Abstract
LncRNA SET-binding factor 2 (SBF2) antisense RNA1 (SBF2-AS1) has been proven to play an oncogenic role in various types of tumors, but the prognostic role of SBF2-AS1 in tumors, especially in diffuse lower-grade glioma (LGG), is still unclear. Here, we aimed to investigate the prognostic value of SBF2-AS1 in LGG. The LGG expression profiles from The Cancer Genome Atlas (TCGA, n = 524) and Chinese Glioma Genome Atlas (CGGA, n = 431) were mined by Kaplan-Meier analysis, Cox regression analysis, Chi-square test and GSEA analysis. Through Kaplan-Meier analysis, we found the prognosis of LGG patients with high expression of SBF2-AS1 were worse than that of patients with low expression (Log Rank P < 0.001). Cox analysis showed SBF2-AS1 was an independent prognostic factor for poorer overall survival in LGG (P < 0.05). SBF2-AS1 was found to be significantly related to IDH mutation status and SBF2-AS1 was highly expressed in IDH wildtype group. GSEA analysis obtained a total of 126 GO terms and 6 KEGG pathways that were significantly enriched in SBF2-AS1 high expression phenotype (NOM P value < 0.05). We found these 126 GO terms and KEGG pathways were mainly related to immunity. In conclusion, lncRNA SBF2-AS1 expression is an immune-related lncRNA associated with unfavorable overall survival in LGG. SBF2-AS1 could be a reliable prognostic biomarker for patients with LGG.
Keywords: diffuse lower-grade gliomas, SBF2-AS1, prognostic biomarker, survival, immunity
Introduction
Glioma is the most common type of intracranial malignant tumor, and can be grade I to IV for diagnosis according to World Health Organization (WHO) classification. Grades II and III gliomas are defined as diffuse lower-grade glioma (LGG). 1,2 Compared with Grade I and IV, LGGs are more heterogeneous, and accurate prognosis judgment is more difficult, so prognostic markers are more needed for clinical guidance. With the application of molecular characteristics (such as isocitrate dehydrogenase (IDH) mutation status) in the grouping of gliomas, 3,4 it is increasingly recognized that molecular biomarkers play a critical role in the prognosis evaluation of LGG.
Long noncoding RNAs (lncRNAs) are a class of non-protein encoded transcripts longer than 200 bp, which play an important role in normal physiological functions such as growth and development and in various human diseases, especially cancer regulate. 5,6 Emerging evidence suggests that lncRNAs have a critical regulatory role in tumour progression and can predict the prognosis of cancer patients. In the field of glioma research, many lncRNAs have been reported to be closely related to tumorigenesis, development, and prognosis. For example, LINC01116 was found to be involved in tumorigenesis of glioma by targeting VEGFA through miR-31-5p. 7 LncRNA HERC2P2 was identified as a tumor suppressor, and its overexpression could significantly inhibit the migration and growth of gliomas in vitro and in vivo. 8 Others have reported that lncRNAs could be prognostic indicators of LGG, such as GAS5, H19, RAB6C-AS1. 9 -11
LncRNA SET-binding factor 2 (SBF2) antisense RNA1 (SBF2-AS1), located on Chromosome 11 p15.4(9,758,268-9,811,335 [GRCh38/hg38]), is widely expressed in various tissues. The tumor-promoting function of SBF2-AS1 was first discovered in non-small cell lung cancer (NSCLC) and was confirmed in various tumors. Meanwhile, SBF2-AS1 was also found to predict the prognosis of cancer patients. For instance, SBF2-AS1 has been reported to promote proliferation of NSCLC cells in vitro or in vivo and regulate the radiosensitivity and apoptosis of NSCLC through microRNA-302a/MBNL3 axis. 12 In cervical cancer, SBF2-AS1 as ceRNA regulates the expression levels of miR-361-5p and FOXM1 to promote the tumour progression. 13 Moreover, up-regulated SBF2-AS1 can promote cell proliferation in esophageal squamous cell carcinoma. 14 As for hepatocellular carcinoma, SBF2-AS1 can promote tumour metastasis and its high expression indicates poor prognosis. 15 By sponging microRNA-143, SBF2-AS1, serving as ceRNA, can accelerate breast cancer tumorigenesis and progression. 16 However, in LGG, it is not known whether SBF2-AS1 has a oncogenic role and can be used as a prognostic marker.
