Abstract
Acinetobacter baumannii has emerged worldwide as a dominant pathogen in a broad range of severe infections, raising an acute need for efficient antibacterials. This is the first report on the resistome and virulome of 33 extended drug-resistant and carbapenem-resistant A. baumannii (XDR CRAB) strains isolated from hospitalized and ambulatory patients in Bucharest, Romania. A total of 33 isolates were collected and analyzed using phenotypic antibiotic susceptibility and conjugation assays, PCR, whole-genome sequencing (WGS), pulsed-field gel electrophoresis (PFGE) and MultiLocus Sequence Typing (MLST). All isolates were extensively drug-resistant (XDR), being susceptible only to colistin. The carbapenem resistance was attributed by PCR mainly to blaOXA-24 and blaOXA-23 genes. PFGE followed by MLST analysis demonstrated the presence of nine pulsotypes and six sequence types. WGS of seven XDR CRAB isolates from healthcare-associated infections demonstrated the high diversity of resistance genes repertoire, as well as of mobile genetic elements, carrying ARGs for aminoglycosides, sulphonamides and macrolides. Our data will facilitate the understanding of resistance, virulence and transmission features of XDR AB isolates from Romanian patients and might be able to contribute to the implementation of appropriate infection control measures and to develop new molecules with innovative mechanisms of action, able to fight effectively against these bugs, for limiting the spread and decreasing the infection rate and mortality.
Subject terms: Microbiology, Infectious diseases
Introduction
Acinetobacter baumannii is one of the most frightening resistant Gram-negative bacteria, being included in both the ESKAPE (later ESCAPE) group and the list of 12 bacteria that represent a threat to human health published in 2017 by the World Health Organization (WHO). The successful development of efficient antibacterials requires a better knowledge of the mechanisms of resistance of this pathogen, to increase the chance to discover new chemical structures with ideally new mechanisms of action1 or different antimicrobial combinations2, 3.
Several mechanisms contribute to A. baumannii strains resistance, such as β-lactamases expression, alteration of cell membrane permeability, increased expression of efflux pumps, mutations in DNA gyrases and topoisomerases encoding genes3–10. The most frequent causes of enzymatic resistance in A. baumannii are represented nowadays by the acquired Carbapenem-Hydrolyzing Class D β-Lactamases (CHDL) (OXA-23, OXA-24, OXA-58, OXA-143, OXA-235) and the overexpression of the intrinsic OXA-51, followed by the presence of class A β-lactamases (i.e. TEM (for Temoneira patient’s name), SHV (Sulfhydryl Variable enzyme), CTX-M (Cefotaxime first isolated in Munich) and KPC (Klebsiella pneumoniae Carbapenemase), GES (Guyana Extended Spectrum β-lactamase), class A extended-spectrum cephalosporinases (usually caused by the over-production of the chromosomal ADCs) and metallo-β-lactamases (MBL) (IMP (active-on-imipenem), VIM (Verona imipenemase), SIM (Seoul imipenemase), SPM (Saõ Paolo metallo-β-lactamase), GIM (German imipenemase) and NDM (New Delhi MBL)11. The expression of OXA genes might be increased by insertion sequences (ISs), such as ISAba1, ISAba4 and ISAba125, which provide an additional strong promoter12, 13. The insertion sequence ISAba1 has been found in A. baumannii isolates upstream of the blaOXA-23,-51,-58 carbapenemase and cephalosporinase blaampC genes14. Among different genetic contexts containing OXA-23 and involving transposons, such as Tn2006, Tn2008, Tn2008B14, Tn2009 and Tn2007, only Tn2006 and Tn2009 are compound transposons, containing two copies of ISAba1 flanking the carbapenemase gene and thus able to regulate its mobility. In the case of the Tn2008 and Tn2008B, there is only one copy of the ISAba1 upstream blaOXA-23 gene12, 15–17.
Carbapenem-resistant A. baumannii (CRAB) is a globally disseminated pathogen with continuously increasing prevalence worldwide. CRAB strains are frequently exhibiting MDR and XDR resistance phenotypes, carrying both intrinsic and acquired resistance genes carried on plasmids, transposons, and integrons18. In Romania, the present knowledge on the distribution and molecular epidemiology of CRAB isolates is scarce and limited to the study of low numbers of clinical strains recovered from the north, west, and south regions of the country. In this context, we aim to investigate the resistance and virulence features of 30 clinical strains and 3 community-acquired XDR CRAB, by phenotypic antibiotic susceptibility assays, PCR, pulsed-field gel electrophoresis (PFGE), MultiLocus Sequence Typing (MLST) and whole-genome sequencing (WGS).
Results
Antimicrobial resistance profiles of CRAB
The great majority of the A. baumannii isolates (96.96%) were resistant to imipenem, meropenem (MIC established by broth microdilution method of 8 mg/L) and ciprofloxacin. High resistance rates were recorded for cefepime (93.93%), trimethoprim-sulfamethoxazole, gentamicin and cefoxitin (90.90% each) and levofloxacin (87.87%). Lower rates were registered for piperacillin-tazobactam (42.42%) and ceftriaxone (39%) (Table 1).
Table 1.
Clinical specimen, carbapenemase, PFGE-type and MLST results, for A. baumannii clinical isolates from Romania.
| Isolate | Source | Clinical Unit/Date | PFGE | MLST | Acquired carbapenemase | Antibiotic Resistance profile |
|---|---|---|---|---|---|---|
| A17 | Blood culture | A, January 2018 | III | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A15 | Pharyngeal exudate | A, January 2018 | II | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| 4542 | Pharyngeal exudate | A, December 2017 | I | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A9 | Stool culture | A, January 2018 | II | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A11 | Anal carriage | A, February 2018 | I | ST2 | OXA23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A07 | Blood culture | A, December 2017 | II | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A6 | Stool culture | A, November 2017 | II | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A12 | Anal carriage | A, February 2018 | I | ST2 | OXA-51, OXA-23 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A14 | Blood culture | A, February 2018 | I | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A10 | Stool culture | A, December 2017 | II | ST2 | OXA-23*, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A13 | Anal carriage | A, February 2018 | V | ST642 | OXA-24, OXA-51 | MEM, IMP, ETP, CEF, CXM, CTX, CAZ, CIP, FOX, FEP, ATM, AMC, PIP-TZP, SXT, AMK, GEN, NIT, LEV, TIG |
| A1 | Blood culture | A, October 2017 | IV | ST312 | OXA-24, OXA-51 | IPM, MEM, AMC, CTX, CAZ, PIP-TZP, CIP, LEV, GEN, AMK |
| A1prim | Blood culture | A, October 2017 | IV | ST312 | OXA-24, OXA-51 | IPM, MEM, AMC, CTX, CAZ, PIP-TZP, CIP, LEV, GEN, AMK |
| 8A | Urine | B, September 2017 | II | ST2 | OXA-23*, OXA-51 | IPM, MEM, PIP-TZP, CAZ, FEP, ATM, AMK, GEN, CIP |
| 7A | Urine | B, September 2017 | IX | ST2 | OXA-24, OXA-51 | IPM, CAZ, FEP, FOX, AMK, GEN, CIP, LEV, SXT, TET |
| 6A | bBood culture | B, September 2017 | VIII | ST636 | OXA-24, OXA-51 | IPM, MEM, CAZ, FEP, FOX, AMK, GEN, CIP, LEV, SXT, TET |
| 22s | Tracheal secretion | C, September 2017 | VI | ST636 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, PIP, IPM, MEM, SXT, FOX, LEV |
| 24s | Tracheal secretion | C, September 2017 | VI | ST636 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 21s | Catheter | C, September 2017 | VI | ST636 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, |
| 18s | Catheter | C, September 2017 | VI | ST1 | OXA-24*, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 19s | Urine | C, September 2017 | VII | ST492 | OXA-24, OXA-51* | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 29s | Tacheal secretion | C, September 2017 | VII | ST492 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 2s | Tracheal secretion | C, September 2017 | VI | ST636 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 33s | Tracheal secretion | C, September 2017 | VII | ST492 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 9s | Tracheal secretion | C, September 2017 | VII | ST492 | OXA-24, OXA-51* | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, TET |
| 14s | Catheter | C, September 2017 | VII | ST492 | OXA-24, OXA-51* | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 01s | Cerebrospinal fluid | C, September 2017 | VI | ST636 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 3s | Catheter | C, September 2017 | VI | ST636 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 4s | Tracheal secretion | C | negative | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, PIP, IPM, MEM, SXT, FOX, LEV, TET | |
| 10s | Tracheal secretion | C, September 2017 | negative | ST492 | OXA-24*, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, TET |
| 26s | Tracheal secretion | C, September 2017 | IV | ST312 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 24s prim | Tracheal secretion | C, September 2017 | VI | ST636 | OXA-24, OXA-51 | AMK, CAZ, FEP, CIP, GEN, PIP, IPM, MEM, SXT, FOX, LEV, TET |
| 4 new | Sputum | B, February 2018 | VI | ST636 | OXA-24, OXA-51 | MEM, FEP, GEN, SXT, LEV, TET |
1 Notes: A = ICU ; B- Ambulatory; C- Children Hospital. (*) indicates the ISAba1 insertion upstream of the gene encoding respective carbapenemase.
