Abstract
Plants and pathogens constantly adapt to each other. As a consequence, many members of the plant immune system, and especially the intracellular nucleotide-binding site leucine-rich repeat receptors, also known as NOD-like receptors (NLRs), are highly diversified, both among family members in the same genome, and between individuals in the same species. While this diversity has long been appreciated, its true extent has remained unknown. With pan-genome and pan-NLRome studies becoming more and more comprehensive, our knowledge of NLR sequence diversity is growing rapidly, and pan-NLRomes provide powerful platforms for assigning function to NLRs. These efforts are an important step toward the goal of comprehensively predicting from sequence alone whether an NLR provides disease resistance, and if so, to which pathogens.
Plant pan-NLRomes aim to fully capture intraspecific diversity of the highly variable NLR immune receptors, enabling systematic analyses of NLR genes and alleles and their roles in disease resistance.
A brief history of plant NLRs
NLRs and disease resistance
Pathogens exert some of the strongest selection pressures on plants, and their prevalence is very dynamic in time and space. It is thus not surprising that the ecological and evolutionary forces resulting from plant–pathogen interactions play an important role in shaping both plant and pathogen genomes, including constant genetic innovation and preservation of long-standing variation on both sides.
Potential pathogens are ubiquitous, but only a few can typically colonize individuals from specific plant species. The first, most common reason for failure to colonize a potential host is that a pathogen lacks the genetic toolkit required for successful establishment and subsequent extraction of resources from this host. A second reason is that a pathogen is recognized by the host because it presents widely distributed, and often highly conserved molecules that mark it as a potential threat. These molecules have been termed microbe- or pathogen-associated molecular patterns (MAMPs/PAMPs), and are detected by pattern recognition receptors (PRRs) that are localized in the plant membrane and that survey extracellular space (Jones and Dangl, 2006). Engagement of these receptors by PAMPs leads to a raft of cellular changes, collectively known as PAMP or pattern triggered immunity (PTI; Jones and Dangl, 2006).
To overcome this first layer of defense, a pathogen has to evade or suppress PTI, which it often does by delivering its own effector molecules into the plant cell. This in turn creates a second opportunity for the plant to detect the presence of the pathogen through intracellular receptors (Chisholm et al., 2006), and their effector-dependent activation leads to what is commonly called effector-triggered immunity (ETI). PTI and ETI, the two branches of the plant immune system, have often been looked at as largely independent from another, but there is increasing evidence of interdependence between PTI and ETI components, which enhances the robustness of the defense response (Hatsugai et al., 2017; Ngou et al., 2020; Yuan et al., 2020).
The central role of nucleotide binding (NB) site leucine-rich repeat (NB-LRR, “nibbler”) proteins in ETI was discovered in the mid-1990s, when several NB-LRR genes were isolated through map-based cloning or transposon tagging. The starting point was plant lines distinguished by presence or absence of disease resistances that followed Flor’s gene-for-gene paradigm (Flor, 1971), in which single pathogen genes trigger a strong defense response in hosts carrying the matching resistance genes (Mindrinos et al., 1994; Bent et al., 1994; Whitham et al., 1994). Such pathogen genes, which are typically found only in some races of a pathogen species, were originally called avirulence genes because their detection by the host makes the pathogen avirulent. Similarly, the host species is polymorphic for the presence of the corresponding resistance genes, resulting in variable gene-for-gene interactions in which a specific group of pathogen strains is recognized only by a subset of similarly distinct host strains (Botella et al., 1998; Ellis et al., 1999).
Cloning of these resistance or R genes revealed several salient facts: (i) Many code for what we now call NLR (instead of NB-LRR) proteins, to emphasize their similarity to animal NOD-like receptor (NLR) immune proteins; (ii) the functional absence of a resistance gene in some cases translates into absence of the entire gene, but in other cases, into the presence of functionally diverged orthologs; and (iii) NLR genes are often organized in clusters that differ greatly between different host strains. Here, we will first describe what has been learned in the past 25 years about inter- and intraspecific NLR diversity. We will then review how much (or how little) progress has been made toward unraveling the known unknowns of intraspecific NLR diversity through pan-genome and pan-NLRome studies, and conclude by discussing challenges for the years ahead.
NLR genes and NLR complexes are (almost) everywhere
NLR proteins are found in plants, animals, fungi, and protists, although the similarities in protein architecture are thought to result from convergent evolution (Yue et al., 2012). Generally, the basic NLR unit follows a modular tri-partite structure. The N terminal domain of NLR proteins can be considered the business end, being the primary structural element of signal transduction (Bentham et al., 2017). In plants, it is usually either a Toll/Interleukin-1 receptor/Resistance protein (TIR) domain, a coiled-coil (CC) domain, or a RESISTANCE TO POWDERY MILDEW 8-like (RPW8 or CC-R) coiled-coil domain. The central and most conserved NLR domain is a signal-transducing ATPase (signal transduction ATPases with numerous domains superfamily, STAND) domain, which likely evolved from a common bacterial ancestor and then gave rise to two different subclades, NB-ARC, typical for plants, and NAIP, CIITA, HET-E, and TP1 (NACHT) domains, typical for animals and fungi (Urbach and Ausubel, 2017). This domain is NB and serves as an ADP–ATP switch that regulates the ON/OFF state of the NLR. The C terminal domain, which is usually composed of repeated units, acts mostly as a ligand binding platform with autoinhibitory function. In plants and some animals, but not in fungi, it is often made up of leucine-rich repeats (LRRs; Soanes and Talbot, 2010; Dyrka et al., 2014).
Plant NLRs recognize pathogen effectors by directly binding to them, or by recognizing effector-mediated modifications of another host component. In the latter scenario, the NLR acts as a guard and its host client is its guardee (Cesari, 2018). Guardees are typically proteins that are targeted by effectors, with some having lost their original function in plant growth or immunity, and only acting as effector decoys (Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008).
Many of the inferences about NLRs came from the study of individual NLR domains and interactions between them and their guardees. The similarity in overall function between animal and plant NLRs was subsequently confirmed by structural analyses. In vertebrates, NLR proteins form wheel-shaped oligomers called inflammasomes, which are assembled upon pathogen recognition and activate a signaling cascade that leads to the formation of pores in the plasma membrane, resulting in localized cell death (Broz and Dixit, 2016). In plants, the CC-NLR HOPZ-ACTIVATED RESISTANCE 1 (ZAR1) and the TIR-NLR Roq1 assemble into oligomeric complexes called resistosomes that are strikingly similar to inflammasomes (Wang et al., 2019a, 2019b; Martin et al., 2020). Elucidation of the ZAR1 structure also revealed that its CC domain is reminiscent of bacterial pore-forming toxins. Since ZAR1 localizes to the plasma membrane, it is tempting to speculate that the oligomerized CC domains directly insert into the membrane, creating pores and thereby potentially changing ion flow and triggering cell death (Wang et al., 2019a). In further support of a common cell death-inducing mechanism across kingdoms is the observation that the CC-R (RPW8) domain can be homology modeled on structures of the mammalian MLKL and the fungal HELo and HELL domains, all of which also form membrane pores and induce cell death (Seuring et al., 2012; Daskalov et al., 2016; Bentham et al., 2018). However, it must be noted that while the idea that CC-NLRs, TIR-NLRs, and RPW8-NLRs can directly make holes in plasma membranes and thereby initiate further cell damage-associated signaling and/or cell death is attractive, it is unclear whether a one-size-fits-all model applies, since plant NLRs can apparently act in different cellular compartments, including the nucleus and the cytoplasm (Zhang et al., 2017a), with resistosomes potentially targeting diverse membranes (Adachi et al., 2019).
If NLRs in plants, animals, and fungi are the product of convergent evolution, what is the advantage of having this particular multi-domain structure and mode of action? Immune receptors should ideally act as hair triggers, such that any threat is immediately met, but at the same time, they should also be robust to inadvertent activation, since inappropriate immunity can have devastating consequences (Bomblies et al., 2007; van Wersch et al., 2016). The multi-domain structure allows for self-inhibition through intramolecular interactions, providing a primary safeguard against spurious activation. The formation of higher order complexes in turn may serve to amplify the triggering signal, but also help to prevent mis-regulation. Lastly, the modular NLR architecture may allow for facile reshuffling of individual domains, endowing them with versatility in recognition specificity, as well as allowing for different selection pressures to act on individual domains.
NLR gene numbers vary greatly between plant species
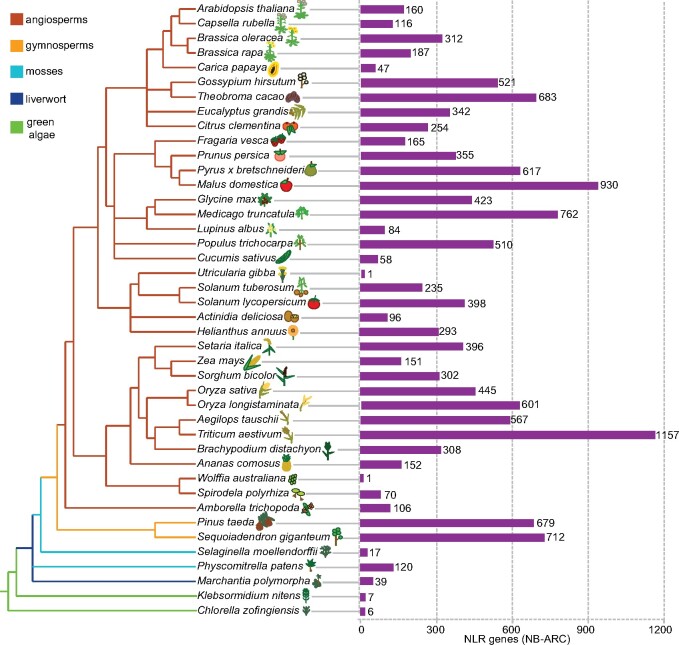
After plants colonized land, plant genomes experienced a massive expansion of NLR genes, going from fewer than a dozen in green algae, where plant NLRs are thought to originate, to many hundreds in land plants, likely as a consequence of adaptation to new pathogen pressures (Shao et al., 2019; Figure 1). NLR numbers across different species are highly variable; among all coding genes, the percentage of NLR genes ranges over many orders of magnitude, from 0.003% in bladderwort (Utricularia gibba) to 2% in apple (Malus domestica; Jia et al., 2015; Baggs et al., 2020). NLR genes appear to turn over rapidly, with frequent births and deaths (Michelmore and Meyers, 1998). Many of the NLR expansion and contraction events are lineage-specific; for example, TIR-NLRs, although present in some mosses and green algae, are largely absent from monocots (Tarr and Alexander, 2009; Gao et al., 2018), and in the Solanaceae, CC-NLRs are greatly expanded (Seo et al., 2016), while Rosales and conifers have experienced independent expansions of RPW8-NLRs (Jia et al., 2015; Van Ghelder et al., 2019; Figure 1). NLR numbers are generally low in the Cucurbitaceae, likely as a result of frequent gene losses and few subsequent duplication events (Lin et al., 2013; Figure 1). Low NLR numbers may result from their functional dispensability; for example, Wolffia australiana, a duckweed with just over 15,000 genes that potentially represent a minimum set of genes necessary for survival in an aquatic environment, has only one canonical NLR (Michael et al., 2020; Figure 1). Bladderwort, another aquatic plant, has at most one, and perhaps no NLRs at all (Baggs et al., 2020). This raises the question of whether evolutionary innovations were required in these plants to compensate for the loss of NLRs. In support of such a scenario, another duckweed with a highly reduced NLR complement, Spirodela polyrhiza, appears to have more components for antimicrobial signaling than other plants (An et al., 2019).
