Abstract
Epigenetic mechanisms play diverse roles in the regulation of genome stability in eukaryotes. In Arabidopsis thaliana, genome stability is maintained during DNA replication by the H3.1K27 methyltransferases ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 (ATXR5) and ATXR6, which catalyze the deposition of K27me1 on replication-dependent H3.1 variants. The loss of H3.1K27me1 in atxr5 atxr6 double mutants leads to heterochromatin defects, including transcriptional de-repression and genomic instability, but the molecular mechanisms involved remain largely unknown. In this study, we identified the transcriptional co-activator and conserved histone acetyltransferase GCN5 as a mediator of transcriptional de-repression and genomic instability in the absence of H3.1K27me1. GCN5 is part of a SAGA-like complex in plants that requires the GCN5-interacting protein ADA2b and the chromatin remodeler CHR6 to mediate the heterochromatic defects in atxr5 atxr6 mutants. Our results also indicate that Arabidopsis GCN5 acetylates multiple lysine residues on H3.1 variants, but H3.1K27 and H3.1K36 play essential functions in inducing genomic instability in the absence of H3.1K27me1. Finally, we show that H3.1K36 acetylation by GCN5 is negatively regulated by H3.1K27me1 in vitro. Overall, this work reveals a key molecular role for H3.1K27me1 in maintaining transcriptional silencing and genome stability in heterochromatin by restricting GCN5-mediated histone acetylation in plants.
The histone post-translational modification H3.1K27me1 maintains transcriptional silencing and genome stability by preventing histone acetylation mediated by the histone acetyltransferase GCN5.
Introduction
Genome and epigenome instability have been implicated in many human diseases, including cancer and neurodegenerative disorders. In proliferating cells, key mechanisms are required to properly copy DNA and different epigenetic states of the genome in the context of ongoing transcription and DNA repair. Chromatin replication is therefore a complex molecular operation that can lead to genomic rearrangements and other types of deleterious mutations in the absence of mechanisms preserving genome stability (Weinert et al., 2009; Chen et al., 2010).
Epigenetic information plays multiple regulatory roles during S phase of the cell cycle that are required to maintain genome stability in eukaryotes. In plants, one of the most well-studied genome maintenance pathways involves the histone post-translational modification (PTM) H3K27me1. The loss of H3K27me1 results in transcriptional de-repression at heterochromatic loci and defects in the structural organization of heterochromatin (Jacob et al., 2009; Stroud et al., 2012). In addition, decreased levels of H3K27me1 induce genome instability characterized by the presence of an excess of repetitive DNA (e.g. transposons) in heterochromatin, hereafter referred to as heterochromatin amplification (Jacob et al., 2010). In Arabidopsis thaliana, H3K27me1 is catalyzed by the plant-specific histone methyltransferases (HMTs) ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 (ATXR5) and ATXR6 (abbreviated ATXR5/6 hereafter), which are recruited to replication forks during DNA replication (Raynaud et al., 2006; Jacob et al., 2009; Davarinejad et al., 2019). Biochemical and structural studies have revealed that the SET domains of ATXR5/6 can methylate replication-dependent H3.1 variants, but not replication-independent H3.3 variants (Jacob et al., 2014). These observations indicate that ATXR5/6 maintain H3K27me1 by methylating newly synthesized H3.1 variants (H3.1K27me1) during DNA replication, which protects against transcriptional de-repression and heterochromatin amplification. The precise molecular mechanism responsible for heterochromatin amplification in the absence of H3.1K27me1 remains unknown. However, a previous study suggested that transcriptional de-repression in the heterochromatin of atxr5 atxr6 double mutant plants (hereafter atxr5/6) is the cause of the genomic instability phenotype, potentially by inducing collisions between the transcription machinery and replication forks, and/or through R-loop formation (Hale et al., 2016). Based on this model, it is predicted that ATXR5/6-catalyzed H3.1K27me1 plays a key role in preventing the transcriptional activity in the heterochromatin of plants.
Many PTMs on histones function as recruitment signals for chromatin reader proteins, which promote specific cellular activities (such as transcription) at genomic regions enriched in these histone PTMs (Musselman et al., 2012). Multiple studies have shown that methylation at H3K27 regulates transcriptional activity through various mechanisms, which are related to the specific methylation level (i.e. me1, me2, or me3) at K27. For example, H3K27me3 is involved in the recruitment of the repressive PRC1 complex in animals (Fischle et al., 2003), and this role is conserved in plants (Huang et al., 2019). H3K27me3 is also directly recognized by the PRC2 complex, which catalyzes K27me3 on histone H3, thus allowing for a “read–write” propagation mechanism that contributes to maintaining H3K27me3 levels in vivo (Hansen et al., 2008; Margueron et al., 2009; Xu et al., 2010). In contrast to H3K27me3, H3K27me1 and H3K27me2 are not as well characterized in animals, but they have specific effects on the regulation of transcriptional activity that do not appear to involve recruitment of chromatin readers. In mouse embryonic stem cells (ESCs), H3K27me2 is present on the majority of total histone H3 in chromatin and safeguards against unintended transcription by preventing CBP/p300-mediated H3K27 acetylation (H3K27ac) at noncell-type-specific enhancers (Ferrari et al., 2014). In contrast, H3K27me1 is present at <5% of total H3s in ESCs, is associated with transcriptionally active genes, and contributes to their expression (Ferrari et al., 2014). However, the mechanism by which H3K27me1 performs this function remains unknown. Predicting the role of ATXR5/6-catalyzed H3K27me1 in plants based on comparative analysis with H3K27me1/me2 in animals is challenging, as it shares the same methylation level of transcriptionally permissive H3K27me1, but its function in heterochromatin silencing in plants suggests properties related to H3K27me2. An additional similarity between plant H3K27me1 and animal H3K27me2 is that these histone PTMs are widely distributed and very abundant in their respective genomes. In Arabidopsis, H3K27me1 was estimated to be present on >50% of total H3 in inflorescence tissues (Johnson et al., 2004), and it is enriched in transcriptionally silent regions of the genome (Jacob et al., 2010). These observations suggest that H3.1K27me1 in plants prevents H3.1K27ac, thus providing a molecular mechanism for the role of ATXR5/6 in protecting against transcriptional de-repression and genomic instability in plants.
In this work, we identify the conserved histone acetyltransferase (HAT) GCN5 as a mediator of transcriptional de-repression and heterochromatin amplification in the absence of H3.1K27me1 in Arabidopsis. GCN5 cooperates with the transcriptional co-activator ADA2b and the chromatin remodeler CHR6 to induce these heterochromatic phenotypes. Our results also show that H3.1K36 plays a key role in inducing genome instability and transcriptional de-repression in the absence of H3.1K27me1, and that H3.1K27me1 interferes with GCN5-mediated acetylation at both H3.1K27 and H3.1K36. Overall, these results demonstrate the key role played by GCN5-mediated histone acetylation in contributing to the heterochromatin phenotypes observed in the absence of ATXR5 and ATXR6 in plants.
Results
Transcriptional de-repression and heterochromatin amplification in the absence of H3.1K27me1 are suppressed in gcn5 mutants
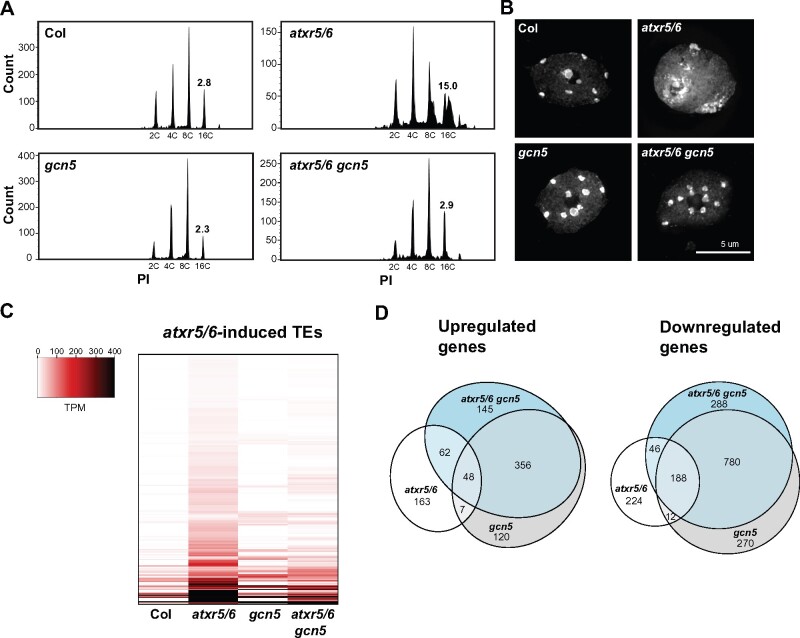
One mechanism by which H3.1K27me1 might interfere with transcription in heterochromatin of plants is by preventing the deposition of H3.1K27ac, as methylation and acetylation at H3K27 have been shown to act antagonistically in other biological systems (Tie et al., 2009; Pasini et al., 2010). H3K27ac is catalyzed by multiple HATs in eukaryotes, including the widely conserved protein GCN5 (Kuo et al., 1996; Suka et al., 2001; Kuo and Andrews, 2013; Cieniewicz et al., 2014; Chen et al., 2017). The Arabidopsis genome contains a single gene encoding a GCN5 homolog (Pandey et al., 2002). To assess if Arabidopsis GCN5 mediates the heterochromatin phenotypes associated with loss of H3.1K27me1, we created an atxr5/6 gcn5 triple mutant by crossing a T-DNA insertion allele of gcn5 (SALK_030913) into the hypomorphic atxr5/6 mutant background (Jacob et al., 2009). This T-DNA mutant allele of gcn5 results in the production of a truncated transcript lacking sequence coding for the C-terminus of the GCN5 protein (Supplemental Figure 1, A and B). Flow cytometry analyses showed strong suppression of heterochromatin amplification in the triple mutant, as represented by the loss of the characteristic broad peaks corresponding to 8C and 16C endoreduplicated nuclei in atxr5/6 mutants (Figure 1A; Supplemental Figure 1C). We also observed by microscopy that the heterochromatin decondensation phenotype of atxr5/6 plants is suppressed in the atxr5/6 gcn5 triple mutant (Figure 1B; Supplemental Figure 1D). A role for GCN5 in inducing genomic instability in atxr5/6 was confirmed by observing suppression of heterochromatin amplification using different mutant alleles of gcn5 (i.e. small indels that change the reading frame of GCN5 downstream of the start codon in the first exon) generated by temperature-optimized CRISPR/Cas9 (Supplemental Figure 1, A, E, F, G, and H; LeBlanc et al., 2018).
Figure 1.
