Abstract
Immunogenetic studies in the past three decades have uncovered a broad range of human genetic factors that seem to influence heterosexual HIV-1 transmission in one way or another. In our own work that jointly evaluated both genetic and non-genetic factors in two African cohorts of cohabiting, HIV-1-discordant couples (donor and recipient pairs) at risk of transmission during quarterly follow-up intervals, relatively consistent findings have been seen with three loci (IL19, HLA-A and HLA-B), although the effect size (i.e., odds ratio or hazards ratio) of each specific variant was quite modest. These studies offered two critical lessons that should benefit future research on sexually transmitted infections. First, in donor partners, immunogenetic factors (e.g., HLA-B*57 and HLA-A*36:01) that operate directly through HIV-1 viral load or indirectly through genital co-infections are equally important. Second, thousands of single nucleotide polymorphisms previously recognized as “causal” factors for human autoimmune disorders did not appear to make much difference, which is somewhat puzzling as these variants are predicted or known to influence the expression of many immune response genes. Replicating these observations in additional cohorts is no longer feasible as the field has shifted its focus to early diagnosis, universal treatment and active management of comorbidities.
Keywords: Africa, epidemiology, genetics, HIV-1 transmission, statistical modeling
Introduction
Sub-Saharan African countries have borne the brunt (60–70%) of the global HIV/AIDS burden for nearly four decades now [1], with heterosexual HIV-1 transmission being a chief driver. Before the implementation of life-long combination antiretroviral therapy (cART) that suppresses viral load, minimizes viral pathogenesis and substantially reduces further spread to at-risk individuals, two study cohorts that began with couple-based voluntary counseling and testing (CVCT) provided a unique opportunity for studying immunogenetic determinants of heterosexual HIV-1 transmission in countries representing eastern and southern Africa. Based on a longitudinal study design, hundreds of initially HIV-1 discordant couples enrolled in Kigali, Rwanda and Lusaka, Zambia had intra-couple transmission events during quarterly follow-up visits, which were confirmed by phylogenetically related viruses. When these epidemiologically linked couples were compared with non-transmission couples for immunogenetic profiles, including variants at HIV-1 coreceptor genes, human major histocompatibility complex (MHC) and other (genome-wide) loci that mediate immune responses, several consensus findings imply the importance of three loci: IL19, HLA-A and HLA-B. Meanwhile, cohort-specific findings are abundant, which might reflect contrasting genetic backgrounds and different HIV-1 subtypes in these study populations.
To highlight advantages and challenges in these immunogenetic studies, this review will summarize robust evidence and lessons learned since 2004, when cumulative sample sizes were adequate enough to allow meaningful analyses of major factors in individuals or couples (with each donor-recipient pair as a single entity) [2–4]. Consistent observations on three loci uncover both direct and indirect mechanisms related to inflammation and immune control, some of which can be verified experimentally using in vitro systems [5–9]. In contrast, thousands of functionally relevant SNPs (single nucleotide polymorphisms), including those known as gene expression quantitative trait loci (eQTLs), have not shown any appreciable impact on heterosexual HIV-1 transmission, suggesting that these eQTLs might not be critical to HIV-1-host interactions at the genital mucosa.
Why do we bother with immunogenetic factors?
Identifying immunogenetic (heritable) correlates of heterosexual HIV-1 transmission is expected to enhance our understanding of host-virus interactions at the genital mucosa and provide much-needed directives for tailoring timely and targeted interventions to suitable subjects or populations. During the course of our cohort studies, the “Fiscal Year 2008 Trans-NIH Plan for HIV-Related Research” [10] made several explicit gestures to encourage the investigation of host factors in HIV/AIDS: (i) “evaluate sexual … transmission and acquisition in relation to...host factors such as sex, age…and host genetic factors,” (ii) “… investigate the contribution of innate host characteristics…and mechanisms for these effects (including host genetic factors…),” (iii) “acquire clinical specimens from populations relevant to HIV vaccine trials…; and (iv) “explore the molecular epidemiology, humoral, and cell-mediated immune responses to HIV-1 and their relationship to class I and class II MHC alleles.” The “Fiscal Year 2012 Trans-NIH Plan”[10] called for strategy that can “delineate the mechanisms by which innate and adaptive immunity, and the effects of genetic or environmental factors on innate and adaptive immunity, influence HIV/SIV replication throughout acute and chronic infection.” By 2019, most of the priorities have shifted to (a) developing next-generation HIV-1 therapies, (b) curing HIV-1 infection and (c) addressing HIV-1-associated comorbidities, coinfections and complications, but reducing the incidence of new HIV-1 infection has remained a critical component [10].
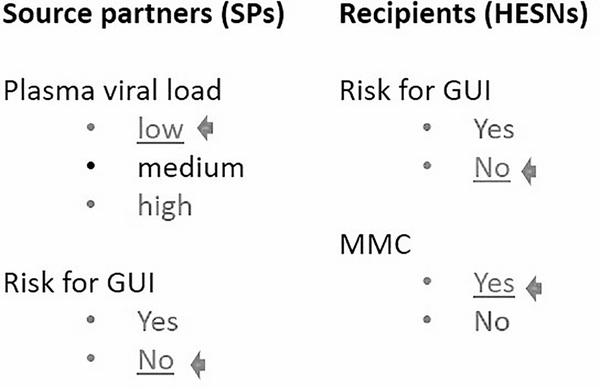
In our search for immunogenetic factors, a multidisciplinary approach was necessary as many non-genetic factors are clearly important to heterosexual HIV-1 transmission (Figure 1). To offer “added values,” our research must first and foremost demonstrate that these gene variants operate independently of other prominent co-factors, especially viral load in the donor partner (Figure 2), genital co-infections in both HIV-1 source partner (SP) and HIV-1-exposed, seronegative partner (HESN), and male medical circumcision in HESNs. Confirmation of these well-known, multifaceted factors can provide further assurance that our study cohorts are valid and can fulfill three main objectives: (i) to advance our understanding of host genetic factors in HIV/AIDS and co-infections; (ii) to identify causal variants of biological importance, through fine-mapping (dense coverage) of target loci and in vitro assays; (iii) to gather an integrated dataset for meta-analyses and other population-based studies.
Figure 1. Several scenarios in the setting of heterosexual HIV-1 transmission among discordant couples.