MEM = meropenem, IPM = imipenem, ETP = ertapenem, CEF = cephalotin, CTX = ceftriaxone, CXM = cefuroxime, FOX = cefoxitin, CAZ = ceftazidime, ATM = aztreonam, FEP = cefepime, AMC = amoxicillin-clavulanic acid, PIP-TZP = piperacillin-tazobactam, CIP = ciprofloxacin, LEV = levofloxacin, GEN = gentamicin, AMK = amikacin, NIT = nitrofurantoin, SXT = trimethoprim-sulfamethoxazole, TET = tetracycline, TIG = tigecycline.
Detection of CHLDs genes
The A. baumannii isolates were investigated for the presence of OXA-type carbapenemases (Table 1). All isolates harbored the intrinsic blaOXA-51-like gene, 66.66% of A. baumannii revealed blaOXA-24 and 33.33% of A. baumannii isolates carried blaOXA-23. We did not detect any MBL gene. The presence of ISAba1 was confirmed in 15 isolates (45.45%) and 10 strains (30.30%) had ISAba1 immediately upstream of the blaOXA-23 gene. No ISAba1 was detected downstream of the blaOXA-23 gene, ruling out the hypothesis that this gene could be part of the composite transposon Tn2006.
Epidemiological typing of isolates by PFGE and MLST
Nine major genotypes were encountered in the analyzed A. baumannii strains using a cut-off of 85% genetic similarity (Table 1): pulsotype I (n = 4), II (n = 6), III (n = 1), IV (n = 3), V (n = 1), VI (n = 9), VII (n = 5), VIII (n = 1) and IX (n = 1), while 2 strains were non-typeable. No correlation could be established between the origin of the analyzed strains (a certain clinical unit) and a specific pulsotype. The MLST analysis performed for the selected clones showed that only 12 of the 33 isolates belonged to the most prevalent global Clonal Complex CC2 (ST2) and 5 singleton STs were identified (ST636, ST492, ST312, ST642, and ST1) (Table 1). The PFGE results have shown that the most common clones were VI (n = 9), II (n = 6), VII (n = 5), I (n = 4) (Table 1).
The PFGE results revealed that there was no outbreak or spread of one single genotype in the clinical strains that were analyzed. However, clones I, II, III, IV and V were associated with hospital A, clones II, VIII and IX with the ambulatory unit B and clones IV, VI and VII were found only in hospital C.
Genomic analyses
The genomes of 7 selected CRAB strains encoded 01s, 14s, 10s, 18s, 24s, A07 and A14 were fully sequenced and analyzed to have a complete picture of the antibiotic resistance and virulence genes repertoires. The analyzed strains were selected for WGS based on the isolation source, all being isolated from healthcare-associated infections, respectively catheter-associated bloodstream infections, ventilator-associated pneumonia and central nervous system infections. Draft-genome sequencing analysis revealed that the chromosome size varied, as expected, between 3.86 and 4.071 Mbp. Instead, the analyzed strains harbored diverse mobile genetic elements. More than half of the CDS were functionally annotated by the RAST program. The general features of the genomes are presented in Table 2.
Table 2.
General features of A. baumannii strains genomes.
| Strain | 01s | 14s | 10s | 18s | 24s | A07 | A14 |
|---|---|---|---|---|---|---|---|
| Size | 3,968,270 | 3,897,785 | 3,860,242 | 3,977,351 | 3,925,255 | 3,970,732 | 4,071,708 |
| GC Content (%) | 39.04 | 39.03 | 38.98 | 39.14 | 39.0 | 39.07 | 38.88 |
| Contig N50 | 161,906 | 162,094 | 132,810 | 81,560 | 107,477 | 127,142 | 135,471 |
| Contig L50 | 9 | 8 | 10 | 16 | 12 | 10 | 10 |
| Number of Contigs (with PEGs) | 166 | 128 | 130 | 232 | 181 | 158 | 205 |
| Number of Coding Sequences | 3908 | 3784 | 3770 | 3931 | 3836 | 3907 | 4061 |
| Genes assigned to COs | 2173 | 2148 | 2165 | 2158 | |||
| Number of RNAs | 71 | 65 | 87 | 70 | 70 | 78 | 69 |
To analyze the acquired antibiotic resistance genes (ARGs), ResFinder version 3.0 with an ID threshold of 90%, and the minimum length set at 60% were used. The analysis revealed that all strains harbor acquired ARGs to aminoglycosides (i.e. aph(3')-VIa, ant(3'')-IIa, aph(6)-Id, aph(3'')-Ib, aadA1, aadA2, armA, aph(3')VIj, ant(3'')-IIa, aph(3')-Ia, aadA1, aac(3)-Ia), β-lactams (blaOXA-66, blaOXA-72, blaOXA-92, blaOXA-23, blaADC-25, blaADC-30, blaADC-81, blaADC-74, blaTEM-1D) and sulphonamides (sul1/ sul2 or both of them), while 4 strains carry ARGs for tetracyclines [tet(B) and tetR encountered in 3 strains from hospital settings A and C and tet(A) gene revealed in one strain]. The predictions were carried with multiple tools as described above, to increase the confidence degree in the predicted genes. The catA1 gene associated with resistance to chloramphenicol was located between TnAs3 and IS1R in 2 strains. ARGs for streptogramin B [msr(E)], mph(E)] have been revealed also in 2 strains from the hospital settings A and C (Table 3).
Table 3.
AR and virulence genetic markers in the analyzed A. baumannii strains.
| 01s | 14s | 10s | 18s | 24s | A07 | A14 | ||
|---|---|---|---|---|---|---|---|---|
| Antibiotic resistance | Aminoglycosides | aph(3')-Ia, aph(3')-VIa, ant(3'')-IIa | aph(6)-Id, ant(3'')-IIa, aph(3'')-Ib, aadA2, armA | aph(3'')-Ib, aph(6)-Id, ant(3'')-IIa, | aph(3')-VIa, ant(3'')-IIa, aac(6')-Ib, aac(3)-I, aadA1, ant(3'')-IIa | aadA1, aph(3')-Ia, aph(3')-VIa, ant(3'')-IIa, | aph(3')VIb, ant(3'')-IIa, aph(3')-Ia, aadA1, aac(3)-Ia | aph(6)-Id, ant(3'')-IIa, aph(3'')-Ib, armA |
| β-lactams | blaOXA-66, blaOXA-72, blaADC-74 | blaOXA-66, blaOXA-72, blaADC-25 | blaOXA-66, blaOXA-72, blaADC-30 | blaOXA-92, blaADC-81, blaOXA-72, blaTEM-84, blaTEM-12, | blaOXA-72, blaOXA-66, blaADC-74, | blaOXA-66, blaADC-11, blaTEM-12, blaPER-1 | blaOXA-66, blaOXA-23, blaADC-11, blaPER-1 | |
| Macrolides | msr(E), mph(E) | msr(E), mph(E) | ||||||
| Tetracycline | tet(B), tetR | tet(B), | tet(A) | tet(B), | ||||
| Phenicol | catA1 | catA1 | catI | |||||
| Folate inhibitors | sul1 | dfrA12, sul1, sul2 | sul2 | sul1 | sul1 | sul1 | sul1 | |
| Quinolones resistance | gyrA (S83L), parC (S84L) | gyrA (S83L), parC (S84L), parC (S467G) | gyrA (S83L), parC (S84L), parC (S467G) | gyrA (S83L), parC (S84L), parC (S467G) | gyrA (S83L), parC (S84L) | gyrA (S83L, parC (S84L), parC (S467G) | gyrA (S83L), parC (S84L), parC (S467G) | |
| antibiotic efflux | abeM, -S; adeABC, -FGH, -IJK, -L, -N, -R, -S | abeM, -S; adeABC, -FGH, -IJK, -L, -N, -R, -S | abaF, -Q; adeK, -I, -S, -L, -G, -A, -C, -N, -F, -H, -R, amvA | abaQ; adeF, -S, -G, -I, -K, -N, -H, -L, amvA | adeA, -C, -S, -L, -G, -K, -I, -R, -F, -H, -N; amvA; AbaF, -Q, | abeM, -S; adeABC, -FGH, -IJK, -L, -N, -R, -S | abeM, -S; adeABC, -FGH, -IJK, -L, -N, -R, -S | |
| Virulence factors | Adherence | ompA | ompA | ompA | ompA | ompA | ompA | ompA |
| Biofilm formation | adeFGH, bap, csuA/BABCDE, pgaABCD, | adeFGH, bap, csuA/BABCDE, pgaABCD |
adeFGH, bap, csuA/BABCDE, pgaABCD |
adeFGH, bap, csuD, csuE, pgaABCD | adeFGH, bap, csuA/BABCDE, pgaABCD | adeFGH, bap, csuA/BABCDE, pgaABCD | adeFGH, bap, csuA/BABCDE, pgaABCD | |
| Enzyme | plc, plcD | plc, plcD | plc, plcD | plc, plcD | plc, plcC, plcD | plc, plcD | plc, plcD | |
| Pore forming toxins | apkA, apkB | apkA, apkB | apkA, apkB | apkA, apkB | apkA, apkB | apkA, apkB | apkA, apkB | |
| Iron uptake | tonB | tonB, hemO | tonB | tonB | tonB | tonB | tonB, hemO | |
