Abstract
At the end of 2019, the World Health Organization (WHO) reported pneumonia that started in Wuhan, China, as a global emergency problem. Researchers quickly advanced in research to try to understand this COVID-19 and sough solutions for the front-line professionals fighting this fatal disease. One of the tools to aid in the detection, diagnosis, treatment, and prevention of this disease is computed tomography (CT). CT images provide valuable information on how this new disease affects the lungs of patients. However, the analysis of these images is not trivial, especially when researchers are searching for quick solutions. Detecting and evaluating this disease can be tiring, time-consuming, and susceptible to errors. Thus, in this study, we aim to automatically segment infections caused by COVID19 and provide quantitative measures of these infections to specialists, thus serving as a support tool. We use a database of real clinical cases from Pedro Ernesto University Hospital of the State of Rio de Janeiro, Brazil. The method involves five steps: lung segmentation, segmentation and extraction of pulmonary vessels, infection segmentation, infection classification, and infection quantification. For the lung segmentation and infection segmentation tasks, we propose modifications to the traditional U-Net, including batch normalization, leaky ReLU, dropout, and residual block techniques, and name it as Residual U-Net. The proposed method yields an average Dice value of 77.1% and an average specificity of 99.76%. For quantification of infectious findings, the proposed method achieves results like that of specialists, and no measure presented a value of ρ < 0.05 in the paired t-test. The results demonstrate the potential of the proposed method as a tool to help medical professionals combat COVID-19. fight the COVID-19.
Keywords: COVID-19, CT findings, Infection quantification, Infection segmentation, Lung segmentation, Medical imaging
Introduction
By the end of 2019, several cases of pneumonia of unknown etiology were reported in Wuhan, China [59]. In December, the World Health Organization (WHO) declared it a global health crisis [70]. Initially, the transmission of the virus causing the disease was restricted to bats in China. However, eventually human-to-human transmission began, and since then this fatal virus has rapidly spread worldwide [6].
The virus is named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the disease is called coronavirus disease (COVID-19). COVID-19 was reported to be the cause of these various cases of pneumonia [4, 42]. Compared with other pandemics such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), COVID19 has accounted for the largest number of contagions in the least amount of time. The WHO declared the SARS outbreak on July 5, 2003. A total of 8096 SARS cases and 774 deaths across 29 countries were reported. MERS, which has still not been contained, has thus far been responsible for 2494 confirmed cases and 858 deaths across 27 countries [44]. In contrast, by February 2020, as reported by Wu and McGoogan [62], there have been 72,528 confirmed COVID-19 cases and 1870 related deaths in only China.
The most common symptoms of COVID-19 include fever, cough, and fatigue, and other symptoms include sputum production, headache, hemoptysis, diarrhea, dyspnea, and lymphopenia [49]. Social isolation is one of the most recommended measures to combat COVID-19. An individual experiencing the symptoms is advised to self-isolate for a 14 day at home. This significantly reduces the chances of the individual passing on the virus to other people. Social isolation is also aimed at preventing the overcrowding of hospitals. Considering how rapidly the number of infections is increasing, if all symptomatic individuals seek emergency medical care at the same time, there would be an overcrowding of hospitals, which would consequently lead to far more deaths [5].
The report published by the WHO on June 10, 2020 is worrisome. As per the report, there are already 7,145,539 cases with 408,025 deaths worldwide. Out of this, Europe and America together account for almost 6,000,000 cases, with 185,537 deaths in Europe and 185,863 in America [41]. Another disturbing data point presented in the report is the number of cases identified as dead in the last 24 h: Out of a total of 105,621 cases, 3629 new deaths were recorded. These statistics are highly alarming, as this number is believed to be significantly higher considering the patients who have not yet been confirmed as COVID-19 positive [40].
The key trends observed in most studies on COVID-19 are the presence of parenchymal involvement due to the formation of lesions with consolidation characteristics, ground glass opacity (GGO), often with rounded morphology and peripheral lung distribution [50]. The consolidation formation occurs on days 4–7 after the patient is infected, whereas the GGO starts in the initial days, and with the evolution of the consolidation until the patient is cured, it may be “dissolved” in GGO [40]. Thus, measuring the degree of infections of both types can guide medical decision-making. Other studies have observed the evolution (both in texture and volume) of these lesions over time [4].
Computed tomography (CT) can present interesting artifacts to medical specialists. For example, a CT can help confirm COVID-19 in an asymptomatic patient, and after a few days, a new follow-up CT can help observe the behavior of the previous findings. Another practical example would be for a patient in a severe condition to undergo an imaging examination. Then, the same patient can undergo treatment, and a new CT can be conducted to evaluate the new tomographic findings.
However, a specialist’s evaluation of tomographic findings can get tiring, and hence susceptible to interpretation errors. The patterns of COVID-19 tomographic findings are different, and evaluating them is a challenging task, especially for doctors who need to diagnose several CT images on the same day. Moreover, the doctor needs to check existing standards and quantify pulmonary impairment due to COVID-19 infection, in addition to diagnosing and suggesting a possible course of treatment. Another significant point of consideration is the verification of the disease’s evolution. The specialist must quantify the findings before and after treatment, mainly to analyze the effectiveness of the therapy. The use of computer-aided diagnosis (CADx) tools is a fundamental task, both for the segmentation of the infectious findings of COVID-19 and for the volumetric quantification of the various types of lesions.
Another significant point, in a verification of the disease ‘s evolution. The specialist must quantify the findings before and after treatment, mainly to analyze the therapy’s effectiveness. The use of Computer Aid-Diagnosis (CADx) tools proves to be a fundamental task, both for the segmentation of the infectious findings of COVID-19 and for the volumetric quantification of the various types of lesions, to be an ally of specialists.
The effectiveness of CADx tools is proportional to the techniques that constitute them. Currently, deep learning techniques have shown promising results in several domains of imaging, mainly in the field of medical imaging. Owing to their power of generalization, methods using deep learning have been applied for organ segmentation [16, 56], lesion detection [14], and disease classification [9].
Thus, the main objective of the present work is to develop an automatic method for the segmentation and volumetric quantification of COVID-19 infection CT findings using deep learning techniques. Our findings will assist clinicians in defining and quantifying results, offering a comprehensive framework for the study of medical images.
Among the various contributions of this work, we highlight the following: (a) an automatic lung segmentation model based on deep learning, (b) segmentation and extraction of pulmonary vessels from the lung parenchyma, (c) volumetric quantification showing the percentage of involvement of each lung and parenchyma, and (d) a fully automatic method to aid detection and diagnosis based on medical imaging.
With this, we provide a promising CADx tool that segments and quantifies lesions, serving as a crucial tool to aid medical professionals to combat the COVID-19 pandemic.
Related work
Two imaging modalities are used to visualize COVID-19 in patients. One is the use of chest radiography. Some studies have already proposed methods for classifying these [21, 37, 43, 61].
In contrast, for classifying CT images, there is a need to develop methods with a series of procedures. Infection segmentation and quantification go through a series of processes including lung segmentation, segmentation of findings, classification of infections, and volumetric quantification.
The literature presents several works that deal with these procedures. It is worth remembering that, as the COVID-19 is a newly discovered pathology, we still need methods that perform all these functions in one go. In this section, we present works that tackle the use of these processes together or separately.
Lung segmentation overview
Lung segmentation, an important step in an automated method, will serve as an initial process in our work for segmenting and extracting pulmonary vessels. It will also be used to extract measurements as we must determine the lung’s volume to measure the degree of lung involvement.
Lung segmentation in CT has been studied for several years [2, 25, 38, 46]. Semi-automatic and automatic techniques have been developed to assist specialists in segmenting this organ. Lung segmentation plays a fundamental role in various spheres of image processing and serves as the pre-processing step for several subsequent steps. It includes detection of nodules, detection of risk organs in radiotherapy treatment, separation of pulmonary lobes, separation between the left and right lungs, and extraction of pulmonary vessels in the parenchyma, among others [20, 39].
According to Hofmanninger et al. [25], lung segmentation is not a methodological problem, but a database problem. Furthermore, the authors state that the lung segmentation methods, despite being published and consolidated, still present questionable results in the clinical routine. The authors of this paper carry out a broad study with several databases and several deep learning networks and compare two works published in the literature. According to the authors, the use of a simple U-Net can present significant (and even better) results compared with sophisticated methods.
To validate the above-mentioned theory, Hofmanninger et al. [25] performed several experiments where they showed that different results can be achieved depending on different training databases. Their research uses four networks for segmentation: U-Net [48], ResU-Net [31], Dilated Residual Network [65], and Deeplabv3 [7]. The databases used were Routine Random, VISCERAL Anatomy3, LTRC, and LCTSC.
Even conducting different experiments, it always results in a measure of the dice coefficient (or just Dice) [13] greater than 92%. This demonstrates that despite the use of another training database to generate a segmentation model, it presents significant results for lung segmentation in a new database. This fact is needed for clinical practice as there will not always be a ground truth to confirm the results, knowing that training in different databases can result in a high degree of accuracy.
Given the urgency for automatic methods and the lack of time for specialists who are currently facing the pandemic, this study used a database from a hospital where the collection of exams does not have a defined acquisition protocol. With the short time the specialists have in hand, it was decided that they would only work on the manual delineation of the infections. In this way, the database has no lung markings. However, there was a need to segment the lungs for better performance of the proposed method.
Thus, to obtain reliable and practical results, it was decided to train a lung segmentation model on a public and diversified database, with several acquisition protocols and using a state-of-the-art fully convolutional network on a segmentation problem. Based on Hofmanninger et al. [25] that, despite not having a metric to prove the result in our base, produces significant results and is capable of being used in the clinical routine.
Infections segmentation overview
As COVID-19 is a current and global crisis, the race for developing methods that can help specialists has accelerated. However, for the development of computational methods in tomography, it is necessary to observe the patterns that such a pathology may present in CT images. Ng et al. [40] describe the main characteristics of infections caused by COVID-19 on CT. The predominant imaging pattern is ground-glass opacities with occasional consolidation in the peripheries.
Some methods for segmentation of infections have already been developed. Yan et al. [64] suggested a new deep neural network for infection segmentation. The network called COVID-SegNet is composed of a block of feature variation proposed by the authors to better delineate infections. The study uses a broad research database consisting of 21,658 chest CT images. Many images are widely used for deep learning training and provide important results when several samples are used for training. The authors’ process achieves 72% Dice in infection segmentation.
In contrast, Fan et al. [19] reported that collecting a large amount of data is impractical within a short time, thus inhibiting the training of a deep model. They proposed a novel COVID-19 lung infection segmentation deep network (Inf-Net) and its semi-supervised version (Semi Inf-Seg), as there are few cases of annotations in its database. The tests were conducted on a database with only 100 slices of CT images, all with infections (database from [11]). A Dice of 68.2% was achieved for Inf-Net and 73.9.
Zheng et al. [68] presented a defined database acquisition protocol and the use of deep learning for infection segmentation. They performed lung segmentation based on a trained lung segmentation model and excluded false positives manually. For infection segmentation, they used a semi-supervised network called DeCovNet in a database of 630 CTs. The results achieved included an area under the ROC curve of 95.9.
In Wu et al. [63] also proposed a method of infection segmentation using deep learning. The method is applied to a standardized database built for the application, composed of 144,167 CT images of 400 COVID-19 patients and 350 uninfected cases. The network used is called joint classification and segmentation (JCS). The method achieved 95.0% sensitivity and 93.0% specificity on COVID-19 classification, and 78.3% Dice on segmentation. It is worth mentioning that specificity values of 93% at the pixel level for the segmentation of infections generate a high number of false positives when assessing the percentage of infection in the volume can result in erroneous or mistaken diagnoses.
