Summary
• Defects in flagella/cilia are often associated with infertility and disease. Motile male gametes (sperm cells) are an ancestral eukaryotic trait that has been lost in several lineages like flowering plants. Here, we made use of a phenotypic male fertility difference between two moss (Physcomitrella patens) ecotypes to explore spermatozoid function.
• We compare genetic and epigenetic variation as well as expression profiles between the Gransden and Reute ecotype to identify a set of candidate genes associated with moss male infertility. We generated a loss-of-function mutant of a coiled-coil domain containing 39 (ccdc39) gene that is part of the flagellar hydin network.
• Defects in mammal and algal homologs of this gene coincide with a loss of fertility, demonstrating the evolutionary conservation of flagellar function related to male fertility across kingdoms. The Ppccdc39 mutant resembles the Gransden phenotype in terms of male fertility.
• Potentially, several somatic (epi-)mutations occurred during prolonged vegetative propagation of Gransden, causing regulatory differences of e.g. the homeodomain transcription factor BELL1. Probably these somatic changes are causative for the observed male fertility defect. We propose that moss spermatozoids might be employed as an easily accessible system to study male infertility of humans and animals in terms of flagellar structure and movement.
Keywords: Physcomitrella patens, moss, male infertility, flagella, spermatozoid, sperm, cilia
Introduction
Motile plant gametes allow studying sperm cell fertility
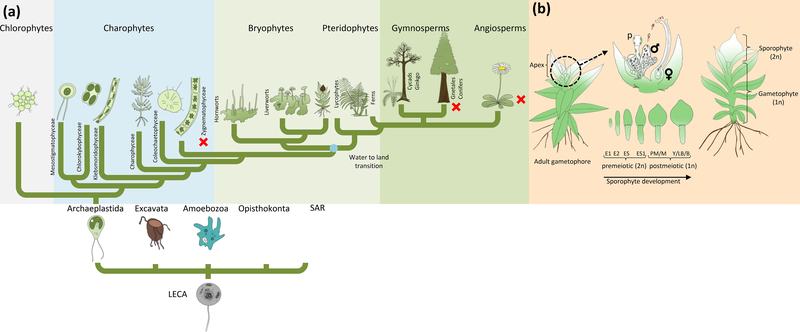
Motile (flagellated) gametes are an ancestral character of eukaryotes (Stewart & Mattox, 1975; Mitchell, 2007) that has been secondarily lost in lineages such as the Zygnematales (Fig. 1A) (Transeau, 1951), which reproduce via conjugation of aplanogametes, and flowering plants, in which pollen transport non-motile gametes to the female (Southworth & Cresti, 1997; Renzaglia & Garbary, 2001). Flagella play an important role in sexual reproduction. In the unicellular algal model organism Chlamydomonas reinhardtii flagella are responsible for motility of the organism and serve during sexual reproduction to mediate a species-specific adhesion between two cells of different mating types (van den Ende et al., 1990). In streptophyte algae (Streptophyta comprise charophycean green algae as well as land plants), male gametes (spermatozoids) are the only motile cells of sessile multicellular algae, swimming to the oogonia to fertilize the egg cell (McCourt et al., 2004; Hackenberg & Twell, 2019). After the water-to-land-transition of plants this system was retained in bryophytes (hornworts, liverworts and mosses) and pteridophytes (lycophytes and monilophytes) in which motile (flagellated) spermatozoids require water to swim to the egg cell. Flagella of spermatozoids show a common architecture (Carvalho-Santos et al., 2011). During land plant evolution, loss of motile sperm occurred twice in seed plants (Fig. 1A), after the evolution of cycads and Ginkgo (both of which have pollen and motile male gametes), probably in the MRCA of conifers plus Gnetales, and in flowering plants (Renzaglia et al., 2000; Renzaglia & Garbary, 2001). Flagellated plants, that have retained motile spermatozoids, are an easily accessible system to study the fertility and other characteristics of male gametes. The moss Physcomitrella patens is particularly attractive in that regard because it develops spermatozoids in superficial male sex organs (antheridia) on an independent generation (gametophyte) that is readily cultured and manipulated (Cove, 2005; Landberg et al., 2013).
Figure 1: Cladogram displaying independent losses of motile sperm during plant evolution and Physcomitrella patens sexual life cycle.
(a): The LECA (last eukaryotic common ancestor) gave rise to eukaryotes of all five kingdoms, in which Archaeplastida harbour the green lineage. Zygnematophyceae, Coleochaetophyceae and Charophyceae (ZCC grade) are sister to land plants (One Thousand Plant Transcriptomes, 2019). Bryophytes are probably monophyletic (see text for references) comprise hornworts, liverworts and mosses, and are sister to vascular plants. Flagella were present in the LECA and were lost three times independently during plant evolution (red cross indicates absence of flagella). Drawn based on DeVries and Archibald 2018, Mast et al., 2014, Puttick et al., 2018.
(b): Upon environmental stimulus, reproductive organs (gametangia) develop on the apex of each gametophore, namely archegonia (female) and antheridia (male). Additionally, paraphyses emerge (p). Mature antheridia release the motile (flagellated) spermatozoids upon watering. The sperm cells swim through the archegonial venter to fertilize the egg cell. After fertilization, embryo development (E1/E2) and sporophyte development (ES-B) occurs. The mature sporophyte is located on the apex of the gametophore and releases haploid spores of the next generation. P. patens is predominantly selfing. Embryo/sporophyte developmental stages according to Hiss et al. (2017).
The moss model Physcomitrella patens
As the probably monophyletic sister clade to vascular plants (Gitzendanner et al., 2018; Puttick et al., 2018; One Thousand Plant Transcriptomes, 2019), bryophytes are important in addressing evolutionary-developmental questions, e.g. (Sakakibara et al., 2008; Aya et al., 2011; Sakakibara et al., 2013; Horst et al., 2016). In particular, the moss P. patens is a well-developed functional genomics model system that is easily accessible for the study of mammalian homologs (Ortiz-Ramirez et al., 2017; Sanchez-Vera et al., 2017). The genome was published in 2008 (Rensing et al., 2008) and recently updated to chromosome scale (Lang et al., 2018). Collections of worldwide accessions are available (Beike et al., 2014), of which some are likely to constitute ecotypes (Lang et al., 2018). The predominantly used ecotype Gransden (Gd) was derived from a single spore isolate collected 1962 in Gransden Wood (UK). The Gd cultures used in most labs worldwide were typically propagated vegetatively and several labs reported fertility issues over the past decades (Ashton & V. S. Raju, 2000; Perroud et al., 2011; Landberg et al., 2013; Hiss et al., 2017), potentially the result of somatic (epi-)mutations through decades of vegetative propagation (Ashton & V. S. Raju, 2000). The ecotype Reute (Re) collected 2006 in Reute (Germany) has a low genetic divergence (low number of SNPs) to Gd, is highly self-fertile and thus has recently been introduced as a Gd alternative to enable studies involving sexual reproduction (Hiss et al., 2017).
P. patens sexual reproduction
P. patens’ sexual reproduction is initiated (under natural conditions in autumn) when day length shifts towards short days and temperature drops (Engel, 1968; Nakosteen & Hughes, 1978; Hohe et al., 2002). On the tip (apex) of the leafy gametophores the apical stem cell produces the antheridium initial stem cell (Kofuji et al., 2018), which subsequently gives rise to a bundle of antheridia, the male reproductive organs (Fig. 1B). Each antheridium consists of a single outer cell layer or jacket surrounding a mass of spermatogenous tissue that produces up to 160 motile spermatozoids. Upon maturity the tip cell of the antheridia swells and bursts (when moistened by water) to release mature, swimming male gametes (spermatozoids). A few days after initiation of antheridia development, the female gametangia (archegonia) start to develop (Kofuji et al., 2009). They comprise a flask shaped egg-containing venter and an elongated neck that opens during maturation via degeneration of the neck canal cells, allowing swimming spermatozoids to reach the egg cell (Landberg et al., 2013; Hiss et al., 2017; Sanchez-Vera et al., 2017). After fertilization, the diploid zygote undergoes embryogenesis and develops into the sporophyte, which eventually generates haploid spores via meiosis (Fig. 1B). Considering the difference in fecundity of Gd and Re (Hiss et al., 2017) we aim here to determine the genes underlying this difference, employing P. patens as a model for studying sperm fertility. For this purpose, we analysed the nearly self-sterile ecotype Gransden (Gd) in comparison with the highly fertile ecotype Reute (Re), using genetic and epigenetic variation data, fertility phenotyping as well as transcript profiling. Based on publicly available array data of developmental stages during sexual reproduction (Ortiz-Ramirez et al., 2017; Fernandez-Pozo et al., 2019) and network analyses, we generated a deletion mutant of a gene exclusively expressed in sexual reproductive organs. The mutant displays a loss of male fertility similar to that observed in Gd, and to that in human/mammal mutants of the ortholog. Our study demonstrates that the moss male germ line can serve as a model for flagellar dysfunction disease.
Material and Methods
Plant material
P. patens Gd (Rensing laboratory strain), Gd Japan (Jp) and Reute (Hiss et al., 2017) were cultivated under the same conditions as described in (Hiss et al., 2017) for sporophyte development, and for crossing analysis as described in (Perroud et al., 2019). For gametophytic stage phenotyping, protonemal cells were placed on 9cm Petri dishes with solid minimal medium (1% w/v agar, Knop’s medium, (Knop, 1868)) enclosed with 3M micropore tape and inoculated 7 days for protonema and in total 10 days for gametophore analysis in long-day (LD) conditions (70 μmol m−2 s−1, 16h light, 8h dark, 22°C). Gametangia harvest and analysis was performed at 21 days after short-day (SD, 20 μmol m−2 s−1, 8h light, 16h dark, 15°C) transfer.
