |
Differentially expressed genes were annotated and analyzed using Ingenuity Pathway Analysis (IPA) to identify key biological processes, pathways, and networks affected by LPS treatment. Top diseases and biological functions associated with LPS treatment in type II cells included immunological disease, cell adhesion, interferon signaling, metabolic disease, immune cell trafficking, cellular movement, inflammasome pathway, and cell to cell signaling and interaction (Table 2).
Table 2.
Enrichment of canonical pathways of the 1064 differentially expressed genes assessed by IPA software. Top 20 pathways are shown.
| Ingenuity Canonical Pathways | −log(p-value) |
|---|---|
| Hepatic Fibrosis / Hepatic Stellate Cell Activation | 15.60 |
| Granulocyte Adhesion and Diapedesis | 13.80 |
| Agranulocyte Adhesion and Diapedesis | 12.90 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 10.20 |
| LXR/RXR Activation | 9.91 |
| Communication between Innate and Adaptive Immune Cells | 8.74 |
| Interferon Signaling | 8.62 |
| Role of Cytokines in Mediating Communication between Immune Cells | 8.05 |
| Altered T Cell and B Cell Signaling in Rheumatoid Arthritis | 7.85 |
| Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis | 7.81 |
| IL-10 Signaling | 7.42 |
| Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza | 6.59 |
| Differential Regulation of Cytokine Production in Intestinal Epithelial Cells by IL-17A and IL-17F | 6.49 |
| TREM1 Signaling | 6.01 |
| VDR/RXR Activation | 5.79 |
| Inflammasome pathway | 5.72 |
| LPS/IL-1 Mediated Inhibition of RXR Function | 5.64 |
| Toll-like Receptor Signaling | 5.35 |
| Dendritic Cell Maturation | 5.33 |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 5.00 |
Of the 1064 genes deregulated by exposure to LPS, 859 were identified as being related to cancer, including genes involved in cell proliferation and cell growth such as matrix metalloproteinase-9 (Mmp9), interferon-γ (Ifn-γ), and nitric oxide synthase 2 (Nos2).52 Another large group of genes (N = 269) included those associated with cell differentiation (Nos2 and Dnmt3l) and proliferation (N = 405), including Dapk1, Cdh13, Il21, and Mmp9. Mmp9 assists in breaking down the extracellular matrix to allow for cell growth.53 NOS2 is an upstream regulator of Mmp9 expression, and an increase in Nos2 expression results in an increase in Mmp9 expression.52 Ifn-γ is an inflammation response gene responsible for signaling and controlling the immune response.54 Dapk1 and Cdh13 are tumor suppressor proteins that are involved in cell survival and adhesion, respectively, and are typically inactivated in lung cancer.55, 56 Il21 is a cytokine that is induced in the inflammation response and signals CD8+-T cells to expand at the site of inflammation.57 Overall, a wide range of genes were affected by exposure to LPS, and our RNA-Seq results for a group of genes (Dapk1, Cdh13, Cyp1a1, Il21, and Ifn-γ) were confirmed using RT-PCR techniques (Supplementary Information, Figure S1).
Discussion
Although immune responses can suppress tumorigenesis, they can also contribute to cancer initiation and progression. COPD patients are more likely to develop lung cancer,6 while patients with chronic inflammatory bowel diseases, such as ulcerative colitis, Lynch syndrome, and Crohn’s disease are at an increased risk of colon cancer.58 In animal models, co-administration of inflammatory agent LPS increases the size and multiplicity of lung tumors induced by the tobacco carcinogen NNK.7 In contrast, anti-inflammatory drugs like aspirin may reduce colorectal cancer risk.59, 60,61 However, the underlying mechanisms of inflammation mediated carcinogenesis are not completely understood.6, 62
One possible link between inflammation and cancer includes inflammation-mediated epigenetic deregulation. Indeed, several studies reported that epithelial cells at sites of chronic inflammation undergo DNA methylation alterations that are similar to those present in cancer cells, suggesting that inflammation may initiate cancer-specific epigenetic changes.5, 63
In the present study, we characterized LPS-associated epigenetic changes in type II alveolar cells, which are thought to be the cells of origin for non-small cell lung cancer due to their high proliferative potential and their stem-cell like nature.9 LPS is an inflammatory agent present in tobacco smoke and known to increase lung tumor size and multiplicity in animal models.7, 27 Our previous studies provided evidence for severe inflammation, macrophage recruitment, and altered gene expression changes in lung tissues and NNK-induced lung tumors of A/J mice treated with LPS.7, 27 However, LPS-induced epigenetic changes in target cells for lung cancer, type II alveolar cells, had not been previously addressed. Since pulmonary inflammation leads to infiltration of inflammatory cells, 7 epigenetic changes observed in earlier studies could have been a result of drastic changes in cell composition.
