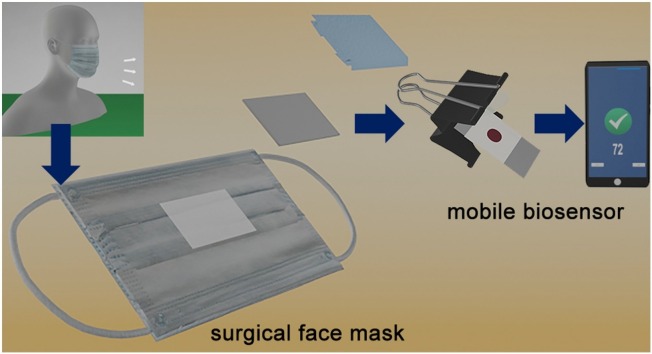
Graphical abstract
Abbreviations: RT-PCR, real-time polymerase chain reaction; IgG, immunoglobulin G; PP, polypropylene; AUC-ROC, area under the ROC curve, where ROC is the receiver operating characteristic curve; COPD, chronic obstructive pulmonary disease
Keywords: SARS-CoV-2, COVID-19, Rapid diagnostic test, Wearable, Biosensor, Smartphone
Abstract
Detecting SARS-CoV-2 antigens in respiratory tract samples has become a widespread method for screening new SARS-CoV-2 infections. This requires a nasopharyngeal swab performed by a trained healthcare worker, which puts strain on saturated healthcare services. In this manuscript we describe a new approach for non-invasive COVID-19 diagnosis. It consists of using mobile biosensors for detecting viral antigens trapped in surgical face masks worn by patients. The biosensors are made of filter paper containing a nanoparticle reservoir. The nanoparticles transfer from the biosensor to the mask on contact, where they generate colorimetric signals that are quantified with a smartphone app. Sample collection requires wearing a surgical mask for 30 min, and the total assay time is shorter than 10 min. When tested in a cohort of 27 patients with mild or no symptoms, an area under the receiving operating curve (AUROC) of 0.99 was obtained (96.2 % sensitivity and 100 % specificity). Serial measurements revealed a high sensitivity and specificity when masks were worn up to 6 days after diagnosis. Surgical face masks are inexpensive and widely available, which makes this approach easy to implement anywhere. The excellent sensitivity, even when tested with asymptomatic patient samples, along with the mobile detection scheme and non-invasive sampling procedure, makes this biosensor design ideal for mass screening.
1. Introduction
The continued spread of COVID-19 has had devastating social and economic repercussions. Mass screening programs have been implemented by many governments in order to detect pre-symptomatic and non-symptomatic patients so that contagions can be reduced through contact-tracing and self-isolation. Currently, the detection of viral nucleic acids with RT-PCR or the immunodetection of viral antigens are the predominant methods for screening early SARS-CoV-2 infections [[1], [2], [3]]. RT-PCR is advantageous because it is sufficiently sensitive to detect the virus in pre-symptomatic and non-symptomatic patients. This method, however, requires expensive reagents and can only be performed by trained personnel in laboratories with immobile instruments, which severely limits the number of tests that can be performed on a daily basis [2]. The other predominant screening method, antigen tests, consist of colorimetric lateral flow immunoassays that detect viral proteins. Although their analytical sensitivity is lower than RT-PCR, their low cost and point-of-need operation have made them popular alternatives for mass screening [2]. In both cases, nasopharyngeal swabs are typically used for sample collection. The drawback of nasopharyngeal swabs is the invasive and uncomfortable sampling procedure that must be performed by a trained healthcare worker [4]. Inadequate sampling may lead to contamination with samples from the upper respiratory tract, which increases the number of false positive results [4,5]. This puts additional pressure on healthcare providers, who need to divert healthcare workers from patient care to perform COVID-19 tests [6]. Saliva specimens are emerging as a non-invasive alternative to sample gathering for nucleic acid testing [[7], [8], [9], [10], [11]]. Unfortunately, the antigen tests currently available yield a high number of false negatives when tested on saliva samples [6]. To overcome these limitations, it would be desirable to find a new, non-invasive antigen test with high sensitivity that can be used at the point of need [12,13]. Such a method could enable mass screening for COVID-19 without overburdening healthcare systems since both sample collection and analysis could be performed by laymen [14,15].
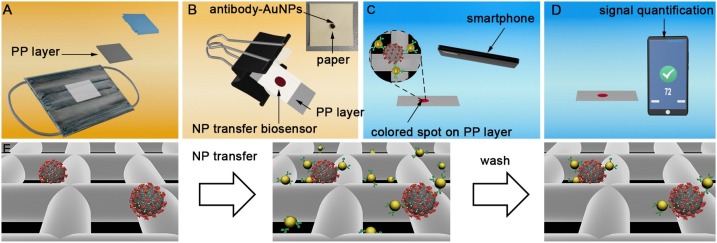
In this manuscript we propose, for the first time in the literature, a non-invasive COVID-19 antigen test based on detecting viral proteins trapped in surgical face masks. Commercial surgical masks contain a hydrophobic polypropylene (PP) layer that provides liquid barrier protection (Fig. 1 A). When wearing a surgical mask, the aerosols and droplets that are produced as the wearer breaths and speaks are captured by this layer [16,17]. We hypothesized that SARS-CoV-2 antigens would therefore accumulate in this layer as well, and that they could subsequently be detected with nanoparticle transfer biosensors. These biosensors consist of a piece of filter paper modified with a polymer in order to store antibody-decorated gold nanoparticles (Fig. 1B, Inset and Fig. S1) [18]. The polymer avoids irreversible interactions between the paper and the nanoparticles in dry conditions, which ensures a complete release of nanoparticles when liquid is added [19]. Paper areas outside the reservoir are modified with wax, which improves the release of nanoparticles from the reservoir compared to our original design [18]. In traditional lateral flow tests, nanoparticles are transferred horizontally from a reservoir to a detection area [[20], [21], [22], [23], [24]]. In contrast, our biosensors transfer nanoparticles vertically upon contact with a receiving substrate [18,19]. While this method was proposed for fabricating paper-only biosensors (e.g. origami biosensors), here we introduce a new application for detecting analytes in exhaled breath by combining the nanoparticle transfer properties of paper immunosensors with the antigen-binding properties of polypropylene face masks (Fig. 1B). Upon contact with the PP layer of the mask, the nanoparticles specifically bind to viral antigens. After 5 min the paper biosensor is removed and, after a quick washing step, the presence of antigens attached to the mask is evaluated by quantifying the colorimetric signal with a smartphone app (Fig. 1C-D). The whole process takes less than 10 min to be completed and can be easily performed at the point of need. The app does not require any smartphone modifications or light boxes to yield consistent readings, which is also advantageous for in-field applications [25]. When tested in a cohort of 27 low- and non-symptomatic patients who wore a surgical mask for 30 min, an area under the receiving operating curve (AUROC) of 0.99 was obtained. Serial measurements revealed an inverse correlation between the days elapsed since diagnosis and the biosensor signal, which indicates that the sensor signal depends on the viral load. Unmodified surgical face masks are used for sample collection, which are inexpensive and widely available. This, along with the rapid assay time and point-of-need detection scheme make our biosensors promising for non-invasive COVID-19 mass testing.
