Abstract
Premelanosome protein (PMEL) is crucial for the formation of melanosomal fibrils through the transition from stage I to stage II melanosomes. It was used as a target antigen in some adoptive T-cell therapy of melanoma. The correlation of PMEL to prognosis and immune cell infiltration level are unknown in melanoma. The PMEL expression was evaluated via Tumor Immune Estimation Resource, Oncomine and Gene Expression Profiling Interactive Analysis (GEPIA). We also evaluate the influence of PMEL on overall survival via GEPIA, PrognoScan, and immunohistochemistry in human tissue microarray. The correlation between PMEL expression level and immune cell or gene markers of immune infiltration level was explored on Tumor Immune Estimation Resource and GEPIA. PMEL expression was significantly higher in skin cutaneous melanoma (SKCM) and SKCM-metastasis in comparison with the other cancers. In SKCM, PMEL expression in high levels was associated with poor overall survival. In both SKCM and SKCM-metastasis patients, PMEL expression is negatively correlated with the infiltration cells of CD8+ T cells, macrophages, and neutrophils. Programmed cell-death protein 1 just showed response rates ranging from 20% to 40% in patients with melanoma, so it is critical to discover a new therapeutic target. PMEL is negatively associated with immune cell infiltration and can be as a negative prognosis marker or new immunotherapy target in SKCM and SKCM-metastasis.
Key Words: PMEL, immune infiltration, prognosis, SKCM
Skin cutaneous melanoma (SKCM) incidence is increasing recent years, with an overall rate of 33% for men and 23% women.1 The programmed cell-death protein 1 (PD-1) immune checkpoints of immunotherapy is the mainly treatment measure for advanced-stage melanoma recently, which have dramatically improved patient outcomes, with the median overall survival (OS) of patients increasing from ~9 months to at least 2 years for those with BRAF-V600-mutant disease.2 The prognosis of early and advanced melanoma are significantly different, such as the 5-year survival rate for early-stage melanoma can reach 99%,3 but large phase I clinical trials of PD-1 also showed response rates ranging from 20% to 40% in patients with melanoma.2 So when and how to apply immunotherapy is crucial for increasing the OS rate. It is urgent to find out a prognostic marker and new immune infiltration related gene to improve the OS for SKCM patients.
Premelanosome protein (PMEL), also recognized as PMEL17, glycoprotein 100 (gp100), silver homologue, Me20, etc. PMEL, is a nonmutated “self” antigen expressed by melanocytes, pigmented retinal cells, and most melanoma cells, plays an essential role in the structural organization of premelanosomes. PMEL is a melanocyte-specific type I membrane protein mainly expressed in normal skin and eye pigment cells,4 and can participate in the formation of melanosomal fibrils through the transition from stage I to stage II melanosomes.5 Compared with normal melanocytes, PMEL is over-expressed at all stages of melanoma progression,6,7 and is a specific marker for melanoma with low-expression in other tissues. Now, PMEL has been focused on a target antigen in adoptive T-cell therapy and proven to be its safety and effectiveness.8 So, PMEL, has represented effective target antigens for the use in adoptive T-cell transfer.9 The latest research demonstrates that the PMEL domain that forms the amyloid core of the melanoma body is essential for the formation of melanoma,10,11 and we can inhibits melanoma cell epithelial to mesenchymal transition, proliferation, migration, invasion, and promotes apoptosis by targeting PMEL through down-regulation of the Wnt signaling pathway.12,13 These finding suggest the PMEL plays a significant role in melanoma formation, invasion, and metastasis of process.
Here, we systematically analyzed PMEL expression and correlation with the prognosis of cancers in the databases including Oncomine, Gene Expression Profiling Interactive Analysis (GEPIA), PrognoScan. Furthermore, we explored the correlation between PMEL expression and immune infiltration immune cells including related markers in the tumor microenvironment via Tumor Immune Estimation Resource (TIMER) and GEPIA. The data in this report reveal that the significant correlation between PMEL expression and OS of melanoma as well as the degree of immune infiltration.
METHODS
Oncomine Database Analysis
The mRNA expression of PMEL in multiple cancers was analyzed via the Oncomine database (www.oncomine.org/resource/main.html).14 The threshold settings were as follows: gene of PMEL, P-value of 1E−4, fold change of 2.0, gene rank of top 10%, and data type of mRNA, respectively.
PrognoScan Database Analysis
The relationship between PMEL expression and OS in multiple types of cancers was analyzed by the PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html), which is a new database for meta-analysis of the prognostic value of genes.15 The Figure 3E and F with cutoff value 75% and 50%, respectively. The OS curves of cancers significantly related to PMEL expression were as follows with P-value<0.05.
FIGURE 3.
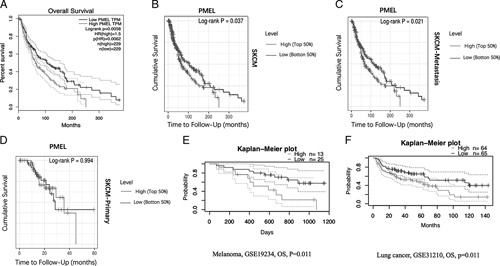
The correlation between PMEL over-expression gene and OS of SKCM in GEPIA and TIMER database (A−D). A, High PMEL expression level corresponded with poor OS prognosis of SKCM in GEPIA database (HR=1.5, log-rank P=0.0058, cutoff value, 50%). B−D, High PMEL expression level correlated with poor OS prognosis of SKCM, SKCM-metastasis and SKCM-primary in TIMER (cutoff value, 50%). The Kaplan-Meier overall survival curves comparing the high and low-expression levels of PMEL in various types of cancer in PrognoScan (E, F). E and F, High PMEL expression level corresponded with poor OS prognosis in melanoma and lung cancer, respectively (P<0.05; cutoff value, 75% and 50%, respectively). GEPIA indicates Gene Expression Profiling Interactive Analysis; HR, hazard ratio; OS, overall survival; PMEL, premelanosome protein; SKCM, skin cutaneous melanoma; TCGA, The Cancer Genome Atlas; TIMER, Tumor Immune Estimation Resource.