In the present study, female A/J mice were intranasally treated with LPS for 4 days, and their lung tissues were harvested and subjected to cell panning to enrich for alveolar type II cells. Isotope dilution capillary HPLC-ESI+-MS/MS was employed to accurately quantify global MeC, hmC, fC levels in genomic DNA, while histone acetylation was assessed by mass spectrometry-based proteomics, and overall changes in gene expression were characterized by RNA-seq. LPS treatment did not affect the global levels of MeC in type II alveolar cells, but a 20% decrease in hmC and 70% decrease in fC was observed (Figure 1A). Although global changes in DNA methylation and hydroxymethylation in mice subjected to acute treatment with LPS were relatively small (Figure 1A), such aggregate analyses cannot reveal methylation and hydroxymethylation changes at individual genomic loci, which may potentially be much greater. Furthermore, a small change in cytosine methylation and hydroxymethylation status can lead to very significant changes in gene expression levels.64 LPS treatment did not affect Histone 4 acetylation, however, Lys 14 and Lys 23 of Histone 3 saw a significant decrease in acetylation (Figure 1C). Acetylation of H3K14 and H3K23 is strongly associated with gene expression, and these changes could have significant implications for gene expression profiles in type II cells.42, 65, 66
RNA-Seq analyses revealed significant LPS induced gene expression changes in type II cells. Specifically, increased expression levels of many proinflammatory genes such as Ifn-γ, Ccl4, Il21, Cxcl9, Cxcl10, and Nos2 were observed (Figure 2B, Table 1). Several epigenetic regulators such as Tet1, Hdac11, and Dnmt3l were affected, with Dnmt3l showing a dramatic increase in expression in LPS treated animals (Figure 2B). Dnmt3l is a catalytically inactive regulatory factor of DNA methyltransferases that can either promote or inhibit DNA methylation depending on the context scaffolding protein for DNA methyltransferases Dnmt3a/b.67, 68 Dnmt3l is essential for the function of de novo DNA methyltransferases: it accelerates the binding of DNA and S-adenosyl-L-methionine (AdoMet) to methyltransferases and dissociates from the complex after DNA binding to the methyltransferases. Furthermore, Dnmt3l recognizes unmethylated histone H3 lysine 4 (H3K4me0) and induces de novo DNA methylation by recruitment or activation of DNMT3. Our observation of dramatically increased expression levels of Dnmt3l in Type II cells upon LPS treatment suggests that the patterns of DNA methylation are likely affected in these cells, despite relatively mild effects of LPS on total cytosine methylation levels.
In contrast, decreased expression of tumor suppressor genes Dapk1, Cdh13, Gata2, and Cdkn2a was observed (Figure 2B). Overall, the list of genes deregulated following LPS treatment was enriched in tumor suppressors, epigenetic regulators, and genes associated with inflammation response (Supplementary Information). A weighted gene co-expression network analysis (WGCNA)38 analysis of data was conducted in an attempt to correlate the changes of DNA modifications, histone modifications, and expressions of genes caused by LPS. These results are included in Supplementary Information. While several modules (e.g. Indianred2, moccasin) were associated with LPS treatment, none of the gene co-expression modules significantly correlated with global 5mC, 5hmC, or fC levels. Therefore, global changes in epigenetic modifications do not explain the observed variations in gene expression.
RNA-Seq data for alveolar type II cells of A/J mice treated with LPS was compared with previously published RNA-Seq data for whole lung tissues of LPS treated mice (see Supplementary Information, Figure S2A).27 We found that many of upstream regulators including Myd88, Nos2, Tnf, interleukins were activated by LPS treatment in both type II cells and whole tissue experiments (Figure S2B). However, some upstream regulators including Tet2, Egf, Mek, and Fox01 were activated in whole lung tissue, but not in in type II cells (Figure S2B). These differences could be due to the inherent dissimilarities between an enriched cell population and the whole tissue, especially since LPS induced lung inflammation increases the number of macrophages and neutrophils that infiltrate the lung tissue as part of immune response.69
Taken together, these results indicate that intranasal exposure of A/J mice to the inflammatory agent LPS induces epigenetic deregulation in alveolar type II cells. LPS treatment reduced the global levels of hmC and fC, led to a loss of H3K14 and H3K23 acetylation, and altered the expression levels of many genes including tumor suppressors and proteins involved in cell adhesion, inflammation, immune response, and epigenetic regulation. To our knowledge, this study provides the first evidence of inflammation –induced epigenetic changes in alveolar type II lung cells, which serve as progenitors of lung adenocarcinoma.