Fig. 1.
Schematic representation of non-invasive detection of SARS-CoV-2 antigens trapped in surgical face masks. (A) The intermediate polypropylene (PP) layer of a face mask worn by a patient is isolated; (B) Nanoparticle transfer biosensors are pressed on top of the PP layer with the aid of a clamp so that antibody-decorated gold nanoparticles (Ab-AuNPs) are transferred to the receiving substrate; Inset: photograph of the biosensor, which consists of a paper reservoir delimited with wax and filled with polystyrene sulfonate (PSS) and Ab-AuNPs (See Fig. S1). (C) After washing, nanoparticles remain attached to the paper mainly through antibody-antigen interactions; (D) The colorimetric signal S (pixel intensity) is calculated in a few seconds with a smartphone app that fixes the imaging conditions with an interactive augmented reality design that compensates for variations in light conditions; (E) Signal generation mechanism based on the specific recognition of masked-trapped antigens by antibody-decorated nanoparticles.
2. Materials and methods
2.1. Materials
Whatman filter paper #1 was purchased from GE Healthcare Life Sciences. Wax paper squares (6″) were purchased from NORPRO. Gold (III) chloride hydrate, sodium citrate tribasic, poly (ethylene glycol) 2-mercaptoethyl ether acetic acid (thiol-PEG-acid) 2100, N-hydroxysulfosuccinimide sodium salt (sulfo-NHS), N-(3-Dimethylaminopropyl)-N′- ethylcarbodiimide hydrochloride (EDAC; EDC), poly (sodium 4-styrenesulfonate, average Mw ∼70.000) 30 % solution, polyethylene glycol 3000 (biotin-PEG), Tween-20, 2-ethanesulfonic acid (MES), and anti-mouse IgG (Fc Specific) (SAB3701021) were obtained from Sigma Aldrich. Mouse monoclonal antibody Anti-SARS-CoV-2 N-protein (35–579) was purchased from ProScience. Recombinant Nucleocapsid protein from SARS-CoV-2 (230−01104) was purchased from Raybiotech. Bovine serum albumin (BSA, protease free) was purchased from VWR chemicals. Aerosol bottles were purchased from Lurrose. Surgical facemasks for experiments in Fig. 3 were purchased from Medline (NONE27377). PBS refers to phosphate buffered saline pH 7.4. PBST refers to PBS supplemented with 0.1 % Tween-20. RT refers to room temperature. PBS-BSA refers to PBS supplemented with 0.5 % BSA.
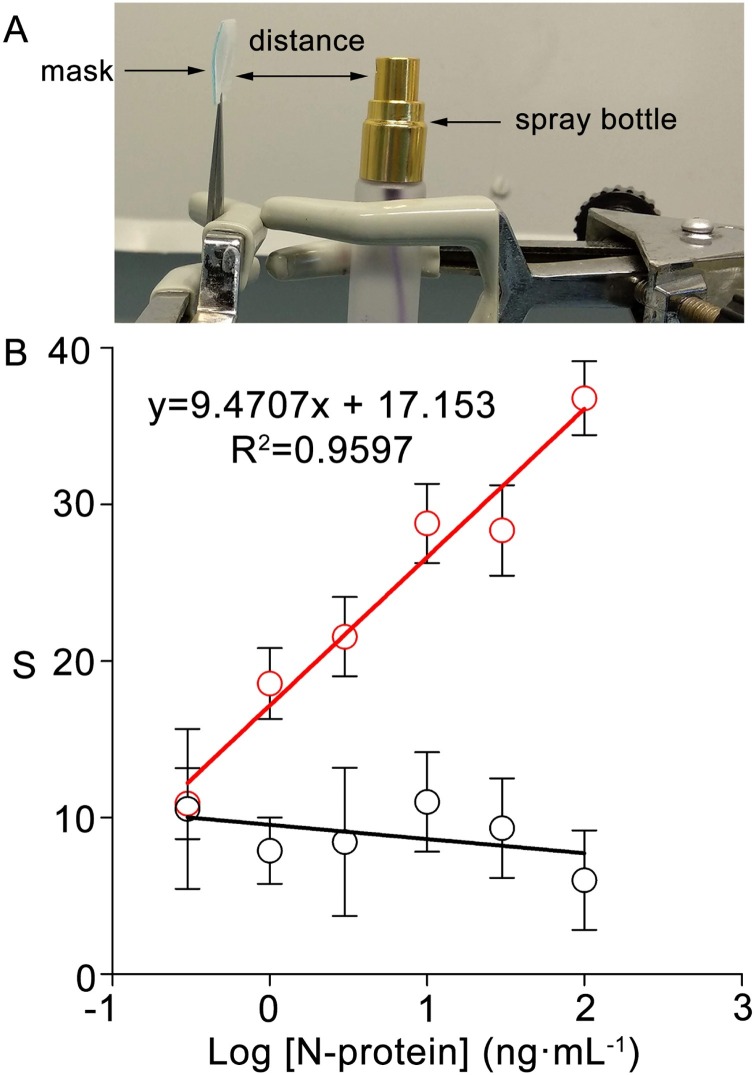
Fig. 3.
Detection of N-antigen in droplets trapped by face masks. (A) Experimental setup for generating droplets; (B) Calibration plot when solutions containing different concentrations of N-antigen (red dots) or BSA (black dots) were dispensed with the setup in (A) (distance = 5 cm, twice). Error bars are the standard deviation of three different biosensors.