Qiu et al. [47] suggest an extremely minimal network for the efficient segmentation of COVID-19 infections . They proposed a framework to provide efficient multi-scale, lightweight learning. The work is applied only to a base with 100 CT images of 60 patients (slices) resulting in approximately 1.6 slices per patient (database available in [11]). The results achieved using their method have 83.62% sensitivity, 97.42% specificity, and 77.28% Dice.
We can observe that the topic of segmenting infections (lesion or COVID19) in CT is already well-studied, and many computational methods have been developed. Among these, the deep learning methods stand out. Virtually all of the literature shows the use of these techniques to target infections Fan et al. [19] and Qiu et al. [47] focused on an image database of only 100 slices, showing the ability to obtain results even on small bases using deep learning methods. However, in clinical practice, a patient does not only have 1.6 slices per exam, and hence the classification of an examination is necessary.
There is, however, a certain standardization in the acquisition of the database, which does not occur in clinical practice, particularly in underdeveloped countries, where tests are carried out with the most diverse types of instruments. Nevertheless, it is not possible for all research to collect such a wide variety of data, primarily due to ethical standards and false diagnoses, among many other factors.
Thus, the segmentation of infections is not a trivial task, presenting at best a Dice of 78.3%. However, in a segmentation system such as these, we must observe the generation of false positives, as it is necessary for evaluating pulmonary impairment. Wu et al. [63] showed that wrong segmentations generate values of wrong commitment, even with 78.3% Dice and a specificity of 93%. The same occurs in Qiu et al. [47], with 77.28% of Dice and 97.42% of specificity.
The present work shows the use of a fully convolutional neural network, aggregating residual blocks [23] for the segmentation of infections in CT. The experiments were conducted on a database acquired without an acquisition process focused on the computational method, with slices ranging from 77 to 585, with patients of various ages, various types of lesions, and varying degrees of commitment. Still, it is a relatively small database in terms of the number of exams, involving only 40 patients, but a total of more than 8000 slices. For infection segmentation, the lung segmentation and vessel extraction processes proposed in our work are crucial for the robustness of the method.
Volumetric quantification overview
Despite playing an important role, segmentation alone does not provide quantitative information for the expert’s assessment. Assessing the volume of the lungs, infections, and the extent of involvement of the infections in the lungs are essential to assess the aggressiveness or stage of COVID-19. For treatment, quantifying impairment is also crucial. Assessing how compromised the lung is before and during treatment can show the effectiveness of the treatment and improve the drug testing process.
Some studies have shown the importance of quantitative assessment of infectious findings in CT. The work of Zhou et al. [69] investigated 62 patients to describe the CT features of COVID-19. They also showed that the CT exam of patients with COVID-19 pneumonia showed a mixed and diverse pattern with both lung parenchyma and the interstitium involvement. The value of assessing the findings is thus seen to suggest measures to the specialist that can be interpreted to monitor the pathology’s development.
The method proposed by Liu et al. [35] aims to quantify pneumonia lesions by computed tomography in the early days to predict progression to severe illness in a cohort of COVID-19 patients. Thus, CT quantification of pneumonia lesions can early and non-invasively predict the progression to severe illness, providing a promising prognostic indicator for clinical management of COVID-19. Three quantitative CT features of pneumonia lesions were automatically calculated, representing the percentages of groundglass opacity volume, semi-consolidation volume, and consolidation volume in both lungs.
Shen et al. [53] showed the importance of some metrics for assessing the degree of the disease: For example, volume (mL), lesion volume (mL), lesion percentage (%), and mean lesion density (HU) of the whole lung, right lung, left lung, and each lobe. Despite the comparison of the results of the computationally calculated values and the values observed by the specialist, it is known that once the lesion is segmented, it is possible to calculate the metrics using mathematical formulations without error.
In addition to the detection/segmentation of infections, the present study presents volumetric quantification of CT findings. Thus, we provide subsidies to specialists for the diagnosis, monitoring, and treatment of patients affected by this disease that has caused deaths of more and more patients every day.
Materials and method
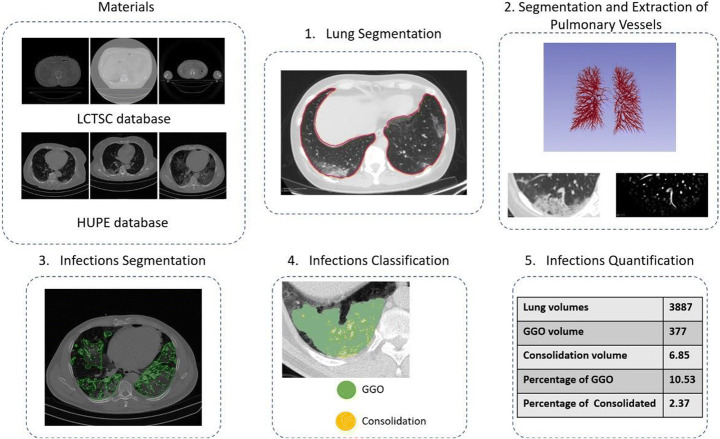
This section describes the materials and the proposed method for segmentation and volumetric quantification of infections caused by COVID-19 in CT. Figure 1 illustrates the steps used in this section. Then, each step is detailed in the subsections that follow.
Fig. 1.
Flowchart of materials and method
Materials
For the development of the method, it was necessary to use two databases retrospectively: a public and a private database. The public database was used for the creation of a lung segmentation model as it has a specialist’s markings for the lungs. The private database was used to perform the experiments on the segmentation of infections caused by COVID-19, to classify infections by texture (GGO or consolidated), and to extract quantitative metrics related to the lung and infection.
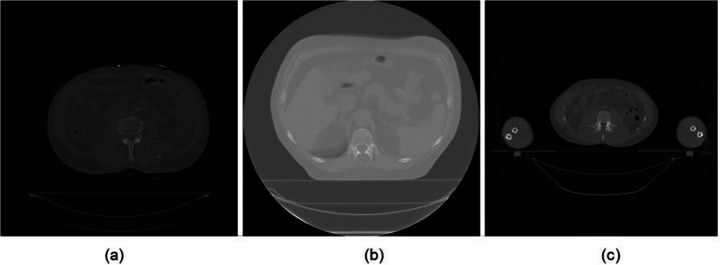
The first database is a public CT database with an expert’s markings for the lungs in 36 exams. It is available in The Cancer Imaging Archive, referenced by the LCTSC database. In this method, there is diversity in the image acquisition process. The images are obtained from three different institutes with several protocols, with the number of slices per exam ranging from 112,288. This database was used only to create a lung segmentation model. Once created, this model is applied to the second database.
The choice of this database for creating a segmentation model is justified by diversity. As the second database has no lung markings, and it was not due to a standard acquisition process due to the practical medical need for speed in the pandemic exams. Thus, a diversified database for the lung segmentation model makes it robust to variations in the acquisition process, generating more effective results when applied to another image database. An example of the diversity of the three institutes in this database is shown in Fig. 2.
Fig. 2.
LCTSC database: (a) slice of institute 1; (b) slice of institute 2; (c) slice of institute 3
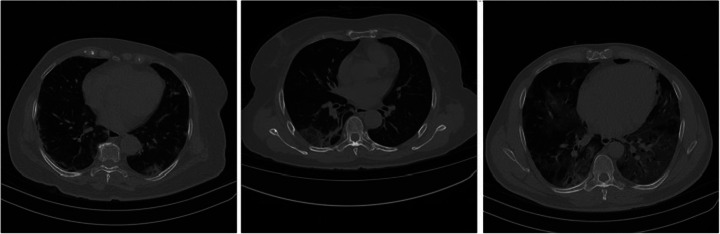
The second database is a private database from the Pedro Ernesto University Hospital of the State of Rio de Janeiro, Brazil, referenced in this work by the HUPE database. This database is composed of CT findings of COVID-19 infections. The appointments were made manually by several specialist doctors for 40 patients who had the most diverse types of lesions and degrees of impairment. There is only the label, without classification by the specialist; that is, it is not given whether the lesions are of a consolidation type or a ground-glass opacity (GGO) type. There is no lung labeling in this database, and thus there is a need to construct a segmentation model for another set of images. Figure 3 presents the examples of patients from this database.
Fig. 3.
HUPE database: Example of three patients with different degrees of infections
In addition, HUPE database patients have a variation in the number of slices, ranging from 77 to 585. The personal information of each patient was discarded to keep his identity confidential.
Therefore, for our experiments, the first database from The Cancer Imaging Archive has 36 patients from three different institutes, and the second database of Pedro Ernesto University Hospital of the State of Rio de Janeiro has 40 CT scans of patients affected by COVID-19 at different stages of contamination.
To analyze the population studying for the creation of the method, in the next subsection we will describe information about the patients of the HUPE database.
Participants of HUPE database
The HUPE database were used to build the infection segmentation, classification and quantification techniques. This database is composed with COVID-19 patients. To develop this database, the inclusion criteria were being ≥18 years of patients age, presenting with acute respiratory symptoms, and having had contact with COVID-19 patients or individuals with symptoms suggestive of the disease, with laboratory confirmation of SARS-CoV-2 infection. Patients in whom the technical standards of the CT scans were deemed unacceptable (movement artifacts or missing sequences) were excluded.
Based on these criteria, the database were composed of 40 different patients with a confirmed diagnosis for COVID-19 from April 17, 2020 to June 23, 2020. Age ranges from 23 to 88 years, with an average age of 49 years. Of the total, 24 (60%) patients were female. All patients had infectious findings in the lung parenchyma.
Image studies were approved by the Institutional Review Board of Pedro Ernesto University Hospital, operated by Rio de Janeiro State University, in the city of Rio de Janeiro, Brazil. Because all the data evaluated were obtained retrospectively from the database of the hospital, the requirement for written informed consent was waived.
Lung segmentation
The lung segmentation step is necessary to assist the next pulmonary vessel extraction step and to extract quantitative metrics related to the lung. For lung segmentation, a model was generated based on a fully convolutional neural network, called U-Net. This network was proposed by Ronneberger et al. [48] and has shown promising results in several domains of images.
First, we define the input images, then the network architecture, after which the training is performed. Finally, we present a segmentation refinement step. This step was necessary because the model generated for the LCTSC database was applied to the HUPE database, which is completely different. Thus, the results achieved for the LCTSC database were not fully reflected for HUPE. The segmentation refinement step tried to exclude some false positives generated for the HUPE database.
Input lung images
As previously mentioned, the training for lung segmentation was performed using the LCTSC database. To build the model, training, validation, and test datasets were defined. However, there are a few examples of the training step. Another significant thing to remember was the large variation in the number of slices. Thus, we opted for slice training (2D). In this way, the number of samples was increased and training with the original size of the x and y axes, which were 512 × 512, was possible.
To preserve database diversity, the three datasets promised patients from the three institutes. Thus, during training, there would be most diverse samples, both concerning protocols from different institutes and the number of slices.
Ensuring this diversity in the training model, the HUPE database acquisition protocol is reflected. In this second database, because the patients included are affected by COVID-19, and need rapid image acquisition, there is no well-defined acquisition protocol. It generates a high amount of diversity in the database. Therefore, ensuring the training of a generalized lung segmentation model is crucial to reflect the results in the HUPE database.
Training lung segmentation model
In the construction of the lung segmentation model, variations were applied to the U-Net model proposed by Ronneberger et al. [48]. U-Net, a convolutional neural network (CNN), uses the concepts and layers of a CNN. Examples include convolutional layers, pooling layers, activation layers, and dropout layers. CNNs are being increasingly used in several problems involving medical imaging [10, 14, 15, 17, 18, 55, 56].