When two main factors in the HIV-1-infected source partners (SPs) are assessed along with two well-documented factors in the recipient partners (also known as HIV-1-exposed, seronegative individuals or HESNs), couples can be divided into various composite risk groups. The group with extremely low risk (having all factors indicated by arrows) is expected to obscure the search for immunogenetic determinants, especially when duration of follow-up is short. GUI, genital ulcer/inflammation (clinical manifestations of genital co-infections); MMC, male medical circumcision (rates of uptake <5% in our cohorts).
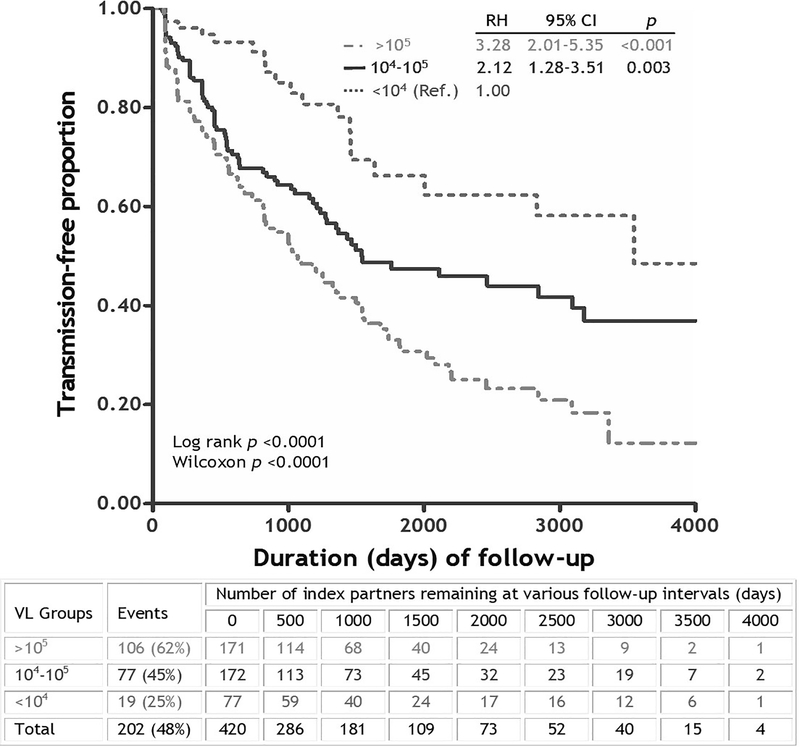
Figure 2. Kaplan-Meier curves showing the impact of HIV-1 viral load (VL) in donor/index partners on heterosexual HIV-1 transmission among 420 HIV-1 discordant Zambian couples with longitudinal follow-up.
The three VL categories followed earlier strategies [83], and subjects with low VL (<10−4 RNA copies/ml of plasma) were treated as the reference (ref.) group. The estimates of relative hazards (RH) and 95% confidence interval (CI) were based on a Cox proportional hazards model. Graph is redrawn from data published in 2008 [17]. Couples that remained eligible for further assessment at various follow-up intervals are indicated below the curves.
The advantages in studying HIV-1 discordant couples
Since 1995, two large cohorts of HIV-1-discordant couples enrolled in Lusaka, Zambia and Kigali, Rwanda served as an optimal platform on which host and viral factors important to heterosexual HIV-1 transmission and co-infections can be evaluated jointly and systematically to inform translational research. By the time (2012) when treatment as prevention (TasP) [11–15] became a new standard of care that outweighed the benefits of CVCT, these projects had already accumulated 1,355 couples who met all four selection criteria: (i) ages between 18 years at enrollment and 65 years at end of follow-up, (ii) donor/source partner viral load >2,000 RNA copies/mL, (ii) epidemiologically linked transmission events or >12 months of transmission-free follow-up (before therapy, drop-out or end of study), and (iv) at least one known risk factor (e.g., unprotected sex or pregnancy) for transmission during study intervals (Table 1).
Table 1.
Assembly of two study cohorts for immunogenetic studies.a
| Overall characteristics | Lusaka, Zambia | Kigali, Rwanda |
|---|---|---|
| Participants | HIV-1 discordant couples | HIV-1 discordant couples |
| Enrollment dates | 1995–2011 | 2002–2011 |
| Intervention | CVCT & cART | CVCT & cART |
| Follow-up visits | Quarterly | Quarterly |
| Genital co-infectionsb | GUI | GUI |
| Total enrollment | 4,759 couples | 1,748 couples |
| Subset for immunogenetic studiesc | 994 pairs (21%) | 361 pairs (21%) |
Whenever applicable, data from other studies pertinent to HIV-1 transmission in African countries are also discussed in the text.
Genital ulcer/inflammation (GUI) was frequent enough for ancillary studies. Other abbreviations: CVCT, couple-based voluntary counseling and testing; cART, combination antiretroviral therapy.
Selection criteria: (i) ages between 18 at enrollment and 65 at end of follow-up, (ii) >12 months of follow-up before therapy or drop-out (while transmission-free), (iii) donor/source partner viral load >2,000 RNA copies/mL, and (iv) at least one known risk factor (e.g., unprotected sex or pregnancy) for transmission during study intervals.
By our study design, immunogenetic factors can be evaluated for SPs and their cohabiting HESNs, with further consideration of covariates (e.g., Figure 1). For SPs, the extent of “HIV-1 control” (as measured by plasma viral load) is critical (Figure 2), so is their susceptibility to genital ulcer/inflammation (GUI) that promote viral shedding at genital mucosa. For HESNs, “HIV-1 susceptibility” (acquisition of infection) can be modulated by GUI as well [16]. When each couple is considered jointly as one entity [17–19], “HIV-1 control” for SP immediately becomes a cofactor for “HIV-1 susceptibility” in HESN (two sides of the same coin), which has been largely overlooked in studies with unknown SPs. Moreover, genetic similarity of each SP-HESN pair can be assessed by allele/haplotype sharing [2, 3] as well as genetic distances defined by principal component analysis (PCA), with further enhancements by a “shared environment” for each SP-HESN pair.