Figure 1.
Plant phylogeny and NLR complement sizes. Adapted from Gao et al. (2018), Munch et al. (2018), and Baggs et al. (2020).
Diversity in NLR genomic organization: from single genes to clusters, pairs, and extras
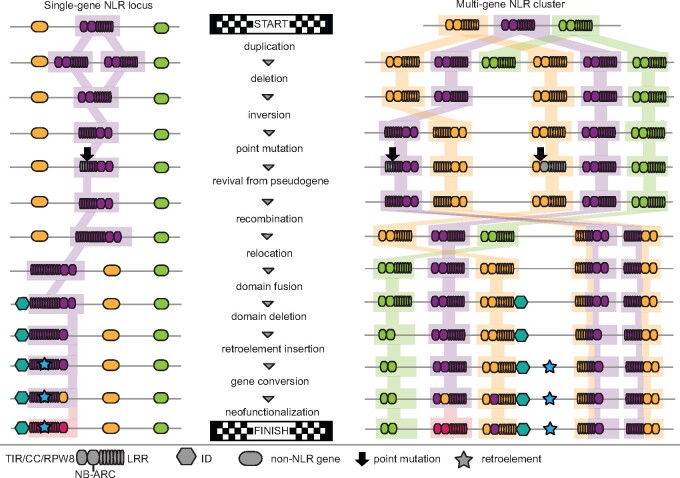
A clustered genomic arrangement is often emphasized as an NLR characteristic. In Arabidopsis thaliana genomes for example, about half of NLR genes are found in clusters (Meyers et al., 2003; Van de Weyer et al., 2019). NLR clusters often appear to be the products of tandem duplication events, sometimes followed by unequal crossing over, as well as intra-cluster rearrangements and gene conversion events (Meyers et al., 1998; Noël et al., 1999; Kuang et al., 2004; Figure 2). Clusters, which are often, but not always, made up of phylogenetically related NLR genes, can range in size from tens of kilobyte, with RPP5 in A. thaliana, which contains five cluster members in the reference accession Col-0, being an example, to several megabytes, with RGC2 in lettuce being a record holder with ∼3.5 Mb and consisting of 24 cluster members (Meyers et al., 1998, 2003).
Figure 2.
Examples of possible changes in sequence and genomic organization in single-gene and clustered NLRs through evolution. Major changes can occur in an NLR locus, and often the reconstruction of the sequence of events will be impossible.
What are the evolutionary advantages and disadvantages of having clusters? Recombination during meiosis is reduced by structural differences, which are particularly high in NLR clusters, as we discuss in more detail below. In agreement, one of the first challenges during efforts to isolate disease resistance genes was often the suppressed recombination across many such loci (to name just a few examples, see Ganal et al. [1989]; Noël et al. [1999]; Wei et al. [1999]; and Chin et al. [2001]). On the other hand, there is evidence of particularly high historical recombination rates around many NLR genes, as measured by linkage disequilibrium (LD) in natural populations (Horton et al., 2012; Choi et al., 2013), and there is only weak evidence for NLR loci as a group to suppress recombination, even for loci that differ in arrangement between parents (Rowan et al., 2019). These apparently contradictory observations likely reflect simply the high variation in recombination rates across NLR loci, with some acting as recombination coldspots, as expected, but others acting actually as recombination hotspots (Choi et al., 2016). This seems to be a function of the extent of structural variation between accessions, and many of the most structurally diverse regions of the A. thaliana genome indeed include NLR clusters with severely suppressed recombination (Jiao and Schneeberger, 2020). As we also discuss below, excessive NLR variation can potentially reduce fitness because of intragenomic immune system conflict, and under reduced recombination, it is more difficult to select for advantageous alleles that are linked to disadvantageous alleles, known as the Hill–Robertson effect (Hill and Robertson, 1966).
Perhaps then the major advantage of clustered gene arrangements comes from several linked, closely related genes providing a means for generating new functional diversity through unequal crossovers (also known as illegitimate recombination) as well as gene conversion involving genes that are not strictly orthologs (Kuang et al., 2004; Wicker et al., 2007). This is further facilitated by the repetitive structure of an important component of NLR exons, the LRR coding sequences.
Importantly, unequal crossover can support both expansion and contraction of sequences. Contraction is perhaps particularly relevant when considering events both at single-gene loci and clusters: At single-gene loci, any deletion will lead to truncation or loss of the gene. In contrast, in a cluster, illegitimate recombination between two genes can simultaneously reduce gene numbers and lead to creation of a new full-length gene (Figure 2). Although new genes resulting from unequal crossovers will often be nonfunctional, they can serve as reservoirs for future evolution. In addition, they might combine the activities of the two original genes, or they could have a different activity all together (Smith and Hulbert, 2005; Figure 2). Furthermore, the more copies of a gene, the higher the chances are that beneficial mutations arise, both because multiple copies constitute a larger mutational target than a single copy, but also because duplicates can undergo relaxed selection due to their functional redundancy (Ohno, 1970; Jiang and Assis, 2017). Gene clusters thus provide a larger and more flexible genetic basis for evolving new resistance specificities through complete or partial domain swaps between closely related homologs.
A special type of clustered NLR genes comprises pairs of phylogenetically unrelated genes arranged in a head-to-head orientation, where one acts as an executor and the other as a sensor, and where the latter often carries a non-canonical integrated domain (ID) that can act as a bait for pathogen effectors (Cesari et al., 2014). In A. thaliana, the sensor TIR-NLR RRS1 features an integrated WRKY domain that is targeted by Ralstonia effector PopP2 and Pseudomonas syringae effector AvrRps4, triggering the formation of an active complex composed of RRS1 and its paired executor NLR RPS4 (Williams et al., 2014; Sarris et al., 2015). In rice (Oryza sativa), RGA4/RGA5 and Pik-1/Pik-2 are two well-characterized head-to-head gene pairs, with RGA5 and Pik-1 encoding HMA domains that are targeted by multiple Magnaporthe oryzae effectors, activating an immune response through their paired NLR partners RGA4 and Pik-2, respectively (Cesari et al., 2013; Maqbool et al., 2015). Minor mutations in IDs can have massive effects on effector recognition; for example, in the aforementioned HMA domain in Pik-1, two adjacent amino-acid changes are sufficient to expand the response profile to different M. oryzae AVR-Pik effector variants (De la Concepcion et al., 2019). Although there is a relatively wide variety of IDs, the most prevalent domains have kinase activity, bind DNA, or mediate protein–protein interactions (Kroj et al., 2016; Sarris et al., 2016; Van de Weyer et al., 2019). Not only are some domains more prevalent than others, it also seems that certain NLR subfamilies are particularly prone to attract IDs, many of which are found in paired arrangements (Bailey et al., 2018). Not all paired NLRs carry IDs; however, for example the TIR-NLR SOC3 is found in a head-to-head orientation with either CHS3 or TN2, both of which are truncated TIR-NLRs. Both pairs monitor SAUL1, an E3 ligase, with SOC3-TN2 responding to overaccumulation of SAUL1, and SOC3-CHS3 to the absence of SAUL1 (Liang et al., 2019). Generally, these paired decoy-containing NLR genes may be evolutionarily advantageous; by being linked, they facilitate co-regulation and co-evolution, leading to effective pathogen recognition and downstream signal transduction.
NLR haplotype diversity at individual loci: the known knowns
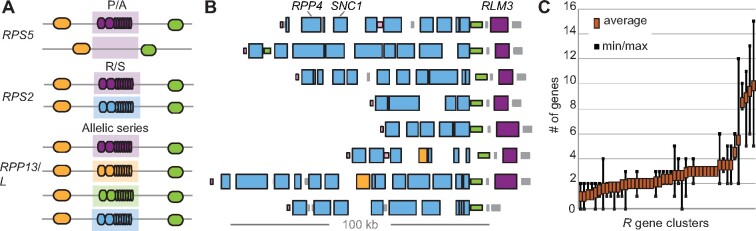
The starting point for the cloning of many NLR loci was naturally occurring resistant and susceptible plants from the same species. An obvious step was therefore to compare the genomic regions between resistant and susceptible individuals, including the underlying NLR genes. At single-gene loci, presence/absence variation (PAV) was found to be common, with entire genes being deleted in susceptible plants. RPS5 in A. thaliana is a classical example of P/A variation at single loci (Henk et al., 1999; Figure 3). The fact that susceptible plants lack the gene completely, instead of simply having functionally divergent alleles, suggested early on that in the absence of the recognized pathogen, the NLR gene imparts a fitness cost, which could indeed be demonstrated experimentally (Tian et al., 2003; Karasov et al., 2014). The two allelic variants in this and other cases, either P/A, or resistant/susceptible as observed for A. thaliana RPS2 (Figure 3), are likely maintained in metapopulations through balancing selection (Caicedo et al., 1999; Stahl et al., 1999; Tian et al., 2002; Mauricio et al., 2003; Bakker et al., 2006; Gos et al., 2012; Rose et al., 2012; Koenig et al., 2019).
Figure 3.
(A) Examples of allelic variation at individual NLR loci. P/A, presence/absence. R/S, resistant/susceptible. (B) RPP4/RPP5 NLR cluster across eight different A. thaliana accessions. Tall rectangles represent NLR genes, and short rectangles non-NLR genes. Colors indicate close sequence relatedness. There is a highly variable number of blue NLR genes in the cluster (including the functionally defined members RPP4, RPP5, and SNC1), while the adjacent RLM3 gene (magenta) shows P/A polymorphism. The RPP4/RPP5 cluster has also been invaded by an unrelated NLR gene (yellow). Finally, some of the non-NLR genes (pink, green) flanking the cluster are duplicated as well. (C) Average number of genes per NLR gene cluster across eight A. thaliana accessions. Most have on average fewer than three genes, while a few are both highly expanded and highly variable in numbers. Data for B and C from Jiao and Schneeberger (2020).
In contrast, RPP13 in A. thaliana and the L locus in flax (Linum usitatissimum) form extensive allelic series, where different functional alleles confer disease resistance to different races of the same pathogen (Figure 3). At the L locus, 10 distinct specificities for recognition of particular races of the rust fungus Melampsora lini are encoded by 11 different alleles, in both cases, recognition differences seem to be mostly due to variation in the LRR domain (Ellis et al., 1999; Bittner-Eddy et al., 2000). To date, the A. thaliana CC-NLR RPP13 remains the most polymorphic single-gene NLR locus known, with 19 different haplotypes initially identified in 24 accessions. RPP13 directly binds to the Hyaloperonospora arabidopsidis (Hpa) effector ATR13, which is also highly polymorphic, making the RPP13/ATR13 system a paradigm for NLR-effector co-evolution (Allen et al., 2004; Hall et al., 2009).
