Abstract
An analytic evaluation of the peak time of a disease allows for the installment of effective epidemic precautions. Recently, an explicit analytic, approximate expression (MT) for the peak time of the fraction of infected persons during an outbreak within the susceptible–infectious–recovered/removed (SIR) model had been presented and discussed (Turkyilmazoglu, 2021). There are three existing approximate solutions (SK-I, SK-II, and CG) of the semi-time SIR model in its reduced formulation that allow one to come up with different explicit expressions for the peak time of the infected compartment (Schlickeiser and Kröger, 2021; Carvalho and Gonçalves, 2021). Here we compare the four expressions for any choice of SIR model parameters and find that SK-I, SK-II and CG are more accurate than MT as long as the amount of population to which the SIR model is applied exceeds hundred by far (countries, ss, cities). For small populations with less than hundreds of individuals (families, small towns), however, the approximant MT outperforms the other approximants. To be able to compare the various approaches, we clarify the equivalence between the four-parametric dimensional SIR equations and their two-dimensional dimensionless analogue. Using Covid-19 data from various countries and sources we identify the relevant regime within the parameter space of the SIR model.
Keywords: Epidemic, SIR model, Peak thresholds, Peak time, COVID-19
1. Introduction
The temporal evolution of COVID-19 (or SARS-CoV-2) pandemic waves has been successfully described, discussed, and forecasted by the mathematical susceptible–infectious–recovered/ removed SIR model [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17] while the model itself had been developed nearly a century ago [18], [19], [20], [21], [22]. There are two noticeable quantities that indicate the occurrence of new pandemic waves: the fraction of infected persons at time , and the fraction of newly infected population per day , where denotes the infection rate, and the fraction of susceptible population. Both indicators and in a pandemic wave first increase with time, undergo a maximum and drop at late times. The two peak times are slightly different. While the peak time in is the one usually reported in the media on the basis of reported number of newly infected persons, the peak time in is the one that determines the peak time of required clinical resources. While an analytic approximant for the usually measurable peak time in exists for both the all-time [18], [23] and semi-time SIR model [19], [20], [21], [24], it is the purpose of this communication to clarify how the several existing approximants for the peak time in compare with each other.
To this end we have collected known approximants for the peak time of the infected compartment in the literature, including the one very recently presented in this journal. We here use “approximant” for an approximate analytic expression whose Taylor expansion about a certain point is not required to exactly coincide with the Taylor expansion of the exact solution about the same point, while such point or points may exist for all approximations to be analyzed. In a first step we are going to explain and prove how the dimensionless and dimensional SIR models are interrelated. The correspondence allows us to compare the existing approximants, as they had been obtained using different notation. We then proceed using a unique language and notation and present all existing approximants without leaving out any single detail in their final expressions. We are not going to repeat all the calculations that lead to these approximants in the several papers we are going to quote, but we are collecting all necessary details in appendices to make this contribution self-contained. The goal of the present study is to find out and clarify, as a service to the readers of Physica D, if the accuracy of the existing approximants supersedes the one presented to them recently by Turkyilmazoglu [25], or not.
There are various papers [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33] that aimed at deriving approximate analytic expressions for various quantities that appear in the all-time or semi-time SIR models [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [34], [35]. And there are several variants [36], [37], [38], [39], [40], [41], [42], [43] of the SIR model, including stochastic variants [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57] or variants that account for vaccination [58], [59], [60], [61], [62] or pulse vaccination [63], [64], [65], [66]. For the semi-time SIR Heng and Althaus [26] provided an analytic approximant for the population fraction of susceptible and infected persons but only for times prior peak time, and they could not derive an approximant for any of the peak times. Harko et al. [67] were able to express time of the semi-time SIR model in terms of an integral, which can only be evaluated numerically, but did not derive an integral or analytic expressions for peak times. Bidari et al. [27] focused on deriving implicit equations for final sizes of compartments and approximate solutions to such equations, but did not obtain an expression for a peak time. An analytic approximate inverse solution of the semi-time SIR model with special initial condition of initially vanishing population fraction of recovered/removed persons was presented by Carvalho and Gonçalves [28]. An explicit series solution of SIR and SIS epidemic models was obtained by making use of the homotopy analysis method by Khan et al. [30]. This approach allows one to study convergence properties of the solution, but in practice, it is inefficient to study the solution of the SIR model using an infinite series. Moreover, this approach does not provide an analytic expression for a peak time. Within the same spirit Barlow and Weinstein [33] derived an accurate closed-form infinite series solution of the SIR model that involves a Vandermonde matrix, whose inversion is explicitly known; however, they do not provide a method for estimating peak time.
In [25] the time -dependent SIR model for population fractions (susceptible), (infected), and (removed/recovered) had been written as follows
| (1) |
subject to the general arbitrary (semi-time) initial conditions
| (2) |
and where and are the assumed stationary infection and recovery rates of population fractions. The initial condition for follows from , because the classical SIR model has only three compartments, and any of the members of the population belongs to one of the three compartments at any time. Allowing for arbitrary initial conditions and restricting the model to future times is known as semi-time SIR model [19], [20], [21], [24]. For the all-time SIR model, and are interrelated [18], [23]. Here we consider the semi-time SIR model for which an analytic expression for the peak time of the infected compartment, defined by
| (3) |
had been derived in [25]. It will be reproduced in Eq. (14). In its classical form Eq. (1) with (2) the dimensional SIR model has four parameters, the rates and , and the initial fractions and .
On the other hand, analytic approximants for the solution of the reduced dimensionless semi-time SIR model are available from [24] and [28]. While the reduced model has only three parameters, it is still equivalent with the original SIR model (the proof of equivalence will be given in the next section). Moreover, one of the three parameters is used to make the time dimensionless, so that one is left with essentially only two parameters and . These parameters appear in the reduced semi-time SIR model [24] as follows
| (4) |
with initial conditions
| (5) |
Here in general a dimensionless reduced time defined by for an arbitrary (including periodic) time-dependent infection rate , but because as in [25] we consider here a stationary infection rate, is simply proportional to time, with a constant rate that is specified in Eq. (7). Moreover, , , and are fractions of the initially unrecovered population. This implies as in [24]. We recall, that the quantities , , and are fractions with respect to the total initial population, including the recovered compartment, and therefore , , and .
Yet another version of a reduced semi-time SIR model was investigated in [28]. They introduced to write
| (6) |
subject to unchanged initial conditions (5). It is important to note here that in the original work Carvalho and Gonçalves [28] formulated their equations in terms of the fractions , , and , did not allow for non-vanishing , and mentioned , which is a correct assignment but only for this special case of . We are here extending their work to allow for non-vanishing so that also has to be revised, as shown next. This extension will allow one to compare all available approximants for arbitrary initial conditions and .
