Abstract
Sea lice (Caligus rogercresseyi) is an ectoparasite which causes major production losses in the salmon aquaculture industry worldwide. Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) are two of the most susceptible salmonid species to sea lice infestation. The objectives of this study were to: (1) identify genomic regions associated with resistance to Caligus rogercresseyi in Atlantic salmon and rainbow trout by performing single-step Genome-Wide Association studies (ssGWAS), and (2) identify candidate genes related to trait variation based on exploring orthologous genes within the associated regions across species. A total of 2626 Atlantic salmon and 2643 rainbow trout were challenged and genotyped with 50 K and 57 K SNP panels, respectively. We ran two independent ssGWAS for sea lice resistance on each species and identified 7 and 13 regions explaining more than 1% of the genetic variance for the trait, with the most important regions explaining 3% and 2.7% for Atlantic salmon and rainbow trout, respectively. We identified genes associated with immune response, cytoskeleton function, and cell migration when focusing on important genomic regions for each species. Moreover, we found 15 common orthogroups which were present in more than one associated genomic region, within- or between-species; however, only one orthogroup showed a clear potential biological relevance in the response against sea lice. For instance, dual-specificity protein phosphatase 10-like (dusp10) and dual-specificity protein phosphatase 8 (dusp8) were found in genomic regions associated with lice density in Atlantic salmon and rainbow trout, respectively. Dusp10 and dusp8 are modulators of the MAPK pathway and might be involved in the differences of the inflammation response between lice resistant and susceptible fish from both species. Our results provide further knowledge on candidate genes related to sea lice resistance and may help establish better control for sea lice in fish populations.
Subject terms: Animal breeding, Genome-wide association studies
Introduction
Sea lice is currently the most harmful parasite for salmon farming worldwide1. Economic losses due to different sea lice species are mainly associated with the reduction of feed conversion rate, growth, indirect mortalities and loss of product value. Furthermore, it has been estimated that the global costs for sea lice control have reached $480 million (USD) annually2. The two most important sea lice species which generate a considerable negative impact in salmon farming are Lepeophtheirus salmonis and Caligus rogercresseyi3.
Caligus rogercresseyi, first described in 2000 by Boxshall and Bravo4, is the main sea lice species affecting salmon aquaculture in Chile5. C. rogercresseyi primarily affects Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss), while coho salmon (Oncorhynchus kisutch) has an innate lower susceptibility to the parasite6. The consequences of the infestation by sea lice include skin lesions, osmotic imbalance and greater susceptibility to bacterial and viral infections through the suppression of the immune response by the damage generated in the skin of the host7. The parasite life cycle is comprised of eight stages of development8: two states of nauplii, one copepod state, four chalimus states and the adult state. The stages of nauplii (I and II) and copepods (infectious stage) are planktonic. The four stages of chalimus (I–IV) are sessile while the adult is a mobile stage9.
Recent studies have estimated low to moderate genetic variation for resistance to sea lice in Atlantic salmon populations, with heritabilities ranging from 0.12 to 0.32 and from 0.13 to 0.33 when resistance was defined as the number of parasites fixed in all the fins7,9,10, or as the logarithm of the parasite density11,12, respectively. A recent study reported a heritability value of 0.09 for sea lice resistance in a rainbow trout breeding population13. These results indicate that it is feasible to improve resistance to sea lice in Atlantic salmon and rainbow trout populations by utilizing selective breeding9,10,14.
Comparative genomic approaches15 allow the identification of genomic similarities between different species, including conserved genes and motifs, traces of genome duplication and gene functions16. Traditionally, these analyses are focused on orthologous genes, which are homologous genes present in different species resulting from direct transmission from a common ancestor16. To date, comparative genomic studies between salmonids have mainly focused on finding evolutionary similarities in the genetic basis of body and sex related traits, including growth, development and sexual differentiation17–19.
A recent study compared results from genome-wide association studies (GWAS) on three salmonid species and identified functional candidate genes involved in resistance to the infection caused by Piscirickettsia salmonis, an intracellular bacterium20. To date, no studies have aimed at comparing genomic regions associated with resistance to sea lice in salmonid species.
The main objectives of the present study were to: (1) identify genomic regions associated with resistance to Caligus rogercresseyi in Atlantic salmon and rainbow trout through GWAS, and (2) identify functional candidate genes potentially related to trait variation through a comparative genomics approach, based on exploring orthologous genes within the associated regions between species.
Results and discussion
The comparative genomics analysis presented here allowed us to identify groups of orthologues genes and several candidate genes among adjacent single nucleotide polymorphisms (SNP) that explained more than 1% of the genetic variance for resistance to C. rogercresseyi. This is the first study aimed at comparing the genomic basis of sea lice resistance in both Atlantic salmon and rainbow trout.
Sea lice challenge test
There was no difference between the average number of sea lice found on Atlantic salmon or rainbow trout in the experimental challenge (Table 1). An average of 5.9 ± 6.6 and 6.1 ± 4.2 parasites per fish was estimated for Atlantic salmon and rainbow trout, respectively. In terms of the maximum number of parasites, this value varied from 106 parasites in Atlantic salmon to 28 in rainbow trout and, for both species, some animals did not present any parasites. The average weight at the end of the experimental challenge was 278.1 ± 90.3 g (ranging from 104 to 569 g) and 173.1 ± 31.4 g (ranging from 86 to 265 g), for Atlantic salmon and rainbow trout, respectively. Although the average number of parasites was not significantly different between the two species, the difference in the average final weight at the end of each challenge could explain the difference in the maximum number of parasites found (~ 4 times more parasites in Atlantic salmon). The number of parasites counted in each species after the challenge is below the range determined in previous studies. For instance, Ødegård et al.11 obtained an average of 20.96 ± 19.68, while Robledo et al.21 reported an average of 38 ± 16 sea lice. The lower number of parasites in comparison to these studies is most likely related to differences in the area of counting (whole body surface versus only fins), parasite species (Lepeophtheirus salmonis versus C.rogercresseyi) and the time of sampling after infestation (8–15 versus 6 days).
Table 1.
Summary statistics for body weight (BW) and lice count (LC) in Atlantic salmon and rainbow trout.
| Species | Mean BW (g) | BW SD (g) | Min BW(g) | Max BW (g) | LC means | LC SD | Min LC | Max LC |
|---|---|---|---|---|---|---|---|---|
| S. salar | 278.1 | 90.3 | 104.0 | 569.0 | 5.9 | 6.61 | 0 | 106 |
| O. mykiss | 173.1 | 31.4 | 86.0 | 265 | 6.1 | 4.22 | 0 | 28 |
SD standard deviation.