These two examples of allelic series, A. thaliana RPP13 and flax L, conform relatively well to the traditional notion of alleles being functionally related (Muller, 1932). In other cases, NLR homologs from different accessions, but in the same genomic location, confer resistance to entirely different pathogens. In A. thaliana, RPP8, HRT, and RCY1 all are at the same genomic location. RPP8 in the Ler accession endows plants with resistance to the Emco5 isolate of Hpa, HRT in the Dijon-17 accession to turnip crinkle virus, and RCY1 in the C24 accession to the yellow strain of cucumber mosaic virus (McDowell et al., 1998; Cooley et al., 2000; Takahashi et al., 2002). Because of the complex nature and copy number variation (CNV) at the small RPP8/HRT/RCY1 cluster, it is unclear whether the different resistances are encoded by different alleles or different genes at the same cluster (MacQueen et al., 2019). In the latter case, the situation would be similar to what has been described for the viral resistance gene Rx1 and the cyst nematode resistance gene Gpa2 in potato (Solanum tuberosum). Both genes are not only located in the same cluster, but also in the same haplotype, and it is therefore clear that they are not allelic (van der Vossen et al., 2000). An unresolved question is whether despite seemingly very different organismal function, such proteins have similar biochemical activity, e.g. if they act as guards, whether they converge on the same guardee that is targeted by effectors from different pathogens.
Finally, even though NLRs are modular proteins where each domain has an assigned function, the absence of some domains does not necessarily render NLR proteins non-functional, with some truncated NLRs being able to confer resistance to pathogens. In A. thaliana for example, the TIR-only protein RBAI can detect the bacterial HopBA1 effector (Nishimura et al., 2017), while the TIR-NB protein TN13 is required for basal resistance (Roth et al., 2017). RLM3, which carries a TIR-NB-X configuration with C-terminal BRX motifs instead of LRRs, confers broad-spectrum resistance to necrotrophic pathogens (Staal et al., 2008). In wheat, YrSP, a CC-NLR that is missing most of its LRR domain, retained the ability to confer resistance to yellow rust, but had different recognition specificities from its highly similar (99.8% identity), apparent full-length variant Yr5 (Marchal et al., 2018). In some cases, truncated NLR may remain functional because they work together with other full-length proteins, including NLRs, to initiate an immune response (Zhao et al., 2015; Zhang et al., 2017b).
Collateral damage of NLR diversity
Extensive sequence diversity found across NLR genes can backfire and lead to genetic incompatibilities, often resulting in hybrid necrosis, a phenomenon formally described as early as 1943 for crosses in wheat (Triticum aestivum; Caldwell and Compton, 1943). Hybrid necrosis cases have since been identified in numerous plants, including rice, lettuce (Lactuca sativa and Lactuca saligna), cotton (Gossypium barbadense and Gossypium hirsutum), tomato (Lycopersicon pimpinellifolium and Lycopersicon esculentum), Capsella spp., monkeyflower (Mimulus caespitosa, M. minor, and M. tilingii), and A. thaliana (Krüger et al., 2002; Bomblies et al., 2007; Alcázar et al., 2009; Jeuken et al., 2009; Yamamoto et al., 2010; Chen et al., 2014; Chae et al., 2014; Todesco et al., 2014; Sicard et al., 2015; Deng et al., 2019; BarragAn et al., 2019; Sandstedt et al., 2020). The genetic architecture underlying these incompatibilities is usually simple, involving one or two mismatching loci that typically encode components of the plant immune system, very often NLR proteins. These incompatibilities lead to deleterious hyperimmunity in the hybrid, thereby limiting the combinations of beneficial alleles that can be assembled in a single genotype (Chae et al., 2014). Conversely, mismatches among pyramided NLR genes can lead to the suppression of disease resistance. In wheat for example, different Pm3 alleles can suppress each other’s ability to confer powdery mildew resistance (Stirnweis et al., 2014), and Pm3 can also suppress resistance provided by its rye (Secale cereale) ortholog Pm8 (Hurni et al., 2014).
The age of NLR pan-genomes: discovering the known unknowns
Why pan-genomes?
From what we have discussed so far, several important points emerge. Foremost is that there is tremendous variation in NLR genes between and within species, and particularly at NLR clusters. In recent years, it has become abundantly clear that one must not speak of “the genome” of a species. In plants, one of the earliest indications that the concept of a single linear sequence as “the genome” was misleading came indeed from comparing haplotypes of individual NLR clusters (Botella et al., 1998; McDowell et al., 1998; Noël et al., 1999; Kuang et al., 2004; Srichumpa et al., 2005). Even from studies of just a few haplotypes, we already learned that there is extensive variation at all levels: in coding sequences, in copy numbers, and in genomic location. However, while we in principle know the molecular processes that can create the observed differences (point mutations, transposon insertions, deletions, duplications, other types of chromosomal rearrangements, gene conversion, illegitimate recombination events, etc.), we still largely do not know the true extent of NLR diversity within a species, both in terms of presence and absence of individual genes nor in terms of haplotype diversity, let alone knowing the underlying evolutionary forces generating and differentially maintaining this diversity.
A decisive factor in answering these questions will be how often, if ever, a sufficient number of haplotypes has survived in a population, so that we can begin to reconstruct the evolutionary history of all, or at least of most, NLR loci. The only way to find out is by assembling more and more genomes from the same species and comparing their NLR content. While this sounds straightforward, there are substantial hurdles in making sense of many genomes due to the exponential increase in all-against-all comparisons: with only 10 different genomes, there are already 45 comparisons, but with 100 genomes, this increases to 4,950 comparisons. In order to make such genome comparisons feasible, data structures built on the concept of pan-genomes must be considered. Below we discuss the challenges of pan-genomes and of pan-NLRomes, the NLR component of pan-genomes: How to produce pan-NLRomes (discovering the unknowns), how to estimate their completeness (estimating the known unknowns), and finally, and clearly the most substantial challenge, how to make sense of the pan-NLRomes (going from known unknowns to knowns).
A brief history of plant pan-genomes
A common definition of a pan-genome is the entire repertoire of DNA sequences and sequence variants in a given species. That closely related bacterial strains may differ by complete arrays of genes, such as pathogenicity islands, was already noticed in the 1980s (Groisman and Ochman, 1996), but one of the first formal mentions of the pan-genome concept was in a study where genomes of eight different Streptococcus agalactiae strains were compared, confirming that often genomes from many individuals are needed to capture the entire gene content of a species (Tettelin et al., 2005). In eukaryotes, generating many high-quality whole genome assemblies was until recently difficult due to both technical and monetary constraints associated with sequencing large genomes, especially when compared with bacteria, where most initial pan-genomic studies were performed (Mukherjee et al., 2019). This is changing quickly, but the mapping of short reads to a single reference genome has so far been the most common method to study variation within and across species. This approach can detect small indels, single-nucleotide polymorphisms (SNPs), or larger deletions in well-conserved regions, but it often fails to adequately capture highly structurally divergent regions and CNVs.
Strictly speaking, it is very rarely possible to know the complete pan-genome of a species. Therefore, as a first step, an initial pan-genome is constructed from a few individuals, and with such a sub-pan-genome in hand, one can estimate how much of the species-wide pan-genome is still missing. Although early pan-genome studies in plants relied mostly on assembling contigs from short reads (e.g. Gordon et al., 2017; Golicz et al., 2016; Hurgobin et al., 2018; Zhao et al., 2018), they already led to the discovery of many genes missing from the reference, demonstrating the importance of sequencing more than one genome to capture inter- and intraspecific genetic diversity (Table 1). For example, the first Brachypodium distachyon pan-genome from 54 lines doubled the number of genes known for this species (Gordon et al., 2017). Similarly, in a study of 66 accessions from different rice species, around ∼10,800 novel genes were identified that were missing from the Nipponbare reference (Zhao et al., 2018).
Table 1.