2. Analytical approximants
Starting from the dimensional SIR model with four parameters, the three parameters of the reduced formulation are given by
| (7) |
In turn, for given , , and , the parameters of the dimensional model are given upon direct inversion of Eqs. (7)
| (8) |
just highlighting the fact, that must drop out. Moreover, any characteristic real time of the dimensional SIR model is related to the corresponding reduced time or also as
| (9) |
where the dimensionless times and depend on two parameters only: the inverse basic reproduction number and the initial fraction of infected population among the non-recovered population, .
If we wish to test an approximate solution of the SIR model for arbitrary choices of parameters, or if we want to compare the quality of different approximants as we are going to do here, it is hence sufficient to perform the test in the 2-dimensional parameter space built by and . While by construction, the is semipositive in general. However, a peak time in occurs at positive times only if the following inequality holds
| (10) |
This inequality follows from [24], as the condition (3) for the peak converts with the help of (4) into
| (11) |
within the reduced model, because , and because monotonically decreases with increasing according to Eq. (4).
We have to still prove the equivalence between dimensional and dimensionless forms of the SIR model. To this end it is sufficient to prove Eq. (7), or the equivalent Eq. (8). Inserting and as well as Eq. (8) into Eq. (1) gives
| (12) |
confirming the equation of change for in Eq. (4), as well as
| (13) |
confirming the equation of change for in Eq. (4), which is obvious, if we divide both sides of the Eqs. (12), (13) by . The initial conditions are also equivalent, as and and .
2.1. MT approximant
The MT approximant by Turkyilmazoglu [25] for the peak time of the infected compartment had been formulated using the dimensional SIR formulation (1) with 4 parameters. Using our replacement rule (8), it receives the form of Eq. (9) with the dimensionless peak time (see Appendix A for details)
| (14) |
where - and -dependent coefficients are given by
| (15) |
Note that the term under the square root is positive for all and . For the special case of , where the denominator of vanishes, the expression can still be evaluated using l’Hopital’s rule, i.e., .
2.2. SK approximants SK-I and SK-II
The Schlickeiser & Kröger (SK) approximants for the peak time of the infected compartment, starting from the reduced SIR model Eqs. (4) had not been explicitly written down in their work [24], where they developed an approximate solution to the whole time-dependency of all SIR quantities, but they can be read off from the provided approximate analytic solution of the reduced SIR model, using Eq. (11). To be more specific, one can readily solve with from Eq. (71) of [24] for (for details see Appendix B). In the quoted work, denotes the cumulative fraction of infected persons, thus , and applies within the regime of reduced time where the peak time actually occurs. The resulting time is the reduced peak time (Appendix B)
| (16) |
with
| (17) |
in terms of a number of quantities characterizing the solution, that are all expressed in terms of and . To be specific, one has [24]
| (18) |
where and are the principal and non-principal solutions of Lambert’s equation [23], [68]. They are both available like inverse trigonometric functions or elliptic integrals in common software packages (python, Mathematica, matlab, eventually also excel). For the so-called SK-I approximant, the remaining two parameters and left to be specified are given by (Appendix B)
| (19) |
while the SK-II approximant is defined by
| (20) |
We are not going to interpret all these quantities here, but it may be useful to mention that is the finally infected population fraction, and the maximum dimensional rate of newly infected persons ( is the maximum reduced rate), that occurs at a time that differs from the reduced peak time in Eq. (16) by the two terms.
2.3. CG approximant
Carvalho and Goncalves [28] (CG) obtained, starting from the reduced SIR model (6) with the help of , for the reduced peak time in the approximate
| (21) |
with coefficients given by
| (22) |
and where is the solution of the nonlinear equation . From Schlickeiser and Kröger [24] we know that the nonlinear equation for that remained unsolved in [28] is solved using Lambert’s principal function [23], [68] as follows
| (23) |
As the reduced times and are related by according to Eq. (9), with according to Eq. (7), the reduced peak time for comparison with the remaining approximants is
| (24) |
where is taken from Eq. (21).
2.4. Results and discussion
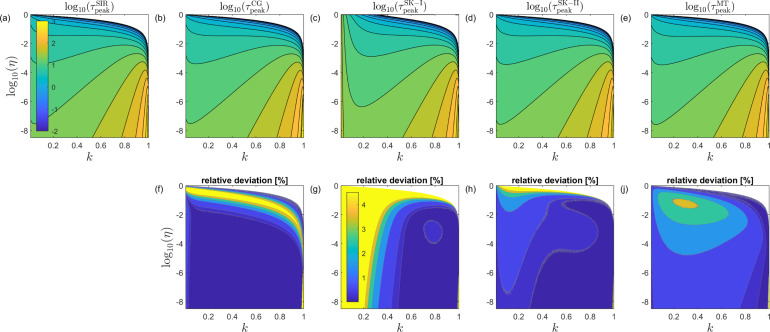
The exact peak time of the infected compartment calculated from the numerical solution of the SIR equations (4) is shown in Fig. 1a over basically the whole admissible (horizontal axis) and (vertical axis) range. Note that we use a semilogarithmic axis and a coloring scheme that reflects to appreciate all details, and that the figure is exactly reproduced if we solve Eqs. (1) or (6) numerically instead. The advantage of (4) or (6) over (1) is, that they have no redundant parameters. The corresponding performance of the four approximants CG, SK-I, SK-II, and MT is shown in Figs. 1b–e, while the relative deviation between approximants and exact solution is given by Fig. 1f–j. In each panel, the two variables of the dimensionless SIR model are thus varied, so that every panel shows the behavior of the peak time over the whole domain of SIR model parameters. While (a) shows the exact result (as a reference), (b)–(e) show the peak time for the four approximate expressions presented in this work. The remaining panels (f)–(j) show the same data (b)–(e) in a different fashion. Shown in (f)–(j) is the relative deviation between the approximate expression and the exact result.
Fig. 1.
Upper row: Decadic logarithm of the reduced peak time of the infected compartment versus and . (a) exact numerical solution of the SIR model, approximants (b) given by Eq. (14), (c) given by Eq. (19), (d) given by Eq. (20), and (e) given by Eq. (24). The dimensional peak time is obtained from upon dividing by the rate . A peak time exists only within the regime . The corresponding relative deviations between approximant and exact solution are shown in the 2nd row (f)–(j). While the performance of the MT approximant is more accurate than 4.3% over the shown --domain, the SK and CG approximants perform better than MT except within the regime of relatively large close to . This is made more precise in Fig. 2. The color bar for all panels of the first row is shown in (a), the single color bar for the 2nd row is shown in (g). The latter goes from blue (high quality, deviation from the exact solution) to yellow (low quality, deviation).