Counting and statistical analysis of sporophytes per gametophore under selfing and crossing conditions, data visualization
To determine the number of sporophytes developed, counting of the selfed F1 of three independent mutant lines (ccdc39#8, ccdc39#41, ccdc39#115, Fig. S1) was performed at 30 days after watering as described in (Hiss et al., 2017) using a Leica S8Apo binocular (Leica, Wetzlar, Germany). Counting of the crosses was performed according to (Perroud et al., 2019) using a fluorescence stereomicroscope SteREO LumarV12 (Carl Zeiss, Oberkochen, Germany). For counting of the selfed F1, at least five independent biological replicates with in total 537 to 742 gametophores per strain were analysed. For the crossings, three biological replicates with in total 300 to 630 gametophores per strain were analysed. For the comparison between Re, Gd and GdJp three replicates with in total 523 to 730 gametophores were analysed. Statistical analysis was carried out using Microsoft Excel 2016 (Microsoft) and R. Plots in R were done using ggplot2 (Wickham, 2016) and ggbeeswarm quasirandom (https://CRAN.R-project.org/package=ggbeeswarm).
Microscopic analysis of gametangia and sporophytes
Harvest and preparation of sporophytes, gametangia and spermatozoids was performed with a Leica S8Apo binocular (Leica, Wetzlar, Germany). Microscopic images were taken with an upright DM6000 equipped with a DFC295 camera (Leica). For both devices the Leica Application Suite version 4.4 was used as the performing software. Images were processed (brightness and contrast adjustment) using Microsoft PowerPoint.
Spermatozoid microscopy, DAPI staining and counting
Preparation for 4’,6-diamidino-2-phenylindole (DAPI) staining/flagellar analysis and counting of spermatozoids per antheridium was performed after 21 days of SD inoculation. Per replicate, a single antheridium was harvested in 4 μl sterile tap water applied to an objective slide. For Gd and Re, 10 antheridia each were analysed. The spermatozoids were released using two ultra-fine forceps (Dumont, Germany) and the sample was dried at room temperature (RT). Samples were fixed with 3:1 ethanol/acetic acid and after drying, spermatozoid nuclei were stained applying 0,7 ng/μl DAPI (Roth, Germany) in tap water. The samples were sealed with nail polish (www.nivea.de). Microscopic images were taken with an upright DM6000 equipped with a DFC295 camera (Leica). Brightness and contrast of microscopy images was adjusted using Microsoft PowerPoint.
Spermatozoid motility analysis
Mature apices (21 d of SD inoculation; n >= 3) were prepared in sterile tap water. The spermatozoids’ motility was recorded using the Leica LAS 4.4 Movie Module (Leica). Analysis of the velocity (μm/s) was performed in ImageJ (Schindelin et al., 2012) using the plugin ManualTracking (Fabrice P. Cordelières).
Electron microscopy
For SEM images, mature spermatozoids (21 d after SD transfer) were harvested on an objective slide covered with polylysine. No differences in the timing of sexual development had previously been observed between Gd and Re ecotypes (Hiss et al., 2017). Chemical fixation was performed with 2.5% glutaraldehyde and an ascending alcohol preparation using 30, 50, 70, 90, and two times 100 % of EtOH was performed. Subsequently, the samples were critical point dryed (Tousimis, Rockville, USA) and coated with 20nm gold using a Leica Sputter (Leica, Wetzlar, Germany). The samples were then analysed with a SEM (Philips XL30, Hamburg, Germany). For TEM, adult apices (21 d after SD transfer) were used. Sample preparation followed that of (Renzaglia et al., 2017) with slightly modified infiltration (25% Epon (Fluka® Analytical, USA) in propylene oxide for 2h; 50% Epon in propylene oxide overnight; next day pure Epon and polymerization). 35–50 antheridia were examined each in Gd, Re and ccdc39 plants, representing all stages of development. The substructure of flagella demonstrated herein in Gd and Re spermatozoids is visible in mature gametes, thus image data were derived only from antheridia with mature or released spermatozoids (2–5 per ecotype). One antheridium in section contains 7–25 spermatozoids in various planes of section.
Nucleic acid isolation
Genomic DNA for genotyping was isolated with a fast extraction protocol, using one to two gametophores as described in (Cove et al., 2009). To determine candidate gene expression levels in juvenile (5 weeks LD growth) and adult (21 days after SD transfer) apices, each at least 25 apices were harvested in 20μl RNA later (Qiagen, Hilden, Germany). Tissue was stored at −20°C, until RNA was extracted using the RNeasy plant mini Kit (Qiagen). For RNA-seq RNA was isolated from 40 antheridia bundles, comprising 4–6 antheridia each, harvested at 21 day after SD transfer and directly stored in 20μl RNAlater. RNA was extracted with a combination of RNeasy plant mini and RNeasy micro kit (Qiagen). The RNeasy plant mini kit was used until step 4. Flowthrough of the QIAshredder spin column was diluted in 0,5 volume 100% EtOH and then transferred to the RNEasy spin columns of the micro kit for further treatment with the micro kit. RNA concentration and quality was analysed with Agilent RNA 6000 Nano Kit at a Bioanalyzer 2100 (Agilent Technologies).
Real-time PCR (RT-PCR) & quantitative real-time PCR (qPCR)
To determine gene expression in juvenile vs. adult, respectively Gd vs. Re adult apices, RNA was extracted as described above. cDNA synthesis was performed using the Superscript III (SSIII) kit (ThermoFisher Scientific) according to the manufacturers` protocol but using ½ of the SSIII amount. Primers were designed manually with an annealing temperature +/− 60°C and a product length of ca. 300 bp. Single genomic locus binding properties were tested by using BLAST against the V3.3 genome of P. patens. Real-time PCR was performed using 5 ng of cDNA as input for PCR reaction with OneTaq from New England Biolabs). PCR products were visualized via gel electrophoresis using peqGREEN from (VWR, Germany). As size standard, the 100 bp ladder (NEB) was used. Real-time qPCR was carried out with two (for juvenile vs. adult comparison) or three (expression determination of ccdc39) biological replicates as published in Hiss et al., 2017. As reference gene, act5 (Pp3c10_17070V3.1, (Le Bail et al., 2013) was chosen, due to homogenous expression in juvenile and adult apices in Gd and Re (Fig. S2). For primer sequences see Table S1.
Antheridia bundle RNA-seq & analysis
RNA-library preparation and RNA-seq was performed at the MPI genome center Cologne (mpgc.mpipz.mpg.de). For each ecotype, three libraries were prepared with the ultra low input RNA-seq protocol followed by sequencing with Illumina HiSeq3000 (150 nt, single ended). For gene expression and DEG analysis for Gd 81,7 mio. and for Re 82,4 mio. uniquely mapped reads were used. The analysis was performed as previously described for the Physcomitrella patens gene atlas project (Perroud et al., 2018), using the strict consensus of three DEG callers, defining transcripts as expressed if their RPKM was greater than or equal to 2. For RPKM data see Table S2 and for DEGs Table S3.
Methylated and differentially methylated positions (DMP) and overlap with single nucleotide polymorphisms (SNP)
The Re data was generated and treated as described for Gd in (Lang et al., 2018). One biological and one technical replicate was prepared. 69,998,270 paired end reads of 90 bp length were used. Methylation levels were called using the R package methylkit (Akalin et al., 2012) and for further analysis only positions with a coverage equal to 9 or above and a minimal mapping quality of 20 were used. DMPs between Gd and Re were called using methylKit using a q-value < 0.01 and a methylation difference of at least 25% (Akalin et al., 2012). Differentially methylated positions were associated with gene models by using bedtools intersect (Quinlan & Hall, 2010). Hyper- / hypo-methylation was determined by comparing the number of DMPs per gene (Fisher’s exact text, q < 0.01, difference = 25). For GO bias calculation all genes within all contexts (CHG, CHH, CG) were used that were detected to be affected by DMPs. This gene list was used to identify genes also affected by a SNP between Gd and Re (Hiss et al., 2017) to generate GObias data of the intersection.
Vector construction and preparation
Deletion constructs were built using pBHRF (Schaefer et al., 2010) as backbone. PCR reactions were performed with OneTaq (NEB), restriction enzymes were purchased from NEB as well as the T4 ligase and 100bp respectively 1kb ladder. For primer sequences see Table S1. 5’- and 3’- homologous regions of Ppccdc39 were amplified from gDNA using primer pairs ccdc39_HR1_for/rev and ccdc39HR2_for/rev. Flanks were subsequently cloned into pME-TA(Miles & Verkade, 2014) via TA cloning to generate 5’- and 3’ flank containing vectors. The 5’ flank was cut and cloned into pBHRF using the restriction enzymes PmeI and XhoI. The 3’ flank was cut and cloned using the restriction enzymes NarI and AscI. The final deletion cassette consists out of the two homologous regions flanking the 35S:Hygromycin:CamVter selection marker (Fig. S3). Plasmid amplification was performed in E. coli Top10 cells, plasmid extraction was performed using the NucleoBond Xtra midi kit (Machery-Nagel, Düren, Germany). For stable transfection, the plasmid was cut using PmeI and AscI, and after ethanol-precipitation solved in sterile TE.
Moss transfection
Moss transfection was carried out in the Re wild type background (Hiss et al., 2017) with small modifications as published in (Cove et al., 2009): after protoplast regeneration all subsequently used medium was Knop medium supplemented with 5mM di-ammonium tartrate (Sigma-Aldrich, Germany). For selection, the antibiotic hygromycin B (Roth, Germany) was used as an additive, with a concentration of 20mg/l.
Genotyping
To identify correctly integrated mutants a set of PCR controls was performed (Fig. S4). WT locus absence was tested by using the primer pair ccdc39_ingDNA_for/rev located in the coding sequence of ccdc39. Proper insertion of the 5’ flank was tested using ccdc39_HR1in (located upstream of the 5’ flank) and p35S_rev (located in the resistance cassette promotor). The 3’ flank was tested using ccdc39_HR2in (located downstream of the 3’ flank) and tCMV_for (located in the terminator of the resistance cassette). Full length PCR was performed using ccdc39_HR1in and ccdc39_HR2in.