GEPIA
The GEPIA database (http://gepia.cancer-pku.cn) is an interactive web which can provides key interactive and customizable functions via the analysis of tumors and normal samples based on TCGA and Genotype-Tissue Expression (GTEx) database.16 And GEPIA was also applied to generate the OS and disease-free survival curves, based on the top-10 over-expressed mRNA levels and log-rank test with group cutoff of median in melanoma.
Using GEPIA to analyze the different expression levels of PMEL between the melanoma samples and normal samples in TCGA and GTEx. The threshold settings were as follows: Expression DIY (do it yourself) of boxplot, cancer name of SKCM, match TCGA normal and GTEx data, Jitter size of 0.4, with |Log2FC| cutoff of 50% and P-value cutoff being 1 and 0.01, respectively.
TIMER Database Analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a web-accessible resource,17 which was used to analysis the correlation of PMEL expression profiles with the abundance of immune cell infiltration including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, via gene modules in melanoma. We also further study the interconnections between PMEL expression and immune related markers of tumor-infiltrated immune cells through correlation modules in TIMER. All of the immune infiltrated markers have been reported in previous reports.18,19 And we also analyzed PMEL expression in different types of cancer and compared their different expression between SKCM-tumor and SKCM-metastasis. The left-most panel shows the levels of PMEL expression against the degree of tumor purity.20 The levels of PMEL expression with 50% cutoff value were demonstrated by log2 RSEM.
Immunohistochemistry (IHC) Analysis of Human Tissue Microarray (TMA) and Scoring
The expression of PMEL in the human TMA was determined by IHC assay according to the manufacturer’s instructions. Primary antibodies were diluted as follows: a rabbit monoclonal (EPR4864) to melanoma gp100, and rabbit monoclonal (SP86; Abcam, Hong Kong; 1:100 dilution, cytoplasmic and nuclear staining). The SKCM TMA (Liao Ding Biotechnology Co. Ltd, Shanghai, China) contains 50 cases/100 cores with histopathologic data, including age, sex, tumor location, Clark, Breslow, tumor size, ulcer, nerve invasion, vascular tumor thrombus, TNM grade, and American Joint Committee on Cancer (AJCC) stage.
Briefly, sections 4-μm thick were placed on slides coated with 3-aminopropyltriethoxysilane. Paraf_x0002_fin sections were deparaffinized in xylene and rehydrated through decreasing concentrations of ethanol (100%, 95%, and 85%, 5 min each). Antigens were unmasked by micro_x0002_wave irradiation for 3 min in pH 6.0 citric buffer and cooled at room temperature for 60 minutes. Endogenous peroxidase activity was blocked by incubation of the slides in 3% H2O2/phosphate-buffered saline, and nonspecific binding sites were blocked with goat serum. Melanoma gp100 staining was observed in the cytoplasm and nucleus of melanoma cells. Staining of melanoma gp100 was scored according to its extent and intensity (extent×intensity), and cytoplasm (extent×intensity)+nucleus (extent×intensity), similar to the methods described previously.21 And high PMEL expression (>0.1875) or low PMEL expression (≤0.1875) was determined by receiver operating characteristic curve.
Statistical Analysis
Survival curves were depicted using the Kaplan-Meier method and compared using the log-rank test. For calculating the best cutoff points for OS, the X-tile statistical package (version 3.5.0; Yale University, New Haven, CT) was used. X-tile plot illustrates the presence of substantial tumor subpopulations and shows the robustness of the relationship between a biomarker and outcome by construction of a 2-dimensional projection of every possible subpopulation.
RESULTS
The mRNA Expression of PMEL in Multiple Cancer Types
To explore the different expression of PMEL in tumor and normal samples of multiple cancer types, the TIMER database was used for analysis. PMEL expression in bladder urothelial carcinoma, head and neck cancer HPV positive, lung adenocarcinoma, lung squamous cell carcinoma, prostate adenocarcinoma, stomach adenocarcinoma were higher compared to adjacent normal tissues (P<0.0001) (Fig. 1). To better understand the mRNA expression of PMEL between tumor and adjacent normal tissues in various human tumors, Oncomine database was adopted. Compared with the normal tissues, higher expression was observed in breast cancer, lung cancer, and melanoma (Fig. 2C). Furthermore, in the TIMER database, we can demonstrate that PMEL expression was significantly higher in SKCM and SKCM-metastasis than other cancers, and much higher in SKCM than in SKCM-metastasis (P<0.01) (Fig. 1). To further explore the expression difference of PMEL between tumors and normal tissues in melanoma, we analyzed the data in TCGA and GTEx based on the GEPIA database and found that the expression level in SKCM (461 samples) was significantly higher than that in normal tissues (558 samples) with statistics difference (P<0.05) (Figs. 2A, B).
FIGURE 1.
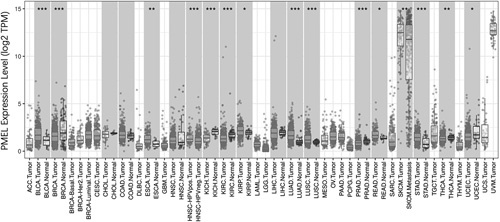
PMEL expression levels in multiple types of cancers. Compare the expression data of PMEL from TCGA using TIMER (*P<0.05, **P<0.01, ***P<0.001). ACC indicates adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical and endocervical cancer; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck cancer; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PMEL, premelanosome protein; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; TCGA, The Cancer Genome Atlas; TIMER, Tumor Immune Estimation Resource; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
FIGURE 2.
