Abstract
Vitamin A deficiency remains prevalent in parts of Asia, Latin America, and sub-Saharan Africa where maize (Zea mays) is a food staple. Extensive natural variation exists for carotenoids in maize grain. Here, to understand its genetic basis, we conducted a joint linkage and genome-wide association study of the US maize nested association mapping panel. Eleven of the 44 detected quantitative trait loci (QTL) were resolved to individual genes. Six of these were correlated expression and effect QTL (ceeQTL), showing strong correlations between RNA-seq expression abundances and QTL allelic effect estimates across six stages of grain development. These six ceeQTL also had the largest percentage of phenotypic variance explained, and in major part comprised the three to five loci capturing the bulk of genetic variation for each trait. Most of these ceeQTL had strongly correlated QTL allelic effect estimates across multiple traits. These findings provide an in-depth genome-level understanding of the genetic and molecular control of carotenoids in plants. In addition, these findings provide a roadmap to accelerate breeding for provitamin A and other priority carotenoid traits in maize grain that should be readily extendable to other cereals.
Eleven genes were identified for grain carotenoids by integrating joint-linkage, genome-wide association, RNA-seq, pleiotropy, and epistasis analyses of the US maize nested association mapping panel.
Introduction
Carotenoids are lipid-soluble isoprenoids (typically C40) that are synthesized by plants, algae, and some fungi, bacteria, and yeast (reviewed in Li et al., 2016). Most carotenoids have yellow, orange, or red colors that are a function of the length of their conjugated double bond system and functional groups (Khoo et al., 2011). Carotenoids containing oxygen functional groups are termed xanthophylls, and those without such groups are termed carotenes. In plants, carotenoids are biosynthesized and localized in plastids, where they play numerous roles in photosystem structure and light harvesting and in photoprotection through their scavenging of singlet oxygen and dissipation of excess excitation energy via the xanthophyll cycle (Jahns and Holzwarth, 2012). Additionally, 9-cis isomers of violaxanthin and neoxanthin are precursors for the biosynthesis of abscisic acid (ABA), a plant hormone with critical roles in embryo dormancy and abiotic stress responses (Kermode, 2005; Tuteja, 2007), while 9-cis β-carotene is the substrate for synthesis of strigolactones; these recently discovered plant hormones are involved in branching and in attracting beneficial arbuscular mycorrhizae (reviewed in Al-Babili and Bouwmeester, 2015; Jia et al., 2018).
Provitamin A carotenoids are an essential micronutrient in human and animal diets, as they are converted to vitamin A (retinol) in the body via oxidative cleavage (reviewed in Eroglu and Harrison, 2013). The most abundant provitamin A carotenoids in the human diet are β-carotene, which yields two molecules of retinol, and β-cryptoxanthin and α-carotene, which yield one (Stahl and Sies, 2005; Combs and McClung, 2017). Clinical vitamin A deficiency affects an estimated 127.2 million preschool children and 7.2 million pregnant women in countries determined to be at risk (West, 2002). Symptoms can include xerophthalmia (“dry eye”), which often progresses to night blindness, as well as increased morbidity and mortality from infections (reviewed in West and Darnton-Hill, 2008). It is estimated that vitamin A deficiency is responsible for the deaths of approximately 650,000 preschool children per year (Rice et al., 2004). Two nonprovitamin A xanthophylls, lutein and zeaxanthin, also play important roles as macular pigments in the human fovea (Beatty et al., 1999; Krinsky et al., 2003; Bernstein and Arunkumar, 2020). Elevated dietary intake of these xanthophylls has been associated with decreased risk of age-related macular degeneration (AMD; Abdel-Aal el et al., 2013; Bernstein and Arunkumar, 2020), which affected an estimated 170 million adults in 2014 and is projected to increase as the global population ages (Wong et al., 2014).
Maize (Zea mays) is a primary food staple in much of Latin America, sub-Saharan Africa, and Asia, where vitamin A deficiency remains highly prevalent (West, 2002). There is extensive natural variation in the levels of maize grain carotenoids, which are most highly concentrated in the vitreous (hard) portion of the endosperm (Blessin et al., 1963; Weber, 1987; Harjes et al., 2008). However, as a dietary staple, the average provitamin A carotenoid levels of diverse yellow maize lines provide less than 20% of the target level established based on recommended dietary allowances (Harjes et al., 2008; Bouis and Welch, 2010; Owens et al., 2014). Genetic improvement of maize grain carotenoid (provitamin A) levels through breeding, an example of biofortification, has been proposed as a cost-effective approach for ameliorating vitamin A deficiency in at-risk populations (Graham et al., 2001; Welch and Graham, 2004; Bouis and Welch, 2010; Diepenbrock and Gore, 2015).
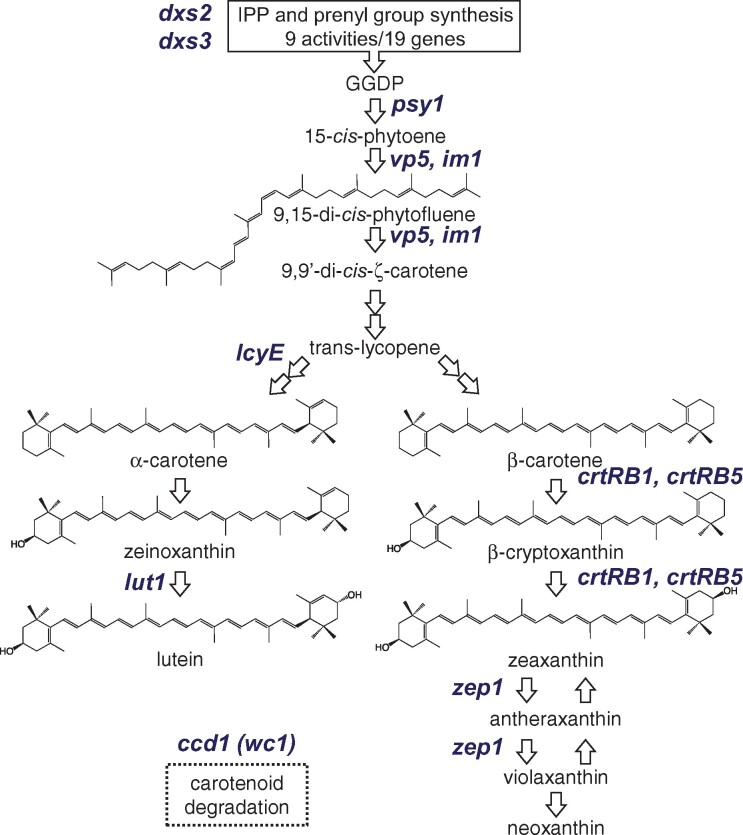
Carotenoids are derived from the five-carbon central intermediate isopentenyl pyrophosphate (IPP) produced by the plastid-localized methyl-d-erythritol-4-phosphate (MEP) pathway (Figure 1). The committed step toward carotenoid biosynthesis is the head-to-head condensation of two 20-carbon geranylgeranyl diphosphate (GGDP) molecules by the enzyme phytoene synthase to form phytoene. Phytoene is sequentially desaturated and isomerized to form lycopene, which is then cyclized with two β-rings to form β-carotene or with one β-ring and one ε-ring to form α-carotene. α- and β-carotenes are hydroxylated twice to form lutein and zeaxanthin, respectively, and zeaxanthin is further modified to yield violaxanthin and neoxanthin. The MEP and carotenoid biosynthetic pathways are well characterized in Arabidopsis (Arabidopsis thaliana), and the genes are highly conserved across plants, enabling the straightforward identification of homologs in other species. In maize, the MEP and carotenoid pathways are encoded by 59 genes, which can be considered a priori candidates for influencing natural variation in maize grain carotenoid levels (Supplemental Data Set S1). Four of these a priori candidate genes—lycopene epsilon cyclase (lcyE), β-carotene hydroxylase 1 (crtRB1), zeaxanthin epoxidase 1 (zep1), and ε-ring hydroxylase (lut1)—have been shown to have genome-wide associations with various carotenoid traits in maize grain (Harjes et al., 2008; Yan et al., 2010; Owens et al., 2014; Suwarno et al., 2015; Azmach et al., 2018; Baseggio et al., 2020). zep1 encodes zeaxanthin epoxidase, which converts zeaxanthin to violaxanthin via antheraxanthin and was associated with zeaxanthin and total β-xanthophyll levels, with large explanation of phenotypic variance for these traits (Owens et al., 2014; Suwarno et al., 2015). lut1 encodes a plastid-localized cytochrome P450 that hydroxylates the ε-ring of α-carotene to form zeinoxanthin and was associated with zeinoxanthin levels and ratios of α-branch compounds (Owens et al., 2014). lcyE catalyzes ε-ring cyclization of lycopene and is a key branch point enzyme: alleles with low expression in grain result in higher flux to β-carotene and β-xanthophylls (Harjes et al., 2008). Finally, crtRB1 encodes a nonheme diiron hydroxylase that sequentially hydroxylates β-carotene to produce β-cryptoxanthin and zeaxanthin; weakly expressed alleles result in higher levels of β-carotene at the expense of β-xanthophylls (Yan et al., 2010).
Figure 1.
Carotenoid biosynthetic pathway in maize; grain carotenoids are primarily produced in endosperm. Precursor pathways are summarized in black boxes. The a priori genes identified in this study are denoted in blue italics and are placed at the pathway step(s) executed by the enzyme that they encode. Gene abbreviations: 1-Deoxy-d-xylulose-5-phosphate synthase (dxs2 and dxs3); phytoene synthase (psy1); phytoene desaturase (vp5); plastid terminal oxidase (im1); lycopene ε-cyclase (lcyE); ε-ring hydroxylase (lut1); β-carotene hydroxylase (crtRB1); zeaxanthin epoxidase (zep1); carotenoid cleavage dioxygenase (ccd1), whitecap1 (wc1; a locus containing a varying number of copies of ccd1).
Provitamin A breeding efforts in maize focusing on marker-assisted selection of favorable lcyE and crtRB1 alleles have been successfully ongoing for more than a decade at two CGIAR centers, in coordination with HarvestPlus and numerous public and private partners (Saltzman et al., 2013; Dhliwayo et al., 2014; Suwarno et al., 2014). Maize hybrid and synthetic varieties that accumulate 40%–70% of the target provitamin A level have been released, and others with higher levels are in national performance trials (Pixley et al., 2013; Menkir et al., 2017). However, introgression of favorable alleles of lcyE and/or crtRB1 can have dramatically different effects depending on the genetic background (Babu et al., 2013; Menkir et al., 2017; Gebremeskel et al., 2018), suggesting that further investigation of these and other involved genes may facilitate and expedite the consistent achievement of target provitamin A levels. Furthermore, simultaneously enhancing and balancing several other priority carotenoid traits (e.g. lutein, zeaxanthin, and total carotenoids) will require a more comprehensive understanding of the genetics underlying natural variation in maize grain carotenoid content and composition.
While substantial insight into the carotenoid pathway has been obtained from studies in Arabidopsis, its seed are green and photosynthetic whereas those of most major crops, including maize, are nonphotosynthetic. Thus, maize grain is both inherently of interest as a key target crop for biofortification efforts and also potentially provides a more suitable model system for carotenoid accumulation in other major crops. In the current study, we used the US maize nested association mapping (NAM) panel to dissect, with high power and resolution, the quantitative trait loci (QTL) and underlying genes responsible for natural variation in grain carotenoid levels.