| Resistance to toxic compounds | Copper | MmcO, CtpA, Bcr/CflA, cueR, pcoB, cutE, CorC | MmcO, CtpA, Bcr/CflA, cueR, pcoB, cutE, CorC | MmcO, CtpA,, Bcr/CflA, cueR, pcoB, cutE, CorC | MmcO, CtpA, Bcr/CflA, cueR, pcoB, pcoD, CusR, CopC, cutE, CorC | MmcO, CtpA,, Bcr/CflA, cueR, pcoB, cutE, CorC | MmcO, CtpA, Bcr/CflA, cueR, pcoB, cutE, CorC | MmcO, CtpA, Bcr/CflA, cueR, pcoB, cutE, CorC |
| Cobalt, Zinc, Cadmium | CzcABCD, MFP, merR, CusA | CzcABD, MFP, merR, CusA | CzcABD, MFP, merR, CusA | CzcABCD, MFP, merR, CusA, CusR, CusS | CzcABCD, MFP, merR, CusA | MFP, czcABD, merR, CusA | MFP, czcABD, merR, CusA | |
| Chromium | ChrA | ChrA | ChrA | ChrA | ChrA | ChrA | ChrA | |
| Mercury | merA, merC, merR | |||||||
| Arsenic | ACR3, arsC, arsR | ACR3, arsC, arsR | ACR3, arsC, arsR | ACR3, arsC, arsH, arsR | ACR3, arsC, arsR | ACR3, arsC, arsH, arsR | ACR3, arsC, arsR | |
| Integron | Incomplete (3’-CS region only) | 5’-CS – dfrA12 – aadA2 – 3’-CS | – | 5’-CS – aac(3)-I – aadA1 – 3’CS | 5’-CS – aac(3)-I – aadA1 – 3’CS | 5’-CS – ac(3)-I – aadA1 – 3’CS | – | |
| Pathogen Finder | 0.853 | 0.852 | 0.852 | 0.845 | 0.852 | 0.852 | 0.854 | |
| Plasmid replicons and carried antibiotic resistance genes | GR6 (pACICU2-like – AN: CP031382.1) - aph(3')-VIa | pMAL-1 like (AN: NZ_KX230793.1) - blaOXA-72, | pMAL-1 like - blaOXA-72 |
pMAL-1 like - blaOXA-72 pESBL234 like (AN: NZ_MT230226.1) – tet(A) |
GR6 (pACICU2-like: ) - aph(3')-VIa | Plasmid unnamed1-like (strain VB1190 AN: VB1190) aph(3')-Ia | Plasmid unnamed (strain T5-67 AN: NZ_CP043569.1 ) - msr(E), mph(E), armA pMAL-1 like | |
| MLST profiles | ST636 | ST492 | ST492 | ST1 | ST636 | ST2 | ST2 |
The XDR phenotype of these strains could be correlated with a large number of multidrug efflux pumps genes, identified in all A. baumannii genomes (Table 3). Genes associated with resistance to copper (MmcO, CtpA, copper chaperon, Bcr/CflA, cueR, pcoB, pcoD, CusR, CopC, cutE, CorC), cobalt, zinc, cadmium (CzcABCD, MFP, merR, CusA, CusR, CusS), chromium (chrA), arsenic compounds (ACR3, arsC, arsH, arsR) and mercury (merA, merC, merR) have been also detected. Also, class 1 integrons, harboring mainly aminoglycoside resistance genes (Table 3) were detected. It is well known that class 1 integrons represent major vehicles enabling the development, accumulation and dissemination of ARGs through horizontal transfer. The association of class 1 integrons with successful A. baumannii clones (such as ST2, ST636 and ST492) could explain their high prevalence and potential of transmission among patients. The GR6 plasmid replicon (pACICU2-like) was detected in 2 strains from community-acquired infections (1S, 24S). Another plasmid, untypable by the PCR-based replicon typing, similar to pMAL-1 plasmid (99% similarity) was detected in all but one clinical strains and one ambulatory strain (A07 and 01S). This pMAL-1-like plasmid sequence harbors blaOXA-72. In the A14 clinical strain, however, the blaOXA-23 gene was truncated, having a deletion of 658 nucleotides (Table 3).
In 2 strains (10s and 14s), 3 intact prophages (70.8 Kb; 37.5 Kb; 13.5 Kb in strain encoded 10s and 40.3 Kb; 52.6 Kb; 41.7 Kb in strain 14s) were also found, without any relation with the presence of ARGs.
According to the virulence factors database all strains harbor a wide range of virulence genes, encoding for: iron acquisition (e.g. tonB), pore-forming toxins (apkA and apkB), adherence, invasion of epithelial cells (ompA), phospholipids degradation (plc, plcC, plcD) and apoptosis induction (Table 3). Two of the strains (14s and A14) contain the hemO (hemoxigenase) gene, which was associated with the hypervirulent phenotype of A. baumannii LAC 419. The ability to produce various virulence factors of these A. baumannii strains could explain their ability to persist and colonize the human host leading to a serious threat for hospitalized patients.
The predicted serological typing scheme for the A. baumannii strains analyzed in this study shows some aspects related to their virulence. It is known that the major immunogenic polysaccharide which is produced A. baumannii as in important virulence factor is the capsular one (K) and not the somatic antigen O, since the non-capsulated strains don’t cause infections20. The Supplementary Fig. 1 shows the genetic variability in the K and O loci, the metrics related to the match confidence, coverage, identity and number of genes, as well the locus type for each of the 7 strains that were sequenced in this study, and their relationships with regards to the subtype.
Pangenome, phage and genomic islands analysis
Genomic Islands
Genomic islands (GI) represent proof of horizontal gene transfer in a population; these DNA segments may integrate into the chromosome of the host and undergo transformation, conjugation or transduction21. The GI predictions showed that only 4 of the 7 strains belonging to ST1, ST2 and ST636, harbored genomic islands: 3 with putative type IV secretion system (T4SS) ICEs and one with putative IME.
The strain 01s has a putative T4SS-type ICE 266 kb (~ 38% GC) region, containing 273 ORFs among which some encode for the following components of T4SS (AAA_10, TrbC, TraL, TraE, TraK, TrbI, TraV, TraU, TraN, TraH, TraG), while others for: a Prim-Pol primase-polymerase, Phage integrase, Pfam-B_3022 (which may be an mRNA interferase toxin of the MqsR-MqsA toxin-antitoxin system, and biofilm/motility regulator), TrwB_AAD_bind type IV coupling protein, TrwC relaxase.
The strain 18s has a putative IME region of 31 kb (41%GC) containing 39 ORFs, some of them encoding for TrbI T4SS component, Rep_trans relaxase and rve integrase.
The strain 24s has a putative T4SS-type ICE 282 kb (~ 38% GC) region containing 280 ORFs among which some encode for the following T4SS components (TrbH, traP_typeI, traK_typeI, AAA_10, T2SSE, Plasmid_RAQPRD)—traP and traK could be hints for an F-like plasmid in A. baumannii. Other ORFs encode for T4CP proteins (such as FtsK_SpoIIIE), DUF1525, relaxases and phage integrases.
The strain A14 has a putative T4SS-type ICE 131 kb (~ 35% GC) region containing 156 ORFs among which some code for the following T4SS components (TraH, traP_typeI, traK, AAA_10, T2SSE, TraG, TraF, TraN, TrbC, TraU, TrbI, TraE, TraL, TrwC, Pfam-B_1474, Plasmid_RAQPRD), rve integrases, T4CP TrwB_AAD_bind, relaxases.
Further, considering the reference genomes used for phylogenetic analysis and the same GI prediction method, the GI were encountered in 19 out of 69 genomes, with sets of genes similar to the selected sequenced samples from the same phylogenetic cluster. While most of the genomes have only one GI region predicted, 3 of the reference genomes had 2 GI regions (Suppl. Table 1).
Most of the genomes with predicted GI belong to the ST1 cluster. Compared with the references from the same subtype, 18s GI exhibits: (i) the lowest number of ORFs, thus fewer known protein types, even though its length is similar with other Putative IME GI from different references; (ii) the highest GC percentage (41.19%) in the genomic island (Suppl. Table 1).