As the development of next generation sequencing technology, massive high-throughput data and bioinformatics methods have facilitated a comprehensive understanding of tumorigenesis and finding novel prognostic markers or therapeutic drugs. 17 -19 Here, lncRNA expression data from a total of 955 patients with LGG were collected from the Cancer Genome Atlas (TCGA) database and the Chinese Glioma Genome Atlas (CGGA). We aimed at mining the large queue of expression profiles and clinical information to reveal the prognostic indicator efficacy of SBF2-AS1 and to investigate the potential role of SBF2-AS1 in the LGG.
Materials and Methods
Data Collection of Diffuse Lower Grade Glioma (LGG) Patients
The gene expression profile of patients with LGG from The Cancer Genome Atlas Program (TCGA) (Release Date: December 18, 2019; Embargo Release Date: August 17, 2018) was measured experimentally using the Illumina HiSeq 2000 RNA Sequencing platform by the University of North Carolina TCGA genome characterization center, which could be obtained from the UCSC Xena browser (https://xenabrowser.net/). 20 Another dataset was downloaded from Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/). 21,22 The BIGD accession number of LGG dataset from CGGA is PRJCA001747 (https://bigd.big.ac.cn/bioproject/browse/PRJCA001747). The corresponding clinical data of these LGG patients were also collected. Clinical details of patients were shown in Table 1.
Table 1.
Clinical Information of LGG Patients From TCGA and CGGA Datasets.
| Characteristic | TCGA | CGGA |
|---|---|---|
| Age | ||
| ≤ 40 | 261 | 221 |
| > 40 | 263 | 210 |
| Sex | ||
| Female | 237 | 193 |
| Male | 287 | 238 |
| Grade | ||
| Unknown | 1 | 0 |
| WHO II | 257 | 180 |
| WHO III | 266 | 251 |
| IDH mutation status | ||
| Unknown | 399 | 38 |
| Mutant | 91 | 297 |
| Wildtype | 34 | 96 |
| Radio status | ||
| Unknown | 67 | 31 |
| NO | 173 | 86 |
| YES | 284 | 314 |
| Histology | ||
| Astrocytoma | 194 | 124 |
| Mixed glioma | 133 | 226 |
| Oligodendroglioma | 197 | 81 |
| 1p19q codeletion status | ||
| Unknown | 38 | |
| Codel | 128 | |
| Non-codel | 265 |
Exploring the Association Between SBF2-AS1 and Clinicopathologic Parameters
X-tile software was used to select optimal cutoff value of the SBF2-AS1 expression value in TCGA and CGGA datasets. 23,24 Then LGG patients were classified into high-expressed or low-expressed group by SBF2-AS1. Survival analysis including Kaplan-Meier, Cox regression analysis was used to explore the prognosis performance of SBF2-AS1. Chi-square test was performed to obtain the relationship between SBF2-AS1 and clinical parameters.
Bioinformatics Analysis of Function Prediction
The above group information of LGG according to the SBF2-AS1 expression was treated as a phenotype label. Then GSEA was performed to analyze the GO and KEGG pathways based on molecular signatures database (c2.cp.kegg.v5.2.symbols.gmt and c5.bp.v5.2.symbols.gmt). 25 The GO terms and KEGG pathways were selected by Nominal P value and Normalized enrichment score (NOM P < 0.05).