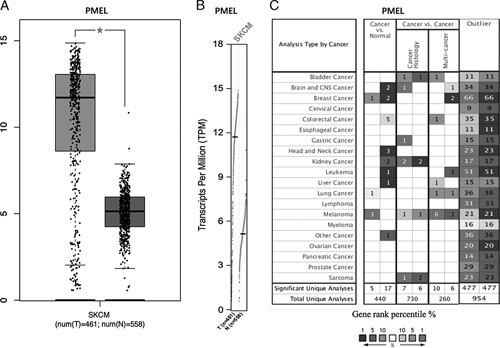
A and B, Human PMEL expression levels in melanoma from TCGA database were determined by GEPIA. C, The expression levels of PMEL in data sets of different cancers between cancer and normal tissues in the Oncomine database. *P<0.01. CNS indicates central nervous system; GEPIA, Gene Expression Profiling Interactive Analysis; PMEL, premelanosome protein; SKCM, skin cutaneous melanoma; TCGA, The Cancer Genome Atlas.
Over-expressed PMEL Predicts the Poor OS of SKCM-metastasis Patients
In the GEPIA database, we found that high PMEL expression was correlated with poor OS (Fig. 3A). Furthermore, In the Cox analysis of PMEL in TIMER database, SKCM-tumor (364 samples, 178 dying) consists of SKCM-primary (95 samples, 25 dying) and SKCM-metastasis patients (269 samples, 153 dying), we found that higher PMEL expressions were also dramatically consistent with the poor OS in SKCM and SKCM-metastasis patients (P<0.05) (Figs. 3B, C). Comparing the high and low-expression levels of PMEL in various types of cancer with Kaplan-Meier OS curves in PrognoScan, we found that high PMEL expression level also corresponded with poor OS prognosis in melanoma and lung cancer (P<0.05) (Figs. 3E, F). So, we could conclude that over-expressed PMEL predicts the poor OS of SKCM-metastasis patients. These results lead that PMEL can be recognized as a prognostic marker for SKCM patients.
PMEL Expression Levels Correlate With the Clinicopathologic Characteristics of SKCM Patients
To further demonstrate the clinical significance of PMEL expression for predicting prognosis in SKCM patients, we tested the expression levels of PEML in a human melanoma TMA by IHC, and evaluated the correlation between PMEL expression and the clinicopathologic characteristics of the SKCM patients. The expression levels of PMEL were graded based on the sum of cytoplasm (extent×intensity) and nucleus (extent×intensity) with score ≥0.1875 considered high PMEL expression. We can demonstrate that PMEL immunoreactivity is mainly in the cytoplasm and nucleus of melanoma cells from the IHC results. Of the 50 TMA cases of SKCM, 45 (90%) of the tissues express detectable PMEL expression.
As is shown in the Table 1, following evaluation of the clinical history of the human SKCM specimens, no significant correlations are found between the expression of PMEL and patient sex, age, tumor location, Breslow, tumor diameter, nerve invasion or vascular tumor thrombus. In addition, we find that PMEL expression is significantly associated with the Clark grade, ulcer and AJCC stage of the SKCM (both P<0.05) (Table 1). As shown, PMEL expression is significantly higher in the higher Clark grade (≥III) compared with those in the lower Clark grade (<III). Moreover, PMEL expression is significantly higher in the higher AJCC stage (III and IV), compared to the lower AJCC stage (I and II).
TABLE 1.
The Analysis of Correlation Between the Expression of PMEL and Clinicopathologic Characteristics of SKCM Based on TMA
| PMEL Expression [n (%)] | ||||
|---|---|---|---|---|
| Clinicopathologic Characteristics | Numbers | Low | High | P |
| Sex | ||||
| Male | 31 | 23 (46.0) | 8 (16.0) | 0.230* |
| Female | 19 | 11 (22.0) | 8 (16.0) | |
| Age (y) | ||||
| ≤60 | 30 | 21 (42.0) | 9 (18.0) | 0.710* |
| >60 | 20 | 13 (26.0) | 7 (14.0) | |
| Tumor location | ||||
| Limbs | 27 | 19 (38.0) | 8 (16.0) | 0.364* |
| Trunk | 5 | 2 (4.0) | 3 (6.0) | |
| Head and neck | 18 | 13 (26.0) | 5 (10.0) | |
| Clark grade | ||||
| I−II | 18 | 16 (32.0) | 2 (4.0) | 0.026 † |
| III−V | 32 | 18 (36.0) | 14 (28.0) | |
| Breslow (mm) | ||||
| ≤11 | 24 | 17 (34.0) | 7 (14.0) | 0.680* |
| >11 | 26 | 17 (34.0) | 9 (18.0) | |
| Tumor diameter (cm) | ||||
| ≤2 | 28 | 19 (38.0) | 9 (18.0) | 0.981* |
| >2 | 22 | 15 (30.0) | 7 (14.0) | |
| Ulcer | ||||
| Yes | 20 | 10 (20.0) | 10 (20.0) | 0.026 * |
| No | 30 | 24 (48.0) | 6 (12.0) | |
| Nerve invasion | ||||
| Yes | 20 | 14 (28.0) | 6 (12.0) | 0.804* |
| No | 30 | 20 (40.0) | 10 (20.0) | |
| Vascular tumor thrombus | ||||
| Yes | 26 | 17 (34.0) | 9 (18.0) | 0.680* |
| No | 24 | 17 (34.0) | 7 (14.0) | |
| AJCC stage (TNM) | ||||
| I−II | 29 | 23 (46.0) | 6 (12.0) | 0.044 * |
| III−IV | 21 | 11 (22.0) | 10 (20.0) | |
Bold values indicate statistical significance (P<0.05).
Pearson χ2 test.
Fisher exact test.
AJCC indicates American Joint Committee on Cancer; PMEL, premelanosome protein; SKCM, skin cutaneous melanoma; TMA, tissue microarray.
To analyze the correlation between the PMEL expression levels and the clinical prognosis of the SKCM. The patients’ follow-up and clinical outcomes of the SKCM TMA were collected. At the time of last follow-up, among 50 patients in the TMA, 28 had died, the median OS was 50 months (range, 9−59 mo).
