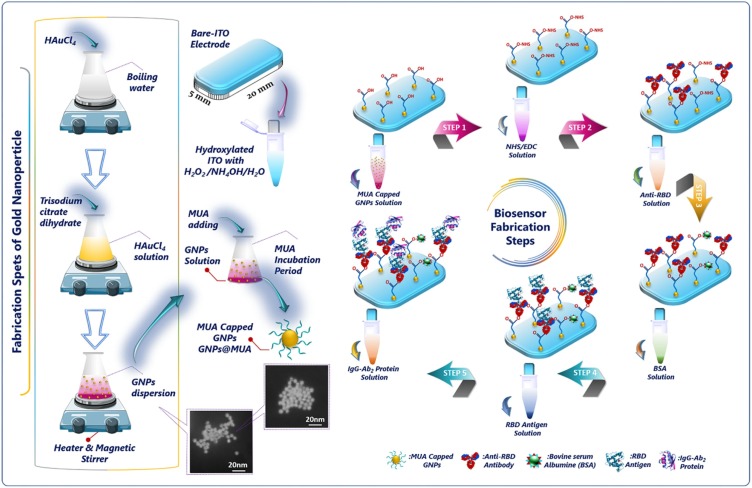
Graphical abstract
Keywords: Spike receptor binding domain, Sandwich immunosensor, MUA-capped gold nanoparticles, Single-use biosensor
Abstract
A label-free electrochemical biosensing system as a suitable analysis technique for COVID 19 specific spike receptor-binding domain protein (RBD) was developed with an aim to facilitate the diagnosis of coronavirus. A novel production procedure for the fabrication of gold nanoparticles (GNPs)-capped 11-mercaptoundecanoic acid (MUA) modified bioelectrode was presented and its application potential for RBD biosensing was examined. The bioelectrode fabrication protocol was based on covalent ester linking formation between hydroxylated ITO electrode and GNPs-capped MUA (GNPs@MUA) with carboxyl ends. For this aim, spherical GNPs were prepared and characterized with scanning-transmission electron microscopy (S-TEM), UV–vis, and Raman spectroscopy. The synthesized GNPs were functionalized with MUA yielding Au-S bonds. Then, covalent immobilization of anti-RBD antibodies on the GNPs@MUA was performed with the help of carbodiimide coupling chemistry. The assembly processes of GNPs@MUA, anti-RBD antibodies and RBD antigens were characterized electrochemical, chemical and morphological techniques. GNPs@MUA was used as immobilization environment and provided the most effective surface design for target immunosensor. The resulting immunosensor is further applied to the impedimetric detection of RBD and it displayed a linear response to RBD antigen in the linear range of 0.002−100 pg mL−1 with a limit of detection of 0.577 fg mL−1 and sensitivity of 0.238 kohmpgmL−1 cm−2. The fabricated immunosensor had a good repeatability, long storage, stability and a reusable property after simple regeneration process. Furthermore, it was successfully employed for selective determination of RBD in artificial nasal secretion samples.
1. Introduction
In late 2019, a new coronavirus called severe acute respiratory syndrome coronavirus (SARS-CoV-2) was first discovered and started to spread around the world [1]. Afterwards, the World Health Organization (WHO) categorized this coronavirus illness 2019 (COVID 19) outbreak as a global pandemic. As of 4 January 2020, there were more than 83 million confirmed COVID 19 cases and more than 1.835 million COVID-119 related deaths globally (http://covid19.who.int/). There are various vaccines for this outbreak; however, not everyone living in the world has been vaccinated. In addition, early diagnosis of COVID 19 disease can have an important role in making the right decision for the isolation of infected humans, and consequently, the spread of the COVID 19 outbreak will slow [2].
SARS-COV-2 is a class of the genus β-COronavirus that contains crown-like, enveloped, positive sense single-stranded RNA viruses [3,4]. The dimension of the whole virus changes from 50 nm to 200 nm. SARS-COV-2 virus encodes at least four structural protein called nucleocapsid (N), spike (S), small envelope (E) and membrane (M) and also several accessory proteins. The N protein holds the viral genome. The M and S glycoproteins are essential for virus binding. The S glycoprotein is important for connecting to host cells, where RBD and S proteins interact with angiotensin-COnverting enzyme 2 (ACE-2) [1,5]. S and N proteins are precious antigen markers for identification of COVID 19 disease. Therefore, screening of RBD as a target of COVID 19 virus is a priority [6,7].
At present, the reverse transcription-polymerase chain reaction (RT-qPCR)-based method has been usually utilized as the gold key diagnostic tool for diagnosis of COVID 19 [8,9]. This technique is sensitive and requires a small amount of input RNA. However, the analysis procedure of this method takes 2 h to get results. In addition, this method requires specialized equipment, specialized handling and transportation, well-trained staff. Furthermore, the result of RT-qPCR can give false-negative results in persons with unapparent clinical findings and false-positive results in recovered patients [2]. Therefore, the development of a fast, ultrasensitive and specific technique will facilitate the identifying the patients with COVID 19 and thus, the spread rate of disease is controlled [6,10,11].
Biosensor have gained the attention throughout the world in recent years. They are utilized in various fields like medical diagnosis, food analysis, environmental measurements and biotechnology because of their rapid response and ease of fabrication [12,13]. Electrochemical immunosensors are a significant class of biosensors due to their simple usage procedure, fast response and possibility of portability and miniaturization [14]. Owing to their excellent sensitivity, high selectivity and low cost, they have utilized to quantify the level of biomarkers, food contaminants, dangerous chemicals for environment and biowarfare agents [15,16]. EIS is a versatile and sensitive technique which provides non-destructive characterization of the electron transfer between modified electrode surfaces and electrolyte solution [17]. This technique is carried out in the presence of a redox couple and it is a proper method to measure molecular interactions of electrochemically inactive compounds taking place on the working electrode surface. Conventional applications of EIS include protein-protein interactions, DNA-DNA interactions and dsDNA-drug interactions. This tool is highly sensitive and can be utilized for a “label-free” detection of a wide range of biorecognition events happening on the working electrode surface. Although this method is quite sensitive, it has some limitations such as inability to discriminate between specific and nonspecific linking. This limitation may be originated from initial electrode contamination and repetitive analyses [18,19].
Nanomaterials such as nanoparticles, nanotubes, graphene and fullerene have gained interest in several research fields because of their promising chemical and physical properties containing large specific surface area, and suitable biocompatibilities [20]. Metal nanoparticles with their outstanding features including large surface area for volume ratio and high surface activity have become an intensive research area and they have been usually utilized in fabrication of biosensors [16]. Especially, gold nanoparticles (GNPs) have been widely employed for construction of immobilization platforms in the development of electrochemical biosensors. The high surface area and a biocompatible microenvironment supported the successful immobilization of biorecognition elements and the bioactivity of biomolecules. In addition, the excellent conductivity of GNPs makes them suitable matrix material and they enhance electron transport between working electrode surface and electrolyte solution [21]. In biosensor fabrication, self-assembled monolayer functionalized GNPs are usually utilized, and this modification method increases the area of the electrode surface. Sulphur containing materials like alkane thiol have high affinity to metals [22]. For the modification of an electrode surface by using SAM functionalized GNPs, silanization agents, polymers and chitosan have been utilized [[23], [24], [25]]. In all these techniques, an extra electrode modification material is necessary and therefore, in this study, we select a simple way for modification of electrode.