Supplementary Material
Acknowledgments
Funding Information
This work was supported by the U.S. National Cancer Institute [2R01 CA-095039].
Abbreviations
- fC
5-formyl-dC
- hmC
5-Hydroxymethylcytosine
- COPD
chronic obstructive pulmonary disease
- FWHM
full-width at half-maximum
- IPA
Ingenuity Pathway Analysis
- IFN-γ
interferon-γ
- LPS
Lipopolysaccharide
- MMP9
matrix metalloproteinase-9
- nanoLC-ESI-MS/MS
nanoflow liquid chromatography electrospray tandem mass spectrometry
- NOS2
nitric oxide synthase 2
- NSCLC
non-small cell lung cancer
- TET 1–3
ten eleven translocation family of proteins
- TLR4
toll-like receptor 4
Footnotes
Supporting Information
Table of primers used for RNA-Seq validation, RNA-Seq validation, Ingenuity pathway analysis summary.
Bibliography
- (1).Siegel RL, Miller KD, and Jemal A. (2019) Cancer statistics, 2019. CA Cancer J Clin 69, 7–34. [DOI] [PubMed] [Google Scholar]
- (2).Ronai ZA, Gradia S, Peterson LA, and Hecht SS (1993) G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4- (methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis 14, 2419–2422. [DOI] [PubMed] [Google Scholar]
- (3).Hasday JD, Bascom R, Costa JJ, Fitzgerald T, and Dubin W. (1999) Bacterial endotoxin is an active component of cigarette smoke. Chest 115, 829–835. [DOI] [PubMed] [Google Scholar]
- (4).Lu YC, Yeh WC, and Ohashi PS (2008) LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151. [DOI] [PubMed] [Google Scholar]
- (5).Gomes M, Teixeira AL, Coelho A, Araujo A, and Medeiros R. (2014) The role of inflammation in lung cancer. Adv Exp Med Biol 816, 1–23. [DOI] [PubMed] [Google Scholar]
- (6).Durham AL, and Adcock IM (2015) The relationship between COPD and lung cancer. Lung Cancer 90, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Melkamu T, Qian X, Upadhyaya P, O’Sullivan MG, and Kassie F. (2013) Lipopolysaccharide enhances mouse lung tumorigenesis: a model for inflammation-driven lung cancer. Vet. Pathol 50, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Alberg AJ, Brock MV, Ford JG, Samet JM, and Spivack SD (2013) Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 143, e1S-e29S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lin C, Song H, Huang C, Yao E, Gacayan R, Xu SM, and Chuang PT (2012) Alveolar type II cells possess the capability of initiating lung tumor development. PLoS One 7, e53817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BL (2013) Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Prince LS, Okoh VO, Moninger TO, and Matalon S. (2004) Lipopolysaccharide increases alveolar type II cell number in fetal mouse lungs through Toll-like receptor 4 and NF-kappaB. Am J Physiol Lung Cell Mol Physiol 287, L999–1006. [DOI] [PubMed] [Google Scholar]
- (12).Warnecke PM, and Bestor TH (2000) Cytosine methylation and human cancer. Curr. Opin. Oncol 12, 68–73. [DOI] [PubMed] [Google Scholar]
- (13).Song J, Teplova M, Ishibe-Murakami S, and Patel DJ (2012) Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335, 709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, and Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, and Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Song CX, and He C. (2013) Potential functional roles of DNA demethylation intermediates. Trends Biochem. Sci 38, 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, and Pfeifer GP (2011) 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71, 7360–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Das PM, and Singal R. (2004) DNA methylation and cancer. J Clin Oncol 22, 4632–4642. [DOI] [PubMed] [Google Scholar]
- (19).Ji S, Shao H, Han Q, Seiler CL, and Tretyakova NY (2017) Reversible DNA-Protein Cross-Linking at Epigenetic DNA Marks. Angew Chem Int Ed Engl 56, 14130–14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Belinsky SA, Swafford DS, Finch GL, Mitchell CE, Kelly G, Hahn FF, Anderson MW, and Nikula KJ (1997) Alterations in the K-ras and p53 genes in rat lung tumors. Environ. Health Perspect 105 Suppl 4, 901–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Klose RJ, and Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31, 89–97. [DOI] [PubMed] [Google Scholar]
- (22).Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, and Crowell RE (2002) Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res 62, 2370–2377. [PubMed] [Google Scholar]
