Abstract
Current eukaryotic replication models postulate that leading and lagging DNA strands are replicated predominantly by dedicated DNA polymerases. The catalytic subunit of the leading strand DNA polymerase ε, Pol2, consists of two halves made of two different ancestral B-family DNA polymerases. Counterintuitively, the catalytically active N-terminal half is dispensable, while the inactive C-terminal part is required for viability. Despite extensive studies of yeast Saccharomyces cerevisiae strains lacking the active N-terminal half, it is still unclear how these strains survive and recover. We designed a robust method for constructing mutants with only the C-terminal part of Pol2. Strains without the active polymerase part show severe growth defects, sensitivity to replication inhibitors, chromosomal instability, and elevated spontaneous mutagenesis. Intriguingly, the slow-growing mutant strains rapidly accumulate fast-growing clones. Analysis of genomic DNA sequences of these clones revealed that the adaptation to the loss of the catalytic N-terminal part of Pol2 occurs by a positive selection of mutants with improved growth. Elevated mutation rates help generate sufficient numbers of these variants. Single nucleotide changes in the cell cycle-dependent kinase gene, CDC28, improve the growth of strains lacking the N-terminal part of Pol2, and rescue their sensitivity to replication inhibitors and, in parallel, lower mutation rates. Our study predicts that changes in mammalian homologs of cyclin-dependent kinases may contribute to cellular responses to the leading strand polymerase defects.
Keywords: DNA replication, leading strand, DNA polymerase ε, cyclin-dependent kinase, CDC28, mutation rates, illegitimate mating, yeast
Introduction
The current model of eukaryotic replication postulates that three essential B-family DNA polymerases have different tasks at the fork (Figure 1A): DNA polymerase α (pol α) in a complex with primase synthesizes short RNA-DNA hybrid primers; pol δ synthesizes the lagging strand and ∼20% of the leading strand; pol ε synthesizes most of the leading DNA strand (Guilliam and Yeeles 2020b). In view of the profound role of pol ε, it is surprising that genetic defects leading to a severe depletion of this polymerase are tolerated during human development, although they lead to a severe disorder: facial dysmorphism, immunodeficiency, livedo, and short stature syndrome (Pachlopnik Schmid et al. 2012), suggesting that the cell can respond to the anomaly of replication. In the present work in the yeast model, we identified suppressors mitigating the replication problems caused by defective leading strand polymerase.
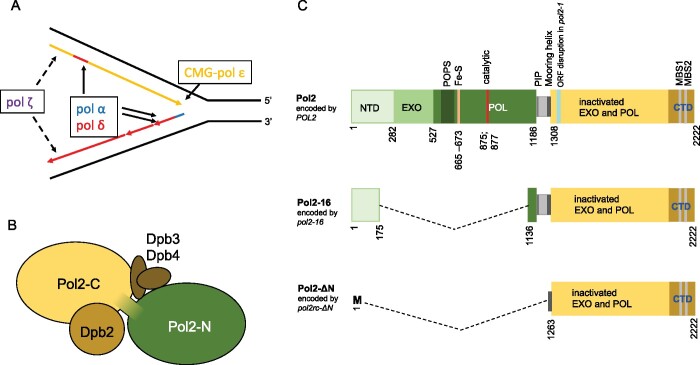
Figure 1.
DNA polymerase ε in replication in eukaryotes. (A) Current view on roles of B-family pols in replication (Guilliam and Yeeles 2020b). CMG complex travels along leading strand and unwinds DNA. Pol ε, a part of CMG, synthesizes most of the leading DNA strand. Primase/pol α and pol δ synthesize lagging strand and almost 20% of the leading strand. Pol ζ assists replication when it is impeded by DNA damage, unusual DNA structures, or impairment of the regular replication machinery (Northam et al. 2006, 2010; Garbacz et al. 2015). (B) Subunit structure of yeast pol ε. The catalytic subunit consists of two halves, active N-terminal pol (green) and inactive C-terminal pol (yellow). The second subunit, Dpb2, is indispensable. Two small subunits, Dpb3 and Dpb4, have a role in the organization of the holoenzyme (rigidity of the complex of two domains of Pol2) (Yuan et al. 2020) but are dispensable although their absence causes genome instability (Aksenova et al. 2010). (C) Top: primary structure of Pol2 protein with the location of critical elements. NTD (N-terminal domain) of unknown function, exonuclease domain, and pol domain of the active N-terminal half are shown in shades of green. Several regions critical for the function are marked. Dark green: POPS (POl2 family-specific catalytic core Peripheral Subdomain) is necessary for proper replication timing and fork progression (Meng et al. 2020), yellow bar: Fe-S cluster is necessary for pol activity (Ter Beek et al. 2019) as well as catalytic carboxylates (red) (Dua et al. 1999). The linker between two halves (gray) has a PCNA binding motif (PIP, thin black line) required for efficient stimulation of Pol ε by PCNA (Devbhandari and Remus 2020) and residues 1270-1308 that form mooring helix for binding Dpb3 and Dpb4 (Yuan et al. 2020). The yellow rectangle is the inactive C-terminal pol with metal-binding sites in the CTD. Teal bar is the site of integration of the URA3 gene in a pol2-1 allele that is viable but confers slow growth phenotype (Morrison et al. 1990). Middle: Pol2-16 protein encoded by allele pol2-16 (Kesti et al. 1999) lacks elements necessary for exonuclease and polymerase activity but possess part of the NTDand elements necessary for interaction with Dpb2, Dpb3, and Dpb4 subunits and PCNA (Ohya et al. 2002). The other name of this allele is pol2-M (Dua et al. 1999). Bottom: Pol2-ΔN encoded by pol2rc-ΔN lacks the NTD and PIP (PCNA-interaction protein box) motif but retains amino acids that can form mooring helix and C-terminal part and thus possesses the ability to interact with Dpb3 and Dpb4 subunits (Devbhandari and Remus 2020), Dpb2 and CMG (Zhou et al. 2017). The allele is also called pol2-Δ1262 or pol2-Δcat (Yeeles et al. 2017; Devbhandari and Remus 2020).
Early studies suggested that two DNA polymerases, pol α and pol δ, perform replication in eukaryotes (Waga and Stillman 1998). However, an unusual “large” form of pol δ purified from human cells had distinct biochemical properties (Nishida et al. 1988; Syvaoja and Linn 1989) and appeared to be the third essential eukaryotic pol of the B family, named pol ε (Syvaoja et al. 1990). This enzyme has four subunits (Figure 1B) (Pospiech and Syvaoja 2003; Pursell and Kunkel 2008). Its catalytic subunit (cs-pol ε), encoded by the POL2 gene in yeast (Morrison et al. 1990), is unusually large and evolved as a fusion of two unrelated ancestral pols of the B family (Figure 1C) (Tahirov et al. 2009; Kazlauskas et al. 2020). The N-terminal part of cs-pol ε contains a proofreading exonuclease (Morrison et al. 1991) and a catalytically active Fe-S cluster-dependent DNA polymerase (Jain et al. 2014; Ter Beek et al. 2019). The N-terminal part of cs-pol ε differs from other members of the B-family polymerases and possesses inserts of stretches of amino acids (Shcherbakova et al. 2003; Hogg et al. 2014) necessary for pol ε-specific functions, for example, for the integration into the replisome, as exemplified by the POPS motif (Meng et al. 2020). The linker between the N-terminal and C-terminal parts of cs-pol ε harbors the PIP motif necessary for interaction with PCNA (Yuan et al. 2020).
Based on the biochemical properties of pol ε, Morrison et al. (1990) proposed a model of eukaryotic replication, where pol α and pol δ synthesize the lagging DNA strand and pol ε synthesizes the leading strand. The model proved to be viable, supported by genetic and biochemical experiments, and is currently widely accepted, with some adjustments (Burgers 2009; Pavlov and Shcherbakova 2010; Stillman 2015; Burgers and Kunkel 2017; Aria and Yeeles 2018; Guilliam and Yeeles 2020b; Lewis et al. 2020; Pavlov et al. 2020) (Figure 1A). The hint against the complete generality of the model was already present in the first paper by Morrison et al. (1990), where yeast strains with the insertion of the URA3 reporter into the middle of the POL2 open reading frame (mutation pol2-1, Figure 1C) were slow growers but viable. Nine years later, it was sensationally discovered that yeast strains with the deletion of the whole N-terminal half of cs-pol ε with all functional elements were viable (mutation pol2-16, Figure 1C), though they grew poorly (Kesti et al. 1999). Notably, deletions leading to the absence of the whole POL2 gene or its C-terminal part were lethal (Morrison et al. 1990; Dua et al. 1999; Kesti et al. 1999). When the active polymerase part of pol ε is deleted, it is likely substituted by pol δ during DNA replication in vivo, as proposed by Kesti et al. (1999). In a reconstituted replication fork in vitro, pol δ in combination with pol α can support replication fork progression when pol ε is devoid of the N-terminal half, albeit at a reduced rate (Yeeles et al. 2017; Devbhandari and Remus 2020). The observation that pol δ can access the 3ʹOH end of the DNA synthesized by pol ε and proofread leading strand errors (Pavlov and Shcherbakova 2010; Bulock et al. 2020) supports the feasibility of such pol replacement. Also, the pattern of mutations in strains without the N-terminal catalytic part of Pol2 is consistent with synthesis of the leading strand by pol δ (Garbacz et al. 2018, 2019). In contrast to the viability of the N-terminal deletion mutant, point mutations in the N-terminus impairing the activity cs-pol ε (Figure 1C) are lethal (Araki et al. 1992; Dua et al. 1999; Pavlov et al. 2001), presumably because the presence of the inactivated part does not allow for the substitution by pol δ.
The critical importance of the C-terminal half of cs-pol ε, primarily the C-terminal domain (CTD), was first demonstrated by mutational analysis of the CTD (Dua et al. 1999). It was later found that this domain participates in the formation of a complex with the second subunit of pol ε and the CMG (Cdc45, Mcm2-7, GINS) helicase; thus, its absence prevents the initiation and progression of DNA synthesis (Tanaka and Araki 2013; Bai et al. 2017). The eukaryotic helicase travels along the leading strand; therefore, pol ε is well-positioned to participate in its replication (Yuan et al. 2020). The C-terminal part of cs-pol ε alone is sufficient for CMG assembly essential for replication (Georgescu et al. 2017; Goswami et al. 2018; Hizume et al. 2018), and this role appears to be more important for replication than the function of pol ε as a polymerase. Excessive DNA unwinding by CMG in strains without pol ε activity is proposed to be suppressed by a Rad53 kinase-dependent mechanism (Devbhandari and Remus 2020).
Several groups confirmed and extended the observation that the deletion of the N-terminal part of cs-pol ε in pol2-16 strains does not stop replication and does not lead to cell death (Dua et al. 1999; Ohya et al. 2002; Garbacz et al. 2018). These studies showed that strains with pol2-16 grow very slowly, are spontaneous mutators, and rapidly acquire the ability to grow at a nearly regular rate. It remained unknown what changes in cells led to the restoration of growth. Besides, several observations in these studies were varying. Some studies revealed the temperature sensitivity of corresponding strains, but others did not see it (Dua et al. 1999; Kesti et al. 1999; Ohya et al. 2002). A recent study questioned even the ability of pol2-16 to support yeast growth (Devbhandari and Remus 2020). It is likely that the slow progress in finding the mechanism of recovery of cells without the N-terminal cs-pol ε half and discrepancies between various studies resulted from (1) difficulties in creating mutant alleles in the chromosomal location and (2) low stability of truncated protein encoded by the pol2-16 allele (Garbacz et al. 2018) leading to a possible defect in CMG. Typically, the pol2-16 strains lacking the N-terminal part of Pol2 need 2 weeks to form visible colonies instead of 2 days for wild-type yeast, and faster-growing isolates possess various undefined adaptations making the allele impractical (Devbhandari and Remus 2020).
Here, we aimed to characterize the biological consequences of the absence of the catalytic half of Pol2 separate from the protein stability defects conferred by pol2-16, as well as to investigate the fork recovery mechanisms. To this end, we designed a robust protocol for making better-growing strains lacking the N-terminal half of Pol2, with a codon-optimized galactose-regulatable allele (named pol2rc-ΔN) encoding for the clear-cut C-terminal part of cs-pol ε (Figure 1C). The colonies of strains bearing pol2rc-ΔN appear three times sooner than the colonies of pol2-16. The strains with pol2rc-ΔN are spontaneous mutators and frequently yield fast-growing variants. Whole-genome sequencing and genetic analysis of pol2rc-ΔN strains provided clues to the nature of genetic events that restore near-normal yeast growth in the absence of the N-terminal half of cs-pol ε. The high proportion of nonsynonymous mutations implied that the emergence of strains was driven by positive selection and supported by elevated mutation rates. We show that recurrent single nucleotide changes in the cell cycle-dependent kinase gene, CDC28, can restore near-normal growth of pol2rc-ΔN strains lacking the N-terminal half of cs-pol and suppress their HU-sensitivity and the mutator phenotype.
Materials and methods
Plasmids
To create strains with galactose-regulatable alleles of POL2, we used integrative plasmids with a TRP1 selectable marker: pJA6, called pRS304(POL2+DPB4-CBP) with a full-length codon-optimized gene that we call POL2rc (Yeeles et al. 2015); and pJA8, called pRS304(POL2-Δ1262+DPB4-CBP) with a POL2 allele with a nucleotide sequence encoding for amino acid residues 1263-2222 of Pol2 flanked by an ATG codon and a nonsense codon (Yeeles et al. 2017). The resulting protein is analogous to the protein encoded by pol2-16 because it similarly lacks the N-terminal catalytic active half of Pol2. However, it is structurally different because pol2-16 encodes for Pol2 variant with amino acid residues 1-174 fused to 1135-2222 (Figure 1C) (Yeeles et al. 2017; Devbhandari and Remus 2020). We renamed pol2Δ1262 (also known as pol2-Δcat) as pol2rc-ΔΝ, where “rc” stands for the recoded gene with codon usage optimized for high expression, and ΔΝ stands for the variant of the gene encoding Pol2 without the first 1262 amino acids. The plasmid without DPB4 (pJA8w/oDPB4) was created by blunt-end ligation of the original pJA8 cut by NotI and SgrAI with ends filled by Klenow fragment of DNA pol I. All plasmids used in this study were sequenced to verify their identity.