Candidate loci
Since the early 1990s, immunogenetic findings have highlighted the importance of a broad range of host genetic factors to HIV-1 transmission, especially those that regulate the expression of HIV-1 co-receptor CCR5 [20–25], as well as prominent candidates highly enriched in the human major histocompatibility complex (MHC) [23–25]. Some of these are “causal variants” (e.g., the CCR5-Δ32 mutation) with well-defined biological pathways, while others are pure “association hits” because of difficulties and uncertainties with fine-mapping [26, 27]. More recently, “unbiased” approach using genome-wide coverage has gained popularity, but most of the findings from genome-wide association studies (GWAS) simply reiterated the importance of MHC genes as the primary quantitative trait locus (QTL) for HIV-1 viral load [28–30].
To provide a systematic and thorough dissection of immunogenetic factors in heterosexual HIV-1 transmission, including those that can help explain “elite controllers” whose persistent viral suppression is the ultimate goal of therapeutic and prophylactic intervention [31–34], our projects followed some of the earlier leads [4, 35, 36], while also considering statistical power within our evolving cohorts (variable sample sizes due to rolling enrollment and continuous updates of clinical findings), with an emphasis on candidate loci (e.g., HLA and cytokine genes) that are (i) amenable to biological interventions (e.g., vaccines and immune adjuvants), (ii) suitable for high-resolution genotyping (to fully resolve alleles and local haplotypes), (iii) related to early events in heterosexual HIV-1 transmission, and (iv) broad implications, especially genital co-infections that facilitate HIV-1 transmission. These efforts further recognized the notion that immunogenetic factors mediate HIV-1 acquisition (by the recipient partners) and immune control of established infection (in the index/source partners) through distinct mechanisms [17, 19, 37], all of which can be altered by inflammation and virus shedding in the genital mucosa.
Genotyping and bioinformatics
In the early phase of our studies, a variety of PCR-based techniques were used to resolve individual SNP genotypes and alleles within each target locus, followed by the evaluation of local and extended haplotypes (resolved by HaploView [38]). The availability of high-throughput assays, including next-generation sequencing and SNP arrays (the ImmunoChip), broadened the search to genome-wide signals or specific gene clusters (e.g., those encoding killer cell immunoglobulin-like receptors/KIRs) that are not adequately covered by other genotyping platforms. In particular, fine-mapping for hundreds of immune response genes was made possible by the ImmunoChip bead arrays (versions 1 and 2) [39–41], although the actual coverage for signal-rich regions (e.g., MHC) remained suboptimal [42, 43]. Occasionally, targeted NGS was able to fill in various gaps through phased allele and haplotype assignments (GenDx, Utrecht, The Netherlands).
Aided by bioinformatics tools, allelic variants of classic human leukocyte antigen (HLA) genes in the human MHC could be analyzed for supertypes [44], functional motifs (e.g., HLA-B Bw4/Bw6 and HLA-B signal peptide) [37, 45], as well as individual amino acid residues [27, 46, 47]. Noncoding SNPs resolved by NGS, including those in the 5’ and 3’ untranslated regions, provide further coverage of regulatory variants, and their functional attributes can be readily surveyed in public databases like HaploReg [48], the ENCODE project [49, 50], dbGaP [51, 52], NCBI Global Cross-database, http://www.ncbi.nlm.nih.gov/), eQTL hits [53], and PhenoScanner [54].
Key findings that are consistent between cohorts or withstood the test of time
When comparable datasets from the two study cohorts were analyzed, consensus findings were limited to three loci, i.e., IL19 (rs12407485-A), HLA-A (allele A*68:02) and HLA-B (alleles in the B*57 group and alleles carrying a Met residue at the P2 position of the leader peptide (Table 2). The functional attributes of HLA-A*68:02 and HLA-B*57 in antigen presentation have been well documented (Robinson, 2020 #777), while rs12407485 has been recognized as an expression QTL for four genes (CD55, IL10, IL19 and IL24). Lack of similar cohorts in other regions has precluded the possibility of further validation (beyond southern and eastern Africa) to fully justify a search for mechanisms underlying these associations.
Table 2.
Summary of consensus immunogenetic findings from two study cohorts.
| Locus | Variant (allele or aa residue) | Setting | Association | Reference(s) |
|---|---|---|---|---|
| IL19 | rs12407485-Aa | HESNs | Resistance to HIV-1 acquisition | [43] |
| HLA-A | A*68:02b | HESNs | Susceptibility to HIV-1 acquisition | [19] |
| HLA-B | B*57b | SPs | HIV-1 transmission | [17, 78] |
| HLA-B | Alleles carrying P2-Metc | HESNs | Susceptibility to HIV-1 acquisition | [6] |
Within a predicted enhancer region. According to a recent (March 2021) search in PhenoScanner (version 2), rs12407485 is a well-recognized eQTL, being associated with the expression of four genes (CD55, IL10, IL19 and IL24).
Confirmative results from the Rwandan cohort have not been published.
Mostly B*57:03 and often reflected by its persistent impact on viral load
Methionine at position 2 of a leader peptide that binds to HLA-E; also supported by in vitro assays [6].
Robust, cohort-specific associations are also worth noting. For example, data from the Zambian cohort suggest that additional variants at HLA-A (allele *A*36:01), HLA-B (allele sharing) and KIR2DS4 (allele *001, corresponding to a functional, full-length receptor) might be informative as well (Table 3). In SPs, A*36:01 promoted HIV-1 transmission in several rounds of data analyses as the number of eligible couples increased over time (Figure 3), and its association with elevated HIV-1 viral load was persistent over time [29]. Epidemiological evidence from an African-American cohort and experimental data supported the association of HLA-B allele sharing and KIR2DS4*001 with unfavorable outcomes in Zambians) [7, 8].
Table 3.
Cohort-specific immunogenetic findings with various supporting evidence.
| Locus | Variant (allele or aa residue) | Setting | Association | Reference(s) |
|---|---|---|---|---|
| HLA-A | A*36:01a | Zambian SPs | Promoting HIV-1 transmission | Ref. [17] |
| HLA-B | Alleles sharingb | Both partners | Promoting HIV-1 transmission | Ref. [2] |
| KIR2DS4 | Full-length allele *001c | Zambian SPs | Promoting HIV-1 transmission | Ref. [18] |
Observation was confirmed multiple times in expanded cohorts (e.g., Figure 2), with clear impact on viral load in the SP partners.
Supported by experimental evidence [8].
The only allele encoding a functional, full-length receptor, with implications for inflammation mediated through natural killer cells [7].