A mutation in GCN5 suppresses transcriptional de-repression and heterochromatin amplification associated with H3.1K27me1 depletion. (A) Flow cytometry profiles of Col, atxr5/6, gcn5, and atxr5/6 gcn5 nuclei stained with propidium iodide (PI) with 2,000 gated events. The numbers below the peaks indicate ploidy levels of the nuclei. The numbers above the 16C peaks indicate the robust coefficient of variation (CV). (B) Leaf interphase nuclei of Col, atxr5/6, gcn5, and atxr5/6 gcn5 stained with DAPI. (C) Heat map showing the relative expression levels of 486 atxr5/6-induced TEs (Supplemental Data Set 1) as measured by TPM (transcripts per million) in Col, atxr5/6, gcn5, and atxr5/6 gcn5. (D) Euler diagrams showing the number of upregulated and downregulated genes (two-fold change) in atxr5/6, gcn5 and atxr5/6 gcn5 compared to Col plants (Padj <0.05).
To measure the impact of GCN5 on transcriptional de-repression in atxr5/6 mutants, we performed RNA-seq analyses and observed widespread suppression of transposable element (TE) reactivation in the atxr5/6 gcn5 triple mutant compared to atxr5/6, although some TEs remained de-repressed compared to Col (Figure 1C; Supplemental Data Set 1). Although GCN5 has a genome-wide impact on transcription, as shown by the 1781 misregulated genes in gcn5 single mutants (Figure 1D; Supplemental Data Set 2), none of the known transcriptional suppressors of atxr5/6 mutants [SERRATE {SE}, AtTHP1, AtSAC3B, AtSTUbL2, AtMBD9, and DDM1] are downregulated in gcn5 mutants or atxr5/6 gcn5 triple mutants (Supplemental Figure 1I; Stroud et al., 2012; Hale et al., 2016; Ma et al., 2018), indicating that suppression of the heterochromatin phenotypes in atxr5/6 gcn5 is not the result of decreased expression levels of these genes.
GCN5 functions with ADA2b and CHR6 to disrupt heterochromatin in the absence of H3.1K27me1
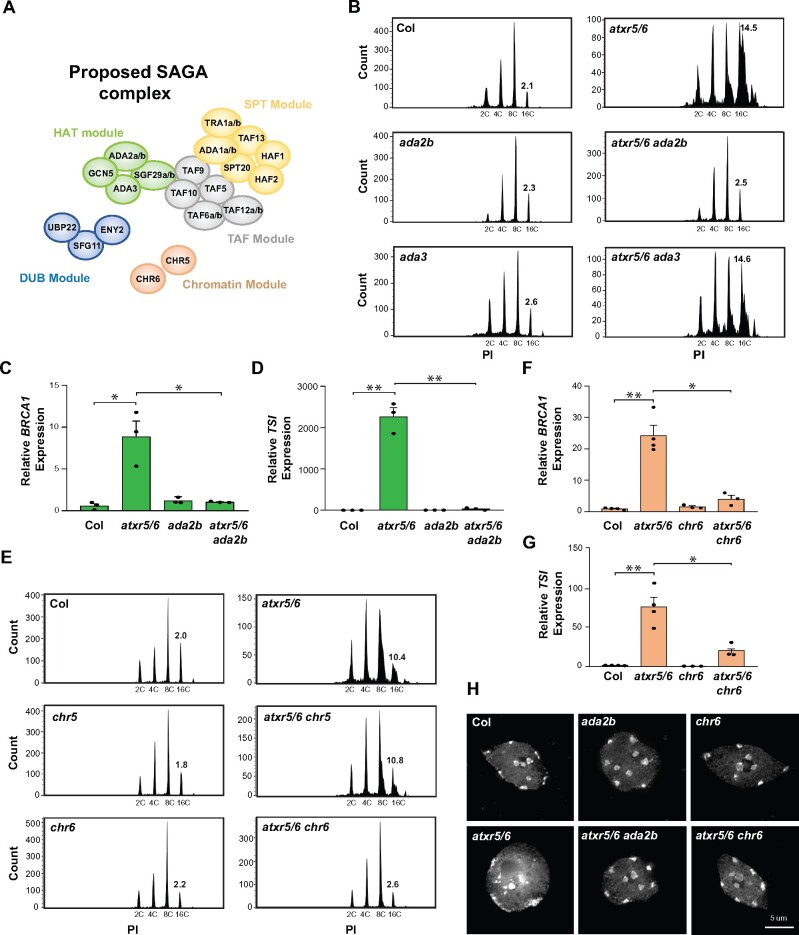
GCN5 is a member of the multi-subunit SAGA complex, which acts as a transcriptional coactivator in yeast and animals, in part by modifying chromatin (Spedale et al., 2012). Key components of this complex are the proteins GCN5, ADA2, ADA3, and SGF29, which form the histone acetylation module of SAGA (Figure 2A). The Arabidopsis genome contains single genes encoding GCN5 and ADA3 and two genes each encoding ADA2 (ADA2a and ADA2b) and SGF29 (SGF29a and SGF29b; Moraga and Aquea, 2015). gcn5 and ada2b (SALK_019407; Kornet and Scheres, 2009) single mutants show pleiotropic phenotypes, which are also shared by the atxr5/6 gcn5 and atxr5/6 ada2b mutants, respectively (Supplemental Figure 2A; Vlachonasios et al., 2003). To test if ADA2b is also required for inducing the heterochromatin phenotypes of atxr5/6 mutants, we generated an atxr5/6 ada2b triple mutant. The results from flow cytometry experiments show that genomic instability is suppressed in the atxr5/6 ada2b triple mutant (Figure 2B; Supplemental Figure 2B). This finding is supported by the altered expression of BRCA1, which functions in eukaryotes as a DNA-damage response gene involved in maintaining genome stability (Prakash et al., 2015; Savage and Harkin, 2015). As previously reported, BRCA1 levels are upregulated in atxr5/6 (Stroud et al., 2012), and our results show that both ADA2b and GCN5 are required for this induction (Figure 2C; Supplemental Figure 2C). Like gcn5, introducing the ada2b mutation into the atxr5/6 background suppressed transcriptional de-repression of the heterochromatic TSI DNA repeat (Figure 2D; Supplemental Figure 2D).
Figure 2.
GCN5, ADA2b, and CHR6 are required to induce heterochromatic defects in atxr5/6 mutants. (A) Proposed subunits of the Arabidopsis SAGA complex; adapted from (Moraga and Aquea, 2015). HAT: histone acetylation module; DUB: deubiquitination module; SPT: recruiting module; TAF: coactivator architecture module. (B) Flow cytometry profiles of Col, atxr5/6, ada2b, atxr5/6 ada2b, ada3, and atxr5/6 ada3. The numbers above the 16C peaks indicate the robust CV. (C and D) RT-qPCR analyses of BRCA1 (C) and the repetitive element TSI (D) in Col, atxr5/6, ada2b, and atxr5/6 ada2b. Data represent the mean of three biological replicates, and error bars indicate the standard error of the mean (sem). Unpaired t-test: * p < 0.05, ** p < 0.001. (E) Flow cytometry profiles of Col, atxr5/6, chr5, atxr5/6 chr5, chr6, and atxr5/6 chr6. (F and G) RT-qPCR analyses of BRCA1 (F) and the repetitive element TSI (G) in Col, atxr5/6, chr6 and atxr5/6 chr6. Data represent the mean of three biological replicates, and error bars indicate sem. Unpaired t-test: *p < 0.05, **p < 0.001. (H) Leaf interphase nuclei of Col, atxr5/6, ada2b, atxr5/6 ada2b, chr6, and atxr5/6 chr6 stained with DAPI.
Next, we generated an atxr5/6 ada3 triple mutant using a T-DNA insertion (SALK_042026C) that prevents expression of a full-length ADA3 transcript (Supplemental Figure 2, E–F), but unlike atxr5/6 ada2b, it did not suppress the genome instability phenotype associated with the atxr5/6 double mutant (Figure 2B). The reported ADA3 protein in Arabidopsis displays low similarity to the ADA3 homologs from yeast and human (26.3 and 16.3%, respectively, compared to >35% similarity for GCN5 and ADA2b; Srivastava et al., 2015) and might therefore have diverged and not be required for GCN5 and ADA2b to acetylate histones in plants. To further investigate whether another module of SAGA mediates the heterochromatin phenotypes associated with the loss of H3.1K27me1, we created triple mutant combinations between atxr5/6 and T-DNA mutant alleles of chr5 or chr6. The chr5 allele (SAIL_504_D01) was characterized in a previous study (Zou et al., 2017), and we performed experiments demonstrating that the chr6 allele (GK_273E06) contains a T-DNA in an exon that results in a late-flowering phenotype also observed for other mutant alleles of chr6 (Supplemental Figure 2, G–K; Ogas et al., 1997, 1999; Henderson et al., 2004). CHR5 and CHR6 are both chromatin-remodeling enzymes that have been proposed to be present in the SAGA complex in plants (Figure 2A). CHR5 is the most closely related plant protein to CHD1-type chromatin remodelers that are part of the SAGA complex in yeast and mammals (Moraga and Aquea, 2015; Srivastava et al., 2015), while CHR6 (also known as CHD3/PICKLE) has been shown to co-purify with SAGA subunits from Arabidopsis tissue (Pfab et al., 2018). Our results show that heterochromatin amplification is suppressed in the atxr5/6 chr6 triple mutant, but not in atxr5/6 chr5 (Figure 2E; Supplemental Figure 2, G–L), thus suggesting an integral function for CHR6 within SAGA in plants. Like mutations in GCN5 and ADA2b, inactivating CHR6 in atxr5/6 mutants suppressed the transcriptional activation of BRCA1 and TSI, and chromatin decondensation (Figure 2, F–H; Supplemental Figure 2M). Overall, these results support an essential role for SAGA-mediated histone acetylation in mediating the heterochromatic phenotypes observed in the absence of H3.1K27me1.
GCN5-mediated H3.1K27ac induces the heterochromatin defects associated with loss of H3.1K27me1
The GCN5 homologs in yeast and mammals have been shown to acetylate multiple lysine residues of histone H3 (i.e. K9, K14, K18, K23, K27, and K36) in vitro; however, the substrate specificity in the context of different histone H3 variants for GCN5 homologs has been unclear (Kuo and Andrews, 2013; Cieniewicz et al., 2014). In addition, while Arabidopsis GCN5 has been shown to acetylate H3K9 and H3K14 on H3 peptides in vitro (Earley et al., 2007), we wanted to examine the role of this protein in acetylation at H3K27 using histone peptides or nucleosomal substrates to better reflect in vivo chromatin.