Results
The Performance of SBF2-AS1 in Predicting the OS of LGG Patients in TCGA and CGGA Datasets
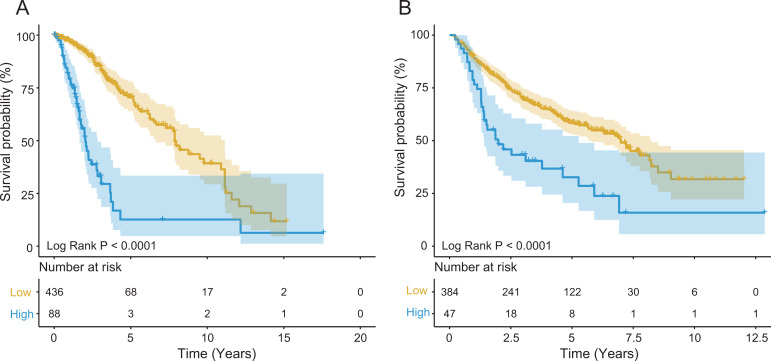
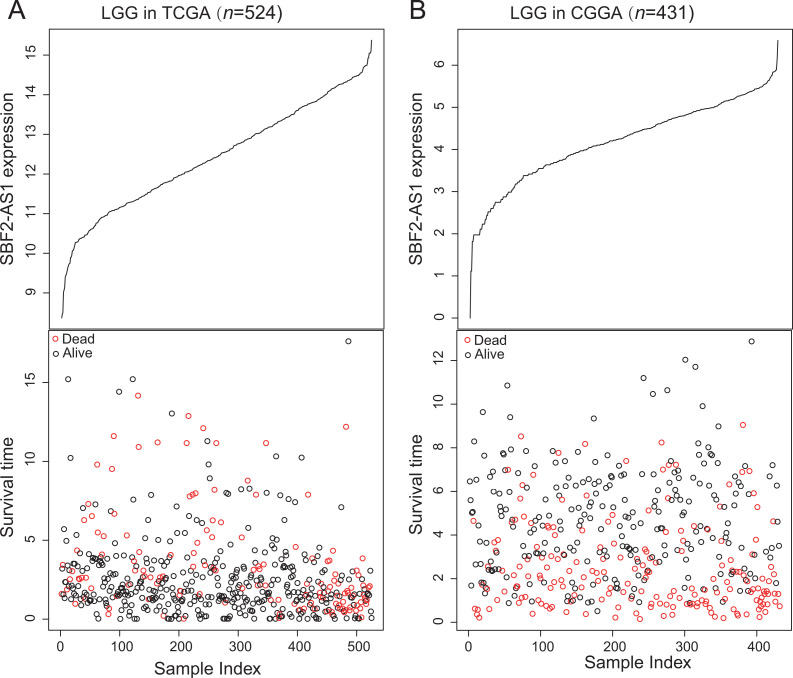
There were 955 LGG patients obtained from the TCGA (n = 524) and CGGA (n = 431) with lncRNAs expression profiles and the corresponding follow-up information for next analysis (Table 1). To investigate the clinical value of SBF2-AS1 in predicting OS of LGG patients, we classified patient with LGG into high- or low-expressed group based on the cutoff value of SBF2-AS1 expression selected by X-tile software, and performed Kaplan–Meier analysis. In TCGA dataset, Kaplan–Meier showed that the survival of high-expressed SBF2-AS1 group (n = 88, median survival time: 2.03 years) was shorter than the low-expressed group (n = 436, median survival time: 7.88 years), and the expression of SBF2-AS1 could distinguish LGG patients with different survival (log-rank P < 0.001; Figure 1A). Then we tested the predictive ability of clinical outcome for SBF2-AS1 in another independent LGG dataset (CGGA, n = 431). The cutoff value of SBF2-AS1 selected by X-tile in CGGA separated LGG patients into 2 groups and Kaplan–Meier analysis verified that the SBF2-AS1 could also assess the prognostic risk of LGG patients (5-year survival: 25.58% vs. 58.43%, log-rank P < 0.001; Figure 1B). Subsequently, in order to describe the relationship between the expression of SBF2-AS1 and survival information, we put them in Figure 2. LGG patients with low expression of SBF2-AS1 lived longer than patients with high expression in the TCGA dataset (Figure 2A) and in the CGGA group (Figure 2B).