In the designing of an electrochemical biosensor, the working electrode is the basic part of the biosensing system. Progress in modification procedures of working electrode can increase the biosensor response, thus more sensitive biosensors can be produced. ITO electrode was a low-cost substrate compared to other carbon-based electrodes and therefore, it was selected as an immobilization platform for immunosensor fabrication. In addition, it had promising properties such as low surface resistivity, good electrical conductivity and optical transparency, high chemical stability, wide electrochemical working area and stable electrochemical features for biosensor fabrication. All these properties make ITO substrate a convenient platform to develop a biosensor. In order to develop a biosensor including ITO substrate as a working electrode, firstly, ITO substrate should be modified for biorecognition element immobilization. In this study, the working ITO electrodes are hydroxylated by using by using NH4OH/H2O2/H2O mixed solution and the GNPs@MUA are bound to these hydroxyl groups with ester bond. As a consequence, the focus of the current study is to develop a suitable and easy fabrication method for RBD biosensor with improved stability and sensitivity.
In this study, a low-cost, easy-to-prepare and easy-to-use biosensor was fabricated by using conductive metal gold nanoparticles and cost-effective ITO working electrode platform for RBD detection. GNPs@MUA were used as an immobilization matrix material and MUA provided attached points for biomolecules. MUA was a bifunctional molecule which contained both thiol and carboxyl functional groups. Thiol ends served as connecting points which bound to GNPs through Au-S bond. Carboxylic acid groups of MUA linked to amino groups of biorecognition elements after a carbodiimide coupling reaction with N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) and N-hydroxysuccinimide (NHS). This procedure provided a stable biosensing interface and high sensitivity. The fabrication of the prepared biosensor was characterized electrochemical techniques by EIS and CV using [Fe(CN)6]3−/4− as a redox probe and SEM microscopic technique. Furthermore, the produced GNPs and the resultant modified GNPs were characterized with UV–vis absorption spectroscopy. The immunosensor was applied for determination of RBD concentrations in artificial samples.
2. Experimental
All utilized materials and reagents in RBD biosensor development and instruments used for characterization of RBD biosensor are available in Supplementary Material File.
2.1. Preparation of gold nanoparticles and MUA-capped gold nanoparticles
Nano-sized GNPs were synthesized employing a reported procedure with minor modifications [26]. The size and surface morphology of gold nanoparticles can be checked during the preparation steps. In a typical preparation route, GNPs were synthesized through Na-citrate reduction of HAuCl4.3H2O [27]. Briefly, gold (III) chloride trihydrate salt (1 mM) was added into 20 mL of boiling and stirring ultra-pure water; then, 2 mL of 1% Na-citrate was rapidly poured out to the prepared solution. After that, the resulting solution was boiled for 10 min, meanwhile, the color of the solution varied from pale yellow to deep red. The prepared GNPs solution cooled down to ambient conditions and then, it was stored at 4 °C until use.
MUA-capped GNPs was prepared by addition of 0.10 mL of MUA (0.05 M prepared in absolute ethanol) into 10 mL of GNPs solution during stirring. After that, the mixed solution was stored in refrigerator overnight. Before fabrication of the proposed biosensor, GNPs@MUA was prepared freshly. The functionalization of nanoparticles was investigated with UV–vis absorption spectroscopy.
2.2. Fabrication of RBD specific bioelectrodes
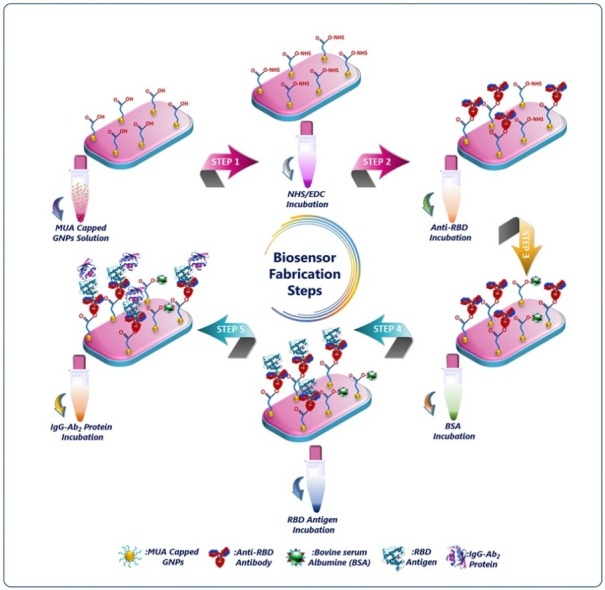
Prior to modification, ITO covered PET plates were sequentially cleaned in ultrasonic bath with acetone, soap solution and ultra-pure water for 10 min each. After that, they were washed with ultra-pure water and finally argon dried. The clean ITO electrodes were dipped into the mixed solution of ultra-pure water/hydrogen peroxide/ammonium hydroxide (5:1:1, v/v) for 90 min. Thus, hydroxylated active ITO surface was produced. These activated surfaces were rinsed with ultra-pure water and dried under a stream of using argon gas. Then, the hydroxylated ITO electrode was immersed in GNPs@MUA solution overnight and ester bond formed between hydroxyl groups of ITO electrodes and carboxyl groups of GNPs@MUA in the presence of EDC (3%) and 4-dimethylaminoantipyrine (DMAP, 0.05 %). To activate the carboxylic ends of GNPs@MUA modified ITO electrodes, they were immersed in a 50 mM PBS solution (pH 7.4) containing EDC (0.4 mM) and NHS (0.1 mM) for 60 min. In the following stage, ITO/GNPs@MUA/NHS-EDC modified surfaces were dipped in anti-RBD solution for 1 h, followed by rinsing with ultra-pure water and drying with argon flow. Anti-RBD antibodies were covalently bound through its amino groups to the carboxyl groups present on the GNPs@MUA modified ITO electrode. After rinsing the electrodes, they were incubated in the BSA solution (0.5 %, for 1 h) to block the unbound ends present on the electrodes. Anti-RBD antibody immobilized electrodes can be utilized directly as immunosensor or kept dry at 4 °C for a few weeks without any decline in the activity. The procedure of the GNPs@MUA decorated RBD biosensor fabrication is illustrated in Scheme 1 .
Scheme 1.
Stepwise fabrication protocol of the novel impedimetric immunosensor for spike RBD antigen.