- (23).Liu F, Killian JK, Yang M, Walker RL, Hong JA, Zhang M, Davis S, Zhang Y, Hussain M, Xi S, Rao M, Meltzer PA, and Schrump DS (2010) Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 29, 3650–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shen H, and Laird PW (2013) Interplay between the cancer genome and epigenome. Cell 153, 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).You JS, and Jones PA (2012) Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Vaz M, Hwang SY, Kagiampakis I, Phallen J, Patil A, O’Hagan HM, Murphy L, Zahnow CA, Gabrielson E, Velculescu VE, Easwaran HP, and Baylin SB (2017) Chronic Cigarette Smoke-Induced Epigenomic Changes Precede Sensitization of Bronchial Epithelial Cells to Single-Step Transformation by KRAS Mutations. Cancer Cell 32, 360–376 e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Qian X, Khammanivong A, Song JM, Teferi F, Upadhyaya P, Dickerson E, and Kassie F. (2015) RNA-sequencing studies identify genes differentially regulated during inflammation-driven lung tumorigenesis and targeted by chemopreventive agents. Inflamm Res 64, 343–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Vuillemenot BR, Hutt JA, and Belinsky SA (2006) Gene promoter hypermethylation in mouse lung tumors. Mol. Cancer Res 4, 267–273. [DOI] [PubMed] [Google Scholar]
- (29).Kerr KM, Galler JS, Hagen JA, Laird PW, and Laird-Offringa IA (2007) The role of DNA methylation in the development and progression of lung adenocarcinoma. Dis. Markers 23, 5–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lieber M, Smith B, Szakal A, Nelson-Rees W, and Todaro G. (1976) A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer 17, 62–70. [DOI] [PubMed] [Google Scholar]
- (31).Kotandeniya D, Seiler CL, Fernandez J, Pujari SS, Curwick L, Murphy K, Wickramaratne S, Yan S, Murphy D, Sham YY, and Tretyakova NY (2018) Can 5-methylcytosine analogues with extended alkyl side chains guide DNA methylation? Chem Commun (Camb) 54, 1061–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, and Carell T. (2011) The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed Engl 50, 7008–7012. [DOI] [PubMed] [Google Scholar]
- (33).Shechter D, Dormann HL, Allis CD, and Hake SB (2007) Extraction, purification and analysis of histones. Nat Protoc 2, 1445–1457. [DOI] [PubMed] [Google Scholar]
- (34).Zhou T, Chung YH, Chen J, and Chen Y. (2016) Site-Specific Identification of Lysine Acetylation Stoichiometries in Mammalian Cells. J Proteome Res 15, 1103–1113. [DOI] [PubMed] [Google Scholar]
- (35).Rappsilber J, Mann M, and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- (36).Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- (37).Vazquez-Baeza Y, Pirrung M, Gonzalez A, and Knight R. (2013) EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Langfelder P, and Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Schmittgen TD, and Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- (40).Strahl BD, and Allis CD (2000) The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- (41).Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, and Esteller M. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37, 391–400. [DOI] [PubMed] [Google Scholar]
- (42).van Leeuwen F, and van Steensel B. (2005) Histone modifications: from genome-wide maps to functional insights. Genome Biol 6, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Lee KK, and Workman JL (2007) Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol 8, 284–295. [DOI] [PubMed] [Google Scholar]
- (44).Wang Y, Kallgren SP, Reddy BD, Kuntz K, Lopez-Maury L, Thompson J, Watt S, Ma C, Hou H, Shi Y, Yates JR 3rd, Bahler J, O’Connell MJ, and Jia S. (2012) Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. J Biol Chem 287, 4386–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Mihelic M, Teuscher C, Turk V, and Turk D. (2006) Mouse stefins A1 and A2 (Stfa1 and Stfa2) differentiate between papain-like endo- and exopeptidases. FEBS Lett 580, 4195–4199. [DOI] [PubMed] [Google Scholar]
- (46).Djurec M, Grana O, Lee A, Troule K, Espinet E, Cabras L, Navas C, Blasco MT, Martin-Diaz L, Burdiel M, Li J, Liu Z, Vallespinos M, Sanchez-Bueno F, Sprick MR, Trumpp A, Sainz B Jr., Al-Shahrour F, Rabadan R, Guerra C, and Barbacid M. (2018) Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci U S A 115, E1147–E1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan S, and Swamynathan SK (2016) Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum Genomics 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Li Z, Su D, Ying L, Yu G, and Mao W. (2017) Study on expression of CDH4 in lung cancer. World J Surg Oncol 15, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Du C, Huang T, Sun D, Mo Y, Feng H, Zhou X, Xiao X, Yu N, Hou B, Huang G, Ernberg I, and Zhang Z. (2011) CDH4 as a novel putative tumor suppressor gene epigenetically silenced by promoter hypermethylation in nasopharyngeal carcinoma. Cancer Lett 309, 54–61. [DOI] [PubMed] [Google Scholar]