Examples of plant pan-genome or pan-NRLome studies and key observations on NLRs or R genes
| Plant pan-genome or pan-NLRome studies | |||||
|---|---|---|---|---|---|
| Organism (s) | N | Sequencing technology | Assembly strategy | Reference | Key observations on NLR or R genes |
| Brassica rapa | 3 | Short reads | De novo assemblies | Lin et al. (2014) | – NLRs enriched in dispensable genome. |
| Glycine soja | 7 | Short reads | De novo assemblies | Li et al. (2014) |
– CNVs common in NLRs: candidates of resistance differences between wild and cultivated accessions. – NLRs numbers and domain architectures varying between species. – NLRs enriched in dispensable genome. |
| Zea mays | 503 | Short reads | De novo transcriptome | Hirsch et al. (2014) | |
| Oryza sativa | 3 | Short reads | De novo assemblies | Schatz et al. (2014) | − NLRs enriched in dispensable genome, e.g. 12% shell versus 0.35% core genes are NLRs. |
| O. sativa | 1483 | Short reads | Iterative assembly | Yao et al. (2015) | – NLRs enriched in dispensable genome. |
| Populus clade | 7 | Short reads | Map to reference | Pinosio et al. (2016) | − CNVs and SVs enriched for NLRs. |
| Brassica oleraceae | 10 | Short reads | Iterative assembly | Golicz et al. (2016) |
– NLRs enriched in genes showing PAV. – 43% of NLRs dispensable, 45% in clusters, and 60% absent from reference. |
| Brachypodium distachyon | 54 | Short reads | De novo assemblies | Gordon et al. (2017) | − NLRs enriched in genes showing PAV, likely underlying variation in disease resistance. |
| Triticum aestivum | 19 | Short reads | Iterative assembly | Montenegro et al. (2017) | – NLRs enriched in genes showing PAV. |
| Medicago truncatula | 15 | Short reads | De novo assemblies | Zhou et al. (2017) | − NLRs enriched in dispensable genome show high nucleotide and protein diversity, large effect SNPs, often with PAV, CNVs and differences in domain architectures. |
| Capsicum clade | 383 | Short reads | Iterative assembly | Ou et al. (2018) | |
| Oryza clade | 66 | Short reads | De novo assemblies | Zhao et al. (2018) | − NLRs enriched in dispensable genome. |
| O. sativa | 3010 | Short reads | Map to reference | Wang et al. (2018) | – NLRs enriched in dispensable genome. |
| Brassica napus | 53 | Short reads | Iterative assembly | Hurgobin et al. (2018) |
– NLR enriched in genes showing PAV. – 30.6% core NLRs, 69.4% variable, and ∼50% absent from reference. |
| Juglans clade | 6 | Short reads (long reads for J. regia) | De novo assemblies | Stevens et al. (2018)/Trouern-Trend et al. (2020) | – Overrepresentation of disease resistance genes in rapidly evolving, contracting, and expanding gene families. |
| Helianthus clade | 493 | Short reads | Iterative assembly | Hübner et al. (2019) | − NLRs overrepresented in regions introgressed from wild into cultivated species. |
| Sesamum clade | 5 | Short reads | De novo assemblies | Yu et al. (2019) | – Defense response genes expanded (e.g. RPM1), positively selected for and fast evolving. |
| Lycopersicon clade | 725 | Short reads | Iterative assembly | Gao et al. (2019) |
– Disease resistance genes enriched in genes lost or selected against during domestication and improvement. – Defense response genes show high PAV. |
| Hordeum vulgare | 63 | Short reads | De novo transcriptome | Ma et al. (2019) |
– Higher proportion of NLRs genes in wild versus cultivated genotypes. – NLRs under high selective pressures during domestication, e.g. Mla genes. |
| Zea mays | 4 | Short reads | De novo assemblies | Haberer et al. (2020) | |
| Arabidopsis thaliana | 8 | Short and long reads | De novo assemblies | Jiao and Schneeberger (2020) |
– Low collinearity between accessions in NLR-rich regions. – SVs in NLRs suppressing meiotic recombination. |
| Solanum clade | 14 | Short and ultra-long reads | De novo assemblies | Alonge et al. (2020) | − SV hotspots introgressed from wild into domesticated species enriched for R genes. |
| P. persica and relatives | 4 | Short reads (long reads and Hi-C for P. mira) | De novo assemblies | Cao et al. (2020) |
– NLRs enriched in dispensable genome. – Cross-species SV underlying nematode resistance. – NLR numbers range from 310 to 339 across species. |
| O. sativa | 12 | Short and long reads and optical map | De novo assemblies | Zhou et al. (2020) | |
| B. napus | 9 | Short and long reads (Hi-C and optical map for some) | De novo assemblies | Song et al. (2020) | – Defense response genes enriched for PAV. |
| Glycine clade | 29 | Short and long reads, Hi-C and optical map | De novo assemblies and graph | Liu et al. (2020b) | − NLRs enriched in dispensable genome. |
| A. thaliana | 64 | RenSeq: short and long reads | Map to reference | Van de Weyer et al. (2019) |
– NLR diversity saturation point reached after 40 accessions. – In 64 accessions, four times higher diversity in NLR architecture than in reference. |
| Solanum/N. benthamiana/C. annum | 18 | RenSeq: short and long reads | Map to reference | Seong et al. (2020) |
– 128 of 314 NLR annotations improved. – Preferential expansion of the extended CC-NLR N terminal region. |
Although short reads only allow for highly fragmented assemblies, these attempts confirmed earlier observations made by assembling left-over reads that could not be mapped to the reference (Cao et al., 2011; Gan et al., 2011; Long et al., 2013), namely that NLR genes are overrepresented in the variable fraction of gene content across accessions (Golicz et al., 2016; Montenegro et al., 2017; Hurgobin et al., 2018). For example, while only 19% of the Brassica oleracea pan-genome was composed of genes missing from the reference, the number of NLR genes missing from the reference was almost 60% (Golicz et al., 2016). Similarly, 50% of the 307 NLR genes found across 53 Brassica napus accessions were absent in the Darmor-bzh reference (Hurgobin et al., 2018). These findings have been confirmed with more recent and more complete long-read assemblies of multiple accessions, for example in soybean (Liu et al., 2020b).
In these early studies, there were generally no attempts to estimate what fraction of gene content was not properly assembled, but given the repetitive nature of many NLR genes, it is almost certain that an unknown portion of the NLR complement was missed. This was partially solved by enriching first for regions of the genome carrying NLR-related DNA sequences by hybridization to baits designed based on known NLR sequences, followed by short read sequencing, which reduces the complexity of the assembly task (Jupe et al., 2013; Andolfo et al., 2014; Arora et al., 2019). This approach, called Resistance gene enrichment sequencing (RenSeq), was later improved upon by using long-read sequencing after enrichment (Witek et al., 2016; Van de Weyer et al., 2019; Seong et al., 2020). Most recently, as the costs for long-read sequencing have dropped precipitously, more and more sets of highly contiguous de novo assemblies are being produced for a range of species, most notably rapeseed, soybean, tomato, rice, and A. thaliana (Alonge et al., 2020; Liu et al., 2020b; Song et al., 2020; Zhou et al., 2020; Jiao and Schneeberger, 2020; Table 1).
Genes in the pan-genome can be divided by their representation frequencies across accessions. Following the convention for bacterial pan-genomes, core genes are those genes that are present in all or almost all accessions (sometimes distinguished as core and soft core), shell genes are found at intermediate frequency, and cloud genes are rare, detected only in one or very few accessions (Figure 4). Shell and cloud genes together make up the dispensable genome (Tettelin et al., 2005; Page et al., 2015). The number of known genes in the dispensable genome is obviously highly dependent on the number of individuals analyzed, but even in comparable data sets their contribution to the pan-genome seems to vary greatly. For example, in a recent comparison of nine B. napus high-quality long-read assemblies, only 56% of genes were assigned core status, while in a comparison of eight A. thaliana genome assemblies of similar quality, 80% of genes were identified as core genes, even though the definition of core genes allowed for absence in one of the studied genomes in B. napus, whereas the A. thaliana study considered only genes present in all genomes as core genes (Jiao and Schneeberger, 2020; Song et al., 2020). How much of this is due to differences in annotation pipelines and methods for identifying orthologs are currently unclear.
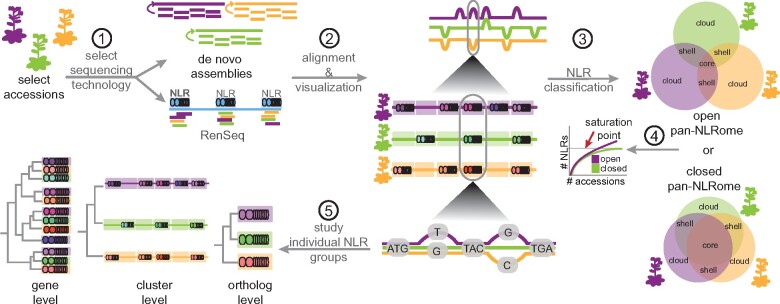
Figure 4.
Potential workflow of the construction, visualization, and analysis of a pan-NLRome. First, accessions to be sequenced are selected in a manner that maximizes diversity, in order to approach a saturated pan-NLRome as quickly as possible (1). Then either the whole genome or the NLR complement is sequenced and assembled (2). The assemblies from each genome are then aligned to each other and potentially combined into a genome graph (3). Core and accessory genes are identified and the saturation point of the NLRome is assessed, to determine whether the NLRome is open or closed (4). Finally, individual NLR groups can be studied, e.g. one can analyze the phylogeny of a certain cluster across accessions, by comparing individual alleles in those accessions that share the same genes, the structure of whole clusters, or the entire set of genes present in all clusters (5).
Key steps in constructing pan-NLRomes
We are interested in the NLR component of pan-genomes, or pan-NLRomes. The first step in constructing a pan-NLRome for a certain species is the same as for a pan-genome: the informed selection of accessions to maximize genetic diversity with as few individuals as possible (Figure 4). Next, one has to decide on the most appropriate approach, either whole-genome assemblies after long-read sequencing or focusing on the assembly of NLR sequences with RenSeq (Witek et al., 2016; Figure 4). The latter, while more cost effective, has several disadvantages: Anchoring assembled NLR contigs to the genome can be difficult, since this depends on the amount of sequences flanking the assembled NLR genes, and because baits are based on known NLR genes, not all NLR genes present in a target genome will be captured equally well. Nevertheless, particularly with very large genomes, this is still the method of choice, especially if the goal is the discovery of a specific resistance gene, rather than producing a complete inventory of all NLR genes in a species (Arora et al., 2019).
De novo assemblies of entire genomes will ideally contain highly contiguous scaffolds that support the facile identification of syntenic sequences. Because RenSeq produces only contigs covering individual NLR genes, the ease with which contigs can be anchored on a reference genome, or linked to larger clusters, will depend on how much flanking sequences these contigs contain, which in turn is a function of the length of the original molecules captured in the enrichment step. As an example, in a recent A. thaliana study, genes that were not clearly orthologous to reference NLR genes could not always be easily anchored to the reference genome (Van de Weyer et al., 2019). The size of captured molecules could potentially be improved using technologies other than hybridization-based sequence capture. One of these is CRISPR/Cas9-based excision of target regions (Gabrieli et al., 2017; Tsai et al., 2017), but this comes with two disadvantages: First, large amounts of very high-molecular weight DNA must be available for size separation of excised molecules from the background. Second, universally conserved regions flanking NLR genes and clusters must be known. An exciting alternative is the capture of large fragments in microdroplets, followed by droplet sorting based on PCR-based detection of NLR-related sequences with fluorescent probes (Madsen et al., 2020), although the design of panels of oligonucleotide primers for comprehensive NLR detection will likely be challenging.
After assembling either whole-genome shotgun or RenSeq reads, NLR genes must be annotated in each individual assembly. Automatic annotation produces a non-negligible number of errors (Meyers et al., 2003; Van de Weyer et al., 2019), even when using a specialized tool such as NLR-annotator (Steuernagel et al., 2020). As with other annotation efforts, RNA-seq data are often very helpful, and in this regard, manipulations such as various stresses that robustly induce the expression of many NLR genes are useful (Lai and Eulgem, 2018; Steuernagel et al., 2020), although manual curation of gene models is still highly recommended. We would like to emphasize, however, that we consider genes or even gene fragments that are not expressed as an important component of the pan-NLRome, because such sequences can still have a role in evolution of the pan-NLRome, as discussed above.
The challenges of analyzing pan-NLRomes
A major unsolved issue in pan-genome and pan-NLRome analyses is how to best compare gene sets and arrangements in multiple genomes. A first step will often be to cluster genes from the different genomes into ortholog groups for further analysis (Enright et al., 2002; Li et al., 2003; Contreras-Moreira et al., 2017; Thanki et al., 2018), which will already reveal NLR genes missing from the initial reference. As discussed above, NLR genes are particularly likely to be insufficiently captured by single reference genomes.
Conventional tools for phylogenetic and population genetic analyses can easily deal with single stand-alone NLR genes. They may show P/A polymorphisms, but if a gene is present, it can in most cases be treated as an orthologous member or true allele of this particular NLR locus. A caveat arises from truncated genes, which may be functional, but might not be correctly identified as an ortholog because orthology is a function of shared sequence.
Many NLR genes are found in clusters, and orthology relationships can be complex, e.g. it may be difficult to discern which, if any, of the multiple copies of an NLR gene in one genome is the true ortholog of that same NLR gene in another genome. This is in turn important if one wants to make inferences about shared functionality and understand the evolution of NLR clusters. In this regard, it is important to remember that there is value not only in discovering new NLR genes and new NLR cluster arrangements, but also in identifying accessions with similar or identical clusters. An excellent criterion for discovering conserved clusters in a principled manner was proposed in the context of comparing eight A. thaliana genomes (Jiao and Schneeberger, 2020). The authors of this study developed a formal measure of pairwise synteny diversity, inspired by the measure of pairwise nucleotide diversity, π. Satisfyingly, many NLR clusters were found to have high synteny diversity across accessions, but some NLR clusters were also remarkably invariant, at least in the number of genes in the cluster (Figure 3).