2.3. Impedimetric measurement of RBD antigen
The electrochemical studies were performed in 10 mL 5.0 × 10−3 M K3[Fe(CN)6]/K4[Fe(CN)6] (1:1) and 1 M KCl solution. EIS signals were recorded with in the frequency range of 0.5 Hz to 50 kHz. The DC and AC voltage were 0 V and 5 mV, respectively. The ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA electrodes were incubated for 60 min in standard RBD antigen solutions with a concentration varying from 0.002 pg mL−1 to 100 pg mL−1, followed by washing thoroughly with ultra-pure water prior to electrochemical measurements. The RBD antigens were captured based a specific interaction between anti-RBD and RBD antigen. This interaction caused a dramatic steric hindrance on the redox probe transfer through electrode surface. Then, the ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA/RBD electrode incubated in the IgG Ab2 solution (250 pg mL−1) to obtain an enhanced signal change. Thus, RBD antigens were detected sensitively and specifically by the proposed sandwich biosensing system.
2.4. Analysis of nasal secretions
Nasal secretions are produced chiefly from submucosal glands and goblet cells. SARS-CoV-2 spike RBD can be determined in nasal and oral cavities. Hence, nasal secretions were chosen a proper analysis matrix for coronavirus. Nasal secretion contains water (95 %), glycoproteins (2%) such as albumin, immunoglobulins, lysozyme, lactoferrin and others, inorganic salts (1%) and lipids (<1%) [28]. In order to test the suggested immunosensor in analytical application, artificial nasal secretion samples were prepared by utilizing water, immunoglobulin G, immunoglobulin M and inorganic salts. Five different levels of RBD were spiked in the prepared solutions and the quantifications of RBD concentrations were performed 3 times.
3. Result and discussion
The synthesis of GNPs and electrochemical RBD immunosensor fabrication strategy are displayed in Scheme 1. Accordingly, GNPs and GNPs@MUA were first prepared through multiple steps. The hydroxylated ITO working electrode incubated in GNPs@MUA overnight to construct the immobilization matrix. Afterwards, the carboxyl ends were activated NHS-EDC crosslinking agents (Step 1). Then, anti‐RBD antibodies were subsequently immobilized on the surface of activated working electrode surface for specific capture of RBD antigen (Step 2). After that, free active carboxyl ends were blocked with BSA protein (Step 3). In the fourth step, RBD antigens were captured by anti-RBD antibodies (Step 4). The resulting RBD antigen/anti-RBD antibody immobilized electrode was dipped in the IgG Ab2 protein to construct a sandwich biosensor (Step 5). On account of specific biorecognition and excellent binding affinity of anti-RBD protein, the sandwich biosensing system was constructed successfully.
3.1. Characterizations of GNPs and MUA-capped GNPs
Spherical and small size distribution gold nanoparticles (GNPs) was successfully prepared by employing a three-step reaction process. During the synthesis of gold nanoparticles, the control of the size and surface morphology of GNPs was possible. This procedure was chosen instead of the other methods because we want to obtain acceptable small size distribution of gold nanoparticles. The detailed GNPs fabrication procedure is given in Scheme 1.
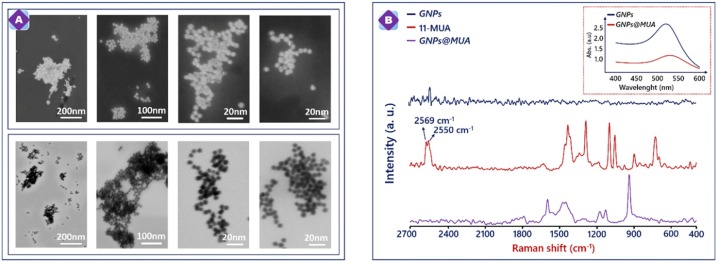
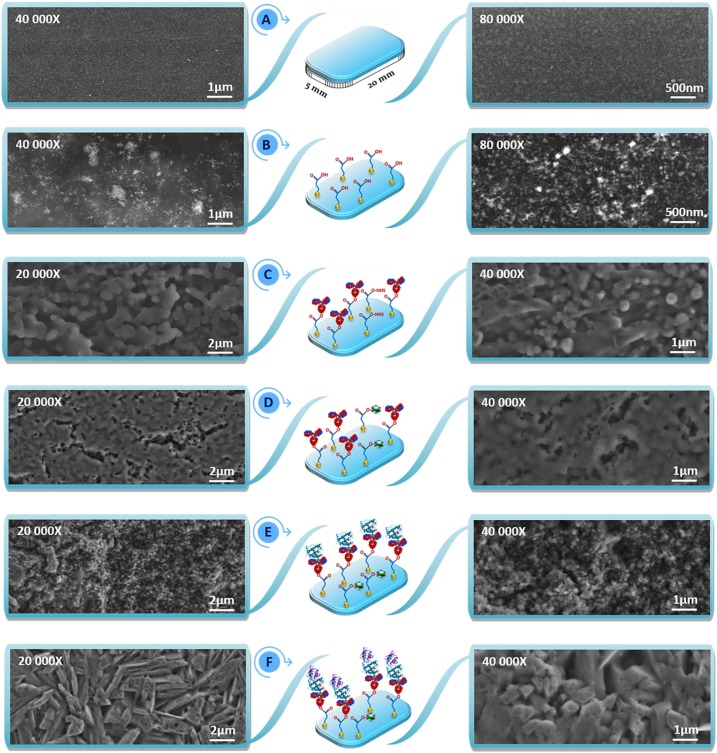
Small size of gold nanoparticles was investigated with S-TEM, UV–vis. and Raman spectroscopy. STEM has emerged as a suitable characterization technique for nanoparticles. In this study, two modes of S-TEM were utilized to characterize GNPs: bright field (BF) and dark field (DF) STEM. Gold nanoparticles was synthesized for morphological monitoring on 200 mesh copper grids with formvar and carbon supports. S-TEM analysis was performed to investigate to particle size of GNPs. Fig. 1 A shows that uniformly dispersed spherical DF (sub-series) and BF images (top-series) of gold nanoparticles. Spherical size diameter of GNPs was average 7–15 nanometers.
Fig. 1.
S-TEM image of GNPs (A). Raman spectra of GNPs (blue line), 11-MUA (red line) and GNPs@MUA (purple line) (B) UV–vis absorption spectrum of the GNPs and GNPs@MUA (Top Corner). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
In order to show the successfully synthesized GNPs and functionalized GNPs with MUA, different MUA amounts were utilized and optimum amount of MUA was selected by following RBD immunosensor signals. The UV–vis absorption spectrum of the GNPs and GNPs@MUA are illustrated in Fig. 1B-Top Corner. It can be observed that the typical absorbance of spherical GNPs appeared at 530 nm (blue line) [29]. After functionalization of the GNPs with MUA, a decline in intensity of characteristic absorbance was seen, and this decrease illustrated successful modification of GNPs through Au-S bond.
Also, GNPs@MUA were characterized with Raman spectroscopy (Fig. 1B). Unlike FT-IR analysis, Raman measurement can directly monitor molecules present in solution because the Raman scattering signal of water is very weak [30]. Raman spectroscopy also supplies similar data about the chemical structure of target molecule like FTIR spectroscopy. Raman spectra of GNPs (blue line), 11-MUA (red line) and GNPs@MUA (purple line) are illustrated in Fig. 1B. The Raman spectrum in 11-MUA presented the bands at 2550 and 2569 cm−1 which was assigned to S—H stretching modes in structure. This bands did not observe spectrum of GNPs@MUA [31]. This indicating that the MUA molecule was adsorbed on the gold nanoparticle by deprotonation of the sulfur atom as a thiolate [32]. Based on these Raman results, it assumed that the gold nanoparticles are covered by amounts of MUA molecules.