In short, this network can be trained end-to-end, where U-Net simply concatenates the encoder feature maps to map decoder feature maps at all stages to form a ladder structure. This design, with its concatenation links, allows the decoder to learn the related features that are lost when grouped into the encoder at each point [48].
Based on the literature, we decided to use U-Net because of its promising results in medical imaging segmentation. However, instead of using the standard U-Net, we propose the following changes to improve its performance: a different number of layers, batch normalization, leaky ReLU, dropout, residual connections, and Dice loss function. We will explain these changes in detail below.
According to Ioffe and Szegedy [27], the training of deep neural networks is complicated, mainly because the distribution of the inputs changes during the training as the parameters of the previous layers are modified. Thus, a normalization was proposed for each layer. One of the problems shown during the development of this work is the variability of our database. As it consists of data from three different institutes, they tend to have high variability in their distribution, and thus, by using batch normalization, we have addressed this problem. Thus, each convolution is followed by batch normalization.
In addition to batch normalization, each convolution is followed by a Leaky ReLU activation [33], and dropout [3].]. The activation functions are used to correctly reset the weight parameters of the networks [33]. Commonly, convolutional neural networks are trained using the ReLU function as activation [32]. However, the ReLU function has problems with negative data during training, without being able to reactivate them. According to Maas et al. [36], Leaky ReLU solved this issue by assigning a constant variable in the negative region of the ReLU activation function. The main criterion for using dropout is to address the problem of overfitting in training. During each dropout operation, some training units are disabled, requiring the network to use other connections to better predict the generated model. In this way, we force the network to calibrate the connections between other neurons and avoid any overfitting caused during training.
According to He et al. [23], when deeper networks start converging, a degradation problem is exposed. With increasing network depth, accuracy becomes saturated (which might be unsurprising) and then degrades rapidly. Unexpectedly, such degradation is not caused by overfitting, and adding more layers to a suitably deep model leads to a higher training error. For this reason, He et al. [23] proposed the use of residual connections. They addressed the degradation problem by introducing a deep residual learning framework. Instead of hoping every few stacked layers directly fit a desired underlying mapping, we explicitly let these layers fit a residual mapping. Therefore, despite the use of Leaky ReLU functions, the proposed network addresses the problem of degradation by using residual connections.
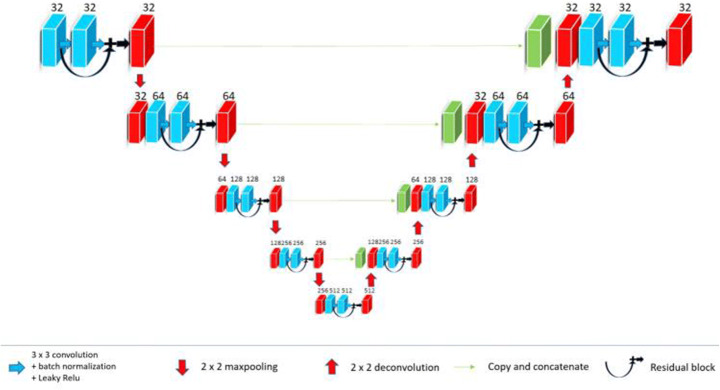
In summary, the RB-U-Net contains four pooling layers, and each convolution layer is composed of a Leaky ReLU activation, batch normalization, dropout, and finally, shortcut connections between the input and output by using residual blocks. All convolution layers used filters of 3 × 3. In the pooling layers, we use an average pooling of 2 × 2. In the deconvolution layers, we used a transposed convolution of 2 × 2. All dropout operations remove 20% of the connections. In this way, it is believed that the proposed network adds a lot of improvements compared to existing U-Nets and addresses major problems such as data variability and overfitting.
An example of the proposed architecture can be seen in the Fig. 4.
Fig. 4.
Architecture of residual U-Net
Also, the network is trained with the dice loss function [57], represented by Eq. 1.
| 1 |
where TP is the true positive, FP is the false positive, and FN is the false negative.
We chose to use the dice loss because we want to optimize the segmentation results based on the specialist’s marking. Thus, the use of this function as a loss helps the network to improve this metric during network training.
In segmentation methods, one of the metrics that reflects how accurate the method is is the Dice Index. This metric, according to Taha and Hanbury [58], is the most used to verify the effectiveness of segmentation methods, and the most compared for this purpose in the literature. As we need to develop a method that accurately delimits infections, the Dice was considered to be used as a loss function, in this way we managed produce a better segmentation result.
After training and creating the model, a test is performed with separate images for testing the LCTSC database. When obtaining satisfactory metrics in this database (compared with the state of the art of lung segmentation), we apply the model to the HUPE database. Although there is no markings to confirm the results achieved, the results visually show promise. However, there were some false positives. Thus, a segmentation refinement step is necessary.
Segmentation refinement
The segmentation refinement process consists of removing false positives, smoothing the contour, and filling holes inside the binary lung mask. The linked components are classified from the binary image to eliminate false positives. We measured the volume of each one. Therefore, we have marked the object with the largest number. Finally, we removed objects with a volume of less than one-fifth of the largest.
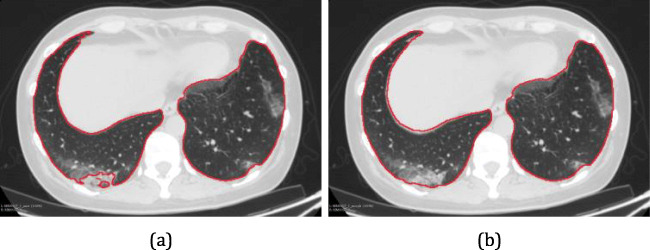
Lung segmentation can result in irregularities near the contour of the lungs. As it is common to have COVID-19 lesions on the walls of the parenchyma because they are denser, they have low contrast concerning the tissue outside the parenchyma. The outline of the lung mask is regulated using morphological filtering techniques of opening and closing by reconstruction. The opening operation can separate connected objects in a binary image and soften the inside of the object’s outline. The closing operation can fill small holes and soften the outside of the object’s outline. Finally, holes not connected to the boundaries of objects are eliminated [34]. Figure 5 presents an example of the result of this step.
Fig. 5.
Segmentation refinement: Contour of the lungs segmented by Residual U-Net (a) and result of segmentation refinement
Segmentation and extraction of pulmonary vessels
The process of marking infections is carried out comprehensively by a specialist. Owing to the difficulty of circumventing existing pulmonary vessels with infections (lesions) during this process, several vessels are incorporated into the markings. When building a model for segmentation, this pattern can end up hindering the training. Therefore, there is a need to exclude these vessels. Thus, the vessels present in the lungs are segmented and subsequently extracted from the specialist’s markings.
Segmentation of pulmonary vessels is performed using filters Hessian Recursive Gaussian Image Filter, and the Hessian 3D To Vesselness Measure Image Filter implemented using the Insight Toolkit (ITK) [30]. The first filter calculates the 3D Hessian matrix of an image by convolution with the second and cross derivatives of a Gaussian. An important parameter of this filter is the sigma, which determines the amount of smoothing applied during the Hessian matrix estimate. A greater sigma will decrease the identification of small vessels.
The second filter highlights tubular structures, such as vessels and bronchi, in three-dimensional medical images. After Hessian matrix eigenvalue decomposition, the bright tubular structures (blood vessels in CT images) have an eigenvalue close to zero and two negative eigenvalues (positive values correspond to dark tubular structures). This filter has been described in more detail in Sato et al. [52]. TThe method produces Hessian matrices as the first stage of the filter, and then highlights the tubular structures in the second stage. Figure 6 presents an example of the segmentation of pulmonary vessels.
Fig. 6.
Enhancement of pulmonary vessels: (a) Result of lung segmentation used to demarcate the region of application of the vessel enhancement filter, which results in (b)
The next step is to create a binary image from the image resulting from blood vessel enhancement filters using thresholding. Experienced radiologists gave us a range of values that could segment blood vessels. Therefore, after several tests, the threshold of 20 was ideal for the base used in this work. It is defined as vessel voxels with a value above a threshold, and otherwise, not vessels. We performed a visual analysis of each image in the bank, subtracting the parenchyma and the binary mask of the vessels generated with a value of 20. Thus, we proved that this value was ideal for our problem. Once the mask of the vessels is obtained, it is extracted from the marking of the lesions made by specialists.
The extraction of blood vessels from the lesions is also important for a better assessment of lesion consolidation. The vessels have high densities and are usually classified as consolidated regions of the lesions, biasing the proposed assessment. The classification of lesions is explained later in Section 3.5. Figure 7 presents an example of the extraction of pulmonary blood vessels from an appointment made by a specialist.
Fig. 7.
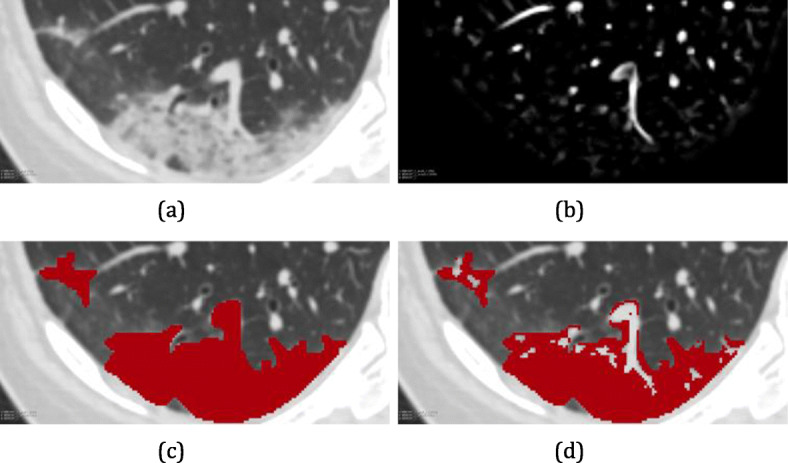
Extraction of pulmonary vessels from the markings made by the specialist: (a) original image; (b) image with emphasis on pulmonary vessels; (c) ground truth built by a specialist; and (d) ground truth without pulmonary vessels
Infections segmentation
After extraction of pulmonary vessels from the specialist’s marking, the next phase is to build a lesion segmentation model. Because the HUPE database has no indication of the type of infection found, the model must be robust enough to segment the most diverse types of infection. The pulmonary vessel exclusion strategy will assist the network in learning lesion patterns without considering vessels and bronchi.
Input infections images
For the input images of the lesion segmentation network, a 2D approach was used. It was passed as an input to the network and slices of patients from the HUPE database. Once again, this approach was chosen for both the diversity of the database and the number of samples. The HUPE database had patients ranging from 77 to 585 slices, showing again the complexity of working with this database.
Thus, in the training phase, the database was divided into three datasets (training, validation, and testing) to ensure that the greatest number of diverse slices was inserted in the training. Examples of slices with several types of CT findings of COVID-19 infections are shown in Fig. 8.
Fig. 8.