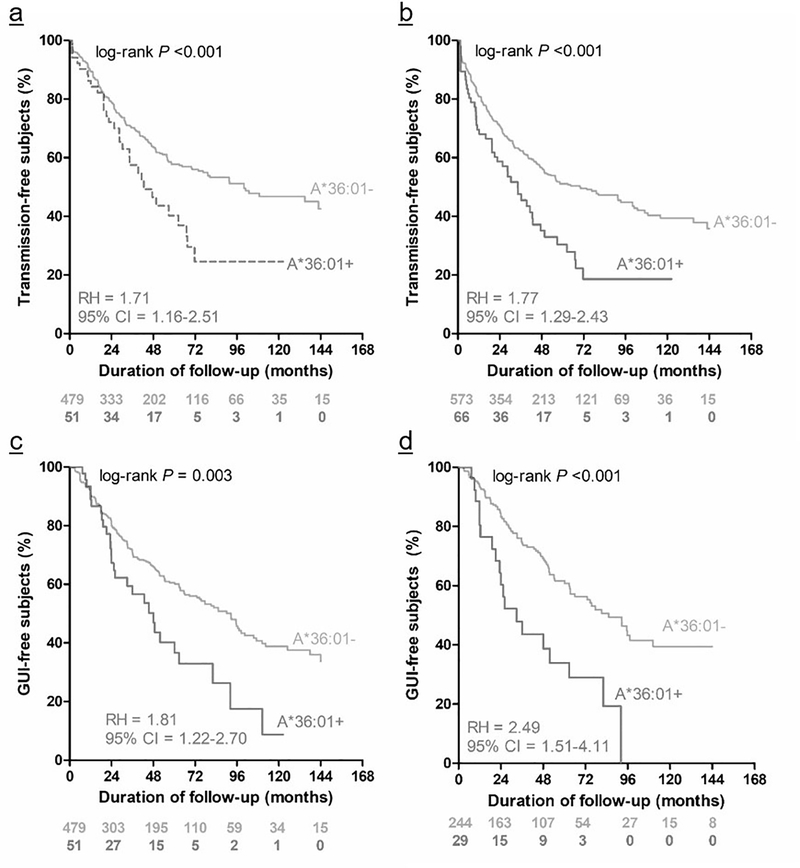
Figure 3. Kaplan-Meier curves showing the impact of HLA-A*36:01 in donor/index partners on heterosexual HIV-1 transmission and genital ulcer/inflammation (GUI) during longitudinal follow-up.
Prompted by our original report on 429 HIV-1 discordant Zambian couples [17], several analyses of expanded datasets (530 couples in panel a versus 639 couples in panel b) confirmed that HLA-A*36:01 in donor/index partners was persistently an unfavorable factor for HIV-1 transmission. This association was partially explained by elevated HIV-1 viral load setpoint in donor partners, as well as presence of GUI over time (panel c, 530 couples), the association with GUI was mostly seen in female-to-male transmission (panel d, with 273 couples) (combined data from two publications [17, 19], including recent updates). The estimates of relative hazards (RH) and 95% confidence interval (CI) were based on Cox proportional hazards models.
In multivariable models, two unfavorable genetic factors found in the Zambian cohort had similar effect sizes in an expanded dataset (re-analyses of data from two publications [17, 19], including recent updates) (Table 4), before and after stratification by the direction of heterosexual transmission (male-to-female or female-to-male). The difference made by the HLA-A*36:01 allele in donor partners was almost identical to that of viral load (per 1.0 log10 or 10-fold change), but its rarity in the Rwandan cohort precluded a cross-cohort validation.
Table 4.
Multivariable models that jointly assess the relative impact of three risk factors (host and viral) on heterosexual HIV-1 transmission among Zambian couples.
| Factors in modela | All subjects (638 couples) | MTF subset (312 couples) | FTM subset (316 pairs) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | RH | 95% CI | p | n | RH | 95% CI | p | n | RH | 95% CI | p | |
| Donor VLc | 634 | 1.8 | 1.6–2.2 | <0.0001 | 312 | 1.67 | 1.3–2.2 | <0.0001 | 316 | 1.9 | 1.5–2.5 | <0.0001 |
| A*36:01 (donor) | 63 | 1.6 | 1.2–2.2 | 0.004 | 28 | 1.44 | 0.9–2.3 | 0.130 | 35 | 1.8 | 1.2–2.9 | 0.009 |
| A*68:02 (recipient)b | 99 | 1.5 | 1.1–2.0 | 0.006 | 58 | 1.51 | 1.0–2.2 | 0.029 | 41 | 1.5 | 0.9–2.4 | 0.099 |
Data from an expanded Zambian cohort (638 couples) (combined data from two publications [17, 19], including recent updates). Results were consistent with earlier findings based on analyses of paired recipient and index/donor partners (each couple was counted as one unit) [17], even when the cohort was split by the direction of transmission (male-to-female/MTF versus female-to-male/FTM).
HIV-1 viral load (VL) was treated as a continuous variable after log10-transformation. The summary statistics, including relative hazards (RH) and 95% confidence interval (CI), are based on Cox proportional hazards models (adjusted for factors in each model).
Genital ulcer/inflammation (GUI) as an important cofactor
HIV/AIDS has a substantial overlap with other infections [55]. As a common manifestation of genital co-infections, GUI appeared to remove a transmission bottleneck such that multiple founder viruses could be transmitted [16, 56]. The importance of GUI to heterosexual HIV-1 transmission was unequivocal in both cohorts, even with a clear additive effect when GUI was observed in both partners (SP + HESN) [17]. In analyses that tested GUI as a secondary outcome (for visits prior to HIV-1 transmission or end of follow-up), KIR2DS4*001 [18] and HLA-A*36:01 (Figure 3) turned out to be unfavorable factors in Zambian SPs, suggesting that both can mediate trans-mucosal infection directly (on viral load) and indirectly (on GUI). Likewise, HLA-A*68:02 as a risk factor for HIV-1 acquisition among Zambian HESNs could be explained in part by high incidence of GUI as well (based on re-analyses of data from two publications [17, 19], including recent updates). Together, these observations revealed two intertwined vicious circles involving distinct immunogenetic pathways. First, unfavorable immunogenetic factors (e.g., KIR2DS4*001 and HLA-A*36:01) in SPs render these subjects susceptible to GUIs, and the spread of genital co-infections (typically more contagious than HIV-1 itself) to HENSs creates a microenvironment that promotes HIV-1 acquisition in HESNs. Conversely, the spread of genital co-infections from HESNs to SPs boosts mucosal HIV-1 shedding and compromises mucosal barriers in SPs, also paving way for HIV-1 transmission. Either way, it was evident that immunogenetic factors in both donor and recipient partners played a prominent role in GUI.