Yeast strains
We used two related strains, both derivatives of CG379 (Morrison et al. 1993): an autodiploid form of E134 (MATα ade5-1 lys2::InsEA14 trp-289 his7-2 leu2-3,112 ura3-52) (Tran et al. 1997; Shcherbakova and Kunkel 1999), PSD93, described in Daee et al. (2010), and an autodiploid of LAN201-Δura3 (MATα ade5-1 lys2-Tn5-13 trp1-289 his7-2 leu2-3,112 ura3-Δ) (Stepchenkova et al. 2017) described in Stepchenkova et al. (2018).
Diploid strains heterozygous for a complete deletion of the POL2 gene (pol2Δ::kanMX, Figure 2A, Supplementary Figure S1) were created by one-step gene disruption (Wach et al. 1994). At the initial step, a diploid heterozygous for pol2::kanMX was constructed by transformation with a PCR product obtained on pFA6-kanMX4 using primers with short homology to the regions flanking the POL2 gene: pol2_kanMX_45up GAAAGAGCACATTCTATCAAGATAACACTCTCAGGGGACAAGTATCAGCTGAAGCTTC GTACGC and pol2_kanMX_45downR TTCATGGTAAAGAGGCCA TTGAACCTCGCGTTATATACTGCTTACGCATAGGCCACTAGTGGATCTG. Once the first heterozygous diploid was created and verified, we used its chromosomal DNA as a template for PCR-amplification of the region encompassing kanMX with long homology to the region flanking the POL2: POL2 144F TATGGATCTTGATACAGAG and POL2 7204R GGCTAATTTTTCGGTTATTT. These procedures resulted in the generation of PSD93 derivative heterozygous for the deletion allele of POL2, named PSD93+/ΔPOL2, and LAN201 derivative named YEE302. The strains were transformed by EcoRV-linearized pJA6 or pJA8 to Trp+ to direct the integration of the whole plasmid into the TRP1 locus, thus creating heterozygotes for the POL2rc or pol2rc-ΔN, respectively (Figure 2A, Supplementary Figure S1). PSD93+/ΔPOL2 derivatives were named PSD93+/ΔPOL2 [pJA6] or PSD93+/ΔPOL2 [pJA8]. Derivatives of YEE302 with [pJA6] or [pJA8] were named YEE303 and YEE304, respectively. Tetrad dissection of these sporulating diploids gave haploid segregants without the natural POL2 gene but with the recoded alleles (Supplementary Figure S1). The presence of an extra copy of the DPB4 gene does not affect the growth of pol2rc-ΔN strains (Supplementary Figure S2). Derivatives of LAN303 and LAN304 strains, heterozygous for Δrev3::hph/+ (constructed by one-step gene disruption) were named YEE303r3hΒ/+ and YEE304r3hΒ/+, respectively. For analysis of suppressor mutations, we crossed pol2rc-ΔN segregants of PSD93+/ΔPOL2 [pJA8] to haploid LAN201-Δura3. For analysis of suppressor effects of CDC28 mutations on the growth of strains with the pol2-1 allele, we crossed 22B-YPOM634 (MATa cdc28-186) or 18A-YPOM643 (MATa cdc28-716) to LAN201-Δura3. We transformed the resulting diploids by EcoRI-cut p2A5 (Morrison et al. 1990), creating a diploid heterozygous for the disruption of POL2 by the URA3 gene (Figure 1C). Strain 10D-D925 (MATα ade1Δ his4Δ lys2Δ ura3Δ leu2Δ thr4Δ) from the strain collection of the Department of Genetics and Biotechnology, St. Petersburg University was used in an illegitimate mating assay to score the frequency of whole chromosome III and its right arm loss. The original strain for the overproduction of pol ε variants, yJF1 (MATa leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 bar1::hph pep4::kanMX rad5-G535R) was from the J. Diffley laboratory (Yeeles et al. 2015; Yeeles et al. 2017).
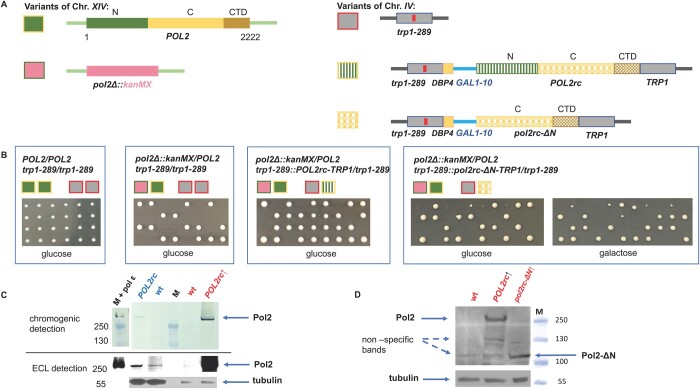
Figure 2.
Generation of the pol2rc-ΔN haploid strains. (A) Diploid used for the construction of pol2rc-ΔN. The genes in chromosomes XIV and IV are color-coded, and pictograms for each allele are shown to the left of the scheme. The POL2 gene is diagramed in green (active polymerase N-half, N) and yellow (inactive polymerases C-terminal part, C). The deletion of the region and substitution with the kanMX cassette is in pink. The TRP1 locus is in grey, and the trp1-289 allele is shown with a red bar inside the ORF representing the mutation. The integration of the optimized POL2rc gene into the TRP1 region is shown by the same colors as the natural POL2 but with a textured fill. The pol2rc-ΔN is in textured yellow. (B) Results of tetrad dissection of the diploids whose genotypes are represented by pictograms above the photograph. Relevant genotypes of the parent diploids are above pictograms. Photos are taken on day 4. (C) Pol2 levels in strain with the wild-type POL2 allele (YEE302, POL2/pol2Δ) and strain with the recoded POL2rc allele (YEE303, POL2/pol2Δ POL2rc, Materials and Methods) grown with or without induction, in YPRAU (raffinose, no induction of GAL1-10 promoter, lanes labeled in blue) or in YPRGAU (raffinose + galactose, induction, lanes labeled in red). Lane labeled “M” is the marker, PageRuler Plus Prestained Protein Ladder covering a range from 10 to 250 kDa (Thermo Fisher Scientific). On lane labeled “M plus pol ε,” in addition to the marker, 2 μl of 2.5 nM purified pol ε were loaded. The upper half represents an image of the membrane developed with the chromogenic substrate (low sensitivity, but the ladder could be seen). The lower part represents the image of the same membrane when the signal is detected by ECL after exposure to film (higher sensitivity). Note that we loaded smaller amounts of extracts prepared from strains grown under induction in YPRGAU. The signal intensity was measured by Image J and adjusted in respect to loading control. (D) Comparable levels of Pol2 (256 kDa) and Pol2-ΔN (110 kDa) proteins in extracts of yeast strain yJF1 (lane 1, labeled wt), or its variants expressing POL2rc (lane 2) or pol2rc-ΔN allele grown in YPRGAU (lane 3). All five repeats of Western blots gave a comparable ratio of normal and truncated proteins, although different detection kits with a wide range of sensitivity were used (Materials and Methods). Under our conditions and antibodies, truncated Pol2-ΔN protein levels in extracts of strains grown without induction were impossible to determine precisely because of nonspecific bands running approximately in the same position.
Yeast media for and growth conditions
We used standard YPD yeast media (Sherman et al. 1986) but supplemented with adenine and uracil for better growth of Ura− and Ade− mutants and prevention of red pigment accumulation in ade2 strains. Yeast extract and peptone were from “ForMedium™” (Norfolk, UK) at UNMC and from Helicon (Moscow, Russia) or DIA-M (Moscow, Russia) in St-Petersburg University. The nomenclature at UNMC is YPDAU (1% yeast extract, 2% peptone, 2% dextrose, 60 mg/L adenine, 62.5 mg/L uracil); YPGAU (same but with 2% galactose instead of glucose), YPRGAU (same, but sugars were 1% raffinose plus 1% galactose); Ccan (synthetic minimal without arginine and with 60 mg/L L-canavanine), HB (minimal with uracil, leucine, threonine, lysine, and histidine). The supplement of adenine and uracil was not needed for YPD or YPRG (named here YPD* and YPRG*) in the St-Petersburg laboratory because of the different sources of yeast extract and peptone. The results between the two laboratories were similar despite the slight difference in media composition. When necessary, we supplemented media with drugs: 100 mM hydroxyurea (HU), 0.65 μM of phleomycin, or 90 μM of benomyl (all from Sigma-Aldrich, USA). Unless stated specifically, yeast strains were grown at 30°. Liquid cultures were grown in 20-ml tubes in a drum rotator.
Molecular genotyping of yeast strains
We routinely did PCR analysis of integration sites using primers complementary to the junctions of the targeted chromosomal region and the vector with recoded genes (the sequences of primers are available upon request). To study the mutations in the CDC28 gene, we amplified the CDC28 gene using yeast chromosomal DNA with primers: CDC28 F242 AGCCAGCACATCAGCTACAGTGG and CDC28 R1414 ACGTCATGGAACACGCCCAGC. The resulting PCR fragment was sequenced with primers: F427 CDC28 GGTGTTCCCAGTACAGCCAT, F947 CDC28 TGAGATCGATCAGATTTTCAAGA, R549 CDC28 GCTTGTGTGCATCAGAGTGAA and R1060 CDC28 GAGGAAAGCTTGGCTTGAAA.
Microscopy
Cells were stained by Vectashield Mounting Medium with DAPI reagent according to the manufacturer's protocol (Vector Laboratories, CA, USA). Imaging was done with a Leica DM 6000B microscope (Leica Microsystems GmBH, Germany) and Leica QWin Standard V3.2.0 software. Fluorescence was detected using the “A” filter component with a 340-nm excitation filter and a 425-nm transmission filter.
Estimating the proportion of dead cells in yeast cultures
Strains of interest were grown in 200 μl of the appropriate liquid media until the stationary phase (48 h). Dead cells were stained with methylene blue as described before (Kwolek-Mirek and Zadrag-Tecza 2014), but using a higher dye concentration. Small aliquots of yeas cultures (1–3 μl) were mixed with 30 μl of staining solution (0.2 mg/ml of methylene blue in 2% sodium citrate) and incubated at room temperature for 5 min. Fifteen microliters of yeast suspension was loaded on a microscope slide. At least 1000 cells for each culture were examined under a bright-field microscope (Zeiss Axio Scope.A1), and the percent of dead cells accumulating the blue stain was determined.
Determination of growth of individual colonies
First, fresh ascospores of the PSD93+/ΔPOL2 [pJA8] strain were obtained by tetrad dissection on complete media with galactose. Four wild-type and four pol2Δ::kanMX trp1-289::pol2rc-ΔN colonies were then transferred to 200 μl of liquid YPRGAU media. The strains were grown for 48 h. Then a small aliquot of each strain was then streaked on thin agar plates with the same media. Using a microscope and micromanipulator, we picked 25 healthy cells with no visible buds for each strain and placed them in a square 5 × 5 pattern. Cells grown at room temperature were monitored every hour during the next 10 h to find the time when first, second, and third buds emerged. The data obtained were used to determine the doubling time or the time at which the visible bud had appeared. After 24 h of incubation, the number of cells that produced microcolonies and the final size of the colonies were recorded.
Antibodies for the C-terminal half of yeast Pol2
We used a custom antibody service from ABclonal (Woburn, MA, USA). Two antigenic peptides, STWEVLQYKDSGEPG and AGAWEGTLPRESIV, corresponding to amino acids 1320–1334 and 2182–2195 of Pol2, respectively, were synthesized. Two rabbits were immunized, boosted, and antiserum further purified by affinity chromatography. The company provided purified antibodies at 2 mg/ml concentration.
Immunoblots
Yeast cells were disrupted by vortexing (twice for 2 min alternated by 1 min chill on ice) with 500 μl glass beads in 500 μl PK lysis buffer (50 mM Tris-Cl pH7.6, 50 mM NaCl, 0.1% Triton X-100, 0.1% Tween 20, 1 mM EDTA, 0.5 mM PMSF) (Pessoa-Brandao and Sclafani 2004) or by alkali method (Kushnirov 2000). Cell debris and beads were pelleted by centrifugation at 14,000 g for 15 min, and the supernatant was transferred to a new tube. Total protein concentration was determined, and the appropriate amount of extract (50–100 μg) was mixed with loading buffer, denatured, and loaded on 4–12% or 10% NuPage Bis-Tris gel. For control of the sizes of separated proteins, PageRuler Plus Prestained Protein Ladder covering a range from 10 to 250 kDa (Thermo Fisher Scientific) and 2.5 nM pol ε purified as described (Xing et al. 2019) were used. After electrophoresis, proteins were transferred to a PVDF membrane (Millipore). The membrane was incubated for 1 h at room temperature in blocking buffer (5% BSA in 1× TBST buffer) and then cut into sections corresponding to larger and to smaller proteins. The section of the membrane with separated 95–300 kDa proteins was incubated with primary antibodies against the C-terminus of Pol2 described in the previous section at 1:500–1:1000 dilution in the diluent provided in the SuperSignal Western Blot Enhancer Kit (Thermo Fisher). The section of the membrane with 50–75 kDa proteins was incubated with rat monoclonal antibodies against yeast α-tubulin (sc-53030, Santa Cruz Biotechnology) at 1:2000–1:4000 dilution. Incubation with primary antibodies was carried out at 4° overnight with gentle rocking. Then, after washing, the membranes were incubated in standard blocking solution with the appropriate horseradish-peroxidase conjugated secondary antibodies diluted at 1:2000–1:4000: goat anti-rabbit IgG, ab97051 (Abcam) or goat anti-rat sc-2006 (Santa Cruz) at room temperature for 1 h. Detection was done by either Western Breeze chromogenic kit or chemiluminescence kit (Pierce™ ECL Western Blotting Substrate, Thermo Fisher) using Kodak BioMax Light film.