While the performance of the MT approximant is rather accurate over the whole --domain, the SK and CG approximants perform better than MT except within the regime of relatively large close to (in the neighborhood of the upper bound of the colored region, where it transits to white in Fig. 1). This is made more precise later below.
To quantify the performance of the approximants at very small (at the bottom of Fig. 1d–e), let us mention the following feature,
| (25) |
Furthermore, both approximants share the following feature at small
| (26) |
At the upper bound the MT approximant correctly vanishes, while the SK approximant does not vanish exactly for all . This feature causes the SK approximant to become poor in the vicinity of (see the yellow regime in Fig. 1g–h).
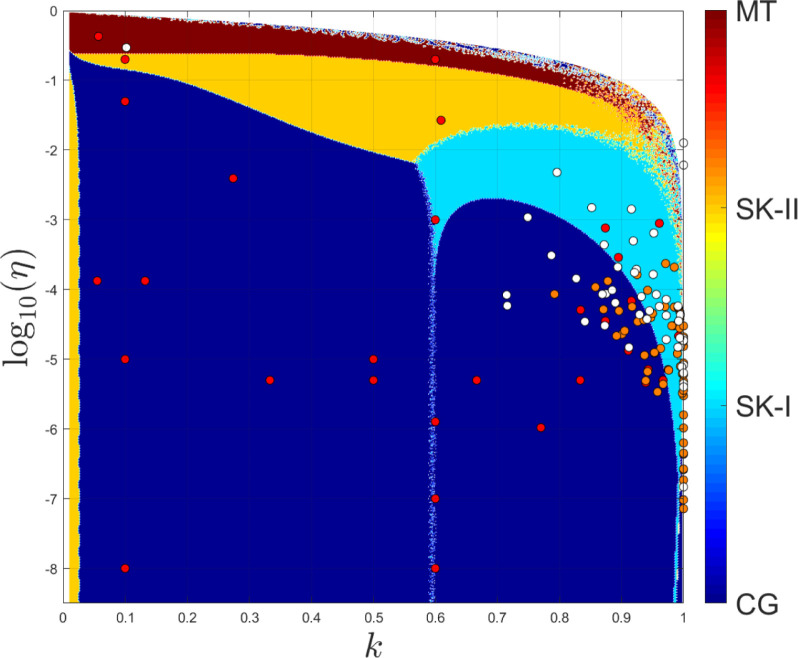
To summarize, Fig. 2 shows the regimes of superior performance of the four approximants. As long as exceeds a critical ,
| (27) |
the MT approximant is more accurate than the other three.
Fig. 2.
Which approximant to use for the peak time of the infected compartment. Within the dark red region at large corresponding to the MT approximant is more accurate than the other approximants. Within the orange area, SK-II performs best, within the light blue and dark blue the SK-I and CG, respectively, exhibit the highest accuracy. There is no peak time (peak lies in the past) within the white area on top, whose border is given by . Red filled circles mark the examples collected in Table 1, Table 2, taken from [25]. The uppermost red circle in the top left of this graph (within the dark red) corresponds to the example quoted by Harko et al. [67] for a population with persons. Orange and white circles correspond to pairs obtained in [17] by analyzing the first and second Covid-19 wave on 2021-04-13 in 60 different countries. Some of them are explicitly listed in Table 1, Table 2 as well. Most of them reside either in the light blue (SK-I) or dark blue (CG) regions. The relative deviation between the approximants and the exact solution was shown in Fig. 1.
For large populations , the fraction of the initially infected compartment is typically of the order of and small compared with the fraction of the initially susceptible compartment, thus is of the order of as well and the inequality (27) holds for , i.e., all not close to unity. Under such circumstances, the other approximants, all involving Lambert’s function, may be used.
For small populations, on the other hand, where is below hundred, say, the initially infected compartment may be a considerable fraction of the whole population. Such as for the example considered by Harko et al. [67], Batiha and Batiha [69]. Here, , , , , and had been considered. This translates into , , and with the help of Eq. (7). The inequality (27) is thus not fulfilled, and the MT approximant should be used.
To get an impression about the relevant regions in – space, Fig. 2 shows in addition many of the published artificial and real data points collected in Table 1, Table 2.
Table 1.
Examples from published literature. The table lists the SIR parameter , , and , as well as their reduced counterparts , , and . Cases for which are marked by (*). Units are omitted from , and , as is common practice. Time is then predicted in the same units (days).
| Ref. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| [67] | 45 | 0.45 | 0.02 | 0.44444 | 0.33333 | 0.057143 | 0.42857 | 0.35 | 1.7143 (*) |
| [70] | 763 | 1.66 | 0.45455 | 0.99476 | 0.0039318 | 0.27418 | 0.003937 | 1.6578 | 0.016273 |
| [71] | 2.87 | 0.91776 | 0.70681 | 1.0442 | 0.77015 | 1.0442 | 0.91776 | 1.3629 | |
| [25] | – | 4.6291 | 2.82 | 0.0268 | 0.60919 | 0.0268 | 4.6291 | 0.20573 | |
| [25] | – | 0.5 | 0.3 | 1.27 | 0.6 | 1.27 | 0.5 | 9.525 | |
| [72] | – | 10 | 1 | 0.05 | 0.1 | 0.05 | 10 | 0.2 | |
| [73] | – | 2 | 1 | 0.9999 | 1 | 0.50005 | 1.0001 | 1.9998 | 6.0011 |
| [74] | 15 000 | 0.13905 | 0.018379 | 0.00013333 | 0.13218 | 0.00013333 | 0.13905 | 0.00053333 | |
| [74] | 15 000 | 0.25695 | 0.014148 | 0.00013333 | 0.05506 | 0.00013333 | 0.25695 | 0.00053333 | |
| [25] | – | 0.3333 | 0.1111 | 5 | 0.3333 | 5 | 0.3333 | 2.2500 | |
| [25] | – | 0.2222 | 0.1111 | 5 | 0.5000 | 5 | 0.2222 | 3.0000 | |
| [25] | – | 0.3333 | 0.1667 | 5 | 0.5002 | 5 | 0.3333 | 3.0009 | |
| [25] | – | 0.1667 | 0.1111 | 5 | 0.6665 | 5 | 0.1667 | 4.4973 | |
| [25] | – | 0.3333 | 0.2222 | 5 | 0.6667 | 5 | 0.3333 | 4.5000 | |
| [25] | – | 0.1333 | 0.1111 | 5 | 0.8335 | 5 | 0.1333 | 9.0068 | |
