Abstract
Administration of convalescent plasma or neutralizing monoclonal antibodies (mAbs) is a potent therapeutic option for coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. However, SARS-CoV-2 variants with mutations in the spike protein have emerged in many countries. To evaluate the efficacy of neutralizing antibodies induced in convalescent patients against emerging variants, we isolate anti-spike mAbs from two convalescent COVID-19 patients infected with prototypic SARS-CoV-2 by single-cell sorting of immunoglobulin-G-positive (IgG+) memory B cells. Anti-spike antibody induction is robust in these patients, and five mAbs have potent neutralizing activities. The efficacy of most neutralizing mAbs and convalescent plasma samples is maintained against B.1.1.7 and mink cluster 5 variants but is significantly decreased against variants B.1.351 from South Africa and P.1 from Brazil. However, mAbs with a high affinity for the receptor-binding domain remain effective against these neutralization-resistant variants. Rapid spread of these variants significantly impacts antibody-based therapies and vaccine strategies against SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, neutralizing antibody, mAb, variant
Graphical abstract
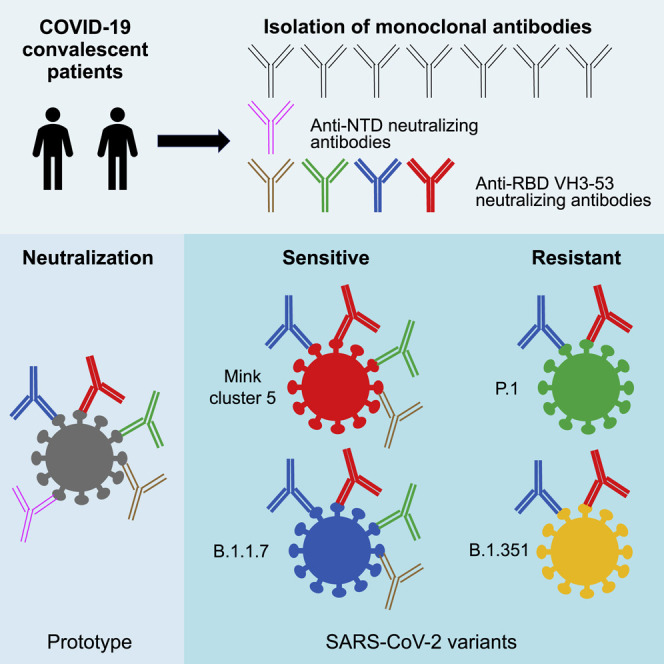
Kaku et al. demonstrate that the efficacy of neutralizing mAbs and convalescent plasma is maintained against SARS-CoV-2 variants B.1.1.7 from the UK and mink cluster 5 but decreases against B.1.351 from South Africa and P.1 from Brazil. Rapid spread of these variants significantly impacts SARS-CoV-2 therapies and vaccine strategies.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), which emerged in late 2019 and became a pandemic (Wu et al., 2020a; Zhou et al., 2020; Zhu et al., 2020). A potent option to treat patients with COVID-19 is the administration of plasma from patients who have recovered from COVID-19 (Duan et al., 2020; Li et al., 2020; Liu et al., 2020b). Neutralization of viruses by antibodies is considered the main mechanism to control COVID-19 by convalescent plasma. Currently, hundreds of monoclonal antibodies (mAbs) against SARS-CoV-2 have been isolated from convalescent COVID-19 patients (Barnes et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Kim et al., 2021; Kreye et al., 2020; Liu et al., 2020a), and potent neutralizing mAbs, which mostly target the receptor-binding domain (RBD) of the SARS-CoV-2 spike (S) protein, have been developed for clinical use (Schäfer et al., 2021; Weinreich et al., 2021).
New SARS-CoV-2 variants with multiple mutations in the S protein have emerged from the D614G variant (Elbe and Buckland-Merrett, 2017; Rambaut et al., 2020). Variants, including B.1.1.7 (also known as VOC-202012/01 or 501Y.V1) and B.1.351 (also known as 501Y.V2), have emerged in the UK and South Africa, respectively, and have since been detected in other countries (Leung et al., 2021; Tegally et al., 2021). P.1, In Brazil, a variant with unique mutations, has emerged in Brazil (Fujino et al., 2021; Maggi et al., 2021). It is important to clarify whether neutralizing antibodies from convalescent patients infected with the prototypic virus are effective against emerging SARS-CoV-2 variants for therapy using plasma or antibodies from convalescent patients. Therefore, we examined the sensitivity of neutralizing mAbs and plasma from convalescent patients to emerging SARS-CoV-2 variants.
Results
Isolation of neutralizing mAbs from two convalescent patients
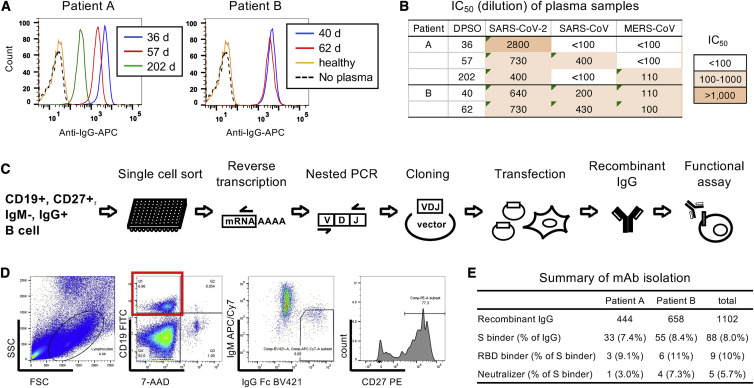
To identify potent neutralizing mAbs against SARS-CoV-2, we selected two patients, A and B, who had recovered from severe COVID-19 (Table S1). These patients were infected in March 2020, during the first wave of SARS-CoV-2 prevalence in Japan. Plasma samples from these patients had high binding activity to the SARS-CoV-2 Wuhan-Hu-1 S protein (Figures 1 A and S1A). Significant binding of plasma samples to S was observed even at a 1:1,000,000 dilution, although the level of binding declined 202 days post-symptom onset (DPSO) in patient A. Neutralizing activity against SARS-CoV-2, which was measured using an HIV-1-based pseudovirus with the SARS-CoV-2 S protein carrying 614G, was detected in these plasma samples (Figure 1B). The potency of plasma sample neutralizing activity correlated with their binding activity, but their neutralizing activity, determined by 50% of the maximal inhibitory concentration (IC50) values ranging from 1:640 to 1:2,800 dilution, was low compared with their strong binding activity (Figures 1A and S1A). Furthermore, these plasma samples had weak cross-neutralization activity against SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) (Figure 1B).
Figure 1.