Phylogeny
A BLAST using the BLOSSUM45 matrix was performed against a database of sequenced eukaryotic genomes. The results were filtered against false positives (Rost, 1999). The alignment was generated using muscle (Edgar, 2004) and manually cropped using Jalview (Waterhouse et al., 2009), to remove columns of low quality as well as sequences representing less than 50% of the alignment. Using ProtTest (Darriba et al., 2011), the best fitting evolutionary model was determined to be LG+G+F which was employed for prior probabilities in MrBayes (Ronquist et al., 2012), run with two hot and two cold chains for 40,000 generations (divergence of split frequencies < 0.01). The tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Gene Ontology analysis and visualization
Gene ontology analysis was performed as described in (Widiez et al., 2014) based on the P. patens V3.3 annotation. Visualization was done using Wordle (http://www.wordle.net/). Green font colors mark over-represented, red font colors mark under-represented GO terms. Size and color correlate with significance. Darker colors and larger font size are correlated with higher significance (q < 0.0001) whereas lighter colors and smaller font size indicate a lower q-value (q > 0.0001 but < 0.05).
Network analysis and gene identifier conversion
For network analysis www.string-db.org (Szklarczyk et al., 2015) was used, using V3.3 protein models. To convert gene IDs from the most recent V3.3 annotation to V1.6 used by string-db.org, the conversion table published by (Perroud et al., 2018) was used.
Results & Discussion
The Gransden ecotype displays a reduced male fertility and a defect in spermatozoid motility
The ecotype Reute (Re) was shown to develop a significantly higher number of sporophytes per gametophore in comparison to the broadly used ecotype Gransden (Gd, (Hiss et al., 2017)). To proof the assumption that the original Gd isolate is not infertile, we analyzed the fertility of GdJp (based on a Gd strain shipped to Japan in the early 90s and propagated involving regular sexual reproduction since then). We found that the sporophyte development was significantly higher in comparison with the predominantly vegetatively propagated Gd, although significantly lower than in Re (t-test, p < 0.05, Fig. S5). To determine whether the reduced fertility of Gd is based on the female or male reproductive apparatus, crossing analyses between Gd and the fluorescent marker line Re-mCherry were performed (Perroud et al., 2011; Perroud et al., 2019). While selfing results in a homozygous sporophyte (inbred line), crossing results in heterozygous sporophytes yielding recombined spores. Re developed on average 98.1% sporophytes per gametophore, of which 3.4% were heterozygous (Fig. 2A). This rate is expected, since P. patens is known to predominantly self-fertilize (Perroud et al., 2011). In comparison, Gd developed 88.33% sporophytes per gametophore of which 97.3% were heterozygous (significantly more than Re: p < 0.01, Chi-square test, Fig. 2A).
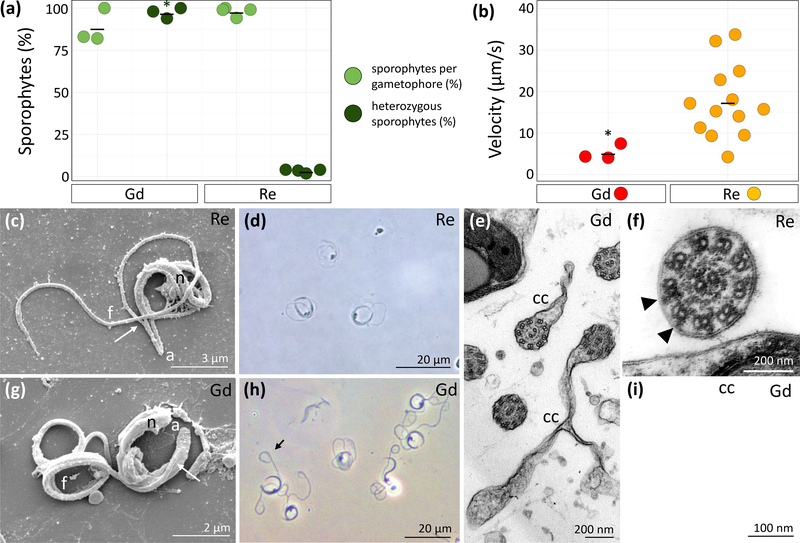
Figure 2: Comparative analysis of Physcomitrella patens ecotypes Gransden (Gd) and Reute (Re) male reproductive apparatus.
(a): Crossing analysis between Re (n = 430), Gd (n = 300) and fluorescent marker line Re-mcherry. Re develops 98.1% of sporophytes per gametophore of which 3.4% are heterozygous (derived from crossing, not selfing). Gd develops 88.33% sporophytes per gametophore of which 97.3% are heterozygous. Mean represented by black line. Significance shown by asterisk (Chi-square test, p < 0.01, four biological replicates each).
(b): Re (n = 10) and Gd (n = 3) spermatozoids differ significantly in velocity (two-sided t-test, p < 0.01,*). Mean represented by black line. Note that only three Gd spermatozoids were measured because the majority shows no motility at all (Fig. S6B), and hence velocity = 0.
(c): Scanning electron micrograph (SEM) of Re sperm cell showing cell architecture. From the cell anterior (a), the two flagella emerge (arrow) at staggered locations from the coiled nucleus (n). One of two long flagella (f) is visible and extends beyond the cell proper.
(d): Phase contrast image of swimming Re spermatozoids.
(e): Transmission electron micrograph (TEM) cross-sections of mature Gd flagella showing cytoplasmic connections (cc) that connect coils.
(f): TEM of a mature spermatozoid of Re. The axoneme exhibits clear outer dynein arms (black arrows) and the plasmalemma is closely associated with the nine doublets.
(g): SEM of Gd spermatozoid with architecture as in Re spermatozoids in C, except the flagella remain coiled.
(h): Phase contrast image of swimming Gd spermatozoids showing loops at the posterior end of the flagella (arrow).
(i): TEM higher magnification of a Gd flagellum in figure 2E showing fewer electron densities associated with the central pair complex (arrow) and microtubule doublets.
Thus, Gd archegonia are fully functional and can develop comparable numbers of sporophytes when fertilized by a male fertile partner. Hence, the Gd male reproductive apparatus is impaired. As previously reported, antheridia development followed the same time frame in both ecotypes (Hiss et al., 2017) and at the time point of the analysis, 1–2 antheridia per bundle were mature (stage 9–10 after (Landberg et al., 2013)), whereas the other antheridia usually varied between stages 6–8. No differences could be seen with regard to the occurrence of axillary hairs in both ecotypes. Analysis of the spermatozoid number per antheridium showed no significant differences between Re (n = 10, median 123) and Gd (n = 10, median 125, Fig. S6A). This is less than determined by an approximation method (Horst & Reski, 2017), counting DAPI stained spermatozoids not released from the antheridium. Motility analysis of 103 Re and 50 Gd spermatozoids showed that only a small number of Gd spermatozoids are motile (i.e., that their velocity is > 0, Fig. S6B). Motile Gd spermatozoids show a significantly reduced velocity (Re mean: 17,58 μm/s; Gd median: 5,32 μm/s; p < 0.01, t-test; Fig. 2B). After release of the spermatozoids through the bursting tip cells of the antheridia, Re spermatozoids started moving a few seconds after release, whereas Gd spermatozoids showed motility only rarely. With regard to their structure, the posterior coils of the flagella in Gd gametes fail to individualize, resulting in a coiled posterior loop significantly more often (90.2%) than in Re (6.6%, t-test, p < 0,01, Fig. 2C, D, G, H, Fig. S6C). Interestingly, coiled flagella similar to those in Gd spermatozoids are also known from mouse and human infertile sperm (Tang et al., 2017; Dong et al., 2018; He et al., 2018). There are further ultrastructural differences between Re and Gd sperm cells. Two cylindrical flagella develop around the outside of the spermatozoid as the nucleus condenses and elongates in both ecotypes (Fig. 2C, D, G, H). Cytoplasmic connections that often extend from flagella and join revolutions of axonemes in Gd spermatozoids are likely responsible for the coiled posterior loop of the flagella (2E). The axonemes of both ecotypes exhibit the typical nine doublets and two central pair microtubules that characterize flagella of most eukaryotes (Fig. 2E, F, I) (Renzaglia et al., 2017). Axonemes in Re mature spermatozoids demonstrate visible dynein arms and protein projections in the central pair complex (Fig. 2F). In contrast, Gd flagella have less clear dynein arms and lack electron densities in the central pair complex (Fig. 2 E, I), indicating loss of associated proteins (Carbajal-Gonzalez et al., 2013).
Genes related to flagellar assembly and motility harbour (epi-)mutations between Gd and Re
Similar to motile sperm, DNA methylation is an ancestral eukaryotic feature (Feng et al., 2010) and is supposed to regulate gene expression on the DNA level (Zemach et al., 2010). In P. patens, methylated gene bodies usually are associated with lower gene expression (Lang et al., 2018) and loss of the DNA methyltransferase PpMET1, which is involved in CG DNA methylation of gene bodies, inhibits sporophyte development (Yaari et al., 2015). Recently, it has been shown, that the male reproductive organs undergo severe changes in DNA methylation in the liverwort M. polymorpha during sexual reproduction (Schmid et al., 2018). Hence, we expected DNA methylation to play a role during regulation of sexual reproduction in moss. Whole genome bisulfite sequencing (bs-seq) of Re and Gd adult gametophores (bearing gametangia; Fig. 1B) was performed. Differentially methylated positions (DMPs) in all three methylation contexts (CHG, CHH, CG) were determined. In total, 671 genes harboring DMPs were found (Table S4). GO bias analysis of the genes containing DMPs showed enriched terms related to cilium movement and motile cilium assembly, as well as protein and macromolecule modification, which includes post-translational modifications like ubiquitination (Fig. S7A). Using the intersect of genes that contain DMPs and single nucleotide polymorphisms (SNPs, Table S5, (Hiss et al., 2017)) the number of GO terms associated with cilia and microtubule based movement was found to be increased (from 3 to 6, Fig. 3A), suggesting genetic and epigenetic effects on the Gd phenotype. In the intersection, additional GO terms related to protein phosphorylation were found to be over-represented (Fig. 3A). In C. reinhardtii, it was shown that protein phosphorylation is a key event of flagellar disassembly (Pan et al., 2011) and that the phosphorylation state of an aurora-like protein kinase coincides with reduced flagellar length (Luo et al., 2011).