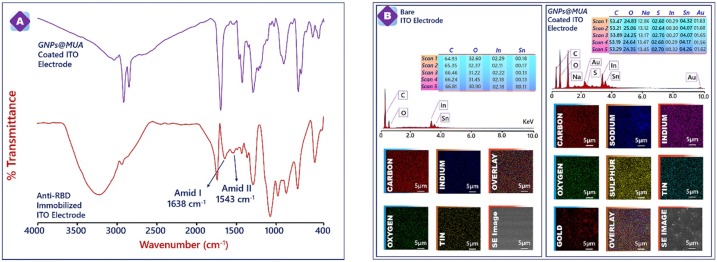
The chemical composition of GNPs@MUA modified electrode surface after and before with anti-RBD immobilization were characterized via FTIR spectroscopy. The FTIR spectra of GNPs@MUA modified electrode surface before (purple line) and after anti-RBD antibody immobilization (red line) are shown in Fig. 2 A, respectively. As for spectrum of GNPs@MUA modified electrode, characteristic peaks were observed at 2915 and 2849 cm−1 (aliphatic CH), 1695 cm— −1 carbonyl groups and 1429 cm−1 (CO—) [33]. As shown in FTIR spectrum of anti-RBD antibody immobilized electrode surface, a broad peak centered at ∼3250 cm-1 is attributed to stretching vibration of O—H, the low-intensity peaks at 1543 cm-1, 1638 cm-1 and sharp peak at 1280 cm-1 are ascribed to the stretching vibrations of amide I, amide II, and amide III, respectively [[34], [35], [36]]. Three main peaks in antibody were proved to presence of anti-RBD protein on the surface.
Fig. 2.
FTIR spectra of MUA capped GNPs and anti-RBD attached surface (A). EDX results of bare and GNPs@MUA covered electrode surface (B).
Furthermore, in order to confirm the presence of GNPs@MUA and the surface homogeneity of the ITO substrate, GNPs@MUA decorated electrode surface was also monitored with Scanning electron microscopy-energy dispersive X-ray analysis (SEM-EDX). The bare ITO electrode had oxygen (O), indium (In) and tin (Sn) elements and after GNPs@MUA fuctionalization, Au and S elements were present on the electrode surface. These elements originated from GNPs@MUA structure. Besides, five different places from ITO/GNPs@MUA electrode were investigated to test the homogeneity of the surface and the percentages of gold elements were similar. This result confirmed that a homogenous GNPs@MUA layer was successfully constructed.
3.2. Electrochemical characterizations of the suggested biosensor fabrication process
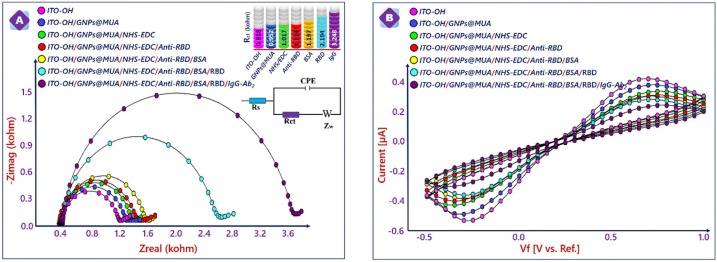
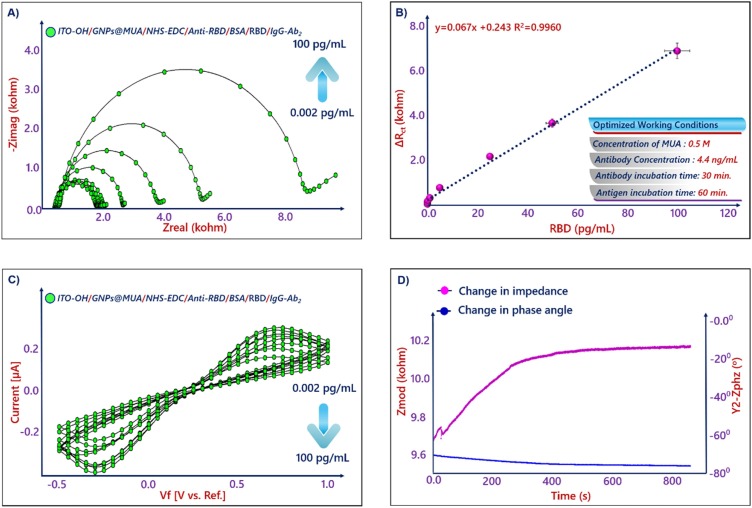
EIS and CV were selected for investigation of electrochemical process happening at the electrode/electrolyte solution interface during the stepwise assembly of the bioelectrode. Impedance alterations on the ITO electrode surface during modification procedure provided useful information about conductivity of formed layers on the electrode surface [27]. Fig. 3 A displays the EIS spectra presented as Nyquist plots obtained on different modified surfaces in a redox solution of 5.0 × 10−3 M [Fe(CN)6]3−/4− and 1 M KCl solution. The EIS signals were fitted by using a Randles equivalent circuit, which contained four elements: (i) Warburg impedance (w), (ii) ohmic resistance of the electrolyte (Rs), (iii) constant phase element (CPE) and (iv) electron transfer resistance (Rct) [37]. The Rct value of the electrode surface was found from diameters of high frequency semicircle. The impedance spectrum of the hydroxylated ITO surface had a small semicircle (Rct = 0.818 kΩ). The Rct of GNPs@MUA modified ITO electrode (0.952 kΩ) was significantly increased compared to hydroxylated ITO electrode. This increase illustrated that the assembled GNPs@MUA layer generated an obstruction to the electron transfer of the redox couple at electrode surface. The carboxyl groups present on the electrode surface was activated NHS-EDC chemistry. Because of electrostatic pushing between negative charged carboxyl groups and redox probe, an electrostatic obstacle, which hindered the ability of the redox couple to access the layer, was formed. Thus, the electron transfer rate decreased, and an increment was observed in electron transfer resistance (1.017 kΩ). When incubating the ITO/GNPs@MUA/NHS-EDC electrode in anti-RBD antibody solution for 1 h, a great increase in semicircle diameter (1.119 kΩ) was observed. The binding of anti-RBD antibodies on the GNPs@MUA decorated electrode surface formed electron transfer-blocking layer. The blocking of free active carboxyl groups with BSA solution increased the Rct value (1.197 kΩ). Afterwards, the Rct value increased to 2.194 kΩ, when the immunosensor was utilized to detect the RBD antigen. The binding between RBD antigen and RBD-antibody formed an additional layer. In the last step of fabrication procedure, IgG Ab2 was utilized to increase the immunosensor signal and a sandwich immunosensing system was constructed. The Rct was measured as 3.248 kΩ after IgG Ab2 immobilization.