Significantly, Kaplan-Meier curves demonstrated that the high PMEL expression was significantly associated with a poor OS (P=0.016) (Fig. 4A), and the 3-year OS rate of high and low PMEL expression group was about 70.6% and 50%, respectively. Furthermore, PMEL expression levels in samples from nonsurvivors are significantly higher than those from survivors (Fig. 4B).
FIGURE 4.
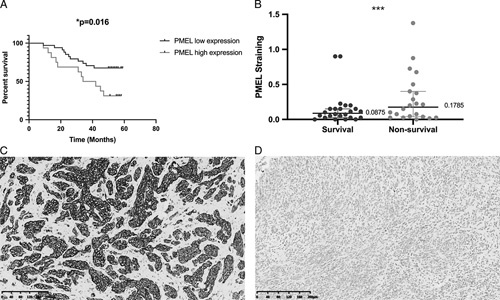
PMEL expression levels associate with clinicopathologic characteristics of SKCM patients. PMEL levels in SKCM tissue microarray were evaluated by immunohistochemistry, and the relationship between PMEL expressions and overall survival was analyzed. A, Kaplan-Meier survival curve of SKCM patients with high PMEL expression (>0.1875) or low PMEL expression (≤0.1875). B, Distribution of PMEL staining scores in the SKCM tissue samples from survival and nonsurvival patients. ***P<0.001. C and D, Representative images of high and low PMEL expression level based on different immunohistochemical staining intensities. PMEL indicates premelanosome protein; SKCM, skin cutaneous melanoma.
PMEL Expression is Negatively Correlated With Immune Infiltration Cells in SKCM and SKCM-metastasis
It has been confirmed that tumor-infiltrating lymphocyte grade is an independent predictor of survival and sentinel lymph node status in patients with melanoma,22,23 which was also demonstrated in other cancers.22,24–26 We assessed the correlations of PMEL expression with immune infiltration levels between SKCM and SKCM-metastasis from TIMER. It was shown that the PMEL expression has significant negative correlations with infiltration cells of CD8+ T cell (r=−0.184, P=1.03e−04), CD4+ T cell (r=−0.138, P=3.59e−03), macrophage (r=−0.307, P=2.47e−11), neutrophil (r=−0.349, P=2.22e−14) and dendritic cell (r=−0.129, P=6.56e−03) in SKCM (Fig. 5A), and CD8+ T cell (r=−0.157, P=3.95e−03), macrophage (r=−0.279, P=1.09e−07) and neutrophil (r=−0.315, P=1.59e−09) in SKCM-metastasis (Fig. 5B). So the result shows that the PMEL expression level have a significant negative correlation with the immune infiltration level, especially in the CD8+ T cell, macrophage, and neutrophil. These findings strongly suggest that PMEL affects patient survival via interacting with immune infiltration in SKCM and SKCM-metastasis.
FIGURE 5.
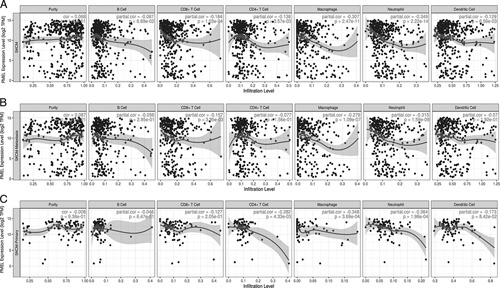
The analysis of correlation between PMEL expression level and immune infiltrates in SKCM (A), SKCM-metastasis (B) and SKCM-primary (C). A, The negative correlations between PMEL expression and immune infiltration of CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell in SKCM. B, The negative correlations between PMEL expression and immune infiltration of CD8+ T cell, macrophage, and neutrophil in SKCM-metastasis. C, The negative correlations between PMEL/SILV expression and immune infiltration of CD4+ T cell, macrophage, and neutrophil in SKCM-primary. PMEL indicates premelanosome protein; SILV, silver homologue; SKCM, skin cutaneous melanoma.
Furthermore, we further demonstrated that the correlation between immune infiltration and survival rate of SKCM and SKCM-metastasis patients in TIMER database. After the adjustment of clinical factors (age, sex, race, stage, tumor purity), we concluded that lower infiltrations of B cell, CD8+ T cell, neutrophil, and dendritic cell were significantly associated with a poor cumulative survival rate in both SKCM and SKCM-metastasis patients (P<0.05) (Figs. 6A, B). So, how can we do to promote the immune infiltration via influencing the PMEL?
FIGURE 6.
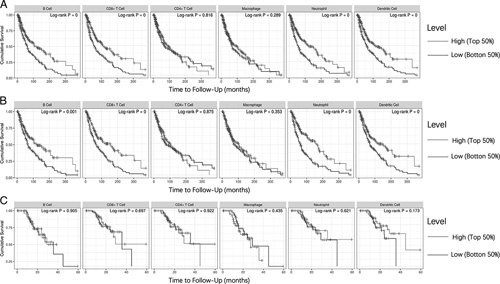
The correlation between immune infiltrates and overall survival rate of SKCM (A), SKCM-metastasis (B) and SKCM-primary (C) patients in TIMER. The indicates were divided into high and low groups with cutoff 50%. SKCM indicates skin cutaneous melanoma; TIMER, Tumor Immune Estimation Resource.
The Analysis of Correlation Between PMEL Expression and Immune Markers
To further illuminate the correlation between PMEL expression and immune infiltration cells, we study the relation between the PMEL expression and immune markers of relative immune infiltration cells of SKCM and SKCM-metastasis in TIMER and GEPIA database. The relative immune infiltration cells are mainly including CD8+ T cell, monocyte, tumor-associated macrophage (TAM), M1 macrophage, M2 macrophage, neutrophils, dendritic cell, Th1, Th2, Tfh, Th17, Treg, exhausted T cell (Table 2). After correlation analysis adjusted by purity, we demonstrated that the expression of PMEL is significantly negative correlated with the amount of immune infiltration relative immune markers with 36 of 48 immune cell markers in SKCM and 25 of 48 immune cell markers in SKCM-metastasis (P<0.05) (Table 2). The difference of correlation between the PMEL expression and immune markers of relative immune infiltration cells demonstrates that PMEL expression may play a negative role in the SKCM.