Isolation of neutralizing mAbs from two convalescent patients
(A) Binding activity of plasma samples from patients A and B to the SARS-CoV-2 S protein was analyzed by flow cytometry. Binding of IgG to cells expressing SARS-CoV-2 S at ×10,000 dilution of plasma samples is shown as a histogram. Dotted line, no plasma control; orange line, plasma from a healthy donor.
(B) Neutralization activity of plasma samples analyzed using pseudoviruses expressing the S protein from SARS-CoV-2, SARS-CoV, and MERS-CoV. IC50 values (dilution) are summarized.
(C) Strategy to isolate neutralizing mAbs is shown schematically.
(D) A representative flow cytometry plot is shown. CD3−CD14−CD8− cells were used to sort memory B cells (7AAD−CD19+IgM−IgG+CD27+ cells), as shown in dotplots.
(E) Numbers of recombinant IgGs, S binders, RBD binders, and neutralizers from patients A and B are summarized.
We sorted IgG+ memory B cells from patients A and B, and mAbs were produced from the amplified immunoglobulin genes (Figures 1C and 1D). The recombinant antibodies were screened for their reactivity to the S protein of SARS-CoV-2 Wuhan-Hu-1 strain (Figure 1E). S-binding antibodies were examined for their neutralizing activity using a pseudovirus expressing SARS-CoV-2 S. Overall, 444 and 658 antibodies were isolated from the immunoglobulin-G-positive (IgG+) memory B cells of patients A and B, respectively (Figure 1E). Among the isolated antibodies, 33 (patient A, 7.4% of total antibodies) and 55 (patient B, 8.4% of total antibodies) were anti-S antibodies. One antibody (3.0% of anti-S antibodies) from patient A and four antibodies (7.3% of anti-S antibodies) from patient B neutralized SARS-CoV-2 pseudovirus (Figures 1E and 2 ).
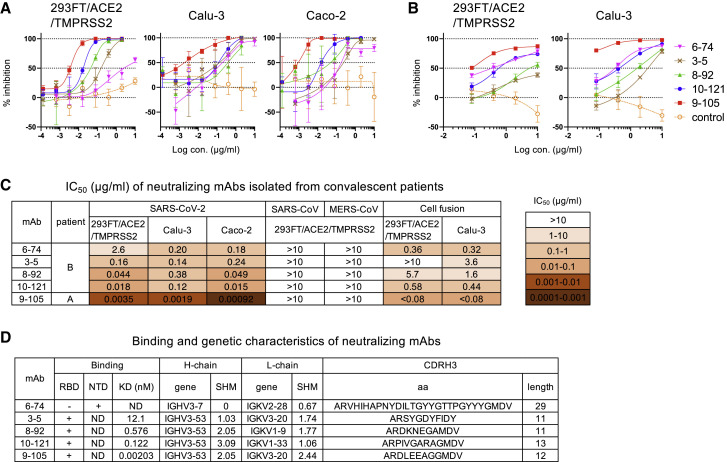
Figure 2.
Characterization of five neutralizing mAbs
(A) Neutralization of SARS-CoV-2 pseudovirus by mAbs 6-74, 3-5, 8-92, 10-121, and 9-105 in 293FT/ACE2/TMPRSS2, Calu-3 (human lung cancer cell line), and Caco-2 (human colon adenocarcinoma cell line) cells is shown. A non-neutralizing mAb, 8-38, was used as a negative control (orange open circle).
(B) Cell fusion of 293FT/DSP8-11/SARS-CoV-2-S cells with 293FT/DSP1-7/ACE2/TMPRSS2 and Calu-3/DSP1-7 cells was measured by luciferase activity at 6 and 20 h after coculture, respectively. A non-neutralizing mAb, 5-76, was used as a negative control (orange open circle).
(C) IC50 values of the five mAbs are summarized.
(D) The binding and genetic characteristics of mAbs are summarized. Binding to RBD and NTD was analyzed by AlphaScreen. KD values were determined by SPR analysis (Figure S1D). Gene usage (gene), somatic hypermutation % (SHM) of heavy and light chains, and CDRH3 amino acids (aa) and length were analyzed by IMGT vquest.
Data shown in (A) and (B) are represented as means ± SD (n = 3). See also Figure S1.
Characterization of five neutralizing mAbs
Among the five neutralizing antibodies, 9-105, which was isolated from patient A 57 DPSO, potently neutralized SARS-CoV-2 pseudovirus at IC50 values of 3.5 ng/mL in 293FT/ACE2/TMPRSS2 cells, 1.9 ng/mL in Calu-3 cells, and 0.92 ng/mL in Caco-2 cells (Figures 2A and 2C). Analysis of their reactivity to RBD and the N-terminal domain (NTD) revealed that four neutralizing mAbs targeted the RBD and one targeted the NTD (Figure 2D). Four RBD-targeting neutralizing antibodies, including 9-105, used the IGHV3-53 gene (Figure 2D), indicating that these mAbs were typical RBD-targeting neutralizing antibodies observed in patients with COVID-19 (Barnes et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Gaebler et al., 2021; Kim et al., 2021; Kreye et al., 2020; Wu et al., 2020b). Similar to the mAbs previously reported, these mAbs had a low somatic hypermutation rate ranging from 1.03% to 3.09% in the VH gene and 1.06% to 2.44% in the VK gene (Figure 2D). One non-RBD-targeting antibody, 6-74, bound to the NTD of SARS-CoV-2 S protein and had an extremely long CDRH3 (Figure 2D). Neutralization activity of 6-74 was low compared with the RBD-targeting mAbs (Figures 2A and 2C), but it had a strong inhibitory activity against SARS-CoV-2 S-mediated cell fusion (Figures 2B and 2C). All the neutralizing mAbs specifically neutralized SARS-CoV-2 pseudovirus but did not cross-neutralize SARS-CoV or MERS-CoV pseudovirus in 293FT/ACE2/TMPRSS2 cells (Figure 2C). No neutralizing activity of mAbs against MERS-CoV was also confirmed by infection of pseudovirus to Caco-2 cells, which express a receptor for MERS-CoV, DPP4 (data not shown).
The neutralization potency of mAbs was proportional to their binding activity to the S protein on the cell surface, although the NTD-targeting mAb, 6-74, bound to the S protein with a greater affinity than the RBD-targeting mAbs with low neutralizing activity (Figure S1C). The binding activity of 6-74 was equivalent to 10-121, which had a 100-fold greater neutralizing activity against SARS-CoV-2 in 293FT/ACE2/TMPRSS2 compared with 6-74 (Figures 2C and S1C). The neutralizing activity of RBD-targeting mAbs was also consistent with the results of surface plasmon resonance (SPR) analysis (Figure S1D). The most potent neutralizing mAb, 9-105, bound to RBD with a dissociation constant (KD) of 2.03 × 10−12 M (Figures 2D and S1D). RBD-targeting mAbs with lower KD values, indicating stronger binding activity, had higher neutralizing activity than RBD-targeting mAbs with higher KD values.