Results
Genetic dissection of carotenoid accumulation in maize grain
We used the US NAM panel—25 families, each comprising 200 recombinant inbred lines (RILs) with B73 as a common parent—to dissect the genetic basis of carotenoid content and composition in maize grain. Seven grain carotenoid compounds were quantified by high-performance liquid chromatography (HPLC) with photodiode array detection. These traits and one summed trait, total carotenoids, had high estimates of line-mean heritability (0.70–0.94, Table 1). These traits exhibited weak negative to strong positive correlations in pairwise relationships (Supplemental Figure S1). Through a joint-linkage (JL) analysis across all 25 NAM families, we identified 117 individual-trait QTL (10–23 for each trait; Tables 1 and 2; Supplemental Data Sets S2 and S3) that each explained 0.7%–40.7% of the phenotypic variance (Supplemental Data Set S4).
Table 1.
Sample sizes, ranges, and heritabilities for carotenoid traits
| Trait | No. of lines | BLUEs |
Heritabilities |
|||
|---|---|---|---|---|---|---|
| Median | SD | Rangea | Estimate | SE | ||
| Phytofluene | 3,521 | 0.39 | 0.54 | −0.43 to 3.02 | 0.72 | 0.02 |
| α-Carotene | 3,556 | 0.91 | 0.84 | −0.70 to 4.77 | 0.70 | 0.03 |
| β-Carotene | 3,538 | 0.96 | 0.97 | −0.58 to 5.49 | 0.78 | 0.03 |
| Zeinoxanthin | 3,465 | 1.44 | 1.99 | −0.95 to 11.19 | 0.89 | 0.01 |
| β-Cryptoxanthin | 3,538 | 1.61 | 1.72 | −0.56 to 10.04 | 0.94 | 0.01 |
| Lutein | 3,581 | 10.68 | 6.74 | −0.72 to 39.47 | 0.94 | 0.01 |
| Zeaxanthin | 3,565 | 7.19 | 7.19 | −0.77 to 42.05 | 0.94 | 0.01 |
| Total carotenoids | 3,581 | 27.33 | 13.69 | −0.89 to 85.40 | 0.94 | 0.01 |
Notes: Medians and ranges (in µg g−1 dry grain) for untransformed BLUEs of eight carotenoid grain traits evaluated in the US maize NAM panel, and estimated heritability on a line-mean basis across 2 years.
SD, standard deviation of the BLUEs; SE, standard error of the heritability estimate.
Negative BLUE values are a product of the statistical analysis. Specifically, it is possible for BLUEs to equal any value from −∞ to +∞.
Table 2.
Genetic association results for carotenoid traits
| Trait | Number of JL-QTL | Median size (SD) of α = 0.01 JL-QTL support interval (Mb) | Number of JL-QTL intervals containing a priori genes | Number of GWAS-associated variants in JL-QTL intervalsa | Maximum RMIP |
|---|---|---|---|---|---|
| Phytofluene | 14 | 5.05 (27.96) | 6 | 65 | 0.67 |
| α-Carotene | 11 | 2.64 (16.36) | 5 | 44 | 0.87 |
| β-Carotene | 10 | 3.86 (6.72) | 8 | 39 | 0.80 |
| Zeinoxanthin | 12 | 2.42 (27.79) | 8 | 40 | 0.86 |
| β-Cryptoxanthin | 23 | 3.52 (26.73) | 11 | 75 | 0.66 |
| Lutein | 14 | 3.17 (19.59) | 7 | 50 | 0.88 |
| Zeaxanthin | 18 | 4.22 (29.42) | 13 | 56 | 0.88 |
| Total carotenoids | 15 | 6.21 (20.77) | 7 | 54 | 0.73 |
| JL-QTL total | 117 | 3.80 (23.58) | 65 | 422 |
Notes: Summary of JL-QTL and GWAS variants identified for eight carotenoid grain traits evaluated in the US maize NAM panel
GWAS variants residing within JL-QTL support intervals for each trait that exhibited a RMIP of 0.05 or greater.
To dissect the identified QTL at higher resolution, we performed a genome-wide association study (GWAS) using ∼27 million HapMap v1 and v2 markers imputed on the ∼3,600 NAM RILs (Table 2 and Supplemental Data Set S5). A total of 983 marker–trait associations (101–142 per trait) had a resample model inclusion probability (RMIP) value ≥ 0.05 (Valdar et al., 2009; Table 2 and Supplemental Data Set S5). Of these, 422 (42.9%) were within a corresponding trait JL interval (Table 2), with 98 of these attributed to 42 markers associated with two or more traits, yielding a total of 366 uniquely associated markers.
Given that individual carotenoid compounds share a biosynthetic pathway (Figure 1), it was not surprising that 80% of overlapping QTL support intervals were significantly pleiotropic (Supplemental Figure S2 and Supplemental Data Set S6). When the 117 individual-trait QTL intervals were merged based on physical overlap, 44 unique QTL were obtained, 21 of which affected multiple traits (Supplemental Data Set S3). We then applied a triangulation approach (Diepenbrock et al., 2017) integrating JL-QTL effect estimates (Supplemental Data Set S7), GWAS marker genotypes, and RNA-sequencing (RNA-seq) expression abundances at six developing kernel stages in the NAM parents (Supplemental Data Set S8) to identify genes underlying the 44 unique QTL (Supplemental Figure S3). The rapid decay of linkage disequilibrium (LD) in proximity to GWAS-detected markers (Supplemental Figure S4), in combination with the high resolving power of the NAM panel (Wallace et al., 2014), supported using a search space spanning ±100 kb of those GWAS signals residing within a unique QTL for gene identification. Based on the confluence of strong triangulation correlations for a single gene within these search spaces, 11 genes were identified as underlying a unique QTL (Figure 2). All 11 genes were contained on the list of 59 a priori genes known from prior studies in various plants to play roles in IPP synthesis and carotenoid biosynthesis and degradation (Supplemental Data Set S1). Five of these 11 genes were in a class that we term correlated expression and effect QTL (ceeQTL), in that their expression levels were significantly associated with the JL allelic effect estimates for the QTL at multiple kernel developmental time points (Figures 2, 3 and Supplemental Figure S3). All QTL with >4% phenotypic variance explained (PVE) were resolved down to an individual gene, with three exceptions: QTL21 for phytofluene (4.68% PVE), QTL24 for α-carotene (6.99%), and QTL34 for α-carotene (5.21% PVE) (Figure 2 and Supplemental Table S1). Of the 33 JL-QTL intervals that could not be resolved to an individual gene (PVE of 0.66%–6.99%), 22 were single-trait intervals (Supplemental Table S1).
Figure 2.
Percent phenotypic variance explained (PVE) by JL-QTL. Blue shading corresponds to the range of PVEs for JL-QTL, with darker blue indicating higher PVEs. ceeQTL indicates significant correlations between expression values and JL-QTL allelic effect estimates at two or more time points for at least one trait. The whitecap1 locus is a macrotransposon insertion-derived tandem-ccd1 gene array located 1.9 Mb away from the progenitor ccd1-r locus (Tan et al., 2017). The high identity of mRNAs from the two loci does not allow ccd1 mRNA from the two loci to be distinguished and therefore locus-specific FDR-corrected P-values could not be calculated. However, a strong correlation was still observed between ccd1 expression (as well as ccd1 copy number) and QTL38 allelic effect estimates for several traits (Figure 3 and Supplemental Table S2). In RefGen_v4, zep1 was erroneously split into the two gene models indicated; in RefGen_v5 they have been correctly merged into a single gene model with identifier Zm00001e007970. PHYF, phytofluene; ACAR, α-carotene; BCAR, β-carotene; ZEI, zeinoxanthin; BCRY, β-cryptoxanthin; LUT, lutein; ZEA, zeaxanthin; TOTCAR, total carotenoids.
Examination of specific variants
We employed two types of variant annotation analyses to provide further functional insight into the 55 unique GWAS variants (i.e. 50 SNPs and 5 indels) involved in one or more marker–trait associations that were within ±100 kb of the 11 identified genes: genomic evolutionary rate profiling (GERP; Davydov et al. 2010) and single-nucleotide polymorphism (SNP) effect analysis (SnpEff; Cingolani et al., 2012). GERP uses multi-species alignments to predict the impact of a variant as it relates to a quantitative measure of evolutionary conservation, with a positive GERP score (>0) indicating that a site may be under evolutionary constraint and mutations with larger scores more likely to be deleterious (Davydov et al., 2010; Rodgers-Melnick et al., 2015). GERP scores from two prior studies (Kistler et al., 2018; Ramstein et al., 2020) were collectively available for 53 of the 55 unique GWAS variants. Of these, 26 variants had positive GERP scores in Kistler et al. (2018) and 12 SNPs had positive GERP scores in Ramstein et al., (2020); (Supplemental Data Set S9). SnpEff, a genomic annotation and functional effect prediction tool (Cingolani et al., 2012), predicted 6 of the 55 SNPs to have low or moderate effects (Supplemental Data Set S9). These results for individual genes are further detailed below.
The role of a priori pathway genes
Of the 11 identified a priori genes, 5 encode enzymes that act upstream of the main carotenoid pathway branch point, where the linear carotene lycopene is cyclized to either α- or β-carotene. All five loci were associated with multiple cyclic carotenoids (Figure 2), including provitamin A compounds (β-carotene, β-cryptoxanthin, and α-carotene), and all but phytoene synthase 1 (psy1) represent novel associations at the genome-wide level with carotenoid variation in maize grain. Consistent with their sequential positions in the upstream portion of the pathway, all of these loci except for phytoene desaturase (PDS) (vp5) showed positive pleiotropy (i.e. having positively correlated QTL allelic effect estimates between pairs of traits) for the downstream cyclic carotenoids with which they were associated (Supplemental File S1). Two of these five genes, dxs2 and dxs3, encode 1-deoxy-d-xylulose 5-phosphate synthase (DXS), the first enzymatic reaction in the plastid-localized MEP pathway that produces IPP for the biosynthesis of carotenoids and other plastidic isoprenoids. dxs2 and dxs3 (QTL 27 and 35, respectively) were associated with eight and six traits, with PVEs of 3.5%–11.3% and 1.2%–4.1%. Of the two genes, dxs2 was found to be a strong ceeQTL in the later stages of kernel development (Figure 3). Six of 15 dxs2 GWAS variants had positive GERP scores (Supplemental Data Set S9). Similarly, 6 of 12 dxs3 GWAS variants had positive GERP scores (Supplemental Data Set S9) with one in the dxs3 genic region (Chr 9: 20, 245, 139 bp) predicted to be a missense variant with moderate effect by SnpEff (Supplemental Data Set S9). The dxs2 and dxs3 homologs were the only MEP pathway genes identified in this study. Phytoene synthase (psy) catalyzes the synthesis of phytoene, the committed biosynthetic intermediate for all carotenoids, from two GGDP molecules (Buckner et al., 1996; Li et al., 2008). The maize genome contains three psy loci: plants with a functional psy1 allele accumulate carotenoids in the endosperm and embryo, whereas those homozygous for a recessive null allele lack carotenoids in endosperm without affecting the carotenoid contents of other tissues (Buckner et al., 1996; Li et al., 2008; reviewed in Gilmore, 1997; Koornneef et al., 2002). The psy1 locus serves as a major genetic controller of quantitative variation for carotenoids in maize endosperm, the major site of carotenoid accumulation in grain (Zhu et al., 2008; Fu et al., 2013b). psy1 (QTL 25) had 5.0%–11.7% PVE for the seven carotenoids analyzed and the largest PVE observed in this study for the sum trait of total carotenoids (24.7%). When considering only the 14 families with nonwhite endosperm parents, indicative of functional psy1 alleles, this locus still showed PVEs of 1.6%–10.8% for the seven measured carotenoids and 22.5% PVE for total carotenoids (Supplemental Data Set S10). psy1 was an extremely strong ceeQTL (Supplemental Figure S3), with positive correlations between expression and effect estimates for several traits throughout kernel development, both across all 25 NAM families and across the 14 families with nonwhite endosperm founders (Supplemental Data Sets S7, S11). Two of the four psy1 GWAS variants had positive GERP scores, with one in the psy1 genic region (Chr 6: 85,064,130 bp) predicted by SnpEff to be a missense variant with medium effect (HapMap_v2). This variant had the second-highest GERP score observed for GWAS variants in this study (Supplemental Data Set S9) and corresponds to a previously identified variant in the fifth exon of psy1 termed SNP7, which was significantly associated with total carotenoid levels (Fu et al., 2013b). SnpEff predicted a Thr to Asn substitution, which is concordant with that identified and suggested to be causal in Fu et al. (2013b).