Mutation rates and mutant frequencies determination
The experimental scheme for determining the mutation rate in TRP1::pol2rc-ΔN pol2Δ segregants possessing only the C-terminal half of Pol2 is presented in Supplementary Figure S3. Colonies of spores grown on media with galactose after tetrad dissection were directly transferred into the glucose-containing broth for mutation rates determination. The spores after tetrad dissections better grow on galactose-containing plates, but after that, the growth of liquid cultures started by material from these colonies is comparable in galactose- or glucose-containing media. The rate of CAN1 forward mutation (all types of mutations, including base pair substitutions, frameshifts, large deletions, and chromosome arm loss) and his7-2 reversion (predominantly +1 insertions and, rarer, −2 deletions) was measured by fluctuation analysis as described (Pavlov et al. 2001; Northam et al. 2010). The median rate was calculated by Drake's formula (Drake 1991), as described before, from at least nine spores of the same genotype for each experiment (repeated five times) (Pavlov et al. 2001). UV light mutagenesis was studied as before (Stepchenkova et al. 2017). Appropriate dilutions of cells were plated on selective and complete plates and irradiated. The mutant frequency in each culture was calculated as a ratio of the number of mutants on selective plates to the number of surviving, colony-forming cells on complete plates multiplied by the dilution factor. We determined mutant frequency at all UV doses for each culture. The UV-induced mutant frequency for each culture was calculated by subtracting the spontaneous background frequency from the mutant frequency in the UV-irradiated cells.
Illegitimate mating assay
The “illegitimate” mating assay, or the alpha-test, was used to score the frequency of several distinct genetic changes in chromosome III. The principles of the assay were described in detail in (Inge-Vechtomov and Repnevskaya 1989) (Kochenova et al. 2011) and presented in short in the Results section. For the assay, aliquots of overnight cultures of nine fresh segregants of MATα POL2 or pol2rc-ΔN strains at an appropriate dilution were plated on YPDAU media to estimate the number of live cells in each culture. In parallel, aliquots of the same cultures were plated together with the 10D-D925 strain (Inge-Vechtomov and Repnevskaya 1989) on HB media for illegitimate hybrid selection. After 3 days of incubation, we counted colonies on HB and YPDAU plates and then determined the frequency of illegitimate hybridization. We tested at least 500 illegitimate hybrids for each wild type and pol2rc-ΔN for the mating type and auxotrophy for markers of the left and right arm of chromosome III to calculate the frequency of chromosome III or its right arm loss.
Genomic DNA sequencing and analysis
We sequenced 96 whole genomes of genotyped segregants (progeny of 28 independent pol2rc-ΔN segregants) as well as the parental strains (PSD93 and its derivatives heterozygous for pol2Δ::kanMX). After tetrad dissection, independent pol2rc-ΔN spores were first streaked on glucose- or galactose-containing media. Among subclones of four spores (2A, 3B, 15C, and 19A), we selected 12 colonies for further sequencing: three big and three small colonies from glucose media and three big and three small colonies from galactose media. For 24 spores (59B to 90B), we selected only two big colonies from glucose media (Supplementary Figure S3). Selected pol2rc-ΔN colonies were grown in 10 ml of the same medium, and chromosomal DNA was isolated by a standard method using phenol extraction (Lada et al. 2013). We sheared DNA in an M220 Focused-ultrasonicator (Covaris, USA), prepared genomic libraries with the TruSeq® Nano DNA LT Prep Kit (Illumina, CA, USA), and performed the whole-genome sequencing using the Illumina HiSeq 2500. We analyzed the raw data using the pipeline described previously (Lada et al. 2015, 2017). The quality of the raw reads was assessed by FastQC. Reads were filtered and trimmed using Trim Galore with base quality equal to or higher than 20. We mapped reads that passed quality control to our reference strain LAN210_v0.10m using Bowtie2. We updated our pipeline using tools from the GATK 4.0.3.0 to call (HaplotypeCaller), sort, and filter variants (SelectVariants, VariantFiltration). For the haploid clones, single-nucleotide variants (SNVs) with AF = 0.5 were manually examined in IGV 2.4.10, confirmed to be false positives, and removed from the final vcf file.
Protein structure modeling
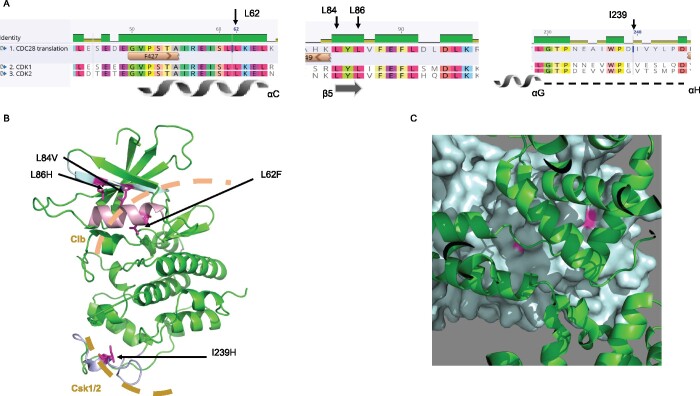
The yeast Cdc28 model (P00546) was built on PDB crystal structure 6gu7.2 that was obtained from the Swiss-Model repository. The yeast Cdc28 model was structurally aligned to Human CDK1 in complex with CsK1 and Clb1 (PDB: 5lqf), and mutations in yeast Cdc28 were visualized using the PyMOL software.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and supplemental materials. The raw NGS data are deposited in Sequence Read Archive (SRA, accession # PRJNA634680, Temporary submission ID: SUB7487615). The BioProject accession number is provided in lieu of SRP and will allow better searching in Entrez. Full scripts used to generate SNV datasets are available upon request. Supplementary files are submitted via GSA figshare portal: https://doi.org/10.25386/genetics.14357234 (accessed 04.28.2021).
Results
Construction of haploid strains without the N-terminal part of cs-pol ε
We and others (Devbhandari and Remus 2020) found that the construction of haploids strains with pol2-16 is challenging (Supplementary Results S1). To investigate the biological effects of the absence of the polymerase with a prominent role in leading strand synthesis, we needed a different allele that leads to the removal of the catalytically active half of Pol2 but does not affect the second role of pol ε as part of CMG helicase. For the reliable construction of such haploid strains, we used a codon-optimized regulatable allele to alleviate the low stability of the truncated protein encoded by pol2-16, as shown by Western blot for C-terminally TAP-tagged Pol2-16 (Garbacz et al. 2018). The complete removal of the N-terminal half of Pol2 results in a protein that is stable in yeast, as demonstrated by Western blot for N-terminally CBP-tagged Pol2Δcat (Pol2-ΔN) and the successful purification of the four-subunit pol ε-Δcat (Yeeles et al. 2017; Goswami et al. 2018; Devbhandari and Remus 2020). The attempts to purify pol ε with Pol2-16 failed (Devbhandari and Remus 2020).
We used the following approach to create yeast strains with the pol2 variant lacking the first half of the gene. In the first step, we created a yeast diploid heterozygous for a complete deletion of the POL2 gene genetically marked by G418 resistance (Materials and Methods, Supplementary Figure S1 and S3, Fig. 2A). Tetrads of the sporulating diploids yielded two viable and two unviable spores (Figure 2B). All viable spores were sensitive to G418, i.e., possessed a wild-type POL2 gene. This result is consistent with the known inviability of strains with the deletion of the whole POL2 gene (Morrison et al. 1990). In the second step, we integrated either POL2rc or pol2rc-ΔΝ alleles into the TRP1 locus of heterozygous diploids using plasmids pJA6 (Yeeles et al. 2015) or pJA8 (Yeeles et al. 2017), respectively, Figure 2A.
Tetrad dissection was done on plates with glucose or raffinose (low levels of expression of pol alleles) or on raffinose plates with galactose (high expression of the optimized genes) (Materials and Methods and Figure 2, B and C). Genetic segregation of the progeny of POL2/pol2Δ strain heterozygous for trp-289::POL2rc::TRP1 (insertion of pJA6) followed a typical digenic scenario with one lethal allele and its unlinked suppressor. Most tetrads had three viable spores (tetratype). All viable spores were of the same size, even on glucose-containing medium, indicating the full complementation of the deletion by the recoded POL2rc allele (Figure 2B) when levels of Pol2 were just twofold higher than in POL2 strains (Figure 2C).
Tetrad analysis of POL2/pol2-Δ strains heterozygous for trp-289::pol2rc-ΔN::TRP1 (Supplementary Figure S1) yielded two classes of haploid colonies, large and small. Representative results are shown in Figure 2B, right half. Small colonies, presumably with pol2rc-ΔΝ, were barely visible on day 3 but were distinctly seen on day 4. The improvement of growth is in stark contrast with the pol2-16 strains that either did not grow (Devbhandari and Remus 2020) or grew very slowly, forming visible colonies in 2 weeks (Kesti et al. 1999; Garbacz et al. 2018). All small colonies were G418-resistant and Trp+; thus, the truncated allele pol2rc-ΔN was the only source of Pol2 (Supplementary Figure S3). Genome sequencing of these segregants confirmed the absence of the normal POL2 gene and duplication of the TRP1 locus (Figure 2A, Supplementary Figure S4). We concluded that the pol2rc-ΔN allele encoding for the Pol2 with the whole N-terminal part deleted (Figure 1C) allows for satisfactory growth in 4 days of the bearer segregants, though colonies are distinctively smaller than the POL2 colonies (Figure 2B, Supplementary Figure S3). The complete absence of the N-terminal half of Pol2, optimization of codon usage, and regulatable expression allow for levels of Pol2-ΔN comparable to Pol2 (Western blot, Figure 2D). It is essential to mention that we have used antibodies detecting untagged Pol2, excluding the possibility that the N-terminal tag is the reason for the stability of the truncated tagged Pol2 (Devbhandari and Remus 2020).
In total, we analyzed 53 tetrads of two heterozygous parent diploids, and the overall ratio of tetrads with four viable spores to tetrads with two and three viable spores was close to 1:1:4 (P = 0.88) predicted by independent segregation of a wild-type POL2 on chromosome XIV and pol2-ΔN on chromosome IV (Supplementary Figure S1). Thus, the truncated Pol2-ΔN lacking the catalytically active part of Pol2 suppresses the lethality caused by the complete deletion of the POL2 gene. We confirmed by sequencing that pol2-ΔN integrated into the chromosome was identical to the original plasmid sequence, ruling out the hypothesis that the truncated inactive pol half acquired polymerase activity by recombination with genes of other active DNA pols. We concluded that the strains with the pol2rc-ΔΝ allele possess sufficient levels of the C-terminal half of Pol2 for the assembly of CMG helicase and can bypass the requirement for the catalytic activity of pol ε on the leading DNA strand.
Characterization of pol2rc-ΔN mutants
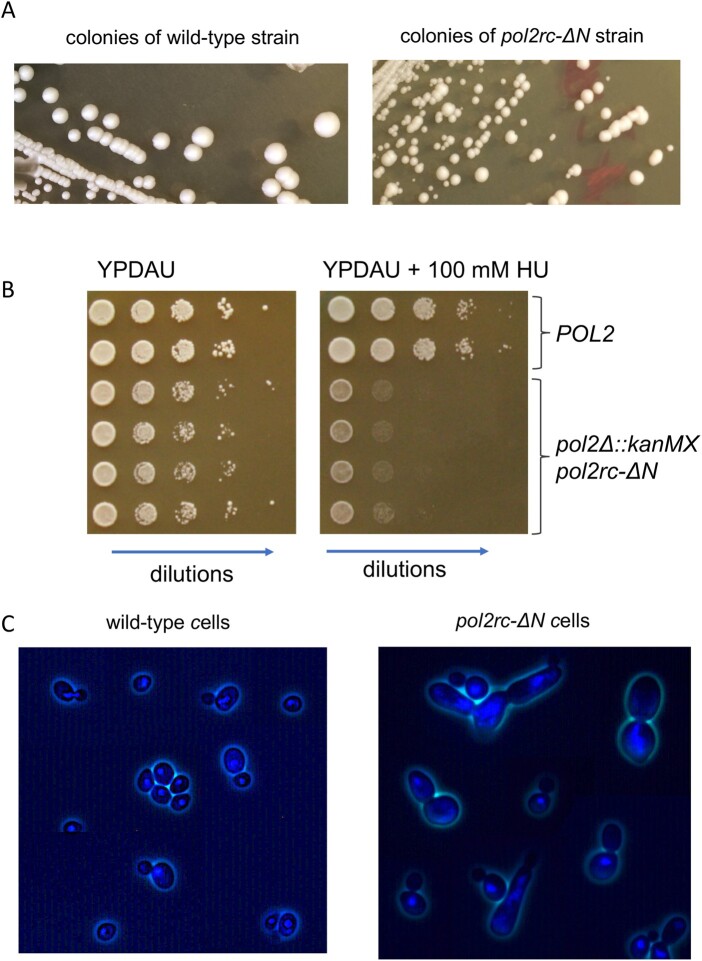
Analysis of the phenotypes of pol2rc-ΔN spores matched most observations made for pol2-16 strains (Dua et al. 1999; Kesti et al. 1999; Garbacz et al. 2018). Strains with pol2rc-ΔN grew slowly (Figure 3A), were sensitive to ribonucleotide reductase inhibitor hydroxyurea (HU) (Figure 3B), and had aberrant nuclear and cell morphology in comparison to wild-type yeast (Figure 3C). Typically, yeast cells are either single or double with smaller buds and larger mother cells. Both types have compact nuclei (Figure 3C, left panel). The pol2rc-ΔN cells (even from relatively normally looking colonies grown on galactose) show several types of anomalies: large cells, buds with no nuclei, abnormally shaped cells, and diffuse nuclei Figure 3C, right panel). The proportion of such defective cells reaches 80% in pol2rc-ΔN cells in drastic contrast to wild-type cells (Supplementary Figure S5). We did not detect cold-sensitivity (to 20°) or thermo-sensitivity (to 37°) of pol2rc-ΔN strains (Supplementary Figure S6), while pol2-16 was described to be temperature sensitive (Ohya et al. 2002). As observed previously for pol2-16, the pol2rc-ΔN strains rapidly produced faster-growing colonies (Figure 3A, right panel).
Figure 3.