2.2. Antibody-decorated gold nanoparticles
A previously described citrate-reducing method was used to obtain gold nanoparticles with a diameter of ca. 40 nm [25]. The citrate around the nanoparticles was substituted for thiol-PEG-acid. The carboxylate moieties were transformed into sulfo-NHS esters by adding EDC (1 mg) and sulfo-NHS (2 mg) in 0.5 M MES buffer pH 5.5 for 30 min. Subsequently, 200 μL of nanoprobes were centrifugated (8000 rpm, 6 min) and anti-mouse IgG (1 mg mL−1) was added and incubated overnight. To cap any remaining sulfo-NHS ester groups, 200 μL of a blocking buffer (glycine 0.1 M, BSA 10 mg mL−1 in phosphate buffer pH 7) was added for 30 min. Nanoparticles were then washed 5 times with PBST (6000 rpm, 6 min) and the pellet resuspended in 200 μL of PBST. The resulting nanoparticles (200 μL) were then modified with 20 μL of monoclonal antibody against the N-protein of SARS-CoV-2 for 1 h followed by washing 3 times with PBST (200 μL, 5500 rpm, 6 min).
2.3. Fabrication of nanoparticle transfer biosensors
Whatman paper #1 was cut into 1.5cm × 1.5cm squares, which were then printed with wax with the following procedure. First, wax paper was cut into 2cm × 2cm squares. A 5 mm circle was removed from the centre with a hole puncher. The remaining wax was aligned with the Whatman paper and the paraffin was transferred by pressing a hot iron against it for 5 s. The procedure was repeated on the other side of the paper to completely seal it. The nanoparticle reservoir was fabricated by adding 3 μL of 30 % PSS in the unwaxed paper area and letting it dry at room temperature (RT) followed by adding 1 μL of antibody-decorated gold nanoparticles and letting it dry at RT.
2.4. Detection of antigens in droplets trapped by face masks
SARS-CoV-2 Nucleocapsid protein solutions at different final concentrations were prepared in PBS. Then each dilution was transferred to a spray bottle. Surgical masks cut into 1.5cm × 2cm squares were sprayed twice with each solution at a fixed 5 cm distance. After drying at RT, the three layers were separated, and the hydrophobic polypropylene layer was hydrated with 1 mL of PBS-BSA. Immediately afterwards, the nanoparticle reservoir was placed onto the surgical facemask filter and held with the aid of a clamp for 5 min. The reservoir was then peeled off and the filter was washed with 1 mL of PBST. The resulting colorimetric signal was measured immediately afterwards with the mobile app as described in previous publications [18,25]. In brief, the biosensor is placed on a gray card that is used to calibrate the camera and compensate varying light conditions. A virtual square on the smartphone screen is aligned with the test in order to control the distance and angle. The app automatically segments the paper biosensors from the background and finds the region of interest. Then it starts calculating the pixel intensity and subtracts it from the background calculated at four points around the region of interest. Inconsistent light conditions (e.g. shadows) are controlled for automatically, i.e. the app stops taking measurements if the lighting becomes uneven. This informs the user that the smartphone needs to be realigned, at which point the measurements resume. The data set is evaluated after 50 valid measurements are captured. The resulting colorimetric signal S ranges from 255 (black) to 0 (white). Measurements are obtained in less than 10 s.
2.5. Analysis of face masks worn by COVID-19 patients
Healthcare professionals that were self-isolated due to a recent positive COVID-19 test were contacted through the risk prevention service from the Balearic Islands healthcare system. All participants gave informed consent (Ethics committee protocol IB 4251/20 PI). Surgical masks were collected and kept at RT for no longer than 48 h before storing them at −80 °C in the Biobank IdISBa and CIBERES Pulmonary Biobank Consortium, a network currently formed by twelve tertiary Spanish hospitals integrated in the Spanish National Biobanks Network. They were processed following standard procedures with the appropriate approval of the Ethics and Scientific Committees and with the collaboration of the healthcare services of the Hospital Universitario Son Espases and Hospital Son Llàtzer. Two 1.5cm × 2cm squares were cut in the centre of each mask. The hydrophobic polypropylene layer was then isolated, and antigens were detected with the method described above.
2.6. Statistical analysis
Statistical analysis was performed using the GraphPad Prism software (San Diego, CA). Correlation between variables was measured using Spearman’s correlation coefficient. Diagnostic test accuracy was evaluated by receiver operating characteristic (ROC) curve analysis. A p value <0.05 was considered statistically significant.
3. Results
3.1. Nanoparticle transfer
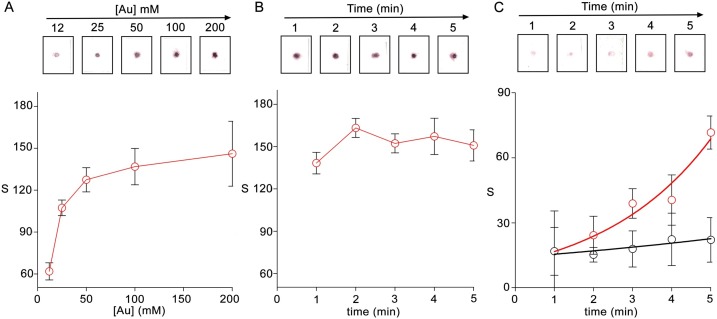
The detection mechanism in the proposed biosensors is based on dispensing nanoparticles to a receiving substrate through contact transfer. While this concept has been previously demonstrated using paper to absorb the nanoparticles [19], it has never been shown to work using the PP layer of a face mask as the receiving substrate. Furthermore, the original design used avidin-modified nanoparticles covered with biotinylated antibodies as nanoprobes. The nanosensors described here used secondary antibodies instead of avidin and therefore have different surface properties. These differences in nanoprobe design and substrate material could have a great impact in nanoparticle transference and immunodetection, which are crucial to detect antigens trapped on face masks. Fig. 2 summarizes the main experiments performed to demonstrate nanoparticle transfer and immunodetection. In Fig. 2A, biosensors containing nanoparticles at different concentrations were pressed against PP substrates for 5 min. After removing the biosensors, the PP layer was scanned, and the colorimetric signal was measured by calculating the pixel intensity with Image J. In this Fig., the characteristic red-mauve hue of the nanoprobes can be seen in the PP layer of the face mask, which demonstrates that the nanoparticles have been transferred. The colorimetric signal increases as the concentration of nanoparticles in the reservoir increases in the range between 12 and 200 mM (calculated as the total concentration of gold, [Au]). Increasing the concentration between 100 and 200 mM raised the variability without substantially increasing the signal magnitude. Therefore, a concentration of 100 mM ([Au]) was used in subsequent experiments. In Fig. 2B, the nanoparticle transfer efficiency was studied as a function of time. In this Fig., a high colorimetric signal is already achieved after 1 min, which indicates that the vast majority of the nanoparticles have been transferred. The signal slightly increases between 1 and 2 min and then plateaus, which shows that no additional nanoparticles are being delivered after 2 min. Finally, we sought to determine the impact of contact time in a model antibody-antigen interaction. To this end, nanoparticles decorated with anti-rabbit IgG were delivered to PP substrates modified with rabbit IgG. Control experiments were performed with substrates modified with PBS only. In Fig. 2C the colorimetric signal exponentially increases as the contact time increases in the 1−5 min range. Control experiments show a much lower increase, which demonstrates that specific antibody-antigen interactions are the main contribution to the colorimetric signal. These experiments show that, although nanoparticles are fully transferred within 2 min, antigen binding by antibody-decorated nanoparticles requires at least 3–5 min. A contact time of 5 min was chosen for subsequent experiments as this value yielded the highest signal-to-noise ratio while still ensuring a total assay time within 10 min.