Figure 3.
Master summaries for selected identified genes. Marker colors indicate the pathway branch of the associated trait: yellow for α-branch compounds; orange for β-branch compounds; purple for phytofluene; and black for total carotenoids. Marker shapes correspond to the hydroxylation state of the associated trait: squares for α- and β-carotene; triangles for β-cryptoxanthin and zeinoxanthin; circles for lutein and zeaxanthin; diamonds for phytofluene; and an X for total carotenoids. Left panels: Directional gene models are depicted as black arrows and the identified gene as a green arrow. Lines with trait names above gene models indicate RMIP of significant GWAS hits ±100 kb of the peak RMIP variant. Lines below gene models indicate pairwise LD (r2) of each GWAS variant with the peak RMIP variant (dark blue line). The blue ribbon depicts the highest LD, per 200-bp window, to the peak RMIP variant, while black ribbons indicate the density of variants tested in GWAS in the 200-bp window (log2 scale). Darker colors correspond to higher values. Right panels: Correlations (r) between JL-QTL allelic effect estimates and expression of the identified gene across six developing kernel time points. Significant correlations are indicated by trait abbreviations above the respective time point. Traits with both JL and GWAS associations appear in black text to the right of the graph and have solid trend lines and symbols, while those with only JL associations are in gray with dashed trend lines and open symbols.
The next step in the pathway involves the sequential desaturation of phytoene to phytofluene and then ζ-carotene by PDS (encoded by vp5; QTL 1), which requires plastoquinone (PQ) as an electron acceptor (Mayer et al., 1990; Norris et al., 1995; Brausemann et al., 2017). The vp5 locus was associated with phytofluene (a desaturation intermediate) as well as α-carotene, β-carotene, and β-cryptoxanthin, with PVEs of 1.4%–1.8% (Supplemental Data Set S3). One of the two vp5 GWAS variants (1.6 kb upstream) had a positive GERP score (Supplemental Data Set S9). Another a priori gene (im1, QTL 5) encodes a homolog of the Arabidopsis plastid terminal oxidase (PTOX, IMMUTANS) gene (Carol et al., 1999; Aluru et al., 2001), which transfers electrons from PQH2 to molecular oxygen to replenish PQ in the absence of an active photosynthetic electron transport chain, i.e. prior to seedling greening (reviewed in Foudree et al., 2012). This locus was associated with phytofluene, α-carotene, β-cryptoxanthin, and zeinoxanthin, with PVEs of 1.2%–2.5%. The three im1 GWAS variants had positive GERP scores, one of which (6.6-kb distal of the gene) had the highest GERP score observed in this study (Supplemental Data Set S9).
Four biosynthetic genes downstream of the pathway branch point were associated with carotenoid traits, and all had major effects (Figure 2). lcyE is the committed step in α-carotene synthesis (Cunningham et al., 1996; Bai et al., 2009; Cazzonelli and Pogson, 2010) and is the major genetic controller of relative flux into the α- or β-carotene pathway branches (Harjes et al., 2008). Accordingly, lcyE (QTL 33) showed negative pleiotropy (i.e. having negatively correlated QTL allelic effect estimates between pairs of traits) for compounds between the two pathway branches and positive pleiotropy for compounds within the same branch (Supplemental File S1). This locus had the largest PVE observed in this study for each of the six α- and β-branch compounds (PVE = 19.6%–40.7%) and—as also reported in Harjes et al. (2008)—had no significant impact on total carotenoids (Figure 2). Of six lcyE GWAS variants, three had positive GERP scores (Supplemental Data Set S9). Two of these GERP positive SNPs were in the genic region, one of which (Chr8: 143,021,025 bp) was associated with five traits in GWAS and was predicted to be a synonymous variant with low effect by SnpEff. This SNP was located 386 bp downstream of a LCYE-5′ transposable element and 347 bp upstream of LCYE-SNP216, two significant variants identified in a prior study (Harjes et al., 2008; Owens et al., 2014). The other GERP positive genic variant (Chr8: 143,025,685 bp) was predicted by SnpEff to cause an intron splice variant with low effect and is located 100 bp downstream of a previously reported significant variant, LCYE-3′ Indel (Harjes et al., 2008; Owens et al., 2014).
Subsequent hydroxylations of the ε- and β-rings of α- and β-carotenes are performed by P450s or nonheme dioxygenases, encoded by two and six genes, respectively, in the maize genome. Of these eight genes, three were associated with carotenoid traits in maize grain: β-carotene hydroxylase 1 (crtRB1) and β-carotene hydroxylase 5 (crtRB5), which preferentially convert β-carotene to β-cryptoxanthin and then zeaxanthin, and lut1, encoding CYP97C, a cytochrome P450-type monooxygenase that preferentially hydroxylates the ε-ring of α-carotene to yield zeinoxanthin (Tian et al., 2004; Quinlan et al., 2012). The primary impact of lut1 (QTL 2) was on the levels of zeinoxanthin (PVE = 6.1%) and lutein (PVE = 2.9%, Figure 2). All three distal lut1 GWAS variants (35.6- and 3.2-kb upstream and 3.6-kb downstream) had positive GERP scores (Supplemental Data Set S9). crtRB1 (QTL 43, also known as hyd3) had PVEs of up to 8.4% for zeaxanthin, β-carotene, and β-cryptoxanthin, with negative pleiotropic effects between its substrate, β-carotene, and its products, β-cryptoxanthin and zeaxanthin (Supplemental File S1). Four of the six crtRB1 GWAS variants had positive GERP scores (Supplemental Data Set S9), with one in the genic region (Chr10: 137,260,105 bp) predicted by SnpEff to cause an intron splice variant with low effect. This variant is located 110- and 1.0-kb upstream of the previously characterized crtRB1-5′TE and crtRB1-3′TE markers, respectively (Yan et al., 2010; Owens et al., 2014). crtRB5 (QTL39, also known as hyd5) was identified with PVE of 1.4% for only a single trait, zeaxanthin (Figure 2), and its single GWAS variant had a positive GERP score (Supplemental Data Set S9). The gene encoding the subsequent β-branch enzyme, zep1, had positive pleiotropic effects between β-cryptoxanthin, zeaxanthin, and total carotenoids (Supplemental File S1). zep1 (QTL 7) primarily had a large effect on its substrate, zeaxanthin, with PVE of 17.9% (Figure 2). Concordant with this large PVE for the highly abundant compound zeaxanthin, this locus also had 5.1% PVE for the sum trait of total carotenoids (Figure 2). One of two zep1 GWAS variants, which was genic, had a positive GERP score (Supplemental Data Set S9).
Finally, the maize genome encodes 12 different carotenoid cleavage enzymes involved in ABA and strigolactone synthesis and carotenoid degradation. However, only one, carotenoid cleavage dioxygenase1 (ccd1), was associated with carotenoid levels in this study. CCD1 has been shown to be active toward multiple cyclic and linear carotenoids in vitro (Vogel et al., 2008). A single copy of ccd1 exists in all lines at the progenitor ccd1-r locus, and in a limited number of lines containing the dominant white cap1 (wc1) locus, a variable number of tandem ccd1 copies (n = 1–11 in the NAM founders) are found within a Tam3L transposon inserted 1.9 Mb proximal to ccd1-r (Tan et al., 2017). The QTL identified at wc1 had PVEs of 2.3%–11.1% for four compounds and 11.1% PVE for total carotenoids, making it the second-largest effect QTL for this trait after psy1 (Figure 2). This locus showed positive pleiotropy for all traits detected (Supplemental File S1) and was a strong ceeQTL (Figure 3). Correlations of ccd1 copy numbers in wc1 alleles from Tan et al. (2017) with NAM JL-QTL allelic effect estimates from our study were particularly strong for the most abundant carotenoids, lutein, zeaxanthin, and total carotenoids (r = −0.76 to −0.85), and moderate for α-carotene (−0.56) (Supplemental Table S2). Additionally, there was a strong, positive correlation detected between ccd1 copy number and ccd1 expression level (log2-transformed FPKM) (r = 0.80–0.84) across all six stages of kernel development.
Epistatic interactions play a minor role in determining most carotenoid traits
We tested all pairs of JL-QTL peak markers for epistasis in a joint analysis of all 25 NAM families, including 11 families that each segregated for a recessive allele of psy1 that conditions extremely low levels of carotenoids in the endosperm. In total, 14 significant epistatic interactions were found across the 25 families, but with relatively small PVEs. For instance, psy1 has main-effect PVEs ranging from 5.0% to 24.7% for seven of the eight traits (Figure 3), but was an epistatic partner in only three interactions, all with PVEs <1% (Supplemental Figure S5 and Supplemental Data Set S12). Only 3 of the 14 interaction terms had PVEs greater than 1%: zep1 with wc1 for zeaxanthin (1.09%), vp5 with QTL24 for α-carotene (6.06%), and dxs2 with QTL34 for α-carotene (1.12%).
Discussion
Here, we performed an extensive dissection of the quantitative genetic basis of maize grain carotenoid levels. With 11 of 44 carotenoid QTL resolved to their underlying genes, the key precursor and core biosynthetic pathway genes underpinning these traits are now largely elucidated (Figure 2 and Supplemental Table S1). With the exception of phytofluene, these 11 genes explain 70.3%–90.9% of all PVE attributed to QTL for each trait (Supplemental Figure S6). Notably, 8 of the 11 genes have large effects (PVE ≥4% for one or more trait), are highly pleiotropic (Supplemental File S1), and six of the eight were also ceeQTL (Figure 2). Taken together, these findings indicate that pleiotropy within the carotenoid pathway may be predominantly regulated (directly or indirectly) at the level of gene expression. A previous examination of kernel gene expression in a 508-line maize association panel supports this conclusion: Fu et al. (2013a) identified expression QTL (marker–gene expression associations) for 4 of our 11 identified genes (lcyE, crtRB1, ccd1, and vp5), 2 of which (lcyE and crtRB1) are represented in the six ceeQTL that we identified. Fu et al. (2013a) also reported significant correlations between lcyE and crtRB1 expression and grain provitamin A concentrations, providing additional support for their designation as ceeQTL.