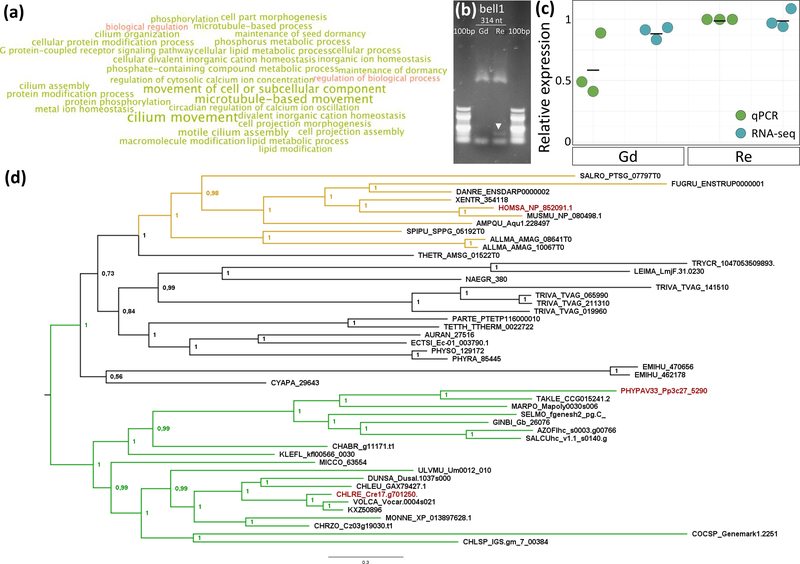
Figure 3: Physcomitrella patens bell1 and ccdc39 expression, GO bias of DMP/SNP overlap and ccdc39 phylogeny.
(a): Gene Ontology bias analysis of genes differentially methylated and SNP- containing in comparison of Gd vs. Re. The intersect of DMP and SNP affected genes shows over-representation of GO terms associated with cilia motility. Over-represented terms are shown in green, whereas under-represented terms are shown in red. Larger font size correlates with a higher significance level.
(b): RT-PCR expression analysis of genes expressed in Gd and Re adult apices shows expression of bell1 (314 nt) in the Re (white arrow) but not in the Gd background.
(c): Relative expression of ccdc39 in qPCR (green, n = 3) and RNA-seq (blue, n = 3) data shows higher expression in Re (mean qPCR: 1, mean RNA-seq: 1) in comparison to Gd (mean qPCR: 0.6, mean RNA-seq: 0.89). Mean marked by black bars.
(d): Phylogenetic analysis of CCDC39 shows clear correlation with the presence of flagella and generally reflects species relationships. Planta are shown in green, opisthokonts in yellow, protozoa, SAR and fungi in black. Homo sapiens, Chlamydomonas reinhardtii and Physcomitrella patens are marked in red.
The transcriptome of antheridia
To gain deeper insight into gene expression differences, cDNA sequencing (RNA-seq) of Gd and Re antheridial bundles was performed. In total 19,481 genes were found to be expressed (RPKM >= 2) in Gd and 19,613 in Re (Table S6/S7). 566 genes were uniquely expressed in Gd and 698 genes in Re (Table S8/S9). GO bias analysis of all genes expressed in antheridia of Gd (Fig. S7B) and Re (Fig. S7C) showed an over-representation of terms like cellular metabolic process, and cellular component organization as compared to the overall distribution in the P. patens genome. Spermine and spermidine belong to the amides (included in the metabolic process and cellular process GO terms) and play a major role in male fertility in mammals (Lefevre et al., 2011) which, based on the RNA-seq GO analysis, also appears to be the case in P. patens. The few under-represented terms found are connected to mRNA capping, potentially indicating that during gamete development mRNA capping could be performed by less characterized capping enzymes, or that translation occurs independently of mRNA capping. It was postulated that the first eukaryotic mRNA translation was probably driven by internal ribosomal entry sites (IRES) and that the cap-dependent (CD) mechanism evolved later. The former mechanism, according to this hypothesis, was retained as an additional regulation of the translation in specific situations like stress responses (Hernandez, 2008; Godet et al., 2019). Also, cap-independent (CI) translation was recently suggested to be a robust partner of CD translation and to appear prevalently in germ cells (Keiper, 2019), which matches with the presented results. The cellular component GO-terms represent the large number of reorganization events taking place during spermatogenesis. Such changes affect the cellular and chromatin organization in e.g. human sperm cells (Lie et al., 2009; Champroux et al., 2016). Interestingly, for Gd the GO term reproduction appears to be under-represented, whereas for Re the GO term reproduction is over-represented in comparison to the reference genome (Gd) indicating less genes with the GO term to be expressed in Gd and more genes to be expressed in the Re background (Fig. S7B, C).
Differential expression between Gd and Re
In human, transcriptional regulation is important to control spermatogenesis (Bettegowda & Wilkinson, 2010). Within the genes expressed in both ecotypes, in total 1,688 transcription associated proteins (TAPs; comprising transcription factors, TFs, and transcriptional regulators, TRs) could be identified using TAPscan (Wilhelmsson et al., 2017). Within the Re uniquely expressed genes, in total 59 TAPs of 25 different TAP families could be identified, including HD_BEL and MADS_MIKCc type proteins (Table S10). Interestingly, bell1 (HD_BEL) previously was described not to be expressed in antheridia (Horst et al., 2016), which indeed is true for Gd (used in the study by Horst et al.), but not for Re (Fig. 3B), suggesting that BELL1 might play a role in male fertility in P. patens. MADS-box genes are important for flower, pollen and fruit development (Theißen & Gramzow, 2016) and have previously been associated with sexual reproduction in P. patens (Hohe et al., 2002; Quodt et al., 2007; Singer et al., 2007). P. patens MIKCc-type genes are important for motile flagella and external water conduction (Koshimizu et al., 2018). In the latter study, PPM1, PPM2 and PPMC6 are the key players for flagellar motility, but since the triple KO did not completely phenocopy the sextuple KO (ppm1, ppm2, ppmads1, ppmc5, ppmc6, ppmadss), the other genes apparently also play a role. While ppm1, ppm2 and ppmc6 are expressed in both, Re and Gd antheridia, mads1 (Hohe et al., 2002) and ppmc5 are exclusively expressed in Re, suggesting a putative involvement of these two genes in flagellar motility. Among the Gd uniquely expressed genes (Table S8), 25 TAPs belonging to 14 different TAP families were found, including MADS and HD proteins (Tab. S10). TIM1 (Pp3c17_24040V3.1) is a type I MADS-box protein, in a sister relationship to all other type I MADS-box proteins (Gramzow & Theissen, 2010), whereas the HD gene lhx4-like (Pp3c9_22420V3.1) was shown to be hyper-methylated in sperm cells of infertile men, which might support a potential function in cilia/flagella (Yu et al., 2015).
From all expressed genes in Gd and Re, a set of 110 differentially expressed genes (DEGs) could be identified using a stringent intersect of three tools. Detailed analysis of the DEGs showed many spermatozoid/sperm and pollen related genes (Table S3). Of the genes expressed significantly higher in Re, many showed no expression in Gd, but moderate to high expression in Re, e.g. the arl13b homolog (Pp3c1_40600V3.1) (He et al., 2018), which plays a role in flagella and cilia stability and signaling via axonemal poly-glutamylation in human, and the protein phosphatase hydin homolog Pp3c3_14230V3.1 (Fig. S8A,B). The central microtubule pair protein hydin is a well analysed gene in human and algae, and connected to the human disease Primary Ciliary Dyskinesia (PCD). Mutations in this gene lead to the loss of flagellar motility and occasionally to loss of one or both central pair microtubules due to missing C2b projections (Lechtreck & Witman, 2007; Olbrich et al., 2012). Interestingly, the Gd ultrastructure showed missing protein projections (Fig. 2E,I), which might be a result from the missing hydin expression. Additionally, the MIKC* MADS box protein MADS3 (Pp3c3_31770V3.1) was found to be differentially expressed, and showed no expression in Gd but in Re. The A. thaliana homolog with the highest BLAST similarity is AGL 104, required for pollen maturation and pollen tube growth (Adamczyk & Fernandez, 2009), suggesting that MADS3 might influence P. patens male gamete development and maturation. We also analysed the TFs discussed in the text with regard to their DMP, SNP and InDel status. No DMPs could be identified, but bell1, ppm1 and ppmc5 show InDels in their UTRs, and bell1, ppm1, ppmc6 and ppmads1 show SNPs. Bell1 contains two synonymous SNPs, ppmads1 features two intron variants and a 5’-UTR SNP, while ppm1 and ppmc6 both show missense variants within their transcript. In addition, ppm1 shows several intron and 3’-UTR variants. These results indicate a potential role of these SNPs in the different expression of the TFs and thus, also a putative contribution to the Gd phenotype (Table S11).
Among the annotated genes significantly higher expressed in Gd were e.g. the membrane associated ring finger march1 homolog (Pp3c18_16700V3.1), which is known to negatively affect the sperm quality and quantity in Chinese Holstein bulls (Liu et al., 2017), and an arabinogalactan 31 homolog (Pp3c5_9210V3.1, Fig. S8B,C). Arabinogalactan proteins are known to be part of mucilage (Lord & Sanders, 1992). In the streptophytic alga Chara vulgaris polysaccharide secretion into the antheridial venter by so called manubria cells could be shown during spermatogenesis, but not during normal antheridial cell proliferation, probably as a part of the complex mucilage production (Pickett-Heaps, 1968; Kwiatkowska & Maszewski, 1987). After maturation of the spermatozoids the mucilage content was reduced, compared to the mucilage content during antheridia development (Gosek & Kwiatkowska, 1991), suggesting that mucilage is required for spermatogenesis, but not for the release of the mature spermatozoids. Arabinogalactan proteins are abundant in the matrix around fern spermatozoids during development and are virtually negligible when antheridia release spermatozoids (Lopez & Renzaglia, 2018). Thus, an overexpression of an arabinogalactan gene could represent an impairment of the spermatozoid maturation process in the Gd background.