TABLE 2.
Correlation Analysis Between PMEL/SILV and Relate Gene Markers of Immune Cells in TIMER Database
| SKCM | SKCM-metastasis | ||||
|---|---|---|---|---|---|
| Description | Gene Markers | Correlation | P | Correlation | P |
| CD8+ T cell | CD8A | −0.139 | ** | −0.106 | 0.0412 |
| CD8B | −0.116 | 0.012 | −0.078 | 0.134 | |
| Monocyte | CD86 | −0.283 | *** | −0.249 | *** |
| CD115 (CSF1R) | −0.335 | *** | −0.307 | *** | |
| CD14 | −0.302 | *** | −0.272 | *** | |
| TAM | CCL2 | −0.305 | *** | −0.287 | *** |
| CD68 | 0.085 | 0.072 | 0.088 | 0.093 | |
| IL10 | −0.238 | *** | −0.205 | *** | |
| M1 macrophage | NOS2 | 0.045 | 0.328 | 0.075 | 0.154 |
| IRF5 | −0.076 | 0.102 | −0.033 | 0.526 | |
| PTGS2 | −0.281 | *** | −0.308 | *** | |
| M2 macrophage | CD163 | −0.297 | *** | −0.269 | *** |
| VSIG4 | −0.277 | *** | −0.246 | *** | |
| MS4A4A | −0.297 | *** | −0.267 | *** | |
| Neutrophils | CEACAM8 | −0.075 | 0.105 | −0.09 | 0.084 |
| CD11b (ITGAM) | −0.244 | *** | −0.208 | *** | |
| CCR7 | −0.035 | 0.443 | −0.033 | 0.534 | |
| Dendritic cell | HLA-DPB1 | −0.172 | ** | −0.139 | * |
| HLA-DQB1 | −0.126 | * | −0.092 | 0.078 | |
| HLA-DRA | −0.21 | *** | −0.177 | ** | |
| HLA-DPA1 | −0.143 | * | −0.099 | 0.057 | |
| BDCA-1 (CD1C) | −0.219 | *** | −0.205 | *** | |
| BDCA-4 (NRP-1) | −0.522 | *** | −0.539 | *** | |
| CD11C (ITGAX) | −0.075 | 0.104 | −0.024 | 0.639 | |
| Th1 | T-bet (TBX21) | −0.144 | * | −0.117 | 0.025 |
| STAT4 | −0.34 | *** | −0.298 | *** | |
| STAT1 | −0.105 | 0.023 | −0.065 | 0.215 | |
| IFN-γ (IFNG) | −0.135 | * | −0.11 | 0.035 | |
| TNF-α (TNF) | −0.107 | 0.020 | −0.115 | 0.027 | |
| Th2 | GATA3 | −0.194 | *** | −0.212 | *** |
| STAT6 | 0.192 | *** | 0.237 | *** | |
| STAT5A | 0.243 | *** | 0.305 | *** | |
| IL13 | −0.174 | ** | −0.184 | ** | |
| Tfh | BCL6 | −0.39 | *** | −0.365 | *** |
| IL21 | −0.133 | * | −0.1 | 0.055 | |
| Th17 | STAT3 | −0.16 | ** | −0.146 | * |
| IL17A | 0.078 | 0.091 | 0.036 | 0.497 | |
| Treg | FOXP3 | −0.101 | 0.028 | −0.088 | 0.094 |
| CCR8 | −0.219 | *** | −0.186 | ** | |
| STAT5B | −0.091 | 0.048 | −0.053 | 0.311 | |
| TGFβ (TGFB1) | −0.303 | *** | −0.31 | *** | |
| T cell exhaustion | PD-1 (SNCA) | 0.539 | *** | 0.582 | *** |
| CTLA4 | −0.061 | 0.184 | −0.079 | 0.128 | |
| LAG3 | −0.13 | * | 0.098 | 0.060 | |
| TIM-3 (HAVCR2) | −0.226 | *** | −0.191 | ** | |
| GZMB | −0.118 | 0.010 | −0.111 | 0.038 | |
None indicates without adjustment; Purity, adjusted by tumor purity; PMEL, premelanosome protein; SILV, silver homologue; SKCM, skin cutaneous melanoma; SKCM-metastasis, the metastasis of skin cutaneous melanoma; TAMs, tumor-associated macrophages; Tfh, follicular helper T cell; Th1, T helper cell 1; Th2, T helper cell 2; Treg, regulatory T cell; TIMER, Tumor Immune Estimation Resource.
P<0.01.
P<0.001.
P<0.0001.
We can see a negative correlation between PMEL expression and monocyte, macrophage and TAM of SKCM and SKCM-metastasis in TIMER database (Table 2). And we also find a low immune infiltration level of macrophage was associated with high PMEL expression level (Figs. 4B, 5A). But we did not find macrophage associated with a poor survival rate of SKCM, SKCM-primary and SKCM-metastasis patients. So, those results can tell us that PMEL influences more function of macrophage than quantity or PMEL on macrophage infiltration might be coordinated by some signals which have less effect on the OS.
Moreover, high PMEL expression in SKCM also relates to low infiltration level of CD8+ T cell (CD8A, CD8B), neutrophils (CD11b), dendritic cell (HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DPA1, BDCA-1, BDCA-4), Th1 (T-bet, STAT4, IFN-γ), Th2 (GATA3, STAT6, STAT5A, IL13), Tfh (BCL6, IL21), Th17 (STAT3), Treg (CCR8, STAT5B, TGFβ) and exhausted T-cell markers (LAG3, TIM-3, GZMB), and the PD-1 is the only 1 positive immune marker correlated to PMEL in SKCM and SKCM-metastasis (P<0.05) (Table 2). In the SKCM-metastasis, compared with the SKCM, the correlation between the PMEL and immune marker including CD8+ T cell (CD8A, CD8B), dendritic cell (HLA-DPB1, HLA-DQB1, HLA-DPA1), Th1 (T-bet, IFN-γ), Tfh (IL21), Treg (STAT5B), and exhausted T-cell markers (LAG3, GZMB) is not confirmed again (P>0.05) (Table 2). Compared with the difference, we guess that PMEL may influence the metastasis of SKCM via interacting the partical function of CD8+ T cell, dendritic cell, Th1, Tfh, Treg, and exhausted T-cell markers.