Importantly, in addition to confirming previously reported gene/trait associations, we identified new and major breeding targets in the IPP pathway and early steps of the carotenoid pathway. Four of the five genes we identified that reside upstream of lycopene cyclization (vp5, im1, dxs2, and dxs3) had not previously been associated with natural variation in maize grain carotenoids at the genome-wide level (Harjes et al., 2008; Yan et al., 2010; Owens et al., 2014; Suwarno et al., 2015; Azmach et al., 2018; Baseggio et al., 2020). Most notably, dxs2 had the second-largest PVEs for four traits in this study, including two provitamin A carotenoids, β-cryptoxanthin and α-carotene, at 11.3% and 7.7%, respectively. dxs2 and dxs3 were also associated with natural variation for tocochromanol (vitamin E-related) traits (Diepenbrock et al., 2017), which also utilize IPP in their synthesis. As was observed for carotenoids, the PVEs of dxs2 for tocochromanol traits were greater than those of dxs3, and dxs2 was also a ceeQTL (FDR < 0.05), while dxs3 was not (Diepenbrock et al., 2017; Figures 2, 3 and Supplemental Figure S3). Engineering and overexpression studies in Arabidopsis and Escherichia coli have shown DXS is a limiting activity in the MEP pathway (Harker and Bramley, 1999; Estevez et al., 2001), and maize dxs2, a likely past target of selection (Fang et al., 2020), appears to be the major genetic control point for IPP synthesis for both carotenoids and tocotrienols, and likely other plastidic isoprenoids in maize grain.
Phytoene synthase catalyzes the committed step in carotenoid synthesis and was previously associated with variation for five carotenoids in maize kernels (Fu et al., 2013b). We confirmed that psy1 has large PVEs for these five carotenoid traits and two others (Figure 2). Considerable haplotype-level variation is still present at the psy1 locus in both temperate and tropical maize (Swarts et al., 2017), and an allelic series was seen in the present study (Supplemental Data Set S7). This suggests that explicit attention to selecting or fixing favorable haplotypes of both psy1 and dxs2, which exhibited large PVEs for many of the same traits, should increase the overall flux of IPP into the carotenoid pathway and further enhance gains obtained from selection on other downstream genes (e.g. lcyE and crtRB1) for provitamin A biofortification of maize.
Genes encoding PDS (vp5) and the plastid alternative oxidase (PTOX, im1), both of which are necessary for the sequential desaturation of phytoene to ζ-carotene via phytofluene, were identified in this study. PDS introduces double bonds into its carotenoid substrates and transfers the electrons to PQ, an essential co-factor for carotenoid biosynthesis, reducing it to PQH2 (Norris et al., 1995). In photosynthetic tissues, PQH2 is efficiently re-oxidized by the photosynthetic electron transport chain (Rosso et al., 2006; Shahbazi et al., 2007; Rosso et al., 2009, reviewed in Foudree et al., 2012). However, in nonphotosynthetic tissues that have an underdeveloped photosynthetic electron transport chain—e.g. germinating seedlings and developing maize grain—PTOX transfers electrons from PQH2 directly to molecular oxygen to regenerate PQ for additional desaturation cycles (Beyer et al., 1989; Mayer et al., 1990; Carol et al., 1999; Wu et al., 1999, reviewed in Rodermel, 2002; Foudree et al., 2012). PTOX loss-of-function mutations in Arabidopsis negatively affect carotenoid synthesis in developing seedlings, resulting in albino sectors of vegetative tissue that hyper-accumulate the PDS substrate phytoene (Wetzel et al., 1994; Carol et al., 1999; Wu et al., 1999; Rosso et al., 2009). Maize vp5 and im1 were associated with the PDS desaturation intermediate phytofluene (Figure 2) but also with three downstream provitamin A active carotenoids, suggesting that vp5 and im1 could be used as targets for breeding or metabolic engineering efforts to enhance kernel provitamin A content.
While biosynthesis is an important control point for carotenoid levels, degradation and the susceptibility to degradation must also be considered. For example, CAROTENOID CLEAVAGE DIOXYGENASE4 (CCD4) in Arabidopsis is a large-effect contributor to natural variation in seed carotenoids (Gonzalez-Jorge et al., 2013), primarily due to its preferential cleavage of β-carotene and epoxy-xanthophylls, whose levels are elevated 3–7-fold in ccd4 null mutants. Zeaxanthin epoxidase (ZEP) has an even greater impact on total carotenoid levels, primarily because ZEP-mediated epoxidation increases susceptibility to degradation by CCD4. A zep null mutant increased zeaxanthin levels by 40-fold, lutein, the most abundant carotenoid in wild-type Arabidopsis seed, by 2.2-fold, and total carotenoids by 5.7-fold (Gonzalez-Jorge et al., 2016). In maize, zep1 and wc1 (encoding CCD1) had analogous impacts on natural variation in zeaxanthin, lutein, and total carotenoids, and both were ceeQTL in developing kernels. Unlike CCD4, maize CCD1 is cytosolic (Tan et al., 2003) and only has access to the outer plastid envelope but is thought to have increased access to plastid-localized carotenoids as plastid membranes lose integrity during kernel desiccation (Tan et al., 2017).
A major finding of this work is that although 10–23 QTL were detected per trait, a large percentage of variance could be explained by only two to five major-effect QTL that contributed 4%–41% PVE, depending on the trait (Figure 2). These major-effect QTL are high-impact targets for genomics-enabled biofortification and/or metabolic engineering strategies. For example, just four genes—lcyE, crtRB1, dxs2, and psy1—explained the majority of variation for the three provitamin A carotenoids. These four genes explained 75% of β-carotene variation attributed to QTL; three of the four (lcyE, dxs2, and psy1) explained 52% for β-cryptoxanthin; and just two (lcyE and dxs2) explained 64% for α-carotene (Figure 2 and Supplemental Data Set S4). These findings suggest that simultaneous increases in both β-carotene and β-cryptoxanthin levels, and potentially α-carotene levels, may be achievable by incorporating the appropriate alleles of this set of four major-effect genes in selection decisions.
Currently, only crtRB1 and lcyE alleles are used in marker-assisted selection efforts (Prasanna et al., 2020). While the combination of two favorable haplotypes of these genes was previously found to decrease total carotenoid levels (Babu et al., 2013), neither crtRB1 or lcyE (separately or in interaction) were associated with total carotenoid levels in the present study (Figure 3). Rather, total carotenoids were positively affected by dxs2 and psy1 alleles and negatively conditioned by specific zep1 and ccd1 alleles (Figure 3). Thus, the previously reported decreases in total carotenoid levels observed when combining favorable lcyE and crtRB1 alleles in the same background can likely be circumvented by conditioning on the presence of the appropriate psy1, ccd1, and zep1 alleles. Enabling tandem selection of lcyE and crtRB1 in this manner should afford considerable gains in both total and provitamin A carotenoid levels, given the large PVEs and allelic effect estimates of lcyE observed in this study in families with both temperate and tropical founders (Figure 3 and Supplemental Data Set S7). In addition to provitamin A carotenoids, lutein and zeaxanthin are the most abundant carotenoids in maize grain and are themselves of direct interest in breeding for human health given their roles in eye health. Three genes—lcyE, psy1, and wc1 (ccd1)—explained 77.3% of lutein variation attributed to QTL, and these three genes, along with zep1, explained 74.1% for zeaxanthin. The data in this study indicate that the simultaneous improvement and balancing of provitamin A and nonprovitamin A carotenoids for human should be feasible.
The high power and resolution of the US maize NAM population has allowed the majority of the phenotypic variation of maize grain carotenoids to be dissected, in some cases approaching the upper limits of the high heritabilities for these traits (Table 1 and Supplemental Data Set S4). The number and PVE of epistatic interaction terms were found to be minor (Supplemental Figure S5)—including compared with those observed for tocochromanol (vitamin E-related) traits in the US maize NAM panel (Diepenbrock et al., 2017)—further suggesting that largely additive variation can be exploited for carotenoid traits in maize grain. The extensive information provided here should focus and accelerate genomics-enabled breeding and/or metabolic engineering efforts to simultaneously achieve provitamin A targets (Bouis and Welch, 2010) and improve the levels of other health-beneficial carotenoids in human populations consuming maize as a staple. The identified genes specifically provide direct, high-value targets for maize biofortification breeding programs. An important next step will be allele mining and examination of allele and haplotype frequencies for the identified genes in germplasm pools and breeding populations. The identified genes are also logical candidates to be assessed as potential controllers of carotenoid variation in seeds of other abundantly consumed monocot crops.
Materials and methods
Field environments and plant materials for genetic mapping
The design of the maize (Z. mays) NAM population has been previously described (Yu et al., 2008; Buckler et al., 2009; McMullen et al., 2009). The experimental field design in 2009 and 2010 for this study—which included the NAM panel, the intermated B73 × Mo17 (IBM) family (Lee et al., 2002), and a 281-line inbred diversity panel (Flint-Garcia et al., 2005)—was conducted as described in Chandler et al. (2013) and Diepenbrock et al. (2017). In brief, 5,000 RILs—25 families, with approximately 200 RILs per family—were generated by crossing 25 diverse inbred lines with B73, a common parent. The 25 families of the NAM population, along with the IBM family (Lee et al., 2002) and a 281-line inbred diversity panel designed for association mapping (Flint-Garcia et al., 2005) were evaluated in West Lafayette, IN. These evaluations were conducted at the Purdue University Agronomy Center for Research and Education in the summers of 2009 and 2010 (i.e. two environments), using standard agronomic practices. The field design for these experiments has been previously described (Chandler et al., 2013). In brief, a sets design was used in each environment, with a given set containing all ∼200 RILs of a family or the 281-line association panel. Each set that contained a NAM family was planted in an augmented 10 × 20 incomplete block α-lattice design, with the two parental lines included as checks in each incomplete block. Each set that contained the 281-line association panel was planted in an augmented 14 × 20 incomplete block α-lattice design, with maize inbred lines B73 and Mo17 included as checks in each incomplete block. One replicate of the entire experiment of 5,481 lines from the 25 NAM families, the IBM family, and the 281-member association panel, plus repeated check lines as described above (for the family sets and 281-line panel), was grown in each of the two environments. Two fields were used in 2009 to grow this entire single replicate, and one field was used in 2010. A single experimental unit was composed of a single inbred line planted in a one-row (3.05 m) plot, which would contain 10 plants on average. At least four plants in each plot were self-pollinated by hand. Grain from these self-pollinated ears was harvested at physiological maturity, dried to ∼15% moisture content, shelled, and bulked (within the plot) to form a representative sample for carotenoid quantification.