Analysis of differential methylation among the DEGs showed that 14 genes are hyper-methylated in Gd (Tab. S12). All of them are expressed in Re but not in Gd, indicating their potential repression through methylation (Lang et al., 2018). Among these genes hydin and arl13b could be detected, re-enforcing the notion that their malfunction in Gd is connected to DNA-methylation.
RNA-seq expression data of hydin and march1 have been confirmed by RT-PCR (Fig. S8A,C), and SNP and DNA methylation data have been analysed. The hydin gene body and potential promoter region (2kbp upstream of the coding sequence) displays six SNPs and one insertion or deletion (InDel) between Gd and Re. One SNP is located in the putative promoter region, three are intron variants and two are located within an exon, causing moderate amino acid changes (Tab. S13). In Gd, CHG and CHH context DMPs are present in the promoter and gene body, while a single CG mark is present in Re (Fig. S9). The lack of expression in Gd matches the presence of SNPs in the promotor region and gene body methylation, shown to coincide with lack of expression (Lang et al., 2018). March1 does show very low levels of DNA methylation but two SNPs between Gd and Re in the 5’-UTR and five SNPs and one InDel in the putative promoter region, which could affect a potential regulatory function of the UTR (Fig. S10, Table S13). Network analysis was performed to gain more insights into putative protein interaction partners. The March1 network revealed proteins known to be involved in degradation of mis-folded proteins via ubiquitination, and modification of proteins via phosphorylation (Fig. S11A). Since protein phosphorylation and ubiquitination pathways are known to act together (Hunter, 2007), march1 disregulation in Gd might be involved in the determined phenotype. The hydin network revealed many proteins involved in ciliary function and motility e.g. radial spoke head protein 9 (RSPH9, (Castleman et al., 2009), glutamylase TLL6-like (He et al., 2018) and CCDC39 (Merveille et al., 2011; Oda et al., 2014), Fig. S11B.
Ccdc39 is essential for proper flagellar development in P. patens
CCDC39 (Pp3c27_5290V3.1) was also detected via a candidate gene approach that screened for expression only in adult gametophores (bearing gametangia). It is part of the hydin network and is a coiled-coil domain containing protein which, as well as hydin and rsph9, belongs to the genes that cause PCD when mutated (Antony et al., 2013; Horani & Ferkol, 2018). In human it is required for the assembly of inner dynein arms and the dynein regulatory complex (Merveille et al., 2011) and in C. reinhardtii it acts as a molecular ruler for the determination of the flagellar length (Oda et al., 2014). Phylogenetic analysis showed the presence of ccdc39 orthologs in all major kingdoms, providing evidence that it was already present in the MRCA of all eukaryotes (Fig. 3D). Interestingly, all species with ccdc39 orthologs also possess flagella, implicating a gene function unique to these motile organelles. Ccdc39 does not show amino acid sequence-affecting SNPs between Gd and Re in the gene body, and no SNPs in the promoter region (Table S13). The promoter region and gene body of ccdc39 display low levels of DNA methylation (Fig. S12). Interestingly, CCDC39 and MARCH1 network analysis showed connections to the same two cGMP-dependent protein kinases (Pp3c16_18360V3.1, Pp3c25_7050V3.1, Fig. S11A,C), implicating involvement of kinases in spermatogenesis of P. patens. Homologs of these kinases were shown to affect mammalian spermatozoa e.g. in terms of capacitation and motility (Miraglia et al., 2011; Cisneros-Mejorado et al., 2014). Analysis of their expression profile via the Physcomitrella efp browser (Ortiz-Ramirez et al., 2016) showed putative function of both in different tissues, whereas Pp3c16_18360V3.1 shows higher expression in tissues related to sexual reproduction and Pp3c25_7050V3.1 in mature sporophytes indicating a putative broad function for these kinases during the P. patens development (Fig. S13).
Expression analysis of ccdc39 via RNA-seq and qPCR in P. patens showed no expression in any tissue except for the antheridia (Fig. S16), and a consistently reduced expression level of all Gd samples as compared to Re (Fig. 3C), suggesting a functional connection to the observed fertility phenotype. To elucidate the function of ccdc39 and to separate the phenotype from hydin function, a loss-of-function mutant was generated, hereafter referred to as ccdc39. Protoplast regeneration, protonemal and gametophytic growth and gametangia development did not reveal an obvious aberrant phenotype (Fig. S14, S15). However, no sporophytes were formed under selfing conditions (Fig. 4A yellow, C). 30 days after watering Re had developed mature brown sporophytes (Fig. 4B), whereas ccdc39 apices showed several unfertilized archegonia and bundles of antheridia in a cauliflower-like structure (Fig. 4C). When no fertilization takes place, P. patens is known to continue gametangiogenesis, leading to the accumulation of multiple archegonia per gametophore (Landberg et al., 2013; Sanchez-Vera et al., 2017). Under crossing conditions, using the male fertile Re-mCherry line (Perroud et al., 2019), a close to normal amount of sporophytes (94%) could be observed, with 100% of them being crosses (Fig. 4A, light and dark green), suggesting a defect in the male reproductive apparatus. Variable phenotypes of flagella could be detected in ccdc39. Some spermatozoids developed very short flagella, others normal length flagella, but with ends that remained in coils, comparable to the Gd phenotype (Fig. 4D–F). Ppccdc39 hence showed a somewhat different phenotype compared to the alga C. reinhardtii, in which a severe reduction of the flagellar length occurred (Oda et al., 2014), an observation only occasionally seen in P. patens ccdc39. No ccdc39 null mutant has been reported for H. sapiens yet, but cilia of epithelial cells of persons bearing mutations in ccdc39 showed a reduction of length in 1/5 cases (Merveille et al., 2011), implying flagellar length variations to be part of the phenotype, as in P. patens ccdc39. Interestingly, all three species share the loss of the inner dynein arms upon mutation of ccdc39 ((Merveille et al., 2011; Oda et al., 2014); Fig. 4 H–I), indicating a conserved function. Ccdc39 gametes undergo cellular morphogenesis into a streamlined coiled architecture, including nuclear compaction/elongation (Fig. 4F). The locomotory apparatus develops normally and the two flagella elongate around the cell perimeter. However, axonemal organization is completely disrupted in the KO spermatozoids. Of the over 300 mature ccdc39 spermatozoids examined in the TEM, all exhibited some degree of disruption of the flagellar axoneme, which was more pronounced along the posterior coils. The most common defect that was visible in virtually every gamete was an anomalous 9+2 organization (compare Re: Fig. 4G and ccdc39: Fig. 4H–J). In cross section this phenotype typically expressed as nine doublet microtubules randomly arranged within an abnormally-shaped flagellar shaft that was rarely cylindrical (Fig. 4H–J). The central pair complex of microtubules with some electron densities remained intact but it was not commonly anchored in the center of the axoneme (Fig. 4F, H,I, J). Floating doublet microtubules show evidence of outer dynein but inner dynein was unclear (Fig. 4G, H). Typically, only a single complement of eight or nine doublets and one central pair occured within an axoneme, but occasional two complements were visible within a single flagellar shaft (Fig. 4F, I). Cytoplasmic connections across coils provided evidence of incomplete separation of cellular coils that is likely responsible for the posterior loop in flagella evident in the light microscope (Fig. 4D). The cytoplasmatic connections observed in the ccdc39 background are similar to those displayed by Gd (Fig. 2E), which also showed a reduced expression of ccdc39 in comparison to Re (Fig. 3C). Together these observations suggest a functional connection between ccdc39 expression and the Gd phenotype.
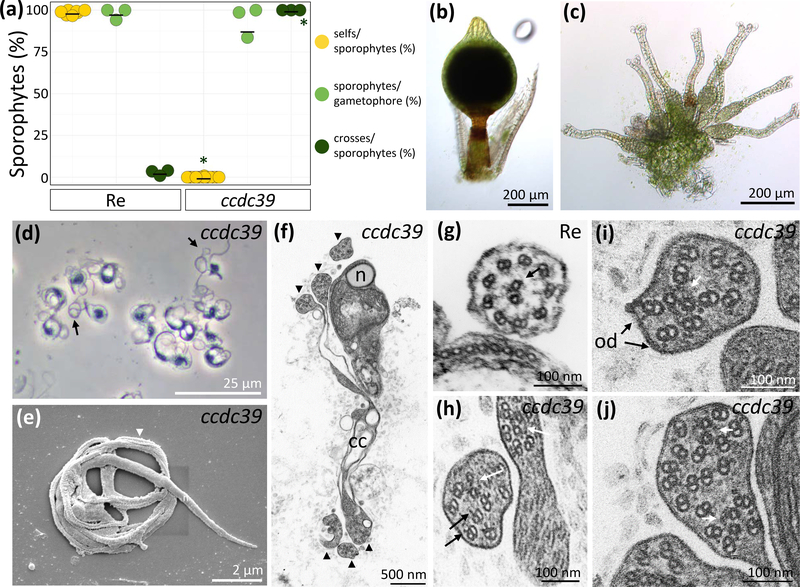
Figure 4: Analysis of Physcomitrella patens ccdc39.
(a): Selfing and crossing analysis between ccdc39 (n = 630), Reute (Re) (n = 331) and fluorescent marker line Re-mcherry. ccdc39 develops 0% and Re develops 98.6% sporophytes per gametophore under selfing conditions. Under crossing conditions, Re develops 98.1% of sporophytes per gametophore of which 2,8% are crosses. ccdc39 develops 94.1% sporophytes per gametophore of which 100% are crosses (asterisk shows significant deviation, Chi-square test, p < 0.01). Mean marked by black line.
Re (b) and ccdc39 (c) apices 30 days after watering show properly developed sporophytes on Re apices and bundles of gametangia on ccdc39 apices.
d: Phase contrast image of fixed mature ccdc39 spermatozoids displaying posterior flagellar loops (arrows).