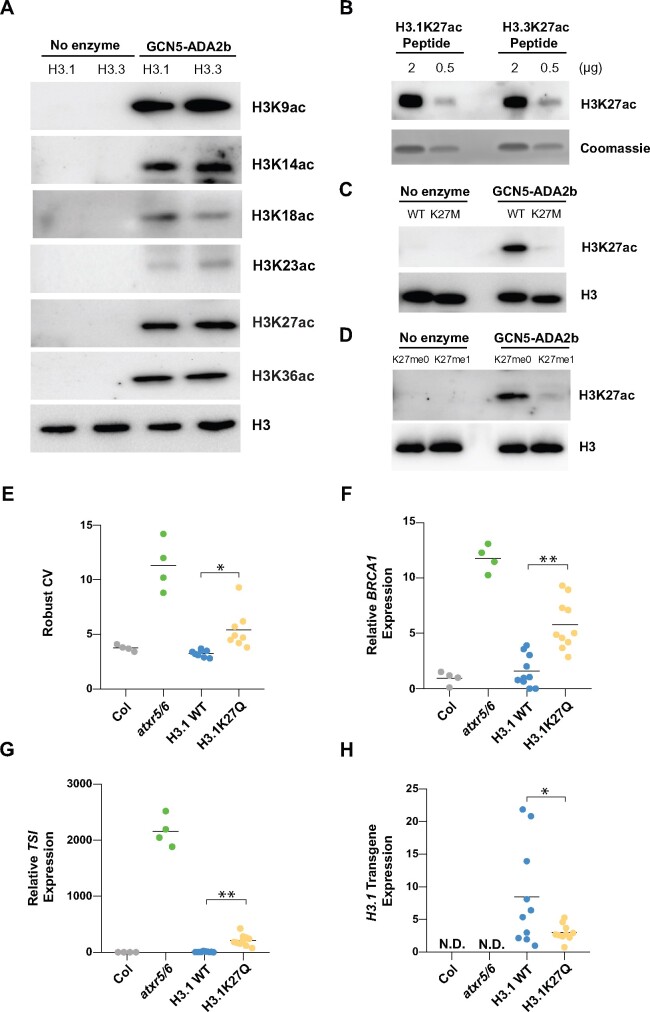
To investigate the substrate specificity of GCN5, we performed in vitro HAT assays using recombinant nucleosomes containing either plant histone H3.1 or H3.3 variants. We recombinantly expressed and purified an Arabidopsis protein complex composed of GCN5 and ADA2b (Supplemental Figure 3). Our results show that GCN5 has HAT activity at K9, K14, K18, K23, K27, and K36 of histone H3 (Figure 3A). Previous studies have shown that GCN5 is involved in the acetylation of H3K9, H3K14, H3K27, and H3K36 in vivo in plants (Chen et al., 2017; Mahrez et al., 2016; Kim et al., 2020), and we validated that it also mediates H3K18ac and H3K23ac by chromatin immunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR; Supplemental Figure 4, A–D). In contrast to ATXR5/6, the enzymatic activity of GCN5 at H3K27 is not regulated by H3 variants, as H3.1 and H3.3 nucleosomes showed equivalent acetylation levels in our HAT assays (Figure 3A). As controls for these results, we used H3.1K27ac and H3.3K27ac peptides to validate that the H3K27ac antibody used did not show preference for H3.1 or H3.3 (Figure 3B), and we validated the specificity of this antibody using H3K27M nucleosomes (Figure 3C). Similar to H3K27, we did not observe any major difference in HAT activity between H3.1 and H3.3 nucleosomes at the other lysine substrates of Arabidopsis GCN5 (Figure 3A). We also confirmed that H3.1K27me1 prevents acetylation by GCN5 at K27 using recombinant nucleosomes mono-methylated at K27 (Figure 3D). To assess if H3.1K27ac mediates the heterochromatin phenotypes present in atxr5/6 mutants in vivo, we introduced a transgene encoding an H3 variant harboring a glutamine residue (Q) instead of K27 (H3K27Q) into wild-type plants. Replacement of lysine with glutamine in histones has been used in in vivo chromatin studies to partially mimic the acetylated state of histone lysine residues (Megee et al., 1990; Zhang et al., 1998; Wang and Hayes, 2008). Our analyses of first-generation transformed (T1) plants showed that expression of H3.1K27Q in wild-type plants is sufficient to induce defects in genome stability, transcriptional activation of the genome instability marker BRCA1, and de-repression of the heterochromatic TSI repeat (Figure 3, E–H). Overall, these results suggest a role for GCN5-mediated H3.1K27ac in inducing the heterochromatic phenotypes associated with the loss of H3.1K27me1 in atxr5/6 mutants.
Figure 3.
Arabidopsis GCN5 acetylates H3.1K27 and induces the heterochromatic defects associated with atxr5/6. (A) In vitro HAT assays with the GCN5–ADA2b complex and H3.1 and H3.3 nucleosomes using anti-H3K9ac, anti-H3K14ac, anti-H3K18ac, anti-H3K23ac, anti-H3K27ac, anti-H3K36ac, and anti-H3 antibodies for detection. (B) Immunoblot of H3.1K27ac and H3.3K27ac peptides using H3K27ac antibody. (C) In vitro HAT assay with the GCN5–ADA2b complex and H3K27M nucleosomes using H3K27ac and H3 antibodies for detection. (D) In vitro HAT assays with the GCN5–ADA2b complex and H3K27me0 and H3K27me1 nucleosomes using anti-H3K27ac and anti-H3 antibodies for detection. (E) Robust CV values for 16C nuclei obtained by flow cytometry analysis. For Col and atxr5/6, each dot represents an independent biological replicate. For the H3.1 replacement lines, each dot represents one T1 plant. Horizontal bars indicate the mean. Unpaired t-test: *p < 0.01. (F, G, and H) RT-qPCR for the genome stability marker BRCA1 (F), the heterochromatic transcriptional reactivation marker TSI (G), and the H3.1 transgene (H) in Col, atxr5/6 and first-generation transformed (T1) plants expressing WT H3.1 or H3.1K27Q. At least eight independent T1 plants were used in the experiments. ND = not detected. Unpaired t-test: *p < 0.05, **p < 0.0001.
H3.1K36 is required to induce genome instability in the absence of H3.1K27me1
Our in vitro results suggest that, in addition to K27, other lysine residues on H3.1 could contribute to GCN5-mediated genomic instability in the absence of H3.1K27me1. To assess this hypothesis, we set up a suppressor screen based on in vivo replacement of histone H3.1 with the point mutant H3.1S28A. Replacement of serine with alanine on H3.1 variants at position 28 (H3.1S28A) generates H3.1 substrates that cannot be methylated by ATXR5/6 (Figure 4A; Bergamin et al., 2017). In contrast, H3.1S28A can still be methylated at K27 by plant PRC2-type complexes and acetylated by the GCN5–ADA2b complex, albeit at lower efficiencies (Supplemental Figure 5, A and B). We transformed the H3.1S28A transgene into a mutant Arabidopsis background expressing a reduced amount of endogenous histone H3.1 (i.e. h3.1 quadruple mutant; Jacob et al., 2014). In T1 plants, we observed phenotypes associated with the loss of H3.1K27me1, including genomic instability (as detected by flow cytometry), increased levels of the genome instability marker gene BRCA1 (Figure 4, B and C), and transcriptional de-repression of the heterochromatic TSI DNA repeat (Figure 4D). Attenuated heterochromatic phenotypes in H3.1S28A lines compared to atxr5/6 mutants are likely due to wild-type H3.1 histone still being present in the h3.1 quadruple mutant background. These results indicate that expressing H3.1S28A in plants generates phenotypes similar to those of atxr5/6 mutants due to the loss of H3.1K27me1. We then introduced a series of H3.1S28A expression constructs containing a second mutation (Lys to Arg replacement) at a residue known to be acetylated by GCN5 into the h3.1 quadruple mutant background and assessed T1 plants for phenotypes associated with the loss of H3.1K27me1. This targeted screen identified H3.1K36 as being essential for inducing genome instability, as flow cytometry analyses demonstrated that H3.1S28A K36R suppresses heterochromatin amplification, while the other targeted mutations do not (Figure 4B). The H3.1S28A K36R replacement line also rescued the increased expression of BRCA1 (Figure 4C) and the transcriptional de-repression of TSI (Figure 4D). Furthermore, expression of the H3.1S28A K36R mutant did not generate a serrated leaf phenotype, as seen in all the other H3.1S28A lines (Supplemental Figure 6). As mutations at K9, K14, K18, and K23 on the H3.1 variant did not suppress the phenotypes associated with the H3.1S28A mutation, these results indicate a specific role for H3.1K36 in inducing genome instability and transcriptional de-repression in the absence of H3.1K27me1.
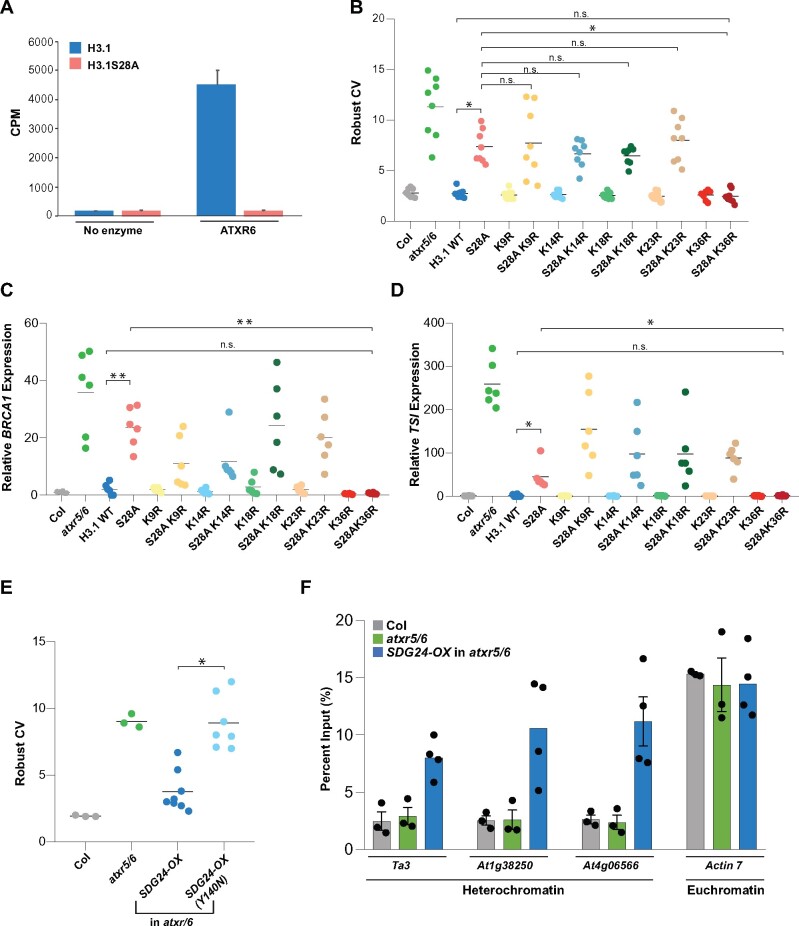
Figure 4.
Heterochromatin amplification in the absence of H3.1K27me1 requires H3.1K36. (A) In vitro histone lysine methylation assays using H3.1 and H3.1S28A peptide substrates and ATXR6. The average of three experiments and SEM are shown. CPM; counts per minute. (B) Robust CV values for 16C nuclei obtained by flow cytometry analysis. For Col and atxr5/6, each dot represents an independent biological replicate. For the H3.1 replacement lines, each dot represents one T1 plant. Horizontal bars indicate the mean. Unpaired t test: * p < 0.00001 and NS = not significantly different. (C and D) RT-qPCR analyses of BRCA1 (C) and the repetitive element TSI (D) in Col, atxr5/6, and H3.1 replacement lines. For Col and atxr5/6, each dot represents an independent biological replicate. For the H3.1 lines, each dot represents one T1 plant. Horizontal bars indicate the mean. Unpaired t-test: * p < 0.01, ** p < 0.0001, and NS = not significantly different. (E) Flow cytometry analyses showing robust CV values for 16C nuclei. For the SDG24-OX lines, each dot represents one T1 plant. Horizontal bars indicate the mean. Unpaired t-test: * p < 0.0001. (F) H3K36me3 ChIP-qPCR at Ta3, At1g38250, At4g06566, and ACTIN7. For Col and atxr5/6, each dot represents an independent biological replicate. For the SDG24-OX lines, each dot represents one T1 plant. Bars indicate the mean. Error bars indicate sem.