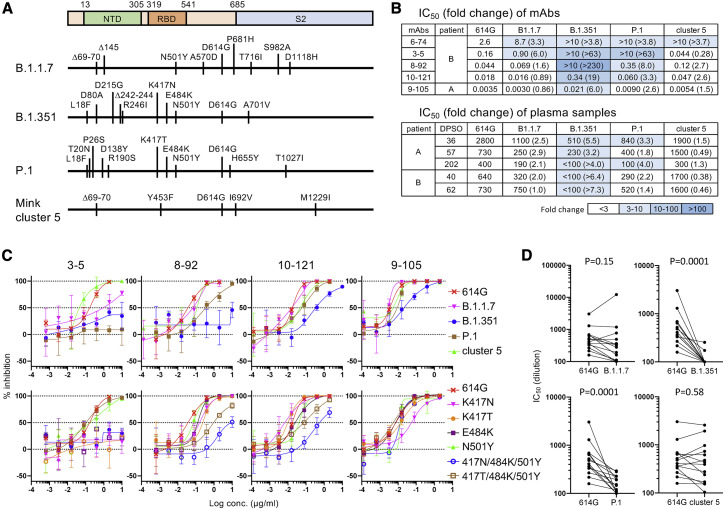
Neutralization activity against SARS-CoV-2 variants
The neutralization potency of mAbs and plasma samples was examined against pseudoviruses expressing S proteins from the emerging SARS-CoV-2 variants, B.1.1.7 from the UK, B.1.351 from South Africa, P.1 from Brazil, and mink cluster 5 from Denmark (Figure 3 ). Most mAbs and plasma samples neutralized B.1.1.7 and mink cluster 5 variants at the same level as the prototypic 614G pseudovirus. B.1.1.7 was slightly resistant to mAbs 6-74 (3.3-fold) and 3-5 (6.0-fold) and was marginally resistant to plasma samples (1.0- to 2.9-fold change). The NTD-targeting mAb 6-74 was not effective against mink cluster 5, but 3-5 and most plasma samples had high potency against this variant. Neutralization resistance was observed for P.1 and especially B1.351 (Figures 3B and 3C). P.1 was not neutralized by mAbs 6-74 and 3-5 and required a high concentration of the other mAbs (2.6- to 8.0-fold) and plasma samples (1.4- to 4.0-fold). B.1.351 was neutralized by mAbs 9-105 and 10-121, but not by mAbs 6-74, 3-5, and 8-92. The potencies of 9-105 and 10-121 were reduced against B.1.351 (6.0- and 19-fold, respectively). B.1.351 was not neutralized by plasma samples from patient B or 202 DPSO from patient A (Figure 3B). Plasma samples at 36 and 57 DPSO from patient A neutralized B.1.351, although the potency decreased 5.5- and 3.2-fold compared with that against the prototypic virus, respectively.
Figure 3.
Neutralizing activity against SARS-CoV-2 variants
(A) Amino acid substitutions in the S protein of SARS-CoV-2 variants B.1.1.7, B.1.351, P.1, and mink cluster 5 are schematically shown.
(B) IC50 values of mAbs (μg/mL) and plasma samples (dilution) against pseudoviruses with SARS-CoV-2 S variants are summarized with fold change (variant IC50 value/614G IC50 value).
(C) Neutralization of pseudoviruses with SARS-CoV-2 S variants (upper panels) and RBD mutants (lower panels) was examined by mAbs in 293FT/ACE2/TMPRSS2 cells. Data are represented as means ± SD (n = 3).
(D) IC50 values of plasma samples from 14 patients with COVID-19 other than patients A and B were compared between the 614G pseudovirus and pseudoviruses carrying the variant S protein. Statistical analysis was performed using a Wilcoxon matched-pairs signed rank test, and p values are shown.
All of the variants tested showed resistance to 6-74, suggesting that variants can easily escape from this mAb targeting the NTD, perhaps due to mutations in the NTD (Figure 3A). The efficacy of the RBD-targeting mAbs was decreased against P.1 and B.1.351, suggesting that the K417N/T, E484K, and N501Y mutations in the RBD region are critical for the resistance of these variants (Figure 3A). Analysis of single mutants revealed that K417N and K417T were critical for escape from 3-5 and slightly decreased the potency of 8-92 (Figure 3C, lower panels). E484K and N501Y single mutation did not confer resistance to these RBD-targeting mAbs. However, pseudoviruses with triple RBD mutations, especially the combination of K417N, E484K, and N501Y, were resistant to 3-5, 8-92, and 10-121. Interestingly, the potency of 9-105 was affected by K417N single mutation but neutralized triple mutants at the same level as prototype pseudovirus (Figure 3C, lower panels).
Neutralization resistance of P.1 and B.1.351 was observed in plasma samples from other patients with COVID-19, who were infected with SARS-CoV-2 before the spread of P.1 and B.1.351 in Japan (Figure 3D; Table S2). Analysis of plasma samples that had IC50 values greater than 1:100 against pseudovirus with SARS-CoV-2 S 614G revealed that neutralization sensitivity was maintained in the B.1.1.7 and mink cluster 5 variants but significantly decreased in the B.1.351 and P.1 variants. Only 2 of 14 plasma samples showed neutralizing activity against B.1.351, suggesting the high resistance of B.1.351 to neutralization by antibodies induced by prototypic SARS-CoV-2 infection (Figure 3D, upper right panel).
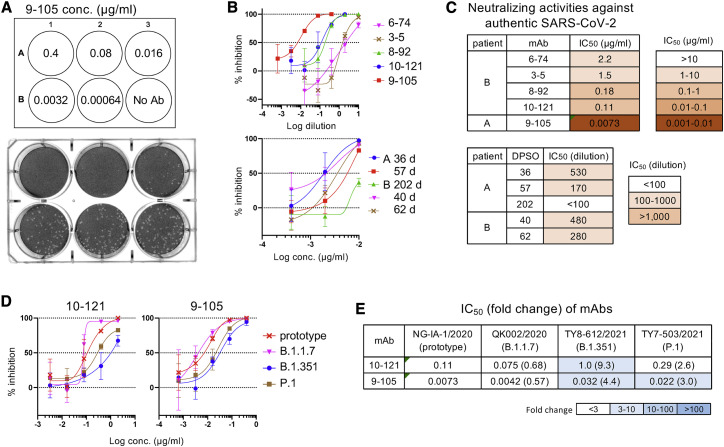
Neutralization of authentic SARS-CoV-2
The neutralizing activity of the five mAbs was also examined by infection of Vero cells with the authentic SARS-CoV-2 Japan/NGS-IA-1/2020 strain, which has an S protein identical to that of the Wuhan-Hu-1 strain. Consistent with the results using pseudovirus, 9-105 was the most potent mAb, and no plaque formation by infection was observed using 0.4 μg/mL 9-105 (Figure 4 A). The neutralization activity of plasma samples and mAbs against authentic virus basically corresponded with those against pseudovirus (Figures 2A, 2C, 4B, and 4C).