DISCUSSION
As a melanocyte-specific type I membrane protein, PMEL, has been considered as a melanocyte biomarker in the diagnosis of primary cutaneous melanoma and in the identification of melanoma metastasis biopsy to sentinel lymph node.27,28 Before the PMEL protein entry into stage I melanosomes, some of fraction of the protein is transiently presented to the cell surface.29–31 It is interesting to note that, the process of PMEL transiently migrating to cell surface makes it possible for immune cells and antibody drug conjugate to target melanoma cell.32 The highly restricted expression of PMEL in normal tissues makes it become a potential drug target for melanoma. Immuno-oncology including the cancer vaccine, immune checkpoints inhibitor is a young and growing field, and melanoma was recognized as a significant cancer model due to its obvious immunogenicity. PMEL, also known as gp100, has been used as a peptide vaccine of antimelanoma for several decades due to the ability to activate the CD8+ T-cell rapidly.33,34 So, we can guess that a significant positive correlation between PMEL expression and immune infiltration cells can be demonstrated. But is this theoretical guess right?
Here, in our present research, we found that the expression of PMEL was negatively associated with the immune infiltration cells such as CD8+ T cell, macrophage, neutrophil, and dendritic cell in the SKCM and SKCM-metastasis. In addition, high expression of PMEL was associated with a poorer OS compared with the lower expression patients in SKCM, which indicated that the PMEL can be recognized as a predictor of poor prognosis. Furthermore, our study showed that immune infiltration levels and various immune infiltration related gene markers are mostly negative correlated with the PMEL expression level. Thus, our study provided a chance to clarify the inner correlation between PMEL expression and immunomelanoma, and the value as a melanoma prognostic biomarker.
Immune infiltration cells of tumor microenvironment are significant determinant for immune response and outcomes in various cancers including melanoma.35,36 Here, we also demonstrated that higher immune infiltration level such as B cell, CD8+ T cell, neutrophil, and dendritic cell were associated with a better OS in the SKCM and SKCM-metastasis patients (Fig. 6), which is consistent with the previous research. But in our study, higher PMEL expression was associated with lower immune infiltration level including CD8+ T cell, macrophage, neutrophil and dendritic cell in the TIMER database (Fig. 5). It was also found that most of the immune infiltration gene marker have a moderate or strong negative correlation with the PMEL expression (Table 2), which implicate the negative role of PMEL in regulating tumor immunology in SKCM. First, gene markers of CD8+ T cell including CD8A and CD8B demonstrated a negative correlation with PMEL expression level, which may explain the phenomenon of poorer prognosis correlated with higher PMEL expression. Interesting, the metastasis of melanoma was most occurred in the lymph nodes, which pooling large numbers of lymphocytes. These may provide an explanation for the negative correlation disappearing in the SKCM-metastasis patients. Second, gene markers of M1 macrophage such as NOS2, IRF5, and PTGS2 showed a weak correlation with PMEL expression but a strong correlation exhibited for the M2 macrophage including CD163, VSIG4, MS4A4A (Table 2), which indicated the potential regulatory function of PMEL in the polarization of TAM. In addition, a moderate to strong negative correlations between PMEL expression level and infiltration level of DCs can be demonstrated in this study, which may be the reason why we should increase cross-presentation of immature dendritic cells to gp100 specific CD4 T cells via modifying glycosylation on gp100.37 However, our study also indicated that PMEL play a significant negative role in activating Tregs and inducing T-cell exhaustion. The higher PMEL expression negatively correlates with the expression of Tregs and exhausted cells gene markers such as CCR8, STAT5B, TGFβ, LAG3, TIM-3, and GZMB (Table 2). The possible hypothesis or explanation about PMEL expression negative correlation of immune cell infiltration is that as a normal, nonmutated “self”-protein, PMEL, presents poor immunogenicity and apparent hyporesponsiveness to the human immune,38,39 so autologous PMEL is challenging to stimulate effective immune cell infiltration. Poor immunogenicity of PMEL antigens is due to the instability of the peptide-major histocompatibility complex,39 and vaccination with autologous PMEL will not trigger these CD8+ T cells unless the vaccine is able to significantly raise the amount of peptide-major histocompatibility complex class I complex on professional antigen presenting cells to a level high enough to surpass the TCR threshold.38
Thus, a positive expression correlation between PMEL and PD-1 was confirmed in our study, and the high expression of PMEL is also associated with the increasing PD-1 expression, which explain the better response for PD-1 in melanoma. Based-PMEL tumor vaccine is one of the important explorations of new treatment methods for melanoma. Studies have found that this type of vaccine can play a better synergistic effect with immune checkpoint inhibitors (PD-1, etc.).40,41 Finally, significant correlations can be found between PMEL expression and several markers of T helper cells including Th1, Th2, Tfh, Th17 in SKCM. These correlations could give us a potential mechanism to explain if and how PMEL regulates the T cells function in SKCM. In conclusion, all these finding indicated that PMEL play a significant role in regulating the immune infiltration function in SKCM.
Melanoma vaccination with various forms combined with PMEL could educate immune system to recognize and eradicate melanoma cells,42,43 in particular to activate PMEL-specific CD8+ T cells against PMEL peptides. PMEL-based melanoma vaccines have been tested extensively in clinical trials.44–48 So high PMEL expression can more efficiently and strongly react with CD8+ T cells stimulated by PMEL-based vaccine, and the effect of antimelanoma immune response will be better.