Unproductive efforts on genome-wide scans
Consistent with earlier work that targeted recipient partners of discordant couples in African countries [57], our recent genome-wide association scan (GWAS) did not reveal any reproducible association hits for the two study cohorts. This work used a relatively recent GWAS array (the Illumina Multi-Ethnic Genotyping Array) that is considered suitable for African cohorts [58] and covers over 1.5 million SNPs, with extra flexibility for accommodating up to 300,000 custom SNPs. In a more focused screening facilitated by two versions of the Illumina ImmunoChip, which is cost-effective and targets a broad range of SNPs mapped to genes that mediate innate and adaptive immune responses [43], ~195,000 ImmunoChip version 1 SNPs passed various quality control measures and were deemed acceptable for association analyses – a discovery phase (in the larger Zambian cohort) was followed by a validation phase (in the smaller Rwandan cohort). Again, not a single SNP met the typical, minimal level of study-wide statistical significance (P <0.05 in both cohorts, after Bonferroni correction for the number of independent tests). Alternatively, no association signals with a false discovery rate <0.05 could be confirmed in both cohorts, although cohort-specific hits have been observed [59]. The addition of ImmunoChip version 2 data (for more recent samples) did not make any difference (Tang et al., unpublished data), neither did alternative analyses for thousands of SNPs previously known as “causal variants” for more than 30 autoimmune disorders [60].
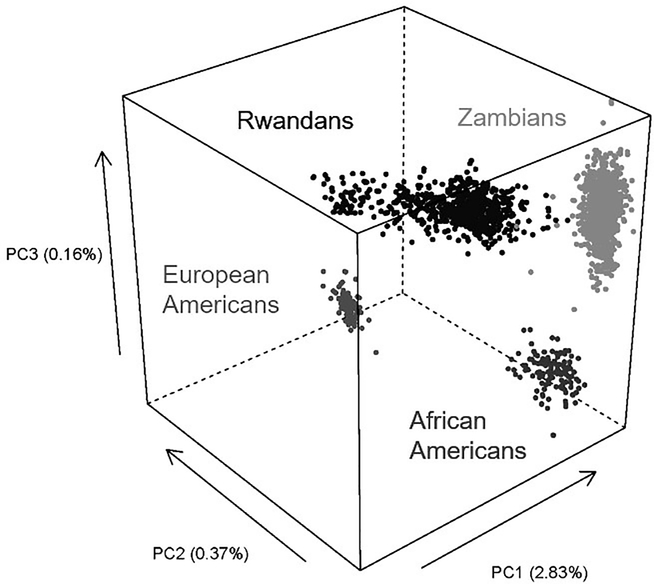
One main byproduct from the use of ImmunoChip bead arrays was the assessment of overall genetic structures. When SNPs with low pairwise LD (r2 <0.20) were used for PCA, the first three components captured most of the genetic variability in each cohort (Figure 4), and these quantitative measures could be kept as covariates in all association analyses. Principal components also formed the basis for measuring genetic distance for each donor-recipient pair, but these metrics did not seem to matter to heterosexual HIV-1 transmission in either cohort. Of note, the Kigali, Rwandan cohort showed a greater genetic diversity than the Zambian cohort (Figure 4), coinciding with a rapid influx of migrants into the Rwandan capital after a massive genocide in 1994.
Figure 4. A 3D graph showing the clustering of subjects from three countries.
In principal component analyses using genome-wide SNPs with low linkage disequilibrium (pairwise r2 <0.20), the first three dimensions (PC1-PC3) captured most of the genetic variations for subjects enrolled from Kigali (Rwanda), Lusaka (Zambia) and Birmingham (United States). The Birmingham cohort consists of African Americans and European Americans. Variance explained by each component is shown in parentheses (Tang et al., unpublished data).
In other applications, genomics data proved useful in determining biological sex, cryptic familial relatedness (kinship) and genetic outliers [43]. In addition, Many SNPs in the extended MHC region helped with the fine-mapping of association signals detected for individual HLA alleles or amino acid residues [9). Indeed, the vast majority of common HLA variants in each cohort, defined at the 1-field (2-digit) and 2-field (4- or 5-digit) specificity levels, could be imputed by SNP data {Wiener, 2018 #770, 42], which may offer a cost-effective approach (<$60/sample) to HLA-based screening of study participants in future studies.
Challenges and limitations
Heterosexual HIV-1 transmission is a highly inefficient process, so our samples sizes remained modest (still the largest to date) despite efforts spanning nearly two decades. As the circulating viruses constantly adapt to immune responses, HIV-1 isolates observed at the beginning of our cohort study may well differ from those relevant to the contemporary populations [61–64]. Analyses of transmitted/founder viruses (TFVs), especially their replicative capacity and pre-adaptation to HLA profiles in the recipient partners, can reflect viral fitness or survival advantages [5, 65–67], but not all TFVs can be readily characterized for linked transmission couples with properly stored specimens. Frequent changes (even interruptions) in research priorities under geopolitical, fiscal and cultural pressures are also unpredictable. Accordingly, the scope of work being discussed here needs to be placed in the right context – that our two study cohorts had multiple high bars to clear. As the global HIV/AIDS landscape evolves, some of the original research objectives are no longer attractive even as we have gained strong footholds in pursuing several complex and intertwined research aims that no other cohorts can achieve.
Another main limitation in these studies is the lack of statistical power for examining gene x gene, gene x environment (e.g., sex and age) and host x virus (HIV-1 subtypes A1 and C) interactions. In theory, clinical outcomes may well depend on biological amplifications (or synergies) under certain microenvironments, but heavy penalty for random testing easily becomes an insurmountable hurdle in these efforts. Perhaps leads from other ongoing studies can gradually justify focused analyses of interactive terms [68].