Figure 4.
Neutralization of authentic SARS-CoV-2
(A) A plaque assay was performed using the authentic SARS-CoV-2 Japan/NGS-IA-1/2020 strain and Vero cells. Representative result of plaque formation in the presence of mAb 9-105 is shown.
(B) Neutralization of authentic SARS-CoV-2 by mAbs and plasma samples are shown.
(C) IC50 values of mAbs and plasma samples are summarized.
(D) Neutralization of authentic SARS-CoV-2 variants B.1.1.7, B.1.351, and P.1 by mAbs 10-121 and 9-105 are shown.
(E) IC50 values of mAbs (μg/mL) against authentic SARS-CoV-2 variants are summarized with fold change (variant IC50 value/prototype IC50 value).
Data shown in (B) and (D) are represented as means ± SD (n = 3). See also Figure S2 for structure of 9-105 and S complex.
The neutralization activity of the potent mAbs 10-121 and 9-105 was also examined against the authentic SARS-CoV-2 variants B.1.1.7 (Japan/QK002/2020), B.1.351 (Japan/TY8-612/2021), and P.1 (Japan/TY7-503/2021). Japan/QK002/2020 has the S protein identical to B.1.1.7 pseudovirus. Japan/TY8-612/2021 has the most S mutations of B.1.351 pseudovirus but lacks L18F and R246I. Japan/TY7-503/2021 has the same S mutations as P.1 pseudovirus and additionally has the V1176F mutation. Consistent with analysis using pseudoviruses, 9-105 and 10-121 neutralized these variants, although the potencies were reduced against B.1.351 and P.1 (Figures 4D and 4E).
Discussion
The emergence of SARS-CoV-2 variants with multiple mutations in the S protein has raised concerns about the efficacy of antibodies elicited by infection or vaccination with prototypic SARS-CoV-2, which emerged in 2019. In this study, we examined the sensitivity of neutralizing mAbs and plasma samples from two convalescent patients to emerging SARS-CoV-2 variants. Four of the five neutralizing mAbs isolated from these patients were RBD-targeting VH3-53 antibodies, which are typically induced in patients with COVID-19 (Barnes et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Gaebler et al., 2021; Kim et al., 2021; Kreye et al., 2020; Wu et al., 2020b). The most potent mAb, 9-105, neutralized SARS-CoV-2 pseudovirus with IC50 values ranging from 0.92 to 3.5 ng/mL in various target cells, indicating that 9-105 is one of the most potent mAbs against SARS-CoV-2 reported to date (Barnes et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Kreye et al., 2020; Liu et al., 2020a).
Previous studies demonstrated that the sensitivity to plasma and mAbs isolated from convalescent patients or vaccinated individuals was moderately decreased against B.1.1.7 and markedly reduced against B.1.351 (Edara et al., 2021; Graham et al., 2021; Muik et al., 2021; Shen et al., 2021; Supasa et al., 2021; Tada et al., 2021; Wibmer et al., 2021; Wu et al., 2021). Consistent with these studies, our study showed a slight and severe decrease in the sensitivity of plasma and mAbs from convalescent patients against B.1.1.7 and B.1.351, respectively. Furthermore, we examined the sensitivity of other variants, P.1 and mink cluster 5, to antibodies. Mink cluster 5 was neutralized by plasma samples from convalescent patients at the same level as prototypic SARS-CoV-2, as reported previously (Tada et al., 2021; Wu et al., 2021). The neutralizing activity of mAbs was similar against prototypic and mink cluster 5 viruses, although 6-74, a mAb against the NTD region, did not neutralize mink cluster 5. In contrast, P.1 was resistant to most mAbs and some of the plasma samples. The resistance level of P.1 was between those of B.1.1.7 and B.1.351.
Variants were reported to be resistant to most NTD-targeting mAbs, including 6-74 examined in this study (Graham et al., 2021; McCallum et al., 2021; Wang et al., 2021). Mutations in the NTD, which are frequently observed in circulating variants, may occur as an escape from NTD-targeting neutralizing mAbs. On the other hand, mutations in the RBD of P.1 and B.1.351 marginally affected the potency of RBD-targeting mAbs when examined by point mutations, except for K417N/T, which resulted in escape from 3-5. The combination of three RBD mutations of P.1 and B.1.351 conferred a significant resistance to 8-92 and 10-121 but did not affect the potency of 9-105. Interestingly, 9-105 potency was slightly decreased by the K417N mutation but recovered to the same level as prototypic virus by the combination of three RBD mutations of P.1 and B.1.351 (Figure 3C). Resistance of B.1.351 to 9-105 may be responsible to the mutations outside the RBD.
The RBD-targeting mAbs tested in this study are typical VH3-53 antibodies, which share structural similarities (Barnes et al., 2020; Cao et al., 2020; Kim et al., 2021; Wu et al., 2020b). Structural analysis suggests that 9-105 binds the RBD region overlapping the ACE2-binding site by interactions similar to other RBD-targeting VH3-53 antibodies (Figure S2) (Barnes et al., 2020; Kaku et al., 2020; Wu et al., 2020b). The combination of RBD-targeting VH3-53 mAbs in this study did not show any synergy in neutralizing activity (data not shown), suggesting that these mAbs recognize the overlapping region in the RBD. The strong binding affinity of 9-105 and 10-121 to RBD may be one reason for cross-neutralizing activity against B.1.351 and P.1 variants. The decrease in RBD binding affinity with mutations may be complemented by interacting with multiple sites of the RBD.