Characteristics of pol2rc-ΔΝ strains. (A) Small colonies after tetrad dissection (Figure 1B) give large and small colonies when re-streaked onto the YPRGAU medium. The photo on the left is a wild-type strain (the photo taken on day 3), and on the right—is the pol2rc-ΔN strain (photo taken on day 4). (B) Sensitivity to hydroxyurea (HU). Serial 10-fold dilutions of cell suspension were plated in horizontal rows by a 48-prong device on control or HU-containing YPDAU plates, and photos were taken on day 2. (C) DAPI-stained cells from small pol2rc-ΔN colonies that were shown in (B) in comparison to wild-type cells. The truncation of POL2 led to aberrant cells with irregular distribution of nuclear material, daughter cells without nuclei, and abnormally large or weird-shaped cells.
To understand the reasons for the slower growth of pol2rc-ΔN cells, we have used two approaches. First, we stained dead cells with methylene blue. The dye penetrates all cells, but only live cells pump the dye out and remain colorless (Kwolek-Mirek and Zadrag-Tecza 2014). We found that the proportion of methylene blue-stained cells in pol2rc-ΔN is 3-5 times higher than in wild-type cells (P < 0.01), Supplementary Figure S7. The proportion of such cells in galactose is higher in both strains because not every cell easily accommodates for galactose utilization, but the trend for a higher proportion of dead cells in pol2rc-ΔN is the same. Interestingly, there is no direct correspondence between atypical cells (80%, Supplementary Figure S5) and dead cells (up to 14%, Supplementary Figure S7), suggesting that cell repair mechanisms could override the anomalies in pol2rc-ΔN mutants with time. Consistent with this, not all abnormally shaped cells are stained by methylene blue (Supplementary Figure S7A). The relatively low number of dead cells only partially explains the slow growth of pol2rc-ΔN strains.
We obtained additional clues on what happens in a second approach where we followed the growth of individual cells (Materials and Methods), Supplementary Figure S8, Table 1. Only a few cells (2%) in wild-type strains do not divide at all in 24 h. The proportion is higher in the pol2rcΔN strain and reaches 8%. The proportion of nondividing cells corresponds to the fraction of inviable cells determined by methylene blue staining. Most wild-type cells divide more than three times, while a large proportion of pol2rcΔN cells complete only one to two divisions in 24 h (Table 1). Interestingly, 76% of pol2rc-ΔN cells divide at a rate similar to wild-type cells, forming microcolonies of the same size but with irregular shapes (Supplementary Figure S8, Table 1). The median of dividing time of pol2rc-ΔN and wild-type cells in the galactose-containing medium is the same 5 h in both strains (P = 0.341, Mann–Whitney U test). These results agree with cell cycle analysis of pol2-16 strains by flow cytometry, where no significant decrease in the rate of DNA synthesis was detected (Kesti et al. 1999). The high heterogeneity of pol2rc-ΔN cells in the ability to divide adds to the colonies’ initial slow growth and abnormal shape of microcolonies.
Table 1.
High proportion of pol2rc-ΔN cells cannot form or have difficulty forming daughter cell buds compared to wild-type cellsa
| Strain | Percent of cells that divided the indicated number of times: (95% confidence intervalb) |
||
|---|---|---|---|
| Zero | One to two | More than three | |
| wild type | 2 (0.5–7) | 1 (0.2–5) | 97 (91.5–99) |
| pol2rc-ΔN | 8 (4–15) | 16 (10–24) | 76 (65–82) |
aThe experiment design is described in Materials and Methods. The photo of the final appearance of the grids is in Supplementary Figure S8.
bThe 95% confidence interval was calculated using the Score method (Breusch and Pagan 1980).
Association between wild type or pol2rc-ΔN and how many times cells divided is statistically significant, estimated by Pearson’s χ2 statistic in a contingency table (χ2 =18.9, P = 0.000079).
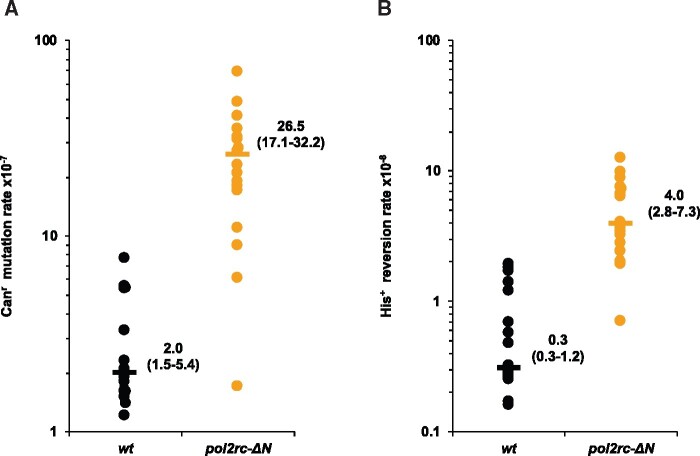
When we analyzed small colonies grown after tetrad dissection without additional subcloning (Materials and Methods, Supplementary Figure S3), segregants with pol2rc-ΔN had a 10- to 100-fold elevated spontaneous forward mutation rate (Figure 4A), as was shown previously for the pol2-1 mutant (Northam et al. 2006) with the insertion of the URA3 gene in the middle of POL2 (Figure 1C) (Morrison et al. 1990) and the pol2-16 mutant (Ohya et al. 2002; Garbacz et al. 2019). The rate of frameshift mutations, as measured by his7-2 reversion, is also increased 10-fold (Figure 4B). The mutation rates in pol2rc-ΔN segregants were highly variable and partially (60%) dependent on the activity of pol ζ (Supplementary Table S1), which corresponds to previous observations with other mutations impairing pol ε (Ohya et al. 2002; Northam et al. 2006; Garbacz et al. 2019).
Figure 4.
pol2rc-ΔN strains are spontaneous mutators. (A) The rate of spontaneous mutations to canavanine-resistance (Canr) is compared in independent POL2 (black dots) and pol2rc-ΔN (yellow dots) segregants. Each dot represents the mutation rate in an independent colony of haploid spore grown on YPRGAU tetrad dissections plates and further cultivated in liquid YPDAU. The procedure minimizes the number of cell divisions and randomizes the effects of putative suppressor mutations (Nick McElhinny et al. 2008). Horizontal lines show the median values. The graph also shows the values of median and 95% confidential interval. The significance of the difference between wild-type and pol2rc-ΔN strains was estimated by Wilcoxon–Mann–Whitney test (P = 1.54E−6). (B) The rate of spontaneous His+ reversion in the descendants of the same spores as in (A). The difference between wild-type and pol2rc-ΔN is statistically significant (P = 3.39E−9).
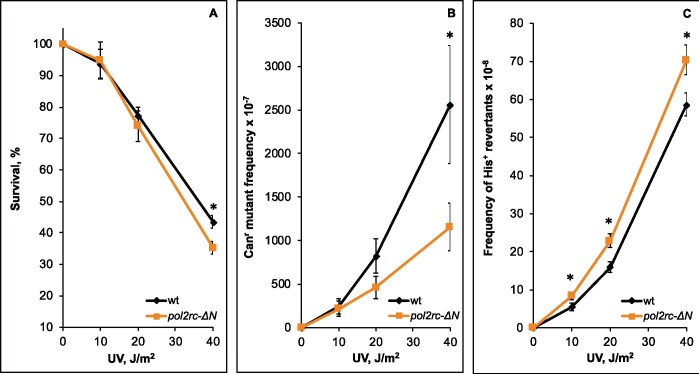
To learn how the absence of cs-pol ε affects the response to DNA damaging agents, we compared the sensitivity of pol2rc-ΔN and wild-type strains to the lethal and mutagenic action of UV light (Figure 5). The slight difference in survival between the strains was seen only at the highest UV light dose (Figure 5A, Supplementary Table S2). This observation goes along with the earlier conclusion that pol2-16 strains are not sensitive to UV (Kesti et al. 1999). We found no differences in the induction of forward mutations at low and intermediate doses of UV light and a twofold decrease at the highest dose (Figure 5B). We observed a small but significant increase in the frequency of induced his7-2 reverse frameshift mutations in pol2-ΔN strains (Figure 5C, Supplementary Table S2). These results suggest that polymerase activity of Pol ε and the Fe-S cluster located in the N-terminal half of Pol2 are not absolutely required for induced mutagenesis, unlike Fe-S clusters in pol δ and pol ζ (Giot et al. 1997; Baranovskiy et al. 2012; Johnson et al. 2012; Makarova et al. 2012; Siebler et al. 2014; Stepchenkova et al. 2017). Further work is required to find what types of mutations are attenuated at high doses of UV.
Figure 5.
Alterations in UV-survival and mutagenesis in pol2rc-ΔN strains. (A) Survival of pol2rc-ΔN (here and in other panels—yellow dots and line) is slightly less than the survival of wild-type strain (here and in other panels—black dots and line) only at the highest UV dose (P = 0.0028). Graphs show the mean and standard error of the mean. Data were obtained in two experiments, each with nine independent measurements for every UV dose. A one-tailed t-test was used to evaluate the significance of the difference between means. Asterisks in this and other panels mark statistically significant differences between wild-type and mutant strains. (B) The frequency of UV-induced forward Canr mutations conferring resistance to L-canavanine in pol2rc-ΔN is similar to the wild-type strain, except for a twofold decrease (P = 0.03) at the highest dose. (C) The pol2rc-ΔN strain has an elevated frequency of reversions to His+ of the frameshift allele his7-2 (10 J/m2: P = 0.02; 20 J/m2: P = 0.0022; 40 J/m2: P = 0.01). The mean ± SEM frequency of spontaneous His+ reversion is 0.28 (± 0.1–1.2) × 10−8 for wild-type clones and 3.9 (± 2.0–7.3) × 10−8 for pol2rc-ΔN clones.
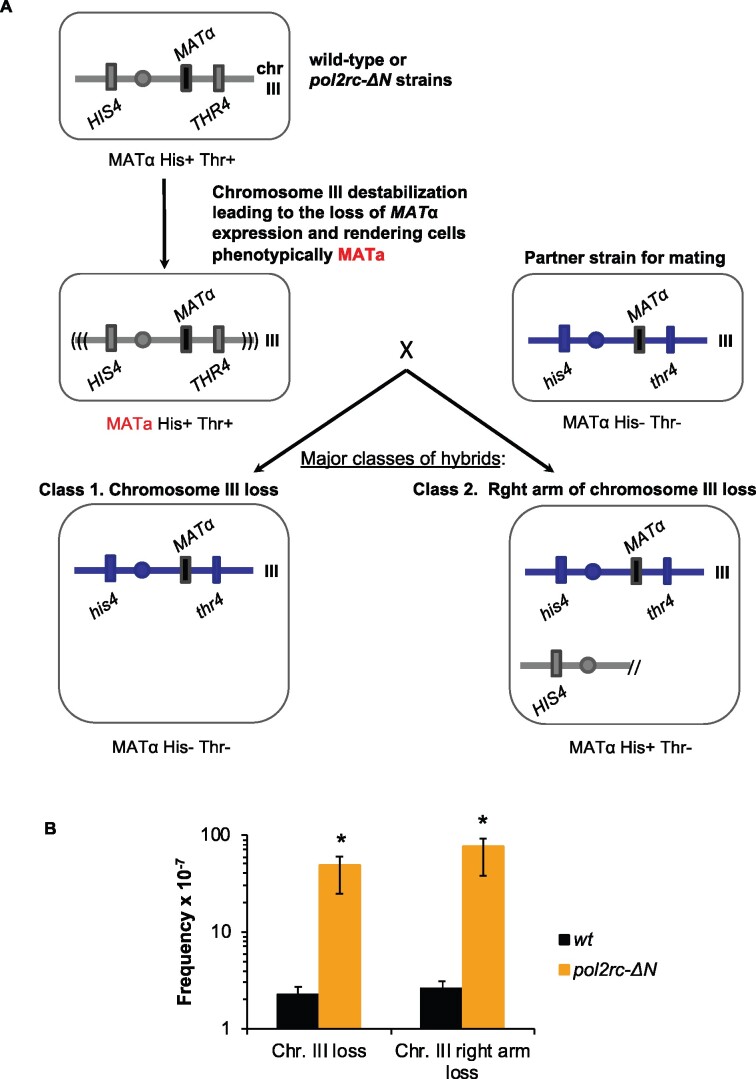
Severe defects in nuclear morphology in pol2rc-ΔN cells indicated difficulties replicating nuclear DNA that may cause chromosomal rearrangements. We used the illegitimate mating test (Inge-Vechtomov and Repnevskaya 1989) to analyze chromosome stability and revealed that the rate of the loss of chromosome III or its arm was elevated 50-fold in pol2rc-ΔN strains (Figure 6). The high frequency of chromosomal abnormalities, together with elevated mutation rates, can contribute to the slow growth and cell death in cultures of pol2rc-ΔN strains. Point mutations or presumed premutation lesions leading to hybridization of strains of the same “α” mating type were elevated fivefold, while recombination events leading to hybridizations were not stimulated in pol2rc-ΔN strains (Supplementary Table S3).
Figure 6.
Increase in illegitimate mating of MATα strains in pol2rc-ΔN strains. (A) Schematic of the prevalent genetic events leading to illegitimate mating. In haploid yeast strains, mating type (α or a) is determined by alternative MATα or MATa alleles of the MAT locus. The MAT locus is located in the right arm of chromosome III. If MATα expression is disturbed, heterothallic yeast strains of the mating-type “α” behave like “a” (marked by red font) and can hybridize with MATα strains (Strathern et al. 1981). The frequency of this illegitimate hybridization reflects the level of genome destabilization. The mating-type changes in heterothallic strains happen when a cell loses the whole chromosome III or its right arm or when the MATα locus is mutated. Also, MATα may be substituted for by an HMRa cassette via recombination/conversion mechanism. It is possible to score the frequency of each of these genetic events when strains have selective markers on both arms of chromosome III, in our case, his4 and thr4 (Inge-Vechtomov and Repnevskaya 1989; Kochenova et al. 2011). Phenotype “α” His− Thr− indicates chromosome III loss; “α” His+ Thr−—for chromosome III arm loss. Minor classes (not shown) are nonmater “n/m” His+ Thr- and “n/m” His+ Thr+. They result from recombination/gene conversion between MATα and HMRa and “α” His+ Thr+ arise by mutations/lesions in the MATα. (B) Fiftyfold-elevation of the frequency of chromosome III or its right arm loss in pol2rc-ΔN strains. Graphs show the median and 95% confidence interval. *The significance of the difference between wild-type and pol2rc-ΔN strains was estimated by Wilcoxon–Mann–Whitney test (P = 0.00002165).