GCN5-mediated acetylation of H3.1K36 could be required to induce the heterochromatin defects of atxr5/6 mutants. One prediction from this model is that increasing histone methylation at H3.1K36 (H3.1K36me) would result in the suppression of the atxr5/6 mutant phenotypes, as H3.1K36me would antagonize H3.1K36 acetylation by GCN5. To test this notion, we constitutively expressed all five Arabidopsis H3K36 methyltransferase genes (SDG4, SDG7, SDG8, SDG24, and SDG26) in atxr5/6 mutants (Baumbusch et al., 2001; Springer et al., 2003). We performed flow cytometry analyses on T1 plants and found that overexpression of SDG24 (SDG24-OX) strongly suppresses the heterochromatin amplification phenotype (Figure 4E; Supplemental Figure 7A). We did not observe a similar effect in T1 lines overexpressing SDG4, SDG7, SDG8, or SDG26 (Supplemental Figure 7B). The ability of SDG24-OX to suppress heterochromatin amplification is dependent on SDG24 having a functional methyltransferase (SET) domain, as overexpression of an SDG24 variant containing a point mutation (Y140N) in a conserved residue essential for SET domain activity did not suppress the phenotype (Figure 4E; Dillon et al., 2005; Jacob et al., 2010). We performed ChIP-qPCR experiments with SDG24-OX plants and detected an increase in H3K36me3 levels at heterochromatic regions (the retrotransposon Ta3, At1g38250, and At4g06566) known to be transcriptionally de-repressed in atxr5/6 mutants (Figure 4F). Taken together, it is likely that H3K36 methylation opposes some features of the atrx5/6 phenotypes, potentially by preventing deposition of H3.1K36ac.
Loss of H3.1K27me1 in plants increases H3K27ac and H3K36ac deposition in heterochromatin
Our results support a model in which GCN5 acetylates both H3K27 and H3K36 in the absence of H3.1K27me1 to induce the heterochromatin phenotypes of atxr5/6 mutants. To assess if H3.1K27me1 depletion leads to an increase in H3K27ac and H3K36ac in vivo, we performed ChIP-Rx (ChIP-seq with reference exogenous genome) for H3K27ac and H3K36ac in Col (WT), atxr5/6, gcn5, and atxr5/6 gcn5 (Orlando et al., 2014). We found that both histone marks are enriched at the 5′-end of protein-coding genes after the transcriptional start site (TSS) in Arabidopsis (Figure 5A) and that this spatial distribution is associated with transcriptional activity, albeit not in a linear relationship (Supplemental Figure 8; Zhang et al., 2015; Mahrez et al., 2016). Comparative analysis of H3K27ac and H3K36ac in Col and gcn5 single mutants demonstrated that the loss of GCN5 results in a decrease in H3K27ac and H3K36ac at euchromatic genes (Figure 5A).
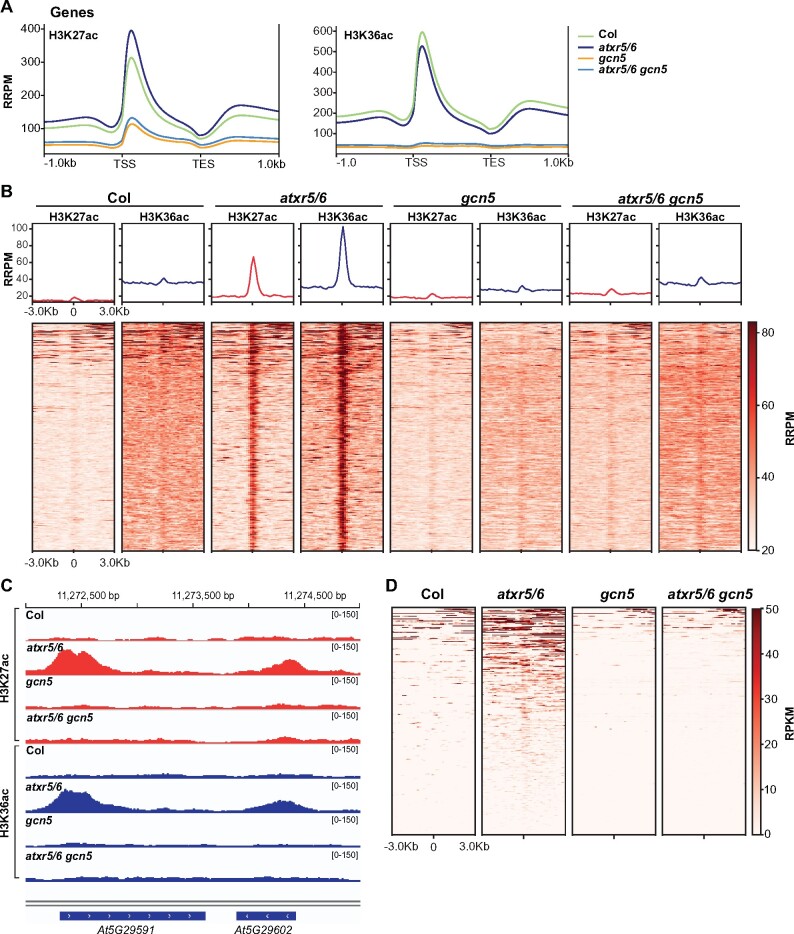
Figure 5.
Mutations in atxr5/6 lead to an increase in H3K27ac and H3K36ac in heterochromatin. (A) Normalized average distribution of H3K27ac and H3K36ac over protein-coding genes for Col, atxr5/6, gcn5, and atxr5/6 gcn5 in reference-adjusted reads per million (RRPM). TSS, transcription start site; TES, transcription end site. (B) Normalized average distribution and heatmap of H3K27ac and H3K36ac normalized reads surrounding the 323 H3K27ac/H3K36ac-enriched heterochromatic regions identified in atxr5/6 compared to Col. The regions are sorted based on levels (RRPM) of H3K27ac/H3K36ac enrichment. (C) Genome browser snapshot showing normalized H3K27ac and H3K36ac ChIP-seq data over a region of chromosome 5 that includes TE genes At5g29591 and At5g29602. The y-axis unit is RRPM. (D) Heatmap showing the RNA-seq reads mapping to the region ±3 kb around the center of the 323 H3K27ac/H3K36ac peaks as measured by RPKM in Col, atxr5/6, gcn5, and atxr5/6 gcn5. The regions are sorted based on expression level (RPKM).
Focusing on heterochromatin, which we defined based on previously identified chromatin states in Arabidopsis (Supplemental Data Set 3; Sequeira-Mendes et al., 2014), we identified 323 regions that were enriched in both H3K27ac and H3K36ac in atxr5/6 but not in Col plants (Figure 5, B and C; Supplemental Data Set 4). H3K27ac and H3K36ac enrichment in heterochromatin was greatly reduced in atxr5/6 gcn5 triple mutants (Figure 5, B and C), suggesting that the higher levels of H3K27ac and H3K36ac in heterochromatic regions of atxr5/6 are almost completely dependent on GCN5. We next tested if the de-repressed TEs identified in atxr5/6 by RNA-seq overlap or are in close proximity (±3 kb) to the 323 genomic regions showing increased levels of H3K27ac and H3K36ac in atxr5/6. We observed a large overlap between transcriptionally de-repressed genomic regions and regions enriched in H3K27ac and H3K36ac in atxr5/6 mutants (Figure 5D; Supplemental Data Set 5). The regions shown in Figure 5D likely represent a low estimate of the total overlap between H3K27ac/H3K36ac regions and transposon reactivation due to the inherent lack of sensitivity of ChIP-seq and RNA-seq experiments in backgrounds showing low-level TE de-repression such as atxr5/6 mutants. For example, we found that a five-fold increase in sequencing depth (75 versus 15 million reads) in our RNA-seq experiments resulted in a 43% increase in the number of de-repressed TEs identified in atxr5/6 (446 TEs versus 312 TEs; Supplemental Data Set 1). To further demonstrate the sensitivity issue associated with low-level de-repression in atxr5/6, we performed RT-qPCR on multiple TEs that showed an increase in H3K27ac in atxr5/6 but were not identified as differently expressed by RNA-seq. For many of these TEs, including At1g36040 and At5g29602 (Supplemental Figure 9), we observed higher expression levels in atxr5/6 compared to wild-type plants, thus confirming the limitations of genome-wide sequencing for detecting low-level TE de-repression in atxr5/6 mutants. Taken together, these results demonstrate that the loss of H3.1K27me1 in atxr5/6 mutants leads to GCN5-dependent increases in H3K27ac and H3K36ac in heterochromatin.
H3.1K27me1 regulates the deposition of H3.1K36ac by GCN5
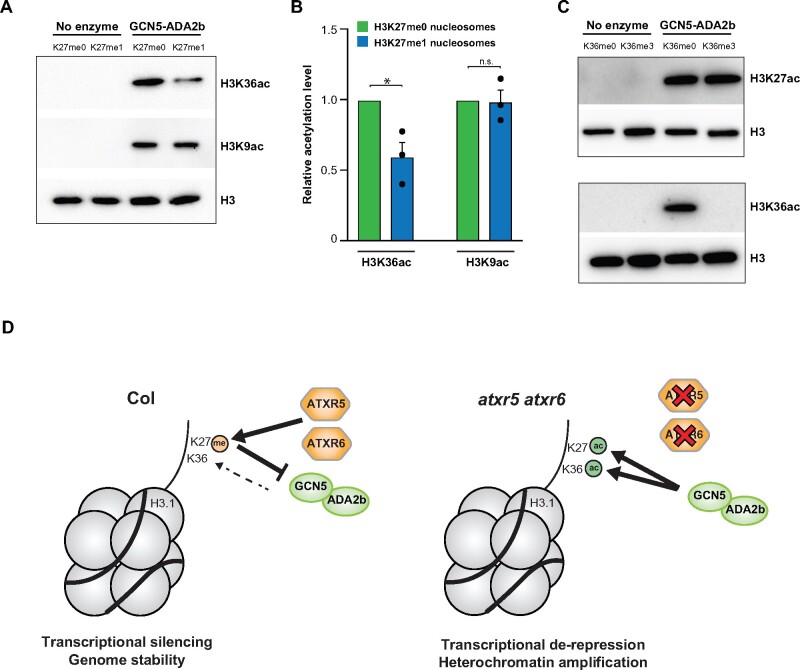
Methylation and acetylation at H3K27 have an antagonistic relationship in the genomes of animals. This relationship is mediated by the interplay between the H3K27 methyltransferase complex PRC2 (H3K27me) and the HATs p300 and CBP, which are responsible for H3K27ac (Tie et al., 2009; Pasini et al., 2010). Our work supports a similar relationship in plants at K27 on H3.1 variants that is mediated by different enzymes, with ATXR5/6-catalyzed H3.1K27me1 preventing the acetylation of H3.1K27 by GCN5. Interactions between PTMs on different histone residues also contribute to chromatin regulation in eukaryotes. One example of this is the inhibition of PRC2 activity towards H3K27 when H3K36 is di- or trimethylated on the same histone (Schmitges et al., 2011; Yuan et al., 2011; Voigt et al., 2012). This suggests that the activity of other chromatin-modifying enzymes may be affected by crosstalk between modified forms of H3K27 and H3K36. To assess if acetylation of H3.1K36 by GCN5 is regulated by H3.1K27me1, we performed in vitro HAT assays using recombinant plant nucleosomes containing either unmodified H3.1 or H3.1K27me1. In these assays, we consistently observed a 40% decrease in the levels of acetylation at H3.1K36 on nucleosomes mono-methylated at H3.1K27 compared to unmodified H3.1 (Figure 6, A and B). This effect of H3.1K27me1 on Arabidopsis GCN5 activity appears to be specific to H3.1K36, as GCN5-mediated acetylation of H3.1K9 was not affected by mono-methylation at K27. Conversely, we also tested if methylation at H3.1K36 would affect acetylation at K27 by GCN5, but we did not observe any difference in acetylation levels at K27 using K36me0 and K36me3 nucleosomes (Figure 6C). Overall, these results suggest that ATXR5/6-catalyzed H3.1K27me1 in plants interferes with GCN5-mediated acetylation at both H3.1K27 and H3.1K36.