When compared with the reference genomes, the strains 24s and 01s belonging to ST636, and A14 from ST2 have the longest GI, with the highest number of ORFs and similar GC.
Beside this prediction, traces of TnAbaR were also found. TnAbaR is a core composite transposon bound by inverted repeats and 2 copies of direct repeats at its ends, forming an AbaR resistance island. Usually, AbaRs are inserted in A. baumannii genes, leading to the loss of their function—in this case, comM (an ATPase-encoding gene known as a hotspot for the integration of AbaR). These islands consist mainly of MGEs, such as transposons or integrons, and various genes that confer MDR22.
Therefore, of the 7 strains, 4 of them (01s, 14s, 24s and A07) fully cover TnAbaR23 (a 50k bp TnAbaR-like island, also containing the partial sequence of comM gene—GenBank: JN676148.1) while 2 of them (10s and A14) partially cover it.
Although they fully cover this resistance island, 14s and 01s strains have the lowest overall identity (20% and 25%, respectively) with TnAbaR23. The contig from 01s has 100% identity to the partial sequence of comM gene, while 14s has the highest local identity (97.1%) only to TnpA IS15DI transposase from Module_I. The strains 24s has an overall 47.1% identity with TnAbaR23, though having 92%, 100% and 100% identity to Module_I,_J and _K, respectively, while A07 has an overall 37.2% identity, though having 100% and 83.6% identity to Module_A and _B, respectively.
The other strains that partially cover TnAbaR23 also have low identities: 10s has an overall 27.1% identity (with 97.1% identity to TnpA gene from Module_I). A14 has an overall 32.8% identity, of which modules A to first half of Module_F have no coverage, the only high similarity region comprising the end of Module_F and the start of Module_G.
On the other hand, the contigs of 18s cover almost entirely TnAbaR23, with 99.97–100% identity to Module_A,_B,_C,_D,_F and _K, 93.4% to Module_G and 36% to Module_I. Almost all of Module_J has no coverage (highlighted in yellow in Suppl. Figure 2): the end part of the CadR (transcriptional regulator of MerR family), whole CadA (heavy metal transport/detoxification protein), whole LspA (prolipoprotein signal peptidase) and almost all of the transposase in this module (protein ID: AFB76410).
The TnAbaR-like island of 18s had most similarities to other TnAbaR islands, mostly on modules A to half of G, and K, given by the backbone transposon Tn6019 (Suppl. Figure 2). The highest similarity is with TnAbaR23 (as it can be seen in the identity distance matrix from Suppl. Figure 2). The region in Module_G (highlighted in cyan in Suppl. Figure 2) is different between 18s/TnAbaR23 and the other transposons. The dissimilarity is increased between 18s and the other transposons on the end part of Module_G and most of Module_I (highlighted in purple in Suppl. Figure 2), where the 18s resistance island may acquire structural variation compared to the others.
Phages
The most phage-abundant regions were found in 2 strains that belong to the ST492 subtype. In these strains we found the highest number of intact phages, phage species and attachment sites for phages compared to the other strains with other subtypes (Suppl. Table 1).
Pangenomes
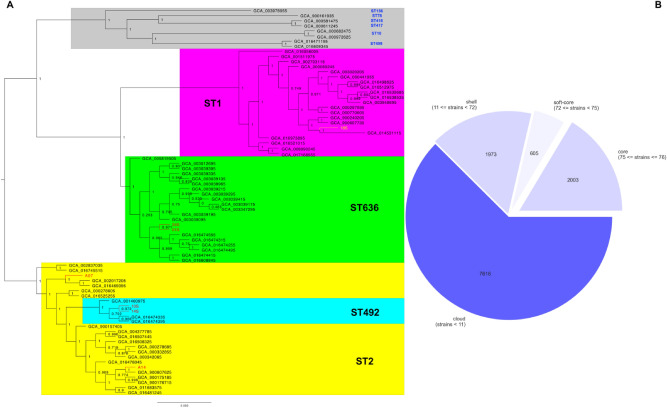
The pangenome analysis shows that the total number of genes found in each of the samples is approximately 3700. Around 43% of these are represented by hypothetical proteins (Fig. 1b). The genomes were subsequently compared one with each other as a matter of unique genes. Then, the lowest numbers taken by subtype belong to ST492 and ST636, while the highest numbers are found in ST2 (6 to 8 times higher than the previous two) and ST1 (almost 3 times higher than in ST2).
Figure 1.
Phylogenetic tree of 76 genomes based on pangenome analysis. (A) Phylogenetic clustering; (B) The distribution of core and accessory genes in all 76 genomes.
The ST492 is of particular interest, considering the subtype is relatively new and not much is known about it in literature. Taking into account that the highest number of phages, but no genomic islands were predicted in two ST492 strains, we took a closer insight into their genome. When compared to the other genomes considered in our phylogenetic analysis, the 2 ST492 strains contain 19 unique genes that were not found in any other genome from the selected ones (including 3 other ST492 reference genomes). Of these 19 genes, 17 are hypothetical and the other 2 are represented by: dnaB2 (a replicative DNA helicase) and bfrD1 (a putative TonB-dependent receptor BfrD). These 2 ST492 strains have 62 proteins in common, of which 57 are hypothetical proteins while the other 5 are represented by: fpvA (ferripyoverdine receptor), pucD (putative xanthine dehydrogenase subunit D), folE (GTP cyclohydrolase 1), bfrD1 and dnaB2. When considering all 5 ST492 genomes from the phylogenetic analysis set, then there are 18 proteins unique only to ST492 of which 11 are hypothetical and the other 7 proteins are: ptk (tyrosine-protein kinase), ptp (low molecular weight protein tyrosine-phosphatase), glxR2 (2-hydroxy-3-oxopropionate reductase), wbpA (UDP-N-acetyl-D-glucosamine 6-dehydrogenase), pglH (GalNAc-alpha-(1- > 4)-GalNAc-alpha-(1- > 3)-diNAcBac-PP-undecaprenol alpha-1,4-N-acetyl-D-galactosaminyltransferase), mshA3 (D-inositol-3-phosphate glycosyltransferase) and pglA (N,N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase).
All 76 samples that were used for the phylogenetic analysis have in common approximatively 2000 core genes and present other 8-9000 accessory genes. This suggests that the selected genomes for the phylogenetic and pangenome analysis have some active MGEs, which may contain various types of resistance genes, integrases, transposases—already predicted and mentioned within the body of this manuscript.
Phylogenetic analysis
The phylogenetic relationships between the selected 76 genomes show a clear clustering between the subtypes of interest in which the studied strains fit in. It also shows the separate evolution of ST492 from ST2 (as previously mentioned, ST492 and ST636 are single and respectively triple locus variants of ST2)23. The other 8 randomly selected subtypes cluster separately (Fig. 1a). Few other studies on ST492 have been mentioned in literature24, 25, but the genomes may have not been uploaded in NCBI database26, although not much is known about this subtype, therefore only 5 genomes for ST492 have been used for the phylogenetic analysis. The 2 ST636 strains from this study (01S and 24S) group together, separately from other ST636, while the ST492 ones (10S and 14S), even if they are a subbranch of ST2, are in the same cluster with the other two ST2 samples (A07 and A14).
Similar to the phylogenetic relationship and MLST predictions, some of the genes are expressed only in the isolates from the same branch (e.g., blaADC-74 predicted to be present only in 01S and 24S isolates (ST636); blaADC-11, blaPER-1 only for A07 and A14 isolates belonging to the ST2 clone; blaADC-30 tet(B), sul2—only for 10S and 14S isolates (ST492 clone). Based on the variant results, approximately 1.25% average of the whole A. baumannii genome length in each of the 7 strains proved to be variants. At the same time, most of the variants predicted by ARIBA are found in the variant list from snippy27, in the same or very similar genes.
Discussions
A. baumannii is one of the most successful pathogens responsible for nosocomial infections, occurred especially in patients admitted to intensive care units (ICUs), but also for community-acquired infections, being able to acquire resistance to carbapenems, fluoroquinolones and aminoglycosides28. Due to the limited options for the antibiotic treatment of the produced infections, CRAB isolates became a significant health problem worldwide29. Although antibiotic resistance may not be considered a traditional virulence factor, in case of A. baumannii it is by far the biggest driver of the clinical outcome by precluding the clinician's ability to eliminate the infecting strain. Our experimental data have shown a high prevalence of imipenem resistance (96.96%) among A. baumannii strains isolated in 2017–2018 from hospitalized and community-acquired infections in patients from Bucharest. This percentage was close to reported rates in Egypt, Turkey, Spain and Italy and higher than from Saudi Arabia30–34.