(e): SEM of ccdc39 spermatozoid that remains in coils. The nucleus (n) is a long coiled cylinder that narrows posteriorly. Irregular flagella coil around the cell (arrowhead).
(f): TEM of ccdc39 whole mature spermatozoid in cross section with compacted nucleus (n) in upper coil. Arrowheads indicate irregular flagella that encircle the cell for over 2 revolutions. Cytoplasmic connections (cc) adjoin axonemes across their length. Bar = 500 nm.
(g): TEM cross-section of a Re immature axoneme showing the regular pattern of nine outer microtubule doublets and two single microtubulis in the central pair complex (white arrow) as well as outer dynein arms (od, black arrows).
(h-j): TEM cross-sections of irregular ccdc39 flagella showing intact acentric central pair complexes (white arrowheads), outer dynein arms (od, black arrows).
(h, j): Double axonemes in single flagellar shafts can be observed.
Conclusions
Our analyses suggest that the long term vegetative propagation of the Gd ecotype lab strain led to accumulation of several (epi-)mutations, affecting the male reproductive apparatus. Unfortunately, the original Gd collection site is not available anymore due to changes in land use. However, the analysis of GdJp (Fig. S5) proves the assumption that the original Gd isolate had not been infertile. Hence, the genetic and epigenetic differences observed between Gd and Re are probably due to somatic changes acquired during vegetative culture as previously proposed (Ashton & V. S. Raju, 2000). To avoid such accumulation of deleterious (epi-) mutations, it appears advisable to regularly include sexual reproduction into the lab strain propagation, and to test the offspring for fertility.
Our network analyses suggest that the components of the protein-protein interaction network of the eukaryotic flagellar structure are well conserved. This network is also supported by the loss-of-function mutant: ccdc39 was found via the network and expression analysis as well as through a candidate gene approach, and resembles the Gd phenotype in terms of male infertility. Most probably, given the lack of SNPs and DMPs in the underlying gene, Ppccdc39 is not the causative master mutation. Rather, it appears probable that one or several upstream regulators are affected, causing (among others) the mis-regulation of Ppccdc39. In this regard it is interesting to note that the homeodomain (HD) TALE TF BELL1, known to be involved in the alternation of generations (haploid to diploid phase transition) in algae and plants (Lee et al., 2008; Horst et al., 2016), shows expression in Re but not in Gd. Intriguingly, the heterodimeric interaction of HD TALE TFs observed in animals and plants has recently been suggested to be an ancestral eukaryotic feature to control the haploid-to-diploid transition only when gametes were correctly fused (Joo et al., 2018). Potentially, master regulators like BELL1 not only control zygote formation, but even earlier processes of sexual reproduction.
Known key players affecting cilia motility in mammal model organisms and human could be identified via their P. patens expression profile as well as through (epi-)mutations between Gd and Re. Some of these were characterized via transcript profiling, demonstrating biased expression between the ecotypes. Primary Ciliary Dyskinesia (PCD) is a heterogeneous genetic defect that results in abnormal structure and function of cilia (Noone et al., 2004). In humans, the condition has a range of biological and clinical phenotypes that include respiratory infections, reduced fertility and situs inversus or developmental left-right inversion of organs. Ultrastructural observations of cilia in patients with PCD reveal perturbations of outer and inner dynein arms, radial spokes and the central pair complex, with 10% exhibiting normal axonemal structure (Papon et al., 2010). The conserved nature of the 9+2 core structure of cilia and flagella is particularly underlined in the present study as a universal phenotype of ccdc39 loss-of-function mutants, since it occurs in moss, dog and human (Merveille et al., 2011). All show axonemal disorganization and inner dynein arm aberrations. Functional analyses indicate that the ccdc39 gene is necessary to assemble the dynein regulatory complex and inner dynein arms. The striking defects evident in ccdc39 moss spermatozoids are even more exaggerated than those in human and dog, perhaps due to the extreme length of flagellar axonemes compared with cilia. Nevers (Nevers et al., 2017) classified motility-associated genes responsible for ciliopathies such as PCD, including ccdc39, in a relatively small subset of ciliary genes necessary to construct a motile flagellum. Hence, flagellated plants (like mosses) apparently harbor male gametes with a flagellar architecture that is conserved with mammals, and thus could serve as easily accessible models for studying human disease or animal foodstock fertility associated with flagella/cilia. Whether only flagella or other elements of the process of motile male gamete development are evolutionarily conserved remains to be determined.
Supplementary Material
Figure S1: Sporophyte per gametophore development for all three independently analysed ccdc39 lines.
Figure S2: Expression level of act5 in juvenile (j) and adult (a) apices of Gd and Re show similar expression.
Figure S3: Vector used for ccdc39 knock-out generation.
Figure S4: Genotyping of ccdc39.
Figure S5: Number of sporophytes per gametophore developed under selfing conditions for P. patens Re, Gd and GdJp.
Figure S6: Detailed analysis of Gd and Re spermatozoid counts and properties.
Figure S7: GO bias analysis and word cloud visualization.
Figure S8: Expression analysis of hydin and march1 in spermatozoids respectively gametangia.
Figure S9: Methylation analysis of the hydin gene.
Figure S10: March1 gene body methylation.
Figure S11: Network analysis of genes associated with proper flagellar functionality.
Figure S12: Percentage of ccdc39 methylated reads per position.
Figure S13: Expression of two cGMP kinases present in the ccdc39 and march1 network analysis.
Figure S14: Vegetative tissue is not affected by ccdc39 mutation.
Figure S15: Archegonia and antheridia of ccdc39 show no morphological differences between Re and ccdc39.
Figure S16: ccdc39 RNA-seq expression throughout different developmental stages.
Table S1: Primer names, sequences, usage and product length for PCRs performed. Orange sequence parts are binding parts for tail-containing primer. Small letters are attached for functionality of the attached restriction sites (blue). In the product column, green fields show product length of Re wild type and red products of Re ccdc39 gDNA template.
Table S2: RPKM values of all biological and technical replicates from the antheridia RNA-seq.
Table S3: Differential expressed genes (DEG) Gd vs Re and their involvement in pollen/sperm development
Table S4: Genes containing DMPs in Gd versus Re and the corresponding over-/under-represented GO terms
Table S5: Genes containing DMPs and DNPs in Gd versus Re and the corresponding over-/under-represented GO terms
Table S6: All genes expressed in Gd antheridial bundles and the corresponding over-/under-represented GO terms
Table S7: All genes expressed in Re antheridial bundles and the corresponding over-/under-represented GO terms
Table S8: Genes uniquely expressed in Gd antheridial bundles
Table S9: Genes uniquely expressed in Re antheridial bundles
Table S10: TAPs found in uniquely expressed genes of Re and Gd antheridial bundles and their family assignment
Table S11: Analysis of the TFs discussed in the text with regard to DMPs, SNPs and INDELs.
Table S12: Genes showing DMPs and are differentially expressed between Gd and Re are hyper-methylated and contain the candidate genes hydin and arl13b.
Table S13: SNPs and InDels between Gd and Re for selected genes.
Acknowledgements
We thank Eva Bieler of the Swiss Nanoscience Institute (SNI), Nano Imaging, University of Basel, for the help taking the SEM pictures, as well as Marion Schön for TEM sample preparation and Rebecca Hinrichs and Jason Backes for assistance. We also thank Karl Ferdinand Lechtreck (University of Georgia) for his helpful comments with regard to the hydin phenotype, Keiko Sakakibara for providing the GdJp strain and Lee Miller for the pME-Ta vector. The authors declare no conflicts of interest. SAR is grateful to DFG for funding (RE 1697/15-1).
Footnotes
State tables and supporting information: Figures S1–16, Tables S1–13
Data availability
All sequencing data has been uploaded to the NCBI SRA. Bs-seq Gd data: SRR4454535 (Lang et al., 2018); bs-seq Re data: SRR9901085. RNA-seq data BioProject PRJNA559055.