Figure 6.
H3K36 acetylation by the GCN5–ADA2b complex is regulated by H3K27me1. (A) In vitro HAT assays with the GCN5–ADA2b complex and H3K27me0 and H3K27me1 nucleosomes using H3K36ac, H3K9ac, and H3 antibodies for detection. (B) Quantification of HAT assay for three technical replicates using independent preparations of nucleosomes and the GCN5–ADA2b complex. Error bars indicate sem. Unpaired t-test: * p < 0.05, and NS = not significantly different. (C) In vitro HAT assays with the GCN5–ADA2b complex and H3K36me0 and H3K36me3 nucleosomes. Data representative of three technical replicates using independent preparations of the GCN5–ADA2b complex. (D) Model depicting the role of H3.1K27me1 in preventing GCN5-mediated acetylation of H3.1K27 and H3.1K36.
Discussion
Previous work had suggested that transcriptional reactivation of heterochromatic regions is responsible for inducing genomic instability in the absence of H3.1K27me1 in plants (Hale et al., 2016). However, the mechanism by which H3.1K27me1 prevents transcriptional de-repression in heterochromatin was unclear. Our study supports a model where ATXR5/6-mediated H3.1K27me1 serves to prevent a SAGA-like complex that includes GCN5, ADA2b, and CHR6 from acetylating the H3.1 variant and initiating transcriptional de-repression (Figure 6D). K27me1 is the most abundant PTM on H3.1K27 in plants (Johnson et al., 2004), and our results suggest that it plays a role analogous to the one proposed for PRC2-catalyzed H3K27me2 in animals, which is present on 50%–70% of total histone H3 in mouse ESCs, interferes with H3K27ac deposition, and prevents spurious transcription (Peters et al., 2003; Jung et al., 2010; Ferrari et al., 2014). In animals, p300 and CBP are the main HATs that contribute to H3K27ac in the absence of PRC2-mediated H3K27 methylation (Tie et al., 2009; Pasini et al., 2010). Our results indicate that in plants, GCN5 plays this role. However, transcriptional de-repression is not completely abolished in gcn5 mutants (Figure 1C), thus suggesting that at least one of the five p300/CBP homologs in Arabidopsis (HAC1/2/4/5/12; Earley et al., 2007; Li et al., 2014) may also contribute to higher histone acetylation levels in the absence of H3.1K27me1.
Our work shows that GCN5-catalyzed histone acetylation plays a key role in mediating transcriptional activation in atxr5/6 mutants. The role of GCN5 as a transcriptional co-activator in other biological systems is well defined, thus supporting a conserved function for GCN5 in all eukaryotes. H3K27ac has been found to be enriched close to the TSS of transcriptionally active protein-coding genes in mammals, maize (Zea mays), rice (Oryza sativa), and Arabidopsis (Wang et al., 2008; Du et al., 2013; Zhang et al., 2015; Yan et al., 2019), a result that we confirmed for Arabidopsis in our ChIP-Rx experiments. H3K36ac has also been shown in multiple biological systems to co-localize with H3K27ac at the TSS of transcriptionally active regions of the genome (Wang et al., 2008; Mahrez et al., 2016). These observations suggest that TSS-localized H3K27ac and H3K36ac play important roles in mediating transcriptional activity. Precisely mapping the H3K27ac and H3K36ac regions in the heterochromatin of atxr5/6 mutants in relation to the TSS of de-repressed TEs is challenging, as TSSs are not well defined for TEs. Nevertheless, we did observe H3K27ac and H3K36ac peaks in atxr5/6 at the 5′ ends of annotated TEs (Figure 5C; Supplemental Figure 8), supporting a similar mode of action for H3K27ac/H3K36ac in regulating the transcription of genes and TEs.
Yeast and animal GCN5 have the ability to acetylate multiple lysines (K9, K14, K18, K23, K27, and K36) in the N-terminal tail of histone H3 (Kuo and Andrews, 2013; Cieniewicz et al., 2014). Our in vitro and in vivo results suggest that the GCN5 homolog in Arabidopsis also has broad substrate specificity. However, the specificity of ATXR5/6 for H3K27 and results from the current study suggest a critical role for K27 over other target sites of GCN5 on H3.1 variants. One observation supporting a unique role for H3.1K27ac over other acetylated lysines of H3 in Arabidopsis comes from experiments showing that increased levels of cytosolic acetyl co-enzyme A (acetyl-CoA; the essential cofactor for protein acetylation) increase H3 acetylation in plants (Chen et al., 2017). Results from these experiments show that H3K27 is predominantly acetylated over other lysine residues of H3 (i.e. H3K9, H3K14, and H3K18; H3K23 and H3K36 were not assessed in that study) in a manner dependent on GCN5. Higher levels of H3K27ac are observed in genic regions, and this correlates with higher transcriptional levels for genes showing gains in H3K27ac (Chen et al., 2017). Like H3.1K27ac, our in vitro and in vivo results implicate H3.1K36ac as playing a key role in mediating the heterochromatin phenotypes of atxr5/6. However, these results do not rule out the possibility that other acetylated sites (e.g. K9, K14, K18, and K23) on H3.1 also help mediate transcriptional de-repression and genomic instability in plants, for example by acting in a functionally redundant manner. Our in vitro HAT assays indicate that deposition of H3K36ac by GCN5 is negatively regulated by H3K27me1, although the molecular mechanism responsible for this crosstalk remains unknown. Previous structural work characterizing a protein complex composed of the HAT domain of GCN5 from the unicellular eukaryote Tetrahymena thermophila and a phosphorylated histone H3 peptide (aa 5–23) showed that the HAT domain interacts with the side chain of glutamine 5 (Q5), located nine amino acids upstream of the target lysine (K14) on the H3 peptide (Clements et al., 2003). As H3K27 is similarly located nine amino acids upstream H3K36, this suggests that the HAT domain of GCN5 in Arabidopsis may interact with the side chain of H3K27 to regulate the catalytic activity of GCN5 at H3K36. Structural studies of the HAT domain of Arabidopsis GCN5 will be needed to validate this model.
The catalytic specificity of ATXR5/6 for replication-dependent H3.1 variants, together with the observation that heterochromatin amplification is suppressed when the H3.1 chaperone CAF-1 is mutated, have led to a model in which the H3.1 variant plays a specific role in maintaining genome stability (Jacob et al., 2014). One possible mechanism that could explain the requirement for H3.1 variants to induce the atxr5/6 mutant phenotypes is that GCN5, like ATXR5/6, specifically modify K27 in H3.1 variants. However, our results show no difference in enzymatic activity for GCN5 on H3.1 versus H3.3 variants (Figure 3A). Therefore, GCN5 is unlikely to be directly involved in mediating the H3.1 requirement for inducing the atxr5/6 mutant phenotypes. An alternative mechanism that could explain the role for H3.1 variants in this process is that downstream chromatin readers that mediate transcriptional de-repression and heterochromatin amplification interact with H3.1K27ac and/or H3.1K36ac, but not H3.3K27ac and/or H3.3K36ac. Another possibility is that transcriptional de-repression mediated through GCN5 is not dependent on H3.1 variants, but heterochromatin amplification is. A previous study showed that expressing an ATXR5/6-resistant H3.1A31T transgene (which partially mimics the N-terminal tail of H3.3 variants) in plants generates low-level transcriptional de-repression in heterochromatin (which is supported by the finding that GCN5 is active on H3.3 variants), but genomic instability in the H3.1A31T lines was not detected (Jacob et al., 2014). Therefore, H3-variant-independent transcriptional de-repression via GCN5 could induce H3.1-dependent genomic instability, or alternatively, these two processes could be uncoupled, although both are regulated by GCN5. Recent work in the yeast Saccharomyces cerevisiae demonstrated that passage through S phase of the cell cycle facilitates epigenetic silencing via the insertion of newly synthesized histones. The insertion of newly synthesized histone H3.1 variants in plants during replication could also be a key step in mediating the epigenetic changes that lead to genomic instability in the absence of H3.1K27me1 (Goodnight and Rine, 2020). More work will be needed to fully understand the relationship between H3 variants, transcriptional de-repression, and genomic instability in plants.
Materials and methods
Plant materials
Arabidopsis thaliana plants were grown in Pro-Mix BX Mycorrhizae soil under cool-white fluorescent lights (approximately 100 μmol m−2 s−1) in long-day conditions (16 h light/8 h dark). The atxr5/6 double mutant was described previously (Jacob et al., 2009). gcn5 (At3g54610, SALK_030913), ada2b (At4g16420, SALK_019407), ada3 (At4g29790, SALK_042026C), chr5 (At2g13370, SAIL_504_D01), and chr6 (At2g25170, GK-273E06) are in the Col-0 genetic background and were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Temperature-optimized CRISPR/Cas9 was used to generate additional mutant alleles of GCN5 (in Col-0 and atxr5/6) used in this study (LeBlanc et al., 2018). The guide RNA transgenes were segregated away from the mutant alleles. The h3.1 quadruple mutant was described previously (Jacob et al., 2014). Transgenic plants expressing WT H3.1 (At5g65360), H3.1K27Q, H3.1S28A, H3.1K9R, H3.1S28A K9R, H3.1K14R, H3.1S28A K14R, H3.1K18R, H3.1S28A K18R, H3.1K23R, H3.1S28A K23R, H3.1K36R, and H3.1S28A K36R were made by transforming plants in the h3.1 quadruple mutant background using the floral dip method (Clough and Bent, 1998). Transgenic plants constitutively expressing (using the 35S promoter) SDG4, SDG7, SDG8, SDG24, and SDG26) were made by transforming plants in the atxr5/6 mutant background.