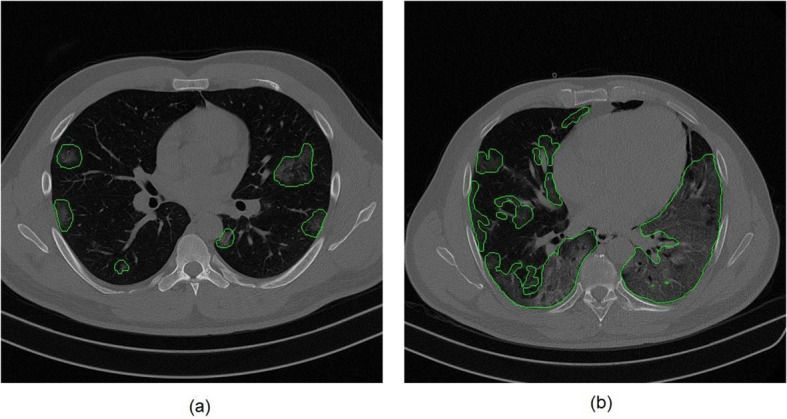
Examples of CT findings of COVID-19 infections: (a) early stage patient and (b) advanced stage patient
Training infections segmentation model
In training the segmentation of infections, once again, we use Residual U-Net, defined in Section 3.2. However, we observed that the use of diceloss was unable to generalize all the examples in the database, without generating significant results. This observation was made by checking the results of the validation dataset. Thus, we decided to use a combination of binary cross-entropy [22] and dice loss as a network loss function.
The loss function is the sum of the binary cross-entropy and dice loss. The network will try to optimize this function in the evolution of the times. An analysis of the influence of the use of these losses is presented in Section 4.
Another important point inserted for infection training was dropout layers [3]. Since the HUPE database is very diversified (in the number of slices and acquisition process) and the number of samples for the various types of existing lesions is low, it was expected that the model would be overfitted. To circumvent this problem, in training the infection segmentation, for each convolution layer followed by batch normalization and leaky ReLU, a dropout layer was inserted.
At the end of the training, the model was applied to separate exams for testing. As segmentation is applied to all slices of the patient, there may be some false positives generated outside the parenchyma region. To achieve this, lung segmentation (Section 3.2) was used to exclude predictions outside the lung region.
Infections classification
CT of the lungs of individuals with COVID-19 shows typical lesions, characterized as ground-glass opacity, air space consolidation, or complete opacity [12, 24]. The level of consolidation found in lung lesions helps to define the stage of the disease. In the initial step, ground-glass opacities located in one or more shapes, in the form of a cloud of light or fine mesh, are common. Occasionally, consolidation is located under the pleura or near the bundles of bronchial blood vessels. In the progressive stage, few patients had increased ground glass shadows, or the affected area enlarged tending to merge, thus presenting with large-scale consolidation. In the severe or critical stage, patients may present with diffuse consolidation of the unilateral or bilateral lungs, accompanied by ground-glass opacities and signs of bronchial infection. In the last step, the absorption stage, the original lesions are completely absorbed and improved [54].
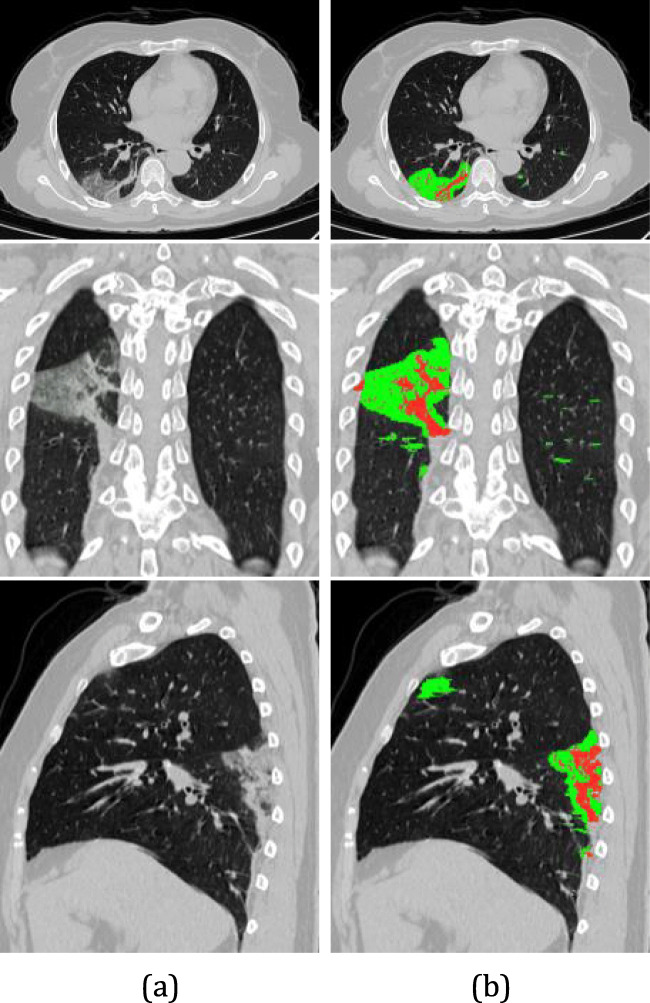
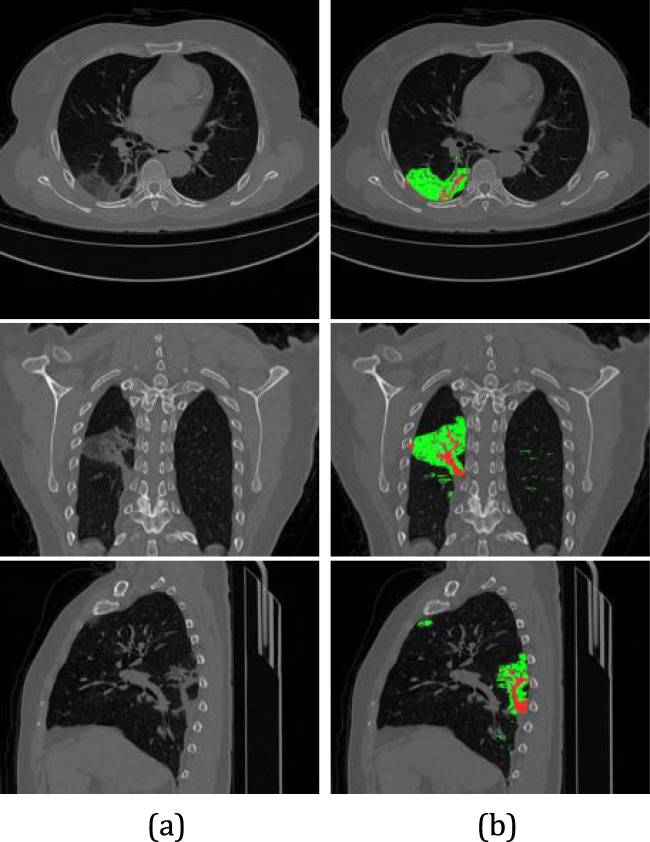
To monitor the level of lung involvement (ratio between the volume of the lesions and the lungs), it is necessary to measure the level of consolidation of the lesions. Thus, sub-regions of the lesions are classified as consolidated if the voxel intensity is greater than 0 Hounsfield units (HU), and in GGO otherwise [40, 66]. Thus, it is possible to evaluate the change in the consolidation of the lesions caused by COVID-19. Figure 9 presents an example of the classification of injuries. Ground-glass lesion regions are marked in green, whereas consolidation regions are red.
Fig. 9.
Infections classification. (a) Original image. (b) GGO (green color) and consolidation (red color) can be seen in the left lung by axial, coronal and sagittal views, respectively
Infections quantification
For the volumetric quantification of infections, it was necessary to calculate the volume of the ground-glass and consolidated subregions of the lesions, the total volume of the parenchyma, the volume of the pulmonary vessels, and the volume of each lung.
In the total volume of the parenchyma, the result of lung segmentation was used (Section 3.2). In the volume of the pulmonary vessels, segmentation of the pulmonary vessels was used (Section 3.3).
The segmentation of each lung was based on the analysis of the vertical projection histogram. For the volume resulting from the lung segmentation, described in Section 3.2, as the lungs are located in the central region of the exam, the vertical projection histogram was calculated only in that region.
Therefore, an interval was defined based on the central value of the exam width with a gap of 50 pixels on both sides. Therefore, the vertical projection histogram shows 101 bins on the horizontal axis and the respective number of lung pixels on the vertical axis. Then, the cut-off point in the width of the exam responsible for separating the lungs was defined based on the bin that has the lowest voxel value, that is, the valley point of the histogram. The segmentation of each lung is shown Fig. 10.
Fig. 10.
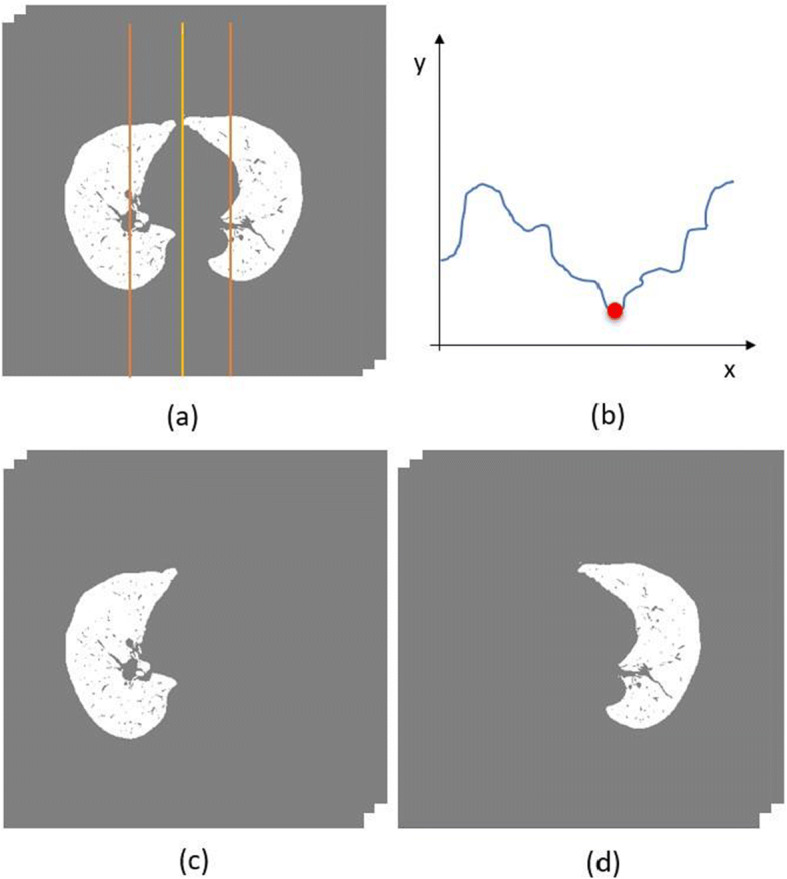
Separation of the lungs. (a) Volume resulting from the lung segmentation step with the range of the projection histogram calculation illustrated by orange, (b) resulting histogram with the cutoff point illustrated by red, (c) left lung, and (d) right lung. Ground glass (GgV), and consolidated (CdV) region volume of the lesions and lung volume (LV). The volume of the lung was calculated by subtracting the volume of the pulmonary vessels. All measures were calculated for the left lung (LL), right lung (RL), and both lungs (BL) in milliliters (mL)
The volumes of the lesions and lungs were calculated from the number of voxels of their respective segmentation and the spacing information from the CT image. Thus, we have the following measures: lesion volume (LesV),
From the volumetric measurements of these structures, some relative measures were calculated and evaluated, such as the proportion of ground glass (PggLes) and consolidation (PcdLes) to the general lesions of a patient, as well as the proportion of ground glass (PggL), consolidation (PcdL), and all lesions (PlesL) to the volume of the patient’s lungs. In addition to volumetric measurements, the average density (LesD) of the lesions was assessed.
At the end of the method, in addition to having visual information giving the location of the infectious lesions, the specialist still has quantitative information from COVID-19 at hand. In this way, the specialist has more subsidies to assess the degree of the disease, the extent of impairment in the lung, or, in the case of treatment, find out temporarily in the TCs if this is the recommended treatment.