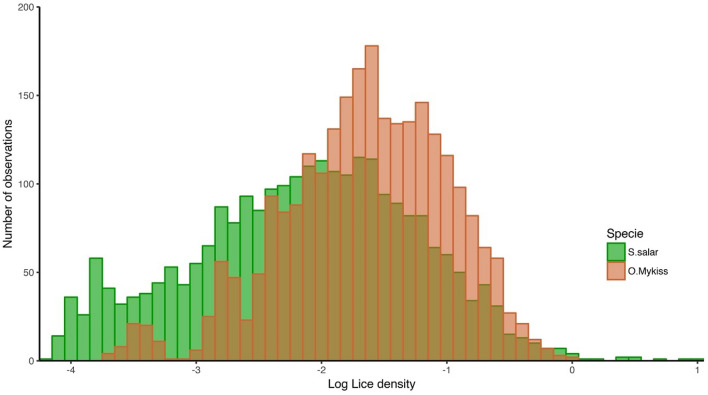
To measure resistance to C. rogercresseyi, lice count values were transformed into lice density on the log scale (LogLD), which allows for correction of the number of parasites based on the body weight of each fish11. The empirical LogLD distribution for both species is shown in Fig. 1. The range of LogLD for Atlantic salmon was greater than for rainbow trout, varying from − 4.60 to 1.02 and from − 4.06 to 0.02, respectively. The average LogLD was − 2.14 ± 0.9 and − 1.66 ± 0.67 for Atlantic salmon and rainbow trout, respectively, which are similar to those reported in a previous study in a different Atlantic salmon population (between − 1.66 ± 0.73 and − 2.55 ± 0.58)11.
Figure 1.
Histogram for log lice density (LogLD) for S. salar (green) and O. mykiss (orange).
Genotyping and genomic heritabilities
A total of 2040 (77.6%) animals, and 45,117 (96.7%) SNPs passed the genotyping quality control for Atlantic salmon. In the case of rainbow trout, 2466 (93.3%) fish and 27,146 (67.4%) SNPs remained for subsequent analyses. In both species, significant genetic variation for resistance to C. rogercresseyi was estimated by using genomic information, with heritability values of 0.19 ± 0.03 and 0.08 ± 0.01 for Atlantic salmon and rainbow trout, respectively (Table 2).
Table 2.
Summary of heritability (h2) for the LogLD trait in both species.
| Specie | Additive variance | Residual variance | h2 |
|---|---|---|---|
| S. salar | 0.09 ± 0.01 | 0.41 ± 0.01 | 0.19 ± 0.03 |
| O. mykiss | 0.02 ± 0.005 | 0.27 ± 0.008 | 0.08 ± 0.01 |
In previous studies in Atlantic salmon populations, Tsai et al.12, estimated genomic heritability values for resistance against L. salmonis of 0.22 ± 0.08 and 0.33 ± 0.08, while Ødegård et al.11, found heritability values of 0.14 ± 0.03 and 0.13 ± 0.03. Similarly, Yañez et al.10 and Correa et al.7 estimated heritability values ranging from 0.10 to 0.12 when defining resistance as the total number of parasites found on all fins using pedigree and genomic information, respectively, and Lhorente et al.9 estimated heritability values of 0.22 ± 0.06 in Atlantic salmon for total count of sessile sea lice per fish, corrected by body weight in the statistical analysis.
GWAS
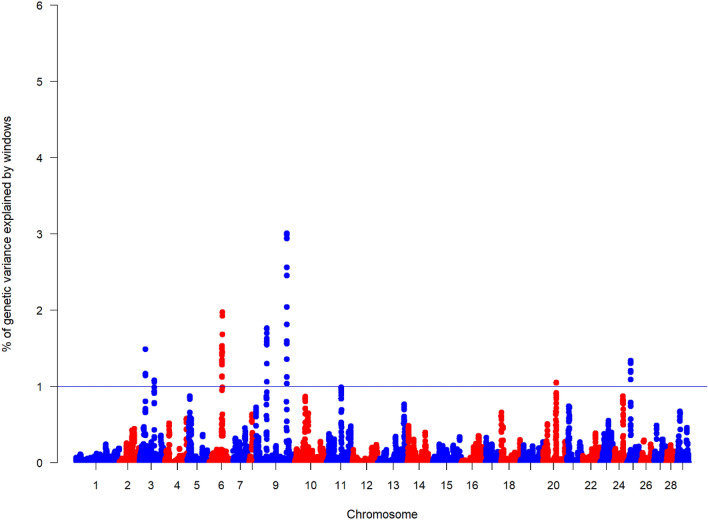
In Atlantic salmon, we found genomic regions explaining more than 1% of the genetic variance for sea lice resistance in five different chromosomes (Fig. 2). Two of these chromosomes (Ssa3 and Ssa9) showed two QTL peaks associated with the trait. Only four SNPs located in Ssa3, Ssa11, Ssa14 and Ssa23 were shown to be significantly associated with the trait at a chromosome-wide level. However, none of these QTLs explained more than 1% of the genetic variance of the trait (Supplementary Figure S1). In general, these regions explained a low percentage of the total genetic variation with a maximum of 3% explained by a single locus. The two SNP windows in Ssa3 explained 1% and 1.4% of the genetic variance while those found in Ssa9 explained 1.7% and 3%. Other QTLs found in Ssa6, Ssa20, and Ssa25 explained 1.9%, 1.05% and 1.33% of the genetic variance, respectively. Supplementary Table S1 shows the variance explained by each window of SNP in both species.
Figure 2.
Weighted single-step GWAS (wssGWAS) results for Log lice density (LogLD) in Atlantic salmon. The Manhattan plot represents the genetic variance explained by windows of 20 SNP and the blue horizontal line indicates 1% threshold.
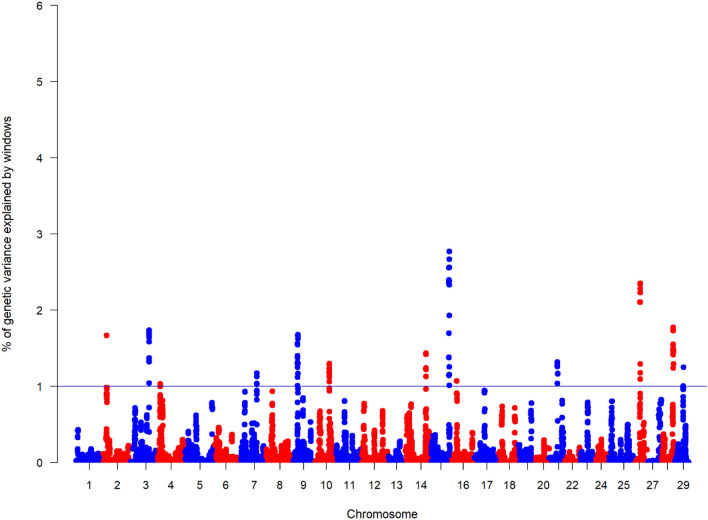
In the case of rainbow trout, the wssGWAS for LogLD identified 13 regions located in different chromosomes that exceeded 1% of the total genetic variance for the trait (Fig. 3). Similar to Atlantic salmon, these windows explained a low percentage of the total variance with a minimum of 1% and a maximum of 2.7%, for QTLs located in Omy17 and Omy15, respectively. In addition, three SNPs located in Omy3, Omy6, Omy9 showed chromosome-wide significant association with the resistance trait in rainbow trout (Supplementary Figure S2).