| [25] | – | 0.3333 | 0.2778 | 5 | 0.8335 | 5 | 0.3333 | 9.0081 | |
| [25] | – | 0.3333 | 0.3222 | 5 | 0.9667 | 5 | 0.3333 | 4.5041 | |
| [25] | – | 0.1149 | 0.1111 | 5 | 0.9669 | 5 | 0.1149 | 4.5355 | |
| [25] | – | 10 | 1 | 0.2 | 0.1 | 0.2 | 10 | 0.8 | |
| [25] | – | 10 | 1 | 1 | 0.1 | 1 | 10 | 4 | |
| [25] | – | 10 | 1 | 1 | 0.1 | 1 | 10 | 4 | |
| [25] | – | 0.5 | 0.3 | 0.2 | 0.6 | 0.2 | 0.5 | 1.5 (*) | |
| [25] | – | 0.5 | 0.3 | 0.001 | 0.6 | 0.001 | 0.5 | 0.0075 | |
| [25] | – | 0.5 | 0.3 | 1 | 0.6 | 1 | 0.5 | 7.5 | |
| [25] | – | 0.5 | 0.3 | 1 | 0.6 | 1 | 0.5 | 7.5 | |
| [25] | – | 0.5961 | 0.8023 | 0.013 | 1.3459 | 0.013 | 0.5961 | −0.113 | |
| [25] | – | 0.5961 | 0.8023 | 0.133 | 1.3459 | 0.133 | 0.5961 | −1.154 | |
| [25] | – | 0.5961 | 0.8023 | 0.333 | 1.3459 | 0.333 | 0.5961 | −2.888 | |
| [24] | 82.7 | 9.94 | 9.8605 | 2.155 | 0.992 | 2.1548 | 9.94 | 0.008080 | |
| [24] | 82.7 | 0.51 | 0.4646 | 1.330 | 0.911 | 1.3301 | 0.51 | 0.000448 | |
| [24] | 11.3 | 1.62 | 1.4839 | 6.876 | 0.916 | 6.8761 | 1.62 | 0.002456 | |
| [24] | 11.3 | 0.54 | 0.4833 | 2.883 | 0.895 | 2.8832 | 0.54 | 0.008238 | |
| [24] | 36.3 | 3.36 | 3.2760 | 7.107 | 0.975 | 7.1074 | 3.36 | 0.000853 | |
| [24] | 36.3 | 0.95 | 0.9130 | 8.889 | 0.961 | 8.8972 | 0.95 | 0.068440 | |
| [24] | 65.6 | 1.52 | 1.4273 | 4.619 | 0.939 | 4.6189 | 1.52 | 0.000227 | |
| [24] | 65.6 | 0.44 | 0.3846 | 3.538 | 0.874 | 3.5381 | 0.44 | 0.000842 | |
| [24] | 32.3 | 1.08 | 1.0292 | 1.226 | 0.953 | 1.2262 | 1.08 | 0.000783 | |
| [24] | 32.3 | 0.24 | 0.2098 | 7.624 | 0.874 | 7.6241 | 0.24 | 0.018153 | |
Table 2.
For all entries of Table 1, where the parameters , and of the reduced SIR system can be found, we here compare the performance of the approximants (14), (19), (20), and (24) against the exact numerical solution of the SIR model.
| Ref. | Best | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| [67] | 0.45 | 0.02 | 0.44444 | 0.33333 | 3.330 | 3.1120 | 12.3148 | 3.2297 | 3.2637 | MT |
| [70] | 1.66 | 0.45455 | 0.99476 | 0.0039318 | 9.123 | 9.1168 | 9.7898 | 9.0485 | 8.8777 | CG |
| [71] | 0.91776 | 0.70681 | 1.0442 | 51.046 | 51.0456 | 50.9659 | 50.9572 | 50.6890 | CG | |
| [75] | 4.6291 | 2.82 | 0.0268 | 6.612 | 6.4521 | 6.6576 | 6.5952 | 6.4352 | SK-II | |
| [76] | 0.5 | 0.3 | 1.27 | 32.115 | 32.1149 | 32.1119 | 32.0545 | 31.7928 | CG | |
| [77] | 10 | 1 | 0.05 | 5.886 | 5.8505 | 10.0904 | 5.7802 | 5.7228 | CG | |
| [73] | 2 | 1 | 0.9999 | 1 | 22.753 | 22.7532 | 22.8343 | 22.7015 | 22.4514 | CG |
| [74] | 0.13905 | 0.018379 | 0.00013333 | 12.671 | 12.6695 | 15.3960 | 12.5626 | 12.4721 | CG | |
| [74] | 0.25695 | 0.014148 | 0.00013333 | 12.627 | 12.6498 | 22.0289 | 12.5291 | 12.4819 | CG | |
| [25] | 0.3333 | 0.1111 | 5 | 19.464 | 19.4635 | 19.8693 | 19.4007 | 19.1987 | CG | |
| [25] | 0.2222 | 0.1111 | 5 | 24.138 | 24.1383 | 24.2195 | 24.0866 | 23.8366 | CG | |
| [25] | 0.3333 | 0.1667 | 5 | 24.144 | 24.1439 | 24.2249 | 24.0921 | 23.8421 | CG | |
| [25] | 0.1667 | 0.1111 | 5 | 33.062 | 33.0618 | 33.0222 | 32.9919 | 32.7266 | CG | |
| [25] | 0.3333 | 0.2222 | 5 | 33.077 | 33.0774 | 33.0376 | 33.0074 | 32.7420 | CG | |
| [25] | 0.1333 | 0.1111 | 5 | 56.791 | 56.7907 | 56.6945 | 56.6914 | 56.4213 | CG | |
| [25] | 0.3333 | 0.2778 | 5 | 56.798 | 56.7973 | 56.7010 | 56.6979 | 56.4279 | CG | |
| [25] | 0.3333 | 0.3222 | 5 | 183.153 | 183.0858 | 183.0371 | 183.037 | 182.7609 | CG | |
| [25] | 0.1149 | 0.1111 | 5 | 183.999 | 183.9297 | 183.8824 | 183.8824 | 183.6062 | CG | |
| [25] | 10 | 1 | 0.2 | 4.078 | 3.9343 | 8.2932 | 3.9712 | 3.9601 | SK-II | |
| [25] | 10 | 1 | 1 | 15.444 | 15.4462 | 19.6447 | 15.3365 | 15.2635 | CG | |
| [25] | 10 | 1 | 1 | 23.120 | 23.1215 | 27.3200 | 23.0118 | 22.9389 | CG | |
| [25] | 0.5 | 0.3 | 0.2 | 1.319 | 1.2842 | 1.4291 | 1.3028 | 1.3130 | MT | |
| [25] | 0.5 | 0.3 | 0.001 | 15.388 | 15.3800 | 15.3907 | 15.3329 | 15.0734 | SK-I | |
| [25] | 0.5 | 0.3 | 1 | 38.469 | 38.4690 | 38.4660 | 38.4086 | 38.1470 | CG | |
| [25] | 0.5 | 0.3 | 1 | 44.226 | 44.2255 | 44.2225 | 44.165 | 43.9034 | CG | |
| [25] | 0.5961 | 0.8023 | 0.013 | 0 | −9.3110 | −7.3538 | −7.3569 | −7.5970 | SK-I | |
| [25] | 0.5961 | 0.8023 | 0.133 | 0 | −2.2972 | −2.1974 | −2.2258 | −2.3032 | SK-I | |
| [25] | 0.5961 | 0.8023 | 0.333 | 0 | −1.4875 | −1.3931 | −1.4747 | −1.6031 | SK-I | |
| [24] | 9.94 | 9.8605 | 2.1548 | 199.096 | 192.1291 | 199.0694 | 199.0694 | 198.9656 | SK-I | |
| [24] | 0.51 | 0.46461 | 1.3301 | 80.306 | 80.2964 | 80.1951 | 80.1947 | 79.9212 | CG | |