Constructs
Cloning of the catalytic fragment of ATXR6 (aa 25–349) and genes for the plant PRC2 complexes for protein expression and in vitro methyltransferase assays was described previously (Jacob et al., 2009, 2014). The histone H3.1 gene (At5g65360) and its promoter (1167-bp upstream of the start codon) were cloned into pENTR/D-TOPO (ThermoFisher Scientific, Waltham, MA, USA) and then sub-cloned using Gateway Technology into the plant binary vectors pB7WG (Karimi et al., 2002). Site-directed mutagenesis to generate the different H3.1 point mutant constructs was performed using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). PCR products corresponding to the genomic sequences of SDG4, SDG7, SGD8, SDG24, and SDG26 (from start to stop codons) were directly cloned into the pMDC32 vector (Curtis and Grossniklaus, 2003) using the AscI and PacI restriction sites. Site-directed mutagenesis was used to create the Y140N point mutation in SDG24. The ADA2b coding sequence was cloned into the pETDuet-1 (Millipore, Burlington, MA, USA) vector using the SalI and NotI restriction sites, yielding pETDuet-1-ADA2b. The GCN5 coding sequence was cloned into the pETDuet-1-ADA2b plasmid using the EcoRV and PacI restriction sites, yielding pETDuet-1-ADA2b-GCN5. The cloning procedure used to make the CRISPR construct targeting GCN5 in Arabidopsis was performed as described previously (Yan et al., 2015).
Protein expression and purification
Expression and purification of the ATXR6 protein and the plant PRC2 complexes CURLY LEAF and MEDEA were described previously (Jacob et al., 2009, 2014). Briefly, the GST-tagged ATXR6 protein was expressed in Escherichia coli BL21 DE3 cells. Protein expression was induced by adding IPTG to a concentration of 0.1-mM, and induction was allowed to proceed overnight at 20°C. The FLAG-tagged PRC2 complexes CLF and MEA were expressed in SF9 insect cells. To purify the complexes, the SF9 cells were resuspended in lysis buffer (50-mM Tris pH 8.0, 150-mM NaCl, 1-mM PMSF, and 0.1% Triton X-100) and sonicated 10 × 20 s on ice. The cell lysate was centrifuged at 20,000g for 40 min at 4°C, and the complexes were purified with anti-FLAG M2 Affinity Gel (ThermoFisher Scientific). The FLAG fusion complexes were eluted from the columns by competition with 100-µg·mL-1 FLAG peptide (ThermoFisher Scientific) in TBS (50-mM Tris–HCl, 150-mM NaCl, pH 7.4).
For the GCN5–ADA2b protein complex, pETDuet-1-ADA2b-GCN5 was transformed into BL21 (DE3) E. coli (Millipore), cultured in LB, and induced to express proteins by adding 1-mM IPTG. The cells were pelleted by centrifugation, resuspended in NPI-10 buffer (50-mM NaH2PO4, 300-mM NaCl, 10-mM Imidazole, pH 8), and lysed by sonication. After centrifugation to remove cell debris, Ni-NTA agarose (Qiagen, Hilden, Germany) was added to the supernatant and rotated at 4°C for 2 h. The Ni-NTA agarose was washed three times using NPI-20 buffer (50-mM NaH2PO4, 300-mM NaCl, 20-mM imidazole, pH 8), and the protein complex was eluted in NPI-250 buffer (50-mM NaH2PO4, 300-mM NaCl, 250-mM imidazole, pH 8). The buffer was changed to 1×PBS (137-mM NaCl, 10-mM phosphate, 2.7-mM KCl, pH 7.4) containing 10% glycerol using an Amicon Ultra-0.5 Centrifugal Filter Unit (30-kDa cutoff). The proteins were aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C.
The protocols to generate the H3K27me1 and H3K36me3 methyl-lysine analog-containing histones and to make the recombinant chromatin used in the in vitro histone modification assays (methylation and acetylation) were described previously (Voigt et al., 2012).
HMT and HAT assays
The general procedure used to perform the in vitro histone modification assays presented in this study were described in detail in a previous publication (Jacob and Voigt, 2018). For the radioactive HMT assays, 0.5 μg of ATXR6, 1.5 μg of MEA, or 1.5 μg of CLF (PRC2) complexes were incubated with 1 μg of Histone H3 peptides (GenScript, Piscataway, NJ, USA) and 1.5 μCi of 3H-SAM (Perkin Elmer, Waltham, MA, USA) in a 25 μL reaction. The HMT buffer contained 50-mM Tris pH 8.0, 2.5-mM MgCl2, and 4-mM DTT. The methylation reactions were incubated at 22°C for 2 h. The samples were pipetted onto Whatman P-81 filter paper and dried for 15 min. The free 3H-SAM was removed by washing 3 × 30 min in 50-mM NaHCO3 pH 9.0. The filter paper was dried and added to a vial containing Opti-Fluor® O (Perkin Elmer). Radioactivity on the filter papers was determined using a liquid scintillation counter (Perkin Elmer).
For the HAT assays with antibody detection, 1 µg of recombinant nucleosomes and 2 µg of the GCN5–ADA2b complex were incubated in 50-µL HAT buffer (1 mM HEPES pH 7.3, 0.02% BSA) containing 50-mM acetyl-CoA (Sigma) at 23°C for 3 h (wild-type H3.1, H3.1K27M, and H3.3 nucleosomes) or 5 h (H3K27me0, H3K27me1, H3K36me0, and H3K36me3 nucleosomes). The reactions were stopped by adding 4X Laemmli Sample Buffer (Bio-Rad) and boiling at 95°C for 5 min. The samples were resolved by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel, transferred to polyvinylidene fluoride membrane, and immunoblot analysis was performed using anti-H3K9ac (Cell Signaling Technology: Danvers, MA, USA: 9649), anti-H3K14ac (Active Motif, Carlsbad, CA, USA: 39698), anti-H3K18ac (Active Motif: 39588), anti-H3K23ac (Active Motif: 39132), anti-H3K27ac (Active Motif: 39135), anti-H3K36ac (Active Motif: 39379), or anti-H3 antibodies (Abcam: ab1791) and a secondary anti-Rabbit HRP-labeled antibody (Sigma).
For the radioactive HAT assays, 1 µg of peptides and 1 µg of GCN5–ADA2 complex were incubated in 25-µL HAT buffer containing 0.625-µCi 3H-acetyl-CoA (PerkinElmer) at 23°C for 2 h. Reactions were stopped by pipetting onto Whatman P-81 filter paper and dried for 15 min. The free 3H-SAM was removed by washing 3 × 30 min in 50-mM NaHCO3 pH 9.0. The filter paper was dried, added to a vial containing Opti-Fluor® O (Perkin Elmer) and activity (c.p.m.) was measured using a liquid scintillation counter (Perkin Elmer). No enzyme controls in the HMT and HAT assays consisted of reactions containing buffer, cofactor and chromatin substrate, but no enzyme.
ChIP
ChIP was performed as described previously (Villar and Kohler, 2010), with some modifications. Briefly, rosette leaves from 3-week-old plants were fixed for 15 min in 1% formaldehyde. For SDG24-OX ChIP experiments, each biological replicate consisted of an independent T1 plant. For ChIP experiments in Figure 5 and Supplemental Figure 4 three plants growing in the same flat were pooled for each biological replicate. After fixation, leaves were flash frozen in liquid nitrogen and ground using a mortar and pestle. Approximately 0.8 g of tissue was added to 10 mL of extraction buffer 1 [0.4-M sucrose, 10-mM Tris–HCl {pH 8.0}, 10-mM MgCl2, 0.1-mM PMSF, 1× protease inhibitors {Roche}] and filtered successively through 70 µm and 40 µm meshes. Samples were centrifuge at 3,000g for 20 min. The pellets were resuspended in 1 mL of extraction buffer 2 [0.25-M sucrose, 10-mM Tris–HCl {pH 8.0}, 10-mM MgCl2, 1% Triton X-100, 0.1-mM PMSF, 1× protease inhibitors] and centrifuged at 12,000g for 10 min. The pellets were then resuspended in 400 µL of extraction buffer 3 [1.7-M sucrose, 10-mM Tris–HCl {pH 8.0}, 0.15% Triton X-100, 0.1-mM PMSF, 1× protease inhibitors]. Extraction buffer 3 (400 µL) was added to fresh tubes. The samples were carefully layered over the buffer and centrifuged for 1 h at 16,000g. The pellets were resuspended in nuclei lysis buffer [50-mM Tris–HCl {pH 8.0}, 10-mM EDTA, 1% SDS, and 1× protease inhibitors], and chromatin was sheared using a Bioruptor 200 sonicator (20 times on a 30-s ON, 30-s OFF cycle). The supernatants were centrifuged at 16,000g for 5 min. ChIP dilution buffer [1.1% Triton X-100, 1.2-mM EDTA, 16.7-mM Tris–HCl {pH 8.0}, 167-mM NaCl, and 1× protease inhibitors] was added to samples to bring to 10× volume. Antibodies were added to 750 μL of diluted sample and incubated at 4°C overnight (while rotating). About 2 μL of Histone H3 antibody (Abcam: ab1791), 2.5 μL of H3K9ac antibody (Cell Signaling Technology: 9649), 2.5 μL of H3K18ac antibody (Active Motif: 39588), 5 μL of H3K23ac antibody (Active Motif: 39132), 2.5 μL of H3K27ac antibody (Active Motif: 39135), 5 μL of H3K36ac antibody (Active Motif: 39379), or 2.5 μL of H3K36me3 (Abcam: ab9050) was used per immunoprecipitation (750 μL of chromatin solution). Immunoprecipitation was performed using protein A magnetic beads (New England BioLabs, Ipswich, MA, USA). The beads were washed twice in each of the following buffers: low salt wash buffer [150-mM NaCl, 0.1% SDS, 1% Triton X-100, 2-mM EDTA, and 20-mM Tris–HCl {pH 8.0}, high salt wash buffer (500-mM NaCl, 0.1% SDS, 1% Triton X-100, 2-mM EDTA, and 20-mM Tris–HCl {pH 8.0}], LiCl wash buffer [0.25-M LiCl, 1% Igepal CA-630, 1% sodium deoxycholate, 1-mM EDTA, and 10-mM Tris–HCl {pH 8.0}], and TE [10-mM Tris–HCl {pH 8.0} and 1-mM EDTA]. The beads were resuspended in 500 µL of elution buffer (1% SDS and 0.1-M NaHCO3) and incubated at 65°C for 15 min. A total of 20 µL of 5-M NaCl was added and samples were incubated at 65°C for 5 h. Around 10 μL of 0.5-M EDTA, 20 μL of 1-M Tris-HCl (pH 6.5), and 2 μL of 10-mg·mL−1 proteinase K were added to each sample and incubated for 2 h at 45°C. Immunoprecipitated DNA was purified using a ChIP DNA Clean & Concentrator kit (Zymo Research, Irvine, CA, USA). For the H3K27ac and H3K36ac ChIP experiments, ChIP with exogenous genome (ChIP-Rx) was performed in order to properly normalize the data (Orlando et al., 2014). For each sample, an equal amount of drosophila chromatin (Active Motif #53083) was added prior to chromatin shearing.