Figure 3.
Weighted single-step GWAS (wssGWAS) results for Log lice density (LogLD) in rainbow trout. Manhattan plot represents the genetic variance explained by windows of 20 SNP and the blue horizontal line indicates 1% significance threshold.
Our results suggest that resistance against C. rogercresseyi is mainly under polygenic control (i.e., influenced by several genes with small effects) in both species. These results are in agreement with previous studies on sea lice resistance, where a similar genetic architecture was suggested for resistance against L. salmonis and C. royercresseyi resistance7,12,22. Recently Robledo et al.23 described and characterized three QTLs related to sea lice resistance in Atlantic salmon by using GWAS and RNA-sequencing approaches. Since sea lice resistance is a polygenic trait, the acceleration of genetic improvement will most likely be best accomplished by employing genomic selection instead of marker-assisted selection or pedigree-based genetic evaluations. For instance, Correa et al.12 and Tsai et al.14 described an increase of over 22% in the accuracy of estimated breeding values (EBVs) using genomic selection over the use of pedigree-based models in Atlantic salmon24.
Candidate genes
The exploration of the genes within the windows that explained over 1% of the genetic variance for LogLD showed several potential candidate genes that were classified into three groups: related to the immune response, cytoskeleton or metalloproteases. The genes are listed in Tables 3 and 4 for Atlantic salmon and rainbow trout, respectively.
Table 3.
Candidate genes for sea lice resistance in Atlantic salmon.
| Protein name | Gene IDa | Location (BP)b | Chrc | Function |
|---|---|---|---|---|
| Tenascin R | tnr | 22,113,558–22,312,716 | 3 | Immune response |
| T-cell activation Rho GTPase-activating protein | tagap | 47,344,667–47,350,357 | 6 | Immune response |
| Forkhead box protein N1-like isoform X1 | LOC106612922 | 111,631,268–111,668,978 | 9 | Immune response |
| Immunoglobulin superfamily member 11-like | igsf11 | 47,453,435–47,570,948 | 20 | Immune response |
| Tripartite motif-containing 45 | trim45 | 31,699,662–31,705,320 | 25 | Immune response |
| pre-B-cell leukemia transcription factor 1-like | LOC106607421 | 43,977,204–44,037,235 | 6 | Immune response |
| Bromodomain-containing protein 4-like isoform | LOC106600922 | 53,482,098–53,526,053 | 3 | Immune response |
| PAPPALYSIN-2 | pappa2 | 21,972,287–22,050,408 | 3 | Metalloprotease |
| Carboxypeptidase D | cdpa | 111,778,091–111,828,613 | 9 | Metalloprotease |
| GEM-interacting protein-like isoform X3 | LOC106600913 | 53,672,635–53,709,985 | 3 | Metalloprotease |
| Epidermal growth factor | egf | 55,552,926–55,584,698 | 9 | Cytoskeleton |
| Procollagen galactosyltransferase 1 | LOC106607427 | 43,820,342–43,851,337 | 6 | Cytoskeleton |
| Heat shock protein HSP 90-beta | hs90b | 48,362,362–48,374,359 | 6 | Cytoskeleton |
| Collagen alpha-1(XXVIII) chain-like | col28a1 | 13,872,240–13,923,778 | 25 | Cytoskeleton |
| Gap junction alpha-4 protein-like | LOC106611294 | 39,373,024–39,373,931 | 9 | Cytoskeleton |
| Pleckstrin homology domain-containing family H member 1-like | LOC106611291 | 39,101,628–39,165,084 | 9 | Cytoskeleton |
| Serine/threonine-protein kinase OSR1-like | LOC106600551 | 44,726,186–44,781,350 | 3 | Cytoskeleton |
| Rho-related GTP-binding protein RhoB-like | LOC106607565 | 46,401,032–46,403,034 | 6 | Cytoskeleton |
| Pleckstrin homology and RhoGEF domain containing G1 | plekhg1 | 64,024,693–64,178,709 | 6 | Cytoskeleton |
aGene identification (Gene ID) from the NCBI database GenBank assembly accession: GCA_000233375.4.
bLocation in base pair (bp).
cChromosome (Chr).
Table 4.
Candidate genes for sea lice resistance in rainbow trout.
| Protein name | Gene ID1 | Location (BP)b | Chrc | Function |
|---|---|---|---|---|
| Nuclear factor of activated T-cells, cytoplasmic 1-like | LOC110518416 | 35,501,191–35,592,262 | 3 | Immune response |
| Thymocyte selection-associated family member 2 | themist2 | 33,704,133–33,726,370 | 4 | Immune response |
| C–C motif chemokine receptor 10 | ccr10 | 21,961,454–21,965,777 | 16 | Immune response |
| T-box 21 | tbx21 | 15,560,804–15,578,932 | 16 | Immune response |
| Leucine-rich repeat-containing protein 15-like | LOC110539182 | 11,715,000–11,717,281 | 2 | Immune response |
| Toll-like receptor 13 | LOC110490289 | 48,117,536–48,120,927 | 15 | Immune response |
| Adhesion G protein-coupled receptor L3 | LOC110531730 | 14,279,162–14,524,070 | 9 | Immune response |
| Inhibin Beta A Chain | inhba | 36,734,061–36,745,506 | 28 | Immune response |
| Interferon gamma 2 | ifng2 | 153,268–183,367 | Unplaced scaffold | Immune response |
| Heat shock protein family A (Hsp70) member 8 | hspa8 | 17,691,060–17,715,579 | 10 | Immune response |
| Fibroblast growth factor | fgf13 | 26,660,561–26,715,195 | 29 | Cytoskeleton |
| Fibroblast growth factor | fgf11 | 38,417,625–38,489,806 | 10 | Cytoskeleton |
| Alpha-actinin-3 | 3a, 3b | 43,817,039–43,841,681 | 10 | Cytoskeleton |
| Dipeptidyl peptidase 3 | ddp3 | 33,026,452–33,044,218 | 29 | Cytoskeleton |
| ELMO/CED-12 domain-containing 1 | elmod1 | 16,419,827–16,441,415 | 10 | Cytoskeleton |
| Cysteine-rich protein 2-like | LOC106611283 | 38,639,636–38,675,313 | 9 | Cytoskeleton |
| Coagulation factor IX-like | LOC110534144 | 43,984,179–43,991,091 | 10 | Cytoskeleton |
| Lysyl oxidase homolog 1 isoform X2 | LOC110526332 | 59,312,382–59,352,343 | 26 | Cytoskeleton |
| Putative ferric-chelate reductase 1 | LOC110490455 | 53,972,784–53,985,982 | 15 | Metalloprotease |
| AFG3 Like Matrix AAA Peptidase Subunit 2 | afg3l2 | 35,488,672–35,515,514 | 28 | Metalloprotease |
| Afg3-Like Protein 1 | LOC110506600 | 19,946,590–19,959,716 | 26 | Metalloprotease |
aGene identification (Gene ID) from the NCBI database GenBank assembly accession: GCA_002163495.1.