Fig. 3.
EIS (E) and CVs (F) responses obtained after fabrication stages.
CVs of fabricated bioelectrodes were recorded in ferro-ferro solution to monitor variations in their electrochemical property at different stages of electrode modifications (Fig. 3B). The CV at the hydroxylated ITO electrode had well-defined redox peaks. After the ITO electrode was decorated with GNPs@MUA, the peak currents decreased dramatically in comparison with those of the hydroxylated ITO electrode. This decrease was originated from the slow electron transfer kinetics through GNPs@MUA SAMs. The activation of carboxyl groups with NHS-EDC crosslinking agent caused slow electron transfer kinetics through the negative charged carboxyl groups. The negatively charged carboxyl groups repulsed the negatively charged redox couple and promoted the electron transfer for the electrode surface and thus, peak currents of ITO/GNPs@MUA/NHS-EDC were less than ITO/GNPs@MUA. After binding of anti-RBD on the ITO surface, decreases were monitored in peak currents. These increases showed that the anti-RBD antibody layer promoted electron transfer. CV of ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA electrode had lower peak currents, which displayed that an insulating layer of BSA was generated. After incubating ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA electrode in RBD antigen solution, decreases were seen in peak currents because of insulating behavior of the protein molecules.
3.3. Morphological characterizations of the constructed bioelectrodes
SEM analyses were carried out to investigate the morphological variations on the working electrode surface obtained after immobilization stages of electrode construction (Fig. 4 ). Fig. 4A shows bare ITO electrode surface and the electrode surface was smooth and pure. The self-assembly of GNPs@MUA on the ITO electrode provided uniformly dispersed spherical GNPs and numerous clusters of GNPs@MUA (Fig. 4B). When anti-RBD antibodies attached on the MUA-capped GNPs modified electrode surface, the morphological image of electrode was changed, and the carboxyl groups of GNPs@MUA provided effective area for attachment of biomolecules. Anti-RBD immobilized surface had globular structures due to morphological shapes of antibodies (Fig. 4C). The SEM image of ITO/GNPs@MUA/NHS-EDC/BSA electrode illustrated that BSA biomolecules were immobilized directly on the free active carboxyl ends and therefore, the surface morphology of electrode was changed (Fig. 4D). The specific immune-interaction between anti-RBD and RBD caused variations in electrode surface. As seen Fig. 4E, RBD antigens were immobilized on the anti-RBD antibody attached electrode surface and the change on the electrode surface was evident of successful fabrication of the proposed biosensor. Lastly, with the immobilization of IgG Ab2, a sandwich system was produced, and this step changed the electrode surface morphology (Fig. 4F).
Fig. 4.
Morphological images obtained after RBD biosensing system fabrication procedure.
3.4. Effect of important parameters for RBD detection
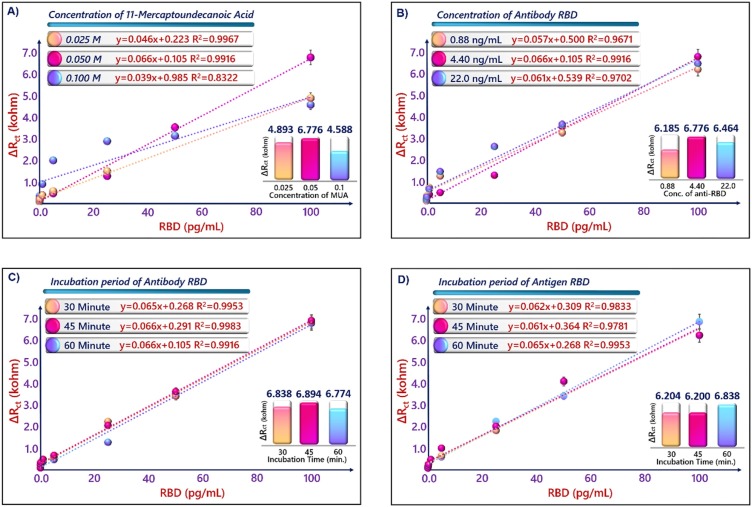
In order to analyze the effects of several parameters (concentrations of MUA-capped GNPs and anti-RBD antibodies, and incubation time of anti-RBD antibody and RBD antigen) on the capacity of immunosensor, different bioelectrodes were prepared and the signals of them were recorded. Firstly, the amount of GNPs@MUA was optimized, because the carboxyl ends present on the MUA provided attachment points for anti-RBD antibodies. Three different MUA concentrations (0.1 M, 0.05 M and 0.025 M) was utilized and different electrodes were prepared with GNPs@MUA. During the optimization of this parameter, the antibody amount, incubating times of antibody-antigen were constant. As seen Fig. 5 A, the high MUA concentration (0.1 M) did not form a smooth SAMs on the GNPs and thus, a low impedimetric signal was obtained. The low MUA level (0.025 M) usage resulted in a low impedimetric response because insufficient number of carboxyl groups were generated for the antibodies to bind. The immunosensor signal, which was measured after 0.05 GNPs@MUA usage had the highest response. Therefore, 0.05 M GNPs@MUA level was the optimum amount (Fig. 5A). Then, the effect of anti-RBD concentration on the RBD immunosensor signal was investigated. During the biosensor construction, different amounts of antibodies (0.88 ng mL−1, 4.4 ng mL−1 and 22 ng mL−1) were employed and maximum response was measured at 4. 4 ng mL−1. Therefore, this concentration was selected as the optimum (Fig. 5B). After optimization the amount of MUA and anti-RBD antibodies, the incubation times of anti-RBD and RBD were optimized. ITO/GNPs@MUA electrodes were incubated in anti-RBD solution (4.4 ng mL−1) for three times (30, 45 and 60 min). The obtained biosensor signals after incubation these durations were similar, and 30 min was sufficient for binding of anti-RBD antibody (Fig. 5C). Lastly, the RBD incubation time was optimized. ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA electrodes were incubated in RBD antigen solutions for three times (30, 45 and 60 min). As seen Fig. 5D, 30 and 45 min were not enough for immunoreaction and after 60-min incubation, the immunosensor had maximum signal (Fig. 5D).
Fig. 5.
Effects of experimental parameters.
3.5. Detection of RBD antigen with the proposed immunosensor
The specific bioreaction between anti-RBD antibody and RBD antigen formed an antibody-antigen immunocomplex. This protein layer created a kinetic barrier that perturbed the interfacial electron transfer at the immunoelectrode and electrolyte solution. Thus, Rct increases and also peak currents decreases were observed. Fig. 6 A and C display the Nyquist curves and CVs after incubation of ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA electrodes in increasing concentrations of target protein, respectively. The fitting Rct values are illustrated in Table SI-1. The calibration curve in Fig. 6B displayed that the impedimetric signal increased linearly with the increment in RBD antigen level in the detection range of 0.002−100 pg mL−1, the detection limit, the quantification limit, and the sensitivity were calculated as 0.577 fg mL−1 1.9 fg mL−1, and 0.284 kΩpg−1mLcm-2, respectively.
Fig. 6.