Experimental setup and results
This section describes our experiments. First, we explain the experimental setup, metrics used to evaluate the method, and training environment used. Then, the results of each step are presented.
Experimental setup
For the proposed method, five validation metrics were used in the model construction steps. The next section describes these metrics.
Evaluation
The metrics used in this work were the most common metrics used in problems with images. They are Dice [13], sensitivity (SEN) [67], specificity (SPEC) [67], accuracy (ACC) [67], area under the curve ROC (AUC) [45], Jaccard index [28], and Hoursdoff distance [26].
Training and testing environment
All tests were implemented in the Python programming language with the help of the Keras deep learning library [8] with TensorFlow-GPU as a backend. For image processing the library Insight Segmentation and Registration Toolkit (ITK) was used [29, 30]. For training and testing of lung segmentation and infection models, a computer with modest configurations with an Intel I7 processor, 128 GB of RAM, 11 GB GeForce RTX 2080, and Windows 10 operating system was used.
The training of the lung segmentation model lasted approximately 360 s per epoch, and the classification for each patient lasted for approximately 12 s. On the other hand, the training of the infection segmentation model lasted for an average time of 419 s per epoch, and the classification per patient lasted approximately 17 s. With trained models, the system lasts about two min from the entry of the CT to the exit of the segmented infections with quantification metrics.
After several training sections, and hyperparameters tuning, the chosen hyperparameters in the training phase were: number of epochs equal to 200, size of batch equal to 3, Adam optimizer with initial learning rate equal to 0.0001, decay equal to 0.000000199, and 10% of volumes for validation.
In the experiments, we tried to use data augmentation to verify improvements in the training phase of the method. However, increasing the sample set by zoom, flip, etc. did not improve the method in terms of infection segmentation. It is worth noting that the changes presented by COVID-19 appear to be more related to texture variations, without having a characteristic behavior of changing geometry; thus, augmentation operations (zoom, flip, rotation, and translation) probably cannot simulate the effects of infections.
Results
In this section, the results obtained by executing the proposed method are presented. To verify all the steps of the method, results and validation metrics were collected at each step. Thus, we divided the experiments into the following:
Lung segmentation.
HUPE database division
Infection segmentation
-
Classification and quantification of infections per patient.
The following subsections describe these experiments.
Results of lung segmentation
As mentioned in Section 3.2, the HUPE database patients did not have lung markings. However, the literature shows that models built for one database can be easily used for another, producing significant and robust results [25]. Thus, tests were performed for the LSTSC database until we found a robust model that could be applied for HUPE. Thus, we divided the LSTSC database into three datasets: training, validation, and test.
The training dataset consisted of 27 patients; as this database is composed from three different institutes, we ensured that nine patients from each institute made up this training dataset. For validation dataset, we used one patient from each institute. The model was applied to the test dataset, which had two patients from each institute. The results can be seen in Table 1, along with the application of the standard U-Net [48] and two U-Net variations [31].
Table 1.
Result of Lung Segmentation in the LSTSC Databasee
We can observe that although U-Net and its variations produce promising results, the proposed variation with residual blocks manages to surpass this result in all metrics, producing a Dice of 96%.
Although the database is varied, consisting of several acquisition protocols from three different institutes, the model was able to generalize the problem and achieve good results for this purpose. The model achieved in the LSTSC database is applied to the entire HUPE database for lung segmentation. In addition, because it is a different database, segmentation refinement processes were applied (Section 3.2.3).
HUPE database division
To validate the results of the proposed method up to the infection segmentation step, several experiments were conducted. First, the database was divided into training, validation, and testing databases. The HUPE database included 40 patients, and thus, we used 30 CTs for training, four for validation, and six for testing. This division was carried out randomly. The only premise adopted was to use as many patients as possible for training, as it is from the variability of training that robust and effective models are created, mainly for models that use deep learning.
As mentioned in Section 3.4, the slices of the training database were used to create the model, using validation to monitor the best set of parameters in the training. With the model in hand, it was possible to apply the prediction on the test database.
Several experiments were then carried out to validate the steps proposed throughout the method. Next, we show the results achieved for the segmentation of infections based on our experiments.
Result of infection segmentation
Validation of the pulmonary vessel segmentation step is not possible because the databases used in this work do not have vessel markings. In addition, works in the literature either do not have vessel marking [1] or use private data sets [51]. Thus, the first experiment was the segmentation of infections with and without the pulmonary vein extraction stage. To validate this experiment, the segmentation and extraction of pulmonary veins were excluded (Section 3.3) and we conducted training with the same training, validation, and testing patients. Table 2 presents the results of these experiments.
Table 2.
Result of Infection Segmentation with and without Pulmonary Vessels Extraction Step
| Pulmonary Vessels | Dice(%) | SEN (%) | SPEC (%) | ACC (%) | AUC(%) | JAC(%) | HD(mm) |
|---|---|---|---|---|---|---|---|
| Without extraction | 48.49 | 47.97 | 99.73 | 99.45 | 73.84 | 51.31 | 11.3102 |
| With extraction | 77.10 | 72.44 | 99.76 | 99.37 | 86.11 | 66.86 | 4.5295 |
We can observe that with the extraction of pulmonary vessels, the method achieves significant improvement for all validation metrics. This is because when the model is trained taking into account the vessels marked by the specialist, the network tends to learn the patterns of the vessels in the model. Thus, when classifying the test images, the model considers several vessels as infection, thus negatively impacting all metrics. The addition of the vessel extraction step improved the index of segmentation of infections by 28.61%.
When inserting the pulmonary vessel extraction step, the method can reach a Dice of 77.1%, resulting in an improvement of more than 25% for this metric.
The same occurs with the sensitivity metric, indicating how robust the method is in finding lesion voxels.
The second experiment was conducted to compare our result with the infection segmentation step using the U-Net proposed by Ronneberger et al. [48]. This experiment intends to show how important the use of the proposed improvements in the network was to achieve the results.
Table 3 shows how promising the improvements applied to our Residual U-Net are. When inserting the residual blocks and layers of dropout and batch normalization, the results compared with U-Net have an improvement of more than 20% in terms of sensitivity Even the ResU-Net and DenseU-Net produce good results; again the proposed method overpassed Dice in more than 10% in terms of sensitivity. It appears in the Dice as a positive factor. We assume that these changes address the issue of the availability of databases, making the network learn more about the patterns found in infections to help describe the COVID-19 lesions.
Table 3.
Result of Infection Segmentation using U-Net and Residual U-Net
To validate the results obtained using the Residual U-Net with the proposed cost function, we conducted another experiment. In this case, we use the same proposed network, changing only the cost function. First, we use binary cross entropy (BCE), dice loss (DL), and finally, the combination of the two (BCE + DL). The results are presented in Table 4.
Table 4.
Result of Infection Segmentation using Different Types of Loss
| Loss Function | Dice(%) | SEN (%) | SPEC (%) | ACC (%) | AUC(%) | JAC(%) | HD(mm) |
|---|---|---|---|---|---|---|---|
| BCE | 39.65 | 25.84 | 99.97 | 99.52 | 62.90 | 45.61 | 16.2633 |
| DL | 66.69 | 53.17 | 99.73 | 99.67 | 76.54 | 56.83 | 8.2578 |
| Proposed Method(BCE + DL) | 77.10 | 72.44 | 99.76 | 99.37 | 86.11 | 66.86 | 4.5295 |
As can be observed, when using only the BCE, the network cannot learn the standards of the training database, presenting a Dice value of 39.65% and sensitivity of 25.84%. When only DL was used, there was a significant improvement in the results, going even beyond the experiment with the U-Net, presenting a Dice of 66.69%. However, on using the combination of the two cost functions, the training performed better, learning in fact the variability of the database. Once again, we show that all the improvements proposed for the proposed method show a significant result (77.1% of Dice) for the task of infection segmentation caused by COVID-19.
To confirm the variability of the database, Table 5 describes the metrics achieved for each of the trained patients. The patient column shows the code name given to each patient in the database.
Table 5.
Result of infections segmentation by test patient
| Patient | Number of slices | Dice(%) | SEN (%) | SPEC (%) | ACC (%) | AUC(%) |
|---|---|---|---|---|---|---|
| MM0001 01 | 425 | 76.72 | 70.45 | 99.89 | 99.65 | 85.17 |
| MM0001 02 | 285 | 72.29 | 72.78 | 99.85 | 99.73 | 86.32 |
| MM0015 01 | 212 | 79.09 | 73.93 | 99.96 | 99.87 | 86.94 |
| MM0016 01 | 256 | 68.19 | 73.28 | 99.87 | 99.80 | 86.58 |
| MM0017 01 | 301 | 76.29 | 77.46 | 99.72 | 99.49 | 88.60 |
| MM0065 01 | 160 | 75.04 | 66.73 | 99.29 | 97.68 | 83.01 |
| Average ± StandardDeviation | 273.17 ± 82.47 | 77.10 ± 3.92 | 72.44 ± 3.29 | 99.76 ± 0.22 | 99.37 ± 0.76 | 86.11 ± 1.71 |
We can observe that the image database used (HUPE database) presents a great diversity concerning the number of slices. This non-standardization makes it necessary to develop a robust model for segmenting infections. By using the various techniques proposed in this methodology, we could present significant Dice metrics for all patients, with the best Dice reaching 79.09% and the worst 67.19%. This produces a standard deviation of only 3.92%, which shows that despite the diversity of the database, the method is capable of presenting significant metrics during the task of infection segmentation.
Result of classification and volumetric quantification of infections by patient
To validate the method and how promising is, we extracted the same measurements from the specialist’s markings to assess how the method performance.
Table 6 presents a comparison of the measures extracted from the marking made by the specialist and the segmentation method. A paired t-test was used to assess the consistency of the results of the distribution of lesions. It was performed using Python’s scipy.stats [60] library, which contains a large number of probability distributions and statistical functions. No measure showed a significant difference (ρ < 0.05).
Table 6.