Suppressors of pol2rc-ΔN “slow growth” phenotype
We were intrigued by the fast accumulation of near-normal growing healthy clones in pol2rc-ΔN strains (Figure 3A). To detect suppressors enabling strains without the active part of Pol2 to grow, we analyzed genomes of pol2rc-ΔN segregants (Supplementary Figure S3). In the first sequencing run, we analyzed differently sized clones of four segregants grown on glucose or galactose plates (Materials and Methods). We found approximately the same number of mutations in the genomes of small and large clones (Supplementary Tables S4 and S5). Mutations present in every genome of all subclones of large and small colonies grown on glucose and galactose (e.g., in GSP1, or RPS1A, or OXA1) may have happened during the early divisions of the spore after tetrad analysis. Mutations present only in genomes of descendants of the large clones likely contributed to the improvement of growth. In some cases, new mutations were found only in genomes obtained from the analysis of small colonies consistent with elevated mutation rates in pol2rc-ΔN strains. We cannot rule out that at least some mutations resulted from the rapid accumulation of fast-growing genetic variants in small clones during their growth for 2 days in the liquid medium before the isolation of genomic DNA.
Because we did not observe drastic differences in the number of mutations between small and large initial colonies and growth conditions, we next sequenced only two independent clones of 24 pol2rc-ΔN segregants. The summary of the overall results of genomic sequencing is presented in Supplementary Tables S4–S6. Overall, we sequenced genomes of 96 strains that originated from 28 independent pol2rc-ΔN segregants (Materials and Methods) and found 292 base pair-substitution mutations. On average, the genome of each analyzed strain possessed three new mutations. Three genomes had no mutations, and nine genomes had no base substitutions in coding regions, though they may have genome rearrangements or epigenetic changes not detected by our analysis. Eighty-four genomes had from one to seven mutations, Supplementary Table S5. Several mutations were observed repeatedly in different genomes. Identical mutations found in genomes of common origin (Supplementary Tables S4–S6, offspring of a single segregant) likely occurred early during spore growth and were recorded as one mutational event. Altogether, we found 197 independent, unique mutations.
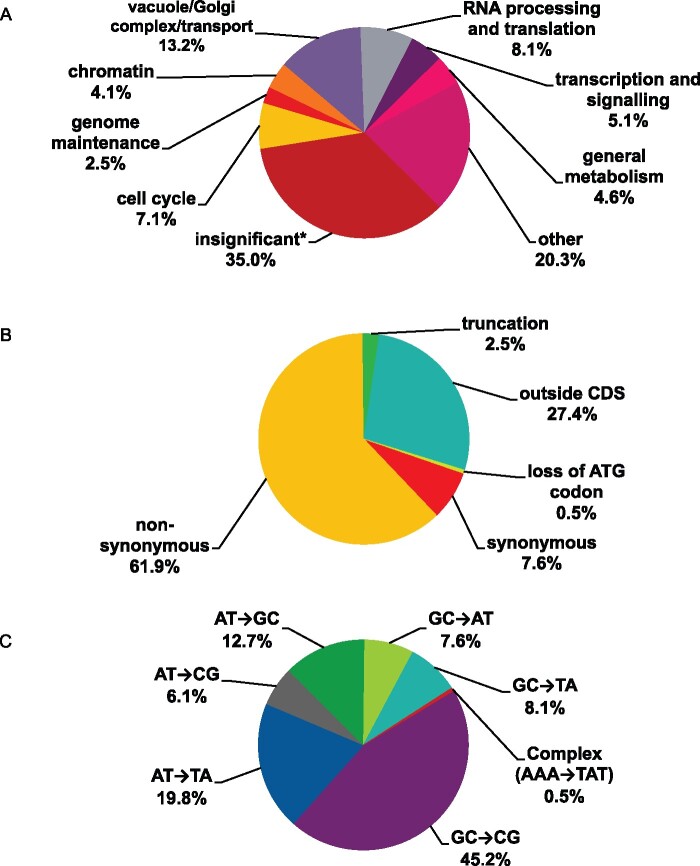
We found mutations in genes associated with cell cycle regulation, DNA metabolism, chromatin, transcription, splicing, translation, cytoskeleton, and other cellular processes (Figure 7A). Two characteristics of these mutations are relevant to the understanding of their origin. First, the proportion of synonymous changes was less than 8% (Figure 7B), indicating that the appearance of clones with these mutations results from positive selection for mutations improving growth. Typically, in collections of unselected genomic mutations, the proportion of synonymous to nonsynonymous changes does not deviate from a 1:1 ratio by more than 15% (Lujan et al. 2014). Second, the prevalent type of mutations was CG to GC transversions that constitute the signature of pol ζ (Northam et al. 2010; Grabowska et al. 2014; Stodola et al. 2016) (Figure 7C, Supplementary Table S6), consistent with the participation of pol ζ in mutagenesis in pol2rc-ΔN strains (Supplementary Table S1). We also found one complex mutation (AAA->TAT), also a type of mutation characteristic of pol ζ-dependent synthesis (Harfe and Jinks-Robertson 2000; Stone et al. 2009; Northam et al. 2010; Kochenova et al. 2017). For most mutations, it is not immediately obvious how certain changes or combinations of changes helped mitigate the growth defect of pol2rc-ΔN. The most relevant to the restoration of typical cell division rates are genes that contribute to cell cycle control, DNA repair and recombination, and cell metabolism regulation. Recurring mutations are the most attractive candidates. Among these, which stood out, were five independent, nonsynonymous mutations in the CDC28 gene, encoding for a cyclin-dependent kinase. In the previous work on the genomic landscape of mutations induced by a base analog or APOBEC deaminases in yeast, we found only one CDC28 mutation (synonymous) among 17,048 mutations (Lada et al. 2013, 2015, 2017). Therefore, we focused further on clarifying the role of changes in CDC28 in the rescue of the growth defect of pol2rc-ΔN strains.
Figure 7.
Overview of mutations found in genomes of pol2rc-ΔN strains. (A) Functional groups of genes where mutations were found. *The group named “insignificant” includes synonymous mutations and mutations outside ORFs. (B) Functional significance of mutations. (C) Types of mutational changes.
Mutations in the CDC28 gene suppress the growth defect of the pol2rc-ΔN
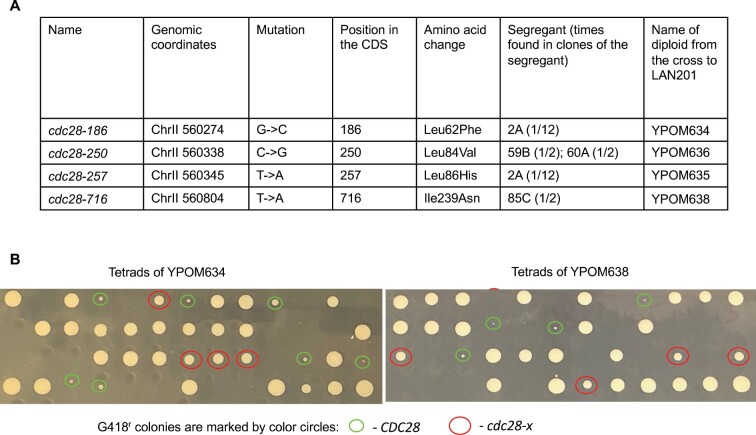
As detailed in the previous subsection, genome sequencing of fast-growing pol2rc-ΔN strains (Figure 3A) revealed five independent occurrences of mutations in the CDC28 gene (Supplementary Table S4, Figure 8A). Two independent clones of spore 2A possessed different mutations, cdc28-186 and cdc28-257. The mutation cdc28-250 was found in clones of two different independent segregants, 59B and 60A. Mutation cdc28-716 was present in segregant 85C. The probability of observing five independent mutations in the same gene in our collection of sequenced genomes by chance is 1.2 × 10−6 (Supplementary Results S2). The length of the CDC28 open reading frame (ORF, 298 codons) is much shorter than the mean ORF length for the yeast genome (491.1 codons; (Marín et al. 2003), thus the probability value is likely to be underestimated (a conservative estimate of the P-value).
Figure 8.
Mutations in CDC28 suppress the growth defect of pol2rc-ΔN. (A) cdc28 alleles found during genomic sequencing. (B) Representative analysis of co-segregation of “good growth” phenotype and alleles of CDC28. The photo shows the appearance of colonies after tetrad analysis after 3 days of growth. The CDC28 status determined by sequencing of G418r colonies is marked by circles of a different color.
To examine the functional significance of CDC28 mutations and their contribution to the improvement of growth of yeast strains on YPDAU without induction, we crossed each segregant to the wild-type strain LAN201 (described in Materials and Methods, illustrated in Supplementary Figure S9, the names of the hybrids are in Figure 8A). We reasoned that half of the G418r segregants bearing the pol2rc-ΔN allele would get wild-type CDC28 from LAN201, while the other half would possess the mutated cdc28 allele (Supplementary Figure S9). The results of the experiments are presented in Table 2 and illustrated in Figure 8B. Two alleles exhibited almost 100% co-segregation of slow growth with wild-type CDC28 status and good growth with cdc28 mutation as determined by sequencing of G418r Trp+ clones. All barely growing colonies (labeled “extra small” in Table 2) with pol2rc-ΔN possessed the wild-type CDC28, and all medium-sized pol2rc-ΔN colonies contained either cdc28-186 or cdc28-716 (Figure 8B). Two mutations, encoding for L84V and L86H, also segregated with the “good growth” phenotype most of the time, with only a few exceptions (Table 2). The effect of these alleles in the rescue of pol2rc-ΔN is apparent. Additional genetic factors may lead to rare outlier well-growing CDC28 clones [one additional mutation in clone S3 of segregant 2A, three additional mutations in clone SR1 of 59B, and one mutation in SR4 clone of segregant 60A (Supplementary Table S4)].
Table 2.
Genetic analysis reveals that mutations in CDC28 efficiently suppress the growth defect of pol2rc-ΔN segregants
| Diploids: assortment of POL2 and CDC28 alleles |
Genotyped segregants with defective POL2 |
||||||
|---|---|---|---|---|---|---|---|
|
pol2Δ::kanMX/POL2
trp1-289::pol2rc-ΔN-TRP1/trp1-289 |
G418r Trp+ segregants (extra small/medium sized) |
||||||
| Tetrads dissected | Total viable spores | Colony size | Wild-type CDC28 allele | Mutant cdc28 allele | |||
| YPOM634 (CDC28/cdc28-186) | 76 | 208 |
71 (34/37) |
Extra small | 31 | 31 | 0 |
| Medium | 32 | 0 | 32 | ||||
| YPOM635 (CDC28/cdc28-250) | 60 | 165 |
49 (29/20) |
Extra small | 23 | 20 | 3 |
| Medium | 11 | 1 | 10 | ||||
| YPOM636 (CDC28/cdc28-257) | 51 | 143 |
49 (23/26) |
Extra small | 14 | 13 | 1 |
| Medium | 11 | 1 | 10 | ||||
| YPOM638 (CDC28/cdc28-716) | 78 |
214 |
67 (36/29) |
Extra small | 36 | 36 | 0 |
| Medium | 26 | 1 | 25 | ||||
|
POL2/pol2-1: |
Ura+ segregants (extra small/medium sized) |
|
|||||
|
YPOM661[p2A5] (CDC28/CDC28) |
10 | 38 |
19 (19/0) |
Extra small Medium |
19 0 |
19 0 |
n/a n/a |
| YPOM664[p2A5] (CDC28/cdc28-186) | 10 | 38 |
19 (7/12) |
Extra small Medium |
12 7 |
12 0 |
0 7 |
| YPOM663[p2A5] (CDC28/cdc28-716) | 20 | 72 | 34 | Extra small | 15 | 15 | 0 |
| (15/19) | Medium | 19 | 0 | 19 | |||
We found recurring mutations (observed at least twice) in several other genes (Supplementary Tables S4 and S5): in MCM4 encoding for a subunit of CMG helicase (Tye and Sawyer 2000) in segregants 59B and 73A; in TRE2, involved in the regulation of metal transporters (Stimpson et al. 2006) in segregants 79B and 82C; in MNN10, encoding for a mannosyltransferase (Dean and Poster 1996) in segregants 3B and 15C; in RPS1A, encoding for a ribosomal protein (Mager et al. 1997) in segregants 3B and 19A; and in BEM2, encoding for a GTPase activator involved in cytoskeleton organization (Wang and Bretscher 1995) in segregants 73A and 75D. These mutations are prospects for further investigation. Some mutations were found only once, but their occurrence adds to the vital role of CDC28-related transactions in the recovery of pol2-ΔN strains because they are in genes encoding for proteins that physically or genetically interact with Cdc28 or regulate the cell cycle (e.g., SIT4, CLB4, CDC5, and CLB2, Supplementary Table S4).
To approach understanding the suppression mechanisms, we compared the growth of wild-type strains, single pol2rc-ΔN, single cdc28-x, and the double pol2rc-ΔN cdc28-x mutants in the presence of different drugs and at different temperatures (Table 3, Supplementary Figure S10, A–D). The pol2rc-ΔN mutation, besides sensitivity to HU, leads to sensitivity to free radical producing drug phleomycin (Enserink et al. 2009) and anti-microtubule drug benomyl (Stearns et al. 1990). Interestingly, none of the cdc28 alleles, when present in POL2 segregants, conferred phenotypes associated with alterations of replication: these clones had the same growth parameters as CDC28 wild-type clones at all conditions examined (compare rows 1 and 7 to 10 of Table 3). All cdc28 alleles partially rescued HU-sensitivity of pol2rc-ΔN at 30° (compare row 2 to rows 3–6). In respect to other factors, different cdc28 alleles showed different properties when combined with pol2rc-ΔN (highlighted by bold in Table 3). The cdc28-186 mutation did not suppress the sensitivity of pol2rc-ΔN to benomyl (compare rows 2 and 3), the cdc28-250 pol2rc-ΔN double combination was even more sensitive to phleomycin than single pol2rc-ΔN (rows 2 and 4), and cdc29-716 did not rescue pol2rc-ΔN sensitivity to HU at high temperature (rows 2 and 6).