Actually, the PMEL is not only highly expressed in melanoma, but also found in other tumor tissues such as glioblastoma multiforme primary cell line,49 breast cancer.50 In our study, we examined the expression level of PMEL mRNA and its correlation with the prognosis of different cancers in Oncomine, TIMER and GEPIA databases. Compared with the normal tissues or the other cancers, the PMEL expression level was highly expressed in melanoma including SKCM on the Oncomine database. From the results of the PrognoScan database, the high PMEL expression level can be used as an independent potential factor for poor prognosis in in melanoma and lung cancer (P<0.05) (Figs. 3E, F). The unique high expression level of PMEL in melanoma and the obvious negatively correlation between PMEL mRNA expression level and prognosis indicate that PMEL will be a potentially better prognostic biomarker in melanoma.
Recent studies demonstrated that the critical PMEL domain has a significant effect on the formation of melanoma, and high PMEL expression plays a positive role in melanoma invasion and metastasis of process.10,12,13 In summary, higher PMEL expression level correlates with poorer prognosis and lower immune cell infiltration level in CD8+ T cell, macrophage, neutrophil, and dendritic cell in the SKCM and SKCM-metastasis. Thus, the correlation between PMEL expression level and immune infiltration markers in SKCM, may provide an explanation that PMEL influence the prognosis of SKCM through inhibiting the infiltration level of macrophage, TAMs, DCs, Tregs, exhausted T cells, and so on. Therefore, PMEL is associated with immune cell infiltration and can be as a prognosis marker or new immunotherapy target in SKCM and SKCM-metastasis.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Shengji Yu and Chunfeng Qu for statistical expertise and critical review of the manuscript. Medical writing and editorial assistance were provided by the other authors.
Conflicts of Interest/Financial Disclosures
Supported by Department of Orthopedics, National Cancer Center/Cancer Hospital, Chinese Academy of Medical sciences and Peking Union Medical College, Beijing, 100021, China and the State Key Lab of Molecular Oncology and Immunology Department, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
All authors have declared that there are no financial conflicts of interest with regard to this work.
Footnotes
Funded by: (1) CAMS Innovation Fund for Medical Sciences (CIFMS) (no. 2017-I2M-1-005). (2) Capital characterized clinical application research Fund of Beijing Municipal Science and Technology Commission of China (no. Z171100001017210). (3) Beijing Hope Run Special Fund of Cancer Foundation of China (no. LC2016L01). (4) Special Fund for Clinical Research of Wu Jieping Medical Foundation (no. 320.6750.14298). (5) Chinese Academy of Medical Sciences Innovative Medicine (No. 2016-I2M-1-007).
Contributor Information
Shuguang Zhang, Email: zhangshuguang02@163.com.
Kun Chen, Email: chenkun_cams@163.com.
Huanmei Liu, Email: 543625412@qq.com.
Changyou Jing, Email: jingchangyou1@163.com.
Xinxin Zhang, Email: xxzhang1983@163.com.
Chunfeng Qu, Email: quchf@cicams.ac.cn.
Shengji Yu, Email: shengjiyu@126.com.
REFERENCES
- 1. Jemal A, Saraiya M, Patel P, et al. Recent trends in cutaneous melanoma incidence and death rates in the United States, 1992-2006. J Am Acad Dermatol. 2011;65(suppl 1):S17–S25. [DOI] [PubMed] [Google Scholar]
- 2. Luke JJ, Flaherty KT, Ribas A, et al. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14:463–482. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 4. Rochin L, Hurbain I, Serneels L, et al. BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc Natl Acad Sci USA. 2013;110:10658–10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoashi T, Muller J, Vieira WD, et al. The repeat domain of the melanosomal matrix protein PMEL17/gp100 is required for the formation of organellar fibers. J Biol Chem. 2006;281:21198–21208. [DOI] [PubMed] [Google Scholar]
- 6. Wagner SN, Wagner C, Schultewolter T, et al. Analysis of PMEL17/gp100 expression in primary human tissue specimens: implications for melanoma immuno- and gene-therapy. Cancer Immunol Immunother. 1997;44:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawakami Y, Eliyahu S, Delgado CH, et al. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci USA. 1994;91:6458–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson LA, Morgan RA, Dudley ME, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simon B, Harrer DC, Schuler-Thurner B, et al. Arming T cells with a gp100-specific TCR and a CSPG4-specific CAR using combined DNA- and RNA-based receptor transfer. Cancers (Basel). 2019;11:693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burgoyne T, O’Connor MN, Seabra MC, et al. Regulation of melanosome number, shape and movement in the zebrafish retinal pigment epithelium by OA1 and PMEL. J Cell Sci. 2015;128:1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bissig C, Rochin L, van Niel G. PMEL amyloid fibril formation: the bright steps of pigmentation. Int J Mol Sci. 2016;17:1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang JJ, Li ZF, Li XJ, et al. Effects of microRNA-136 on melanoma cell proliferation, apoptosis, and epithelial-mesenchymal transition by targetting PMEL through the Wnt signaling pathway. Biosci Rep. 2017;37:BSR20170743. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Hee JS, Mitchell SM, Liu X, et al. Melanosomal formation of PMEL core amyloid is driven by aromatic residues. Sci Rep. 2017;7:44064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;4:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pan JH, Zhou H, Cooper L, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X, Xiong H, Liang D, et al. The role of SRGN in the survival and immune infiltrates of skin cutaneous melanoma (SKCM) and SKCM-metastasis patients. BMC Cancer. 2020;20:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi S, Ye S, Mao J, et al. CMA1 is potent prognostic marker and associates with immune infiltration in gastric cancer. Autoimmunity. 2020;53:210–217. [DOI] [PubMed] [Google Scholar]