Comparison between the Measures Evaluated by the Experts and a Computer using the Proposed Method
| Measures | Method | Specialist | t* | ρ’ |
|---|---|---|---|---|
| LVBL | 3887.91 ± 1214.80 | – | – | – |
| LVLL | 2044.44 ± 572.04 | – | – | – |
| LVRL | 1843.46 ± 647.00 | – | – | – |
| LesVBL | 383.67 ± 429.18 | 460.42 ± 562.55 | 1.38 | 0.23 |
| LesVLL | 207.31 ± 153.84 | 242.44 ± 197.83 | 1.85 | 0.12 |
| LesVRL | 176.36 ± 276.00 | 217.98 ± 366.70 | 1.11 | 0.32 |
| GgVBL | 377.82 ± 420.56 | 455.60 ± 557.66 | 1.36 | 0.23 |
| GgVLL | 203.52 ± 149.51 | 238.58 ± 194.49 | 1.81 | 0.13 |
| GgVRL | 174.30 ± 271.79 | 217.02 ± 365.31 | 1.10 | 0.32 |
| CdVBL | 5.85 ± 9.09 | 4.82 ± 5.31 | −0.64 | 0.55 |
| CdVLL | 3.79 ± 4.99 | 3.86 ± 4.08 | 0.15 | 0.89 |
| CdVRL | 2.06 ± 4.24 | 0.96 ± 1.44 | −0.94 | 0.39 |
| PggLesBL | 98.37 ± 1.59 | 98.32 ± 1.74 | −0.16 | 0.88 |
| PggLesLL | 98.20 ± 1.70 | 98.08 ± 1.81 | −0.40 | 0.71 |
| PggLesRL | 99.36 ± 0.70 | 83.01 ± 40.67 | −0.98 | 0.37 |
| PcdLesBL | 1.63 ± 1.59 | 1.68 ± 1.74 | 0.16 | 0.88 |
| PcdLesLL | 1.80 ± 1.70 | 1.92 ± 1.81 | 0.40 | 0.71 |
| PcdLesRL | 0.64 ± 0.70 | 0.33 ± 0.34 | −1.05 | 0.34 |
| PlesLBL | 10.80 ± 11.90 | 12.78 ± 15.34 | 1.33 | 0.24 |
| PlesLLL | 10.63 ± 7.78 | 12.23 ± 9.76 | 1.75 | 0.14 |
| PlesLRL | 11.25 ± 16.83 | 13.73 ± 22.04 | 1.11 | 0.32 |
| PggLBL | 10.63 ± 11.68 | 12.64 ± 15.22 | 1.31 | 0.25 |
| PggLLL | 10.43 ± 7.59 | 12.03 ± 9.61 | 1.71 | 0.15 |
| PggLRL | 11.12 ± 16.58 | 13.66 ± 21.95 | 1.11 | 0.32 |
| PcdLBL | 0.17 ± 0.24 | 0.14 ± 0.14 | 0.16 | 0.88 |
| PcdLLL | 0.20 ± 0.24 | 0.20 ± 0.20 | 0.40 | 0.71 |
| PcdLRL | 0.13 ± 0.25 | 0.07 ± 0.09 | −1.05 | 0.34 |
| LesDBL | −536.59 ± 95.98 | −545.58 ± 90.63 | −1.01 | 0.36 |
| LesDLL | −536.70 ± 90.60 | −544.33 ± 87.45 | −0.74 | 0.49 |
| LesDRL | −586.23 ± 108.19 | −500.26 ± 262.56 | 0.70 | 0.51 |
| GgDBL | −545.47 ± 90.32 | −555.22 ± 85.98 | −1.30 | 0.25 |
| GgDLL | −546.58 ± 84.10 | −555.40 ± 81.83 | −0.99 | 0.37 |
| GgDRL | −589.75 ± 104.94 | −502.28 ± 263.18 | 0.72 | 0.51 |
| CdDBL | 48.34 ± 21.37 | 71.67 ± 36.28 | 1.71 | 0.15 |
| CdDLL | 48.37 ± 21.64 | 73.37 ± 39.60 | 1.66 | 0.16 |
| CdDRL | 30.50 ± 26.09 | 54.02 ± 40.98 | 1.76 | 0.14 |
It is worth mentioning that for patients in advanced stages, the specialist opts for a quickly marking, as the degree of impairment is clearly visible, which results in either ignoring minor infections or adding several lesions in the same marking.
However, as our method classifies voxel-to-voxel, it can leading to the existence of some divergent metrics extracted by the specialist. The abbreviation of the metrics can be found in Section 3.6.
Lung volume (LV), lesion volume (LesV), ground glass volume (GgV) and condensation volume (CdV).
Proportion of GGO (PggLes) and consolidation (PcdLes) to the general lesions of a patient. Proportion of GGO (PggL), consolidation (PcdL) and all lesion (PlesL) to the volume of the patient’s lung.
Mean lesion density of GGO (GgD), consolidation (CdD) and all lesion (LesD). Left lung (LL), right lung (RL) and bilateral lung (BL).
A Paired T-test to assess the consistency of the results of the distribution of lesions obtained by a specialist and proposed method.
Discussion
In this section, we discuss the quantitative and qualitative results of the method through case studies. Existing methods are compared with the proposed method. Finally, the advantages and limitations of the proposed method are highlighted.
Case study
Two test cases are presented. Considering the results presented in Section 4, we present the patient with the highest Dice value (MM0015 01) and the lowest Dice value (MM0015 01).
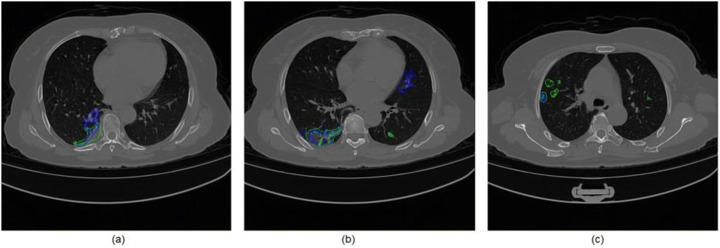
Infection segmentation experiments show a visual and quantitative way to compare our results with a real specialist’s markings. We can see the edges of the mark (blue) and the segmentation of the method (green), where a large part is superimposed, which shows us the robustness of the method to be similar to the specialist’s markings. However, although in some points the specialist’s markings go beyond the borders found using the proposed method, it is worth remembering that the doctor while marking the infections tends to leave a risk margin so as to ensure a certain safety cover, or because it is difficult to define the boundaries and behavior of COVID-19 on CT. Therefore, first, we will show the results concerning infection segmentation compared with the specialist markings. Next, we will show the regions highlighted in GGO and consolidation together with the volumetric quantification for each patient.
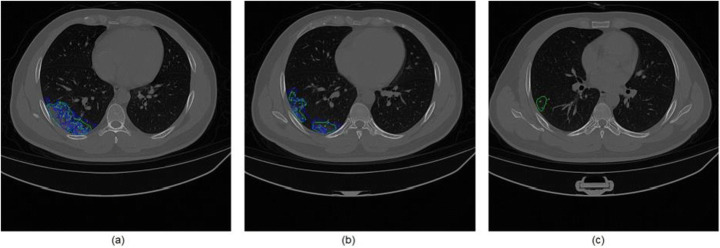
The first case study is patient MM0015 01 (see Table 5), which presented a Dice of 79.09%. The result of the infection segmentation with the marking of the specialist is shown in Fig. 11.
Fig. 11.
Patient case study MM0015 01 in blue marked by the specialist and green marked by the method (a) slice 80, (b) slice 97, and (c) slice 114
We can observe that the specialist’s mark is very faithful to the mark obtained using the proposed method. It is worth mentioning that because it presents a specificity of 99.76%, practically no false positives are generated. In Fig. 11(c) we can see that the proposed method can find regions in GGO that were not marked by the specialist.
After segmenting the infections, the next step is to evaluate the regions of GGO and the condensation of this patient. Table 7 presents the results of the patients MM0015 01 e MM0016 01.
Table 7.
Infections Quantification from Case Studies: patient MM0015 01 and MM0016 01
| Measures | MM0015 1 Method Specialist |
MM0016 1 Method Specialist |
||
|---|---|---|---|---|
| LVBL | 4445.18 | – | 2713.41 | – |
| LVLL | 2268.14 | – | 1432.97 | – |
| LVRL | 2177.04 | – | 1280.44 | – |
| LesVBL | 126.26 | 155.89 | 94.40 | 83.57 |
| LesVLL | 125.19 | 155.89 | 87.89 | 79.66 |
| LesVRL | 1.07 | 0.00 | 6.51 | 3.90 |
| GgVBL | 123.42 | 152.85 | 90.29 | 79.40 |
| GgVLL | 122.35 | 152.85 | 83.87 | 75.50 |
| GgVRL | 1.07 | 0.00 | 6.42 | 3.90 |
| CdVBL | 2.85 | 3.04 | 4.11 | 4.17 |
| CdVLL | 2.85 | 3.04 | 4.02 | 4.17 |
| CdVRL | 0.00 | 0.00 | 0.09 | 0.00 |
| PggLesBL | 97.75 | 98.05 | 95.65 | 95.01 |
| PggLesLL | 97.73 | 98.05 | 95.43 | 94.77 |
| PggLesRL | 100.00 | 0.00 | 98.63 | 99.95 |
| PcdLesBL | 2.25 | 1.95 | 4.35 | 4.99 |
| PcdLesLL | 2.27 | 1.95 | 4.57 | 5.23 |
| PcdLesRL | 0.00 | 0.00 | 1.37 | 0.05 |
| PlesLBL | 2.84 | 3.51 | 3.48 | 3.08 |
| PlesLLL | 5.52 | 6.87 | 6.13 | 5.56 |
| PlesLRL | 0.05 | 0.00 | 0.51 | 0.30 |
| PggLBL | 2.78 | 3.44 | 3.33 | 2.93 |
| PggLLL | 5.39 | 6.74 | 5.85 | 5.27 |
| PggLRL | 0.05 | 0.00 | 0.50 | 0.30 |
| PcdLBL | 0.06 | 0.07 | 0.15 | 0.15 |
| PcdLLL | 0.13 | 0.13 | 0.28 | 0.29 |
| PcdLRL | 0.00 | 0.00 | 0.01 | 0.00 |
| LesDBL | −468.10 | −505.56 | −468.29 | −454.83 |
| LesDLL | −466.20 | −505.56 | −463.01 | −443.86 |
| LesDRL | −690.00 | 0.00 | −539.62 | −678.62 |
| GgDBL | −479.92 | −516.56 | −491.26 | −481.08 |
| GgDLL | −478.09 | −516.56 | −486.96 | −470.85 |
| GgDRL | −690.00 | 0.00 | −547.42 | −678.95 |
| CdDBL | 44.75 | 46.79 | 36.31 | 45.16 |
| CdDLL | 44.75 | 46.79 | 36.65 | 45.17 |
| CdDRL | 0.00 | 0.00 | 21.17 | 21.00 |
On analyzing Fig. 12 and Table 7, we concluded that the method was more accurate compared with the specialist’s markings. Despite the divergence of some metrics, using the proposed method it is possible to assess how compromised the lung is, and thus provide a series of subsidies to specialists.
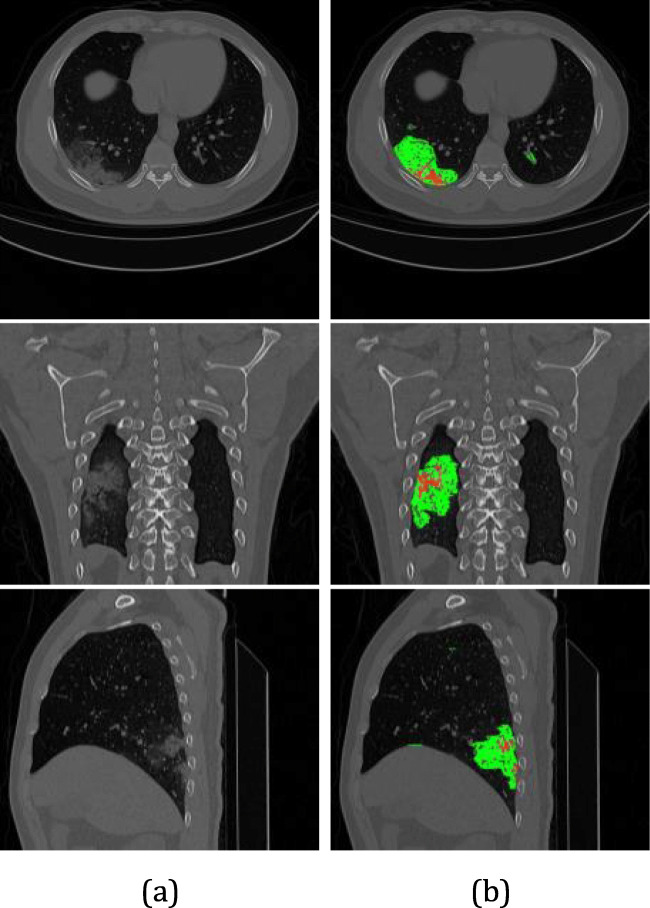
Fig. 12.