Unfortunately, there will be limits to pairwise comparisons, as discussed earlier, and ultimately one would want to move to a unified data structure for multiple genomes. An obvious solution is provided by genome graphs (Herbig et al., 2012; Nguyen et al., 2015; Paten et al., 2017; Computational Pan-Genomics Consortium, 2018). In addition, powerful tools are being developed for variant calling and gene annotation on such genome graphs (Paten et al., 2017; Eggertsson et al., 2019; Kim et al., 2019; Rakocevic et al., 2019), and in principle one could envision how these tools could be extended to support phylogenetic analyses of sequences present in graph genomes (Figure 4).
A major challenge in understanding the evolution of NLR clusters is to reconstruct the entire sequence of events such as duplications, deletions, invasions by non-NLR genes, and phylogenetically unrelated NLR genes (Figure 2), which will be contingent on how many of the individual steps in the evolution of NLR clusters are still represented in a population. Methods for reconstructing the histories of tandem duplications and more complex gene clusters, including illegitimate recombination events, have been proposed (Elemento et al., 2002; Vinař et al., 2009), but they are not yet widely used, most likely because the available data have been too sparse in most cases.
These issues relate to another important question: How many individuals need to be analyzed to capture a substantial fraction of a species’ pan-genome? This will greatly depend on the underlying diversity of the group of accessions considered, as nicely illustrated with a recent analysis of structural variants, which include the absence/presence polymorphisms typical for many NLR genes, in tomato. Near-saturation for common structural variants was achieved much more quickly for domesticated tomato varieties, which have undergone a recent genetic bottleneck, than for wild tomato accessions (Alonge et al., 2020). If additional genomes lead to the addition of fewer and fewer new genes to the pan-genome, one can infer that saturation of pan-genome content is achievable, and the pan-genome is said to be closed. If the number of new genes added by each additional genome analyzed does not diminish, and if it is therefore unclear whether saturation is achievable, a pan-genome is said to be open (Figure 4).
The openness of bacterial pan-genomes is primarily due to horizontal gene transfer (HGT) including uptake of environmental DNA by transformation (Vernikos et al., 2015; Brockhurst et al., 2019). Although HGT exists in plants and should not be dismissed as a contributor to plant pan-genomes (Bock, 2010), a likely more important route for expanding pan-genomes is through homoploid hybridization, that is, hybridization between distant relatives without a change in chromosome number, which is very common across all major eukaryotic lineages including plants (Mallet et al., 2016; Payseur and Rieseberg, 2016; Taylor and Larson, 2019). Often, the genomes from the original two species contribute very unevenly to the extant genomes we observe today, and the sequences coming from the underrepresented genome are considered as introgressed segments. Of particular interest in this context is adaptive introgression, where alien sequences are favored under natural selection because they confer some fitness benefit (Suarez-Gonzalez et al., 2018). It is not surprising that there are many instances of introgressions from wild material intro crops (Janzen et al., 2019), and introgression through wide crosses has a long history in breeding, including breeding for disease resistance (Stevens and Rick, 1988; Niu et al., 2014; Gaiero et al., 2018).
Pan-NLRomes: a step toward fewer unknowns
Two recent RenSeq studies focused specifically on the pan-NLRome, with the goal of capturing a substantial fraction of inter- and intraspecific NLR diversity (Table 1). The first study compared 64 A. thaliana accessions (Van de Weyer et al., 2019), while the second study spanned 16 accessions from 5 different Solanum species, plus single accessions of Nicotiana benthamiana and Capsicum annuum, also from the Solanaceae family (Seong et al., 2020). In both A. thaliana and Solanum, the average number of NLR genes discovered in the different accessions was similar to that in the reference genomes, with some accessions having more NLR genes than the reference, pointing to the completeness of the RenSeq efforts. This was similar in a comparison of four high-quality long read assemblies from domesticated rice accessions (Wang et al., 2019c). In A. thaliana, 26 new IDs in addition to the 10 known from the original reference genome were found, and the universe of different NLR domain architectures was expanded from 22 in the reference genome to 97 in the entire set of 64 accessions (Van de Weyer et al., 2019), highlighting how incomplete our understanding of NLR diversity is with a single reference genome. This dataset also confirmed different modes of evolution for subsets of NLRs (Kuang et al., 2004). While the phylogeny of most groups of apparent orthologs behaved in a manner consistent with allelic series, some groups showed more complex patterns indicative of sequence exchanges and neofunctionalization. This was also recently observed in Lee and Chae (2020), where a distinction between conserved “high fidelity” and highly-variable “radiating” NLR genes was made. Notably, radiating NLRs included all three known cases in A. thaliana where genes at the same locus confer resistance to very different pathogens, such as the RPP8/HRT/RCY1 example discussed above (Van de Weyer et al., 2019).
Both the A. thaliana and the Solanaceae pan-NLRome analyses showed that there are apparently core NLR genes that are present in all or almost all accessions, but that these account only for a minority of all NLR genes (Van de Weyer et al., 2019; Seong et al., 2020). Furthermore, although the length of sequencing reads in these studies was substantially below what would be possible in 2020 with current long-read sequencing technology, the assembled contigs often contained more than one NLR, and also often neighboring non-NLR genes, allowing at least for partial ordering of NLR genes in clusters and anchoring of a fraction of contigs to the corresponding place in the reference genome. A more conclusive analysis of variation in NLR cluster size was presented in a comparison of eight completely assembled A. thaliana genomes, which confirmed substantial CNV in most clusters, and also identified small clusters of up to three genes that appeared to be size invariant in this set (Jiao and Schneeberger, 2020; Figure 3).
As alluded to earlier, a key question is how extensive NLR diversity within the target group is and how many genome assemblies are needed to capture most of the variation. The increase in known diversity in the A. thaliana study with 64 accessions—more than quadrupling the number of NLR domain architectures known from the reference—may appear daunting, but it was also observed that a set of about 40 maximally diverse accessions would have been sufficient to discover the majority of NLR genes, as defined by phylogenetic orthology (Van de Weyer et al., 2019; Table 1). This might perhaps appear surprising at first glance, but it likely reflects strong selection for NLR genes, which are therefore more likely to spread faster throughout the entire population, and are therefore more evenly distributed in the population. With pan-NLRome information, it should become possible to test such hypotheses, including whether long-range linkage among NLR genes due to population structure is lower than for other genes.
Turning Pan-NLRome sequence knowledge into functional knowledge
The ultimate goal of pan-NLRome studies is to accelerate the discovery of NLR specificity in disease resistance. Such knowledge has practical value in breeding, but it is equally interesting to understand the diversification of NLR function in an evolutionary and ecological context. The first step is the generation of species-wide pan-NLRomes, which is clearly within reach. The second step, using the pan-NLRomes to better understand how NLR diversity is distributed and to discern the evolutionary history of NLRs in a species, is more challenging, but we are optimistic that this can be solved as well. A highly innovative approach to parsing NLR diversity has recently been introduced, with a focus on revealing potential functional sites in NLRs that will aid in the rational design of novel or broad-spectrum disease resistance (Prigozhin and Krasileva, 2020).
What remains is to comprehensively assign function to all existing NLRs. The most common known molecular activities of NLRs are (i) detecting a specific pathogen, or a specific pathogen effector, and (ii) enabling the functioning of other, pathogen-detecting NLRs (helper function). An additional activity for which there is less direct evidence so far is NLR expression being part of broad-spectrum disease resistance after a pathogen attack.
Most NLRs with a known function in disease resistance were discovered through linkage mapping in experimental crosses, using resistant and susceptible parents, often from natural populations, and also from mutant screens (Kourelis and van der Hoorn, 2018). Similar to other genes underlying traits that vary in natural or crop populations, the identification of disease resistance genes has benefited from advances with genome-wide association studies (GWASs). Disease resistance lends itself particularly well to GWAS because it is often controlled by loci of large effect (Atwell et al., 2010; Liu et al., 2020a), although this advantage is partially reduced, at least in wild plant species, by genetic heterogeneity, where different loci may underlie resistance to the same pathogen race (Nemri et al., 2010). The identification of NLRs responsible for disease resistance by GWAS has been accelerated by focusing on the NLR complement of genomes with RenSeq and using alignment-free approaches for identifying the causal genes (Arora et al., 2019). Similarly, RenSeq without assembly is a powerful technique to search for evolutionary signals of selection in NLR genes, which can help to prioritize specific genes for functional follow-up studies (Stam et al., 2016, 2019). In this regard, there is a virtuous circle: The better the pan-NLRome of a species is known, the better strategies can be designed for focused interrogation of NLR genes.
Despite the advances with genetic mapping in experimental crosses or in natural populations, these approaches do not easily scale. As an example, a review of the literature indicated that only 88 NLR genes in A. thaliana had some assigned function in the middle of the year 2020 (Kourelis and Kamoun, 2020), yet there are on the order of 500 orthologous groups of NLR genes that are represented in multiple accessions (Van de Weyer et al., 2019). Of course, a major unknown is how many NLR genes have a positive function in the sense that they can detect effectors that pose an actual threat to the NLR carrier, and how many no longer or not yet have such a function, but it seems likely that many more than 88 NLR genes in this species play a role in disease resistance in nature.
For comprehensive assignment of NLR function, clearly more direct functional tests need to be pursued. The simplest cases for direct functional tests are those where a function is already known for a particular NLR, and based on this knowledge a system can be devised for rapid investigation of sequence-related NLRs. An example is the RPP13 allelic series in A. thaliana, where transient expression was used to determine which RPP13 alleles recognized which allele of the matching effector ATR13 (Allen et al., 2004, 2008; Krasileva et al., 2011).
A more general approach introduces NLR genes into susceptible plant backgrounds followed by extensive testing with different races of the pathogen of interest (Yang et al., 2013; Zhang et al., 2015; Wang et al., 2019c). Direct functional testing of NLRs by expressing them in a foreign background may, however, not always be so straightforward. Many NLRs act as sensor NLRs and rely on helper NLRs for conferring disease resistance (Wu et al., 2017; Jubic et al., 2019; Feehan et al., 2020). In the case of paired NLRs, this limitation could be circumvented by always introducing both members of a pair, which are easily found due to their characteristic arrangement in the genome. More challenging are NLR networks that include unlinked NLRs, such as those found in the complex NRC immune network in the Solanaceae (Wu et al., 2017). While the sensor NLRs in this case are massively expanded, they converge on relatively few, conserved helper NLRs, although it cannot be excluded that some sensor NLRs require co-evolved helper NLRs. A clear opportunity that emerges from pan-NLR network studies is to identify combinations of interacting NLRs that do not currently exist in available breeding material, essentially generating a pan-NLR network, to extend both qualitative aspects (new resistances) or quantitative aspects (fine-tuning existing resistances).