Other implications and future directions
Immunogenetic factors that help shape the natural history of HIV-1 infection, especially during the pivotal, early phases of HIV-1-host interactions, may further impact therapeutic outcomes [69]. As the HIV/AIDS field shifts its focus on immune restoration after cART or eradication of viral reservoirs (“functional cure”) [70], a “genetic profile” that predicts immunologic deterioration early in the course of infection [71] or atypical response to treatment could alert caregivers to the need for more aggressive versus standard care. The demand for prognostically useful information and its translation into the clinic will accelerate as several phenomena occur in the era of universal cART: (i) options for early and targeted intervention increase, (ii) “genetic profile” or “causal variant” reliably predicts outcome; (iii) pressure intensifies to tailor individual treatment to a predicted disease trajectory (including non-AIDS morbidities) or risk for further transmission (threat to public health); and (iv) barriers (including costs) to routine genetic profiling and counseling diminish.
HLA genotyping has been well received in clinics where there is a need to promptly identify HLA-B*57:01 patients who are destined to develop life-threatening hypersensitivity reactions to abacavir [72]. Beyond this immediate application, prognostic genetic profiling can benefit clinical trials by improving trial efficiency (pre-treatment stratification) and reducing subgroup heterogeneity (as a covariate in analysis of efficacy) [73], as reflected by data from a promising AIDS vaccine trial in Thailand (RV144) [74, 75] and in certain nonhuman primate models [76]. Various research activities related to the two study cohorts in Rwanda and Zambia are being integrated into the International AIDS Vaccine Initiative [77, 78] and may prove to have far-reaching significance in the African epicenter, although follow-up research will certainly require some strategic adjustments, under the new premises of the 90:90:90 initiative – to rid the world free of AIDS by substantially reducing the further spread of HIV-1 infection, through concerted efforts for active diagnosis, early treatment and targeted prevention [79, 80]. A new generation of investigators, who have been trained by several research teams involved in these cohort studies, are now armed with well-documented expertise in epidemiology, genetics, immunology, virology and biostatistics. Their contributions to scientific research will undoubtedly go beyond HIV/AIDS.
Acknowledgements
The original data included in this review came primarily from an R01 project (AI071906) funded by the National Institute of Allergy and Infectious Disease, with further funding from R01-AI64060. Unpublished genomics data for African-Americans and European Americans came from a third project (R21-AG051309). I am grateful to Dr. Richard A. Kaslow, Dr. Susan Allen and Dr. Eric Hunter for their leadership during these projects. Members of the Rwanda-Zambia HIV-1 Research Group were essential to patient enrollment and data management. Refinements of earlier findings based on updated datasets were made possible by statistical models prepared by Mr. Xuelin Li, Dr. Heather A. Prentice, and Dr. Howard Wiener.
Footnotes
Conflict of Interest
The author has no potential conflict of interest to declare.
References
- 1.Tebit DM and Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011; 11: 45–56. [DOI] [PubMed] [Google Scholar]
- 2.Dorak MT, Tang J, Penman-Aguilar A, Westfall AO, Zulu I, Lobashevsky ES, et al. Transmission of HIV-1 and HLA-B allele-sharing within serodiscordant heterosexual Zambian couples. Lancet. 2004; 363: 2137–2139. [DOI] [PubMed] [Google Scholar]
- 3.Tang J, Tang S, Lobashevsky E, Zulu I, Aldrovandi G, Allen S, et al. HLA allele sharing and HIV type 1 viremia in seroconverting Zambians with known transmitting partners. AIDS Res Hum Retroviruses. 2004; 20: 19–25. [DOI] [PubMed] [Google Scholar]
- 4.Tang J, Penman-Aguilar A, Lobashevsky E, Allen S, Kaslow R and Zambia-UAB HIV Research Project. HLA-DRB1 and -DQB1 alleles and haplotypes in Zambian couples and their associations with heterosexual transmission of human immunodeficiency virus type 1. J Infect Dis. 2004; 189: 1696–1704. [DOI] [PubMed] [Google Scholar]
- 5.Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009; 206: 909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merino AM, Sabbaj S, Easlick J, Goepfert P, Kaslow RA and Tang J. Dimorphic HLA-B signal peptides differentially influence HLA-E- and natural killer cell-mediated cytolysis of HIV-1-infected target cells. Clin Exp Immunol. 2013; 174: 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merino AM, Dugast AS, Wilson CM, Goepfert PA, Alter G, Kaslow RA, et al. , KIR2DS4 promotes HIV-1 pathogenesis: new evidence from analyses of immunogenetic data and natural killer cell function. PLoS ONE. 2014; 9: e99353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabbaj S, Scanlon N, Du VY, Wang Y, Tang J, Hunter E, et al. Enhanced allogeneic cellular responses to mismatched HLA-B antigens results in more efficient killing of HIV infected cells. J Acquir Immune Defic Syndr. 2016; 71: 493–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claiborne DT, Scully EP, Palmer CD, Prince JL, Macharia GN, Kopycinski J, et al. Protective HLA alleles are associated with reduced LPS levels in acute HIV infection with implications for immune activation and pathogenesis. PLoS Pathog. 2019; 15: e1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Institutes of Health (NIH). NIH Strategic Plan for HIV and HIV-Related Research Archive. 2020. [cited 2021 April 4]; Available from: https://www.oar.nih.gov/hiv-policy-and-research/strategic-plan/archive.