bLocation in base pair (bp).
cChromosome (Chr).
A recent study on gene expression with C. rogercresseyi infestation in susceptible and resistant Atlantic salmon indicated that several components of the immune system (inflammatory response, cytokine production, TNF and NF-kappa B signaling and complement activation) and tissue repair are upregulated during infestation21. In salmonids, the main response of the immune system to parasites is mediated by T-Helper 1 and T-Helper 2 cells25. Thus, genes related to the immune response, either by promoting leukocyte growth or favoring migration or activation are strong candidate genes. For instance, in Atlantic salmon we found, T-cell activation Rho GTPase-activating protein (tagap) in Ssa6, which participates in the activation and recruitment of T cells by cytokines26, and tenascin R (tnr) in Ssa3, which is an extracellular matrix protein, present in bone marrow, thymus, spleen and lymph nodes27. The latter has been described as having an adhesin function favoring the mobility of lymphocytes and lymphoblasts27,28. In rainbow trout, we found candidate genes with similar functions, such as T-box 21 (tbx21), also known as T-bet (T-box expressed in T cells), found in Omy16. This gene belongs to the sub-Tbr1 family29, and generates type 1 immunity and participates in the maturation and migration of T-helper 1 (Th1) cells, which in turn produce interferon-gamma (IFN-γ). Studies have described T-bet expression in NK cells (natural killer), dendritic cells and T CD8+ cells30,31.
Forkhead box protein N1-like (foxn1) present on Ssa09 of Atlantic salmon is part of a family of genes widely studied in humans, which are related to various functions including cell growth, lymph node development, and T cell differentiation32. It has been proposed that foxn1 has a role in the activation of fibroblast growth factor receptors32.
Meanwhile, in rainbow trout on Omy21, serine/threonine-protein phosphatase 2A 56 kDa was identified, which is described as having participated in cell growth and signaling33. Robledo et al.21 recently found that in Atlantic salmon, this protein showed the most significant change in the expression differences between healthy skin and skin where sea lice were attached21. In Atlantic salmon, we identified Tripartite motif-containing protein 45 (trim45) on Ssa25 which belongs to a large family of proteins present in diverse organisms that can function as a ligase and can modify ubiquitin and proteins stimulated by interferon of 15 kDa (isg15)34.
Several metalloproteases were found in genomic regions associated with resistance in both species, but for the interest of this study, we focused on GEM-interacting protein which interacts with rab27a or its effector in leukocytes. Rab is a large family of GTPases responsible for vesicle cellular transport35. Deficiencies of this molecule is correlated with immune deficiencies due to the malfunction of cytotoxic activity of T-lymphocytes, natural killer cells and neutrophils36.
Considering the importance of cell growth and movement in response to sea lice infestation, the cytoskeleton may play a considerable role in sea lice resistance as well. For Atlantic salmon, genes related to the cytoskeleton, such as epidermal growth factor (egf) in Ssa9, were identified. This gene is part of a superfamily of receptors with tyrosine kinase activity that have been described in a variety of organs with growth promoter functions, cellular differentiation38 and could participate in tissue repair by promoting cell growth29. In rainbow trout, fibroblast growth factors (fgf11, fgf13) located in Omy10 and Omy29 respectively, are involved in angiogenesis and pro-inflammatory responses, and were identified as important genes in sea lice resistance in previous transcriptomic studies by Skugor et al. (2009) and Robledo et al. (2018) in Atlantic salmon21,39. In addition, ELMO/CED-12 domain-containing prot 1 was identified in Omy10 in rainbow trout. This protein participates in phagocytosis of apoptotic cells, and in mammals, it also has a role in cell migration40. Other cytoskeleton related candidate genes include: Procollagen galactosyltransferase 1 present in Ssa6, collagen alpha-1 (XXVIII) chain-like on Ssa25 and pleckstrin homology domain-containing family H member 1-like on Ssa941. The top ten SNPs that explained the highest variance for sea lice resistance are located on Ssa9 in Atlantic salmon, representing the most important QTL in this species. This QTL is harboring the breast carcinoma-amplified sequence 3 (bcas3) gene, which in Atlantic salmon codes for a cell migration factor associated with microtubules that favor cellular mobility42. Cell migration is generally induced in response to chemotactic signals, which induces changes in the cytoskeleton and extracellular matrix43. We also found, the tripartite motif-containing protein 16-like on Omy15, which is part of the trim superfamily and has functions related to cell differentiation, apoptosis, regulation of transcription and signaling pathways34. This gene is similar to Tripartite motif-containing protein 45 present on Ssa25. In this region, we also found a locus that codes for interferon-γ 2 (ifng2), which is a cytokine that participates in type 1 immune responses and that favors the presentation of antigens and activation of macrophages44. On this same chromosome (Omy15), we also identified putative ferric-chelate reductase 1 (frrs1), which functions in the fixation of iron in teleosts 45. Robledo et al.21 identified heme-binding protein 2 (hebp2) as a gene involved in Atlantic salmon sea lice resistance, which has an iron-binding function. Different authors46,47 have stated that decreasing the availability of iron can be part of a nutritional defense mechanism against sea lice infestation.