Carotenoid quantification
Extraction of carotenoids was conducted on ∼50 ground kernels per plot, and seven carotenoid compounds—α-carotene, β-carotene, β-cryptoxanthin, lutein, phytofluene, zeaxanthin, and zeinoxanthin—as well as the sum trait of total carotenoids were quantified via HPLC as previously described (Owens et al., 2014), with units of μg g−1 seed. Carotenoids were assessed based on HPLC data passing internal quality control measures that were collected on 9,411 grain samples from 4,871 NAM and 198 IBM RILs, as well as the 850 repeated parental check lines. HPLC-generated measurements of carotenoid compounds and their respective isomers were combined for zeinoxanthin and β-cryptoxanthin to obtain an overall value for the level of the compound. For technical replicates of the same sample, the mean value of the replicate measurements was recorded for each of the seven carotenoids; i.e. α-carotene, β-carotene, β-cryptoxanthin, lutein, phytofluene, zeaxanthin, and zeinoxanthin. Initial total carotenoid values were calculated as the sum of the quantified levels for these seven compounds.
Phenotypic data processing
The HPLC data set was further cleaned to standardize sample genotype names, as associated with experimental field location, and any samples that lacked proper field data were removed. For each NAM family dataset, we retained only samples with genotype assignments belonging to that family or the family’s parental genotypes (which were used as checks). Samples from each NAM family and their parental genotypes were categorized as “yellow-to-orange-grain family” (Y) or “white-grain family” (W) based on the grain endosperm color phenotype of the non-B73 parent of that family. For all samples in the “W” class for which it was available, the genotype at the psy1 locus, defined as the genomic region spanning chromosome 6 position 82,017,148 to 82,020,879 bp on the AGPv2 maize reference genome, was obtained from GBS SNP marker data downloaded from MaizeGDB (Portwood et al., 2019). To eliminate possible sample contamination and select the subset of samples expected to have a functional core carotenoid pathway, samples in the “W” class were further classified into “low” and “high” carotenoid classes using Gaussian decomposition applied to the total carotenoids values for each NAM family × year combination. Gaussian decomposition was performed using the R package “mclust,” specifying two mixture components and one-dimensional variable variance (Scrucca et al., 2016; R Core Team, 2018). Samples with classification uncertainty ≥10% were assigned to the “ambiguous” class. Any samples in the “W” class that switched carotenoid class assignments between years were removed from further analysis, as well as those that were assigned to the “low” or “ambiguous” carotenoid class in both years or that had a homozygous alternate genotype call at the psy1 locus. Finally, samples in both the “W” and “Y” classes were subjected to outlier analysis. Specifically, for each NAM family × year combination, a quartile analysis was performed on the total carotenoid values using the “boxplot.stats” function in R, and any samples with total carotenoids values at least 1.25 * interquartile range (IQR) smaller than the first quartile were marked as low total carotenoids outliers and removed from further analysis.
Following the sample-level filtering, compound measurements were set to missing for any NAM family × year × compound measurement combination that did not have at least 20 samples measured for that compound, and all samples were removed for any NAM family × year combination that did not have at least 40 remaining samples. For the remaining samples, any missing data for a given compound was assigned a value generated by random uniform sampling from the interval (0, min_measured), where min_measured is the lowest (minimum) value measured for that compound in the corresponding NAM family × year group of samples. Following this process, the total carotenoids values were recalculated to include the assigned random uniform values.
As in Diepenbrock et al. (2017), the IBM RILs were not included in JL analysis or the GWAS, as they exhibit a differential recombination rate (due to being intermated). However, IBM was still included in the model fitting process to generate best linear unbiased estimators (BLUEs) along with the 25 NAM families to provide additional information regarding spatial variation within environments and potential interactions of genotype and environment.
To examine the data for phenotypic outliers, mixed linear model selection was conducted using a custom R script that calls ASRemlR version 4 (Butler et al., 2017). This model selection process was conducted separately for each of the eight traits. To conduct model selection, a mixed linear model was fit where the grand mean was the only fixed effect, and random effects automatically included in the base model (i.e. without being tested in model selection) were the genotypic effects (family and RIL nested within family) and baseline spatial effects (year and field nested within year). The best random structure was then identified using the Bayesian information criterion (BIC; Schwarz, 1978). The random structures that were tested included all combinations of the following, thus representing one to five terms (or zero, if the base model were to exhibit the most favorable BIC value) to be fit as additional random effects: a laboratory effect (HPLC auto-sampler plate) and certain additional spatial effects (set nested within field within year, block nested within set within field within year, family nested within year, and RIL nested within family within year). The best residual structure was then also identified using BIC, after the best random structure had been identified and included in the mixed linear model. The residual structures that were tested, to account for potential spatial variation across rows and/or columns within each environment, were identity by year; autoregressive for range and identity for row, by field-in-year; identity for range and autoregressive for row, by field-in-year; and autoregressive (first-order, AR1 × AR1) for range and row, by field-in-year. Field-in-year represents a new factor that combines the field name and year to enable fitting a unique error structure for each of the three fields.
From the final fitted model for each trait, phenotypic outliers with high influence were detected using the DFFITS (“difference in fits”) criterion (Neter et al., 1996; Belsley et al., 2005) as previously described in Diepenbrock et al. (2017), and observations were set to NA if they exceeded a conservative DFFITS threshold previously suggested for this experimental design (Hung et al., 2012). Following outlier removal, the model-fitting process described above was conducted again to estimate BLUEs for the RILs, but with the genotypic effects of family and RIL within family now included as sparse fixed effects rather than random effects. Note that the model was first fit in this step with these genotypic effects as random, then updated with these effects as sparse fixed. All terms except for the grand mean were again fitted as random effects to estimate variance components for the calculation of line-mean heritabilities. These heritability estimates were calculated only across the 25 NAM families (Hung et al., 2012), and the delta method was used to obtain standard errors (Holland et al., 2003).
The BLUEs generated for each trait were then examined to detect any remaining statistical outliers. Specifically, the Studentized deleted residuals (Kutner et al., 2004) were examined using PROC MIXED in SAS version 9.3 (SAS Institute 2011). These residuals were obtained as in Diepenbrock et al. (2017) from a parsimonious linear model that contained the grand mean and a single randomly sampled, representative SNP (PZA02014.3) from the original genetic map for the NAM panel (McMullen et al., 2009) as fixed effects. The BLUE value of a given RIL for a given trait was set to NA if its corresponding Studentized deleted residual had a magnitude greater than the Bonferroni critical value of The significance level (α) used in this step was 0.05; n was the sample size of 3,585 RILs; and p was the number of predictors.
The Box–Cox power transformation (Box and Cox, 1964) was then performed separately on BLUEs for each trait as in Diepenbrock et al. (2017). Briefly, the same parsimonious model used to generate Studentized deleted residuals was also used to identify the most appropriate Box–Cox transformation (with λ tested between −2 and 2, step of 0.05) that corrected for heteroscedasticity and error terms that were not normally distributed. PROC TRANSREG within SAS version 9.3 (SAS Institute 2011) was used to find the optimal λ for each trait (Supplemental Table S3) and apply the transformation. Note that the Box–Cox power transformation requires positive input values. Each of the traits had some number of negative BLUE values (ranging from 1 to 283 RILs per trait); these are a reasonable result of the BLUE-fitting process (Burkschat, 2009). The lowest possible integer needed to make all values positive for a given trait was added as a constant across that trait vector for all of the RILs before applying the transformation; this constant had a value of 1 for all traits.
JL analysis
A 0.1-cM consensus genetic linkage map (14,772 markers) was used for JL analysis, as in Diepenbrock et al. (2017). This map was generated by imputing SNP genotypes at 0.1-cM intervals in a process previously described (Ogut et al., 2015), using genotyping-by-sequencing (GBS) data for ∼4,900 NAM RILs as anchors (Elshire et al., 2011; Glaubitz et al., 2014). JL analysis was then conducted as previously described (Diepenbrock et al., 2017) across the 25 families of the NAM population to map QTL for natural variation in one or more maize grain carotenoid traits. Briefly, joint stepwise regression was implemented using modified source code in TASSEL version 5.2.53 (Bradbury et al., 2007; modified source code provided on GitHub), with transformed BLUEs as the response variable and the family main effect forced into the model first as an explanatory variable. The effects of each of the 14,772 markers in the 0.1-cM linkage map, nested within family, were then tested for inclusion in the final model as explanatory variables. The significance threshold for model entry and exit of marker-within-family effects was based on conducting JL analysis on 1,000 permutations of transformed BLUEs for each trait and selecting the entry P-value thresholds (from a partial F-test) that control the Type I error rate at α = 0.05. The permutation-derived entry thresholds are listed in Supplemental Table S3. Exit thresholds were set to be twice the value of these empirically derived entry thresholds, so that a marker could not enter and exit the model in the same step.
Upon examining the results of initial JL analysis conducted via the above-described procedure, peak markers in the vicinity of psy1 for various traits, which is the only locus in this genomic interval expected to control for the presence/absence of endosperm carotenoids in white-grain families, were found to have low minor allele counts exclusively among white-grain families (with fewer than 40 individuals across all 25 families having a genotypic state score greater than zero; zero represents homozygosity for the major allele), resulting in apparent inflation of the PVE by these markers. The RILs that had a genotypic state score greater than zero at the psy1-proximal peak marker for a given trait were removed from the data set for all traits (prior to the DFFITs step and BLUE generation), comprising 54 unique RILs removed in total. The analytical pipeline was then re-conducted from the mixed linear model selection step to re-generate JL models and for use of the results in all downstream analyses. After this additional removal step, it was confirmed that no markers in the vicinity of psy1 exhibited significant JL signal in any white-grain families. The sample sizes per population in this final data set used in all 25-family analyses are listed in Table 1. The permutation procedure was also applied within the 14 NAM families with parents with nonwhite endosperm color to enable an additional, separate JL analysis within those families using the appropriate thresholds, e.g. due to the reduced sample size.
Some multicollinearity between markers in the consensus map was expected, and indeed for two traits (zeaxanthin and zeinoxanthin), two pairs and one pair of markers present in the final JL model, respectively, had a Pearson’s correlation coefficient (r) with magnitude greater than 0.8 between their SNP genotype states. In these cases, the marker with the smaller sum of squares within the JL model was removed. A re-scan procedure was then conducted in the vicinity of any of the remaining peak markers for a given trait to test whether the removal of the multicollinear marker(s) meant a shift in the association signal. Specifically, if another marker within the support interval now had a larger sum of squares than the original peak marker, that marker would replace the original peak marker in the model, and this process was repeated (including re-calculation of the support interval) until a local maximum in the sum of squares was found. The final peak JL markers following re-scan, along with the family term, were then re-fitted to obtain final statistics from the JL analysis of each trait. Allelic effect estimates for each QTL, nested within family, were generated as in Diepenbrock et al. (2017) by fitting final JL models with the “lm” function (R, lme4 package), which also evaluates the significance of these QTL-within-family terms in two-sided independent t tests. In this step, the false discovery rate was controlled at 0.05 via the Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995).
Support intervals (α = 0.01) were calculated for the JL-QTL in each final model as previously described (Tian et al. 2011). Logarithm of the odds scores were calculated (R, “logLik” base function). The PVE by each joint QTL was calculated using previous methods (Li et al., 2011), with some modifications as described in Diepenbrock et al. (2017) to account for segregation distortion across the families. While transformed data were used throughout the analyses in the present study, including in all steps that required statistical inference, it was also desired to more closely examine the signs and magnitudes of QTL allelic effect estimates on the original trait scale and in directly interpretable units of nutrition. For this single purpose, the final JL model determined using transformed BLUEs was refit with untransformed BLUEs without further model selection or re-scan.