SARS-CoV-2 has evolved, and new variants continue to emerge. Adaptation to humans, such as the conformational change in the S protein by the D614G mutation (Gobeil et al., 2021; Weissman et al., 2021; Yurkovetskiy et al., 2020), may enhance its transmission in humans. Moreover, variants that mutate to escape from antibody neutralization, which occurs during chronic infection (Kemp et al., 2021; Starr et al., 2021), as well as selection by the prototypic SARS-CoV-2-based vaccine, are a major concern for the treatment and prevention of SARS-CoV-2. It is important to monitor the emergence of new variants and identify the mutations associated with immune escape.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Biotin CD3 Monoclonal Antibody (UCHT1) | eBioscience | Cat#13-0038-82, RRID:AB_466323 |
| Biotin Mouse Anti-Human CD8 (RPA-T8) | BD | Cat#555365; RRID:AB_395768 |
| Biotin CD14 Monoclonal Antibody (61D3) | eBioscience | Cat#13-0149-82; RRID:AB_466373 |
| Anti-human CD19 Antibody FITC (HIB19) | BioLegend | Cat#302206; RRID:AB_314236 |
| Anti-human IgM Antibody APC/Cy7 (MHM-88) | BioLegend | Cat#314520; RRID:AB_10900422 |
| Anti-Human IgG BV421 (G18-145) | BD | Cat#562581; RRID:AB_2737665 |
| anti-human CD27 Antibody PE (M-T271) | BioLegend | Cat#356406; RRID:AB_2561825 |
| APC-conjugated AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG (H + L) | Jackson ImmunoResearch | Cat#109-136-088; RRID:AB_2337691 |
| Bacterial and virus strains | ||
| SARS-CoV-2 Japan/NGS-IA-1/2020 | Sakurai et al., 2021 | GISAID ID number: EPI_ISL_481251 |
| SARS-CoV-2 Japan/TY7-503/2021 | National Institute of Infectious Diseases, Japan | GISAID ID number: EPI_ISL_877769 |
| SARS-CoV-2 Japan/TY8-612-P1/2021 | National Institute of Infectious Diseases, Japan | GISAID ID number: EPI_ISL_1123289 |
| SARS-CoV-2 Japan/QK002/2020 | National Institute of Infectious Diseases, Japan | GISAID ID number: EPI_ISL_768526 |
| Stbl2 Competent Cells | Invitrogen | Cat#10268019 |
| Biological samples | ||
| Blood of SARS-CoV-2 infected patient A and B, See Table S1 | This paper | N/A |
| Plasma of SARS-CoV-2 infected individuals, See Table S2 | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| MojoSort Magnet | BioLegend | Cat#480019 |
| 7AAD | BD | Cat#559925 |
| guanidine thiocyanate | Invitrogen | Cat#AM9422 |
| dNTP-Mix (10mM) | Invitrogen | Cat#18427088 |
| Recombinant RNase inhibitor | Takara | Cat#2313B |
| Superscript III reverse transcriptase | Invitrogen | Cat#18080085 |
| Sensor Chip CM5 | cytiva | Cat#BR100030 |
| Human Antibody Capture Kit | cytiva | Cat#BR100839 |
| Amine Coupling Kit | cytiva | Cat#BR100050 |
| G418 | Millipore | Cat#345812-20ML |
| Hygromycin B | Nacalai Tesque | Cat#09287-84 |
| RBD | This paper | N/A |
| NTD | This paper | N/A |
| Protein A AlphaScreen Donor Beads | PerkinElmer | Cat#AS102D |
| Anti-His AlphaLISA Acceptor beads | PerkinElmer | Cat#AL178C |
| Carboxymethyl cellulose | Sigma | Cat#C9481-500G |
| Methylene blue | Nacalai Tesque | Cat#22412-14 |
| Critical commercial assays | ||
| FectoPRO | Polyplus-transfection | Cat#116-010 |
| FectoCHO Expression System | Polyplus-transfection | Cat#716-06LKIT |
| Luciferase assay system | Promega | Cat#E4550 |
| EnduRen | Promega | Cat#E6481 |
| Gibson Assembly Master Mix | New England Biolabs | Cat#E2611L |
| HiTrap rProtein A FF Column | cytiva | Cat#17507901 |
| COSMOGEL His-Accept | Nacalai Tesque | Cat#09277-56 |
| Superdex 75 Increase 10/300 GL | cytiva | Cat#29148721 |
| Deposited data | ||
| Antibody nucleotide sequence | This paper | GenBank accession number: MZ089617 to MZ089626 |
| Structure of antibody (COVA2-04 or COVA2-39)-RBD complex | Wu et al., 2020b | PDB: 7JMO and 7JMP |
| Structure of 9-105-RBD complex | This paper | N/A |
| Experimental models: cell lines | ||
| 293T | ATCC | CRL-3216 |
| 293A | Invitrogen | Cat#R70507 |
| 293FT/DSP1-7/ACE2/TMPRSS2 | Yamamoto et al., 2020 | N/A |
| 293FT/DSP8-11/SARS-CoV-2-S | Yamamoto et al., 2020 | N/A |
| Calu3-DSP1-7 | Yamamoto et al., 2020 | N/A |
| Calu-3 | ATCC | HTB-55 |
| Caco-2 | ATCC | HTB-37 |
| Vero | ATCC | CCL-81 |
| ExpiCHO-S | GIBCO | Cat#A29127 |
| Oligonucleotides | ||
| Primer: 3′ Cγ CH1: GGAAGGTGTGCACGCCGCTGGTC | Tiller et al., 2008 | N/A |
| Primer: HcnestU: GG(ACTAGT)TCTTGTCCACCTTGGTGTTG | Coronella et al., 2000 | N/A |
| Primer: P1-Cκ (modified): CAGCAGGCACACAACAGAGGCAGTTCC | He et al., 2014 | N/A |
| Primer: 3′ Cλ: CACCAGTGTGGCCTTGTTGGCTTG | Tiller et al., 2008 | N/A |
| Primer: Lnest: GCTCTAGAACTAATGCGTGACCTGGCAGCTGT | Coronella et al., 2000 | N/A |
| Recombinant DNA | ||
| pIgGH | Ramirez Valdez et al., 2015 | N/A |
| pKVA2 | Ramirez Valdez et al., 2015 | N/A |
| pLSH | Ramirez Valdez et al., 2015 | N/A |
| SARS-CoV-2 Spike ORF expression plasmid | Sino Biological | Cat#VG40589-UT |
| pSARS-CoV-2-S-IRES-EGFP | This paper | N/A |
| psPAX2-IN/HiBiT | Ozono et al., 2020 | N/A |
| pWPI-Luc2 | Ozono et al., 2020 | N/A |
| pC-SARS2-S-D614G | Ozono et al., 2021 | N/A |
| pSARS-CoV-2-S-D614G-19del | This paper | N/A |
| SARS-CoV S-expressing plasmid | Sino Biological | Cat#VG40150-G-N |
| MERS-CoV S-expressing plasmid | Sino Biological | Cat#VG40069-G-N |
| Plasmids to express pSARS-CoV-2 S variants | This paper | N/A |
| Plasmids to express pSARS-CoV-2 S mutants | This paper | N/A |
| Software and algorithms | ||
| IMGT vquest | Brochet et al., 2008 | http://imgt.org/ |
| PyIR | Soto et al., 2020 | https://github.com/crowelab/PyIR |
| FlowJo | TreeStar | Version 10.7.1 https://www.flowjo.com |
| BIAevaluation | GE healthcare | Version 4.1 http://www.cytivalifesciences.com/country-selection?originalItemPath=%2f |
| Prism | GraphPad Software | Version 8.4.3 https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shuzo Matsushita (shuzo@kumamoto-u.ac.jp).