To date, in Romania, there has been reported a high prevalence of CRAB strains in different parts of the country including Bucharest, the capital city35. Even though blaOXA-58 has been identified in A. baumannii (ST1 clone) recovered from patients in our country36, the ST2 associated with blaOXA-23 remains the most common among the CRAB isolates. A pilot study from 3 Romanian hospitals—from Iași and Târgu-Mureș, performed between 2014 and 2015—showed the presence of carbapenemases OXA-23 and OXA-24/72 in A. baumannii nosocomial isolates37.
In our study, the MLST analysis has revealed that the strains belonged to 6 different clones: ST2 (pulsotypes I, II, III and IX), ST636 (pulsotypes VI and VIII), ST492 (pulsotype VII), ST312 (pulsotype IV), ST642 (pulsotype V) and ST1 (pulsotype VI); ST2 was the most frequently encountered clone (12/33; 36.36%) followed by ST636 (9/33; 27.27%) and ST492 (6/33; 18.18%) (Table 1).
Regarding the distribution of the carbapenemases genes among the identified clones, it has been observed that the ST2 clone encountered in hospital A was associated with the production of blaOXA-23 gene (10/33; 30.30%) carbapenemase and revealed, in most cases, the presence upstream the carbapenemase gene, of the ISAba1 element (9/33; 27.27%). The presence of ISAba1 upstream blaOXA-23 and blaOXA-51 is required to confer resistance to carbapenems38. Although the relationship between ISAba1 upstream blaOXA-51 and carbapenem resistance was confirmed, this might not be enough to confer resistance, as A. baumannii isolates susceptible to carbapenems with the association ISAba1/ blaOXA-51 have already been described39. The clone harboring 2 carbapenemases: OXA-23 and OXA-51 was identified in the ambulatory sector (clinical unit B) (Table 1); the ST636 clone was related with the presence of OXA-24 carbapenemase, most of the strains being isolated from hospital C (7/33; 21.21%) and 2 strains from the ambulatory. The other isolates belonging to STs 492, 312, 642 and ST1 were associated with blaOXA-24 gene (Table 1). The distribution of these clones per hospital unit revealed that ST492 and ST1 were found exclusively in hospital C; ST642 in hospital A, while ST312 was identified in both hospital units (A and C) (Table 1).
Currently, worldwide carbapenem resistant strains are mostly associated with international clone II, with blaOXA-23 as the main carbapenem resistance mechanism12. In Greece, it has been observed that the ST2 was the most common clone circulating in Greek hospital settings40. With regards to MDR, several other authors have demonstrated the association of blaOXA-23, blaOXA-58, blaOXA-72 and ST224, 41–44. Furthermore, the international clone ST2 was found to be broadly spread among our country45. Very recently Lukovik et al., have reported the same association of CRAB circulating in Serbia: blaOXA-66/blaOXA-23/ST2 blaOXA-66/blaOXA-72/ST492, and blaOXA-66/ blaOXA-72/ST63625. These data confirm the multidirectional evolution of the CRAB clones in neighboring countries. A particular case has been noticed in Serbia by Misic et al. in 2018 which have demonstrated the presence of Aci1 carrying blaOXA-72 belonging to ST1 in a companion animal emphasizing the importance for both animal and public health46. Jakovac et al. demonstrated the presence of the international clone II carrying blaOXA-23 and blaOXA-72 in nosocomial CRAB recovered from the southwestern part of Bosnia and Herzegovina47. A sporadic CRAB clone harboring a unique blaOXA-72 carrying plasmid have been reported in China from a patient with community-acquired pneumonia48.
Similar to our results, the presence of multiple ARGs for β-lactams, aminoglycosides, sulfonamide and tetracyclines was evidenced in clinical MDR A. baumannii isolates from Spain and Switzerland49, 50.
Some of the identified plasmids (e.g. pACICU-2 like ST2 carrying blaOXA-23; pMAL-1like ST492 and ST1 carrying blaOXA-72) were previously described in our country, in A. baumannii strains belonging to different clones35, 51 and in Serbia (GenBank accession no. KX230793.1), while others (e.g. pACICU-2like ST636 carrying blaOXA-72 gene; pACICU-2 like and pMAL-1 like ST2 carrying blaOXA-23) are reported for the first time in community-acquired and intra-hospital A. baumannii strains isolated in Bucharest. In our study, due to sequencing limitations, the blaOXA-23 gene location was not identified. Our data revealed that community-acquired A. baumannii harbored pACICU-2 linked to ST636 producing OXA-72; pMAL-1linked to ST492 and ST1 producing OXA-72 and clinical CRAB pACICU-2 like and pMAL-1linked to ST2 OXA-23 producing.
The OC locus prediction shows that the analyzed strains contain 3 locus types: OCL1, OCL2 and OCL4. OCL1, present in 4 out of 7 strains, is known to be the most abundant gene cluster in the major global clone groups GC1 and GC2, of which ST1 and ST2 belong to. In this case the ST1 strain belongs to OCL4 and the ST492 to OCL1 (supporting thus the hypothesis that ST492, although rare, resulted from ST2, as suggested by phylogeny, or vice versa). The major difference is found between the strains belonging to the ST636 subtype, where wecB gene, known to be involved in the biosynthesis of sialic acid, occurs between the glycosyltransferases gtrOC8 and gtrOC952.
On the other hand, the diversity is increased in the K loci, the 7 strains containing 5 different gene clusters: KL1, KL3, KL30, KL40 and KL77.
The major differences appear between the capsular export region and the repeat unit of translocase (wzx), with the presence or absence of various types of genes: UDP-N-acetylglucosamine-2-epimerase (mnaA) and UDP-N-acetylmannosamine dehydrogenase (mnaB) in the ST636 strain; UDP-d-GlcpNAcA epimerase (gne2) in ST1; d-glucosaminate PTS permease components EIIA, EIIB, EIIC (dgaA, dgaB, dgaC) in A14 from ST2; Photosystem I P700 chlorophyll a apoprotein A1 & A2 (psaA, psaB), Photosystem I iron-sulfur center (psaC), Photosystem I reaction center subunit II & IV & III (psaD, psaE, psaF) in A07 from ST2. Actually, A07 strain has the most diverse K locus from the analyzed strains—there is also an acyl/acetyl transferase (e.g. atr20) which is inserted in the simple sugar synthesis region. More than that, the insertion of atr20 may suggest a novel K locus even though it matches the K77 reference, according to the Kaptive documentation. Similarly may happen to the K loci in ST492 strains, since there is no coverage in the respective region for wzy, gtr63 and itrA2; this finding, together with the low matching confidence and missing genes between gna and wzx compared with the other samples, is suggesting a possible novel variant. We also revealed that the p-ACICU-2like and pMAL-1like carrying blaOXA-23, p-ACICU-2like carrying blaOXA-72 and pMAL-1like carrying blaOXA-72 presented virulence-related genes involved in iron acquisition (such as the TonB-dependent receptor), adherence (ompA), biofilm formation (pgaABCD locus), invasion (septicolysin) and haemolytic activity against human erythrocytes, aiding in iron acquisition) (e.g. plc, plcC and plcD)53. TonB-dependent transporters are outer membrane proteins (OMPs) that bind and transport siderophores in addition to vitamin B12, nickel complexes, and carbohydrates and may be involved in the survival of bacteria in the lungs central nervous system and blood54. Septicolysin, on the other hand, is a pore-forming toxin with a cytolytic activity that mediates invasion of tissues or cells55.
Materials and methods
Bacterial isolates
Thirty-three carbapenemase-producing A. baumannii were isolated from hospitalized, ICU patients (n = 30) and ambulatory patients (3) during Sept. 2017–Feb. 2018 and identified by commercial systems (VITEK2, Microscan and Maldi TOF) (Table 1).
Susceptibility testing
The susceptibility of A. baumannii strains to meropenem (MEM—10 µg), imipenem (IMP—10 µg), ertapenem (ETP—10 µg), cephalotin (CEF—30 µg) ceftriaxone (CTX—30 µg), cefuroxime (CXM—30 µg), cefoxitin (FOX—30 µg), ceftazidime (CAZ—30 µg), aztreonam (ATM—30 µg), cefepime (FEP—30 µg), amoxicillin-clavulanic acid (AMC—10 µg), piperacillin-tazobactam (PIP-TZP—10 µg), ciprofloxacin (CIP—5 µg), levofloxacin (LEV—5 µg) gentamicin (GEN—5 µg), amikacin (AMK—5 µg), nitrofurantoin (NIT—300 µg), trimethoprim-sulfamethoxazole (SXT—23.75 µg), tetracycline (TET—30 µg), tigecycline (TIG) and colistin (COL) (Table 1) was tested by disk diffusion and microdilution methods (for MEM, COL and TIG assessed by broth microdilution method with Mueller–Hinton broth), according to Clinical and Laboratory Standards Institute (CLSI), 2018 and 2019 guidelines56, 57. Strains displaying MICs > 8 mg/L for MEM; MICs > 4 mg/L for COL and MICs > 2 mg/L for TIG according EUCAST breakpoints for Enterobacterales were considered resistant58. Pseudomonas aeruginosa ATCC 27,853 was used as a quality control strain. A. baumannii were classified as follows: multidrug-resistant (MDR) [resistant to at least one agent in 3 or more antimicrobial categories], extensively drug-resistant (XDR) [resistant to at least one agent in all, but 2 or fewer antimicrobial categories] and pandrug-resistant (PDR) [resistant to all agents in all antimicrobial categories tested59.