Fig. 2.
Nanoparticle transfer to PP substrates and immunodetection of adsorbed proteins. Photographs and densitometric analysis of PP substrates after transferring antibody-AuNPs from paper reservoirs at different concentrations (A); Transferring antibody-AuNPs at a concentration of 100 mM ([Au]) for different times (B); Transferring nanoparticles to substrates modified with rabbit IgG (red) or BSA (black) (C). Error bars are the standard deviation of three independent experiments. Lines are a guide to the eye.
3.2. Detection of N-antigen in droplets
After demonstrating that nanoparticles can be transferred to PP substrates, we sought to study the detection of SARS-CoV-2 antigens in liquid droplets. This required dispensing droplets of solutions containing a known concentration of N-antigen onto face masks, which were subsequently analyzed as schematically shown in Fig. 1. The setup in Fig. 3A was devised in order to generate the droplets. It consisted of placing a piece of face mask (containing all three layers) at a fixed distance of 5 cm from a spray bottle (Fig. S2). After spraying face masks twice, the PP layer was isolated and tested with the procedure schematized in Fig. 1. In Fig. 3B, there is a linear relationship between the logarithm of the antigen concentration and the colorimetric signal in the concentration range between 0.3 and 102 ng mL−1. The limit of detection, expressed as the first sample that yields a signal above two times the standard deviation of the blank, is 3 ng mL−1. Control experiments performed by spraying BSA instead of N-antigen yielded lower signals, which demonstrated that color is mainly originated by specific antibody-antigen interactions. These experiments demonstrate that nanoparticle transfer biosensors can detect SARS-CoV-2 antigens present in droplets that have been captured on a surgical face mask. Other proteins were detected with the same procedure in a dose-dependent manner, which suggests that nanoparticle transfer biosensors could be used to detect other relevant biomarkers in droplets trapped by face masks (Fig. S3). It should be noted that the limit of detection and sensitivity of this calibration method are only indicative, as it could change depending on the number of times that antigen solutions are sprayed on the face masks, among other factors. Likewise, in real applications, the sensitivity may depend on the time that the patient has worn the mask. Below we explore these points further by analyzing face masks worn by volunteers for different amounts of time.
3.3. Detection of N-antigen trapped in face masks worn by patients
To study the detection of SARS-CoV-2 antigens trapped in face masks, healthcare workers and their family members who had been recently diagnosed as COVID-19 positive with either RT-PCR or a commercial antigen test were contacted to participate in our study. All participants were self-isolated in their homes with mild (21) or no (6) symptoms (N = 27), and all had been diagnosed within the previous 6 days. Ages were between 7 and 60 years old. Participants were asked to wear a new surgical face mask for 30 min, and another new one for 120 min. No control was exerted over the brand of the face masks other than ensuring that they were surgical masks with an intermediate PP layer. A subpopulation of 15 patients agreed to repeat the experiment 2 days and 3–4 days afterwards. Control experiments were performed on healthy volunteers with ages ranging from 25 to 92 years old. Some had potentially confounding respiratory diseases such as asthma or COPD, which were duly recorded.
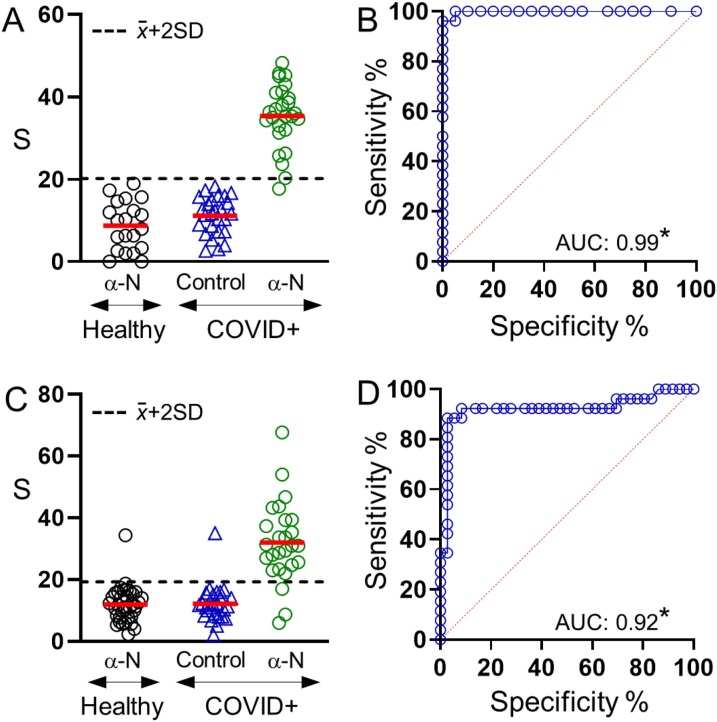
Two central squares of the face mask were cut out and analyzed with either anti-N antigen biosensors or control biosensors containing nanoparticles modified with anti-mouse IgG. Control biosensors were used to define the limit of detection, which was established as two times the standard deviation above the average control signal. Fig. 4 A shows the results obtained after analyzing masks worn for 30 min. In this Fig., only one COVID-19 positive sample yielded a signal below the limit of detection. Masks worn by COVID-19 negative subjects always yielded signals below the limit of detection as expected from volunteers without SARS-CoV-2 infection. In Fig. 4B, comparing signals from COVID-19 positive and negative samples by receiver operating characteristic (ROC) curve analysis yields an area under the ROC curve (AUC) of 0.99, with a sensitivity of 96.2 % and a specificity of 100 % for a threshold S value of 19.65 (negative likelihood ratio 0.04). In Fig. 4C face masks worn by the same volunteers on the same day for 120 min yielded three samples below the limit of detection, as well as one COVID-19 negative sample above the limit of detection. The average colorimetric signal for positive patients changed from 35 ± 8 for 30 min to 31 ± 13 for 120 min, which means that measurements were less precise (relative standard deviation RSD 22 % and 41 %, respectively). Control experiments did not change substantially (11 ± 5 vs 12 ± 6). In Fig. 4D, the AUC also decreased to 0.92 with an associated sensitivity of 88.5 % and specificity of 97.2 % for S = 20.3 (positive and negative likelihood ratios are 31.9 and 0.12, respectively). These experiments demonstrate that SARS-CoV-2 antigens can be detected in surgical face masks worn by COVID-19 patients with high sensitivity and specificity using nanoparticle transfer biosensors. Higher sensitivity and specificity values are obtained when the patients wear the mask for only 30 min.