- 11.Montaner JS, Lima VD, Barrios R, Yip B, Wood E, Kerr T, et al. Association of highly active antiretroviral therapy coverage, population viral load, and yearly new HIV diagnoses in British Columbia, Canada: a population-based study. Lancet. 2010; 376: 532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delva W, Eaton JW, Meng F, Fraser C, White RG, Vickerman P, et al. HIV treatment as prevention: Optimising the impact of expanded HIV treatment programmes. PLoS Med. 2012; 9: e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen MS, Gamble T and McCauley M. Prevention of HIV Transmission and the HPTN 052 Study. Annu Rev Med. 2020; 71: 347–360. [DOI] [PubMed] [Google Scholar]
- 14.Herce ME, Hoffmann CJ, Fielding K, Topp SM, Hausler H, Chimoyi L, et al. Universal test-and-treat in Zambian and South African correctional facilities: a multisite prospective cohort study. Lancet HIV. 2020; 7: e807–e816. [DOI] [PubMed] [Google Scholar]
- 15.Coburn BJ, Okano JT and Blower S. Using geospatial mapping to design HIV elimination strategies for sub-Saharan Africa. Sci Transl Med. 2017; 9: eaag0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009; 5: e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, Allen S, et al. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol. 2008; 181: 2626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merino A, Malhotra R, Morton M, Mulenga J, Allen S, Hunter E, et al. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J Infect Dis. 2011; 203: 487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song W, He D, Brill I, Malhotra R, Mulenga J, Allen S, et al. Disparate associations of HLA class I markers with HIV-1 acquisition and control of viremia in an African population. PLoS ONE. 2011; 6: e23469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrington M and O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003; 54: 535–51. [DOI] [PubMed] [Google Scholar]
- 21.Tang J and Kaslow RA. The impact of host genetics on HIV infection and disease progression in the era of highly active antiretroviral therapy. AIDS. 2003; 17: S51–S60. [DOI] [PubMed] [Google Scholar]
- 22.Nolan D, Gaudieri S, John M and Mallal S. Impact of host genetics on HIV disease progression and treatment: new conflicts on an ancient battleground. AIDS. 2004; 18: 1231–40. [DOI] [PubMed] [Google Scholar]
- 23.Fellay J, Shianna KV, Telenti A and Goldstein DB. Host genetics and HIV-1: the final phase? PLoS Pathog. 2010; 6: e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentice HA and Tang J. HIV-1 dynamics: A reappraisal of host and viral factors, as well as methodological issues. Viruses. 2012; 4: 2080–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goulder PJ and Walker BD. HIV and HLA class I: an evolving relationship. Immunity. 2012; 37: 426–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorak MT, Shao W, Machulla HK, Lobashevsky ES, Tang J, Park MH, et al. Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun. 2006; 7: 450–67. [DOI] [PubMed] [Google Scholar]
- 27.McLaren PJ, Ripke S, Pelak K, Weintrob AC, Patsopoulos NA, Jia X, et al. Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum Mol Genet. 2012; 21: 4334–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009; 5: e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang J, Malhotra R, Song W, Brill I, Hu L, Farmer PK, et al. , Human leukocyte antigens and HIV type 1 viral load in early and chronic infection: Predominance of evolving relationships. PLoS ONE. 2010; 5: e9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereyra F et al. and International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010; 330: 1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Autran B, Descours B, Avettand-Fenoel V and Rouzioux C. Elite controllers as a model of functional cure. Curr Opin HIV AIDS. 2011; 6: 181–7. [DOI] [PubMed] [Google Scholar]
- 32.Goulder P and Deeks SG. HIV control: Is getting there the same as staying there? PLoS Pathog. 2018; 14: e1007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kayongo A, Gonzalo-Gil E, Gumusgoz E, Niwaha AJ, Semitala F, Kalyesubula R, et al. Brief Report: Identification of Elite and Viremic Controllers From a Large Urban HIV Ambulatory Center in Kampala, Uganda. J Acquir Immune Defic Syndr. 2018; 79: 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phetsouphanh C, Aldridge D, Marchi E, Munier CML, Meyerowitz J, Murray L, et al. Maintenance of Functional CD57+ Cytolytic CD4+ T Cells in HIV+ Elite Controllers. Front Immunol. 2019; 10: 1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002; 76: 8276–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu L, Song W, Brill I, Mulenga J, Allen S, Hunter E, et al. Genetic variations and heterosexual HIV-1 infection: analysis of clustered genes encoding CC-motif chemokine ligands. Genes Immun. 2012; 13: 202–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merino AM, Song W, He D, Mulenga J, Allen S, Hunter E, et al. , HLA-B signal peptide polymorphism influences the rate of HIV-1 acquisition but not viral load. J Infect Dis. 2012; 205: 1797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett JC, Fry B, Maller J and Daly MJ, Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21: 263–5. [DOI] [PubMed] [Google Scholar]
- 39.Cortes A and Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther, 2011. 13: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet. 2013; 45: 664–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowes J, Budu-Aggrey A, Huffmeier U, Uebe S, Steel K, Hebert HL, et al. Dense genotyping of immune-related susceptibility loci reveals new insights into the genetics of psoriatic arthritis. Nat Commun. 2015; 6: 6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prentice HA, Pajewski NM, He D, Zhang K, Brown EE, Kilembe W, et al. Host genetics and immune control of HIV-1 infection: fine mapping for the extended human MHC region in an African cohort. Genes Immun. 2014; 15: 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Zhang K, Pajewski NM, Brill I, Prentice HA, Shrestha S, et al. Immunogenetic influences on acquisition of HIV-1 infection: consensus findings from two African cohorts point to an enhancer element in IL19 (1q32.2). Genes Immun. 2015; 16: 213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazaryan A, Lobashevsky E, Mulenga J, Karita E, Allen S, Tang J, et al. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J Virol. 2006; 80: 6056–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, Leung JY, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci USA. 2001; 98: 5140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kloverpris HN, Harndahl M, Leslie AJ, Carlson JM, Ismail N, van der Stok M, et al. HIV control through a single nucleotide on the HLA-B locus. J Virol. 2012; 86: 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiener HW, Shrestha S, Lu H, Karita E, Kilembe W, Allen S, et al. Immunogenetic factors in early immune control of human immunodeficiency virus type 1 (HIV-1) infection: Evaluation of HLA class I amino acid variants in two African populations. Hum Immunol. 2017; 79: 166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward LD and Kellis M, HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012; 40: D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010; 38: D620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ENCODE Project Consortium, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wooten EC and Huggins GS. Mind the dbGAP: the application of data mining to identify biological mechanisms. Mol Interv. 2011; 11: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meesters C, Leber M, Herold C, Angisch M, Mattheisen M, Drichel D, et al. Quick, “imputation-free” meta-analysis with proxy-SNPs. BMC Bioinformatics. 2012; 13: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012; 44: 502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019; 35: 4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corey L, Wald A, Celum CL and Quinn TC. The effects of herpes simplex virus-2 on HIV-1 acquisition and transmission: a review of two overlapping epidemics. J Acquir Immune Defic Syndr. 2004; 35: 435–45. [DOI] [PubMed] [Google Scholar]