References
- Adamczyk BJ, Fernandez DE. 2009. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol 149(4): 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. 2012. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13(10): R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony D, Becker-Heck A, Zariwala MA, Schmidts M, Onoufriadis A, Forouhan M, Wilson R, Taylor-Cox T, Dewar A, Jackson C, et al. 2013. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum Mutat 34(3): 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton NW, Raju MVS 2000. The distribution of gametangia on gametophores of Physcomitrella (Aphanoregma) patens in culture. Journal of Bryology,22:1,9–12. [Google Scholar]
- Aya K, Hiwatashi Y, Kojima M, Sakakibara H, Ueguchi-Tanaka M, Hasebe M, Matsuoka M. 2011. The Gibberellin perception system evolved to regulate a pre-existing GAMYB-mediated system during land plant evolution. Nat Commun 2: 544. [DOI] [PubMed] [Google Scholar]
- Beike AK, von Stackelberg M, Schallenberg-Rudinger M, Hanke ST, Follo M, Quandt D, McDaniel SF, Reski R, Tan BC, Rensing SA. 2014. Molecular evidence for convergent evolution and allopolyploid speciation within the Physcomitrium-Physcomitrella species complex. BMC Evol Biol 14: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda A, Wilkinson MF. 2010. Transcription and post-transcriptional regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci 365(1546): 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajal-Gonzalez BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D. 2013. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken) 70(2): 101–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol 194(2): 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castleman VH, Romio L, Chodhari R, Hirst RA, de Castro SC, Parker KA, Ybot-Gonzalez P, Emes RD, Wilson SW, Wallis C, et al. 2009. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet 84(2): 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champroux A, Torres-Carreira J, Gharagozloo P, Drevet JR, Kocer A. 2016. Mammalian sperm nuclear organization: resiliencies and vulnerabilities. Basic Clin Androl 26: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros-Mejorado A, Hernandez-Soberanis L, Islas-Carbajal MC, Sanchez D. 2014. Capacitation and Ca(2+) influx in spermatozoa: role of CNG channels and protein kinase G. Andrology 2(1): 145–154. [DOI] [PubMed] [Google Scholar]
- Cove D 2005. The moss Physcomitrella patens. Annu Rev Genet 39: 339–358. [DOI] [PubMed] [Google Scholar]
- Cove DJ, Perroud PF, Charron AJ, McDaniel SF, Khandelwal A, Quatrano RS. 2009. The moss Physcomitrella patens: a novel model system for plant development and genomic studies. Cold Spring Harb Protoc 2009(2): pdb emo115. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8): 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong FN, Amiri-Yekta A, Martinez G, Saut A, Tek J, Stouvenel L, Lores P, Karaouzene T, Thierry-Mieg N, Satre V, et al. 2018. Absence of CFAP69 Causes Male Infertility due to Multiple Morphological Abnormalities of the Flagella in Human and Mouse. Am J Hum Genet 102(4): 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5): 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel PP. 1968. The induction of biochemical and morphological mutants in the moss Physcomitrella patens. American Journal of Botany 55(4): 438–446. [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A 107(19): 8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Haas FB, Meyberg R, Ullrich KK, Hiss M, Perroud PF, Hanke S, Kratz V, Powell AF, Vesty EF., et al. 2019. PEATmoss (Physcomitrella Expression Atlas Tool): a unified gene expression atlas for the model plant Physcomitrella patens. Plant J. doi: 10.1111/tpj.14607. [DOI] [PubMed] [Google Scholar]
- Gitzendanner MA, Soltis PS, Wong GK, Ruhfel BR, Soltis DE. 2018. Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. Am J Bot 105(3): 291–301. [DOI] [PubMed] [Google Scholar]
- Godet AC, David F, Hantelys F, Tatin F, Lacazette E, Garmy-Susini B, Prats AC. 2019. IRES Trans-Acting Factors, Key Actors of the Stress Response. Int J Mol Sci 20(4),924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosek A, Kwiatkowska M. 1991. Cytochemical studies on the antheridial mucilage and changes in its concentration and amount during the spermatogenesis in Chara vulgaris L. Folia Histochem Cytobiol 29(3): 91–99. [PubMed] [Google Scholar]
- Gramzow L, Theissen G. 2010. A hitchhiker’s guide to the MADS world of plants. Genome Biol 11(6): 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, Twell D. 2019. The evolution and patterning of male gametophyte development. Curr Top Dev Biol 131: 257–298. [DOI] [PubMed] [Google Scholar]
- He K, Ma X, Xu T, Li Y, Hodge A, Zhang Q, Torline J, Huang Y, Zhao J, Ling K, et al. 2018. Axoneme polyglutamylation regulated by Joubert syndrome protein ARL13B controls ciliary targeting of signaling molecules. Nature Communications 9(1): 3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G 2008. Was the initiation of translation in early eukaryotes IRES-driven? Trends Biochem Sci 33(2): 58–64. [DOI] [PubMed] [Google Scholar]
- Hiss M, Meyberg R, Westermann J, Haas FB, Schneider L, Schallenberg-Rudinger M, Ullrich KK, Rensing SA. 2017. Sexual reproduction, sporophyte development and molecular variation in the model moss Physcomitrella patens: introducing the ecotype Reute. Plant J 90(3): 606–620. [DOI] [PubMed] [Google Scholar]
- Hohe A, Rensing S, Mildner M, Lang D, Reski R. 2002. Day Length and Temperature Strongly Influence Sexual Reproduction and Expression of a Novel MADS-Box Gene in the Moss Physcomitrella patens. Plant Cell Rep 20, 1135–1140. [Google Scholar]
- Horani A, Ferkol TW. 2018. Advances in the Genetics of Primary Ciliary Dyskinesia: Clinical Implications. Chest 154(3): 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst NA, Katz A, Pereman I, Decker EL, Ohad N, Reski R. 2016. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat Plants 2: 15209. [DOI] [PubMed] [Google Scholar]
- Horst NA, Reski R. 2017. Microscopy of Physcomitrella patens sperm cells. Plant Methods 13: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 28(5): 730–738. [DOI] [PubMed] [Google Scholar]
- Joo S, Wang MH, Lui G, Lee J, Barnas A, Kim E, Sudek S, Worden AZ, Lee JH. 2018. Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida. BMC Biol 16(1): 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper BD. 2019. Cap-Independent mRNA Translation in Germ Cells. Int J Mol Sci 20(1):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop W 1868. Der Kreislauf des Stoffs: Lehrbuch der Agriculture-Chemie. Leipzig: H. Haessel. [Google Scholar]
- Kofuji R, Yagita Y, Murata T, Hasebe M. 2018. Antheridial development in the moss Physcomitrella patens: implications for understanding stem cells in mosses. Philos Trans R Soc Lond B Biol Sci 373(1739). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R, Yoshimura T, Inoue H, Sakakibara K, Hiwatashi Y, Kurata T, Aoyama T, Ueda K, Hasebe M 2009. Gametangia Development in the Moss Physcomitrella patens. The moss Physcomitrella patens: Blackwell Publishing Ltd, 167–181, editors: Knight Celia D., Pierre-François Perroud, Cove David J.. [Google Scholar]
- Koshimizu S, Kofuji R, Sasaki-Sekimoto Y, Kikkawa M, Shimojima M, Ohta H, Shigenobu S, Kabeya Y, Hiwatashi Y, Tamada Y, et al. 2018. Physcomitrella MADS-box genes regulate water supply and sperm movement for fertilization. Nat Plants 4(1): 36–45. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska M, Maszewski J. 1987. Ultrastructural and autoradiographic studies of non-generative cells in the antheridium of Chara vulgaris. I. Manubria. Folia Histochem Cytobiol 25(3–4): 215–223. [PubMed] [Google Scholar]
- Landberg K, Pederson ER, Viaene T, Bozorg B, Friml J, Jonsson H, Thelander M, Sundberg E. 2013. The MOSS Physcomitrella patens reproductive organ development is highly organized, affected by the two SHI/STY genes and by the level of active auxin in the SHI/STY expression domain. Plant Physiol 162(3): 1406–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Ullrich KK, Murat F, Fuchs J, Jenkins J, Haas FB, Piednoel M, Gundlach H, Van Bel M, Meyberg R, et al. 2018. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J 93(3): 515–533. [DOI] [PubMed] [Google Scholar]
- Le Bail A, Scholz S, Kost B. 2013. Evaluation of reference genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS ONE 8(8): e70998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Witman GB. 2007. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J Cell Biol 176(4): 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lin H, Joo S, Goodenough U. 2008. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133(5): 829–840. [DOI] [PubMed] [Google Scholar]
- Lefevre PL, Palin MF, Murphy BD. 2011. Polyamines on the reproductive landscape. Endocr Rev 32(5): 694–712. [DOI] [PubMed] [Google Scholar]
- Lie PP, Cheng CY, Mruk DD. 2009. Coordinating cellular events during spermatogenesis: a biochemical model. Trends Biochem Sci 34(7): 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yin H, Li C, Qin C, Cai W, Cao M, Zhang S. 2017. Genetic effects of PDGFRB and MARCH1 identified in GWAS revealing strong associations with semen production traits in Chinese Holstein bulls. BMC Genet 18(1): 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez RA, Renzaglia KS. 2018. The Ceratopteris (fern) developing motile gamete walls contain diverse polysaccharides, but not pectin. Planta 247(2): 393–404. [DOI] [PubMed] [Google Scholar]
- Lord EM, Sanders LC. 1992. Roles for the extracellular matrix in plant development and pollination: a special case of cell movement in plants. Dev Biol 153(1): 16–28. [DOI] [PubMed] [Google Scholar]
- Luo M, Cao M, Kan Y, Li G, Snell W, Pan J. 2011. The phosphorylation state of an aurora-like kinase marks the length of growing flagella in Chlamydomonas. Curr Biol 21(7): 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt RM, Delwiche CF, Karol KG. 2004. Charophyte algae and land plant origins. Trends Ecol Evol 19(12): 661–666. [DOI] [PubMed] [Google Scholar]
- Merveille AC, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataille G, Belmont J, Beydon N, Billen F, Clement A, et al. 2011. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet 43(1): 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles LB, Verkade H. 2014. TA-cloning vectors for rapid and cheap cloning of zebrafish transgenesis constructs. Zebrafish 11(3): 281–282. [DOI] [PubMed] [Google Scholar]
- Miraglia E, De Angelis F, Gazzano E, Hassanpour H, Bertagna A, Aldieri E, Revelli A, Ghigo D. 2011. Nitric oxide stimulates human sperm motility via activation of the cyclic GMP/protein kinase G signaling pathway. Reproduction 141(1): 47–54. [DOI] [PubMed] [Google Scholar]
- Mitchell DR. 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv Exp Med Biol 607: 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakosteen PC, Hughes KW. 1978. Sexual Life Cycle of Three Species of Funariaceae in Culture. The Bryologist 81(2): 307–314. [Google Scholar]