In addition, focusing only on pathogens that cause disease on the focal plant species may not reveal the function of all of its NLRs, because some might provide resistance to microbes or other enemies that are not considered pathogens of that species (Cevik et al., 2019). Even if one can assign function to NLRs in terms of recognizing a specific pathogen, the question remains which effectors they detect. Knowledge of both the NLR and matching effector in turn is required for predicting the spectrum of pathogen strains that is recognized by different plant accessions in the field. An alternative therefore is to identify the effector proteins from pathogens or the plant proteins with which NLRs interact, an approach taken in two large yeast-two-hybrid interactome studies (Mukhtar et al., 2011; Weßling et al., 2014). These studies identified interactors for a large fraction of tested NLR fragments; having defined such NLR partners, one should be able to easily test how the interactions are affected by sequence variation mined from pan-NLRome efforts. In principle, this approach could be scaled up to include all members of a pan-NLRome. As pan-genome efforts take off also for pathogens (Badet and Croll, 2020), pan-NLRomes could then be tested in this manner against pan-effectoromes and their host targets. Importantly, pan-NLRome resources will help to choose alleles that provide an agriculturally optimal response to a specific effector or pathogen race, with the best compromise between yield penalty in the absence of the pathogen, effective pathogen detection and minimal collateral damage from too strong an immune reaction (Harris et al., 2013). Finally, in order to derive predictive models for NLR function, functional information must be captured in a structured manner; RefPlantNLR is a recent initiative that aims to do just that (Kourelis and Kamoun, 2020).
The ultimate goal is to arrive at a point where NLR function can be predicted from sequence alone. By combining knowledge of protein–protein interactions with other sources of information such as structures of NLRs and their interactors, as well as knowledge of host protein modification by pathogen effectors, we foresee that predictive models of NLR function will come into reach in the next decade. Finally, with global knowledge of NLRs and matching effectors in hand, mapping the co-occurrence of such pairs in hosts and pathogens in time and space should reveal the true extent of past and ongoing arms races between hosts and their pathogens.
Acknowledgments
The authors thank Jonathan Jones, Sophien Kamoun, Jeff Ellis, Jeff Dangl, Oliver Furzer, and Gautam Shirsekar for comments on the manuscript as well as the anonymous reviewers for their valuable comments. They apologize to those authors whose work they could not cite due to space limitations.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft through Collaborative Research Center SFB1101 and by the Max Planck Society (to D.W.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement. None declared.
Contributor Information
A Cristina Barragan, Department of Molecular Biology, Max Planck Institute for Developmental Biology, 72076 Tübingen, Germany.
Detlef Weigel, Department of Molecular Biology, Max Planck Institute for Developmental Biology, 72076 Tübingen, Germany.
A.C.B. and D.W. outlined, wrote, and edited this review.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Detlef Weigel (weigel@tue.mpg.de).
References
- Adachi H, Kamoun S, Maqbool A (2019) A resistosome-activated “death switch.” Nat Plants 5: 457–458 [DOI] [PubMed] [Google Scholar]
- Alcázar R, García AV, Parker JE, Reymond M (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci 106: 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RL, Bittner-Eddy PD, Grenville-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL (2004) Host–parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Allen RL, Meitz JC, Baumber RE, Hall SA, Lee SC, Rose LE, Beynon JL (2008) Natural variation reveals key amino acids in a downy mildew effector that alters recognition specificity by an Arabidopsis resistance gene. Mol Plant Pathol 9: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, et al. (2020) Major Impacts of Widespread Structural Variation on Gene Expression and Crop Improvement in Tomato. Cell 182: 145–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo G, Jupe F, Witek K, Etherington GJ, Ercolano MR, Jones JDG (2014) Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol 14: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Zhou Y, Li C, Xiao Q, Wang T, Zhang Y, Wu Y, Li Y, Chao D-Y, Messing J, Wang W (2019) Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proc Natl Acad Sci U S A 116: 18893–18899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O, Johnson R, Enk J, Periyannan S, Singh N, et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol 37: 139–143 [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badet T, Croll D (2020) The rise and fall of genes: origins and functions of plant pathogen pangenomes. Curr Opin Plant Biol 56: 65–73 [DOI] [PubMed] [Google Scholar]
- Baggs E, Monroe JG, Thanki AS, O’Grady R, Schudoma C, Haerty W, Krasileva KV (2020) Convergent loss of an EDS1/PAD4 signaling pathway in several plant lineages reveals co-evolved components of plant immunity and drought response. Plant Cell 32: 2158–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey PC, Schudoma C, Jackson W, Baggs E, Dagdas G, Haerty W, Moscou M, Krasileva KV (2018) Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol 19: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EG, Toomajian C, Kreitman M, Bergelson J (2006) A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18: 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragan CA, Wu R, Kim S-T, Xi W, Habring A, Hagmann J, Van de Weyer A-L, Zaidem M, Ho WWH, Wang G, Bezrukov I, et al. (2019) RPW8/HR repeats control NLR activation in Arabidopsis thaliana. PLoS Genet 15: e1008313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860 [DOI] [PubMed] [Google Scholar]
- Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B (2017) Animal NLRs provide structural insights into plant NLR function. Ann Bot 119: 827–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham AR, Zdrzalek R, De la Concepcion JC, Banfield MJ (2018) Uncoiling CNLs: structure/function approaches to understanding CC domain function in plant NLRs. Plant Cell Physiol 59: 2398–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J 21: 177–188 [DOI] [PubMed] [Google Scholar]
- Bock R (2010) The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci 15: 11–22 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D (2007) Autoimmune response as a mechanism for a Dobzhansky–Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JD (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10: 1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Harrison E, Hall JPJ, Richards T, McNally A, MacLean C (2019) The ecology and evolution of pangenomes. Curr Biol 29: R1094–R1103 [DOI] [PubMed] [Google Scholar]
- Broz P, Dixit VM (2016) Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16: 407–420 [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Schaal BA, Kunkel BN (1999) Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc Natl Acad Sci U S A 96: 302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell RM, Compton LE (1943) Complementary lethal genes in wheat causing a progressive lethal necrosis of seedlings. J Hered 34: 67–70 [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Cao K, Peng Z, Zhao X, Li Y, Liu K, Arus P, Zhu G, Deng S, Fang W, Chen C, et al. (2020) Pan-genome analyses of peach and its wild relatives provide insights into the genetics of disease resistance and species adaptation. bioRxiv:200204
- Cesari S (2018) Multiple strategies for pathogen perception by plant immune receptors. New Phytol 219: 17–24. [DOI] [PubMed] [Google Scholar]
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN (2014) A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front Plant Sci 5: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik V, Boutrot F, Apel W, Robert-Seilaniantz A, Furzer OJ, Redkar A, Castel B, Kover PX, Prince DC, Holub EB, et al. (2019) Transgressive segregation reveals mechanisms of Arabidopsis immunity to Brassica-infecting races of white rust (Albugo candida). Proc Nat Acad Sci U S A 116: 2767–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim S-T, Karelina D, Zaidem M, Ossowski S, Martín-Pizarro C, Laitinen RAE, Rowan BA, Tenenboim H, et al. (2014) Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Lin Y-S, Shen J-B, Shan J-X, Qi P, Shi M, Zhu M-Z, Huang X-H, Feng Q, et al. (2014) A two-locus interaction causes interspecific hybrid weakness in rice. Nat Commun 5: 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin DB, Arroyo-Garcia R, Ochoa OE, Kesseli RV, Lavelle DO, Michelmore RW (2001) Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157: 831–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Choi K, Serra H, Reinherd C, Ziolkowski PA, Underwood CJ, Zhao X, Hardcastle TJ, Yelina NE, Griffin C, Jackson M, et al. (2016) Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLoS Genet 12: e1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FC, et al. (2013) Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Computational Pan-Genomics Consortium (2018) Computational pan-genomics: status, promises and challenges. Brief Bioinform 19: 118–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Moreira B, Cantalapiedra CP, García-Pereira MJ, Gordon SP, Vogel JP, Igartua E, Casas AM, Vinuesa P (2017) Analysis of plant pan-genomes and transcriptomes with GET_HOMOLOGUES-EST, a clustering solution for sequences of the same species. Front Plant Sci 8:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley MB, Pathirana S, Wu HJ, Kachroo P, Klessig DF (2000) Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Sabaté R., Berbon M, Martinez D, Chaignepain S, Coulary-Salin B, Hofmann K, Loquet A, Saupe SJ (2016) Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proceedings of the National Academy of Sciences 113: 2720–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Concepcion JC, Franceschetti M, MacLean D, Terauchi R, Kamoun S, Banfield MJ (2019) Protein engineering expands the effector recognition profile of a rice NLR immune receptor. Elife 8: e47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Fang L, Zhu X, Zhou B, Zhang T (2019) A CC-NBS-LRR gene induces hybrid lethality in cotton. J Exp Bot 70: 5145–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrka W, Lamacchia M, Durrens P, Kobe B, Daskalov A, Paoletti M, Sherman DJ, Saupe SJ (2014) Diversity and variability of NOD-like receptors in fungi. Genome Biol Evol 6: 3137–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson HP, Kristmundsdottir S, Beyter D, Jonsson H, Skuladottir A, Hardarson MT, Gudbjartsson DF, Stefansson K, Halldorsson BV, Melsted P (2019) GraphTyper2 enables population-scale genotyping of structural variation using pangenome graphs. Nat Commun 10: 5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Gascuel O, Lefranc M-P (2002) Reconstructing the duplication history of tandemly repeated genes. Mol Biol Evol 19: 278–288 [DOI] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehan JM, Castel B, Bentham AR, Jones JD (2020) Plant NLRs get by with a little help from their friends. Curr Opin Plant Biol 56: 99–108 [DOI] [PubMed] [Google Scholar]
- Flor HH (1971) Current status of the gene for gene hypothesis. Annu Rev Phytopathol 9: 275–295 [Google Scholar]
- Gabrieli T, Sharim H, Michaeli Y, Ebenstein Y (2017) Cas9-assisted targeting of CHromosome segments (CATCH) for targeted nanopore sequencing and optical genome mapping. bioRxiv: 110163
- Gaiero P, Speranza P, de Jong H (2018) Introgressive hybridization in potato revealed by novel cytogenetic and genomic technologies. Am J Potato Res 95: 607–621 [Google Scholar]