Table 3.
Mutations in CDC28 relieve growth defects and sensitivity to cell-damaging agents of pol2rc-ΔN segregantsa
| Relevant genotype | 30o | 37o | HU, 100 mM | HU, 100 mM, 37o | Phleomycin, 0.65 μM | UV light, 120 J/m2 | Benomyl, 90 μM |
|---|---|---|---|---|---|---|---|
| Wild type | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | +++ |
| pol2rc-ΔN | +++ | +++ | ++ | ++ | ++ | ++ | ++ |
| pol2rc-ΔN cdc28-186 | ++++ | ++++ | ++++ | ++++ | +++ | +++ | ++ |
| pol2rc-ΔN cdc28-250 | ++++ | ++++ | ++++ | ++++ | ++ | +++ | +++ |
| pol2rc-ΔN cdc28-257 | ++++ | ++++ | +++ | +++ | +++ | +++ | +++ |
| pol2rc-ΔN cdc28-716 | ++++ | ++++ | ++++ | ++ | +++ | +++ | +++ |
| cdc28-186 | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | +++ |
| cdc28-250 | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | +++ |
| cdc28-257 | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | +++ |
| cdc28-716 | +++++ | +++++ | +++++ | +++++ | ++++ | +++ | +++ |
The number of plus signs is an estimate of robustness of growth and corresponds to the number of dilutions where colonies are readily visible (Supplementary Figure S10). Bold and larger font marks outliers in the behavior of the double pol2rc-ΔN cdc28-x strains.
Mutations in the CDC28 gene suppress the mutator effect of pol2rc-ΔN
Mutations in CDC28 mitigated most phenotypes of pol2-ΔN caused by abnormal replication. If elevated mutation rates in strains with pol2-ΔN are the result of replication stress, we expected that cdc28 alleles would also suppress the mutator phenotype. The analysis of CAN1 forward and his7-2 reverse mutations is summarized in Table 4. Mutations in CDC28 alone did not affect mutation rates (compare row 1 and rows 2–5). pol2-ΔN leads to a 50-fold elevation of Canr mutations and a 4-fold increase of his7-2 frameshift reversions. All cdc28 alleles led to a decrease of mutation rates in pol2-ΔN strains; the effect varied from complete suppression (no statistical difference from wild type for cdc28-716) to an 8- to 15-fold reduction in comparison to pol2rc-ΔN for cdc28-186 to a 2-fold reduction for Canr mutation in strains with cdc28-250 (statistically significant, P = 0.0083).
Table 4.
cdc28 mutations suppress mutator effects of pol2rc-ΔN
| Strain | Relevant genotype | Canr × 10-7, median (95% confidence interval) | His+ × 10-8, median (95% confidence interval) |
|---|---|---|---|
| LAN201 | Wild type | 1.4 | 1.8 |
| (0.7–5.7) | (0.8–5.1) | ||
| 25A-YPOM639 | cdc28-186 | 1.3 | 1.8 |
| (0.6–2.4) | (0.6–2.5) | ||
| SEGS-YPOM641a | cdc28-250 | 4.7 | 1.7 |
| (1.5–5.8) | (1.1–6.1) | ||
| 18B-YPOM640 | cdc28-257 | 1.1 | 1.1 |
| (0.5–6.2) | (0.4–1.7) | ||
| 16D-YPOM643 | cdc28-716 | 1.4 | 0.9 |
| (0.5–1.8) | (0.2–1.7) | ||
| 33D-YPOM634 | pol2rc-ΔN | 69 b | 8 d |
| (30–96.5) | (2.1–14) | ||
| 31C-YPOM639 | pol2rc-ΔN cdc28-186 | 3.4 | 3.1 |
| (1.5–15.7) | (1.5–4.5) | ||
| SEGS-YPOM639a | pol2rc-ΔN cdc28-186 | 7.6c | 4.1 |
| (1.6–15.1) | (2.2–5.6) | ||
| 21C-YPOM639 | pol2rc-ΔN cdc28-186 | 4.6 | 2.5 |
| (3.4–7.2) | (0.6–5.1) | ||
| SEGS-YPOM641a | pol2rc-ΔN cdc28-250 | 25.5c | 3.0 |
| (16–46.5) | (1.7–7.6) | ||
| 26A-YPOM640 | pol2rc-ΔN cdc28-257 | 8.2d | 2.7 |
| (4.2–9.6) | (1.3–5.6) | ||
| 10A-YPOM643 | pol2rc-ΔN cdc28-716 | 3.6 | 2.1 |
| (2.1–16) | (0.6–3.9) |
Analysis of six independent segregants of the same genotype.
Statistically significant difference from the wild type, P = 0.00512, Wilcoxon–Mann–Whitney test.
Statistically significant difference from the wild type, P = 0.0164.
Statistically significant difference from the wild type, P = 0.0083.
The suppression effect of cdc28 mutations is not confined to only the pol2rc-ΔN allele
The expression of pol2rc-ΔN at the unusual chromosomal location is driven by the GAL1-10 promoter, and it is formally possible that the effect of cdc28 mutations is confined to that particular allele and depends on the nature of the artificial promoter or position in another chromosome. To exclude this possibility, we studied the interaction of cdc28 alleles with another POL2 allele, pol2-1 (Figure 1C, disruption of natural POL2 by the URA3 gene) that also confers slow growth and high mutator effect (Morrison et al. 1990; Northam et al. 2006). For this purpose, we created a diploid heterozygous for cdc28 mutations and pol2-1 as described in Materials and Methods. After sporulation and tetrad dissection, we evaluated the size of growing Ura+ colonies and determined the status of the CDC28 gene (Table 2, last three rows). All small Ura+ colonies were CDC28, and all medium-sized colonies carried one of the cdc28 alleles. The drastic effect of cdc28 mutations on the restoration of growth of pol2-1 is illustrated in Figure 9. We conclude that cdc28 mutations suppress the various defects of pol ε, not the specific pol2rc-ΔN allele.
Figure 9.
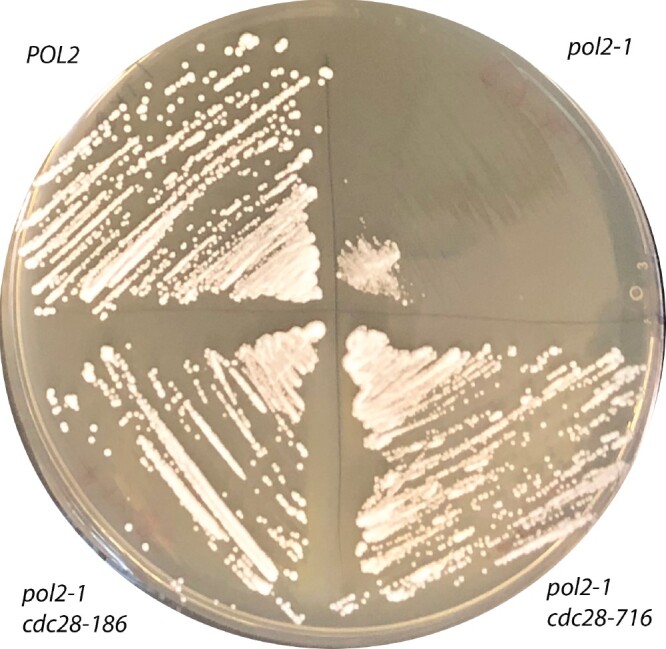
The suppression of slow growth of pol2-1 by cdc28 alleles. Genotyped colonies (Table 2) were streaked onto YPDAU, and the photo was taken on day 2 of growth at 30°.
Discussion
Current models of eukaryotic replication assert that the leading DNA strand is synthesized almost entirely by DNA pol ε (Burgers and Kunkel 2017; Li and O'Donnell 2018; Guilliam and Yeeles 2020b). The participation of pol α and pol δ in the leading strand was thought to be mostly limited to origins (Aria and Yeeles 2018; Garbacz et al. 2018) and termination zones (Zhou et al. 2019). However, Pol δ is unconstrained and, in addition to proofreading errors made by pol α (Pavlov et al. 2006), can proofread errors made by pol ε on leading strand (Pavlov and Shcherbakova 2010; Bulock et al. 2020) and thus can participate in the synthesis of other parts of the leading strand. Pol δ also operates on the leading DNA strand after replication restart when DNA is damaged (Guilliam and Yeeles 2020a). The current view, taking into account all available information, is that pol ε synthesizes more than 80% of the leading DNA strand (Guilliam and Yeeles 2020b). Surprisingly, yeast manages to endure the absence of one-half of the catalytic subunit of pol ε responsible for DNA synthesis, although growth is poor, at least at first (Kesti et al. 1999). Other polymerases, most likely pol δ, can substitute for the lost catalytic part (Kesti et al. 1999; Garbacz et al. 2018; Devbhandari and Remus 2020). Moreover, cells quickly adapt to the loss of the leading strand polymerase, and colonies start growing almost normally after a relatively short period of struggle ((Kesti et al. 1999; Garbacz et al. 2018), and this study). In the current work, we examined the genetic factors ensuring the resilience of replication and the ability to adapt to the loss of the essential replisome component.
Previous studies with the pol2-16 allele could not unequivocally determine the consequence of the loss of the Pol2N-terminus because it disrupted two functions of pol ε, polymerase activity due to the loss of the N-terminal half and participation in the assembly of the CMG helicase as a result of low stability of the Pol2-16 (Garbacz et al. 2018). Our study overcomes this limitation and allows us to assess the in vivo effects of Pol2-ΔN reliably. We developed a robust protocol for the generation of yeast strains without the catalytic half of Pol2 (Materials and Methods, Figure 2, Supplementary Figures S1 and S3). Expression of the recoded pol2rc-ΔN from the GAL1-10 promoter (Figure 1C) resulted in an improvement of growth not only on galactose- but also on glucose-containing medium (Figure 2B). The level of untagged Pol2-ΔN protein detected by antibodies against the C-terminal part of Pol2 under induction conditions was similar to that of the wild-type Pol2 (Figure 2D). This result suggests that it is not the truncation of Pol2 itself but the low stability of Pol2-16 (the C-terminal essential part of the protein fused to the first 175 amino acids of the N-terminal part, Figure 1C) that makes pol2-16 strains sick or inviable (Garbacz et al. 2018; Devbhandari and Remus 2020). It appears that when the C-terminal half of Pol2 necessary for the proper assembly of the CMG complex is stable and available [complete deletion of the N-terminal half encoded by pol2rc-ΔN (Devbhandari and Remus 2020) and present work], growth is improved. This observation argues that the 4Fe-4S cluster present in the N-terminal part of Pol2 (Jain et al. 2014; Ter Beek et al. 2019), the POPS insert (Meng et al. 2020), and PIP motifs (Figure 1C) are not critically involved in the regulation of the function of pol ε in the replisome, although they are needed for the maintenance of the proper structure of the active polymerase part. UV-mutability of pol2rc-ΔN strains argues that the 4Fe-4S cluster in the catalytic part of pol ε does not play a significant role in processes regulating induced mutagenesis, unlike the Fe-S cluster in pol δ and pol ζ (Giot et al. 1997; Baranovskiy et al. 2012, 2018; Stepchenkova et al. 2017). The difference in UV induced forward mutagenesis (predominantly base-pair substitutions) and frameshift reversion (+1 and −2 in poly AT run) (Shcherbakova and Kunkel 1999; Pavlov et al. 2001) (Figure 5) is consistent with the different mechanisms of processing of UV damage at regular vs repeated sequences, as suggested by less pronounced and variable dependence of UV-induced frameshift mutations on pol ζ (Lawrence et al. 1984; Abdulovic and Jinks-Robertson 2006).
The absence of the catalytic function of pol ε is not benign for the cells, even for the descendants of pol2rc-ΔN strains with near-normal growth. The strains experience difficulty starting divisions (Supplementary Figure S8), accumulate dead cells ((Supplementary Figure S7), are sensitive to hydroxyurea (Figure 3B), and have an increased rate of spontaneous mutation (Figure 4) that partially depends on pol ζ (Supplementary Table S1). The latter dependence is not specific to misfunction of pol ε and is characteristic of a broad spectrum of replication defects caused by mutations in genes for the catalytic subunits of all B family pols (Pavlov et al. 2001; Northam et al. 2006); accessory subunits (Northam et al. 2006; Jaszczur et al. 2008; Aksenova et al. 2010), parts of the CMG complex (Grabowska et al. 2014; Garbacz et al. 2015), by high GC content of the templates (Kiktev et al. 2018), and by replication stress caused by HU (Northam et al. 2010). The improvement of growth on galactose-containing medium did not suppress the high level of mutagenesis. Notably, the effect of REV3 deletion on mutagenesis in pol2-16 (Garbacz et al. 2019) or pol2rc-ΔN strains (Supplementary Table S1; 40% decrease of mutagenesis) is less pronounced than for other mutations leading to a defective replisome, including the pol2-1 allele (>80% reduction of mutagenesis) (Northam et al. 2006). The reasons for these peculiarities are still unclear and might reflect the multifaceted role of Pol2 and its catalytically active domain in the replication fork (Meng et al. 2020). In addition to the elevation of mutation rate, pol2rc-ΔN strains exhibit a high rate of illegitimate mating, resulting from chromosomal rearrangements and losses, point mutations, and “primary” DNA lesions temporarily affecting MAT locus transcription (Inge-Vechtomov and Repnevskaya 1989; Kochenova et al. 2011). These events are signs of replication stress in yeast strains lacking the critical part of Pol2, likely because of CMG helicase progression without a corresponding amount of DNA synthesis (Devbhandari and Remus 2020).