- Nevers Y, Prasad MK, Poidevin L, Chennen K, Allot A, Kress A, Ripp R, Thompson JD, Dollfus H, Poch O, et al. 2017. Insights into Ciliary Genes and Evolution from Multi-Level Phylogenetic Profiling. Mol Biol Evol 34(8): 2016–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noone PG, Leigh MW, Sannuti A, Minnix SL, Carson JL, Hazucha M, Zariwala MA, Knowles MR. 2004. Primary ciliary dyskinesia: diagnostic and phenotypic features. Am J Respir Crit Care Med 169(4): 459–467. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, Kikkawa M. 2014. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 346(6211): 857–860. [DOI] [PubMed] [Google Scholar]
- Olbrich H, Schmidts M, Werner C, Onoufriadis A, Loges NT, Raidt J, Banki NF, Shoemark A, Burgoyne T, Al Turki S, et al. 2012. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet 91(4): 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes I. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574(7780): 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Hernandez-Coronado M, Thamm A, Catarino B, Wang M, Dolan L, Feijo JA, Becker JD. 2016. A Transcriptome Atlas of Physcomitrella patens Provides Insights into the Evolution and Development of Land Plants. Mol Plant 9(2): 205–220. [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Michard E, Simon AA, Damineli DSC, Hernandez-Coronado M, Becker JD, Feijo JA. 2017. GLUTAMATE RECEPTOR-LIKE channels are essential for chemotaxis and reproduction in mosses. Nature 549(7670): 91–95. [DOI] [PubMed] [Google Scholar]
- Pan J, Naumann-Busch B, Wang L, Specht M, Scholz M, Trompelt K, Hippler M. 2011. Protein phosphorylation is a key event of flagellar disassembly revealed by analysis of flagellar phosphoproteins during flagellar shortening in Chlamydomonas. J Proteome Res 10(8): 3830–3839. [DOI] [PubMed] [Google Scholar]
- Papon JF, Coste A, Roudot-Thoraval F, Boucherat M, Roger G, Tamalet A, Vojtek AM, Amselem S, Escudier E. 2010. A 20-year experience of electron microscopy in the diagnosis of primary ciliary dyskinesia. Eur Respir J 35(5): 1057–1063. [DOI] [PubMed] [Google Scholar]
- Perroud PF, Cove DJ, Quatrano RS, McDaniel SF. 2011. An experimental method to facilitate the identification of hybrid sporophytes in the moss Physcomitrella patens using fluorescent tagged lines. New Phytol 2(10): 1469–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud PF, Haas FB, Hiss M, Ullrich KK, Alboresi A, Amirebrahimi M, Barry K, Bassi R, Bonhomme S, Chen H, et al. 2018. The Physcomitrella patens gene atlas project: large-scale RNA-seq based expression data. Plant J 95(1): 168–182. [DOI] [PubMed] [Google Scholar]
- Perroud PF, Meyberg R, Rensing SA. 2019. Physcomitrella patens Reute mCherry as a tool for efficient crossing within and between ecotypes. Plant Biol (Stuttg) 21 Suppl 1: 143–149. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J 1968. Ultrastructure and Differentiation In Chara Sp. III. Formation of the Antheridium. Australian Journal of Biological Sciences 21(2): 255–274. [Google Scholar]
- Puttick MN, Morris JL, Williams TA, Cox CJ, Edwards D, Kenrick P, Pressel S, Wellman CH, Schneider H, Pisani D, et al. 2018. The Interrelationships of Land Plants and the Nature of the Ancestral Embryophyte. Curr Biol 28(5): 733–745 e732. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6): 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quodt V, Faigl W, Saedler H, Munster T. 2007. The MADS-domain protein PPM2 preferentially occurs in gametangia and sporophytes of the moss Physcomitrella patens. Gene 400(1–2): 25–34. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319(5859): 64–69. [DOI] [PubMed] [Google Scholar]
- Renzaglia KS, Duff RJT, Nickrent DL, Garbary DJ. 2000. Vegetative and reproductive innovations of early land plants: implications for a unified phylogeny. Philos Trans R Soc Lond B Biol Sci 355(1398): 769–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzaglia KS, Garbary DJ. 2001. Motile Gametes of Land Plants: Diversity, Development, and Evolution. Critical Reviews in Plant Sciences 20(2): 107–213. [Google Scholar]
- Renzaglia KS, Lopez RA, Henry JS, Flowers ND, Vaughn KC. 2017. Transmission Electron Microscopy of Centrioles, Basal Bodies and Flagella in Motile Male Gametes of Land Plants. Bio-protocol 7(19): e2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology 61(3): 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B 1999. Twilight zone of protein sequence alignments. Protein Eng 12(2): 85–94. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Ando S, Yip HK, Tamada Y, Hiwatashi Y, Murata T, Deguchi H, Hasebe M, Bowman JL. 2013. KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 339(6123): 1067–1070. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Deguchi H, Hasebe M. 2008. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol Dev 10(5): 555–566. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vera V, Kenchappa CS, Landberg K, Bressendorff S, Schwarzbach S, Martin T, Mundy J, Petersen M, Thelander M, Sundberg E. 2017. Autophagy is required for gamete differentiation in the moss Physcomitrella patens. Autophagy 13(11): 1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer DG, Delacote F, Charlot F, Vrielynck N, Guyon-Debast A, Le Guin S, Neuhaus JM, Doutriaux MP, Nogue F. 2010. RAD51 loss of function abolishes gene targeting and de-represses illegitimate integration in the moss Physcomitrella patens. DNA Repair (Amst) 9(5): 526–533. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MW, Giraldo-Fonseca A, Rovekamp M, Smetanin D, Bowman JL, Grossniklaus U. 2018. Extensive epigenetic reprogramming during the life cycle of Marchantia polymorpha. Genome Biol 19(1): 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SD, Krogan NT, Ashton NW. 2007. Clues about the ancestral roles of plant MADS-box genes from a functional analysis of moss homologues. Plant Cell Rep 26(8): 1155–1169. [DOI] [PubMed] [Google Scholar]
- Southworth D, Cresti M. 1997. Comparison of Flagellated and Nonflagellated Sperm in Plants. American Journal of Botany 84(9): 1301–1311. [PubMed] [Google Scholar]
- Stewart KD, Mattox KR. 1975. Comparative cytology, evolution and classification of the green algae with some consideration of the origin of other organisms with chlorophylls A and B. The Botanical Review 41(1): 104–135. [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(Database issue): D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Wang X, Li W, Yang X, Li Z, Liu W, Li C, Zhu Z, Wang L, Wang J, et al. 2017. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am J Hum Genet 100(6): 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theißen G, Gramzow L 2016. Structure and Evolution of Plant MADS Domain Transcription Factors. Plant Transcription Factors, Academic Press:127–138 Editor: Gonzalez Daniel H.. [Google Scholar]
- Transeau EN. 1951. The Zygnemataceae (fresh-water Conjugate Algae) with Keys for the Identification of Genera and Species: And Seven Hundred Eighty-nine Illustrations: Ohio State University Press. [Google Scholar]
- van den Ende H, Musgrave A, Klis FM. 1990. The Role of Flagella in the Sexual Reproduction of Chlamydomonas Gametes. Springer, Boston, MA. [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9): 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. [Google Scholar]
- Widiez T, Symeonidi A, Luo C, Lam E, Lawton M, Rensing SA. 2014. The chromatin landscape of the moss Physcomitrella patens and its dynamics during development and drought stress. Plant J 79(1): 67–81. [DOI] [PubMed] [Google Scholar]
- Wilhelmsson PKI, Mühlich C, Ullrich KK, Rensing SA. 2017. Comprehensive Genome-Wide Classification Reveals That Many Plant-Specific Transcription Factors Evolved in Streptophyte Algae. Genome Biology and Evolution 9(12): 3384–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari R, Noy-Malka C, Wiedemann G, Auerbach Gershovitz N, Reski R, Katz A, Ohad N. 2015. DNA METHYLTRANSFERASE 1 is involved in (m)CG and (m)CCG DNA methylation and is essential for sporophyte development in Physcomitrella patens. Plant Mol Biol 88(4–5): 387–400. [DOI] [PubMed] [Google Scholar]
- Yu B, Zhou H, Liu M, Zheng T, Jiang L, Zhao M, Xu X, Huang Z. 2015. Epigenetic Alterations in Density Selected Human Spermatozoa for Assisted Reproduction. PLoS One 10(12): e0145585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328(5980): 916–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Sporophyte per gametophore development for all three independently analysed ccdc39 lines.
Figure S2: Expression level of act5 in juvenile (j) and adult (a) apices of Gd and Re show similar expression.
Figure S3: Vector used for ccdc39 knock-out generation.
Figure S4: Genotyping of ccdc39.
Figure S5: Number of sporophytes per gametophore developed under selfing conditions for P. patens Re, Gd and GdJp.
Figure S6: Detailed analysis of Gd and Re spermatozoid counts and properties.
Figure S7: GO bias analysis and word cloud visualization.
Figure S8: Expression analysis of hydin and march1 in spermatozoids respectively gametangia.
Figure S9: Methylation analysis of the hydin gene.
Figure S10: March1 gene body methylation.
Figure S11: Network analysis of genes associated with proper flagellar functionality.
Figure S12: Percentage of ccdc39 methylated reads per position.
Figure S13: Expression of two cGMP kinases present in the ccdc39 and march1 network analysis.
Figure S14: Vegetative tissue is not affected by ccdc39 mutation.
Figure S15: Archegonia and antheridia of ccdc39 show no morphological differences between Re and ccdc39.
Figure S16: ccdc39 RNA-seq expression throughout different developmental stages.
Table S1: Primer names, sequences, usage and product length for PCRs performed. Orange sequence parts are binding parts for tail-containing primer. Small letters are attached for functionality of the attached restriction sites (blue). In the product column, green fields show product length of Re wild type and red products of Re ccdc39 gDNA template.
Table S2: RPKM values of all biological and technical replicates from the antheridia RNA-seq.
Table S3: Differential expressed genes (DEG) Gd vs Re and their involvement in pollen/sperm development
Table S4: Genes containing DMPs in Gd versus Re and the corresponding over-/under-represented GO terms
Table S5: Genes containing DMPs and DNPs in Gd versus Re and the corresponding over-/under-represented GO terms
Table S6: All genes expressed in Gd antheridial bundles and the corresponding over-/under-represented GO terms
Table S7: All genes expressed in Re antheridial bundles and the corresponding over-/under-represented GO terms
Table S8: Genes uniquely expressed in Gd antheridial bundles
Table S9: Genes uniquely expressed in Re antheridial bundles
Table S10: TAPs found in uniquely expressed genes of Re and Gd antheridial bundles and their family assignment
Table S11: Analysis of the TFs discussed in the text with regard to DMPs, SNPs and INDELs.
Table S12: Genes showing DMPs and are differentially expressed between Gd and Re are hyper-methylated and contain the candidate genes hydin and arl13b.
Table S13: SNPs and InDels between Gd and Re for selected genes.
Data Availability Statement
All sequencing data has been uploaded to the NCBI SRA. Bs-seq Gd data: SRR4454535 (Lang et al., 2018); bs-seq Re data: SRR9901085. RNA-seq data BioProject PRJNA559055.