Infections classification from case studies MM0015 01: (a) original image. (b) GGO (green color) and consolidation (red color) can be seen by axial, coronal and saggittal views, respectively
The second case study concerns the patient MM0016 01 (see Table 5), which presented a Dice equal to 67.19%, the worst result presented by the method. The result of the infection segmentation together with the markings by the specialist is shown in Fig. 13.
Fig. 13.
Patient case study MM0016 01 in blue marked by the specialist, green marked by the method (a) slice 93, (b) slice 107, and (c) slice 160
In Fig. 13(a), the method marks a condensed region outside the markings of the specialist, which can be considered as an error.
In fig. 13(b), there is a region of infection that was not found by the method but marked by the specialist. However, Fig. 13(c) manages to demarcate more regions than the one marked by the specialist. All of these scenarios negatively impact the value of Dice, and as a result, its value is only 67%.
As in the first case study, segmented infections, the regions of GGO, and condensation of this patient were evaluated (Fig. 14) and the metrics are evaluated (Table 7). Once again, despite the low Dice value, when comparing the metrics extracted from the method and the specialist’s markings, it did not have a significant impact on the evaluation of the results. It is possible to analyze lung impairment from the extracted metrics.
Fig. 14.
Infections classification from case studies MM0016 01: (a) original image. (b) GGO (green color) and consolidation (red color) can be seen by axial, coronal and sagittal views, respectively
Comparison with related works
Even though it is a recent problem, considering how aggravating it is, some studies have already been begun trying to address the segmentation of infections and volumetric quantification. Although there is no way to compare the results of volumetric quantification, the works already developed served as a basis for our study. When observing the opportunity to not only segment but also to measure the degree of infection caused by COVID-19, the works in the literature gave us subsidies to extract metrics after our segmentation.
In contrast, some works deal only with the segmentation of infections. Although they do not use the same database, they serve as a comparison to our method. In Section 2.2 some works are presented that do the segmentation of infections.
All the works presented use deep learning models, which highlights the promise these techniques hold. Some of the works presented propose semisupervised models, that is, models that need some degree of human interaction to achieve desired results [19, 68]. It is worth mentioning that our method is completely automatic, which in addition to targeting infections on a remarkably diverse database also extracts metrics related to infections to assist specialists.
Fan et al. [19], Qiu et al. [47] used a limited database, presenting only 100 slices of patients. Although training with few slices and good results are relevant, the possibility of an overfitted model is great. In clinical practice, usually, 100 slices correspond to the CT of a single patient. Our work uses a database with 40 patients with more than 8000 slices and achieves a Dice of 77.1% with approximately 100% specificity.
Yan et al. [64] and Wu et al. [63] used a very extensive image database with fixed acquisition protocols. In deep learning approaches, the greater the number of images for training, the more robust the model becomes. Despite using 21,658 chest CT, the work of Yan et al. [64] achieve 72% Dice. Wu et al. [63] used 144,167 chest CT, presenting a Dice of 78.3%; however, the specificity is 93%. This causes a series of false positives, which in our approach would be a problem because we deliver measures related to infections. Compared with these two works, our method is used in fewer images; however, it achieves promising results, as we reach 77.1% Dice with a specificity of 99.76%. In our method, we value consistent values of Dice and specificity, mainly due to the existence of the quantification step, to generate a minimum of false positives.
Table 8 shows the results of the presented and the proposed method.
Table 8.
Comparison with related works
We can observe that although a faithful comparison is not possible, our method has a prominent place among the related works. Our metrics, when not superior, are comparable to other methods in the literature.
However, it is worth mentioning that methods for segmenting infections are very important in clinical practice to assist specialists. Thus, it is believed that our method is promising in the task of infection segmentation, reaching an average Dice of 77.1% and specificity of 99.76%.
Finally, we highlight that this is despite the comparable results concerning segmentation. Our method also presents a step of classification of infection in GGO and condensation areas and quantification of these findings to provide more subsidies to the specialist.
Advances and limitations
Infection segmentation is not a trivial task. Developing a method capable of addressing this problem requires a robust model that circumvents all adversities encountered by patients and procurement protocols. By achieving an effective method for this task, several advances are achieved, such as:
A fully automatic method is capable of segmenting infections and quantifying them to provide subsidies to specialist doctors. The use of a public database to build a lung segmentation model has proved to be very promising. Due to the lack of specialists’ time, it would be very difficult to find a database with both lung marking and infections. By creating a robust model on a diversified database, we were able to generalize to other databases.
For the lungs segmentation, a model based on U-Net [48] was proposed, which evaluated the results, showed to be quite promising, achieving better results than the U-Net itself.
By observing that the markings comprised a series of unnecessary information for the training of the network (pulmonary vessels), by excluding this from the mark, our method became even more robust. In this way, we managed to solve the problem of generating many false positives
All techniques proved to be fundamental for the creation of an infection segmentation model. From these results, it was possible to find a Dice of 77.1% with only 99.76% specificity.
As there is no classification of the types of infections, we propose to identify the condition of the infection tissue by analyzing the HU, classifying them as GGO, or condensed. This is important because the evolution of COVID19 changes the structure of the lesion, which may progress between GGO and condensation.
From the segmentation and classification of infections, it was possible to extract metrics, and such metrics show the degree to which each lung and the whole parenchyma is compromised. This in clinical practice can be essential, both to detect disease, diagnose, prevent, and even monitor whether a particular treatment is reducing the impairment.
Finally, all the steps made the proposed method very promising. For the segmentation of infections, it reached 77.1% Dice, 72.44% sensitivity, 99.76% specificity, 99.37% accuracy, and 86.11% AUC. As for quantification, the method showed a value of ρ < 0.05, which shows that the extracted metrics were faithful to the specialist’s marking.
Despite being a promising method with a series of advances, it has some limitations, such as:
As it is a national emergency, it has not yet been possible to find a database composed of numerous cases marked by specialists. Thus, building or obtaining a large database can be fundamental for creating 3D models of networks.
One of the main problems when it comes to deep learning is the number of parameters, being able to find these parameters automatically can provide better results for the method.
Another important point is the metrics, since the specialist uses them to interpret the degree of the disease, providing more measures than those already proposed can be important for medical interpretation.
Thus, we show that the proposed method is promising in clinical practice allied with specialist expertise. The use of a computer-supported method can assist all spheres of healthcare to detect disease, diagnosis, prevent, and even monitor whether a particular treatment.
Conclusion
This study presented a method for segmenting infections caused by COVID-19 and quantifying these findings. The method was created using two databases: one public (LSTSC database) and one private (HUPE database). Furthermore, the proposed method was composed of five stages: lung segmentation, segmentation and extraction of pulmonary vessels, infection segmentation, infection classification, and quantification of infections.
The proposed method proved to be promising, and all steps involved were crucial to achieving outstanding results. For infection segmentation, we achieved a Dice of 77.1% with 99.56% specificity. This indicates how accurate the proposed method was in finding infections by generating as few false positives as possible. As our approach also proposes measures for the infections found, having few false positives is important in order not to tend to quantify these infections.
In addition, when proposing an analysis of pulmonary impairment through volumetric quantification, the proposed method achieved results similar to that of the specialist, where no measure presented a value of ρ < 0.05 in the paired t-test.
Thus, it is believed that the proposed method may play a fundamental role in clinical practice as it can point out both infections and measures related to them to the specialists.
Finally, we believe that some improvements could be introduced to produce even better results. First, new architectures for the U-Net used could be tested or created, as the literature has several publications related to this. Second, a database with several marked cases could be acquired, which would allow 3D training for these patients. Finally, other metrics can be used to provide even more subsidies to specialists.
Acknowledgements
This research was carried out with the support of the institutions Applied Computing Group (NCA) from Federal University of Maranhão (UFMA), Federal Institutes of Maranhão (IFMA) and Amazonas (IFAM), Pedro Ernesto University Hospital (HUPE) from State University of Rio de Janeiro (UERJ), Fundação de Amparo à Pesquisa e ao Desenvolvimento Científico e Tecnológico do Maranhão (FAPEMA), and Tecgraf Institute for Technical and Scientific Software Development (Tecgraf/PUC-Rio).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
João O. B. Diniz, Email: joao.bandeira@ifma.edu.br
Darlan B. P. Quintanilha, Email: dquintanilha@nca.ufma.br
Antonino C. Santos Neto, Email: antoninocalisto@nca.ufma.br
Giovanni L. F. da Silva, Email: giovannilucca@nca.ufma.br
Jonnison L. Ferreira, Email: jonnison@nca.ufma.br
Stelmo M. B. Netto, Email: stelmo@nca.ufma.br
José D. L. Araújo, Email: josedenes@nca.ufma.br
Luana B. Da Cruz, Email: luana.b.cruz@nca.ufma.br
Thamila F. B. Silva, Email: thamila.fontenele@nca.ufma.br
Caio M. da S. Martins, Email: caiomanfredini@nca.ufma.br
Marcos M. Ferreira, Email: marcos.melo@nca.ufma.br
Venicius G. Rego, Email: venicius.gr@nca.ufma.br
José M. C. Boaro, Email: boaro@nca.ufma.br
Carolina L. S. Cipriano, Email: carol@nca.ufma.br
Aristófanes C. Silva, Email: ari@nca.ufma.br
Anselmo C. de Paiva, Email: paiva@nca.ufma.br
Geraldo Braz Junior, Email: geraldo@nca.ufma.br.
João D. S. de Almeida, Email: jdallyson@nca.ufma.br
Rodolfo A. Nunes, Email: rodolfoacatauassu@yahoo.com.br
Roberto Mogami, Email: pq242001@hotmail.com.