EIS (A) and CVs (C) of the RBD immunosensor towards different levels of RBD antigen, the calibration plot drawn as function of RBD levels of EIS response (B), SFI (D) results.
The kinetic behavior of the immunoreaction between anti-RBD and RBD proteins was appraised by following the variations in impedance and phase angle at a single frequency (SFI) in a nonfaradaic environment. The determination of the single frequency (44 Hz) was performed with the help of the Bode plot. The Bode plot give information concerning the kinetic behaviors formed on the electrode-electrolyte solution interface for various frequencies. The impedance of this system at 44 Hz is demonstrated in Fig. 4D. As seen in the Fig. 6D, the binding between RBD antigen and anti-RBD antibody is quite high for the first 400 s. The increment in impedance value was an evidence of specific biointeraction and the developed immunosensing system demonstrated the satisfactory selectivity for the determination of RBD antigen.
Table 1A displays the characteristic features of some reported RBD biosensors. The developed biosensors had high performance, low detection limit and wide linear range. As can be figured out in Table 1A, the suggested RBD sensor had a lower detection limit than other RBD sensing system stated in Table 1A. Furthermore, the developed biosensor was a simple and low-cost technique for determination of RBD antigen. Thus, it can be concluded that the constructed RBD immunosensor had a good performance and it was effective in measurement of RBD antigen.
Table 1.
Comparison of RBD sensing systems (A), results of nasal secretions by suggested RBD biosensor (B).
| Analysis method | Detection method | Detection Range (pg/mL) | Detection limit (pg/mL) | Reference |
|---|---|---|---|---|
| Silanized microfluidic device | Fluorescence | – | 1.6 × 105 | [38] |
| Co-functionalized TiO2 nanotubes | Electrochemical | 0.721−72.1 | 36 | [39] |
| Anti-S1 cell-based biosensor | Colorimetric | 10−2-106 | 1 | [40] |
| Paper based ePAD | Colorimetric | 103-106 | 11 × 105 | [41] |
| Lateral flow assay | Colorimetric | 5 × 103-5 × 105 | 106 | [42] |
| GNPs@MUA decorated ITO platform | Electrochemical | 0.002−100 | 0.577 | Present study |
| B) Sample | Added RBD (pg/mL) | Found by biosensor (pg/mL) | *SD and *CV (%) | Recovery (%) | Relative difference (%) |
|---|---|---|---|---|---|
| 1 | 0.10 | 0.12 | 0.01−14.43 | 100.47 | 0.47 |
| 1 | 0.10 | 0.10 | 100.07 | 0.07 | |
| 1 | 0.10 | 0.09 | 99.67 | −0.33 | |
| 2 | 0.25 | 0.24 | 0.01−6.28 | 99.63 | −0.37 |
| 2 | 0.25 | 0.22 | 99.25 | −0.75 | |
| 2 | 0.25 | 0.25 | 100.02 | 0.02 | |
| 3 | 1.00 | 1.06 | 1.14−7.13 | 101.41 | 1.41 |
| 3 | 1.00 | 1.23 | 104.95 | 4.95 | |
| 3 | 1.00 | 1.14 | 103.02 | 3.02 | |
| 4 | 25.00 | 23.36 | 0.34−1.43 | 94.27 | −5.73 |
| 4 | 25.00 | 23.52 | 94.84 | −5.16 | |
| 4 | 25.00 | 24.01 | 96.54 | −3.46 | |
| 5 | 50.00 | 51.03 | 0.48−0.96 | 101.92 | 1.92 |
| 5 | 50.00 | 50.16 | 100.29 | 0.29 | |
| 5 | 50.00 | 50.96 | 101.78 | 1.78 |
*SD: Standard Deviation, *CV: Coefficient Variation.
3.6. Repeatability, reproducibility, long-time stability, specificity and regeneration of the biosensor
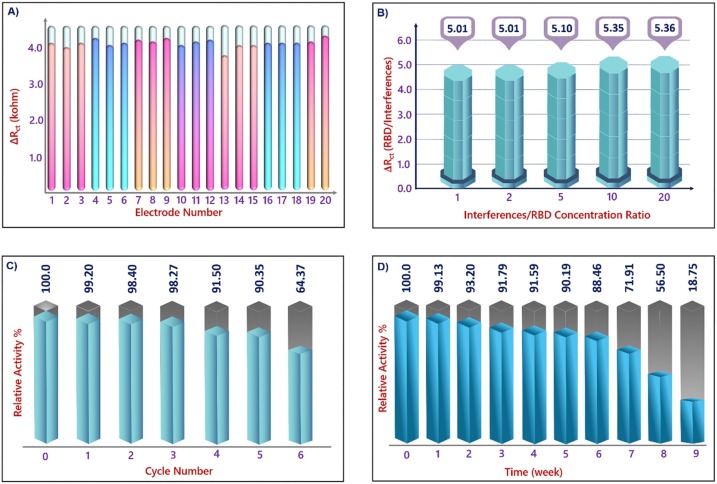
Repeatability and reproducibility are important parameters which provided information about precision of the immunosensor. The repeatability of this sensor was tested by impedimetric measurement of 2.5 pg mL−1 RBD antigen with ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA bioelectrodes and a low relative standard deviation (RSD 6.47 %) was obtained (Fig. 7 A). The reproducibility of the suggested sensor was studied by preparing 10 different biosensors. These biosensors were prepared under identical conditions and used for RBD antigen detection. The RSD of the reproducibility test was found as 1.49 % (Fig. SI-1).
Fig. 7.
Repeatability (A), specificity (B), reusability (C) and long-time stability (D) results.
Considering in terms of application, an immunosensor should be not only sensitive but also selective [43]. To measure the specificity of the fabricated RBD biosensor, control experiments were carried out using nucleocapsid and other biological markers found in nasal secretions (interleukin 1β, interleukin 8 and tumor necrosis factor α) [44]. ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA electrodes were incubated 60 min in different PBS solutions containing 5 different levels of interferents. As seen in Fig. 7B, incubation with interferents did not generate important variations in ΔRct ratios. These obtained results proved that the suggested immunosensor had excellent specificity to the RBD antigen.
Although ITO is cheap material, being reusable is very important in practice. Therefore, the reusability feature of the prepared electrode was investigated. First of all, the prepared ITO/GNPs@MUA/NHS-EDC/anti-RBD/BSA electrode was incubated in RBD solution and then, the impedimetric signal of this prepared electrode was recorded. After measurement, the electrode was regenerated with acid solution (10 mM HCl) and then, it was dipped in RBD antigen solution. After incubation in RBD solution, the impedimetric signal was re-measured. This treatment contained until the electrode was damaged. As shown in Fig. 7C, the prepared electrode could be regenerated 6 times with good activity.
The long-term stability of the bioelectrode was also studied. Firstly, 10 different bioelectrodes (ITO/GNPs@MUA/NHS-EDC/BSA) were prepared and stored at 4 °C. At each week one of them were used for RBD antigen (10 pg mL−1) detection and this measurement procedure was contained ten weeks. After 5 weeks, there was no obvious variation in impedimetric response and the measured EIS responses retained about 71.91 % of its original signals after seven weeks, illustrating acceptable stability (Fig. 7D). The high stability, repeatability and reproducibility implied the excellent biocompatibility of GNPs@MUA interface. The role of covalent attachment between hydroxylated ITO electrode and GNPs@MUA in increment the stability of the bioelectrode should not be-ignored.