The most exciting result came to the fore during the analysis of the remarkable ability of pol2rc-ΔN strains to overcome the initial growth defect (Figure 3A). Genome sequencing of independent isolates with this mutation revealed a high prevalence of nonsynonymous changes implying positive selection forces, which, in combination with a mutator effect (with the domination of CG to GC transversions that typically require the involvement of pol ζ (Kochenova et al. 2017), lead to a rapid acquisition of changes allowing for the near-normal growth of pol2rc-ΔN strains. Mutations in the genes controlling various pathways, including cell cycle, DNA metabolism, and chromatin remodeling, might have contributed to the effect (Figure 7A), and further analysis will elaborate on the role of these mutations. The most striking case of recurrent mutations has been analyzed in the current work. Using genetic analysis, we showed that mutations in the CDC28 gene, the primary regulator of the cell cycle, mediate the improvement of the growth of the pol2rc-ΔN strains. Better growth was apparently connected to the alleviation of replication stress caused by the absence of the N-terminal half of Pol2 because cdc28 alleles suppressed HU-sensitivity (Table 3) and dramatically lowered mutation rates in pol2rc-ΔN strains (Table 4). The effect on mutation rates was stronger than the effect of deletion of REV3 (compare Table 4 and Supplementary Table S1). The suppression of mutator effects of pol δ alleles was seen previously for the checkpoint dun1 mutants (Datta et al. 2000; Mertz et al. 2015).
Cdc28 in complex with various cyclins inactivates/activates more than 70 cellular targets during the cell cycle (Enserink and Kolodner 2010; Mertz et al. 2015). One function of Cdc28 is the phosphorylation of the second subunit of pol ε, Dpb2, during the initiation of replication (Kesti et al. 2004). The complete understanding of how the CDC28 mutations revive strains without cs-pol ε is a complicated task because of the large number of Cdc28 targets and partners (Pines 1996; Wood and Endicott 2018). The amino acid residues changed by mutations improving survival of pol2rc-ΔN are located on the two different surfaces of the protein far from the kinase domain (Figure 10, A–C), and most likely modulate interactions with a subunit of the Cdc28 kinase, Csk1/2, with cyclins and other proteins, which, together with Cdc28, regulate the cell cycle (Enserink et al. 2009; Malumbres 2014). Notably, one group of mutations is at the surface of Cdc28 interacting with Clbs, while one mutation affects the surface necessary for interaction with Csk1/2 (Figure 10). We observed that different cdc28 mutations differentially affected the drug sensitivity of pol2rc-ΔN (Supplementary Figure S10, Table 3). The plethora of properties of Cdc28 variants found in our study suggests several ways of suppressing the pol2rc-ΔN defect. One idea stems from the observation that the proper level of Cdc28 is required to complete the cell cycle when replication continues beyond S-phase (Ivanova et al. 2020). In the pol2rc-ΔN cells, the proportion of under-replicated genomic regions might be higher than in normal cells, as indicated by aberrant cell and nucleus morphology (Figure 3C) and chromosome instability (Figure 6B). The cdc28 mutations may alter this balance of Cdc28 activity and other replication factors resulting in better completion of replication in pol2rc-ΔN cells. The deficiency of Cks in budding yeast leads to G2 arrest, and elevated levels lead to G2 delay (Pines 1996). Of interest, in our collection, there are several mutations in genes encoding those cyclins that participate in the late stages of the cell cycle (Supplementary Table S4). Another possible explanation of cdc28 mutations-mediated suppression of pol ε deficiency is that cdc28 mutations allow for the robust remodeling of the replication fork and easier access of pol δ to the replication of the leading DNA strand. Pol δ can bind the leading DNA strand during replication restart after damage (Guilliam and Yeeles 2020a). It is possible that alterations in the cell cycle or phosphorylation of DNA pols caused by the cdc28 mutations facilitate pol δ recruitment.
Figure 10.
Location of amino acid changes on the modeled structure of Cdc28 in complex with Csk1 and Clb1. (A) Amino acid residues that were changed in cdc28 mutants are conserved between yeast Cdc28 and human CDK1 and CDK2. A screenshot of amino acid sequence alignment done by Geneious (Biomatters, Ltd) is shown with secondary structures [α-helical regions as spirals, β-strands as arrows, and loops as dotted lines (Wood and Endicott 2018)]. (B) Amino acid residues that were changed in cdc28 mutants (magenta) are on protein surfaces of modeled Cdc28 (colored green) that interact with Csk1 and 2 (sand-like dashed arc) and cyclins (light brown dashed arc). (C) Amino acid residues (pink) that were changed in Cdc28 (teal) at the surface interacting with cyclin B (green).
Based on our results, we envisage the following mechanism leading to the appearance of fast-growing clones in pol2rc-ΔN strains. The initial defect causes a delay in replication and activation of cell-cycle checkpoint (Garbacz et al. 2019), followed by remodeling of the replication fork with the aid of Rad53 kinase and the remaining B-family polymerases replicating both lagging and leading DNA strands (Devbhandari and Remus 2020). The unusual arrangement of pols leads to increased mutagenesis and genome instability, providing sufficient material for rapid selection against slow-growing cells. It is likely that mutations and, as hypothesized in Garbacz et al. (2019), epigenetic changes may lead to the appearance of fast-growing cells. Changes in many different genes (Figure 7A) can potentially suppress the growth defect of pol2rc-ΔN, thus contributing to the ability of cells to adapt to the defect of the leading DNA strand replication. In this study, we revealed the prominent role of Cdc28 alterations in the resilience of the replication machinery.
Our studies in yeast are relevant to phenotypic manifestation of pol ε dysfunction in higher eukaryotes. In humans, homozygous mutations severely decreasing levels of pol ε are surprisingly not lethal but cause immunodeficiency, facial dysmorphism, livedo, and short stature called FILS syndrome (Pachlopnik Schmid et al. 2012). Bi-allelic mutations in POLE, encoding the catalytic subunit of human pol ε, cause IMAGe syndrome (Logan et al. 2018). Mutations in POLE cause disorders similar to some DNA breakage/instability syndromes (Thiffault et al. 2015). In mice, destabilization of pol ε by a deletion of the gene of the fourth subunit causes growth defects, leukopenia, and a predisposition to cancer (Bellelli et al. 2018). Our work establishing the role of mutations in the CDC28 in the suppression of growth defects caused by pol ε deficiency in yeast suggests that variations in its mammalian homologs, CDKs, might also play a role in suppressing the effects of pol ε. Multiple alignment of CDK protein kinase orthologs (Supplementary Figure S11) suggested that all four amino acid residues changed in our mutants are located in evolutionarily conserved positions, although the Leu84 site shows some limited variability of residues, and instead of Ile, the 239 site has Val in most species. Mutations in human CDK1 and CDK2, resulting in amino acid changes in the same region that is affected by yeast cdc28-716, are found in head and neck tumors and breast invasive ductal carcinoma (CBioPortal, http://www.cbioportal.org/). In this respect, a new important observation is that the levels of CDK2 modulate the response of breast cancer cells to low levels of POLE (Sviderskiy et al. 2020). We expect that further studies of the suppression of leading DNA strand replication defects will lead to novel approaches to alleviate the biological consequences of replication stress. It is also likely that studying the genetic consequences of the absence of the catalytic half of Pol2 will help understand the overwhelming predominance of mutations affecting this part of pol ε in human cancers (Rayner et al. 2016; Barbari and Shcherbakova 2017; Pavlov et al. 2020).
Acknowledgments
The authors want to thank John Diffley for sharing the plasmids and strains used in the study and Lucy Drury for helping us to understand the details of the constructs. The authors thank C. Wittenberg for plasmids with the pol2-16 allele. The authors thank Tahir Tahirov, Andrey Baranovskiy, Tom Petes, Sue Jinks-Robertson, and Julian Sale for helpful and stimulating discussions of the work. The authors thank Dmitry Gordenin for a suggestion on how to better abbreviate the catalytic subunit of DNA polymerase ε. The authors thank the reviewers of the manuscript for their helpful suggestions. The authors thank Elizabeth Moore and Irina Zotova for their excellent technical assistance. The authors thank the Research Park of St. Petersburg State University for next-generation sequencing and UNMC facilities for the sequencing of CDC28.
Y.I.P. designed the study, executed genetic and biochemical experiments, and wrote the first draft; E.I.S. contributed to the study design, did genetic experiments, and participated in writing on the first draft. A.S.Z. created plasmids and was one of the main contributors to the bioinformatics and statistical part of the work; J.C. participated in the verification of antibodies, and Western blots, and the analysis of mutants; E.R.T. did genetic experiments; S.R.B. participated in strain construction and analysis of proteins; P.V.S. participated in the discussion of work strategy, generated reagents and critically edited the text; D.E.P. supervised next-generation sequencing; R.F. did estimations of the probability of recurrent mutations; E.P. did molecular modeling and created structural images; I.B.R. supervised the statistical analysis and contributed to the discussion of the significance of the study; A.G.L. led the analysis of the genomic sequences. All authors participated in writing parts of the manuscript and its editing.
Funding
The genome sequencing, microscopy, and mutant frequency tests were done with the financial support of RSF grant # 20-15-00081 to E.I.S. The genome analysis work was supported by the Government of the Russian Federation through the ITMO Fellowship and Professorship Program to A.S.Z. The National Institutes of Health grants ES015869 and CA239688 to P.V.S. supported work done by S.R.B and P.V.S. EP and I.B.R. were supported by the Intramural Research Programs of the National Eye Institute and National Library of Medicine, National Institutes of Health. S.R.B. was supported by the University of Nebraska Medical Center Graduate Studies Fellowship and by the Cancer Biology Training Grant T32CA009476 from the National Cancer Institute. Y.I.P. and J.C. were supported by the Eppley Institute for Research in Cancer Pilot grant. The University of Nebraska Medical Center Genomics Facility and other facilities are administrated through the Office of the Vice-Chancellor for Research and supported by state funds from the Nebraska Research Initiative (NRI), the University of Nebraska Foundation, the Nebraska Banker's Fund, and by the NIH-NCRR Shared Instrument Program. The facilities also receive partial support from the National Institute for General Medical Science (NIGMS) INBRE—P20 GM103427 and COBRE—P30 GM106397 and P30GM110768 grants, as well as support from the National Cancer Institute (NCI) for The Fred & Pamela Buffett Cancer Center Support Grant—P30 CA036727, The Center for Root and Rhizobiome Innovation (CRRI) 36-5150-2085-20, and the Nebraska Research Initiative. The contents and interpretations of this publication are the sole responsibility of the authors.
Conflicts of interest
None declared.