M. Gattass, Email: mgattass@inf.puc-rio.br
References
- 1.Agam G, Armato SG, Wu C. Vessel tree reconstruction in thoracic CT scans with application to nodule detection. IEEE Trans Med Imaging. 2005;24(4):486–499. doi: 10.1109/TMI.2005.844167. [DOI] [PubMed] [Google Scholar]
- 2.Armato SG, III, Sensakovic WF. Automated lung segmentation for thoracic ct: impact on computer-aided diagnosis1. Acad Radiol. 2004;11(9):1011–1021. doi: 10.1016/j.acra.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Baldi P, Sadowski PJ (2013) Understanding dropout. In: Advances in neural information processing systems, pp. 2814–2822
- 4.Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, et al. (2020) Chest ct findings in coronavirus disease-19 (covid-19): relationship to duration of infection. Radiology p 200463 [DOI] [PMC free article] [PubMed]
- 5.Boccia S, Ricciardi W, Ioannidis JP (2020) What other countries can learn from Italy during the covid-19 pandemic. JAMA Internal Med [DOI] [PubMed]
- 6.Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, Xing F, Liu J, Yip CCY, Poon RWS, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European conference on computer vision (ECCV), pp 801–818
- 8.Chollet F, et al. (2018) Keras: the python deep learning library. Astrophysics Source Code Library
- 9.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Cruz LB, Araujo JDL, Ferreira JL, Diniz JOB, Silva AC, de Almeida JDS, de Paiva AC, Gattass M (2020) Kidney segmentation from computed tomography images using deep neural network Computers in Biology and Medicine p 103906 [DOI] [PubMed]
- 11.[dataset] (2020) Covid-19 ct segmentation dataset. URL http://medicalsegmentation.com/covid19/
- 12.de Barros Machado DJ, Ianhez LE (2020) Covid-19 pneumonia in kidney transplant recipients-where we are? Transplant infectious disease p e13306 [DOI] [PMC free article] [PubMed]
- 13.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. doi: 10.2307/1932409. [DOI] [Google Scholar]
- 14.Diniz JOB, Diniz PHB, Valente TLA, Silva AC, de Paiva AC, Gattass M. Detection of mass regions in mammograms by bilateral analysis adapted to breast density using similarity indexes and convolutional neural networks. Comput Methods Prog Biomed. 2018;156:191–207. doi: 10.1016/j.cmpb.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Diniz PHB, Valente TLA, Diniz JOB, Silva AC, Gattass M, Ventura N, Muniz BC, Gasparetto EL. Detection of white matter lesion regions in mri using slic0 and convolutional neural network. Comput Methods Prog Biomed. 2018;167:49–63. doi: 10.1016/j.cmpb.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Diniz JOB, Diniz PHB, Valente TLA, Silva AC, Paiva AC. Spinal cord detection in planning ct for radiotherapy through adaptive template matching, imslic and convolutional neural networks. Comput Methods Prog Biomed. 2019;170:53–67. doi: 10.1016/j.cmpb.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Diniz JOB, Ferreira JL, Diniz PHB, Silva AC, de Paiva AC (2020) Esophagus segmentation from planning ct images using an atlas-based deep learning approach Computer Methods and Programs in Biomedicine p 105685 [DOI] [PubMed]
- 18.Diniz JOB, Ferrreira J, Diniz P, Serejo B, Ribeiro N, Santos O, Silva A, Paiva A (2020) Spinal cord segmentation as oar in planning ct for radiotherapy using histogram matching, template matching, and u-net. Revista de Sistemas e Computac¸˜ao-RSC 10(1)
- 19.Fan DP, Zhou T, Ji GP, Zhou Y, Chen G, Fu H, Shen J, Shao L (2020) Inf-net: automatic covid-19 lung infection segmentation from ct scans. arXiv preprint arXiv:200414133 [DOI] [PubMed]
- 20.Firmino M, Morais AH, Mendonca RM, Dantas MR, Hekis HR, Valentim R. Computer-aided detection system for lung cancer in computed tomography scans: review and future prospects. Biomed Eng Online. 2014;13(1):41. doi: 10.1186/1475-925X-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao T (2020) Chest x-ray image analysis and classification for covid-19 pneumonia detection using deep cnn. medRxiv
- 22.Godoy D (2019) Understanding binary cross-entropy/log loss: a visual explanation
- 23.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778
- 24.He W, Chen L, Chen L, Yuan G, Fang Y, Chen W, Wu D, Liang B, Lu X, Ma Y, et al. (2020) Covid-19 in persons with haematological cancers. Leukemia pp 1–9 [DOI] [PMC free article] [PubMed]
- 25.Hofmanninger J, Prayer F, Pan J, Rohrich S, Prosch H, Langs G (2020) Automatic lung segmentation in routine imaging is a data diversity problem, not a methodology problem. arXiv preprint arXiv:200111767 [DOI] [PMC free article] [PubMed]
- 26.Huttenlocher DP, Klanderman GA, Rucklidge WJ. Comparing images using the hausdorff distance. IEEE Trans Pattern Anal Mach Intell. 1993;15(9):850–863. doi: 10.1109/34.232073. [DOI] [Google Scholar]
- 27.Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:150203167
- 28.Jaccard P. The distribution of the flora in the alpine zone. 1. New Phytol. 1912;11(2):37–50. doi: 10.1111/j.1469-8137.1912.tb05611.x. [DOI] [Google Scholar]
- 29.Johnson H, McCormick M, Ibañez L (2009) Insight segmentation and registration toolkit (itk)
- 30.Johnson HJ, McCormick MM, Ibanez L (2015) The ITK software guide book 2: design and functionality-volume 2. Kitware, Inc.
- 31.Kolărík M, Burget R, Uher V, Ríha K, Dutta MK. Optimizedˇ high resolution 3d dense-u-net network for brain and spine segmentation. Appl Sci. 2019;9(3):404. doi: 10.3390/app9030404. [DOI] [Google Scholar]
- 32.Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp. 1097–1105
- 33.Lau MM, Lim KH (2018) Review of adaptive activation function in deep neural network. In: 2018 IEEE-EMBS conference on biomedical engineering and sciences (IECBES), IEEE, pp 686–690
- 34.Lehmann G. Label object representation and manipulation with itk. Insight J. 2007;8:1–31. [Google Scholar]
- 35.Liu F, Zhang Q, Huang C, Shi C, Wang L, Shi N, Fang C, Shan F, Mei X, Shi J, Song F, Yang Z, Ding Z, Su X, Lu H, Zhu T, Zhang Z, Shi L, Shi Y. Ct quantification of pneumonia lesions in early days predicts progression to severe illness in a cohort of covid-19 patients. Theranostics. 2020;10(12):5613–5622. doi: 10.7150/thno.45985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maas AL, Hannun AY, Ng AY (2013) Rectifier nonlinearities improve neural network acoustic models. In: proc. icml, vol 30, p 3
- 37.Mangal A, Kalia S, Rajgopal H, Rangarajan K, Namboodiri V, Banerjee S, Arora C (2020) Covidaid: Covid-19 detection using chest x-ray. arXiv preprint arXiv:200409803
- 38.Mansoor A, Bagci U, Xu Z, Foster B, Olivier KN, Elinoff JM, Suffredini AF, Udupa JK, Mollura DJ. A generic approach to pathological lung segmentation. IEEE Trans Med Imaging. 2014;33(12):2293–2310. doi: 10.1109/TMI.2014.2337057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mittal A, Hooda R, Sofat S. Lung field segmentation in chest radiographs: a historical review, current status, and expectations from deep learning. IET Image Process. 2017;11(11):937–952. doi: 10.1049/iet-ipr.2016.0526. [DOI] [Google Scholar]
- 40.Ng MY, Lee EY, Yang J, Yang F, Li X, Wang H, Lui MM, CSY L, Leung B, Khong PL, et al. Imaging profile of the covid-19 infection: radiologic findings and literature review. Radiology: Cardiothoracic Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Organization WH, et al. (2020) Coronavirus disease (covid-19): situation report - 142. Tech. rep., World Health Organization
- 42.Organization WH, et al. (2020) Coronavirus disease (covid-19) technical guidance: the unity studies: early investigations protocols. Tech. rep., World Health Organization
- 43.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of covid-19 cases using deep neural networks with x-ray images. Computers in biology and medicine p 103792 [DOI] [PMC free article] [PubMed]
- 44.Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E (2020) Covid19, sars and mers: are they closely related? Clinical Microbiology and Infection [DOI] [PMC free article] [PubMed]
- 45.Provost F, Domingos P. Tree induction for probability-based ranking. Mach Learn. 2003;52(3):199–215. doi: 10.1023/A:1024099825458. [DOI] [Google Scholar]
- 46.Pu J, Roos J, Chin AY, Napel S, Rubin GD, Paik DS. Adaptive border marching algorithm: automatic lung segmentation on chest ct images. Comput Med Imaging Graph. 2008;32(6):452–462. doi: 10.1016/j.compmedimag.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu Y, Liu Y, Xu J (2020) Miniseg: an extremely minimum network for efficient covid-19 segmentation. arXiv preprint arXiv:200409750 [DOI] [PubMed]
- 48.Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention, Springer, pp. 234–241
- 49.Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (covid-19) outbreak. Journal of autoimmunity p 102433 [DOI] [PMC free article] [PubMed]
- 50.Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A (2020) Coronavirus disease 2019 (covid-19): a systematic review of imaging findings in 919 patients. American Journal of Roentgenology pp 1–7 [DOI] [PubMed]
- 51.Samet R, Yildirim Z (2016) A new methodology for blood vessel segmentation on lung CT images. In: Proceedings – NICOGRAPH International 2016, NicoInt 2016, Institute of Electrical and Electronics Engineers Inc., pp 1–7, DOI 10.1109/NicoInt.2016.1
- 52.Sato Y, Nakajima S, Shiraga N, Atsumi H, Yoshida S, Koller T, Gerig G, Kikinis R. Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal. 1998;2(2):143–168. doi: 10.1016/S1361-8415(98)80009-1. [DOI] [PubMed] [Google Scholar]
- 53.Shen C, Yu N, Cai S, Zhou J, Sheng J, Liu K, Zhou H, Guo Y, Niu G (2020) Quantitative computed tomography analysis for stratifying the severity of coronavirus disease 2019. Journal of Pharmaceutical Analysis [DOI] [PMC free article] [PubMed]
- 54.Shen KL, Yang YH, Jiang RM, Wang TY, Zhao DC, Jiang Y, Lu XX, Jin RM, Zheng YJ, Xu BP, et al. (2020) Updated diagnosis, treatment and prevention of covid-19 in children: experts’ consensus statement (condensed version of the second edition). World Journal of Pediatrics pp 1–8 [DOI] [PMC free article] [PubMed]
- 55.Sousa JA, Paiva A, Silva A, Almeida JD, Braz Junior G, Diniz JO, Figueredo WK, Gattass M. Automatic segmentation of retinal layers in oct images with intermediate age-related macular degeneration using u-net and dexined. PLoS One. 2021;16(5):e0251591. doi: 10.1371/journal.pone.0251591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Souza JC, Diniz JOB, Ferreira JL, da Silva GLF, Silva AC, de Paiva AC. An automatic method for lung segmentation and reconstruction in chest x-ray using deep neural networks. Comput Methods Prog Biomed. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, Springer, pp 240–248 [DOI] [PMC free article] [PubMed]
- 58.Taha AA, Hanbury A. Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC Med Imaging. 2015;15(1):1–28. doi: 10.1186/s12880-015-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan W, Zhao X, Ma X, Wang W, Niu P, Xu W, Gao GF, Wu G A novel coronavirus genome identified in a cluster of pneumonia cases—wuhan, china 2019–2020. China CDC Weekly 2(4):61–62 [PMC free article] [PubMed]
- 60.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. Scipy 1.0: fundamental algorithms for scientific computing in python. Nat Methods. 2020;17(3):261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Lin ZQ, Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (covid-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. Jama. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 63.Wu YH, Gao SH, Mei J, Xu J, Fan DP, Zhao CW, Cheng MM (2020) Jcs: An explainable covid-19 diagnosis system by joint classification and segmentation. arXiv preprint arXiv:200407054 [DOI] [PubMed]
- 64.Yan Q, Wang B, Gong D, Luo C, Zhao W, Shen J, Shi Q, Jin S, Zhang L, You Z (2020) Covid-19 chest ct image segmentation–a deep convolutional neural network solution. arXiv preprint arXiv:200410987
- 65.Yu F, Koltun V, Funkhouser T (2017) Dilated residual networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 472–480
- 66.Yu Q, Wang Y, Huang S, Liu S, Zhou Z, Zhang S, Zhao Z, Yu Y, Yang Y, Ju S. Multicenter cohort study demonstrates more consolidation in upper lungs on initial ct increases the risk of adverse clinical outcome in covid-19 patients. Theranostics. 2020;10(12):5641–5648. doi: 10.7150/thno.46465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng C, Zhu Z, Liu G, Hu W, Wang X, Yang C, Wang H, He D, Tan J. Randomized, double-blind, placebo-controlled trial of oral enalapril in patients with neurally mediated syncope. Am Heart J. 1998;136(5):852–858. doi: 10.1016/S0002-8703(98)70131-0. [DOI] [PubMed] [Google Scholar]
- 68.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Liu W, Wang X (2020) Deep learning-based detection for covid-19 from chest ct using weak label. medRxiv
- 69.Zhou S, Wang Y, Zhu T, Xia L (2020) Ct features of coronavirus disease 2019 (covid-19) pneumonia in 62 patients in Wuhan, China. Am J Roentgenology pp 1–8 [DOI] [PubMed]
- 70.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team China novel coronavirus investigating and research team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]