Fig. 4.
Detection of SARS-CoV-2 antigens trapped in face masks worn by volunteers for 30 min (A, B) or 120 min (C, D); (A, C) colorimetric signal S obtained after analyzing COVID-19 positive and negative samples with biosensors modified with anti-N antigen (α-N); or after analyzing COVID-19 samples with control biosensors modified with anti-mouse IgG; (B, D) ROC analysis for each data set. AUC is the area under the ROC curve. *p < 0.001.
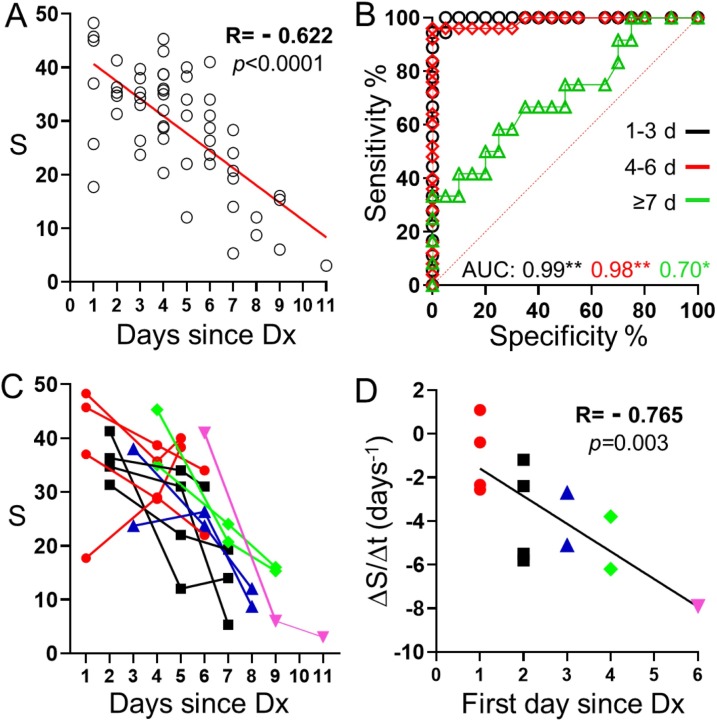
Next, we studied serial measurements over time with nanoparticle transfer biosensors. In Fig. 5 A the colorimetric signal S is plotted as a function of the time elapsed since the official diagnostic with RT-PCR or a commercial antigen test when masks were worn for 30 min. In this Fig., there is a statistically significant negative correlation between both parameters, as expected from the fact that the viral load decreases as time passes for patients with mild or asymptomatic COVID-19 [26]. The same analysis showed a poor correlation that was not statistically significant when the analysis was made with masks worn by the same patients for 120 min (Fig. S4). Fig. 5B shows the AUC analysis for face masks worn by patients within the 3 first days after the first diagnosis, 4–6 days afterwards, and 7 or more days afterwards. In these plots the AUC stays high within the first 6 days (AUC 0.99 and 0.98) but drops to 0.70 after 7 days. Analysis of masks worn for 120 min yielded poorer results (AUC 0.87, 0.55 and 0.52, respectively, Fig. S4). These experiments show that nanoparticle transfer biosensors can diagnose COVID-19 with high sensitivity and specificity when the masks are worn within the first 6 days of confirmed infection. They also show that wearing the masks for 120 min is detrimental to the diagnosis. Fig. 5C shows the time evolution of the S signal for each participant. In this Fig., S decreases slowly, or even increases, when patients had been recently diagnosed, but sharply decreases if several days have passed since diagnosis. This is summarized in Fig. 5D, where the variation of signal S with time was calculated as the slope of a linear regression plot crossing the three points shown in Fig. 5C. The slope was then plotted as a function of the first day that the mask was worn since diagnosis. In this Fig., there is a statistically significant negative correlation between both parameters. S decreases faster for patients that are diagnosed at a later stage, in agreement with the observation that viral replication is the highest during the first stages of infection [27]. These experiments confirm that there is a relationship between time elapsed since diagnosis and the sensor signal, which is presumably linked to variations in the viral load.
Fig. 5.
Impact of time elapsed since first diagnosis (Dx) on the colorimetric signal (S) obtained after analyzing masks worn during 30 min. (A) Correlation plot; (B) ROC analysis for data points corresponding to masks during the first 3 days since the official diagnosis (black, *p < 0.001), 4-6 days afterwards (red, *p < 0.001) or more than 7 days afterwards (green, **p < 0.05). AUC is the area under the ROC curve. (C) Biosensor signal evolution with time for all participants firstly diagnosed 1 (red), 2 (black), 3 (blue), 4 (green), or 6 (pink) days ago. (D) Signal variation rate as function of time passed since diagnosis for the first sample in the series.
4. Discussion
Several aspects of the experiments described here deserve special consideration.
The results shown in Fig. 3 demonstrate that the signal is specific and dose-dependent when the only parameter that changes is the concentration of the antigen. The amount of antigen trapped by the mask depends on the distance between the spray and the mask, as well as the number of times that the solution is sprayed onto the mask. As such, the Figs. of merit reported in Fig. 3 are indicative only. The setup does not pretend to fully mimic the way that aerosols and droplets are generated in the human body. A recent paper describes the airborne transmission of SARS-CoV-2 as aerosols and/or droplets [16]. Droplets are larger and emitted when the patient sneezes or coughs, whereas aerosols consist of smaller particles generated by exhaled breath. It should be noted that none of the participants in the study reported cough or sneezing as a symptom, although 26 % did report rhinorrhea or nasal congestion.