- Ganal MW, Young ND, Tanksley SD (1989) Pulsed field gel electrophoresis and physical mapping of large DNA fragments in the Tm-2a region of chromosome 9 in tomato. Mol Gen Genet 215: 395–400 [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gonda I, Sun H, Ma Q, Bao K, Tieman DM, Burzynski-Chang EA, Fish TL, Stromberg KA, Sacks GL, et al. (2019) The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet 51: 1044–1051 [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang W, Zhang T, Gong Z, Zhao H, Han G-Z (2018) Out of water: the origin and early diversification of plant R-genes. Plant Physiol 177: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz AA, Bayer PE, Barker GC, Edger PP, Kim H, Martinez PA, Chan CKK, Severn-Ellis A, McCombie WR, Parkin IAP, et al. (2016) The pangenome of an agronomically important crop plant Brassica oleracea. Nat Commun 7:13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, Stritt C, Roulin AC, Schackwitz W, Tyler L, et al. (2017) Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat Commun 8: 2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gos G, Slotte T, Wright SI (2012) Signatures of balancing selection are maintained at disease resistance loci following mating system evolution and a population bottleneck in the genus Capsella. BMC Evol Biol 12: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Ochman H (1996) Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87: 791–794 [DOI] [PubMed] [Google Scholar]
- Haberer G, Kamal N, Bauer E, Gundlach H, Fischer I, Seidel MA, Spannagl M, Marcon C, Ruban A, Urbany C, et al. (2020) European maize genomes highlight intraspecies variation in repeat and gene content. Nat Genet 52: 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SA, Allen RL, Baumber RE, Baxter LA, Fisher K, Bittner-Eddy PD, Rose LE, Holub EB, Beynon JL (2009) Maintenance of genetic variation in plants and pathogens involves complex networks of gene-for-gene interactions. Mol Plant Pathol 10: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci U S A 110: 21189–21194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Igarashi D, Mase K, Lu Y, Tsuda Y, Chakravarthy S, Wei H-L, Foley JW, Collmer A, Glazebrook J, Katagiri F (2017) A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J 36: 2758–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henk AD, Warren RF, Innes RW (1999) A new Ac-like transposon of Arabidopsis is associated with a deletion of the RPS5 disease resistance gene. Genetics 151: 1581–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig A, Jäger G, Battke F, Nieselt K (2012) GenomeRing: alignment visualization based on SuperGenome coordinates. Bioinformatics 28: i7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG,, Robertson A (1966) The effect of linkage on limits to artificial selection. Genet Res 8: 269–294 [PubMed] [Google Scholar]
- Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, Peñagaricano F, Lindquist E, Pedraza MA, Barry K, et al. (2014) Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RAL, Kamoun S (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton M, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. (2012) Genome-wide pattern of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner S, Bercovich N, Todesco M, Mandel JR, Odenheimer J, Ziegler E, Lee JS, Baute GJ, Owens GL, Grassa CJ, et al. (2019) Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants 5: 54–62 [DOI] [PubMed] [Google Scholar]
- Hurgobin B, Golicz AA, Bayer PE, Chan C-KK, Tirnaz S, Dolatabadian A, Schiessl SV, Samans B, Montenegro JD, Parkin IAP, et al. (2018) Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol J 16: 1265–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurni S, Brunner S, Stirnweis D, Herren G, Peditto D, McIntosh RA, Keller B (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J 79: 904–913 [DOI] [PubMed] [Google Scholar]
- Janzen GM, Wang L, Hufford MB (2019) The extent of adaptive wild introgression in crops. New Phytol 221: 1279–1288 [DOI] [PubMed] [Google Scholar]
- Jeuken MJW, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, Michelmore RW, Visser RGF, Niks RE (2009) Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21: 3368–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Assis R (2017) Natural selection drives rapid functional evolution of young Drosophila duplicate genes. Mol Biol Evol 34: 3089–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W-B, Schneeberger K (2020) Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat Commun 11:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Yuan Y, Zhang Y, Yang S, Zhang X (2015) Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet 16: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL (2019) Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol 50: 82–94 [DOI] [PubMed] [Google Scholar]
- Jupe F, Witek K, Verweij W, Śliwka J, Pritchard L, Etherington GJ, Maclean D, Cock PJ, Leggett RM, Bryan GJ, et al. (2013) Resistance gene enrichment sequencing (R en S eq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J 76: 530–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov TL, Kniskern JM, Gao L, DeYoung BJ, Ding J, Dubiella U, Lastra RO, Nallu S, Roux F, Innes RW, et al. (2014) The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D, Hagmann J, Li R, Bemm F, Slotte T, Neuffer B, Wright SI, Weigel D (2019) Long-term balancing selection drives evolution of immunity genes in Capsella. Elife 8: e43606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30: 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, Kamoun S (2020) RefPlantNLR: a comprehensive collection of experimentally validated plant NLRs. bioRxiv:193961. [DOI] [PMC free article] [PubMed]
- Krasileva KV, Zheng C, Leonelli L, Goritschnig S, Dahlbeck D, Staskawicz BJ (2011) Global analysis of Arabidopsis/downy mildew interactions reveals prevalence of incomplete resistance and rapid evolution of pathogen recognition. PLoS ONE 6: e28765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T, Chanclud E, Michel-Romiti C, Grand X, Morel J-B (2016) Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol 210: 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JDG (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747 [DOI] [PubMed] [Google Scholar]
- Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16: 2870–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Eulgem T (2018) Transcript-level expression control of plant NLR genes. Mol Plant Pathol 19: 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RRQ, Chae E (2020) Variation patterns of NLR clusters in Arabidopsis thaliana Genomes. Plant Commun 1: 100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, van Wersch S, Tong M, Li X (2019) TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol 221: 2054–2066 [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Zhang N, Severing EI, Nijveen H, Cheng F, Visser RGF, Wang X, de Ridder D, Bonnema G (2014) Beyond genomic variation—comparison and functional annotation of three Brassica rapa genomes: a turnip, a rapid cycling and a Chinese cabbage. BMC Genomics 15:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Zhang Y, Kuang H, Chen J (2013) Frequent loss of lineages and deficient duplications accounted for low copy number of disease resistance genes in Cucurbitaceae. BMC Genomics 14: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Kang H, Xu Y, Peng Y, Wang D, Gao L, Wang X, Ning Y, Wu J, Liu W, et al. (2020a) Genome-wide association study identifies an NLR gene that confers partial resistance to Magnaporthe oryzae in rice. Plant Biotechnol J 18: 1376–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, Zhou G-A, Zhang H, Liu Z, Shi M, et al. (2020b) Pan-genome of wild and cultivated soybeans. Cell 182: 162–176.e13 [DOI] [PubMed] [Google Scholar]
- Li Y-H, Zhou G, Ma J, Jiang W, Jin L-G, Zhang Z, Guo Y, Zhang J, Sui Y, Zheng L, et al. (2014) De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol 32: 1045–1052 [DOI] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjálmsson BJ, Korte A, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A, Tian D, Chang W, Holub E, Kreitman M, Bergelson J (2019) Population genetics of the highly polymorphic RPP8 gene family. Genes 10: 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Höijer I, Kvist T, Ameur A, Mikkelsen MJ (2020) Xdrop: targeted sequencing of long DNA molecules from low input samples using droplet sorting. Hum Mutat 41: 1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Besansky N, Hahn MW (2016) How reticulated are species? Bioessays 38: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool A, Saitoh H, Franceschetti M, Stevenson CEM, Uemura A, Kanzaki H, Kamoun S, Terauchi R, Banfield MJ (2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 4: 08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Zhang J, Zhang P, Fenwick P, Steuernagel B, Adamski NM, Boyd L, McIntosh R, Wulff BBH, Berry S, et al. (2018) BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants 4: 662–668 [DOI] [PubMed] [Google Scholar]
- Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, Staskawicz BJ (2020) Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ. bioRxiv:46413 [DOI] [PMC free article] [PubMed]
- Mauricio R, Stahl EA, Korves T, Tian D, Kreitman M, Bergelson J (2003) Natural selection for polymorphism in the disease resistance gene RPS2 of Arabidopsis thaliana. Genetics 163: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Liu M, Stiller J, Liu C (2019) A pan-transcriptome analysis shows that disease resistance genes have undergone more selection pressure during barley domestication. BMC Genomics 20: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10: 1861–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Ernst E, Hartwick N, Chu P, Bryant D (2020) Genome and time-of-day transcriptome of Wolffia australiana link morphological extreme minimization with un-gated plant growth. bioRxiv:018291 [DOI] [PMC free article] [PubMed]
- Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8: 1113–1130 [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Montenegro JD, Golicz AA, Bayer PE, Hurgobin B, Lee H, Chan C-KK, Visendi P, Lai K, Doležel J, Batley J, et al. (2017) The pangenome of hexaploid bread wheat. Plant J 90: 1007–1013 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Katta HY, Mojica A, Chen I-MA, Kyrpides NC, Reddy T (2019) Genomes OnLine database (GOLD) v.7: updates and new features. Nucleic Acids Res 47: D649–D659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis A-R, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ (1932) Further studies on the nature and causes of gene mutations. Proc Int Congr Genet 6: 213–255 [Google Scholar]
- Munch D, Gupta V, Bachmann A, Busch W, Kelly S, Mun T, Andersen SU (2018) The Brassicaceae family displays divergent, shoot-skewed NLR resistance gene expression. Plant Physiol 176: 1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemri A, Atwell S, Tarone AM, Huang YS, Zhao K, Studholme DJ, Nordborg M, Jones JDG. (2010) Genome-wide survey of Arabidopsis natural variation in downy mildew resistance using combined association and linkage mapping. Proc Natl Acad Sci U S A 107: 10302–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Jones JDG (2020) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. bioRxiv:034173 [DOI] [PubMed]
- Nguyen N, Hickey G, Zerbino DR, Raney B, Earl D, Armstrong J, Kent WJ, Haussler D, Paten B. (2015) Building a pan-genome reference for a population. J Comput Biol 22: 387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Anderson RG, Cherkis KA, Law TF, Liu QL, Machius M, Nimchuk ZL, Yang L, Chung E-H, Kasmi FE, et al. (2017) TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc Natl Acad Sci U S A 114: E2053–E2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z, Klindworth DL, Yu G, Friesen TL, Chao S, Jin Y, Cai X, Ohm J-B, Rasmussen JB, Xu SS (2014) Development and characterization of wheat lines carrying stem rust resistance gene Sr43 derived from Thinopyrum ponticum. Theor Appl Genet 127: 969–980 [DOI] [PubMed] [Google Scholar]
- Noël L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099–2112 [PMC free article] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer, Berlin, Heidelberg [Google Scholar]
- Ou L, Li D, Lv J, Chen W, Zhang Z, Li X, Yang B, Zhou S, Yang S, Li W, et al. (2018) Pan-genome of cultivated pepper (Capsicum) and its use in gene presence-absence variation analyses. New Phytol 220: 360–363 [DOI] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31: 3691–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B, Novak AM, Eizenga JM, Garrison E (2017) Genome graphs and the evolution of genome inference. Genome Res 27: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Rieseberg LH (2016) A genomic perspective on hybridization and speciation. Mol Ecol 25: 2337–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinosio S, Giacomello S, Faivre-Rampant P, Taylor G, Jorge V, Le Paslier MC, Zaina G, Bastien C, Cattonaro F, Marroni F, et al. (2016) Characterization of the poplar pan-genome by genome-wide identification of structural variation. Mol Biol Evol 33: 2706–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhin DM, Krasileva KV (2020) Intraspecies diversity reveals a subset of highly variable plant immune receptors and predicts their binding sites. bioRxiv:190785 [DOI] [PMC free article] [PubMed]