Materials availability
Further information and requests for resources and reagents should be directed to Shuzo Matsushita (shuzo@kumamoto-u.ac.jp).
Data and code availability
-
•
Sequence data for antibodies have been deposited at GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Sample collection from patients with COVID-19
Peripheral blood samples were obtained from donors hospitalized in Kumamoto City Hospital and Kyushu Medical Center, who recovered from COVID-19 disease. Peripheral blood samples for antibody isolation were obtained from patients A (male) and B (male), and the information of patients were in Table S1. Age of these patients are undisclosed due to privacy policy. Information of patients with COVID-19 other than patients A and B is in Table S2. The study protocol was approved by the institutional ethical review boards of Kumamoto City Hospital (546), Kyushu Medical Center (20C120) and Kumamoto University (2013 and 2066). All the study participants provided their written informed consent for the collection of the samples and their subsequent analysis. All the patient samples were collected before the emergence of variants tested in this study.
Cell lines
Human embryonic kidney cells, 293T, 293A, 293FT/DSP1-7/ACE2/TMPRSS2 and 293FT/DSP8-11/SARS-CoV2-S, were maintained in high glucose DMEM (Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich). Calu-3 (human lung cancer cell line), Calu-3-DSP1-7 and Caco-2 (human colon adenocarcinoma cell line) cells were maintained in EMEM with non-essential amino acids (Fujifilm) supplemented with 20% FBS. Vero cells were maintained in EMEM (Fujifilm) supplemented with 5% FBS. ExpiCHO-S cells were maintained in ExpiCHO Expression Medium (Invitrogen).
Method details
Isolation of IgG+ memory B cells from patients with COVID-19
Samples collected from patient A 36 and 57 DPSO and patient B 40 DPSO were used for isolation of mAbs. Peripheral blood mononuclear cells were isolated by Ficoll density gradient centrifugation and B cells were enriched by negative selection using antibodies against CD3 CD8 and CD14 nd MojoSort Magnet. The enriched B cells were stained with 7AAD, CD19-FITC, IgM-APC/Cy7, IgG-BV421 and CD27-PE. IgG+ memory B cells (7AAD-CD19+IgM-IgG+CD27+ cells) were sorted at single cell density into 5 μl/well of 50 μM ice-cold guanidine thiocyanate.
Cloning of immunoglobulin variable genes
Reverse transcription was performed in a total volume of 20 μl/well in a 96-well sorting plate containing 1.4 μL of 100 μM gene specific primers (Coronella et al., 2000; He et al., 2014; Tiller et al., 2008), 2 μL of 10 mM each dNTP-Mix, 1 μL of 0.1 M DTT, 20 U recombinant RNase inhibitor, 4 μL of 5X First-Strand Buffer and 50 U Superscript III reverse transcriptase as follows: 42°C for 10 min, 25°C for 10 min, 50°C for 60 min and 94°C for 5 min.
Nested PCR was performed for cloning of immunoglobulin variable genes, as previously described (Ramirez Valdez et al., 2015). IgG heavy and light chain expression plasmids were constructed by the homologous recombination of the nested PCR products with pIgGH, pKVA2 and pLSH, respectively using the Gibson Assembly Master Mix. The nucleotide sequences of the immunoglobulin variable regions were analyzed for germline gene verification, framework and CDR mapping and quantification of percent identity to germline using IMGT vquest (Brochet et al., 2008) and PyIR, which is an IgBLAST wrapper and parser (Soto et al., 2020).
Production and purification of recombinant IgG
Recombinant IgG was produced and purified as previously described (Ramirez Valdez et al., 2015). Briefly, a pair of heavy and light chain plasmids were transfected into 293T cells using FectoPRO and the supernatant 2 days post transfection was used for the screening of antibodies reactive to SARS-CoV-2 S. Many antibodies were produced by the transient transfection of ExpiCHO cells with the FectoCHO Expression Kit or the establishment of 293A cells stably expressing IgG, which were cloned from colonies selected with G418 (1000 μg/ml) and hygromycin (150 μg/ml) from the cells transfected with heavy and light chain plasmids. IgG were purified using a HiTrap rProtein A FF Column.
Analysis of the binding activity of antibodies by flowcytometry
The binding activity of antibodies was analyzed using cells expressing SARS-CoV-2 S and enhanced green fluorescent protein (EGFP). The internal ribosome entry site (IRES) and the EGFP gene were inserted into a SARS-CoV-2 Spike ORF mammalian expression plasmid, and the resultant plasmid, pSARS-CoV-2-S-IRES-EGFP, was used for the transfection of 293T and 293A cells. Transiently or stably transfected cells were stained with primary antibody for 15 min at room temperature (RT). The cells were washed twice with PBS containing 0.2% BSA and incubated with APC-conjugated AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG (H + L) for 15 min at RT. The stained cells were analyzed by FACSCanto II (BD Biosciences). The reactivity of antibodies was analyzed after gating on EGFP+ cells using FlowJo.
Analysis of the binding activity of mAbs by surface plasmon resonance
Surface plasmon resonance (SPR) experiments were performed using a Biacore 3000 system (cytiva). For the mAb 9-105, anti-human IgG antibody was immobilized on a CM5 sensor chip by amine-coupling using a human Antibody capture kit, and then the mAb was immobilized on an anti-human IgG antibody-coupled chip. The RBD-9-105 complex was removed for regeneration because the bound RBD was not released from 9-105 under conventional regeneration conditions. Other mAbs were immobilized on the CM5 sensor chip by amine-coupling using an amine-coupling kit. Purified RBD (2–2500 nM) in HBS-P buffer (10 mM HEPES, 150 mM NaCl, 0.005% surfactant P20, pH 7.4) was injected over the immobilized mAbs. The binding response at each concentration was calculated by subtracting the equilibrium response measured in the control flow cell from the response in each sample flow cell. The data were analyzed using BIAevaluation version 4.1 software and K d values were determined by equilibrium analyses using nonlinear curve fitting of the Langmuir binding isotherm.
Analysis of the binding activity of mAbs to RBD and NTD
Protein expression and purification of the S-protein RBD (amino acid number, P322 to N536) and NTD (amino acid number, S12 to S305) were performed as previously described for other viral glycoproteins (Kubota et al., 2016). Briefly, an expression plasmid encoding RBD or NTD was transiently transfected into 80% confluent HEK293 cells. At 6 days post-transfection, supernatant containing the secreted protein was harvested. Then, RBD or NTD was purified using COSMOGEL His-Accept in purification buffer (50 mM NaH2PO4, 150 mM NaCl, 10 mM imidazole, pH 8.0), and eluted with elution buffer (50 mM NaH2PO4, 150 mM NaCl, 500 mM imidazole, pH 8.0). RBD or NTD was further purified using a gel filtration column Superdex 75 Increase 10/300 GL in buffer containing 100 mM NaCl and 20 mM Tris-HCl (pH 8.0).