| [24] | 1.62 | 1.4839 | 6.8761 | 63.742 | 63.6843 | 63.6397 | 63.6393 | 63.3699 | CG | |
| [24] | 0.54 | 0.4833 | 0.00028832 | 41.376 | 41.2577 | 41.2913 | 41.2905 | 41.0343 | SK-I | |
| [24] | 3.36 | 3.276 | 7.1074 | 205.753 | 205.5298 | 205.6436 | 205.6436 | 205.3713 | SK-I | |
| [24] | 0.95 | 0.91295 | 0.00088972 | 28.987 | 27.7059 | 28.9755 | 28.9752 | 28.9152 | SK-I | |
| [24] | 1.52 | 1.4273 | 4.6189 | 121.758 | 121.7482 | 121.6419 | 121.6417 | 121.3665 | CG | |
| [24] | 0.44 | 0.38456 | 3.5381 | 54.722 | 54.7135 | 54.6197 | 54.6184 | 54.3470 | CG | |
| [24] | 1.08 | 1.0292 | 1.2262 | 125.512 | 125.4527 | 125.3988 | 125.3987 | 125.1243 | CG | |
| [24] | 0.24 | 0.20976 | 0.00076241 | 29.500 | 29.3287 | 29.4311 | 29.4295 | 29.1909 | SK-I |
3. Summary and conclusions
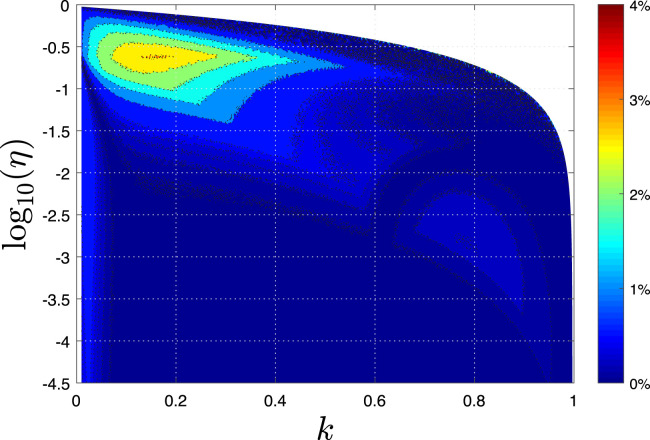
We have compared the quality of the available approximants point-wise, i.e., for each possible case of model parameters separately. Overall, if we limit ourself to the broad regime of and the mean relative deviation between the exact numerical solution and the approximants can be extracted as well. We obtain for the CG approximant, for the SK-II approximant, 2.38% for the MT approximant, and a large value of 24% for the SK-I approximant, as it fails dramatically at large and small , as can be seen in Fig. 1(g). Despite this, each of the approximants fails by more than 50% at least in some of the – region. This fact highlights the necessity to use the best available approximant, given by Fig. 1, depending on the – regime that applies to a real situation at hand. Using the combined approximant, the best approximant depending on the pair value, the relative deviation to the exact result is quantified in Fig. 3.
Fig. 3.
Combined approximant, if the best approximant for each region in - space is used. This combined approximant has an accuracy smaller than 3% for any choice of SIR parameters. The region of worst performance of the combined approximant is located in the vicinity of and , while for or , the performance is just excellent (deviations below 1%).
There are many examples to which the MT approximant had been applied in the original work [25]. We list them in Table 1. Table 2 confirms that the MT approximant is more accurate when , while all approximants are good and work very well. We include in the last three rows of the table a case that is not allowed in the semi-time SIR model, the case of , as it has been discussed in [25] as well.
Examples of relevance for the ongoing Covid-19 pandemics in 60 countries are available from [17]. The authors analyze the first and second Covid-19 wave in real time. Rather than adding all additional pairs from 2021-04-13 to our tables, we have included them all as orange and white circles corresponding to first and second waves to Fig. 2. There is a general trend of a decreasing for the second pandemic wave, even though the fraction of the remaining un-recovered population has diminished during the second wave. All crosses reside either in the yellow or white regions.
This means, that for most cases of relevance for the ongoing pandemics the existing SK-II and CG approximants are the most accurate. Still, we had to clarify here the relationship between different notation, dimensional versus dimensionless versions of the SIR equations, to allow for a direct comparison. Here, we clarified the correspondence between equivalent versions. The MT approximant might be a good choice if one does not want to calculate a Lambert value, or if the Lambert function is not available at all within the computational environment.
One should keep in mind that the peak time of the daily reported new cases does not coincide with the peak time of the infected compartment. While the former solves , the latter solves , and the final fraction of infected population is not related to an integral over , but equals . To be precise, the peak times differ by the two terms in Eq. (16) above. Both terms tend to vanish as approaches unity. The peak time of the newly infected population fraction is given by with and given by Eq. (18), as shown in [24]. This measurable peak time is thus identical for approximants SK-I and SK-II.
CRediT authorship contribution statement
Martin Kröger: Conceptualization, Methodology, Formal analysis, Writing - original draft. Mustafa Turkyilmazoglu: Writing - review & editing. Reinhard Schlickeiser: Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the referees for their constructive and helpful comments.