The patient that yielded a signal below the limit of detection in Fig. 4A reported fever and headache symptoms and had been diagnosed with a commercial antigen test one day prior to wearing the mask. Masks worn by the same patient in the following days yielded a positive result. The antibodies used in this study for detecting N-antigen differentiate between the same protein produced by SARS-CoV-1 and SARS-CoV-2 (Fig. S5), which means that interference from an infection by another virus from the Coronaviridae family is highly unlikely. Since the patient showed COVID-19 symptoms and the patient tested positive afterwards, the false negative was probably caused by a biosensor malfunction rather than by a low viral load or a false positive result in the original test.
Better specificity and sensitivity were always found when masks were worn for only 30 min. It has been proposed that proteins adsorb onto polypropylene fibers following a two-stage mechanism [28]. During the first stage a protein monolayer is formed onto the pristine material, whereas during the second stage more materials adsorb onto the first monolayer. The first layer is strongly bound to PP via hydrophobic interactions, whereas during the second stage additional proteins are bound via weaker non-specific protein-protein interactions. We hypothesized that when masks are worn for long periods of time, antigens attach to the PP layer following the second mechanism. Since antigens are weakly bound, antibody-decorated nanoparticles recognizing them are easily removed during the washing step. This decreases the signal and increases the variability of the assay as observed in Fig. 4 and Fig. S4. Additional experiments will be undertaken to determine whether the mask wearing time can be reduced below 30 min.
In this study, the time elapsed between the first diagnosis and sample gathering was used as a proxy for studying the evolution of the viral load. Participants in the study were self-confined and were not allowed to travel for RT-PCR analysis. Healthcare resources could not be diverted for nurses to visit them and take a sample. Nevertheless, this cohort of patients allowed us to access chronological data from mild or asymptomatic COVID-19 patients, which are the main target population for mass screening. Our results show that the biosensors signal decreases as time evolves, and that when the diagnosis is delayed the chances of obtaining a negative result increase. However, since we do not have data on actual viral loads it is unknown whether the patients are still contagious when the biosensor signal drops.
5. Conclusions
In conclusion, we have reported a new non-invasive method for COVID-19 diagnosis. It is based on using nanoparticle transfer biosensors for detecting SARS-CoV-2 antigens trapped in surgical face masks. The biosensors show an excellent sensitivity and specificity, even when tested with asymptomatic patients, when masks were worn for only 30 min. The biosensor signal decreases chronologically with serial testing. The total turnaround time is fast (under 10 min) and signals can be read with a smartphone, which is ideal for decentralized, point-of-need mass screenings. Circumventing the need for nasopharyngeal swabs in sample collection ensures an uncomplicated screening process where participants wear a face mask for 30 min and receive a test result in 10 min. The antigen-based detection mechanism makes them complementary to recent approaches for nucleic acid detection in saliva with mobile biosensors. In the future, the proposed biosensors could be used for detecting antigens from other pathogens as well as biomarkers of inflammation such as interleukins, which have been shown to be present in exhaled breath [29].
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
CRediT authorship contribution statement
Andreu Vaquer: Methodology, Investigation, Validation. Alejandra Alba-Patiño: Methodology, Investigation, Validation. Cristina Adrover-Jaume: Methodology, Investigation, Validation. Steven M. Russell: Methodology, Investigation. María Aranda: Investigation, Resources. Marcio Borges: Investigation, Resources. Joana Mena: Investigation, Resources. Alberto del Castillo: Investigation, Resources. Antonia Socias: Investigation, Resources. Luisa Martín: Investigation, Resources. María Magdalena Arellano: Investigation, Resources. Miguel Agudo: Investigation, Resources. Marta Gonzalez-Freire: Data curation, Writing - review & editing. Manuela Besalduch: Methodology. Antonio Clemente: Data curation, Supervision, Methodology, Writing - review & editing. Enrique Barón: Methodology, Supervision, Writing - review & editing. Roberto de la Rica: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
R.R., A.C. and C.A. acknowledge a Radix, Folium and Talent Junior fellowship, respectively, from IdISBa/Impost turisme sostenible/Agència d’Estratègia Turística de les Illes Balears/Govern de les Illes Balears. A.A acknowledges a fellowship for doctoral studies from Roche (“Stop fuga de cerebros” program). E.B. acknowledges funding from the Instituto de Salud Carlos III (Sara Borrell contract). A.V. acknowledges funding from European Social Fund (ESF) through JoTReSdOS program. MG-F acknowledges funding by the Miguel Servet Program (MS19/00201), Instituto de Salud Carlos III (ISCIII), Madrid. We want to particularly acknowledge the patients, Biobank IdISBa and CIBERES Pulmonary Biobank Consortium (PT17/0015/0001) member of the Spanish National Biobanks Network financed by the Carlos III Health Institute.
Biographies
Andreu Vaquer obtained his degree in Chemistry from Balearic Islands University in 2017. He is currently on the first year of the MSc Science and Chemical technology at the University of the Balearic Islands. He is a member of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) since April 2019. His research interest focuses into paper microfluidic devices and nanoparticle functionalization.
Alejandra Alba-Patiño In 2018 Alejandra Alba-Patiño obtained her MSc degree in Chemical Science and Technology from Balearic Islands University where she is currently doing her PhD. Thanks to a scholarship funded by Roche pharma, she is performing her research at the Balearic Islands Health Research Institute (IdISBa). As a member of the Multidisciplinary Sepsis Group, she focuses on the development of colorimetric paper-based biosensors for the early diagnosis of infections and sepsis.
Cristina Adrover-Jaume Cristina Adrover obtained her degree in Chemistry in 2019 and her MSc degree in Science and Chemical Technology in 2020, both from the Balearic Islands University (UIB). Currently, she is in the first year of her PhD degree in the Balearic Islands Health Research Institute (IdISBa). She is a member of the Multidisciplinary Sepsis Group (IdISBa) since June 2019. Her research focuses into the development of paper biosensors based on nanoparticles.
Steven M. Russell Steven M. Russell is a researcher in computer vision and machine learning. His work applies real-time image processing to biosensing. He is also interested in the 3D modelling of biochemical interactions and in developing paper-based microfluidic devices.