3.7. Applications of the RBD immunosensor to nasal secretions
In order to evaluate the applicability of the suggested immunosensor for RBD analysis, artificial nasal secretions were tested. Firstly, artificial nasal secretions were prepared by using water, immunoglobulin G, immunoglobulin M and inorganic salts. Five different concentrations of RBD antigen were added these prepared solutions and the determinations of RBD concentrations were carried out 3 times. In these experiments polyclonal IgG for SARS-CoV-2 spike RBD was used as a secondary antibodies as before. As illustrated in Table 1B, this method had good recovery rates from 94.27%–104.95 % and this result indicated that the developed immunosensor could be used to detect RBD antigen with low concentrations.
4. Conclusion
In this study, a simple and different modification strategy was utilized for the fabrication of a RBD immunosensor to achieve an efficient biosensing system. The self-assembled GNPs@MUA on the electrode surface provided a biocompatible environment and a large surface area for antibody binding and improved the electrode surface conductivity. The RBD antigen specific anti-RBD antibodies attached covalently with their amino groups to carboxyl groups of MUA. This covalent linkage resulted in effective binding and increased the bioactivity of antibodies. By this way, the fabricated biosensor had a selective behavior to RBD antigens and at the same time, it maintained this bioactivity for a long time. The developed RBD biosensor had a double functionality, containing specific biorecognition and signal amplification for detection of RBD. This sandwich type impedance based immunosensor achieved a linear detection range from 0.002 pg mL−1 to 100 pg mL-1 and a detection limit of 0.58 fg mL−1. Furthermore, in clinical diagnosis, the biosensing strategy was successfully utilized for detection of RBD protein in artificial nasal secretions. The obtained results demonstrated that this label-free immunosensor illustrated the notable advantages such as highly sensitive, economical, selective and simple. Consequently, the recommended RBD immunosensor approach provided a feasible and sensitive method for detecting spike RBD protein in the early diagnosis of COVID 19 disease.
CRediT authorship contribution statement
Elif Burcu Aydın: Biosensor fabrication, all electrochemical measurements, and writing. Muhammed Aydın: Gold nanoparticle synthesis, electrochemical measurements, SEM, AFM, Raman, and FT-IR experimets, and writing. Mustafa Kemal Sezgintürk: Conceptualization, Methodology, Supervision, Validation, Writing- Reviewing and Editing.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The authors would like to acknowledge the financial support from The Scientific and Technological Research Council of Çanakkale Onsekiz Mart University, (FIA-2020-3304).
Biographies
Elif Burcu AYDIN is an assoc. professor in biochemistry at Scientific and Technological Research Center, Tekirdağ Namık Kemal University. She received his Ph.D. degree in biochemistry from Tekirdağ Namık Kemal University in 2018. Her current research focuses on the fabrication and application of electrochemical immunosensors for cancer biomarkers detection.
Muhammet AYDIN is an assoc. professor in chemistry at Scientific and Technological Research Center, Tekirdağ Namık Kemal University. He received his Ph.D. degree polymer chemistry from Gebze Technical University in 2017. His work is focused on the polymer synthesis for fabrication of different kind of immunosensors.
Mustafa Kemal Sezgintürk is a Professor in the Bioengineering Department at the University of Onsekiz Mart where he has been a faculty member since 2016. Sezgintürk completed his Ph D. at Biochemistry Department of Ege University in 2007, his M.Sc. at Biochemistry Department of Ege Univesity in 2002 and his undergraduate from Chemistry Department of Ege University in 1998. His research interests lie in the area of biochemistry, biosensors, cancer biomarkers, electrochemical impedance spectroscopy, enzymology, protein purification and characterization. Professor Sezgintürk coordinated several scientific projects such as TÜBİTAK (The Scientific and Technological Research Council of Turkey), BAP (Projects of Scientific Investigation), so on and so forth. In 2016, He has won the Awards for Outstanding Young Scientist (Turkish Academy of Sciences-GEBİP), which is one of the prestigious scientific award.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2021.130355.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji T., Liu Z., Wang G., Guo X., Lai C., Chen H., et al. Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushik A.K., Dhau J.S., Gohel H., Mishra Y.K., Kateb B., Kim N.-Y., et al. Electrochemical SARS-CoV-2 sensing at point-of-care and artificial intelligence for intelligent COVID-19 management. ACS Appl. Bio Mater. 2020;3:7306–7325. doi: 10.1021/acsabm.0c01004. [DOI] [PubMed] [Google Scholar]
- 5.Sheikhzadeh E., Eissa S., Ismail A., Zourob M. Diagnostic techniques for COVID-19 and new developments. Talanta. 2020 doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Zhang G., Liu L., Li Z. Emerging biosensing technologies for improved diagnostics of COVID-19 and future pandemics. Talanta. 2020 doi: 10.1016/j.talanta.2020.121986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu L., Li D., Ramadan S., Li Y., Klein N. Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steiner D.J., Cognetti J.S., Luta E.P., Klose A.M., Bucukovski J., Bryan M.R., et al. Array-based analysis of SARS-CoV-2, other coronaviruses, and influenza antibodies in convalescent COVID-19 patients. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laghrib F., Saqrane S., El Bouabi Y., Farahi A., Bakasse M., Lahrich S., et al. Current progress on COVID-19 related to biosensing technologies: new opportunity for detection and monitoring of viruses. Microchem. J. 2020 doi: 10.1016/j.microc.2020.105606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhalla N., Pan Y., Yang Z., Payam A.F. Opportunities and challenges for biosensors and nanoscale analytical tools for pandemics: COVID-19. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R., Mukherjee M.D., Sumana G., Gupta R.K., Sood S., Malhotra B. Biosensors for pathogen detection: a smart approach towards clinical diagnosis. Sens. Actuators B. 2014;197:385–404. [Google Scholar]
- 13.Bahadır E.B., Sezgintürk M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015;478:107–120. doi: 10.1016/j.ab.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Aydın E.B., Aydın M., Sezgintürk M.K. Selective and ultrasensitive electrochemical immunosensing of NSE cancer biomarker in human serum using epoxy-substituted poly (pyrrole) polymer modified disposable ITO electrode. Sens. Actuators B. 2020;306 [Google Scholar]