Communicated by V.M. Perez-Garcia
Appendix A. Derivation of
Our Eq. (14) for the reduced peak time arises from the dimensional peak time denoted by in Eq. (7b) of [25]. In this latter work one of us (MT) had already shown that his approximant supersedes alternate approximants such as their Eq. (7a) so that there is no need to compare with those. Using the abbreviation he obtained
| (A.1) |
where we here introduce abbreviations for the otherwise rather lengthy original expression as follows
| (A.2) |
So far we have just reproduced the existing expression for that had been obtained for the dimensional SIR model (1). To convert this into the dimensionless counterpart, we have to make use of Eq. (8), i.e., we have to replace , , , and we have to use Eq. (9), which states . As a result, the , , should all drop out automatically. Using the transformation rules (8) the quantities in (A.2) become
| (A.3) |
where we have re-used the quantities – defined by (15) to allow for a direct comparison between (A.1), (14). The indeed drops out because only the ratio appears in (A.1), and the remaining sign is taken care of the asymmetry of . The rate still appearing in drops out in the reduced (dimensionless) peak time because according to (9). We have thus shown that the reduced time depends only on and , and that (14) is the reduced peak time that corresponds to the dimensional peak time in [25].
Appendix B. Derivation of
This appendix provides details on how to read off Eq. (16) from the results obtained by Schlickeiser and Kröger [24]. Equation (71) in [24] for the cumulative fraction of infected persons after the occurrence of a peak in the differential rate of newly infected persons, within the so-called ‘decay’ period for reduced times , reads
| (B.1) |
as function of the reduced time . Here, is the final cumulative fraction of infected persons, given by Eq. (18), defined by Eq. (54) of [24] (for the special case of , note also that as stated after Eq. (61) in [24]) and reproduced here in (17). Furthermore as mentioned after Eq. (69) of [24], and is a characteristic reduced time given by Eq. (73) in [24], identical with the first term on the right hand side of (16). The coefficients and for the SK-I and SK-II approximants are given by Eqs. (63)-66) in [24] and reproduced in (19), (20). Analytic results for the characteristic cumulative fraction , the differential rate of infections at peak time , and the cumulative fraction and the peak time of are stated in Eqs. (48), (49), and (62) of [24]. The quantity in (B.1) is the cumulative fraction of infected persons at reduced time , and thus as denotes the susceptible fraction at reduced time . Since we are interested in the peak time of the infected compartment, and because this peak time is delayed with respect to the peak time of the differential rate of newly infected persons, rather than applies; the latter quantity, valid for , had been derived in [24] as well. Making use of (11), one has . To be specific, using (B.1), the equation determining becomes
| (B.2) |
This equation is readily solved for , the result is given by Eq. (16).
Data availability statement
All data generated or analyzed during this study are included in this published article.
References
- 1.Gopagoni D., Lakshmi P.V. Susceptible, infectious and recovered (SIR model) predictive model to understand the key factors of COVID-19 transmission. Int. J. Adv. Comput. Sci. Appl. 2020;11:296–302. [Google Scholar]
- 2.Law K.B., Peariasamy K.M., Gill B.S., Singh S., Sundram B.M., Rajendran K., Dass S.C., Lee Y.L., Goh P.P., Ibrahim H., Abdullah N.H. Tracking the early depleting transmission dynamics of COVID-19 with a time-varying SIR model. Sci. Rep. 2020;10:21721. doi: 10.1038/s41598-020-78739-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmetolan S., Bilge A.H., Demirci A., Peker-Dobie A., Ergonul O. What can we estimate from fatality and infectious case data using the susceptible-infected-removed (SIR) model? A case study of Covid-19 pandemic. Front. Med. 2020;7 doi: 10.3389/fmed.2020.556366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neher R.A., Dyrdak R., Valentin D., Hodcroft E.B., Albert J. Potential impact of seasonal forcing on a SARS-CoV-2 pandemic. Swiss Med. Wkly. 2020;150:w20224. doi: 10.4414/smw.2020.20224. [DOI] [PubMed] [Google Scholar]
- 5.Babajanyan S.G., Cheong K.H. Age-structured SIR model and resource growth dynamics: a COVID-19 study. Nonlinear Dyn. 2021 doi: 10.1007/s11071-021-06384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brugnano L., Iavernaro F., Zanzottera P. A multiregional extension of the SIR model, with application to the COVID-19 spread in Italy. Math. Methods Appl. Sci. 2021;44(6):4414–4427. doi: 10.1002/mma.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X.W., Li J., Xiao C., Yang P.L. Numerical solution and parameter estimation for uncertain SIR model with application to COVID-19. Fuzzy Optim. Decis. Mak. 2021;20:189–208. [Google Scholar]
- 8.Chen Y.C., Lu P.E., Chang C.S., Liu T.H. A time-dependent SIR model for COVID-19 with undetectable infected persons. IEEE Trans. Netw. Sci. Eng. 2020;7:3279–3294. doi: 10.1109/TNSE.2020.3024723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper I., Mondal A., Antonopoulos C.G. A SIR model assumption for the spread of COVID-19 in different communities. Chaos Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaffar A., Alanazi S., Alruwaili M., Sattar M.U., Ali W., Humayun M., Siddiqui S.Y., Ahmad F., Khan M.A. Multi-stage intelligent smart lockdown using SIR model to control COVID 19. Intell. Autom. Soft Comput. 2021;28(2):429–445. [Google Scholar]
- 11.Postnikov E.B. Estimation of COVID-19 dynamics ”on a back-of-envelope?: Does the simplest SIR model provide quantitative parameters and predictions? Chaos Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah S.T.A., Mansoor M., Mirza A.F., Dilshad M., Khan M.I., Farwa R., Khan M.A., Bilal M., Iqbal H.M.N. Predicting COVID-19 spread in Pakistan using the SIR model. J. Pure Appl. Microbiol. 2020;14:1423–1430. [Google Scholar]