María Aranda María Aranda Perez obtained her degree in Medicine from Universidad Complutense de Madrid in 2007. She specialized in Critical Care Medicine at HU Son Llàtzer hospital in 2015 and from 2015she is working at HU Son Llàtzer as intensivist and is member of Sepsis Unit from its creation.
Marcio Borges Coordinator of Multidisciplinary Sepsis Unit. Intensive Care Unit. Son Llàtzer University Hospital. Associated Professor in Infectious Diseases by Balearic Islands University (UIB). Coordinator of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) since January 2017. Director of Sepsis Area by Iberian and Panamerican Federation of Intensive Care. Spanish’s Director of Sepsis Code endorsed by Health Ministry of Spain.
Joana Mena Joana Mena obtained her degree in Nursery from Balearic Islands University in 2009. She is currently working in Hospital Universitari Son Llàtzer of the Balearic Islands. Member of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) since 2018, she collaborates in the different studies of the Multidisciplinary Sepsis Group.
Alberto del Castillo Alberto del Castillo obtained his degree in Medicine from University of Salamanca in 2002 and completed the Intensive Care specialty at the Hospital Dr.Peset (Valencia) in 2008. He is currently working in the Intensive Care Unit of the University Hospital Son Llàtzer. He is a member of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa). His research interest focuses into sepsis.
Antonia Socias Antonia Socias obtained her degree in Medicine from Universitat Autonoma de Barcelona in 2000. She specialized in Critical Care Medicine at Vall d'Hebron hospital and from 2006 she is working at HU Son Llàtzer as intensivist and is member of Sepsis Unit from its creation. She is currently a PhD student in Toxicology.
Luisa Martín Maria Luisa Martin Fajardo has a degree in Medicine and Surgery from Universidad of Seville. She specialized in Internal Medicine and Infectious Diseases at Son Espases Hospital and currently, she is working at the Emergency Department of Son Llàtzer Hospital (Balearic Islands) and is a member of the Sepsis group of the same department.
María Magdalena Arellano Maria Magdalena Arellano graduated in Medicine from the University of Zaragoza in 2000. She specialized in Family and Community Medicine at Son Dureta Hospital in Palma de Mallorca. Since 2007 she has been working in the Emergency Department of Son Llàtzer Hospital as an emergency doctor and is a member of the Sepsis group of the same department.
Miguel Agudo Miguel Agudo is currently working in the Emergency Department of Son Llàtzer Hospital and is also a member of the Multidisciplinary Sepsis group.
Marta Gonzalez-Freire Dr. Marta Gonzalez-Freire completed his PhD at the European University of Madrid in Madrid, Spain. After that she started a postdoctoral fellowship at the National Institutes of Health (NIH), USA, for almost 6 years. She has experience in epidemiology studies and biology of aging and inflammation. She is currently a Miguel Servet Investigator, a tenure track position, at the Balearic Islands Health Research Institute, IdISBa since January 2020.
Manuela Besalduch Manuela Besalduch Vidal has a degree in Medicine and Surgery from Universidad Autonóma of Barcelona. She is a specialist in Occupational Medicine and in Family and Community Medicine. She has developed her work life in the Health Service from Balearic Islands and currently, she performs as head of the Prevention Department.
Antonio Clemente Dr. Antonio Clemente obtained his MSc degree on Microbiology in 2008 and his PhD degree on Immunology from the Autonomous University of Barcelona in 2014. During 1-year postdoctoral position at the August Pi Sunyer Biomedical Research Institute (IDIBAPS) he studied nanotechnology applications to immunological disorders therapies. Currently, he is working at the Health Research Institute of the Balearic Islands (IdISBa) with a 2-year post-doc contract (FOLIUM). His research focuses into biosensor diagnosis of bacterial infections
Enrique Barón Dr. Enrique Barón obtained his MSc degree on Advanced Chemistry in 2012 and his PhD degree on Analytical Chemistry from the University of Barcelona in January 2016. During his PhD he studied the environmental behavior of different halogenated flame retardants. Currently, he is working at the Health Research Institute of the Balearic Islands (IdISBa) with a 3-year post-doc contract (Sara Borrell). His research focuses into the development of biosensors for the detection of different disease biomarkers.
Roberto de la Rica Dr. Roberto de la Rica completed his PhD at the National Center for Microelectronics in Barcelona, Spain. He has worked at the City University of New York, at the University of Twente and at Imperial College London. He has experience in nanolithography, synthesis of nanomaterials, biosensors and bio-enabled nanofabrication. He is the coordinator of the technological division of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) since January 2019.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2021.130347.
Appendix A. Supplementary data
The following files are available free of charge.