Genome-wide association study
Chromosome-specific residuals for each trait were obtained from the final transformed JL models with the family term and any joint QTL located on the given chromosome removed. These residuals were used as the response variable in GWAS, whereas the genetic markers tested as explanatory variables consisted of the 26.9 million variants (SNPs and indels <15 bp) of the maize HapMap v. 1 and 2 projects (Gore et al., 2009; Chia et al., 2012), as previously described (Wallace et al., 2014), that were upliftable to RefGen_v4 coordinates. Uplifting of HapMap markers from the B73 RefGen_v2 to RefGen_v4 assembly was conducted by clipping 50 nucleotides from each side of a given marker in its v2 position (101 nucleotides of flanking sequence in total). These were then aligned to the B73 RefGen_v4 assembly using Vmatch (v2.3.0; Kurtz, 2019), with the options of -d -p -complete -h1. The resulting alignments were then filtered to keep the highest scoring and unique alignment for each marker. If a marker did not have a high-confidence, unique alignment, it was omitted from the set of upliftable markers.
The upliftable markers were projected onto the NAM RILs using the dense 0.1-cM resolution linkage map, and GWAS was conducted in the NAM-GWAS plugin in TASSEL version 4.1.32 (Bradbury et al., 2007) as previously described (Wallace et al., 2014; Diepenbrock et al., 2017). Briefly, a forward selection regression procedure was conducted 100 times for each chromosome, with 80% of the RILs from every family sub-sampled each time. For each trait, the model entry threshold was empirically determined by conducting GWAS on 1,000 permutations of chromosome-specific residuals for each trait and averaging the results across chromosomes (Wallace et al., 2014) to control the genome-wide Type I error rate at α = 0.05 (Supplemental Data Set S12). The significance threshold used for a marker in GWAS was its RMIP value, or the proportion of the 100 final GWAS models in which that marker was included (i.e. meeting the model entry threshold). Markers with an RMIP ≥ 0.05 were considered in downstream analyses.
RNA-seq
Sample collection for RNA-seq—in three biological replicates of the NAM founders at six developing kernel stages, and in root and shoot tissues—and RNA sequencing and sample quality assessment were conducted as described in Diepenbrock et al. (2017). Briefly, one self-pollinated ear per plot was sampled for each developing kernel stage, frozen in liquid nitrogen in the field and held at −80°C until shelling and the removed kernels were stored at −80°C. Thirty kernels were then randomly sampled from across the replicates for a given parent and bulked; for the majority of samples, 10 seeds were used per replicate. For root and shoot samples, seeds were surface sterilized and germinated on wet filter paper for 4–5 days at room temperature under grow lamps. Germinated seedlings were then transplanted into soil in pots and grown in a greenhouse under long-day conditions (supplemental light via high-pressure sodium lamps) for an additional 14 days at 30°C–33°C. Plants were removed from pots, rinsed with water to remove soil, and root and shoot tissue harvested separately and flash-frozen in liquid nitrogen. Samples were stored at −80°C until RNA extraction. Equal weights of shoots and roots were sampled from across the replicates for a given parent and bulked. Total RNA was extracted and sequenced, and raw reads processed and aligned, as previously described with no modifications (Diepenbrock et al., 2017). In the present study, RNA-seq reads from Diepenbrock et al. (2017) were directly downloaded from the National Center for Biotechnology Information Sequence Read Archive (BioProject PRJNA174231) and processed using the same pipeline as described in Hoopes et al. (2018). In brief, read quality was assessed using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and MultiQC (Ewels et al., 2016) and then cleaned using Cutadapt (Martin, 2011) to remove adaptors and low-quality sequences, aligned to AGPv4 of B73 using TopHat2 (Kim et al., 2013) with the parameters -i 5 -I 60000 –library-type fr-unstranded, and expression abundances determined using Cufflinks (Trapnell et al., 2012) in the unstranded mode with a maximum intron length of 60 kb and the AGPv4 annotation.
FPKM filtering
The gene set was filtered (as in Diepenbrock et al., 2017) such that at least one of the kernel developmental samples in at least one sampled founder line had an FPKM greater than 1.0; a total of 30,121 genes remained upon filtering with this criterion. Expression data for genes passing the specified threshold were transformed according to log2(FPKM + 1), where the constant of 1 was added to allow the transformation of “0” values. These log2-transformed values are referred to as “gene expression levels” (Supplemental Data Set S8).
Triangulation analysis
The JL support intervals from two or more individual-trait models that were physical overlapping were merged to form common support intervals, as in Diepenbrock et al. (2017). Physically distinct support intervals detected for a single trait were also retained. Triangulation analyses were conducted as in Diepenbrock et al. (2017), based on all pairwise Pearson correlations between trait JL-QTL effect estimates; marker genotype state for each significant GWAS marker in the interval for the respective trait(s); and log2-transformed expression values of genes within ± 100 kb of any of these significant GWAS markers. The search space of 100 kb was selected based on LD decay (Supplemental Figure S4). For those correlations involving one of the five traits with a negative optimal lambda for the Box–Cox transformation (i.e. an inverse power transformation was applied for these traits), the sign of the correlation was reversed in graphical and tabular representations (Figure 3 and Supplemental Figure S3 for master gene summaries, Supplemental Figure S2 and Supplemental Data Set S6 for pleiotropy) to represent the true directionality of the relationship between traits.
Epistasis
For the peak markers in the final JL model for each trait, each possible additive × additive pairwise interaction was individually tested for significance in a model containing all marker main effects as in Diepenbrock et al. (2017). This procedure was conducted for the 25 families. The model entry thresholds for these interaction terms was determined by modeling 1,000 null permutations of transformed trait BLUEs with only additive terms in the model and selecting the P-value approximating a Type I error rate at α = 0.05. Final epistatic models were then fit with all marker main effects and any significant interactions. PVE was calculated as described above, except that pairwise genotype scores were collapsed into three classes for interaction terms as previously described (Diepenbrock et al., 2017). Significant interactions were graphically depicted using the Circos software package (Krzywinski et al., 2009; Supplemental Figure S5).
Pleiotropy
Pleiotropy was assessed as previously described (Buckler et al., 2009), by applying the JL QTL model for each trait to every other trait. Pearson correlations between the allelic effect estimates for the original trait and the trait to which its model was applied were evaluated for significance at α = 0.01 after FDR correction via the Benjamini–Hochberg method. Significant pleiotropic relationships were visualized using the network R package (Butts, 2008, 2015; Supplemental Figure S2). Pleiotropy was also examined within each common support interval (Supplemental File S1) to validate the merging of individual-trait intervals, a step conducted in previous NAM JL analyses (Tian et al., 2011). In this QTL-level analysis, each peak JL marker within the interval was fit for every other trait that had a peak JL marker in the interval.
LD analysis
The same imputed genotypic data set of 26.9 million segregating markers used in JL-GWAS was used to estimate LD. Specifically, pairwise LD of each significant GWAS marker with all other markers within ±250 kb was estimated using custom Python and R scripts as previously described (Weir, 1996; Wallace et al., 2014; Diepenbrock et al., 2017). A null distribution was generated by performing the same estimation for 50,000 markers selected at random. LD was examined in both v2 and uplifted v4 coordinates; in the latter case, the small minority of flanking markers that moved outside of the ±250-kb region upon uplifting were dropped from the v4 analysis for that given marker.
Variant annotation
Variant annotation was conducted on the GWAS variants in this study; i.e. genetic markers that were significant for one or more traits in GWAS and that were within ±100 kb of an identified gene. Existing GERP scores in maize were downloaded from the publicly available data sets of Ramstein et al., (2020) and Kistler et al. (2018). GERP scores [along with minor allele frequencies, from Ramstein et al., (2020)] were extracted for the GWAS variants in this study based on their AGP_v4 chromosome and position. Annotation of genetic variants and prediction of effects was conducted in SnpEff (Cingolani et al., 2012). First, the input files used in GWAS were converted to VCFs in TASSEL 5.2.65 (Bradbury et al., 2007). The GWAS variants in this study were then annotated using AGP_v4 coordinates in SnpEff 5.0 (build 2020-08-09) (Cingolani et al., 2012), using the following command (and Ensembl Genome release 46; ftp://ftp.ensemblgenomes.org/pub/release-46): java -Xmx8g -jar snpEff.jar Zea_mays [infile].vcf > [outfile]_Annot.vcf.
Accession numbers
The genes that were identified in this study are listed in Figure 2. Their accession numbers (in the form of RefGen_v4 gene IDs) are as available in MaizeGDB (www.maizegdb.org; Portwood et al., 2019): Zm00001d027936, Zm00001d029822, Zm00001d001909, Zm00001d003512/Zm00001d003513, Zm00001d036345, Zm00001d019060, Zm00001d011210, Zm00001d045383, Zm00001d048373, Zm00001d048469, and Zm00001d026056. Sequence data from the maize HapMap v1 and v2 projects (Gore et al., 2009; Chia et al., 2012, respectively) are available in the NCBI Short Read Archive (and at www.panzea.org); accession numbers are SRP001145 (for HapMap v1) and SRA051245 (HapMap v2). Expression data are available via the MSU Maize Genomics Resource (Hoopes et al., 2018; http://maize.plantbiology.msu.edu/index.shtml) via a JBROWSE installation and as a downloadable expression matrix. The data reported herein are made available in the supplemental data sets, and scripts are available on GitHub (https://github.com/GoreLab/Vitamaize_NAM_GWAS_LabVersion/Carot.git).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Pairwise phenotypic correlations between untransformed BLUEs of eight carotenoid grain traits.
Supplemental Figure S2. Pleiotropy of 117 QTL identified in JL analysis for eight grain carotenoid traits in the US maize NAM population.
Supplemental Figure S3. Master summaries for the remaining genes identified in this study.
Supplemental Figure S4. LD estimates between GWAS variants in the US maize NAM population.
Supplemental Figure S5. Genome-wide distribution of carotenoid JL-QTL and their pairwise epistatic interactions in the 25 US NAM families.
Supplemental Figure S6. Relative explanation of phenotypic variance for eight carotenoid grain traits by JL-QTL in which a gene was identified, or that remain unresolved.
Supplemental Table S1. Percent PVE by unresolved QTL (JL-QTL).
Supplemental Table S2. Relationship between ccd1 copy number and JL-QTL allelic effect estimates for the NAM JL-QTL containing wc1.
Supplemental Table S3. Lambda values used in Box–Cox transformation and JL and GWAS entry thresholds determined from permutations for each trait.
Supplemental Data Set S1. Genomic information for the 59 a priori candidate genes.
Supplemental Data Set S2. BLUEs for the US maize NAM population used in the present study. Transformed BLUEs are presented in Supplemental Data Set S2a, and untransformed BLUEs are presented in Supplemental Data Set S2b (Supports Table 1).
Supplemental Data Set S3. Summary of JL-QTL and associated GWAS signals for eight carotenoid grain traits evaluated in the US maize NAM population.
Supplemental Data Set S4. Final JL models for the eight grain carotenoid traits evaluated in the US maize NAM population.
Supplemental Data Set S5. Complete list of GWAS results.
Supplemental Data Set S6. Summary of pleiotropy analyses.
Supplemental Data Set S7. Summary of JL-QTL allelic effect estimates for eight carotenoid traits evaluated in the US maize NAM population.
Supplemental Data Set S8. Tissue description and number of sequences for RNA samples used in this study.