- 13.dos Santos I.F.F., Almeida G.M.A., de Moura F. Adaptive SIR model for propagation of SARS-CoV-2 in Brazil. Physica A. 2021;569 doi: 10.1016/j.physa.2021.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMahon A., Robb N.C. Reinfection with SARS-CoV-2: Discrete SIR (susceptible, infected, recovered) modeling using empirical infection data. Jmir Publ. Health Surveill. 2020;6(4):279–287. doi: 10.2196/21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telles C.R., Lopes H., Franco D. SARS-COV-2: SIR model limitations and predictive constraints. Symmetry-Basel. 2021;13:676. [Google Scholar]
- 16.Venkatasen M., Mathivanan S.K., Jayagopal P., Mani P., Rajendran S., Subramaniam U., Ramalingam A.C., Rajasekaran V.A., Indirajithu A., Somanathan M.S. Forecasting of the SARS-CoV-2 epidemic in India using SIR model, flatten curve and herd immunity. J. Ambient Intell. Hum. Comput. 2021 doi: 10.1007/s12652-020-02641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kröger M., Schlickeiser R. Forecast for the second Covid-19 wave based on the improved SIR model with a constant ratio of recovery to infection rate. R. Soc. Open Sci. 2021 doi: 10.1098/rsos.211379. submitted for publication. URL: https://www.complexfluids.ethz.ch/cgi-bin/covid19-waveII. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kendall D.G. Proc. Third Berkeley Symp. on Math. Statist. and Prob., Vol. 4. Univ. of Calif. Press; Berkeley, United States: 1956. Deterministic and stochastic epidemics in closed populations; pp. 149–165. [Google Scholar]
- 19.Kermack W.O., McKendrick A.G. Contributions to the mathematical theory of epidemics—I. 1927. Bull. Math. Biol. 1991;53:33–55. doi: 10.1007/BF02464423. [DOI] [PubMed] [Google Scholar]
- 20.Kermack W.O., McKendrick A.G. Contributions to the mathematical theory of epidemics—II. The problem of endemicity. 1932. Bull. Math. Biol. 1991;53:57–87. doi: 10.1007/BF02464424. [DOI] [PubMed] [Google Scholar]
- 21.Kermack W.O., McKendrick A.G. Contributions to the mathematical theory of epidemics—III. Further studies of the problem of endemicity. 1933. Bull. Math. Biol. 1991;53:89–118. doi: 10.1007/BF02464425. [DOI] [PubMed] [Google Scholar]
- 22.Britton T., Pardoux E., Ball F., Larédo C., Sirl D., Tran V.C. Stochastic Epidemic Models with Inference. vol. 2255. Springer; Berlin: 2019. (Lecture Notes in Mathematics). [Google Scholar]
- 23.Kröger M., Schlickeiser R. Analytical solution of the SIR-model for the temporal evolution of epidemics. Part A: Time-independent reproduction factor. J. Phys. A. 2020;53 [Google Scholar]
- 24.Schlickeiser R., Kröger M. Analytical solution of the SIR-model for the temporal evolution of epidemics. Part B: Semi-time case. J. Phys. A. 2021;54 [Google Scholar]
- 25.Turkyilmazoglu M. Explicit formulae for the peak time of an epidemic from the SIR model. Physica D. 2021;422 doi: 10.1016/j.physd.2021.132902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heng K., Althaus C.L. The approximately universal shapes of epidemic curves in the susceptible-exposed-infectious-recovered (SEIR) model. Sci. Rep. 2020;10:19365. doi: 10.1038/s41598-020-76563-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bidari S., Chen X.Y., Peters D., Pittman D., Simon P.L. Solvability of implicit final size equations for SIR epidemic models. Math. Biosci. 2016;282:181–190. doi: 10.1016/j.mbs.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho A.M., Gonçalves S. An analytical solution for the Kermack-McKendrick model. Physica A. 2021;566 [Google Scholar]
- 29.Guerrero F., Santonja F.J., Villanueva R.J. Solving a model for the evolution of smoking habit in Spain with homotopy analysis method. Nonlinear Anal. RWA. 2013;14(1):549–558. [Google Scholar]
- 30.Khan H., Mohapatra R.N., Vajravelu K., Liao S.J. The explicit series solution of SIR and SIS epidemic models. Appl. Math. Comput. 2009;215(2):653–669. [Google Scholar]
- 31.Liu J.L., Peng B.Y., Zhang T.L. Effect of discretization on dynamical behavior of SEIR and SIR models with nonlinear incidence. Appl. Math. Lett. 2015;39:60–66. [Google Scholar]
- 32.Van Mieghem P. Approximate formula and bounds for the time-varying susceptible-infected-susceptible prevalence in networks. Phys. Rev. E. 2016;93 doi: 10.1103/PhysRevE.93.052312. [DOI] [PubMed] [Google Scholar]
- 33.Barlow N.S., Weinstein S.J. Accurate closed-form solution of the SIR epidemic model. Physica D. 2020;408 doi: 10.1016/j.physd.2020.132540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yildirim A., Cherruault Y. Analytical approximate solution of a SIR epidemic model with constant vaccination strategy by homotopy perturbation method. Kybernetes. 2009;38:1566–1575. [Google Scholar]
- 35.Cadoni M., Gaeta G. Size and timescale of epidemics in the SIR framework. Physica D. 2020;411 doi: 10.1016/j.physd.2020.132626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chekroun A., Kuniya T. Global threshold dynamics of aninfection age-structured SIR epidemic model with diffusion under the Dirichlet boundary condition. J. Differential Equations. 2020;269(8):117–148. [Google Scholar]
- 37.Imron C., Hariyanto T., Yunus M., Surjanto S.D., Dewi N.A.C., Iop V.C. Stability and persistence analysis on the epidemic model multi-region multi-patches. J. Phys. Conf. Ser. 2019;1218 [Google Scholar]
- 38.Karaji P.T., Nyamoradi N. Analysis of a fractional SIR model with general incidence function. Appl. Math. Lett. 2020;108 [Google Scholar]
- 39.Mohamadou Y., Halidou A., Kapen P.T. A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19. Appl. Intell. 2020 doi: 10.1007/s10489-020-01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samanta S., Sahoo B., Das B. Dynamics of an epidemic system with prey herd behavior and alternative resource to predator. J. Phys. A. 2019;52(42) [Google Scholar]
- 41.Sene N. SIR epidemic model with Mittag-Leffler fractional derivative. Chaos Solitons Fractals. 2020;137 doi: 10.1016/j.chaos.2021.111030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon M. SIR epidemics with stochastic infectious periods. Stochastic Process. Appl. 2020;130(7):4252–4274. [Google Scholar]
- 43.Tian C.R., Zhang Q.Y., Zhang L. Global stability in a networked SIR epidemic model. Appl. Math. Lett. 2020;107 [Google Scholar]
- 44.El Koufi A., Adnani J., Bennar A., Yousfi N. Analysis of a stochastic SIR model with vaccination and nonlinear incidence rate. Int. J. Differ. Equ. 2019;2019 [Google Scholar]
- 45.Houy N. Are better vaccines really better? the case of a simple stochastic epidemic SIR model. Econ. Bull. 2013;33:207–216. [Google Scholar]