PCR for carbapenemase genes
Genes encoding common class D carbapenemases (blaOXA-51-like, blaOXA-58-like, blaOXA-23-like, blaOXA-24-like, blaOXA-143-like and blaOXA-235-like) and class B metallo-β-lactamases (blaIMP, blaVIM, blaNDM, blaOXA-48) were detected by PCR60, 61, followed by sequencing of selected amplicons. The presence of ISAba1 upstream of blaOXA-23-like, blaOXA-24-like was tested by PCR and sequencing using custom-designed primers (OXA-24A: CTCTAAGCCCCAAAATTTCC; OXA-24B: CGCATAAGGCGTATTATGTTA; ISAbaF1: CCTCAGTTTAATGCCAATGCT). The increased expression of blaOXA-51 genes was analyzed by PCR for ISAba1 upstream of the gene35. Sequence analysis was performed using Chromas Lite 2.1 software and compared with sequences deposited in the GenBank database26.
Pulsed-field gel electrophoresis (PFGE)
The clonality of blaOXA-23 and blaOXA-24 carrying A. baumannii was determined with PFGE using the ApaI enzyme and CHEF-DRII apparatus (Bio-Rad). Pulsotypes were defined as isolates with PFGE band patterns of 85% similarity or above62. The isolates were classified according to the criteria described63.
Mating experiments
Transferability of blaOXA-23 by conjugation was tested in solid mating using a rifampicin (RIF)—resistant Acinetobacter baylyi ADP1 as a recipient. Briefly, equal amounts (100 µL) of overnight cultures of the donor (A14; A17; 8A) and recipient strains were mixed and incubated in Brain heart infusion agar plates, cells were resuspended in saline solution and selected in plates containing RIF (300 mg/L) and MEM (0.5 mg/L)64. Characterization of the transconjugants was conducted by PCR to confirm the conjugative transfer.
Multilocus sequence typing (MLST)
MLST was used to establish the clonal relationships in A. baumannii representative strain from each pulsed-field type, using 7 housekeeping loci (elongation factor EF-G [fusA], citrate synthase [gltA], CTP synthase [pyrG], homologous recombination A factor [recA], 60-KDa chaperonin [cpn 60], 50S ribosomal protein L2 [rplB] and RNA polymerase subunit B [rpoB] according to the Institute Pasteur scheme65.
Whole-genome sequencing and bioinformatic protocols
The genomes of 7 A. baumannii strains isolated from different infections were sequenced using HiSeq and MiSeq X Ten, Illumina. The paired-end raw reads have been submitted to the GenBank under accession numbers: SRX4094320, SRX4094321, SRX4094322, SRX4094323, SAMN14525745, SAMN14525746, SAMN14525747 and were subsequently assembled using shovill66 with SPADES 3.12.067 already implemented, while the quality of assembling was checked with QUAST68. The samples underwent a reference mapping step in Geneious Prime 2020.1.169, with trimming in a 5 iteration step and taking into account only the reads with a quality factor higher than Q30 resulting in coverage of approximatively 35x-to-38x on the whole genome. Annotations were performed with RAST70 and Prokka71 while the prediction of resistance and virulence profiles was performed by using the following programs and databases: Abricate (compared against VFDB database for virulence factor prediction), ARIBA (downloading and preparing at first the reference data from NCBI database), ResFinder (compared against implemented NCBI database), PlasmidFinder (compared against implemented plasmidfinder database), PathogenFinder, CARD, PubMLST, ISfinder, PATRIC72–83. The resistance genes and virulence factors were selected based on the highest identity and coverage. Plasmid replicons were detected using AB-PBRT84 by an in silico PCR approach sequences85. The untypable plasmids were compared with the NCBI nucleotide database, while the plasmids integrated within the bacterial chromosome were detected using the BLAST tool86 using a command-line interface. All the other predictions were run by using default parameters.
Integron analysis was performed using command-line blastn86, with 5’CS and 3’CS sequences (Supplementary file 1) as queries against assembled genomes. Integron sequences annotations were performed manually.
Additional plasmids analyses were performed using the PLSDB database Search tool87, using the Mash strategy, with search parameter set as default (max. p-value at 0.1 and min. identity 0.99).
The phylogenetic analysis was performed using the following protocol: (i) taking into account the subtyping information from MLST prediction for the 7 strains discussed in our study, the assembly contigs from 69 reference sequences were selected from the NCBI database26. After repeating the MLST prediction on the majority of A. baumannii genomes from NCBI (that were uploaded as contigs) the selection of the 69 sequences used for phylogeny was done randomly. Approximatively 20 genomes were selected from each subtype corresponding to the subtypes found in our samples (except ST492—only 3 out of ~ 6000 genomes were found) while other 8 genomes belong to different subtypes. (ii) Whole genome annotation with Prokka (using genetic code 11 and Genbank/ENA/DDJB compliance options) was performed for each of the 76 genomes selected for phylogeny. (iii) the output from Prokka was used for pangenome analysis using Roary88. (iv) the phylogenetic tree was generated (on the alignment of core genome gene sequences in Roary with MAFFT89, using FastTree90 with Jukes-Cantor model, Maximum-likelihood and minimum-evolution NNIs and SPRs algorithms, implemented in Geneious Prime 2020.1.1. The phylogenetic tree image was generated using FigTree software (http://tree.bio.ed.ac.uk/software/figtree/). The Genomic Islands predictions were performed with ICEfinder online tool91, while the phage predictions were done with PHASTER online tool92. K and OC locus predictions were performed using the Kaptive software20. TnAbaR sequences have been aligned with MAFFT, implemented in Geneious Prime.
Conclusions
Our results highlight the presence of an impressive armamentarium of ARGs and of mobile elements required to form their mobilization and transmission, such as the plasmids, transposons and ISs in CRAB strains isolated from Romanian patients. These strains belong to widespread clones reported in intrahospital infections or community-acquired patients. The most frequently encountered clones identified in A. baumannii clinical strains were ST2, ST636 and ST492, while the most frequent carbapenemase genes were blaOXA-23 or blaOXA-24. Among resistance determinants, several virulence genes, as well as factors that contribute to the persistence of these bacteria in the hospital environment, have been detected. All sequenced A. baumannii isolates have the genetic equipment conferring them the ability to grow in iron-depleted media and to survive in the presence of desiccation, antimicrobials and toxic compounds. Our data will facilitate the understanding of resistance, virulence and transmission features of XDR AB isolates from Romanian patients and might contribute to the implementation of appropriate infection control measures for limiting the spread and decreasing the infection rate and mortality.
Supplementary Information
Author contributions
Conceptualization, I.G.; I.S.; and Z.Z.; methodology, I.G.; and I.S.; software, I.C.B.; M.S.; and Y.F.; validation, M.C.C.; and Z.Z.; formal analysis, I.G.; and L.-I.P.; investigation, I.G.; I.C.B.; I.S.; S.P.; resources, M.C.C.; and Z.Z.; data curation, M.C.C.; and V.L.; writing—original draft preparation, I.G.; I.C.B.; M.S.; writing—review and editing, I.G.; I.C.B.; M.S.; D.O.; visualization, M.C.C.; V.L., D.O.; supervision, M.C.C.; project administration, I.G.; funding acquisition, M.C.C. All authors have read and agreed to the published version of the manuscript I.G.; I.C.B.; and M.S.; have equally contributed to this paper as main authors.
Funding
This research was funded by the Research Projects PN-III-P1.1-PD-2016-1798 (PD 148/2018), PN-III-P4-ID-PCCF-2016-0114 awarded by UEFISCDI, FDI-2020-0834 awarded by CNFIS and grant no. 20964/30.10.2020 awarded by ICUB. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Date Availability
A. baumannii strains are available from the authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Irina Gheorghe, Ilda Czobor Barbu and Marius Surleac.
Contributor Information
Irina Gheorghe, Email: iryna_84@yahoo.com.
Ilda Czobor Barbu, Email: ilda.barbu@bio.unibuc.ro.
Marius Surleac, Email: marius.surleac@gmail.com.
Ionela Sârbu, Email: ionela24avram@yahoo.com.
Laura Ioana Popa, Email: lpbio_laura@yahoo.com.
Simona Paraschiv, Email: mona@mateibals.ro.
Yu Feng, Email: a1204350@qq.com.