Supplemental Data Set S9. GERP and SnpEff results for all GWAS variants within ±100 kb of 1 of the 11 genes identified in the present study.
Supplemental Data Set S10. Summary of JL-QTL within the 14 nonwhite-endosperm families of the US maize NAM population for eight grain carotenoid traits.
Supplemental Data Set S11. Summary of JL-QTL allelic effect estimates for eight carotenoid traits evaluated within the 14 nonwhite-endosperm families of the US maize NAM population.
Supplemental Data Set S12. Significant pairwise interaction terms between JL-QTL peak markers for eight carotenoid grain traits evaluated in the 25 families of the US maize nested associated mapping (NAM) population.
Supplemental File S1. Pleiotropy for eight maize grain carotenoid traits within each of the 11 US NAM JL-QTL in which an underlying gene was identified in this study.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Arthur Gilmour for expert support in AsREML and Dr. Guillaume Ramstein for advisement related to GERP scores.
Funding
This research was supported by the National Science Foundation (DBI-0922493 to D.D.P., C.R.B., E.S.B., and T.R. and DBI-0820619 and IOS-1238014 to E.S.B.), by the USDA-ARS (E.S.B.), by Cornell University startup funds (M.A.G.), and by the University of California, Davis startup funds (C.H.D.).
Conflict of interest statement. None declared.
Contributor Information
Christine H Diepenbrock, Department of Plant Sciences, University of California, Davis, California 95616.
Daniel C Ilut, Plant Breeding and Genetics Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853.
Maria Magallanes-Lundback, Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, Michigan 48824.
Catherine B Kandianis, Plant Breeding and Genetics Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853; Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, Michigan 48824.
Alexander E Lipka, Institute for Genomic Diversity, Cornell University, Ithaca, New York 14853.
Peter J Bradbury, Institute for Genomic Diversity, Cornell University, Ithaca, New York 14853; United States Department of Agriculture—Agricultural Research Service, Robert W. Holley Center for Agriculture and Health, Ithaca, New York 14853.
James B Holland, United States Department of Agriculture—Agricultural Research Service, Plant Science Research Unit, Department of Crop and Soil Sciences, North Carolina State University, Raleigh, North Carolina 27695.
John P Hamilton, Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824.
Edmund Wooldridge, Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824.
Brieanne Vaillancourt, Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824.
Elsa Góngora-Castillo, Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824.
Jason G Wallace, Department of Crop and Soil Sciences, University of Georgia, Athens, Georgia 30602.
Jason Cepela, Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824.
Maria Mateos-Hernandez, Department of Agronomy, Purdue University, West Lafayette, Indiana 47907.
Brenda F Owens, Department of Agronomy, Purdue University, West Lafayette, Indiana 47907.
Tyler Tiede, Department of Agronomy, Purdue University, West Lafayette, Indiana 47907.
Edward S Buckler, Plant Breeding and Genetics Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853; Institute for Genomic Diversity, Cornell University, Ithaca, New York 14853; United States Department of Agriculture—Agricultural Research Service, Robert W. Holley Center for Agriculture and Health, Ithaca, New York 14853.
Torbert Rocheford, Department of Agronomy, Purdue University, West Lafayette, Indiana 47907.
C Robin Buell, Department of Plant Biology, Michigan State University, East Lansing, Michigan 48824.
Michael A Gore, Plant Breeding and Genetics Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853.
Dean DellaPenna, Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, Michigan 48824.
C.H.D., M.A.G., and D.D.P co-wrote the manuscript. C.H.D., D.C.I., C.B.K., and A.E.L. co-led data analysis. M.M.-L. performed HPLC analyses and metabolite quantifications. B.V., E.G.-C., J.P.H., E.W., and J.C. performed transcriptome analysis. J.P.H. performed uplifting analyses. J.G.W. generated marker data sets. J.G.W. and D.C.I. coded GWAS, GWAS permutation, and figure scripts. J.C. created website/databases. D.C.I. and P.J.B co-led phenotypic data processing. J.B.H. conceptualized and coded spatial model fitting and heritability implementation. P.J.B. modified TASSEL source code and oversaw epistasis calculations. T.R. overall management of NAM population growth. M.M.-H. managed planting, pollination, harvesting, and processing of NAM population. B.F.O. and T.T. generated developing kernels for transcriptome analysis. E.S.B. oversaw germplasm, genotyping, and imputation, and advised on mapping analysis. C.R.B. oversaw RNA-seq and transcriptome analysis, managed all informatics. M.A.G. oversaw data analysis, project management, design, and coordination. D.D.P. overall project management and coordination, oversaw data and metabolite analyses, and biological interpretation.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Christine H. Diepenbrock (chdiepenbrock@ucdavis.edu), Michael A. Gore (mag87@cornell.edu), and Dean DellaPenna (dellapen@msu.edu).
References
- Abdel-Aal el SM, Akhtar H, Zaheer K, Ali R (2013) Dietary sources of lutein and zeaxanthin carotenoids and their role in eye health. Nutrients 5: 1169–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66: 161–186 [DOI] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR (2001) The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmach G, Menkir A, Spillane C, Gedil M (2018) Genetic loci controlling carotenoid biosynthesis in diverse tropical maize lines. G3 8: 1049–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu R, Rojas NP, Gao S, Yan J, Pixley K (2013) Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor Appl Genet 126: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Kim EH, DellaPenna D, Brutnell TP (2009) Novel lycopene epsilon cyclase activities in maize revealed through perturbation of carotenoid biosynthesis. Plant J 59: 588–599 [DOI] [PubMed] [Google Scholar]
- Baseggio M, Murray M, Magallanes-Lundback M, Kaczmar N, Chamness J, Buckler ES, Smith ME, DellaPenna D, Tracy WF, Gore MA (2020) Natural variation for carotenoids in fresh kernels is controlled by uncommon variants in sweet corn. Plant Genome 13: e20008. [DOI] [PubMed] [Google Scholar]
- Beatty S, Boulton M, Henson D, Koh H-H, Murray IJ (1999) Macular pigment and age related macular degeneration. Br J Ophthalmol 83: 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsley DA, Kuh E, Welsch RE (2005) Regression Diagnostics: Identifying Influential Data and Sources of Collinearity, John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Met 57: 289–300 [Google Scholar]
- Bernstein PS, Arunkumar R (2020) The emerging roles of the macular pigment carotenoids throughout the lifespan and in prenatal supplementation. J Lipid Res: jlr.TR120000956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer P, Mayer M, Kleinig H (1989) Molecular oxygen and the state of geometric isomerism of intermediates are essential in the carotene desaturation and cyclization reactions in daffodil chromoplasts. Eur J Biochem 184: 141–150 [DOI] [PubMed] [Google Scholar]
- Blessin CW, Brecher JD, Dimler RJ (1963) Carotenoid of corn and sorghum: V. Distribution of xanthophylls and carotenes in hand-dissected and dry-milled fractions of yellow dent corn. Cereal Chem 40: 582–586 [Google Scholar]
- Bouis HE, Welch RM (2010) Biofortification—a sustainable agricultural strategy for reducing micronutrient malnutrition in the Global South. Crop Sci 50: S-20–S-32 [Google Scholar]
- Box GEP, Cox DR (1964) An analysis of transformations. J R Stat Soc B Met 26: 211–252 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Brausemann A, Gemmecker S, Koschmieder J, Ghisla S, Beyer P, Einsle O (2017) Structure of phytoene desaturase provides insights into herbicide binding and reaction mechanisms involved in carotene desaturation. Structure 25: 1222–1232 [DOI] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Buckner B, Miguel PS, Janick-Buckner D, Bennetzen JL (1996) The y1 gene of maize codes for phytoene synthase. Genetics 143: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkschat M (2009) Linear estimators and predictors based on generalized order statistics from generalized Pareto distributions. Commun Stat Theory Methods 39: 311–326 [Google Scholar]
- Butler DG, Cullis BR, Gilmour AR, Gogel BG, Thompson R (2017) ASReml-R Reference Manual Version 4, VSN International Ltd, Hemel Hempstead
- Butts C (2008) Network: a package for managing relational data. R J Stat Softw 24: 1–36 [Google Scholar]
- Butts C (2015) Network: classes for relational data. The Statnet Project (http://statnet.org)
- Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M (1999) Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Chandler K, Lipka AE, Owens BF, Li H, Buckler ES, Rocheford T, Gore MA (2013) Genetic analysis of visually scored orange kernel color in maize. Crop Sci 53: 189–200 [Google Scholar]
- Chia JM, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, Elshire RJ, Gaut B, Geller L, Glaubitz JC, et al. (2012) Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet 44: 803–807 [DOI] [PubMed] [Google Scholar]
- Cingolani P Platts A Wang LL Coon M Nguyen T Wang L Land SJ Lu X Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92 10.416119695 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs GF, McClung JP (2017) The Vitamins: Fundamental Aspects in Nutrition and Health, 5th edn, Academic Press, London [Google Scholar]
- Cunningham FX Jr, Pogson BJ, Sun Z, McDonald KA, DellaPenna D, Gantt E (1996) Functional analysis of the β and ε lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8: 1613–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhliwayo T, Palacios-Rojas N, Crossa J, Pixley KV (2014) Effects of S1 recurrent selection for provitamin A carotenoid content for three open-pollinated maize cultivars. Crop Sci 54: 2449–2460 [Google Scholar]
- Diepenbrock CH, Kandianis CB, Lipka AE, Magallanes-Lundback M, Vaillancourt B, Gongora-Castillo E, Wallace G, Cepela J, Mesberg A, Bradbury PJ, et al. (2017) Novel loci underlie natural variation in vitamin E levels in maize grain. Plant Cell 29: 2374–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepenbrock CH, Gore MA (2015) Closing the divide between human nutrition and plant breeding. Crop Sci 55: 1437–1448 [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler E, Mitchell S (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez JM, Cantero A, Reindl A, Reichler S, Leon P (2001) 1-Deoxy-d-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276: 22901–22909 [DOI] [PubMed] [Google Scholar]
- Eroglu A, Harrison EH (2013) Carotenoid metabolism in mammals, including man: formation, occurrence, and function of apocarotenoids. J Lipid Res 54: 1719–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Kaller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Fu X, Wang Y, Xu J, Feng HY, Li WY, Xu JT, Jittham O, Zhang X, Zhang LL, et al. (2020) Genetic basis of kernel nutritional traits during maize domestication and improvement. Plant J 101: 278–292 [DOI] [PubMed] [Google Scholar]
- Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES (2005) Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44: 1054–1064 [DOI] [PubMed] [Google Scholar]
- Foudree A, Putarjunan A, Kambakam S, Nolan T, Fussell J, Pogorelko G, Rodermel S (2012) The mechanism of variegation in immutans provides insight into chloroplast biogenesis. Front Plant Sci 3: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Cheng Y, Linghu J, Yang X, Kang L, Zhang Z, Zhang J, He C, Du X, Peng Z, et al. (2013a) RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun 4: 2832. [DOI] [PubMed] [Google Scholar]
- Fu Z, Chai Y, Zhou Y, Yang X, Warburton ML, Xu S, Cai Y, Zhang D, Li J, Yan J (2013b) Natural variation in the sequence of PSY1 and frequency of favorable polymorphisms among tropical and temperate maize germplasm. Theor Appl Genet 126: 923–935 [DOI] [PubMed] [Google Scholar]