Comparative genomics
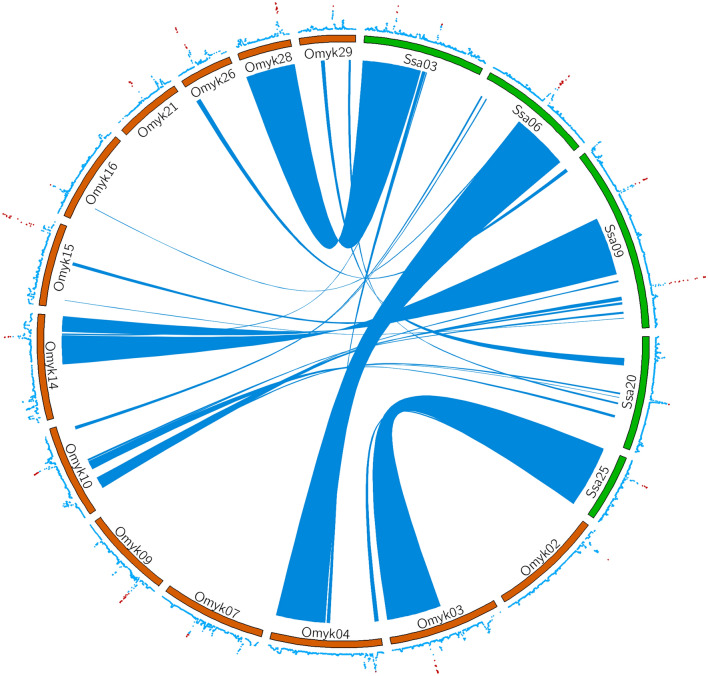
The comparative genomic analyses performed show regions of synteny between chromosomes associated with sea lice resistance in Atlantic salmon and rainbow trout (Fig. 4). Thus, there are homologous regions which are associated with the trait and share similarities between chromosomes from both species. However, there were no obvious shared regions associated to sea lice resistance within species (i.e. homeologous regions). The examined populations shared homeologous regions harboring genes controlling resistance, which might suggest similar genomic regions involved in the regulation of resistance in the two species. For example, Ssa3 (Atlantic salmon) shares extensive homology with Omy28 (rainbow trout) and Ssa25 with Omy3. In addition, when performing the search for orthologue genes (Supplementary Table S2), we found uncharacterized proteins in both species, which shared functionality identified by genomic ontology.
Figure 4.
A circos plot for genomic regions explaining more than 1% of the genetic variance for sea lice resistance in Atlantic salmon and rainbow trout. The inner ribbons mark syntenic regions between Atlantic salmon (green and labeled Ssa) and rainbow trout (orange and labeled Omyk) chromosomes. Genetic variance explained by 20 SNP windows obtained from the wssGWAS analysis are plotted on the outer ring, with the most important windows plotted in red (windows explaining ≥ 1% of the genetic variance).
We determined 15 orthogroups that were present in QTLs for sea lice resistance and were shared both within and between species (Supplementary Table S2). These orthogroups were classified according to gene ontology annotations48. One of the most interesting groups is orthogroup 12 which contained lysophosphatidic acid receptor 2-like (lpa2) in Atlantic salmon, and a G-protein coupled receptor 12-like in rainbow trout. This orthogroup shares the same GO categories (GO: 0004930, GO: 0007186, GO: 0016021, GO: 0070915, GO: 0007165, GO: 0016020) related to the receptor signaling pathway associated with protein G. The activation of LPA2 participates in multiple biological processes, such as cytoskeleton modification via actin fiber formation49 and have a role in the activation of related adhesion focal tyrosine kinase (raftk)50, which in turn participates like a stimulating factor for monocytes and macrophages51. In orthogroup 13, we identified dual-specificity protein phosphatase 10-like (dusp10) in Atlantic salmon and dual-specificity protein phosphatase 8 (dusp8) in rainbow trout. These genes might have a similar function in both species, which is most likely related to modulating p3852 within the MAPK cascade53, a pathway of pro-inflammatory regulators. It has been previously shown that the lice resistant individuals have an up regulated production of pro inflammatory genes than most susceptible fish54. The other orthogroups found here did not show a clear relationship with sea lice resistance (Supplementary Table S2).
Conclusion
The GWAS performed here for Atlantic salmon and rainbow trout made it possible to compare the genetic basis of sea lice resistance in both species. We present novel information about resistance to sea lice in both species. Our results suggest that resistance might be mediated by genes controlling leukocyte response and the cytoskeleton, which promote cell mobility and repair of the wound. The analysis of orthologous proteins provided few characterized proteins. Therefore, further investigations are needed to better annotate genes and generate advances in the elucidation of the genetics behind resistance to Caligus rogercresseyi and other important traits in salmonids. We found uncharacterized common genes classified under similar mechanisms by GO terms that could explain resistance in both species. These results suggest that similar mechanisms may regulate sea lice resistance in Atlantic salmon and rainbow trout. Our results provide further knowledge to help establish better control and treatment measures for one of the most important parasitic diseases affecting Atlantic salmon and rainbow trout aquaculture.
Material and methods
Experimental animals
All experiments were performed under relevant guidelines and regulations and were approved by the Institutional Committee for Animal Care and Use of the University of Chile (Certificate N 17,041-VET-UCH).
Sea lice challenge in rainbow trout
The fish for this study belong to a rainbow trout breeding population established in 1998 by Aguas Claras S.A., at Quetroleufu, IX Region, Chile, and currently owned by EFFIGEN S.A. (Puerto Montt, Chile). The population of this study were from the year-class 2011, which has undergone three generations of selection growth, carcass quality and others traits of interest. The details of the population management and breeding program were described by Yoshida et al.55. For this study, a total of 2588 PIT (Passive Integrated Transponder) -tagged rainbow trout, originated from 105 maternal full-sib families from the 2012 year-class, were used. For the challenge, the fish were separated into three different tanks so that each family was equally represented in each tank. The C. rogercresseyi infestation was conducted with a total of 105,600 copepodites, i.e., an infestation pressure of ~ 40 copepods/fish, which were produced in vitro from ovigerous females. The infestation consisted of depositing the copepodites in each one of the three test tanks, stopping the water flow and keeping the room in darkness for 6 h. On the sixth day after infestation, parasite counting on all fins was performed and caudal fins were sampled for genetic analysis. All fish were euthanized and fins were examined for parasite count using a stereoscopic magnifying glass. Body weight was also recorded for each animal at the end of the challenge.
Sea lice challenge in Atlantic salmon
A total of 2559 Atlantic salmon smolts belonging to 118 maternal full-sib families from the 2010 year-class of Salmones Chaicas S.A. (Puerto Montt, Chile), were challenged with C. rogercresseyi. The fish were PIT-tagged, acclimated and distributed into three tanks as described in previous studies9,10. Infestation with the parasite was carried out using 13–24 copepods per fish, stopping the water flow for 6 h after the infestation. The challenge lasted 6 days, then the fish were euthanized and the sea lice were counted on all of the fins. A sample of the caudal fin was taken for genetic analysis and the body weight of each fish was measured at the end of the challenge.