Literature cited
- Abdulovic AL, Jinks-Robertson S.. 2006. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: insights into Pol ζ- and Pol η-dependent frameshift mutagenesis. Genetics. 172:1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenova A, Volkov K, Maceluch J, Pursell ZF, Rogozin IB, et al. 2010. Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase epsilon. PLoS Genet. 6:e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Ropp PA, Johnson AL, Johnston LH, Morrison A, et al. 1992. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 11:733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aria V, Yeeles JTP.. 2019. Mechanism of bidirectional leading-strand synthesis establishment at eukaryotic DNA replication origins. Mol Cell. 73:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Yuan Z, Sun J, Georgescu R, O'Donnell ME, et al. 2017. Architecture of the Saccharomyces cerevisiae replisome. Adv Exp Med Biol. 1042:207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, et al. 2012. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J Biol Chem. 287:17281–17287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Siebler HM, Pavlov YI, Tahirov TH.. 2018. Iron-Sulfur clusters in DNA polymerases and primases of eukaryotes. Methods Enzymol. 599:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbari SR, Shcherbakova PV.. 2017. Replicative DNA polymerase defects in human cancers: consequences, mechanisms, and implications for therapy. DNA Repair (Amst)). 56:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellelli R, Borel V, Logan C, Svendsen J, Cox DE, et al. 2018. Polε instability drives replication stress, abnormal development, and tumorigenesis. Mol Cell. 70:707–721.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breusch TS, Pagan AR.. 1980. The Lagrange multiplier test and its applications to model specification in econometrics. Rev Econ Stud. 47:239–253. [Google Scholar]
- Bulock CR, Xing X, Shcherbakova PV.. 2020. DNA polymerase delta proofreads errors made by DNA polymerase epsilon. Proc Natl Acad Sci U S A. 117:6035–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM. 2009. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 284:4041–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PMJ, Kunkel TA.. 2017. Eukaryotic DNA replication fork. Annu Rev Biochem. 86:417–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daee DL, Mertz TM, Shcherbakova PV.. 2010. A cancer-associated DNA polymerase delta variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci U S A. 107:157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Schmeits JL, Amin NS, Lau PJ, Myung K, et al. 2000. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol Cell. 6:593–603. [DOI] [PubMed] [Google Scholar]
- Dean N, Poster JB.. 1996. Molecular and phenotypic analysis of the S. cerevisiae MNN10 gene identifies a family of related glycosyltransferases. Glycobiology. 6:73–81. [DOI] [PubMed] [Google Scholar]
- Devbhandari S, Remus D.. 2020. Rad53 limits CMG helicase uncoupling from DNA synthesis at replication forks. Nat Struct Mol Biol. 27:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 88:7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL.. 1999. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 274:22283–22288. [DOI] [PubMed] [Google Scholar]
- Enserink JM, Hombauer H, Huang ME, Kolodner RD.. 2009. Cdc28/Cdk1 positively and negatively affects genome stability in S. cerevisiae. J Cell Biol. 185:423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink JM, Kolodner RD.. 2010. An overview of Cdk1-controlled targets and processes. Cell Div. 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbacz M, Araki H, Flis K, Bebenek A, Zawada AE, et al. 2015. Fidelity consequences of the impaired interaction between DNA polymerase epsilon and the GINS complex. DNA Repair (Amst)). 29:23–35. [DOI] [PubMed] [Google Scholar]
- Garbacz MA, Cox PB, Sharma S, Lujan SA, Chabes A, et al. 2019. The absence of the catalytic domains of Saccharomyces cerevisiae DNA polymerase strongly reduces DNA replication fidelity. Nucleic Acids Res. 47:3986–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbacz MA, Lujan SA, Burkholder AB, Cox PB, Wu Q, et al. 2018. Evidence that DNA polymerase delta contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat Commun. 9:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, et al. 2017. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci U S A. 114:E697–E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Chanet R, Simon M, Facca C, Faye G.. 1997. Involvement of the yeast DNA polymerase δ in DNA repair in vivo. Genetics. 146:1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami P, Abid Ali F, Douglas ME, Locke J, Purkiss A, et al. 2018. Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome. Nat Commun. 9:5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska E, Wronska U, Denkiewicz M, Jaszczur M, Respondek A, et al. 2014. Proper functioning of the GINS complex is important for the fidelity of DNA replication in yeast. Mol Microbiol. 92:659–680. [DOI] [PubMed] [Google Scholar]
- Guilliam TA, Yeeles JTP.. 2020a. Reconstitution of translesion synthesis reveals a mechanism of eukaryotic DNA replication restart. Nat Struct Mol Biol. 27:450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliam TA, Yeeles JTP.. 2020b. An updated perspective on the polymerase division of labor during eukaryotic DNA replication. Crit Rev Biochem Mol Biol. 55:469–481. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S.. 2000. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell. 6:1491–1499. [DOI] [PubMed] [Google Scholar]
- Hizume K, Endo S, Muramatsu S, Kobayashi T, Araki H.. 2018. DNA polymerase epsilon-dependent modulation of the pausing property of the CMG helicase at the barrier. Genes Dev. 32:1315–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, et al. 2014. Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat Struct Mol Biol. 21:49–55. [DOI] [PubMed] [Google Scholar]
- Inge-Vechtomov SG, Repnevskaya MV.. 1989. Phenotypic expression of primary lesions of genetic material in Saccharomyces yeasts. Genome. 31:497–502. [DOI] [PubMed] [Google Scholar]
- Ivanova T, Maier M, Missarova A, Ziegler-Birling C, Dam M, et al. 2020. Budding yeast complete DNA synthesis after chromosome segregation begins. Nat Commun. 11:2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Vanamee ES, Dzikovski BG, Buku A, Johnson RE, et al. 2014. An iron-sulfur cluster in the polymerase domain of yeast DNA polymerase epsilon. J Mol Biol. 426:301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszczur M, Flis K, Rudzka J, Kraszewska J, Budd ME, et al. 2008. Dpb2p, a noncatalytic subunit of DNA polymerase epsilon, contributes to the fidelity of DNA replication in Saccharomyces cerevisiae. Genetics. 178:633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S.. 2012. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc Natl Acad Sci U S A. 109:12455–12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas D, Krupovic M, Guglielmini J, Forterre P, Venclovas Č.. 2020. Diversity and evolution of B-family DNA polymerases. Nucleic Acids Res. 48:10142–10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C.. 1999. DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 3:679–685. [DOI] [PubMed] [Google Scholar]
- Kesti T, McDonald WH, Yates JR 3rd, Wittenberg C.. 2004. Cell cycle-dependent phosphorylation of the DNA polymerase epsilon subunit, Dpb2, by the Cdc28 cyclin-dependent protein kinase. J Biol Chem. 279:14245–14255. [DOI] [PubMed] [Google Scholar]
- Kiktev DA, Sheng Z, Lobachev KS, Petes TD.. 2018. GC content elevates mutation and recombination rates in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 115:E7109–E7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenova OV, Bezalel-Buch R, Tran P, Makarova AV, Chabes A, et al. 2017. Yeast DNA polymerase ζ maintains consistent activity and mutagenicity across a wide range of physiological dNTP concentrations. Nucleic Acids Res. 45:1200–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenova OV, Soshkina JV, Stepchenkova EI, Inge-Vechtomov SG, Shcherbakova PV.. 2011. Participation of translesion synthesis DNA polymerases in the maintenance of chromosome integrity in yeast Saccharomyces cerevisiae. Biochemistry (Mosc)). 76:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV. 2000. Rapid and reliable protein extraction from yeast. Yeast. 16:857–860. [DOI] [PubMed] [Google Scholar]
- Kwolek-Mirek M, Zadrag-Tecza R.. 2014. Comparison of methods used for assessing the viability and vitality of yeast cells. FEMS Yeast Res. 14:1068–1079. [DOI] [PubMed] [Google Scholar]
- Lada AG, Kliver SF, Dhar A, Polev DE, Masharsky AE, et al. 2015. Disruption of transcriptional coactivator Sub1 leads to genome-wide re-distribution of clustered mutations Induced by APOBEC in active yeast genes. PLoS Genet. 11:e1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lada AG, Stepchenkova EI, Waisertreiger IS, Noskov VN, Dhar A, et al. 2013. Genome-wide mutation avalanches induced in diploid yeast cells by a base analog or an APOBEC deaminase. PLoS Genet. 9:e1003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lada AG, Stepchenkova EI, Zhuk AS, Kliver SF, Rogozin IB, et al. 2017. Recombination Is responsible for the increased recovery of drug-resistant mutants with hypermutated genomes in resting yeast diploids expressing APOBEC deaminases. Front Genet. 8:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, O'Brien T, Bond J.. 1984. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants of yeast. Mol Gen Genet. 195:487–490. [DOI] [PubMed] [Google Scholar]
- Lewis JS, Spenkelink LM, Schauer GD, Yurieva O, Mueller SH, et al. 2020. Tunability of DNA polymerase stability during eukaryotic DNA replication. Mol Cell. 77:17–25.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, O'Donnell ME.. 2018. The Eukaryotic CMG helicase at the replication fork: emerging architecture reveals an unexpected mechanism. Bioessays. 40:1700208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CV, Murray JE, Parry DA, Robertson A, Bellelli R, et al. ; SGP Consortium. 2018. DNA polymerase epsilon deficiency causes IMAGe syndrome with variable immunodeficiency. Am J Hum Genet. 103:1038–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Clausen AR, Clark AB, MacAlpine HK, MacAlpine DM, et al. 2014. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 24:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager WH, Planta RJ, Ballesta JG, Lee JC, Mizuta K, et al. 1997. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 25:4872–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM.. 2012. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 40:11618–11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. 2014. Cyclin-dependent kinases. Genome Biol. 15:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín A, Gallardo M, Kato Y, Shirahige K, Gutiérrez G, et al. 2003. Relationship between G+C content, ORF-length and mRNA concentration in Saccharomyces cerevisiae. Yeast. 20:703–711. [DOI] [PubMed] [Google Scholar]
- Meng X, Wei L, Devbhandari S, Zhang T, Xiang J, et al. 2020. DNA polymerase ε relies on a unique domain for efficient replisome assembly and strand synthesis. Nat Commun. 11:2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz TM, Sharma S, Chabes A, Shcherbakova PV.. 2015. Colon cancer-associated mutator DNA polymerase delta variant causes expansion of dNTP pools increasing its own infidelity. Proc Natl Acad Sci U S A. 112:E2467–E2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A.. 1990. A third essential DNA polymerase in S. cerevisiae. Cell. 62:1143–1151. [DOI] [PubMed] [Google Scholar]
- Morrison A, Bell JB, Kunkel TA, Sugino A.. 1991. Eukaryotic DNA polymerase amino acid sequence required for 3’—> 5' exonuclease activity. Proc Natl Acad Sci U S A. 88:9473–9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Johnston AL, Johnston LH, Sugino A.. 1993. Pathway correcting DNA replication errors in S. cerevisiae. EMBO J. 12:1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA.. 2008. Division of labor at the eukaryotic replication fork. Mol Cell. 30:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida C, Reinhard P, Linn S.. 1988. DNA repair synthesis in human fibroblasts requires DNA polymerase delta. J Biol Chem. 263:501–510. [PubMed] [Google Scholar]
- Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV.. 2006. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 25:4316–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV.. 2010. Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 184:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya T, Kawasaki Y, Hiraga S, Kanbara S, Nakajo K, et al. 2002. The DNA polymerase domain of pol(ε) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J Biol Chem. 277:28099–28108. [DOI] [PubMed] [Google Scholar]
- Pachlopnik Schmid J, Lemoine R, Nehme N, Cormier-Daire V, Revy P, et al. 2012. Polymerase epsilon1 mutation in a human syndrome with facial dysmorphism, immunodeficiency, livedo, and short stature ("FILS syndrome"). J Exp Med. 209:2323–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Frahm C, McElhinny SA, Niimi A, Suzuki M, et al. 2006. Evidence that errors made by DNA polymerase a are corrected by DNA polymerase delta. Curr Biol. 16:202–207. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV.. 2010. DNA polymerases at the eukaryotic fork-20 years later. Mutat Res. 685:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Kunkel TA.. 2001. In vivo consequences of putative active site missense mutations in yeast replicative DNA polymerases α, ε, δ and ζ. Genetics. 159:47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Zhuk AS, Stepchenkova EI.. 2020. DNA Polymerases at the eukaryotic replication fork: thirty years after: connection to cancer. Cancers (Basel). 12:3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa-Brandao L, Sclafani RA.. 2004. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics. 167:1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. 1996. Cell cycle: reaching for a role for the Cks proteins. Curr Biol. 6:1399–1402. [DOI] [PubMed] [Google Scholar]
- Pospiech H, Syvaoja JE.. 2003. DNA polymerase ε—more than a polymerase. ScientificWorldJournal. 3:87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Kunkel TA.. 2008. DNA polymerase epsilon: a polymerase of unusual size (and complexity). Prog Nucleic Acid Res Mol Biol. 82:101–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, et al. 2016. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 16:71–81. [DOI] [PubMed] [Google Scholar]
- Shcherbakova PV, Kunkel TA.. 1999. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 19:3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, et al. 2003. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem. 278:43770–43780. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks JB.. 1986. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 180 p. [Google Scholar]
- Siebler HM, Lada AG, Baranovskiy AG, Tahirov TH, Pavlov YI.. 2014. A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differential regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae. DNA Repair (Amst)). 24:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Hoyt MA, Botstein D.. 1990. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 124:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepchenkova EI, Shiriaeva AA, Pavlov YI.. 2018. Deletion of the DEF1 gene does not confer UV-immutability but frequently leads to self-diploidization in yeast Saccharomyces cerevisiae. DNA Repair (Amst). 70:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepchenkova EI, Tarakhovskaya ER, Siebler HM, Pavlov YI.. 2017. Defect of Fe-S cluster binding by DNA polymerase delta in yeast suppresses UV-induced mutagenesis, but enhances DNA polymerase zeta—dependent spontaneous mutagenesis. DNA Repair (Amst). 49:60–69. [DOI] [PubMed] [Google Scholar]
- Stillman B. 2015. Reconsidering DNA polymerases at the replication fork in eukaryotes. Mol Cell. 59:139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson HE, Lewis MJ, Pelham HR.. 2006. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 25:662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stodola JL, Stith CM, Burgers PM.. 2016. Proficient replication of the yeast genome by a viral DNA polymerase. J Biol Chem. 291:11698–11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JE, Kissling GE, Lujan SA, Rogozin IB, Stith CM, et al. 2009. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase zeta. Nucleic Acids Res. 37:3774–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J, Hicks J, Herskowitz I.. 1981. Control of cell type in yeast by the mating type locus: the α1-α2 hypothesis. J Mol Biol. 147:357–372. [DOI] [PubMed] [Google Scholar]
- Sviderskiy VO, Blumenberg L, Gorodetsky E, Karakousi TR, Hirsh N, et al. 2020. Hyperactive CDK2 activity in basal-like breast cancer imposes a genome Integrity liability that can be exploited by targeting DNA polymerase ε. Mol Cell. 80:682–698.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvaoja J, Linn S.. 1989. Characterization of a large form of DNA polymerase delta from HeLa cells that is insensitive to proliferating cell nuclear antigen. J Biol Chem. 264:2489–2497. [PubMed] [Google Scholar]
- Syvaoja J, Suomensaari S, Nishida C, Goldsmith JS, Chui GS, et al. 1990. DNA polymerases alpha, delta, and epsilon: three distinct enzymes from HeLa cells. Proc Natl Acad Sci U S A. 87:6664–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV.. 2009. Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H.. 2013. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 5:a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Beek J, Parkash V, Bylund GO, Osterman P, Sauer-Eriksson AE, et al. 2019. Structural evidence for an essential Fe-S cluster in the catalytic core domain of DNA polymerase ε. Nucleic Acids Res. 47:5712–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiffault I, Saunders C, Jenkins J, Raje N, Canty K, et al. 2015. A patient with polymerase E1 deficiency (POLE1): clinical features and overlap with DNA breakage/instability syndromes. BMC Med Genet. 16:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA.. 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 17:2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye BK, Sawyer S.. 2000. The hexameric eukaryotic MCM helicase: building symmetry from nonidentical parts. J Biol Chem. 275:34833–34836. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P.. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10:1793–1808. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B.. 1998. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 67:721–751. [DOI] [PubMed] [Google Scholar]
- Wang T, Bretscher A.. 1995. The rho-GAP encoded by BEM2 regulates cytoskeletal structure in budding yeast. Mol Biol Cell. 6:1011–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DJ, Endicott JA.. 2018. Structural insights into the functional diversity of the CDK-cyclin family. Open Biol. 8:180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing X, Kane DP, Bulock CR, Moore EA, Sharma S, et al. 2019. A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat Commun. 10:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF.. 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 519:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JTP, Janska A, Early A, Diffley JFX.. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell. 65:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Georgescu R, Schauer GD, O'Donnell ME, Li H.. 2020. Structure of the polymerase ε holoenzyme and atomic model of the leading strand replisome. Nat Commun. 11:3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, et al. 2017. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc Natl Acad Sci U S A. 114:4141–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZX, Lujan SA, Burkholder AB, Garbacz MA, Kunkel TA.. 2019. Roles for DNA polymerase delta in initiating and terminating leading strand DNA replication. Nat Commun. 10:3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and supplemental materials. The raw NGS data are deposited in Sequence Read Archive (SRA, accession # PRJNA634680, Temporary submission ID: SUB7487615). The BioProject accession number is provided in lieu of SRP and will allow better searching in Entrez. Full scripts used to generate SNV datasets are available upon request. Supplementary files are submitted via GSA figshare portal: https://doi.org/10.25386/genetics.14357234 (accessed 04.28.2021).