An AlphaScreen assay was performed as described in the manufacturer’s protocol (PerkinElmer). In brief, 5 μL of recombinant IgG and supernatant from the transfected cells, were mixed with 50 ng of RBD or NTD. After incubation at 37°C for 40 min, 12.5 μg of Protein A AlphaScreen Donor Beads and Anti-6xHis AlphaLISA Acceptor beads were added to the reaction mix, respectively. After incubation at 37°C for 40 min, the AlphaScreen score was measured by EnSpire Multimode Plate Reader (PerkinElmer).
Neutralization assay using pseudovirus
The neutralization activity of antibodies was determined using HIV-1-based pseudovirus with SARS-CoV-2 S, as previously described (Ozono et al., 2020, 2021) In brief, 293T cells were transfected with lentiviral packaging plasmid psPAX2-IN/HiBiT, the firefly luciferase-expressing lentiviral transfer plasmid pWPI-Luc2 and a plasmid expressing SARS-CoV-2 S. We used pSARS-CoV-2-S-D614G-19del expressing S with the D614G mutation and a 19 amino acid deletion in the cytoplasmic tail, which was constructed from an S-expressing plasmid and pC-SARS2-S-D614G (Ozono et al., 2021), as a prototypic S. Plasmids to express S of variants and mutants were constructed by PCR-mutagenesis from pSARS-CoV-2-S-D614G-19del. Supernatant after 48 h of transfection was stored at ˗80°C. The median tissue culture infectious dose (TCID50) of each pseudovirus was determined by infection with serially-diluted viruses. The neutralization assay was performed in triplicate. Serially diluted antibody and virus (200 TCID50) were incubated for 1 h and transferred to cells plated at 1-2x104 cells/well in a 96-well plate 1 day before infection. After incubation for 2 h, medium was changed to fresh medium, and incubated for an additional 48 h. Luciferase activity was measured using luciferase assay system and EnSpire Multimode Plate Reader (PerkinElmer). The relative light units (RLU) were compared to calculate the reduction in infectivity and 50% of the maximal inhibitory concentration (IC50) was calculated using Prism 8.
DSP assay to monitor cell fusion
Cell fusion was analyzed using 293FT/DSP1-7/ACE2/TMPRSS2, 293FT/DSP8-11/SARS-CoV-2-S and Calu-3/DSP1-7 cells, as previously described (Yamamoto et al., 2020). Briefly, 293FT/DSP1-7/ACE2/TMPRSS2 and Calu-3/DSP1-7 cells, which were seeded in a 6-well plates (9 × 105 cells/3 ml) one day before the assay, were treated with 6 μM EnduRen. Serially-diluted mAb or plasma were added to a96-well black plate, in which 293FT/DSP8-11/SARS-CoV-2-S cells (3x104 cells/100 μl) were seeded one day before. After 2-4 h of EnduRen incubation, cells were detached and the 50 μl cell suspension was transferred to a 96-well black plate. Luciferase activity was measured periodically using EnSpire Multimode Plate Reader. The RLU were compared to calculate the reduction in infectivity and IC50 was calculated using Prism 8. Results of cell fusion using 293FT/DSP1-7/ACE2/TMPRSS2 and Calu-3/DSP1-7 cells at 6 h and 20 h after coculture, respectively, are shown.
Neutralization of authentic virus
Plaque assays were performed using Japan/NGS/IA-1/2020 (Sakurai et al., 2021), Japan/TY7-503/2021, Japan/TY8-612-P1/2021, and Japan/QK002/2020 strains of SARS-CoV-2. Vero cells were plated at 5x105 cells/well in a 6-well plate or 2x105 cells/well in a 12-well plate 1 day before experiment. Plasma or mAb was serially diluted in virus dilution buffer containing 1x MEM, 20 mM HEPES, 1x NEAA, and 1x penicillin and streptomycin. The diluted mAb or plasma (110 μl) was added to the same volume as 110 PFU of SARS-CoV-2, and incubated for 1 h. In 6-well plate, 50μl or 150 μl of the mixture was added to Vero cells supplemented with 950 μl or 850 μl of dilution buffer, respectively. In 12-well plate, 40μl or 160 μl of the mixture was added to Vero cells supplemented with 460 μl or 340 μl of dilution buffer, respectively. After incubation for 2 h, 2 and 1 mL PFU buffer containing 1x MEM, 3% FBS, and 1.5% carboxymethyl cellulose was overlaid to 6-well plate and 12-well plate, respectively. After further incubation for 3 days, wells were washed three times with PBS, and cells were fixed with 4% paraformaldehyde in PBS. Wells were washed with water, dried, and stained with 0.1% methylene blue to visualize the plaques. IC50 values were calculated using Prism 8.
Quantification and statistical analysis
Neutralization and fusion inhibition data are processed by Prism 8, and shown by the means ± SD of triplicates. The statistical analysis for comparison of IC50 values were performed using Prism 8 by Wilcoxon matched-pairs signed rank test where p < 0.05 are considered significant.
Acknowledgments
We thank all the clinical staff who provided care for the patients in Kumamoto City Hospital and Kyusyu Medical Center. We thank Dr. Misumi and Dr. Shuto (Kumamoto University) and Dr. Gohda (University of Tokyo) for providing cell lines, Dr. Yasuda (Nagasaki University) for providing authentic SARS-CoV-2, Dr. Ito (National Institute of Infectious Diseases) for providing authentic SARS-CoV-2 variants, and Dr. Tokunaga (National Institute of Infectious Diseases) for providing plasmids for pseudovirus production. We also thank Dr. Oshiumi (Kumamoto University) for cooperation in obtaining authentic SARS-CoV-2 variants and Dr. Croxford (Edanz Group) for editing a draft of this manuscript. This study was supported by the Japan Agency for Medical Research and Development (grant JP20fk0108271 to S.M., T.K., Y. Maeda, and T.T; grant JP19fk0108111 to T.H.; and grant JP20fk0108270 to N.T. and Y. Koyanagi); a grant from the Senshin Medical Research Foundation to S.M.; Ministry of Education, Culture, Sports, Science and Technology KAKENHI grants 20H05773 and 20K20596 to T.H.; a grant from the JST Core Research for Evolutional Science and Technology (20356730) and the JSPS Core-to-Core Program A, the Advanced Research Networks to T.N. and Y. Koyanagi; an intramural grant from Kumamoto University COVID-19 Research Projects (AMABIE) to Y. Maeda, T.I., and C.M.; Kumamoto University International Collaborative Research Grants to T.U.; and the Joint Usage/Research Center program of the Institute for Frontier Life and Medical Sciences Kyoto University (S.M. and Y. Koyanagi).