- 15.Ansari S., Karimi M. Novel developments and trends of analytical methods for drug analysis in biological and environmental samples by molecularly imprinted polymers. TrAC, Trends Anal. Chem. 2017;89:146–162. [Google Scholar]
- 16.Mishra S.K., Srivastava A.K., Kumar D. Bio-functionalized Pt nanoparticles based electrochemical impedance immunosensor for human cardiac myoglobin. RSC Adv. 2014;4:21267–21276. [Google Scholar]
- 17.Aydın M., Aydın E.B., Sezgintürk M.K. Electrochemical immunosensor for CDH22 biomarker based on benzaldehyde substituted poly (phosphazene) modified disposable ITO electrode: a new fabrication strategy for biosensors. Biosens. Bioelectron. 2019;126:230–239. doi: 10.1016/j.bios.2018.10.051. [DOI] [PubMed] [Google Scholar]
- 18.Bogomolova A., Komarova E., Reber K., Gerasimov T., Yavuz O., Bhatt S., et al. Challenges of electrochemical impedance spectroscopy in protein biosensing. Anal. Chem. 2009;81:3944–3949. doi: 10.1021/ac9002358. [DOI] [PubMed] [Google Scholar]
- 19.Zamfir L.-G., Puiu M., Bala C. Advances in electrochemical impedance spectroscopy detection of endocrine disruptors. Sensors. 2020;20:6443. doi: 10.3390/s20226443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurbanoglu S., Ozkan S.A., Merkoci A. Nanomaterials-based enzyme electrochemical biosensors operating through inhibition for biosensing applications. Biosens. Bioelectron. 2017;89:886–898. doi: 10.1016/j.bios.2016.09.102. [DOI] [PubMed] [Google Scholar]
- 21.Arduini F., Micheli L., Moscone D., Palleschi G., Piermarini S., Ricci F., et al. Electrochemical biosensors based on nanomodified screen-printed electrodes: recent applications in clinical analysis. TrAC Trends Anal. Chem. 2016;79:114–126. [Google Scholar]
- 22.Atta N.F., Galal A., Ekram H. A novel sensor of cysteine self-assembled monolayers over gold nanoparticles for the selective determination of epinephrine in presence of sodium dodecyl sulfate. Analyst. 2012;137:2658–2668. doi: 10.1039/c2an16210e. [DOI] [PubMed] [Google Scholar]
- 23.Saxena U., Chakraborty M., Goswami P. Covalent immobilization of cholesterol oxidase on self-assembled gold nanoparticles for highly sensitive amperometric detection of cholesterol in real samples. Biosens. Bioelectron. 2011;26:3037–3043. doi: 10.1016/j.bios.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Charan S., Sanjiv K., Singh N., Chien F.-C., Chen Y.-F., Nergui N.N., et al. Development of chitosan oligosaccharide-modified gold nanorods for in vivo targeted delivery and noninvasive imaging by NIR irradiation. Bioconjugate Chem. 2012;23:2173–2182. doi: 10.1021/bc3001276. [DOI] [PubMed] [Google Scholar]
- 25.Qin A., Fu L.T., Wong J.K., Chau L.Y., Yip S.P., Lee T.M. Precipitation of PEG/carboxyl-modified gold nanoparticles with magnesium pyrophosphate: a new platform for real-time monitoring of loop-mediated isothermal amplification. ACS Appl. Mater. Interfaces. 2017;9:10472–10480. doi: 10.1021/acsami.7b00046. [DOI] [PubMed] [Google Scholar]
- 26.De Souza C.D., Nogueira B.R., Rostelato M.E.C. Review of the methodologies used in the synthesis gold nanoparticles by chemical reduction. J. Alloys Compd. 2019;798:714–740. [Google Scholar]
- 27.Pashai E., Darzi G.N., Jahanshahi M., Yazdian F., Rahimnejad M. An electrochemical nitric oxide biosensor based on immobilized cytochrome c on a chitosan-gold nanocomposite modified gold electrode. Int. J. Biol. Macromol. 2018;108:250–258. doi: 10.1016/j.ijbiomac.2017.11.157. [DOI] [PubMed] [Google Scholar]
- 28.Ramvikas M., Arumugam M., Chakrabarti S., Jaganathan K. Elsevier; 2017. Nasal Vaccine Delivery, Micro and Nanotechnology in Vaccine Development; pp. 279–301. [Google Scholar]
- 29.Kesik M., Kanik F.E., Hızalan G., Kozanoglu D., Esenturk E.N., Timur S., et al. A functional immobilization matrix based on a conducting polymer and functionalized gold nanoparticles: synthesis and its application as an amperometric glucose biosensor. Polymer. 2013;54:4463–4471. [Google Scholar]
- 30.Carden A., Morris M.D. Application of vibrational spectroscopy to the study of mineralized tissues. J. Biomed. Opt. 2000;5:259–268. doi: 10.1117/1.429994. [DOI] [PubMed] [Google Scholar]
- 31.Tripathy S.K., Yu Y.-T. Spectroscopic investigation of S–Ag interaction in ω-mercaptoundecanoic acid capped silver nanoparticles. Spectrochim. Acta Part A. 2009;72:841–844. doi: 10.1016/j.saa.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Love J.C., Estroff L.A., Kriebel J.K., Nuzzo R.G., Whitesides G.M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005;105:1103–1170. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 33.Niu W.-J., Shan D., Zhu R.-H., Deng S.-Y., Cosnier S., Zhang X.-J. Dumbbell-shaped carbon quantum dots/AuNCs nanohybrid as an efficient ratiometric fluorescent probe for sensing cadmium (II) ions and L-ascorbic acid. Carbon. 2016;96:1034–1042. [Google Scholar]
- 34.Miyazawa T., Shimanouchi T., Mizushima Si. Characteristic infrared bands of monosubstituted amides. J. Chem. Phys. 1956;24:408–418. [Google Scholar]
- 35.Miyazawa T., Shimanouchi T., Mizushima Si. Normal vibrations of N‐methylacetamide. J. Chem. Phys. 1958;29:611–616. [Google Scholar]
- 36.Khanmohammadi M., Garmarudi A.B. Infrared spectroscopy provides a green analytical chemistry tool for direct diagnosis of cancer. TrAC Trends Anal. Chem. 2011;30:864–874. [Google Scholar]
- 37.Saxena R., Srivastava S. An insight into impedimetric immunosensor and its electrical equivalent circuit. Sens. Actuators B. 2019;297 [Google Scholar]
- 38.Rodriguez-Moncayo R., Cedillo-Alcantar D.F., Guevara-Pantoja P.E., Chavez-Pineda O.G., Hernandez-Ortiz J.A., Amador-Hernandez J.U., et al. A high-throughput multiplexed microfluidic device for COVID-19 serology assays. Lab Chip. 2021;21:93–104. doi: 10.1039/d0lc01068e. [DOI] [PubMed] [Google Scholar]
- 39.Vadlamani B.S., Uppal T., Verma S.C., Misra M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors. 2020;20:5871. doi: 10.3390/s20205871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., et al. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishra R.K., Hayat A., Catanante G., Ocaña C., Marty J.-L. A label free aptasensor for Ochratoxin A detection in cocoa beans: an application to chocolate industries. Anal. Chim. Acta. 2015;889:106–112. doi: 10.1016/j.aca.2015.06.052. [DOI] [PubMed] [Google Scholar]
- 44.Riechelmann H., Deutschle T., Friemel E., Gross H., Bachem M. Biological markers in nasal secretions. Eur. Respir. J. 2003;21:600–605. doi: 10.1183/09031936.03.00072003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.