4′,6-Diamidino-2-phenylindole (DAPI) staining of nuclei
Leaves from 4-week-old plants were fixed in 3.7% formaldehyde in cold Tris buffer (10-mM Tris–HCl pH 7.5, 10-mM NaEDTA, 100-mM NaCl) for 20 min. Formaldehyde solution was removed, and the leaves were washed twice for 10 min in Tris buffer. The leaves were then finely chopped with a razor blade in 500-μL LB01 buffer (15-mM Tris–HCl pH 7.5, 2-mM NaEDTA, 0.5-mM spermine-4HCl, 80-mM KCl, 20-mM NaCl, and 0.1% Triton X-100). The lysate was filtered through a 30 µm mesh (Sysmex Partec, Gorlitz, Germany). About 5 μL of lysate was added to 10 μL of sorting buffer (100-mM Tris–HCl pH 7.5, 50-mM KCl, 2-mM MgCl2, 0.05% Tween-20, and 5% sucrose) and spread onto a coverslip until dried. Cold methanol was added onto each coverslip for 3 min, and then rehydrated with TBS-Tx (20-mM Tris pH 7.5, 100-mM NaCl, 0.1% Triton X-100) for 5 min. The coverslips were mounted onto slides with Vectashield mounting medium DAPI (Vector Laboratories, Burlingame, CA, USA). Nuclei were imaged under a Nikon Eclipse Ni-E microscope with a 100× CFI PlanApo Lamda objective (Nikon, Minato City, Tokyo, Japan). Digital images were obtained using an Andor Clara camera. Z-series optical sections of each nucleus were obtained at 0.3-μm steps. Images were deconvolved by ImageJ using the deconvolution plugin.
RT-qPCR
Total RNA was extracted from 3-week-old leaf tissue using TRIzol (Invitrogen, Carlsbad, CA, USA). The samples were treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA) at 37°C for 30 min. SuperScript II Reverse Transcriptase (Invitrogen) was used to produce cDNA from 1 µg of total RNA. Reverse transcription was initiated using oligo dT primers. Quantification of cDNA was done by PCR using a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with KAPA SYBR FAST qPCR Master Mix (2×) Kit (Kapa Biosystems, Wilmington, MA, USA). The cycling conditions were the following: 95°C for 3 min; 40 cycles of 95°C for 3 s, 60°C for 25 s, followed by dissociation curve analysis. Each primer pair was assessed for the efficiency of amplification (Supplemental Table 1). Relative quantities were determined by the Ct method (Livak and Schmittgen, 2001). ACTIN was used as the normalizer. At least three biological samples were used for each experiment. Three plants growing in the same flat were pooled for each biological replicate.
Flow cytometry
Rosette leaves from 3-week-old plants were finely chopped in 0.5-mL Galbraith buffer (45-mM MgCl2, 20-mM MOPS, 30-mM sodium citrate, 0.1% Triton X-100, 40-μg·μL–1 RNase A) using a razor blade. The lysate was filtered through a 30 µm mesh (Sysmex Partec, Gorlitz, Germany). Propidium iodide (Sigma, St Louis, MO, USA) was added to each sample to a concentration of 20 µg·mL-1 and vortexed for 3 s. Each sample was analyzed using a BD FACS LSR Fortessa X20 (Becton Dickinson, Franklin Lakes, NJ, USA). Quantification (nuclei counts and robust CV values) was performed using Flowjo 10.0.6 (Tree Star, Ashland, OR, USA). Each biological replicate consisted of a leaf from one plant.
Next-generation sequencing library preparation
RNA samples were prepared from 3-week-old leaf tissue using an RNeasy Plant Mini Kit (Qiagen). Three plants growing in the same flat were pooled for each biological replicate. RNA and ChIP sequencing libraries were prepared at the Yale Center for Genome Analysis. RNA samples were quantified and checked for quality using the Agilent 2100 Bioanalyzer Nano RNA Assay. Library preparation was performed using Illumina’s TruSeq Stranded Total RNA with Ribo-Zero Plant in which samples were normalized with a total RNA input of 1 µg and library amplification with eight PCR cycles. ChIP library preparation was performed using a TruSeq Library Prep Kit (Illumina, San Diego, CA, USA). Libraries were validated using the Agilent Bioanalyzer 2100 High sensitivity DNA assay and quantified using a KAPA Library Quantification Kit for Illumina Platforms kit. Sequencing was done on an Illumina NovaSeq 6000 using the S4 XP workflow.
RNA-seq processing and analysis
Two independent biological replicates for Col, atxr5/6, gcn5, and atxr5/6 gcn5 were sequenced. Paired-end reads were filtered and trimmed using BBTools (version 38.79; Bushnell et al., 2017). Reads with quality scores <20 were removed (Supplemental Table 3). The resulting data sets were aligned against the Arabidopsis genome (TAIR10) using STAR (version 2.7.2a) allowing two mismatches (–outFilterMismatchNmax 2; Dobin et al., 2013). Consistency between biological replicates was confirmed by Pearson correlation using deepTools2 (Supplemental Figure 10; Ramirez et al., 2016). Protein-coding genes and TEs were defined as described in the TAIR10 annotation gff3 file. The program featureCounts (version 1.6.4; Liao et al., 2014) was used to count the paired-end fragments overlapping with the annotated protein-coding genes and TEs. Differential expression analysis of protein-coding genes was performed using DESeq2 version 1.26 (Love et al., 2014) on raw read counts to obtain normalized fold changes (FCs) and Padj-values for each gene. Genes were considered to be differentially expressed only if they showed a log2FC >1 or log2FC < −1 and a Padj-values <0.05. TPM (transcripts per million) values were calculated for TEs. To define TEs as upregulated in the atxr5/6 mutant, they must show two-fold upregulation compared to Col in both biological replicates and have a value of TPM > 5. The heatmap was drawn with the R program (version 3.6.2; Team, 2018).
ChIP-seq processing and analysis
Two independent biological replicates for Col, atxr5/6, gcn5, and atxr5/6 gcn5 were sequenced. In order to properly compare H3K27ac and H3K36ac levels between each genotype, we performed ChIP-Rx (ChIP with reference exogenous genome; Orlando et al., 2014) using equal amounts of Drosophila chromatin in each sample as a reference. Paired-end reads were filtered and trimmed using BBTools (Bushnell et al., 2017). Reads with quality scores <20 were removed (Supplemental Table 3). Data sets were aligned against the combined genomes of A. thaliana (TAIR10) and Drosophila melanogaster (dm6) using bowtie2 (Langmead and Salzberg, 2012) with default parameters. Duplicate reads were removed using Picard toolkit; https://broadinstitute.github.io/picard/faq.html) (MarkesDuplicates with REMOVE_DUPLICATES=true). Consistency between biological replicates was confirmed by Pearson correlation using deepTools2 (Supplemental Figure 11; Ramirez et al., 2016). To calculate the Rx scaling factor of each biological replicate, Drosophila-derived IP read counts were normalized according to the number of input reads. Spike-in normalization was performed as previously described (Nassrallah et al., 2018). We used α = r/Nd_IP from Orlando et al. (2014) to compute the scaling factor α for each replicate, with Nd_IP corresponding to the number of reads (in millions) aligning to the D. melanogaster genome in the IP and with r = 100 * Nd_i / (Na_i + Nd_i), where Nd_i and Na_i are the number of input reads (in millions) aligning to the D. melanogaster or A. thaliana genome, respectively. The Rx factors are presented in Supplemental Table 2. We generated bedgraph files with a bin size of 10 bp using deepTools. The bedgraph files were then scaled by adjusting the number of reads in each bin with the Rx factors and therefore generating reference-adjusted reads per million. H3K27ac- and H3K36ac-enriched regions were identified by computing the differential between each bin (±1 kb) to define local maxima.
The number of reads corresponding to euchromatic regions was much higher than the ones from heterochromatic regions. To best determine the heterochromatic enrichment of H3K27ac in each genotype of interest, we avoided the noise from the euchromatic reads by first defining heterochromatic regions and extracting the corresponding reads from each genotype. We defined the heterochromatic regions based on chromatin states proposed previously (Sequeira-Mendes et al., 2014). The authors defined four different chromatin states enriched in genes (states 1, 3, 6, and 7), three chromatin states enriched in the distinctive polycomb mark H3K27me3 (states 2, 4, and 5), and two types of heterochromatin states (states 8 and 9). We attributed the value of the state number (1–9) for each bin of the Sequeira-Mendes et al. annotation, and averaged them on 100-kb windows along the A. thaliana genome. Only regions with average chromatin state scores >7 were defined as heterochromatic regions (Supplemental Data Set 3). We then generated a bam file with the reads corresponding to the defined heterochromatic regions. We identified heterochromatic H3K27ac- and H3K36ac-enriched regions by calculating the log2 ratio between H3K27ac or H3K36ac IP and H3 input using the heterochromatin bam file. The enriched regions were defined with the following criteria: log2 (IP/H3) >0.3. To compare the H3K27ac- and H3K36ac-enriched regions between Col and our mutant genotypes, we computed log2 (mutant/Col), using the Rx factor normalized bedgraph file. We considered the levels of H3K27ac and H3K36ac to be differential between genotypes when log2 (mutant/Col) >0.8. These regions needed to be detected in both replicate in order to be considered.
Statistical analyses
Statistical analysis data are provided in Supplemental Data Set 6.
Primers
All primers used in this study are listed in Supplemental Table 1.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: ATXR5 (At5g09790), ATXR6 (At5g24330), GCN5 (At3g54610), ADA2b (At4g16420), ADA3 (At4g29790), CHR5, (At2g13370), CHR6 (At2g25170), SDG4 (At4g30860), SDG7 (At2g44150), SDG8 (At1g77300), SDG24 (At3g59960), SDG26 (At1g76710), CLF (At2g23380), MEA (At1g02580), H3.1 (At5g65360), BRCA1 (At4g21070), SE (At2g27100), AtTHP1 (At2g19560), AtSAC3B (At3g06290), AtSTUbL2 (At1g67180), AtMBD9 (At3g01460), and DDM1 (At5g66750).
Sequencing data are available at the Gene Expression Omnibus under accession code GSE146126.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effect of GCN5 on genome stability and transcriptional de-repression.
Supplemental Figure 2. Role of SAGA-related proteins in transcriptional de-repression and genome stability.
Supplemental Figure 3. Purification of the GCN5–ADA2b complex.
Supplemental Figure 4. In vivo acetylation levels at different lysines of H3 are dependent on GCN5.
Supplemental Figure 5. In vitro histone modification assays.
Supplemental Figure 6. Growth and developmental phenotypes of T1 plants expressing different H3.1 transgenes.
Supplemental Figure 7. Analyses of the effects of overexpression of H3K36 methyltransferases on genome stability.
Supplemental Figure 8. Average distribution of H3K27ac and H3K36ac over protein-coding genes grouped by their expression levels.
Supplemental Figure 9. Validation of ChIP-seq and RNA-seq analyses.