Genotyping
Genomic DNA was extracted from the caudal fin of each challenged fish using the DNeasy Blood & Kit tissue kit (Qiagen), following the manufacturer's instructions. The 2628 Atlantic salmon samples were genotyped using a custom Affymetrix® 50 K Axiom® myDesign™ Genotyping Array designed by AquaInnovo and the University of Chile56, while the 2643 rainbow trout samples were genotyped with a 57 K SNP array developed by the United State Department of Agriculture (USDA)57. Quality control of the genotypes was carried out in PLINK v1.90b3.34. SNPs with a call rate ≤ 0.95, a Minor Allele Frequency (MAF) < 0.05 and those that were not in Hardy–Weinberg equilibrium (p < ) were discarded. Individuals were filtered if they had a call rate ≤ 0.95. All the SNPs and fish that passed quality control, were used for downstream analysis.
Genomic association analysis
Resistance to C. rogercresseyi was defined as follows, according to Ødegård et al.11:
where LD is the C. rogercresseyi density defined as the lice count (LC) on each fish at the end of the experimental challenge plus a unity, divided by the cube root of the squared body weight of the fish on the same day (BW), which is an approximation of the surface of the skin of each fish. The logarithm of LD was used as it has an approximately normal distribution.
A weighted single-step genomic association study (wssGWAS)58 was used to identify associations between SNPs and resistance to C. rogercresseyi in both species, using the BLUPF90 family of programs59. This approach uses a combination of both genomic and pedigree matrixes. Genotype and pedigree information were used to generate the H kinship matrix60, as defined in the following equation:
where is the inverse of the relationship matrix for all the animals, constructed from the pedigree, is the inverse of the pedigree relationship matrix for the genotyped animals, and is the inverse of the genomic relationship matrix for the genotyped animals. The SNPs were weighted with equal value and assigned the constant 1 to perform the single-step GWAS method. For the weighted single-step GWAS method, the markers were assigned to weights estimated by the previous method. The association analysis for both species were performed using the following mixed linear model y = Xb + Za + e, where y is the vector of phenotypic values (LogLD); b is the vector of fixed effects (tank); a is the vector of random animal effects, considering the structure of covariance between individuals established by matrix H, and e is the vector of random residuals; X and Z are the incidence matrices for fixed and random animal effects, respectively.
To identify the regions of the genome associated with resistance, we generated windows of 20 adjacent SNPs. Thereafter, if a window explained more than 1% of the genetic variance, it was considered associated with the trait. We also estimated the p-values for individual SNP-trait associations by using BLUPF90 software61.
Genome comparison
The rainbow trout (GCF_002163495.1)62 and Atlantic salmon (GCF_000233375.1)63 genomes were downloaded from the NCBI database and the subset for chromosomes associated with sea lice resistance was aligned with software Samtools 64. Synteny between the chromosomes was identified by aligning sequences using the program Symap v3.465. Circos66 was used to visualize the relationships between genomic regions associated to sea lice resistance in rainbow trout and Atlantic salmon chromosomes.
Candidate genes
The 71 pb flanking sequences surrounding SNPs associated with sea lice resistance were aligned to the most recent reference genomes of Atlantic salmon and rainbow trout using BLASTn67. Sequences covering 1 Mb, flanking the associated SNPs (0.5 Mb downstream and 0.5 Mb upstream), were saved in the FASTA format. BLASTx was then used to identify coding sequences for proteins in these 1 Mb associated windows. Blast2GO68 was used in parallel with the FASTA file to identify proteins and classify them by function. For both species, the reference genome of Danio rerio (GenBank Assembly Accession: GCA_000002035.4) was used to annotate proteins that were not characterized in the rainbow trout or Atlantic salmon reference genomes. To identify orthologous proteins/genes between species, the OrthoFinder69 program was used with the FASTA sequences obtained with BLASTx.
Ethics approval and consent to participate
All the experimental challenges were approved by the Institutional Committee for Animal Care and Use of the University of Chile (Certificate N 17,041-VET-UCH). We also confirm that the study was carried out in compliance with the ARRIVE guidelines.
Supplementary Information
Acknowledgements
We would like to thank to EFFIGEN S.A. (Puerto Montt, Chile) by providing phenotypic and genotypic data from the rainbow trout population used here.
Author contributions
P.C. assessed the analyses and wrote the initial version of the manuscript. A.B. contributed with results interpretation discussion and writing. K.C. contributed to genome comparison analysis, writing, and discussion. L.N.B. and K.C. managed samples, performed DNA extraction and performed the quality control of genotypes. B.G. contributed with writing, revision and discussion of the results. J.P.L. contributed to the study design. J.M.Y. conceived the study, contributed to results interpretation with a discussion. All authors approved the manuscript.
Data availability
Atlantic salmon and rainbow trout phenotype and genotype data are available at https://figshare.com/articles/Compative_genomic_of_O_mykiss_and_S_salar_for_resistance_to_Sea_lice/7676147 and https://figshare.com/projects/Compative_genomic_of_O_mykiss_and_S_salar_for_resistace_to_Sea_lice/70904, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92425-3.