- (50).Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, and Elias JA (2011) Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol 73, 479–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Tan W, Zolotukhin AS, Tretyakova I, Bear J, Lindtner S, Smulevitch SV, and Felber BK (2005) Identification and characterization of the mouse nuclear export factor (Nxf) family members. Nucleic Acids Res 33, 3855–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zergoun AA, Zebboudj A, Sellam SL, Kariche N, Djennaoui D, Ouraghi S, Kerboua E, Amir-Tidadini ZC, Chilla D, Asselah F, Touil-Boukoffa C, Merghoub T, and Bourouba M. (2016) IL-6/NOS2 inflammatory signals regulate MMP-9 and MMP-2 activity and disease outcome in nasopharyngeal carcinoma patients. Tumour Biol 37, 3505–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Mehner C, Hockla A, Miller E, Ran S, Radisky DC, and Radisky ES (2014) Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget 5, 2736–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, and Schreiber RD (2001) IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410, 1107–1111. [DOI] [PubMed] [Google Scholar]
- (55).Drilon A, Sugita H, Sima CS, Zauderer M, Rudin CM, Kris MG, Rusch VW, and Azzoli CG (2014) A prospective study of tumor suppressor gene methylation as a prognostic biomarker in surgically resected stage I to IIIA non-small-cell lung cancers. J Thorac Oncol 9, 1272–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Buckingham L, Penfield FL, Kim A, Liptay M, Barger C, Basu S, Fidler M, Walters K, Bonomi P, and Coon J. (2010) PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int. J. Cancer 126, 1630–1639. [DOI] [PubMed] [Google Scholar]
- (57).Davis MR, Zhu Z, Hansen DM, Bai Q, and Fang Y. (2015) The role of IL-21 in immunity and cancer. Cancer Lett 358, 107–114. [DOI] [PubMed] [Google Scholar]
- (58).Freeman HJ (2008) Colorectal cancer risk in Crohn’s disease. World J Gastroenterol 14, 1810–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Burn J, Gerdes AM, Macrae F, Mecklin JP, Moeslein G, Olschwang S, Eccles D, Evans DG, Maher ER, Bertario L, Bisgaard ML, Dunlop MG, Ho JW, Hodgson SV, Lindblom A, Lubinski J, Morrison PJ, Murday V, Ramesar R, Side L, Scott RJ, Thomas HJ, Vasen HF, Barker G, Crawford G, Elliott F, Movahedi M, Pylvanainen K, Wijnen JT, Fodde R, Lynch HT, Mathers JC, Bishop DT, and Investigators C. (2011) Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet 378, 2081–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Cao Y, Nishihara R, Wu K, Wang M, Ogino S, Willett WC, Spiegelman D, Fuchs CS, Giovannucci EL, and Chan AT (2016) Population-wide Impact of Long-term Use of Aspirin and the Risk for Cancer. JAMA Oncol 2, 762–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Force, U. S. P. S. T. (2017) Final Recommendation Statement: Aspirin Use to Prevent Cardiovascular Disease and Colorectal Cancer: Preventive Medication. [Google Scholar]
- (62).Talikka M, Sierro N, Ivanov NV, Chaudhary N, Peck MJ, Hoeng J, Coggins CR, and Peitsch MC (2012) Genomic impact of cigarette smoke, with application to three smoking-related diseases. Crit Rev. Toxicol 42, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Lee J, Taneja V, and Vassallo R. (2012) Cigarette smoking and inflammation: cellular and molecular mechanisms. J. Dent. Res 91, 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Baylin SB, and Jones PA (2016) Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Cai L, Sutter BM, Li B, and Tu BP (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, and Tora L. (2012) H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Chedin F, Lieber MR, and Hsieh CL (2002) The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A 99, 16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Jia D, Jurkowska RZ, Zhang X, Jeltsch A, and Cheng X. (2007) Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Dagvadorj J, Shimada K, Chen S, Jones HD, Tumurkhuu G, Zhang W, Wawrowsky KA, Crother TR, and Arditi M. (2015) Lipopolysaccharide Induces Alveolar Macrophage Necrosis via CD14 and the P2X7 Receptor Leading to Interleukin-1alpha Release. Immunity 42, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.