- 46.Jornet-Sanz M., Corberan-Vallet A., Santonja F.J., Villanueva R.J. A bayesian stochastic SIRS model with a vaccination strategy for the analysis of respiratory syncytial virus. Stat. Oper. Res. Trans. 2017;41:159–175. [Google Scholar]
- 47.Li X.N., Zhang Q.M. Time to extinction and stationary distribution of a stochastic susceptible-infected-recovered-susceptible model with vaccination under markov switching. Math. Popul. Stud. 2020;27:259–274. [Google Scholar]
- 48.Liu Q., Jiang D.Q. The threshold of a stochastic delayed SIR epidemic model with vaccination. Physica A. 2016;461:140–147. [Google Scholar]
- 49.Liu Q., Jiang D.Q., Shi N.Z., Hayat T. Dynamics of a stochastic delayed SIR epidemic model with vaccination and double diseases driven by levy jumps. Physica A. 2018;492:2010–2018. [Google Scholar]
- 50.Miao A.Q., Zhang J., Zhang T.Q., Pradeep B. Threshold dynamics of a stochastic SIR model with vertical transmission and vaccination. Comput. Math. Methods Med. 2017;2017 doi: 10.1155/2017/4820183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen C., Carlson J.M. Optimizing real-time vaccine allocation in a stochastic SIR model. Plos One. 2016;11 doi: 10.1371/journal.pone.0152950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F.Y., Wang X.Y., Zhang S.W., Ding C.M. On pulse vaccine strategy in a periodic stochastic SIR epidemic model. Chaos Solitons Fractals. 2014;66:127–135. [Google Scholar]
- 53.Wang L., Teng Z.D., Tang T.T., Li Z.M. Threshold dynamics in stochastic SIRS epidemic models with nonlinear incidence and vaccination. Comput. Math. Methods Med. 2017;2017 doi: 10.1155/2017/7294761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Witbooi P.J. Stability of a stochastic model of an SIR epidemic with vaccination. Acta Biotheor. 2017;65:151–165. doi: 10.1007/s10441-017-9308-5. [DOI] [PubMed] [Google Scholar]
- 55.Xu C.Y., Li X.Y. The threshold of a stochastic delayed SIRS epidemic model with temporary immunity and vaccination. Chaos Solitons Fractals. 2018;111:227–234. [Google Scholar]
- 56.Zhang Y., Li Y., Zhang Q.L., Li A.H. Behavior of a stochastic SIR epidemic model with saturated incidence and vaccination rules. Physica A. 2018;501:178–187. [Google Scholar]
- 57.Zhao X., He X., Feng T., Qiu Z.P. A stochastic switched SIRS epidemic model with nonlinear incidence and vaccination: stationary distribution and extinction. Int. J. Biomath. 2020;13 [Google Scholar]
- 58.Priesemann V., Balling R., Brinkmann M.M., Ciesek S., Czypionka T., Eckerle I., Giordano G., Hanson C., Hel Z., Hotulainen P., Klimek P., Nassehi A., Peichl A., Perc M., Petelos E., Prainsack B., Szcurek E. An action plan for pan-European defence against new SARS-CoV-2 variants. Lancet. 2021;397:469–470. doi: 10.1016/S0140-6736(21)00150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Priesemann V., Brinkmann M.M., Ciesek S., Cuschieri S., Czypionka T., Giordano G., Gurdasani D., Hanson C., Hens N., Iftekhar E., Kelly-Irving M., Klimek P., Kretzschmar M., Peichl A., Perc M., Sannino F., Schernhammer F., Schmidt A., Stainers A., Szcurek E. Calling for pan-European commitment for rapid and sustained reduction in SARS-CoV-2 infections. Lancet. 2021;397:92–93. doi: 10.1016/S0140-6736(20)32625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colombo R.M., Garavello M. Optimizing vaccination strategies in an age structured SIR model. Math. Biosci. Eng. 2020;17:1074–1089. doi: 10.3934/mbe.2020057. [DOI] [PubMed] [Google Scholar]
- 61.Schlickeiser R., Kröger M. Analytical modeling of the temporal evolution of epidemics outbreaks accounting for vaccinations. Physics. 2021;3:386–426. [Google Scholar]
- 62.Mungkasi S. Variational iteration and successive approximation methods for a SIR epidemic model with constant vaccination strategy. Appl. Math. Model. 2021;90:1–10. [Google Scholar]
- 63.Meng X.Z., Chen L.S. The dynamics of a new SIR epidemic model concerning pulse vaccination strategy. Appl. Math. Comput. 2008;197:582–597. [Google Scholar]
- 64.Moneim I.A., Greenhalgh D. Threshold and stability results for an SIRS epidemic model with a general periodic vaccination strategy. J. Biol. Syst. 2005;13:131–150. [Google Scholar]
- 65.Terry A.J. PULSE vaccination strategies in a metapopulation SIR model. Math. Biosci. Eng. 2010;7:455–477. doi: 10.3934/mbe.2010.7.455. [DOI] [PubMed] [Google Scholar]
- 66.D’Onofrio A. Pulse vaccination strategy in the SIR epidemic model: global asymptotic stable eradication in presence of vaccine failures. Math. Comput. Model. 2002;36:473–489. [Google Scholar]
- 67.Harko T., Lobo F.S.N., Mak M.K. Exact analytical solutions of the susceptible-infected-recovered (SIR) epidemic model and of the SIR model with equal death and birth rates. Appl. Math. Comput. 2014;236:184. [Google Scholar]
- 68.Abramowitz M., Stegun I.A. Dover Publications; New York: 1972. Handbook of Mathematical Functions with Formulas, Graphs, and Mathematical Tables. [Google Scholar]
- 69.Batiha A.-M., Batiha B. A new method for solving epidemic model. Aust. J. Basic Appl. Sci. 2011;5:3122–3126. [Google Scholar]
- 70.Balkew T.M. East Tennessee State University; Tennessee, USA: 2010. The SIR Model When is a Multi-Exponential Function. [Google Scholar]
- 71.Cano C. Louisiana Tech Digital Commons, Louisiana Tech University; USA: 2020. The SIR Models, their Applications, and Approximations of their Rates. [Google Scholar]
- 72.Kiss I.Z., Miller J.C., Simon P.L. Springer; Berlin: 2017. Mathematics of Epidemics on Networks. [Google Scholar]
- 73.Simon C.M. The SIR dynamic model of infectious disease transmission and its analogy with chemical kinetics. PeerJ Phys. Chem. 2020;2 [Google Scholar]
- 74.Sasaki K. 2021. COVID-19 dynamics with SIR model. URL: https://www.lewuathe.com/covid-19-dynamics-with-sir-model.html. [Google Scholar]
- 75.Sulsky D. The University of New Mexico; Albuquerque, USA: 2012. Using Real Data in an SIR Model. URL: https://www.math.unm.edu/ sulsky/mathcamp/ApplyData.pdf. [Google Scholar]
- 76.Smith D., Moore L. 2021. The SIR model for spread of disease. URL: https://www.math.duke.edu/education/ccp/materials/diffcalc/sir/index.html. [Google Scholar]
- 77.Miller J.C. Mathematical models of SIR disease spread with combined non-sexual and sexual transmission routes. Inf. Dis. Mod. 2017;2:35. doi: 10.1016/j.idm.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.