- Rakocevic G, Semenyuk V, Lee W-P, Spencer J, Browning J, Johnson IJ, Arsenijevic V, Nadj J, Ghose K, Suciu MC, et al. (2019) Fast and accurate genomic analyses using genome graphs. Nat Genet 15: 354–362 [DOI] [PubMed] [Google Scholar]
- Rose L, Atwell S, Grant M, Holub EB (2012) Parallel loss-of-function at the RPM1 bacterial resistance locus in Arabidopsis thaliana. Front Plant Sci 3: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Lüdke D, Klenke M, Quathamer A, Valerius O, Braus GH, Wiermer M (2017) The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-α3 interaction partner required for plant immunity. Plant J 92: 808–821 [DOI] [PubMed] [Google Scholar]
- Rowan BA, Heavens D, Feuerborn TR, Tock AJ, Henderson IR, Weigel D (2019) An ultra high-density Arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features. Genetics 213: 771–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstedt GD, Wu CA, Sweigart AL (2020) Evolution of multiple postzygotic barriers between species in the Mimulus tilingii species complex. bioRxiv:241489 [DOI] [PMC free article] [PubMed]
- Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac SU, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Sarris PF, Cevik V, Dagdas G, Jones JDG, Krasileva KV (2016) Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Maron LG, Stein JC, Wences AH, Gurtowski J, Biggers E, Lee H, Kramer M, Antoniou E, Ghiban E, et al. (2014) Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol 15: 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Kim S, Yeom S-I, Choi D (2016) Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding leucine-rich repeat gene family among Solanaceae plants. Front Plant Sci 7: 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K, Seo E, Witek K, Li M, Staskawicz B (2020) Evolution of NLR resistance genes with noncanonical N-terminal domains in wild tomato species. New Phytol (doi: 10.1101/786194) [DOI] [PubMed] [Google Scholar]
- Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R (2012) The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol 10: e1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z-Q, Xue J-Y, Wang Q, Wang B, Chen J-Q (2019) Revisiting the origin of plant NBS-LRR genes. Trends Plant Sci 24: 9–12 [DOI] [PubMed] [Google Scholar]
- Sicard A, Kappel C, Josephs EB, Lee YW, Marona C, Stinchcombe JR, Wright SI, Lenhard M (2015) Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nat Commun 6: 7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Hulbert SH (2005) Recombination events generating a novel Rp1 race specificity. Mol Plant Microbe Interact 18: 220–228 [DOI] [PubMed] [Google Scholar]
- Soanes DM, Talbot NJ (2010) Comparative genome analysis reveals an absence of leucine-rich repeat pattern-recognition receptor proteins in the kingdom Fungi. PLoS ONE 5: e12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J-M, Guan Z, Hu J, Guo C, Yang Z, Wang S, Liu D, Wang B, Lu S, Zhou R, et al. (2020) Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat Plants 6: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srichumpa P, Brunner S, Keller B, Yahiaoui N (2005) Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139: 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal J, Kaliff M, Dewaele E, Persson M, Dixelius C (2008) RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J 55: 188–200 [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671 [DOI] [PubMed] [Google Scholar]
- Stam R, Scheikl D, Tellier A (2016) Pooled enrichment sequencing identifies diversity and evolutionary pressures at NLR resistance genes within a wild tomato population. Genome Biol Evol 8: 1501–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam R, Silva-Arias GA, Tellier A (2019) Subsets of NLR genes show differential signatures of adaptation during colonization of new habitats. New Phytol 224: 367–379 [DOI] [PubMed] [Google Scholar]
- Steuernagel B, Witek K, Krattinger SG, Ramirez-Gonzalez RH, Schoonbeek H-J, Yu G, Baggs E, Witek AI, Yadav I, Krasileva KV, et al. (2020) The NLR-annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol 183: 468–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KA, Woeste K, Chakraborty S, Crepeau MW, Leslie CA, Martínez-García PJ, Puiu D, Romero-Severson J, Coggeshall M, Dandekar AM, et al. (2018) Genomic variation among and within six Juglans species. G3 8: 2153–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MA, Rick CM (1988) Genetics and breeding. In Atherton JG, Rudich J, eds, The Tomato Crop, Chapman and Hall, London, pp. 35–109 [Google Scholar]
- Stirnweis D, Milani SD, Brunner S, Herren G, Buchmann G, Peditto D, Jordan T, Keller B. (2014) Suppression among alleles encoding nucleotide-binding-leucine-rich repeat resistance proteins interferes with resistance in F1 hybrid and allele-pyramided wheat plants. Plant J 79: 893–903 [DOI] [PubMed] [Google Scholar]
- Suarez-Gonzalez A, Lexer C, Cronk QCB (2018) Adaptive introgression: a plant perspective. Biol Lett 14:20170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miller J, Nozaki Y, Takeda M, Shah J, Hase S, Ikegami M, Ehara Y, Dinesh-Kumar SP (2002) RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J 32: 655–667 [DOI] [PubMed] [Google Scholar]
- Tarr DEK, Alexander HM (2009) TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes 2: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Larson EL (2019) Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat Ecol Evol 3: 170–177 [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Scott Durkin A, et al. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci U S A 102: 13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanki AS, Soranzo N, Haerty W, Davey RP (2018) GeneSeqToFamily: a Galaxy workflow to find gene families based on the Ensembl Compara GeneTrees pipeline. Gigascience 7: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Araki H, Stahl E, Bergelson J, Kreitman M (2002) Signature of balancing selection in Arabidopsis. Proc Natl Acad Sci U S A 99: 11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Todesco M, Kim S-T, Chae E, Bomblies K, Zaidem M, Smith LM, Weigel D, Laitinen RAE. (2014) Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. PLoS Genet 10: e1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouern-Trend AJ, Falk T, Zaman S, Caballero M, Neale DB, Langley CH, Dandekar AM, Stevens KA, Wegrzyn JL (2020) Comparative genomics of six Juglans species reveals disease-associated gene family contractions. Plant J 102: 410–423 [DOI] [PubMed] [Google Scholar]
- Tsai Y-C, Greenberg D, Powell J, Höijer I, Ameur A, Strahl M, Ellis E, Jonasson I, Pinto RM, Wheeler VC, et al. (2017) Amplification-free, CRISPR-Cas9 targeted enrichment and SMRT sequencing of repeat-expansion disease causative genomic regions. bioRxiv: 203919
- Urbach JM, Ausubel FM (2017) The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc Natl Acad Sci U S A 114: 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Weyer A-L, Monteiro F, Furzer OJ, Nishimura MT, Cevik V, Witek K, Jones JDG, Dangl JL, Weigel D, Bemm F (2019) A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178: 1260–1272.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ghelder C, Parent GJ, Rigault P, Prunier J, Giguère I, Caron S, Stival Sena J, Deslauriers A, Bousquet J, Esmenjaud D, et al. (2019) The large repertoire of conifer NLR resistance genes includes drought responsive and highly diversified RNLs. Sci Rep 9: 11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G, Medini D, Riley DR, Tettelin H (2015) Ten years of pan-genome analyses. Curr Opin Microbiol 23: 148–154 [DOI] [PubMed] [Google Scholar]
- Vinař T, Brejová B, Song G, Siepel A (2009) Reconstructing histories of complex gene clusters on a phylogeny. In FD Ciccarelli, I Miklós, eds, Comparative Genomics, Springer, Berlin, Heidelberg, pp. 150–163 [Google Scholar]
- van der Vossen EA, van der Voort JN, Kanyuka K, Bendahmane A, Sandbrink H, Baulcombe DC, Bakker J, Stiekema WJ, Klein-Lankhorst RM (2000) Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J 23: 567–576 [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang H-W, Zhou J-M, Chai J (2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364: 6435. [DOI] [PubMed] [Google Scholar]
- Wang JZ, Wang J, Hu MJ, Wang HW, Zhou JM, Chai JJ (2019b) Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364: eaav5868 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao L, Zhang X, Zhang Q, Jia Y, Wang G, Li S, Tian D, Li W-H, Yang S (2019c). Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc Natl Acad Sci U S A 116: 18479–18487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. (2018) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP (1999) The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153: 1929–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch R, Li X, Zhang Y (2016) Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Front Plant Sci 7: 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Gläßer C, et al. (2014) Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Keller B (2007) Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J 51: 631–641 [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303 [DOI] [PubMed] [Google Scholar]
- Witek K, Jupe F, Witek AI, Baker D, Clark MD, Jones JDG (2016) Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat Biotechnol 34: 656–660 [DOI] [PubMed] [Google Scholar]
- Wu C-H, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S (2017) NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci U S A 114: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Takashi T, Morinaka Y, Lin S, Wu J, Matsumoto T, Kitano H, Matsuoka M, Ashikari M (2010) Gain of deleterious function causes an autoimmune response and Bateson–Dobzhansky–Muller incompatibility in rice. Mol Genet Genomics 283: 305–315 [DOI] [PubMed] [Google Scholar]
- Yang S, Li J, Zhang X, Zhang Q, Huang J, Chen JQ, Hartl DL, Tian D (2013) Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc Natl Acad Sci U S A 110: 18572–18577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Li G, Zhao H, Wang G, Lian X, Xie W (2015) Exploring the rice dispensable genome using a metagenome-like assembly strategy. Genome Biol 16: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, He SY (2020) Pattern-recognition receptors are required for NLR-mediated plant immunity. bioRxiv:031294 [DOI] [PMC free article] [PubMed]
- Yue J-X, Meyers BC, Chen J-Q, Tian D, Yang S (2012) Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol 193: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Yu J, Golicz AA, Lu K, Dossa K, Zhang Y, Chen J, Wang L, You J, Fan D, Edwards D, et al. (2019) Insight into the evolution and functional characteristics of the pan-genome assembly from sesame landraces and modern cultivars. Plant Biotechnol J 17: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang S, Wang J, Jia Y, Huang J, Tan S, Zhong Y, Wang L, Gu L, Chen J-Q, et al. (2015) A genome-wide survey reveals abundant rice blast R genes in resistant cultivars. Plant J 84: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dodds PN, Bernoux M (2017a) What do we know about NOD-like receptors in plant immunity? Annu Rev Phytopathol 55: 205–229 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Liu J, Ding Y, Wang S, Zhang X, Liu Y, Yang S (2017b) Temperature-dependent autoimmunity mediated by chs1 requires its neighboring TNL gene SOC3. New Phytol 213: 1330–1345 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, et al. (2018) Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50: 278–284 [DOI] [PubMed] [Google Scholar]
- Zhao T, Rui L, Li J, Nishimura MT, Vogel JP, Liu N, Liu S, Zhao Y, Dangl JL, Tang D (2015) A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet 11: e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Silverstein KAT, Ramaraj T, Guhlin J, Denny R, Liu J, Farmer AD, Steele KP, Stupar RM, Miller JR, et al. (2017) Exploring structural variation and gene family architecture with de novo assemblies of 15 Medicago genomes. BMC Genomics 18: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Chebotarov D, Kudrna D, Llaca V, Lee S, Rajasekar S, Mohammed N, Al-Bader N, Sobel-Sorenson C, Parakkal P, et al. (2020) A platinum standard pan-genome resource that represents the population structure of Asian rice. Sci Data 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]