- 56.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, et al. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014; 345: 1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lingappa JR, Petrovski S, Kahle E, Fellay J, Shianna K, McElrath MJ, et al. Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PLoS ONE. 2011; 6: e28632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adebamowo SN, Adeyemo AA, Rotimi CN, Olaniyan O, Offiong R, Adebamowo CA, et al. Genome-wide association study of prevalent and persistent cervical high-risk human papillomavirus (HPV) infection. BMC Med Genet. 2020; 21: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prentice HA, Pajewski NM, Porter TR, Zhang K, Borwon EE, Allen SA, et al. Host genetics and susceptibility to HIV-1 infection: Novel MHC associations among serodiscordant couples. Poster presentation at AIDS Vaccine 2013. (Barcelona, Spain: ). Full data are posted online at https://www.researchgate.net/publication/263124868_Host_Genetics_and_Susceptibility_to_HIV-1_Infection_Novel_MHC_Associations_Among_Serodiscordant_Couples. [Google Scholar]
- 60.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015; 518: 337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Manen D, Gras L, Boeser-Nunnink BD, van Sighem AI, Maurer I, Mangas Ruiz MM, et al. Rising HIV-1 viral load set point at a population level coincides with a fading impact of host genetic factors on HIV-1 control. AIDS. 2011; 25: 2217–26. [DOI] [PubMed] [Google Scholar]
- 62.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009; 458: 641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bansal A, Carlson J, Yan J, Akinsiku OT, Schaefer M, Sabbaj S, et al. CD8 T cell response and evolutionary pressure to HIV-1 cryptic epitopes derived from antisense transcription. J Exp Med. 2010; 207: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson JM, Du VY, Pfeifer N, Bansal A, Tan VY, Power K, et al. Impact of pre-adapted HIV transmission. Nat Med. 2016; 22: 606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goepfert PA, Lumm W, Farmer P, Matthews P, Prendergast A, Carlson JM, et al. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008; 205: 1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Claiborne DT, Prince JL, Scully E, Macharia G, Micci L, Lawson B, et al. Replicative fitness of transmitted HIV-1 drives acute immune activation, proviral load in memory CD4+ T cells, and disease progression. Proc Natl Acad Sci USA. 2015; 112: E1480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monaco DC, Dilernia DA, Fiore-Gartland A, Yu T, Prince JL, Dennis KK, et al. Balance between transmitted HLA preadapted and nonassociated polymorphisms is a major determinant of HIV-1 disease progression. J Exp Med. 2016; 213: 2049–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X, Price MA, He D, Kamali A, Karita E, Lakhi S, et al. Host genetics and viral load in primary HIV-1 infection: clear evidence for gene by sex interactions. Hum Genet. 2014; 133: 1187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuniholm MH, Gao X, Xue X, Kovacs A, Anastos K, Marti D, et al. Human leukocyte antigen genotype and risk of HIV disease progression before and after initiation of antiretroviral therapy. J Virol. 2011; 85: 10826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saez-Cirion A, Pancino G, Sinet M, Venet A and Lambotte O. HIV controllers: how do they tame the virus? Trends Immunol. 2007; 28: 532–40. [DOI] [PubMed] [Google Scholar]
- 71.Tang J, Li X, Price MA, Sanders EJ, Anzala O, Karita E, et al. CD4:CD8 lymphocyte ratio as a quantitative measure of immunologic health in HIV-1 infection: findings from an African cohort with prospective data. Front Microbiol. 2015; 6: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saag M, Balu R, Phillips E, Brachman P, Martorell C, Burman W, et al. High sensitivity of human leukocyte antigen-B*5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin Infect Dis. 2008; 46: 1111–8. [DOI] [PubMed] [Google Scholar]
- 73.Kulkarni H, Marconi VC, Agan BK, McArthur C, Crawford G, Clark RA, et al. Role of CCL3L1-CCR5 genotypes in the epidemic spread of HIV-1 and evaluation of vaccine efficacy. PLoS ONE. 2008; 3: e3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paris R, Bejrachandra S, Thongcharoen P, Nitayaphan S, Pitisuttithum P, Sambor A, et al. HLA class II restriction of HIV-1 clade-specific neutralizing antibody responses in ethnic Thai recipients of the RV144 prime-boost vaccine combination of ALVAC-HIV and AIDSVAX(R) B/E. Vaccine. 2012; 30: 832–836. [DOI] [PubMed] [Google Scholar]
- 75.Gartland AJ, Li S, McNevin J, Tomaras GD, Gottardo R, Janes H, et al. Analysis of HLA A*02 association with vaccine efficacy in the RV144 HIV-1 vaccine trial. J Virol. 2014; 88: 8242–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, et al. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci Transl Med. 2011; 3: 81ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang J, Cormier E, Gilmour J, Price MA, Prentice HA, Song W, et al. Human leukocyte antigen variants B*44 and B*57 are consistently favorable during two distinct phases of primary HIV-1 infection in sub-Saharan Africans with several viral subtypes. J Virol. 2011; 85: 8894–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prentice HA, Porter TR, Price MA, Cormier E, He D, Farmer PK, et al. HLA-B*57 versus HLA-B*81 in HIV-1 infection: slow and steady wins the race? J Virol. 2013; 87: 4043–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Granich R, Williams B, Montaner J and Zuniga JM. 90–90-90 and ending AIDS: necessary and feasible. Lancet. 2017; 390: 341–343. [DOI] [PubMed] [Google Scholar]
- 80.de Bree GJ, van Sighem A, Zuilhof W, van Bergen J, Prins M, Heidenrijk M, et al. Is reaching 90–90-90 enough to end AIDS? Lessons from Amsterdam. Curr Opin HIV AIDS. 2019; 14: 455–463. [DOI] [PubMed] [Google Scholar]
- 81.MacDonald KS, Fowke KR, Kimani J, Dunand VA, Nagelkerke NJ, Ball TB, et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J Infect Dis. 2000; 181: 1581–9. [DOI] [PubMed] [Google Scholar]
- 82.MacDonald KS, Embree JE, Nagelkerke NJ, Castillo J, Ramhadin S, Njenga S, et al. The HLA A2/6802 supertype is associated with reduced risk of perinatal human immunodeficiency virus type 1 transmission. J Infect Dis. 2001; 183: 503–6. [DOI] [PubMed] [Google Scholar]
- 83.Fideli US, Allen SA, Musonda R, Trask S, Hahn BH, Weiss H, et al. , Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retroviruses. 2001; 17: 901–10. [DOI] [PMC free article] [PubMed] [Google Scholar]