Veronica Lazăr, Email: veronica.lazar2009@gmail.com.
Mariana Carmen Chifiriuc, Email: carmen.chifiriuc@bio.unibuc.ro.
Dan Oţelea, Email: dotelea@mateibals.ro.
Zong Zhiyong, Email: zongzhiyong@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92590-5.
References
- 1.Duval RE, Grare M, Demoré B. Fight against antimicrobial resistance: We always need new antibacterials but for right bacteria. Molecules. 2019;24:3152. doi: 10.3390/molecules24173152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliva A, Garzoli S, De Angelis M, Marzuillo C, Vullo V, Mastroianni CM, Ragno R. In-vitro evaluation of different antimicrobial combinations with and without colistin against carbapenem-resistant Acinetobacter Baumannii. Molecules. 2019;24:886. doi: 10.3390/molecules24050886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrancianu CO, Gheorghe I, Czobor IB, Chifiriuc MC. Antibiotic resistance profiles, molecular mechanisms and innovative treatment strategies of Acinetobacter baumannii. Microorganisms. 2020;8:935. doi: 10.3390/microorganisms8060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pogue JM, Mann T, Barber KE, Kaye KS. Carbapenem-resistant Acinetobacter baumannii: epidemiology, surveillance and management. Expert Rev Anti- Infect Ther. 2013;11:383–393. doi: 10.1586/eri.13.14. [DOI] [PubMed] [Google Scholar]
- 5.Ramoul A, Loucif L, Bakour S, Amiri S, Dekhil M, Rolain J-M. Cooccurrence of blaNDM-1 with blaOXA-23 or blaOXA-58 in clinical multidrug-resistant Acinetobacter baumannii isolates in Algeria. J. Glob. Antimicrob. Resist. 2016;6:136–141. doi: 10.1016/j.jgar.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Bakour S, Olaitan AO, Ammari H, Touati A, Saoudi S, Saoudi K, et al. Emergence of colistin- and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in Algeria: First case report. Microb. Drug Resist. 2015;21:279–285. doi: 10.1089/mdr.2014.0214. [DOI] [PubMed] [Google Scholar]
- 7.Mathlouthi N, El Salabi AA, Ben Jomàa-Jemili M, Bakour S, Al-Bayssari C, Zorgani AA, et al. Early detection of metallo-β-lactamase NDM-1- and OXA-23 carbapenemase-producing Acinetobacter baumannii in Libyan hospitals. Int. J. Antimicrob Agents. 2016;48:46–50. doi: 10.1016/j.ijantimicag.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Evans BA, Amyes SGB. OXA β-Lactamases. Clin. Microbiol. Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 10.Abdi SN, Ghotaslou R, Ganbarov K, Mobed A, Tanomand A, Yousefi M, Asgharzadeh M, Kafil HS. Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect Drug Resist. 2020;13:423–434. doi: 10.2147/IDR.S228089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammoudi D, Ayoub MC, Hakime N, Houmani M, Barakat A, Najjar Z, Suleiman M, Fayad N, Sarraf R, Sarkis K. Spread of imipenem-resistant Acinetobacter baumannii co-expressing OXA-23 and GES-11 carbapenemases in Lebanon. Int J Inf Dis. 2015;36:56–61. doi: 10.1016/j.ijid.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopes BS, Amyes SGB. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J. Med. Microbiol. 2012;61:1103–1108. doi: 10.1099/jmm.0.044156-0. [DOI] [PubMed] [Google Scholar]
- 14.Martínez P, Mattar S. Imipenem-resistant Acinetobacter baumannii carrying the ISAba1- blaOXA-23, 51 and ISAba1-blaADC-7 genes in Monteria, Colombia. Brazil. J. Microbiol. 2012;2012:1274–1280. doi: 10.1590/S1517-83822012000400006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corvec S, Poirel L, Naas T, et al. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:1530–1533. doi: 10.1128/AAC.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Zong Z, Lu X. Tn2008 is a major vehicle carrying blaOXA-23 in Acinetobacter baumannii from China. Diagn. Microbiol. Infect. Dis. 2011;69:218–222. doi: 10.1016/j.diagmicrobio.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Mugnier PD, Poirel L, Nordmann P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 2009;191:2414–2418. doi: 10.1128/JB.01258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makke G, Bitar I, Salloum T, Panossian B, Alousi S, Arabaghian H, Medvecky M, Hrabak J, Merheb-Ghoussoub S, Tokajian S. Whole-genome-sequence-based characterization of extensively drug-resistant Acinetobacter baumannii Hospital Outbreak. mSphere. 2020;5:00934–1019. doi: 10.1128/mSphere.00934-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou HY, Kuang SN, He X, Molgora BM, Ewing PJ, Deng Z, Osby M, Chen W, Xua HH. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci. Rep. 2015;5:8643. doi: 10.1038/srep08643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyres KL, Cahill SM, Holt KE, Hall RM, Kenyon JJ. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom. 2020;6:e000339. doi: 10.1099/mgen.0.000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulaiman, S., Yusoff, N. S., Mun, N. S., Makmur, H. & Firdaus-Raih, M. Inference of Horizontal Gene Transfer: Gaining Insights Into Evolution via Lateral Acquisition of Genetic Material. In Encyclopedia Bioinformatics and Computational Biology (Academic Press, pp. 953-964). 10.1016/B978-0-12-809633-8.20173-8 (2019).
- 22.https://apps.szu.cz/anemec/Krizova_Thesis.pdf (2014).
- 23.https://ses.library.usyd.edu.au/handle/2123/18156 (2018).
- 24.Dortet, L., Bonnin, R. A., Girlich, D., Imanci, D., Bernabeu, S., Fortineau, N. & Naas, T. Whole-Genome Sequence of a European Clone II and OXA-72-producing Acinetobacter baumannii strain from Serbia. Genome Announc. 3, e01390–15. 10.1128/genomeA.01390-15 (2015). [DOI] [PMC free article] [PubMed]
- 25.Lukovic B, Gajic I, Dimkic I, et al. The first nationwide multicenter study of Acinetobacter baumannii recovered in Serbia: Emergence of OXA-72, OXA-23 and NDM-1-producing isolates. Antimicrob. Resist. Infect Control. 2020;9:10. doi: 10.1186/s13756-020-00769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Center for Biotechnology Information (NCBI) [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988]. Available from: https://www.ncbi.nlm.nih.gov/. Accessed June 01, 2021.
- 27.https://github.com/tseemann/snippy. Accessed February 26, 2020.