- 21. Yu H, Jin GZ, Liu K, et al. Twist2 is a valuable prognostic biomarker for colorectal cancer. World J Gastroenterol. 2013;19:2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oble DA, Loewe R, Yu P, et al. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human melanoma. Cancer Immun. 2009;9:3. [PMC free article] [PubMed] [Google Scholar]
- 23. Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30:2678–2683. [DOI] [PubMed] [Google Scholar]
- 24. Liakou CI, Narayanan S, Ng Tang D, et al. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human bladder cancer. Cancer Immun. 2007;7:10. [PMC free article] [PubMed] [Google Scholar]
- 25. Ohtani H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PMC free article] [PubMed] [Google Scholar]
- 26. Dunn GP, Dunn IF, Curry WT. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human glioma. Cancer Immun. 2007;7:12. [PMC free article] [PubMed] [Google Scholar]
- 27. Yaziji H, Gown AM. Immunohistochemical markers of melanocytic tumors. Int J Surg Pathol. 2003;11:11–15. [DOI] [PubMed] [Google Scholar]
- 28. Rothberg BE, Moeder CB, Kluger H, et al. Nuclear to non-nuclear PMEL17/gp100 expression (HMB45 staining) as a discriminator between benign and malignant melanocytic lesions. Mod Pathol. 2008;21:1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: Driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007;19:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valencia JC, Watabe H, Chi A, et al. Sorting of PMEL17 to melanosomes through the plasma membrane by AP1 and AP2: evidence for the polarized nature of melanocytes. J Cell Sci. 2006;119:1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yasumoto K, Watabe H, Valencia JC, et al. Epitope mapping of the melanosomal matrix protein gp100 (PMEL17): rapid processing in the endoplasmic reticulum and glycosylation in the early Golgi compartment. J Biol Chem. 2004;279:28330–28338. [DOI] [PubMed] [Google Scholar]
- 32. Chen Y, Chalouni C, Tan C, et al. The melanosomal protein PMEL17 as a target for antibody drug conjugate therapy in melanoma. J Biol Chem. 2012;287:24082–24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kamta J, Chaar M, Ande A, et al. Advancing Cancer therapy with present and emerging immuno-oncology approaches. Front Oncol. 2017;7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu Z, Huang H, Grenier JM, et al. Cytomegalovirus-based vaccine expressing a modified tumor antigen induces potent tumor-specific CD8(+) T-cell response and protects mice from melanoma. Cancer Immunol Res. 2015;3:536–546. [DOI] [PubMed] [Google Scholar]
- 35. Selitsky SR, Mose LE, Smith CC, et al. Prognostic value of B cells in cutaneous melanoma. Genome Med. 2019;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iglesia MD, Parker JS, Hoadley KA, et al. Genomic analysis of immune cell infiltrates across 11 tumor types. J Natl Cancer Inst. 2016;108:djw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aarnoudse CA, Bax M, Sanchez-Hernandez M, et al. Glycan modification of the tumor antigen gp100 targets DC-SIGN to enhance dendritic cell induced antigen presentation to T cells. Int J Cancer. 2008;122:839–846. [DOI] [PubMed] [Google Scholar]
- 38. Overwijk WW, Tsung A, Irvine KR, et al. gp100/PMEL 17 is a murine tumor rejection antigen: induction of “self”-reactive, tumoricidal T cells using high-affinity, altered peptide ligand. J Exp Med. 1998;188:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu Z, Theoret MR, Touloukian CE, et al. Poor immunogenicity of a self/tumor antigen derives from peptide-MHC-I instability and is independent of tolerance. J Clin Invest. 2004;114:551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yazdani M, Hatamipour M, Alani B, et al. Liposomal gp100 vaccine combined with CpG ODN sensitizes established B16F10 melanoma tumors to anti PD-1 therapy. Iran J Basic Med Sci. 2020;23:1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yazdani M, Gholizadeh Z, Nikpoor AR, et al. Vaccination with dendritic cells pulsed ex vivo with gp100 peptide-decorated liposomes enhances the efficacy of anti PD-1 therapy in a mouse model of melanoma. Vaccine. 2020;38:5665–5677. [DOI] [PubMed] [Google Scholar]
- 42. Fotaki G, Jin C, Kerzeli IK, et al. Cancer vaccine based on a combination of an infection-enhanced adenoviral vector and pro-inflammatory allogeneic DCs leads to sustained antigen-specific immune responses in three melanoma models. Oncoimmunology. 2018;7:e1397250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Przybyla A, Lehmann AA, Zhang T, et al. Functional T cell reactivity to melanocyte antigens is lost during the progression of malignant melanoma, but is restored by immunization. Cancers (Basel). 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Middleton MR, McAlpine C, Woodcock VK, et al. Tebentafusp, A TCR/Anti-CD3 bispecific fusion protein targeting gp100, potently activated antitumor immune responses in patients with metastatic melanoma. Clin Cancer Res. 2020;26:5869–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cassaday RD, Sondel PM, King DM, et al. A phase I study of immunization using particle-mediated epidermal delivery of genes for gp100 and GM-CSF into uninvolved skin of melanoma patients. Clin Cancer Res. 2007;13:540–549. [DOI] [PubMed] [Google Scholar]
- 47. Powell DJ, Jr, Rosenberg SA. Phenotypic and functional maturation of tumor antigen-reactive CD8+ T lymphocytes in patients undergoing multiple course peptide vaccination. J Immunother. 2004;27:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tarhini AA, Leng S, Moschos SJ, et al. Safety and immunogenicity of vaccination with MART-1 (26-35, 27L), gp100 (209-217, 210M), and tyrosinase (368-376, 370D) in adjuvant with PF-3512676 and GM-CSF in metastatic melanoma. J Immunother. 2012;35:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu G, Ying H, Zeng G, et al. HER-2, gp100, and MAGE-1 are expressed in human glioblastoma and recognized by cytotoxic T cells. Cancer Res. 2004;64:4980–4986. [DOI] [PubMed] [Google Scholar]
- 50. Sellappan S, Grijalva R, Zhou X, et al. Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 2004;64:3479–3485. [DOI] [PubMed] [Google Scholar]