References
- 1.Costello MJ. How sea lice from salmon farms may cause wild salmonid declines in Europe and North America and be a threat to fishes elsewhere. Proc. R. Soc. B Biol. Sci. 2009;276:3385–3394. doi: 10.1098/rspb.2009.0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abolofia J, Asche F, Wilen JE. The cost of lice: Quantifying the impacts of parasitic sea lice on farmed salmon. Mar. Resour. Econ. 2017;32:329–349. doi: 10.1086/691981. [DOI] [Google Scholar]
- 3.Barrett LT, Oppedal F, Robinson N, Dempster T. Prevention not cure: A review of methods to avoid sea lice infestations in salmon aquaculture. Rev. Aquac. 2020;12:2527–2543. doi: 10.1111/raq.12456. [DOI] [Google Scholar]
- 4.Boxshall A, Bravo S. On the identity of the common Caligus (Copepoda: Siphonostomatoida: Caligidae) from salmonid netpen systems in southern Chile. Contrib. Zool. 2000;69:137–146. doi: 10.1163/18759866-0690102015. [DOI] [Google Scholar]
- 5.Lhorente JP, Araneda M, Neira R, Yáñez JM. Advances in genetic improvement for salmon and trout aquaculture: The Chilean situation and prospects. Rev. Aquac. 2019;11:340–353. doi: 10.1111/raq.12335. [DOI] [Google Scholar]
- 6.Hamilton-West C, et al. Epidemiological description of the sea lice (Caligus rogercresseyi) situation in southern Chile in August 2007. Prev. Vet. Med. 2012;104:341–345. doi: 10.1016/j.prevetmed.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Correa K, et al. Genome wide association study for resistance to Caligus rogercresseyi in Atlantic salmon (Salmo salar L.) using a 50K SNP genotyping array. Aquaculture. 2017;472:61–65. doi: 10.1016/j.aquaculture.2016.04.008. [DOI] [Google Scholar]
- 8.Gonzalez L, Juan C. Life cycle of Caligus rogercresseyi, (Copepoda: Caligidae) parasite of Chilean reared salmonids. Aquaculture. 2003;220:101–117. doi: 10.1016/S0044-8486(02)00512-4. [DOI] [Google Scholar]
- 9.Lhorente JP, et al. Quantitative genetic basis for resistance to Caligus rogercresseyi sea lice in a breeding population of Atlantic salmon (Salmo salar) Aquaculture. 2012;324–325:55–59. doi: 10.1016/j.aquaculture.2011.10.046. [DOI] [Google Scholar]
- 10.Yáñez JM, et al. Genetic co-variation between resistance against both Caligusrogercresseyi and Piscirickettsia salmonis, and body weight in Atlantic salmon (Salmo salar) Aquaculture. 2014;433:295–298. doi: 10.1016/j.aquaculture.2014.06.026. [DOI] [Google Scholar]
- 11.Ødegård J, et al. Genomic prediction in an admixed population of Atlantic salmon (Salmo salar) Front. Genet. 2014;5:1–8. doi: 10.3389/fgene.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai HY, et al. Genomic prediction of host resistance to sea lice in farmed Atlantic salmon populations. Genet. Sel. Evol. 2016;48:1–11. doi: 10.1186/s12711-016-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassini N, et al. Genetic parameters for resistance to Caligus rogercresseyi, Piscirickettsia salmonis and body weight in rainbow trout (Oncorhynchus mykiss) Aquaculture. 2017;472:127–127. [Google Scholar]
- 14.Correa K, Bangera R, Figueroa R, Lhorente JP, Yáñez JM. The use of genomic information increases the accuracy of breeding value predictions for sea louse (Caligusrogercresseyi) resistance in Atlantic salmon (Salmo salar) Genet. Sel. Evol. 2017;49:1–5. doi: 10.1186/s12711-017-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardisson RC. Comparative genomics. Brenner’s Encycl. Genet. Second Ed. 2013;1:102–105. [Google Scholar]
- 16.Fang G, Bhardwaj N, Robilotto R, Gerstein MB. Getting started in gene orthology and functional analysis. PLoS Comput. Biol. 2010;6:20. doi: 10.1371/journal.pcbi.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid DP, Szanto A, Glebe B, Danzmann RG, Ferguson MM. QTL for body weight and condition factor in Atlantic salmon (Salmo salar): Comparative analysis with rainbow trout (Oncorhynchus mykiss) and Arctic charr (Salvelinus alpinus) Heredity (Edinb). 2005;94:166–172. doi: 10.1038/sj.hdy.6800590. [DOI] [PubMed] [Google Scholar]
- 18.Moghadam HK, Ferguson MM, Danzmann RG. Evolution of Hox clusters in salmonidae: A comparative analysis between atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) J. Mol. Evol. 2005;61:636–649. doi: 10.1007/s00239-004-0338-7. [DOI] [PubMed] [Google Scholar]
- 19.Woram RA. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 2003;13:272–280. doi: 10.1101/gr.578503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yáñez JM, et al. Comparative genomic analysis of three salmonid species identifies functional candidate genes involved in resistance to the intracellular bacterium Piscirickettsia salmonis. Front. Genet. 2019;10:665. doi: 10.3389/fgene.2019.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robledo D, Gutiérrez AP, Barría A, Yáñez JM, Houston RD. Gene expression response to sea lice in Atlantic salmon skin: RNA sequencing comparison between resistant and susceptible animals. Front. Genet. 2018;9:1–10. doi: 10.3389/fgene.2018.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rochus CM, et al. Genome-wide association analysis of salmon lice (Lepeophtheirussalmonis) resistance in a North American Atlantic salmon population. Aquac. Res. 2018;49:1329–1338. doi: 10.1111/are.13592. [DOI] [Google Scholar]
- 23.Robledo D, et al. Discovery and functional annotation of quantitative trait loci affecting resistance to Sea lice in Atlantic salmon. Front. Genet. 2019;10:1–10. doi: 10.3389/fgene.2019.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallejo RL, et al. Genomic selection models double the accuracy of predicted breeding values for bacterial cold water disease resistance compared to a traditional pedigree-based model in rainbow trout aquaculture. Genet. Sel. Evol. 2017;49:1–13. doi: 10.1186/s12711-017-0293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegertjes GF, Wentzel AS, Spaink HP, Elks PM, Fink IR. Polarization of immune responses in fish: The ‘macrophages first’ point of view. Mol. Immunol. 2016;69:146–156. doi: 10.1016/j.molimm.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Tamehiro N, et al. T-cell activation RhoGTPase-activating protein plays an important role in T H 17-cell differentiation. Immunol. Cell Biol. 2017;95:729–735. doi: 10.1038/icb.2017.27. [DOI] [PubMed] [Google Scholar]