Supplementary material file: Photograph of biosensors (Fig. S1 Set up for dispensing droplets (Fig. S2); Detection of rabbit IgG (Fig. S3); Impact of time elapsed since first diagnosis (Dx) for masks worn during 120 min (fig. S4); cross-reactivity studies (Fig. S5) (PDF)
The following is Supplementary data to this article:
References
- 1.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Il Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 2.Fleurence R.L., Ph D., Collins F.S., Ph D. Rapid scaling up of Covid-19 diagnostic testing in the United States — the NIH RADx initiative. N. Engl. J. Med. 2020;383:1071–1077. doi: 10.1056/NEJMsr2022263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators, B Chem. 2021;329 doi: 10.1016/j.snb.2020.129196. 129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohn M.K., Loh T.P., Bin Wang C., Mueller R., Koch D., Sethi S., Rawlinson W.D., Clementi M., Erasmus R., Leportier M., Grimmler M., Yuen K.Y., Mancini N., Kwon G.C., Menezes M.E., Patru M.M., Gramegna M., Singh K., Najjar O., Ferrari M., Horvath A.R., Lippi G., Adeli K. IFCC interim guidelines on serological testing of antibodies against SARS-CoV-2. Clin. Chem. Lab. Med. 2020;58:2001–2008. doi: 10.1515/cclm-2020-1413. [DOI] [PubMed] [Google Scholar]
- 5.Higgins T.S., Wu A.W., Ting J.Y. SARS-CoV-2 nasopharyngeal swab testing-false-Negative results from a pervasive anatomical misconception. JAMA Otolaryngol. Head Neck Surg. 2020;146(11):993–994. doi: 10.1001/jamaoto.2020.2946. [DOI] [PubMed] [Google Scholar]
- 6.Agulló V., Fernández-González M., Ortiz de la Tabla V., Gonzalo-Jiménez N., García J.A., Masiá M., Gutiérrez F. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. J. Infect. 2021;82(5):186–230. doi: 10.1016/j.jinf.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ning B., Adv S., Ning B., Yu T., Zhang S., Huang Z., Tian D., Lin Z., Niu A., Golden N., Hensley K. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 2020;3703 doi: 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senok A., Alsuwaidi H., Atrah Y., Al Ayedi O., Al Zahid J., Han A., Al Marzooqi A., Al Heialy S., Altrabulsi B., AbdelWareth L., Idaghdour Y., Ali R., Loney T., Alsheikh-Ali A. Saliva as an alternative specimen for molecular COVID-19 testing in community settings and population-based screening. Infect. Drug Resist. 2020;13:3393–3399. doi: 10.2147/IDR.S275152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of sars-cov-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czumbel L.M., Kiss S., Farkas N., Mandel I., Hegyi A., Nagy Á., Lohinai Z., Szakács Z., Hegyi P., Steward M.C., Varga G. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front. Med. 2020;7:1–10. doi: 10.3389/fmed.2020.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112765. 112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharifi M., Hasan A., Haghighat S., Taghizadeh A., Attar F., Bloukh S.H., Edis Z., Xue M., Khan S., Falahati M. Rapid diagnostics of coronavirus disease 2019 in early stages using nanobiosensors: challenges and opportunities. Talanta. 2021;223 doi: 10.1016/j.talanta.2020.121704. 121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivakumar R., Dinh V.P., Lee N.Y. Ultraviolet-induced in situ gold nanoparticles for point-of-care testing of infectious diseases in loop-mediated isothermal amplification. Lab Chip. 2021;21(4):700–709. doi: 10.1039/d1lc00019e. [DOI] [PubMed] [Google Scholar]
- 14.Gowri A., Ashwin Kumar N., Suresh Anand B.S. Recent advances in nanomaterials based biosensors for point of care (PoC) diagnosis of Covid-19 – a minireview. TrAC Trends Anal. Chem. 2021;137 doi: 10.1016/j.trac.2021.116205. 116205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu K., Chugh V.K., di Girolamo A., Liu J., Saha R., Su D., Krishna V.D., Nair A., Davies W., Wang A.Y., Cheeran M.C.J., Wang J.P. A portable magnetic particle spectrometer (MPS) for future rapid and wash-free bioassays. ArXiv. 2021;13(7):7966–7976. doi: 10.1021/acsami.0c21040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo Y.Y., Uspal W.E., Wei T. Airborne transmission of COVID-19: aerosol dispersion, lung deposition, and virus-receptor interactions. ACS Nano. 2020;14:16502–16524. doi: 10.1021/acsnano.0c08484. [DOI] [PubMed] [Google Scholar]
- 17.Shan B., Broza Y.Y., Li W., Wang Y., Wu S., Liu Z., Wang J., Gui S., Wang L., Zhang Z., Liu W., Zhou S., Jin W., Zhang Q., Hu D., Lin L., Zhang Q., Li W., Wang J., Liu H., Pan Y., Haick H. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020;14:12125–12132. doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- 18.Adrover-Jaume C., Alba-Patiño A., Clemente A., Santopolo G., Vaquer A., Russell S.M., Barón E., del M. González del Campo M., Ferrer J.M., Berman-Riu M., García-Gasalla M., Aranda M., Borges M., de la Rica R. Paper biosensors for detecting elevated IL-6 levels in blood and respiratory samples from COVID-19 patients. Sens. Actuators, B Chem. 2021;330 doi: 10.1016/j.snb.2020.129333. 129333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alba-Patiño A., Adrover-Jaume C., La Rica R.D. Nanoparticle reservoirs for paper-only immunosensors. ACS Sens. 2020;5:147–153. doi: 10.1021/acssensors.9b01937. [DOI] [PubMed] [Google Scholar]
- 20.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 21.Yu S., Nimse S.B., Kim J., Song K., Kim T. Development of a lateral flow strip membrane assay for rapid and sensitive detection of the SARS-CoV-2. Anal. Chem. 2020;92:14139–14144. doi: 10.1021/acs.analchem.0c03202. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 23.Feng M., Chen J., Xun J., Dai R., Zhao W., Lu H., Xu J., Chen L., Sui G., Cheng X. Development of a sensitive immunochromatographic method using lanthanide fluorescent microsphere for rapid serodiagnosis of COVID-19. ACS Sens. 2020;5:2331–2337. doi: 10.1021/acssensors.0c00927. [DOI] [PubMed] [Google Scholar]
- 24.Cavalera S., Colitti B., Rosati S., Ferrara G., Bertolotti L., Nogarol C., Guiotto C., Cagnazzo C., Denina M., Fagioli F., Di Nardo F., Chiarello M., Baggiani C., Anfossi L. A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2. Talanta. 2021;223 doi: 10.1016/j.talanta.2020.121737. 121737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alba-Patiño A., Russell S.M., Borges M., Pazos-Perez N., Alvarez-Puebla R.A., de la Rica R. Nanoparticle-based mobile biosensors for the rapid detection of Sepsis biomarkers in whole blood. Nanoscale Adv. 2020;2:1253–1260. doi: 10.1039/d0na00026d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J.-T., Ran R.-X., Lv Z.-H., Feng L.-N., Ran C.-Y., Tong Y.-Q., Li D., Su H.-W., Zhu C.-L., Qiu S.-L., Yang J., Xiao M.-Y., Liu M.-J., Yang Y.-T., Liu S.-M., Li Y. Chronological changes of viral shedding in adult inpatients with COVID-19 in Wuhan, China. Clin. Infect. Dis. an Off. Publ. Infect. Dis. Soc. Am. 2020;71:2158–2166. doi: 10.1093/cid/ciaa631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2020;2:e13–e22. doi: 10.1016/s2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trang H.K., Schadock-Hewitt A.J., Jiang L., Marcus R.K. Evaluation of loading characteristics and IgG binding performance of Staphylococcal protein A on polypropylene capillary-channeled polymer fibers. J. Chromatogr. B-ANALYTICAL Technol. Biomed. LIFE Sci. 2016;1015:92–104. doi: 10.1016/j.jchromb.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 29.Chen H., Li J., Zhang X., Li X., Yao M., Zheng G. Automated in vivo nanosensing of breath-borne protein biomarkers. Nano Lett. 2018;18:4716–4726. doi: 10.1021/acs.nanolett.8b01070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.