Author contributions
Y. Kaku, T.K., and S.M. designed the experiments. H.I., Y.N., R.M., C.M, and M.T. collected patient samples. Y. Kaku isolated B cells. Y. Kaku., H.M.Z., N.K., S.B., K.M. M.S., Y.K., K.S., and C.O. performed antibody cloning. T.H., T.S., J.S., T.N, Y. Muramoto, Y. Kaku, and T.T produced proteins and performed binding assay. T.I., M.T., and T.K. prepared authentic viruses. T.K. performed neutralization assays. Y. Kaku, T.K., Y. Maeda, T.U., Y. Koyanagi, and S.M. interpreted data. Y. Kaku., T.K., and S.M. prepared the figures and wrote the paper. S.M. designed and coordinated the study.
Declaration of interests
Y. Kaku, T.K., and S.M. are listed as inventors on a patent application related to this work. The remaining authors declare no competing interests.
Published: July 13, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109385.
Supplemental information
References
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X., Lefranc M.-P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503-8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronella J.A., Telleman P., Truong T.D., Ylera F., Junghans R.P. Amplification of IgG VH and VL (Fab) from single human plasma cells and B cells. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.20.e85. E85–E85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.V., Floyd K., Lai L., Gardner M., Hudson W., Piantadosi A., Waggoner J.J., Babiker A., Ahmed R., Xie X. Infection and mRNA-1273 vaccine antibodies neutralize SARS-CoV-2 UK variant. medRxiv. 2021 2021.02.02.21250799. [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T., Nomoto H., Kutsuna S., Ujiie M., Suzuki T., Sato R., Fujimoto T., Kuroda M., Wakita T., Ohmagari N. Novel SARS-CoV-2 Variant Identified in Travelers from Brazil to Japan. Emerg. Infect. Dis. J. 2021;27:1243–1245. doi: 10.3201/eid2704.210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil S.M.C., Janowska K., McDowell S., Mansouri K., Parks R., Manne K., Stalls V., Kopp M.F., Henderson R., Edwards R.J. D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction. Cell Rep. 2021;34:108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham C., Seow J., Huettner I., Khan H., Kouphou N., Acors S., Winstone H., Pickering S., Galao R.P., Lista M.J. Impact of the B.1.1.7 variant on neutralizing monoclonal antibodies recognizing diverse epitopes on SARS-CoV-2 Spike. bioRxiv. 2021 doi: 10.1016/j.immuni.2021.03.023. 2021.02.03.429355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Sok D., Azadnia P., Hsueh J., Landais E., Simek M., Koff W.C., Poignard P., Burton D.R., Zhu J. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci. Rep. 2014;4:6778. doi: 10.1038/srep06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku Y., Kuwata T., Gorny M.K., Matsushita S. Prediction of contact residues in anti-HIV neutralizing antibody by deep learning. Jpn. J. Infect. Dis. 2020;73:235–241. doi: 10.7883/yoken.JJID.2019.496. [DOI] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Noh J., Kim S., Choi Y., Yoo D.K., Lee Y., Lee H., Jung J., Kang C.K., Song K.H. Stereotypic neutralizing VH antibodies against SARS-CoV-2 spike protein receptor binding domain in patients with COVID-19 and healthy individuals. Sci. Transl. Med. 2021;13:eabd6990. doi: 10.1126/scitranslmed.abd6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota M., Takeuchi K., Watanabe S., Ohno S., Matsuoka R., Kohda D., Nakakita S., Hiramatsu H., Suzuki Y., Nakayama T. Trisaccharide containing α2,3-linked sialic acid is a receptor for mumps virus. Proc. Natl. Acad. Sci. U S A. 2016;113:11579–11584. doi: 10.1073/pnas.1608383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26:2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., Kong Y., Ren L., Wei Q., Mei H. Effect of Convalescent Plasma Therapy on Time to Clinical Improvement in Patients With Severe and Life-threatening COVID-19: A Randomized Clinical Trial. JAMA. 2020;324:460–470. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu S.T.H., Lin H.-M., Baine I., Wajnberg A., Gumprecht J.P., Rahman F., Rodriguez D., Tandon P., Bassily-Marcus A., Bander J. Convalescent plasma treatment of severe COVID-19: a propensity score-matched control study. Nat. Med. 2020;26:1708–1713. doi: 10.1038/s41591-020-1088-9. [DOI] [PubMed] [Google Scholar]
- Maggi F., Novazzi F., Genoni A., Baj A., Spezia P.G., Focosi D., Zago C., Colombo A., Cassani G., Pasciuta R. Imported SARS-COV-2 Variant P.1 Detected in Traveler Returning from Brazil to Italy. Emerg. Infect. Dis. J. 2021;27:1249–1251. doi: 10.3201/eid2704.210183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Tobiume M., Kishigami S., Tokunaga K. Super-rapid quantitation of the production of HIV-1 harboring a luminescent peptide tag. J. Biol. Chem. 2020;295:13023–13030. doi: 10.1074/jbc.RA120.013887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S., Zhang Y., Ode H., Sano K., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez Valdez K.P., Kuwata T., Maruta Y., Tanaka K., Alam M., Yoshimura K., Matsushita S. Complementary and synergistic activities of anti-V3, CD4bs and CD4i antibodies derived from a single individual can cover a wide range of HIV-1 strains. Virology. 2015;475:187–203. doi: 10.1016/j.virol.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Sakurai Y., Ngwe Tun M.M., Kurosaki Y., Sakura T., Inaoka D.K., Fujine K., Kita K., Morita K., Yasuda J. 5-amino levulinic acid inhibits SARS-CoV-2 infection in vitro. Biochem. Biophys. Res. Commun. 2021;545:203–207. doi: 10.1016/j.bbrc.2021.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:218. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29:529–539.e3. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., Finn J.A., Willis J.R., Day S.B., Sinkovits R.S., Jones T., Schmitz S., Meiler J., Branchizio A., Crowe J.E., Jr. PyIR: a scalable wrapper for processing billions of immunoglobulin and T cell receptor sequences using IgBLAST. BMC Bioinformatics. 2020;21:314. doi: 10.1186/s12859-020-03649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic-Golden M., Herati R.S., Cornelius A., Mulligan M.J., Landau N.R. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. BioRxiv. 2021 2021.02.05.430003. [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., van Gils M.J. An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. BioRxiv. 2021 2021.01.25.427948. [Google Scholar]
- Yamamoto M., Kiso M., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Imai M., Takeda M., Kinoshita N., Ohmagari N., Gohda J., Semba K. The Anticoagulant Nafamostat Potently Inhibits SARS-CoV-2 S Protein-Mediated Fusion in a Cell Fusion Assay System and Viral Infection In Vitro in a Cell-Type-Dependent Manner. Viruses. 2020;12:629. doi: 10.3390/v12060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Sequence data for antibodies have been deposited at GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.