- 28.Blanco N, Harris AD, Rock C, Johnson JK, Pineles L, Bonomo RA, et al. Risk factors and outcomes associated with multidrug-resistant Acinetobacter baumannii upon intensive care unit admission. Antimicrob. Agents Chemother. 2017;62:e01631–e1717. doi: 10.1128/AAC.01631-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempf M, Rolain J-M. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int. J. Antimicrob. Agents. 2012;39:105–114. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Al-Agamy MH, Khalaf NG, Tawfick MM, Shibl AM, El Kholy A. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. Int. J. Infect. Dis. 2014;22:49–54. doi: 10.1016/j.ijid.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Ciftci IH, Asık G, Karakece E, Oksuz L, Yagcı S, Sesli C, E, , et al. Distribution of blaOXA genes in Acinetobacter baumannii strains: a multicenter study. Mikrobiyol Bul. 2013;47:592–602. doi: 10.5578/mb.6388. [DOI] [PubMed] [Google Scholar]
- 32.Villalon P, Valdezate S, Medina-Pascual MJ, Carrasco G, Vindel A, Saez-Nieto JA. Epidemiology of the Acinetobacter-derived cephalosporinase, carbapenemhydrolysing oxacillinase and metallo-b-lactamase genes, and of common insertion sequences, in epidemic clones of Acinetobacter baumannii from Spain. J. Antimicrob. Chemother. 2013;68:550–553. doi: 10.1093/jac/dks448. [DOI] [PubMed] [Google Scholar]
- 33.Principe L, Piazza A, Giani T, Bracco S, Caltagirone MS, Arena F, et al. Epidemic diffusion of OXA-23-producing Acinetobacter baumannii isolates in Italy: Results of the first cross-sectional countrywide survey. J. Clin. Microbiol. 2014;52:3004–3010. doi: 10.1128/JCM.00291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Badawy MF, Abdelwahab SF, Alghamdi SA, Shohayeb MM. Characterization of phenotypic and genotypic traits of carbapenem-resistant Acinetobacter baumannii clinical isolates recovered from a tertiary care hospital in Taif, Saudi Arabia. Infect Drug Resist. 2019;12:3113–3124. doi: 10.2147/IDR.S206691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gheorghe I, Novais A, Grosso F, Rodrigues C, Chifiriuc MC, Lazar V, Peixe L. Snapshop of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in Bucharest hospitals reveals unusual clones and novel genetic surroundings for blaOXA-23. J. Antimicrob. Chemother. 2015;70:1016–1020. doi: 10.1093/jac/dku527. [DOI] [PubMed] [Google Scholar]
- 36.Bonnin RA, Poirel L, Licker M, Nordmann P. Genetic diversity of carbapenem-hydrolysing β-lactamases in Acinetobacter baumannii from Romanian hospitals. Clin. Microbiol. Infect. 2011;17:1524–1528. doi: 10.1111/j.1469-0691.2011.03622.x. [DOI] [PubMed] [Google Scholar]
- 37.Timofte, D., Panzaru, C. V., Maciuca, I. E., Dan, M., Mare, A. D., Man, A., Toma, F. Active surveillance scheme in three Romanian hospitals reveals a high prevalence and variety of carbapenemase-producing Gram negative bacteria: a pilot study, December 2014 to May 2015. Eurosurveillance. 21, pii=30262. 10.2807/1560-7917.ES.2016.21.25.30262 (2016). [DOI] [PubMed]
- 38.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006;258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 39.Pagano M, Martins AF, Barth AL. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 2016;47:785–792. doi: 10.1016/j.bjm.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pournaras S, Dafopoulou K, Del Franco M, et al. Predominance of international clone 2 OXA-23-producing-Acinetobacter baumannii clinical isolates in Greece, 2015: Results of a nationwide study. Int. J. Antimicrob. Agents. 2017;49:749–753. doi: 10.1016/j.ijantimicag.2017.01.028. [DOI] [PubMed] [Google Scholar]
- 41.Mammina C, Palma DM, Bonura C, Aleo A, et al. Epidemiology and clonality of carbapenem-resistant Acinetobacter baumannii from an intensive care unit in Palermo, Italy. BMC Res. Notes. 2012;5:365. doi: 10.1186/1756-0500-5-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izdebski R, Janusz Fiett J, Waleria Hryniewicz W, Marek Gniadkowski M. Molecular analysis of Acinetobacter baumannii isolates from invasive infections in 2009 in Poland. J. Clin. Microbiol. 2012;50:3813–3815. doi: 10.1128/JCM.02271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrova AP, Stanimirova ID, Ivanov IN, Petrov MM, et al. Carbapenemase production of clinical isolates Acinetobacter baumannii and Pseudomonas aeruginosa from a Bulgarian University Hospital. Folia Med. 2017;59:413–421. doi: 10.1515/folmed-2017-0060. [DOI] [PubMed] [Google Scholar]
- 44.Nemec A, Krízová L, Maixnerová M, Diancourt L, van der Reijden TJ, Brisse S, van den Broek P, Dijkshoorn L. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 2008;62:484–489. doi: 10.1093/jac/dkn205. [DOI] [PubMed] [Google Scholar]
- 45.Di Popolo A, Giannouli M, Triassi M, Brisse S, Zarrilli R. Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin. Microbiol. Infect. 2011;17:197–201. doi: 10.1111/j.1469-0691.2010.03254.x. [DOI] [PubMed] [Google Scholar]
- 46.Misic D, Asanin J, Spergser J, Szostak M, Loncaric I. OXA-72-mediated carbapenem resistance in sequence type 1 multidrug (Colistin)-resistant Acinetobacter baumannii associated with urinary tract infection in a dog from Serbia. Antimicrob Agents Chemother. 2018;62:e00219–e318. doi: 10.1128/AAC.00219-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jakovac, S., Goić-Barišić, I., Pirija, M., Kovačić, A., Hrenović, J., Tanja Petrović, T., et al. Molecular characterization and survival of carbapenem resitant Acinetobacter baumannii isolated from hospitalized patients in Mostar, Bosnia and Herzegovina. Microb Drug Res. 27, 383–390. 10.1089/mdr.2020.0163 (2021). [DOI] [PubMed]
- 48.Jia H, Sun Q, Ruan Z, Xie X. Characterization of a small plasmid carrying the carbapenem resistance gene blaOXA-72 from community-acquired Acinetobacter baumannii sequence type 880 in China. Infect Drug Resist. 2019;12:1545–1553. doi: 10.2147/IDR.S202803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labrador-Herrera, G., Álvarez, R., López-Rojas., R, Smani, Y., Cebrero-Cangueiro, T., Rueda, A., Pérez Florido, J., Pachón, J. & Pachón-Ibáñez, M. E. Draft genome sequences of seven multidrug-resistant Acinetobacter baumannii strains, isolated from respiratory samples in Spain. Genome Announc. 4, e00083–16. 10.1128/genomeA.00083-16 (2016). [DOI] [PMC free article] [PubMed]
- 50.Abdelbary, M. H., Prodhom, G., Greub, G., Senn, L. & Blanc, D. S. Draft genome sequences of two carbapenemase-producing Acinetobacter baumannii clinical strains isolated from Albanian and Togolese Patients. Genome Announc. 5, e00115–17. 10.1128/genomeA.00115-17 (2017). [DOI] [PMC free article] [PubMed]
- 51.Gheorghe, I., Czobor, I., Ratiu, A.C., Petrut, S., Avram, I., Zhiyong, Z., Cristea, V.C., Chifiriuc, M.C. & Ecovoiu, A. Draft plasmid sequence of an XDR Acinetobacter baumannii strain reveals linked dissemination of carbapenem resistance and virulence genes. 27th ECCMID Viena, Austria, 22–25 april, EV0486 (2017).
- 52.Chou WK, Hinderlich S, Reutter W, Tanner ME. Sialic acid biosynthesis: stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP-N-acetylglucosamine 2-epimerase. J Am Chem Soc. 2003;125:2455–2461. doi: 10.1021/ja021309g. [DOI] [PubMed] [Google Scholar]
- 53.Fiester, S. E. et al. Iron-regulated phospholipase C activity contributes to the cytolytic activity and virulence of Acinetobacter baumannii. PLoS One.11, e0167068. 10.1371/journal.pone.0167068 (2016). [DOI] [PMC free article] [PubMed]
- 54.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, et al. The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 2008;10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 28th informational supplement. M02, M07 and M11 (2018).
- 57.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 29th informational supplement. M02, M07 and M11 (2019).
- 58.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 8.1 (2018).
- 59.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistaant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 60.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents. 2006;27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Higgins PG, Lehmann M, Seifert H. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. J. Antimicrob Agents. 2010;35:305. doi: 10.1016/j.ijantimicag.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 62.Van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, et al. Guildnes for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 2007;13:1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 63.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng Y, Yang P, Wang X, Zong Z. Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J. Antimicrob. Chemother. 2016;71:71–75. doi: 10.1093/jac/dkv324. [DOI] [PubMed] [Google Scholar]
- 65.http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html. Accessed May 15, 2019.
- 66.https://github.com/tseemann/shovill. Accessed February 4, 2020.
- 67.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, et al. Assembling genomes and mini-metagenomes from highly chimeric. Reads. 2013;7821:158–170. doi: 10.1007/978-3-642-37195-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;15:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.https://www.geneious.com. Accessed February 26, 2020.
- 70.Aziz, R. K. et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics.9, 75. 10.1186/1471-2164-9-75 (2008). [DOI] [PMC free article] [PubMed]
- 71.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 72.https://github.com/tseemann/abricate. Accessed January 10, 2020.
- 73.Hunt, M. et al. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom.3, e000131. 10.1099/mgen.0.000131 (2017). [DOI] [PMC free article] [PubMed]
- 74.Feldgarden M, Brover V, Haft DH, et al. Validating the AMR finder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019;63:e00483–e519. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, et al. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cosentino S, Voldby Larsen M, Møller Aarestrup F, Lund O. PathogenFinder - distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE. 2013;8:77302. doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.http://www.genomicepidemiology.org/. Accessed November 20, 2019.
- 79.https://card.mcmaster.ca/. Accessed October 18, 2019.
- 80.https://pubmlst.org/. Accessed August 12, 2019.
- 81.Siguier, P., Perochon, J., Lestrade, L., Mahillon, J. & Chandler, M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. https://isfinder.biotoul.fr/ (2006). [DOI] [PMC free article] [PubMed]
- 82.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016;4:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;4:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents. Chemother. 2010;54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.https://github.com/egonozer/in_silico_pcr. Accessed September 04, 2020.
- 86.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 87.Galata, V., Fehlmann, T., Backes, C. & Keller, A. PLSDB: A resource of complete bacterial plasmids, Nucleic Acids Res. 47, D195-D202. 10.1093/nar/gky1050 (2018). [DOI] [PMC free article] [PubMed]
- 88.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum-evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu M, Li X, Xie Y, Bi D, Sun J, Li J, et al. ICEberg 20: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019;47(1):660–665. doi: 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arndt, D., Grant, J., Marcu, A., Sajed, T., Pon, A., Liang, Y. & Wishart, D.S. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. 10.1093/nar/gkw387 (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A. baumannii strains are available from the authors.