- 27.Ruegg CR, Chiquet-Ehrismann R, Alkan SS. Tenascin, an extracellular matrix protein, exerts immunomodulatory activities. Proc. Natl. Acad. Sci. 1989;86:7437–7441. doi: 10.1073/pnas.86.19.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark RA, Erickson HP, Springer TA. Tenascin supports lymphocyte rolling. J. Cell Biol. 1997;137:755–765. doi: 10.1083/jcb.137.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fast MD. Fish immune responses to parasitic copepod (namely sea lice) infection. Dev. Comp. Immunol. 2014;43:300–312. doi: 10.1016/j.dci.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Berry G, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 31.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/S1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 32.Coffer PJ, Burgering BMT. Forkhead-box transcription factors and their role in the immune system. Nat. Rev. Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 33.Veerle J, Jozef G. Protein phosphatase 2A: A high regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353:417–439. doi: 10.1042/bj3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr. Opin. Immunol. 2011;23:46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 35.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 36.Johnson JL, Monfregola J, Napolitano G, Kiosses WB, Catz SD. Vesicular trafficking through cortical actin during exocytosis is regulated by the Rab27a effector JFC1/Slp1 and the RhoA-GTPase-activating protein Gem-interacting protein. Mol. Biol. Cell. 2012;23:1902–1916. doi: 10.1091/mbc.e11-12-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valenzuela-Muñoz V, Boltaña S, Gallardo-Escárate C. Comparative immunity of Salmo salar and Oncorhynchus kisutch during infestation with the sea louse Caligus rogercresseyi: An enrichment transcriptome analysis. Fish Shellfish Immunol. 2016;59:276–287. doi: 10.1016/j.fsi.2016.10.046. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, et al. Expression and function of the epidermal growth factor receptor in physiology and disease. Physiol. Rev. 2016;96:1025–1069. doi: 10.1152/physrev.00030.2015. [DOI] [PubMed] [Google Scholar]
- 39.Skugor S, Glover KA, Nilsen F, Krasnov A. Local and systemic gene expression responses of Atlantic salmon (Salmo salar L.) to infection with the salmon louse (Lepeophtheirus salmonis) BMC Genom. 2008;9:1–18. doi: 10.1186/1471-2164-9-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brzostowski JA, Fey P, Yan J, Isik N, Jin T. The Elmo family forms an ancient group of actin-regulating proteins. Commun. Integr. Biol. 2009;2:337–340. doi: 10.4161/cib.2.4.8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemmon MA, Ferguson KM, Abrams CS. Pleckstrin homology domains and the cytoskeleton. FEBS Lett. 2002;513:71–76. doi: 10.1016/S0014-5793(01)03243-4. [DOI] [PubMed] [Google Scholar]
- 42.Wang T, Holland JW, Martin SAM, Secombes CJ. Sequence and expression analysis of two T helper master transcription factors, T-bet and GATA3, in rainbow trout Oncorhynchus mykiss and analysis of their expression during bacterial and parasitic infection. Fish Shellf. Immunol. 2010;29:705–715. doi: 10.1016/j.fsi.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 43.Lamalice L, Le Boeuf F, Huot J. Endothelial cell migration during angiogenesis. Circ. Res. 2007;100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 44.Zou J, et al. Identification and bioactivities of IFN- in rainbow trout Oncorhynchus mykiss: The first Th1-type cytokine characterized functionally in fish. J. Immunol. 2005;175:2484–2494. doi: 10.4049/jimmunol.175.4.2484. [DOI] [PubMed] [Google Scholar]
- 45.Bury N, Grosell M. Iron acquisition by teleost fish. Comp. Biochem. Physiol. C. 2003;135:76490. doi: 10.1016/s1532-0456(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 46.Jones SRM, Fast MD, Johnson SC, Groman DB. Differential rejection of salmon lice by pink and chum salmon: Disease consequences and expression of proinflammatory genes. Dis. Aquat. Organ. 2007;75:229–238. doi: 10.3354/dao075229. [DOI] [PubMed] [Google Scholar]
- 47.Valenzuela-Muñoz V, Gallardo-Escárate C. Iron metabolism modulation in Atlantic salmon infested with the sea lice Lepeophtheirus salmonis and Caligus rogercresseyi: A matter of nutritional immunity? Fish Shellf. Immunol. 2017;60:97–102. doi: 10.1016/j.fsi.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 48.Bateman A, et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moolenaar WH, Kranenburg O, Postma FR, Zondag GCM. Lysophosphatidic acid: G-protein signalling and cellular responses. Curr. Opin. Cell Biol. 1997;9:168–173. doi: 10.1016/S0955-0674(97)80059-2. [DOI] [PubMed] [Google Scholar]
- 50.Park SY, Schinkmann KA, Avraham S. RAFTK/Pyk2 mediates LPA-induced PC12 cell migration. Cell. Signal. 2006;18:1063–1071. doi: 10.1016/j.cellsig.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 51.Hatch WC, et al. The related adhesion focal tyrosine kinase (RAFTK) is tyrosine phosphorylated and participates in colony-stimulating factor-1/macrophage colony-stimulating factor signaling in monocyte-macrophages. Blood. 1998;91:3967–3973. doi: 10.1182/blood.V91.10.3967. [DOI] [PubMed] [Google Scholar]
- 52.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 53.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 54.Núñez-Acuña G, et al. Transcriptome immunomodulation of in-feed additives in Atlantic salmon Salmo salar infested with sea lice Caligus rogercresseyi. Fish Shellf. Immunol. 2015;47:450–460. doi: 10.1016/j.fsi.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida GM, et al. Genomic prediction accuracy for resistance against Piscirickettsia salmonis in farmed rainbow trout. G3 Genes Genomes Genet. 2018;8:719–726. doi: 10.1534/g3.117.300499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yáñez, J. M. et al. Genomewide single nucleotide polymorphism discovery in Atlantic salmon (Salmo salar): validation in wild and farmed American and European populations. Mol Ecol Resour16, 1002-1011 (2016). [DOI] [PubMed]
- 57.Palti Y, et al. The development and characterization of a 57K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 2015;15:662–672. doi: 10.1111/1755-0998.12337. [DOI] [PubMed] [Google Scholar]
- 58.Legarra A, Christensen OF, Aguilar IM. Single step, a general approach for genomic selection. Livest. Sci. 2014 doi: 10.1016/j.livsci.2014.04.029. [DOI] [Google Scholar]
- 59.Misztal I, Aggrey SE, Muir WM. Experiences with a single-step genome evaluation. Poult. Sci. 2013;92:2530–2534. doi: 10.3382/ps.2012-02739. [DOI] [PubMed] [Google Scholar]
- 60.Aguilar I, et al. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score1. J. Dairy Sci. 2010;93:743–752. doi: 10.3168/jds.2009-2730. [DOI] [PubMed] [Google Scholar]
- 61.Misztal, I. et al. Manual for BLUPF90 family of programs. Univ. Georg. Athens, USA 125 (2018).
- 62.Pearse DE, et al. Sex-dependent dominance maintains migration supergene in rainbow trout. bioRxiv. 2018 doi: 10.1101/504621. [DOI] [PubMed] [Google Scholar]
- 63.Lien S, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533:200–205. doi: 10.1038/nature17164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soderlund C, Bomhoff M, Nelson WM. SyMAP v3.4: A turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011;39:20. doi: 10.1093/nar/gkr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Connors J, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madden T. The BLAST sequence analysis tool. In: McEntyre J, Ostell J, editors. The NCBI Handbook. National Center for Biotechnology Information; 2002. [Google Scholar]
- 68.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 69.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:1–14. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atlantic salmon and rainbow trout phenotype and genotype data are available at https://figshare.com/articles/Compative_genomic_of_O_mykiss_and_S_salar_for_resistance_to_Sea_lice/7676147 and https://figshare.com/projects/Compative_genomic_of_O_mykiss_and_S_salar_for_resistace_to_Sea_lice/70904, respectively.