- Gebremeskel S, Garcia-Oliviera AL, Menkir A, Adetimirin V, Gedil M (2018) Effectiveness of predictive markers for marker assisted selection of pro-vitamin A carotenoids in medium-late maturing maize (Zea mays L.) inbred lines. J Cereal Sci 79: 27–34 [Google Scholar]
- Gilmore AM (1997) Mechanistic aspects of xanthophyll cycle-dependent photoprotection in higher plant chloroplasts and leaves. Physiol Plant 99: 197–209 [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9: e90346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S, Mehrshahi P, Magallanes-Lundback M, Lipka AE, Angelovici R, Gore MA, DellaPenna D (2016) ZEAXANTHIN EPOXIDASE activity potentiates carotenoid degradation in maturing seed. Plant Physiol 171: 1837–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S, Ha SH, Magallanes-Lundback M, Gilliland LU, Zhou A, Lipka AE, Nguyen Y-N, Angelovici R, Lin H, Cepela J, et al. (2013) Carotenoid cleavage dioxygenase4 is a negative regulator of beta-carotene content in Arabidopsis seeds. Plant Cell 25: 4812–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Graham RD, Welch RM, Bouis HE (2001) Addressing micronutrient malnutrition through enhancing the nutritional quality of staple foods: principles, perspectives, and knowledge gaps. Adv Agron 70: 77–142 [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, et al. (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker M, Bramley PM (1999) Expression of prokaryotic 1-deoxy-d-xylulose-5-phosphatases in Escherichia coli increases carotenoid and ubiquinone biosynthesis. FEBS Lett 448: 115–119 [DOI] [PubMed] [Google Scholar]
- Holland JB, Nyquist WE, Cervantes-Martínez CT (2003) Estimating and Interpreting Heritability for Plant Breeding: An Update in Plant Breeding Reviews, John Wiley & Sons, Inc., New York [Google Scholar]
- Hoopes GM, Hamilton JP, Wood JC, Esteban E, Pasha A, Vaillancourt B, Provart NJ, Buell CR (2018) An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J 97: 1154–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HY, Browne C, Guill K, Coles N, Eller M, Garcia A, Lepak N, Melia-Hancock S, Oropeza-Rosas M, Salvo S, et al. (2012) The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity 108: 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns P, Holzwarth AR (2012) The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim Biophys Acta 1817: 182–193 [DOI] [PubMed] [Google Scholar]
- Jia KP, Baz L, Al-Babili S (2018) From carotenoids to strigolactones. J Exp Bot 69: 2189–2204 [DOI] [PubMed] [Google Scholar]
- Kermode AR (2005) Role of abscisic acid in seed dormancy. J Plant Growth Regul 24: 319–344 [Google Scholar]
- Khoo HE, Prasad KN, Kong KW, Jiang Y, Ismail A (2011) Carotenoids and their isomers: color pigments in fruits and vegetables. Molecules 16: 1710–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler L, Maezumi SY, de Souza JG, Przelomşka NAS, Costa FM, Smith O, Loiselle H, Ramos-Madrigal J, Wales N, Ribeiro ER, et al. (2018) Multiproxy evidence highlights a complex evolutionary legacy of maize in South America. Science 362: 1309–1313 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33–36 [DOI] [PubMed] [Google Scholar]
- Krinsky NI, Landrum JT, Bone RA (2003) Biologic mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu Rev Nutr 23: 171–201 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra M (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S (2019) The Vmatch large scale sequence analysis software (http://www.vmatch.de)
- Kutner MH, Nachtsheim CJ, Neter J, Li W (2004) Applied Linear Statistical Models, McGraw Hill Irwin, Boston, MA [Google Scholar]
- Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair D, Hallauer A (2002) Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol Biol 48: 453–461 [DOI] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET (2008) The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol 147: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bradbury P, Ersoz E, Buckler ES, Wang J (2011) Joint QTL linkage mapping for multiple-cross mating design sharing one common parent. PLoS ONE 6: e17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yuan H, Zeng Y, Xu Q (2016) Plastids and carotenoid accumulation. Subcell Biochem 79: 273–293 [DOI] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- Mayer MP, Beyer P, Kleinig H (1990) Quinone compounds are able to replace molecular oxygen as terminal electron acceptor in phytoene desaturation in chromoplasts of Narcissus pseudonarcissus L. Eur J Biochem 191: 359–363 [DOI] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Menkir A, Maziya-Dixon B, Mengesha W, Rocheford T, Oladeji E (2017) Accruing genetic gain in pro-vitamin A enrichment from harnessing diverse maize germplasm. Euphytica 213: 105 [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W (1996) Applied Linear Statistical Methods, Irwin, Chicago, IL [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D (1995) Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogut F, Bian Y, Bradbury PJ, Holland JB (2015) Joint-multiple family linkage analysis predicts within-family variation better than single-family analysis of the maize nested association mapping population. Heredity 114: 552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens BF, Lipka AE, Magallanes-Lundback M, Tiede T, Diepenbrock CH, Kandianis CB, Kim E, Cepela J, Mateos-Hernandez M, Buell CR, et al. (2014) A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels. Genetics 198: 1699–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pixley K, Palacios NP, Babu R, Mutale R, Simpungwe E (2013) Biofortification of maize with provitamin A carotenoids. In Tanumihardo SA, ed., Carotenoids, Human Health and Nutrition, Springer Science + Business Media, New York, pp. 271–292 [Google Scholar]
- Portwood JP II, Woodhouse MR, Cannon EK, Gardiner JM, Harper LC, Schaeffer ML, Walsh JR, Sen TZ, Cho KT, Schott DA, et al. (2019) MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Res 47: D1146–D1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanna BM, Palacios-Rojas N, Hossain F, Muthusamy V, Menkir A, Dhliwayo T, Ndhlela T, San Vicente F, Nair SK, Vivek BS, et al. (2020) Molecular breeding for nutritionally enriched maize: status and prospects. Front Genet 10: 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan RF, Shumskaya M, Bradbury LM, Beltran J, Ma C, Kennelly EJ, Wurtzel ET (2012) Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol 160: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2018) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria
- Ramstein GR, Larsson SJ, Cook JP, Edwards JW, Ersoz ES, Flint-Garcia S, Gardner CA, Holland JB, Lorenz AJ, McMullen MD, et al. (2020) Dominance effects and functional enrichments improve prediction of agronomic traits in hybrid maize. Genetics 215: 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice AL, West KP Jr, Black RE (2004) Vitamin A deficiency. In Ezzati M, Lopez AD, Rodgers A, Murray CJL, eds., Comparative Quantification of Health Risks: Global and Regional Burden of Disease Attributable to Selected Major Risk Factors, Vol. 1, World Health Organization, Geneva, Switzerland, pp. 211–256 [Google Scholar]
- Rodermel S (2002) Arabidopsis variegation mutants. Arabidopsis Book 1: e0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Bradbury PJ, Elshire RJ, Glaubitz JC, Acharya CB, Mitchell SE, Li C, Li Y, Buckler ES (2015) Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc Natl Acad Sci USA 112: 3823–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D, Ivanov AG, Fu A, Geisler-Lee J, Hendrickson L, Geisler M, Stewart G, Krol M, Hurry V, Rodermel SR, et al. (2006) IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of Arabidopsis during steady-state photosynthesis. Plant Physiol 142: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D, Bode R, Li W, Krol M, Saccon D, Wang S, Schillaci LA, Rodermel S, Maxwell D, Huner N (2009) Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21: 3473–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman A, Birol E, Bouis HE, Boy E, De Moura FF, Islam Y, Pfeiffer WH (2013) Biofortification: progress toward a more nourishing future. Glob Food Sec 2: 9–17 [Google Scholar]
- SAS Institute Inc. (2011) Base SAS® 9.3 Procedures Guide: Statistical Procedures, SAS Institute Inc., Cary, NC [Google Scholar]
- Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6: 461–464 [Google Scholar]
- Scrucca L, Fop M, Murphy TB, Raftery AE (2016) mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J 8: 289–317 [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M, Gilbert M, Laboure AM, Kuntz M (2007) Dual role of the plastid terminal oxidase in tomato. Plant Physiol 145: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl W, Sies H (2005) Bioactivity and protective effects of natural carotenoids. Biochim Biophys Acta 1740: 101–107 [DOI] [PubMed] [Google Scholar]
- Suwarno WB, Pixley KV, Palacios-Rojas N, Kaeppler SM, Babu R (2014) Formation of heterotic groups and understanding genetic effects in a provitamin A biofortified maize breeding program. Crop Sci 54: 14–24 [Google Scholar]
- Suwarno WB, Pixley KV, Palacios-Rojas N, Kaeppler SM, Babu R (2015) Genome-wide association analysis reveals new targets for carotenoid biofortification in maize. Theor Appl Genet 128: 851–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts K, Gutaker RM, Benz B, Blake M, Bukowski R, Holland J, Kruse-Peeples M, Lepak N, Prim L, Cinta Romay M, et al. (2017) Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America. Science 357: 512–515 [DOI] [PubMed] [Google Scholar]
- Tan B-C, Guan J-C, Ding S, Wu S, Saunders JW, Koch KE, McCarty DR (2017) Structure and origin of the White Cap locus and its role in evolution of grain color in maize. Genetics 206: 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B-C, Joseph LM, Deng W-T, Liu L, Li Q-B, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D (2004) The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid E-ring hydroxylation activity. Proc Natl Acad Sci U S A 101: 402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2: 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Holmes CC, Mott R, Flint J (2009) Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Tan B-C, McCarty DR, Klee HJ (2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J Biol Chem 283: 11364–11373 [DOI] [PubMed] [Google Scholar]
- Wallace JG, Bradbury PJ, Zhang N, Gibon Y, Stitt M, Buckler ES (2014) Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet 10: e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber EJ (1987) Carotenoids and tocols of corn grain determined by HPLC. J Am Oil Chem Soc 64: 1129–1134 [Google Scholar]
- Weir BS (1996) Genetic data analysis II, Sinauer Associates
- Welch RM, Graham RD (2004) Breeding for micronutrients in staple food crops from a human nutrition perspective. J Exp Bot 55: 353–364 [DOI] [PubMed] [Google Scholar]
- West KP, Darnton-Hill I (2008) Vitamin A deficiency. In Semba RD, Bloem M, eds., Nutrition and Health in Developing Countries, 2nd edn, The Humana Press, Inc., Totowa, NJ, pp. 377–433 [Google Scholar]
- West KP Jr (2002) Extent of vitamin A deficiency among preschool children and women of reproductive age. J Nutr 132: 28575–28665 [DOI] [PubMed] [Google Scholar]
- Wetzel CM, Jiang C-Z, Meehan LJ, Voytas DF, Rodermel SR (1994) Nuclear–organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J 6: 161–175 [DOI] [PubMed] [Google Scholar]
- Wong WL, Su X, Li X, Cheung CMG, Klein R, Cheng C-Y, Wong TY (2014) Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Health 2: e106–e116 [DOI] [PubMed] [Google Scholar]
- Wu D, Wright DA, Wetzel C, Voytas DF, Rodermel S (1999) The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, Bai L, Kim E-H, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, et al. (2010) Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat Genet 42: 322–327 [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Naqvi S, Breitenbach J, Sandmann G, Christou P, Capell T (2008) Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Natl Acad Sci U S A 105: 18232–18237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.