Supplemental Figure 10. Scatterplots and Pearson correlation coefficients for RNA-seq replicates of Col, atxr5/6, gcn5, and atxr5/6 gcn5.
Supplemental Figure 11. Scatterplots and Pearson correlation coefficients for H3K27ac and H3K36ac ChIP-seq replicates of Col, atxr5/6, gcn5, and atxr5/6 gcn5.
Supplemental Table 1. Cloning and PCR primers.
Supplemental Table 2. Rx factors for Col, atxr5/6, gcn5, and atxr5/6 gcn5 replicates.
Supplemental Table 3. Statistics for mapping and coverage of the NGS data.
Supplemental Data Set 1. TEs de-repressed in atxr5/6.
Supplemental Data Set 2. Misregulated genes in atxr5/6, gcn5, and atxr5/6 gcn5.
Supplemental Data Set 3. Regions of Arabidopsis genome defined as heterochromatin.
Supplemental Data Set 4. Heterochromatic regions enriched in H3K27ac and H3K36ac in atxr5/6.
Supplemental Data Set 5. TEs that are de-repressed and overlap with heterochromatic regions enriched in H3K27ac and H3K36ac in atxr5/6.
Supplemental Data Set 6. Statistical analysis data.
Supplementary Material
Acknowledgments
We thank members of our lab for discussions and advice during the course of this work. We want to acknowledge Christopher Bolick and his staff at Yale for help with plant growth and maintenance. We also thank Jean-François Couture (University of Ottawa) for sending the K27M nucleosomes used in this study, and Kenneth Nelson (Yale University) for technical help with flow cytometry.
Funding
This project was supported by grant #R35GM128661 from the National Institutes of Health to Y.J. B.M. was supported by a Yale University Brown Fellowship. V.J. is supported by the Fonds de Recherche du Québec-Nature et Technologies (FRQNT) (grant no. 272565). Work in the Voigt lab is supported by the Wellcome Trust (grant no. 104175/Z/14/Z, Sir Henry Dale Fellowship to P.V.) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERC-STG grant agreement No. 639253). The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust [203149]. We are grateful to the Edinburgh Protein Production Facility (EPPF) for their support. The EPPF was supported by the Wellcome Trust through a Multi-User Equipment grant (101527/Z/13/Z).
Conflict of interest statement. None declared.
Contributor Information
Jie Dong, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Chantal LeBlanc, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Axel Poulet, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Benoit Mermaz, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Gonzalo Villarino, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Kimberly M Webb, Wellcome Centre for Cell Biology, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3BF.
Valentin Joly, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Josefina Mendez, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Philipp Voigt, Wellcome Centre for Cell Biology, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3BF.
Yannick Jacob, Yale University, Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, 260 Whitney Avenue, New Haven, CN 06511.
Y.J., J.D., C.L., and A.P. designed the experiments. Y.J. wrote the paper with contributions from J.D., C.L., and A.P. All in vitro assays were performed by J.D. C.L. performed the ChIP experiments. A.P. did the bioinformatics analyses of all ChIP-seq experiments. A.P. and V.J. did RNA-seq analyses. Microscopy was done by C.L. Flow cytometry analyses were performed by C.L., B.M, G.V., and J.M. RNA extractions and RT-qPCR were done by C.L., J.D., and G.V. Crosses were done by G.V. and B.M. Genotyping and plant transformations were performed by G.V., J.M., C.L., and B.M. G.V. made the CRISPR/Cas9 mutants. K.M.W. and P.V. made the modified and unmodified nucleosomes used in the in vitro assays.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yannick Jacob (yannick.jacob@yale.edu).
References
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamin E, Sarvan S, Malette J, Eram MS, Yeung S, Mongeon V, Joshi M, Brunzelle JS, Michaels SD, Blais A, et al. (2017) Molecular basis for the methylation specificity of ATXR5 for histone H3. Nucleic Acids Res 45: 6375–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B, Rood J, Singer E (2017) BBMerge - Accurate paired shotgun read merging via overlap. PLoS One 12: e0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li C, Wang Y, Renaud J, Tian G, Kambhampati S, Saatian B, Nguyen V, Hannoufa A, Marsolais F, et al. (2017) Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat Plants 3: 814–824 [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP (2010) Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol 20: 222–233 [DOI] [PubMed] [Google Scholar]
- Cieniewicz AM, Moreland L, Ringel AE, Mackintosh SG, Raman A, Gilbert TM, Wolberger C, Tackett AJ, Taverna SD (2014) The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3. Mol Cell Proteomics 13: 2896–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A, Poux AN, Lo WS, Pillus L, Berger SL, Marmorstein R (2003) Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol Cell 12: 461–473 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davarinejad H, Joshi M, Ait-Hamou N, Munro K, Couture JF (2019) ATXR5/6 Forms Alternative Protein Complexes with PCNA and the Nucleosome Core Particle. J Mol Biol 431: 1370–1379 [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Li H, Wei Q, Zhao X, Wang C, Zhu Q, Yi X, Xu W, Liu XS, Jin W, et al. (2013) Genome-wide analysis of histone modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. Japonica. Mol Plant 6: 1463–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS (2007) In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 52: 615–626 [DOI] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, Fischle W, Bonaldi T, Pasini D (2014) Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53: 49–62 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnight D, Rine J (2020) S-phase-independent silencing establishment in Saccharomyces cerevisiae. eLife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CJ, Potok ME, Lopez J, Do T, Liu A, Gallego-Bartolome J, Michaels SD, Jacobsen SE (2016) Identification of multiple proteins coupling transcriptional gene silencing to genome stability in Arabidopsis thaliana. PLoS Genet 12: e1006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300 [DOI] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134: 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Jiang L, Liu BY, Tan CF, Chen DH, Shen WH, Ruan Y (2019) Evolution and conservation of polycomb repressive complex 1 core components and putative associated factors in the green lineage. BMC Genomics 20: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Bergamin E, Donoghue MT, Mongeon V, LeBlanc C, Voigt P, Underwood CJ, Brunzelle JS, Michaels SD, Reinberg D, et al. (2014) Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343: 1249–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD (2009) ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Stroud H, Leblanc C, Feng S, Zhuo L, Caro E, Hassel C, Gutierrez C, Michaels SD, Jacobsen SE (2010) Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466: 987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Voigt P (2018) In vitro assays to measure histone methyltransferase activity using different chromatin substrates. Methods Mol Biol 1675: 345–360 [DOI] [PubMed] [Google Scholar]
- Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE (2004) Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res 32: 6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HR, Pasini D, Helin K, Jensen ON (2010) Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics 9: 838–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim S, Piquerez SJM, Ramirez-Prado JS, Mastorakis E, Veluchamy A, Latrasse D, Manza-Mianza D, Brik-Chaouche R, Huang Y, Rodriguez-Granados NY, et al. (2020) GCN5 modulates salicylic acid homeostasis by regulating H3K14ac levels at the 5' and 3' ends of its target genes. Nucleic Acids Res 48: 5953–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornet N, Scheres B (2009) Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383: 269–272 [DOI] [PubMed] [Google Scholar]
- Kuo YM, Andrews AJ (2013) Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PLoS One 8: e54896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y (2018) Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93: 377–386 [DOI] [PubMed] [Google Scholar]
- Li C, Xu J, Li J, Li Q, Yang H (2014) Involvement of Arabidopsis HAC family genes in pleiotropic developmental processes. Plant Signal Behav 9: e28173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Castillo-Gonzalez C, Wang Z, Sun D, Hu X, Shen X, Potok ME, Zhang X (2018) Arabidopsis serrate coordinates histone methyltransferases ATXR5/6 and RNA processing factor RDR6 to regulate transposon expression. Dev Cell 45: 769–784 [DOI] [PubMed] [Google Scholar]
- Mahrez W, Arellano MS, Moreno-Romero J, Nakamura M, Shu H, Nanni P, Kohler C, Gruissem W, Hennig L (2016) H3K36ac is an evolutionary conserved plant histone modification that marks active genes. Plant Physiol 170: 1566–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM (1990) Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247: 841–845 [DOI] [PubMed] [Google Scholar]
- Moraga F, Aquea F (2015) Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses. Front Plant Sci 6: 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG (2012) Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19: 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassrallah A, Rougee M, Bourbousse C, Drevensek S, Fonseca S, Iniesto E, Ait-Mohamed O, Deton-Cabanillas AF, Zabulon G, Ahmed I, et al. (2018) DET1-mediated degradation of a SAGA-like deubiquitination module controls H2Bub homeostasis. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG (2014) Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. (2010) Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 38: 4958–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Pfab A, Bruckmann A, Nazet J, Merkl R, Grasser KD (2018) The adaptor protein ENY2 is a component of the deubiquitination module of the Arabidopsis SAGA transcriptional co-activator complex but not of the TREX-2 complex. J Mol Biol 430: 1479–1494 [DOI] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M (2015) Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 7: a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Sozzani R, Glab N, Domenichini S, Perennes C, Cella R, Kondorosi E, Bergounioux C (2006) Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J 47: 395–407 [DOI] [PubMed] [Google Scholar]
- Savage KI, Harkin DP (2015) BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. Febs J 282: 630–646 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Araguez I, Peiro R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C (2014) The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G, Timmers HT, Pijnappel WW (2012) ATAC-king the complexity of SAGA during evolution. Genes Dev 26: 527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM (2003) Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol 132: 907–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Rai KM, Pandey B, Singh SP, Sawant SV (2015) Spt-Ada-Gcn5-Acetyltransferase (SAGA) complex in plants: genome wide identification, evolutionary conservation and functional determination. PLoS One 10: e0134709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Hale CJ, Feng S, Caro E, Jacob Y, Michaels SD, Jacobsen SE (2012) DNA methyltransferases are required to induce heterochromatic re-replication in Arabidopsis. PLoS Genet 8: e1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Team RC (2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar CB, Kohler C (2010) Plant chromatin immunoprecipitation. Methods Mol Biol 655: 401–411 [DOI] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ (2003) Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, LeRoy G, Drury WJ 3rd, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D (2012) Asymmetrically modified nucleosomes. Cell 151: 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hayes JJ (2008) Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol 28: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T, Kaochar S, Jones H, Paek A, Clark AJ (2009) The replication fork’s five degrees of freedom, their failure and genome rearrangements. Curr Opin Cell Biol 21: 778–784 [DOI] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, et al. (2010) Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc Natl Acad Sci USA 107: 19266–19271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Yan W, Chen D, Schumacher J, Durantini D, Engelhorn J, Chen M, Carles CC, Kaufmann K (2019) Dynamic control of enhancer activity drives stage-specific gene expression during flower morphogenesis. Nat Commun 10: 1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B (2011) H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY (1998) Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. Embo J 17: 3155–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Garcia N, Feng Y, Zhao H, Messing J (2015) Genome-wide histone acetylation correlates with active transcription in maize. Genomics 106: 214–220 [DOI] [PubMed] [Google Scholar]
- Zou B, Sun Q, Zhang W, Ding Y, Yang DL, Shi Z, Hua J (2017) The Arabidopsis chromatin-remodeling factor CHR5 regulates plant immune responses and nucleosome occupancy. Plant Cell Physiol 58: 2202–2216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.