Figure 1.
Kaplan–Meier analysis showed LGG patients could be classified into 2 groups with significantly different survival based on SBF2-AS1 expression in the TCGA (A) and CGGA (B) datasets.
Figure 2.
The expression of SBF2-AS1, survival status and survival time for LGG patients in the TCGA (A) and CGGA (B) datasets. X axis represents the index of samples. Red color means that the survival status of patients was dead, and black color means that the survival status of patients was alive.
The Association Between SBF2-AS1 and Clinical Parameters of LGG
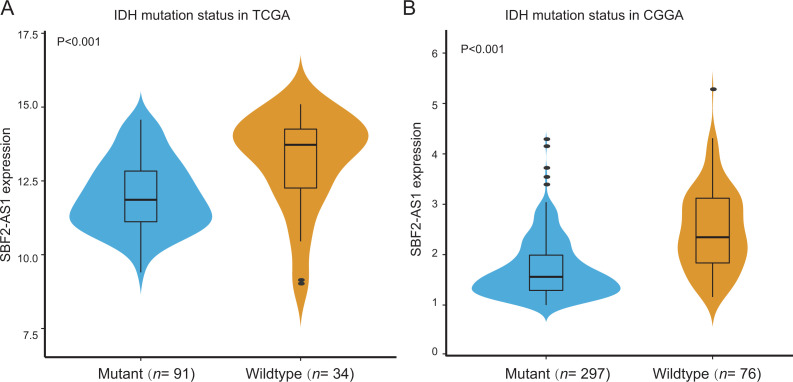
Although the expression of SBF2-AS1 was related to Age, Radio status and Grade in TCGA dataset, the CGGA group could not verify the association (P < 0.05, Table 2). However, as shown in Table 2, Chi-square test found the expression of SBF2-AS1 was related to IDH mutation status in 2 datasets (P < 0.001, Table 2). The SBF2-AS1 expression of IDH mutation group was lower than that of IDH wildtype group in TCGA (Figure 3A) and CGGA (Figure 3B) datasets.
Table 2.
Association of SBF2-AS1 Expression With Clinicopathological Characteristics in LGG Patients.
| Variables | TCGA | P a | CGGA | P a | ||
|---|---|---|---|---|---|---|
| Low | High | Low | High | |||
| Age | <0.001 | 0.266 | ||||
| ≤ 40 | 242 | 19 | 201 | 20 | ||
| > 40 | 194 | 69 | 183 | 27 | ||
| Sex | 0.181 | 0.158 | ||||
| Female | 191 | 46 | 177 | 16 | ||
| Male | 245 | 42 | 207 | 31 | ||
| IDH mutation | <0.001 | <0.001 | ||||
| Unknown | 333 | 66 | 35 | 3 | ||
| Mutant | 83 | 8 | 284 | 13 | ||
| Wildtype | 20 | 14 | 65 | 31 | ||
| Radio | 0.010 | 0.625 | ||||
| Unknown | 50 | 17 | 26 | 5 | ||
| NO | 155 | 18 | 77 | 9 | ||
| YES | 231 | 53 | 281 | 33 | ||
| 1p19q codeletion status | 0.001 | |||||
| Unknown | 38 | 0 | ||||
| Codel | 121 | 7 | ||||
| Non-codel | 225 | 40 | ||||
| Grade | 0.002 | 0.724 | ||||
| Unknown | 1 | 0 | ||||
| WHO II | 229 | 28 | 162 | 18 | ||
| WHO III | 206 | 60 | 222 | 29 | ||
a The Chi-squared test, P value < 0.05 was considered significant.
Figure 3.
SBF2-AS1 expression patterns in IDH wildtype and mutant groups in the TCGA (A) and CGGA (B) datasets.
SBF2-AS1 Is an Independent Risk Factor for LGG
In TCGA and CGGA dataset, Cox regression analyses were performed. The univariate Cox analysis results indicated Grade, IDH mutation status and the SBF2-AS1 expression were risk factors of LGG; The multivariable Cox analysis obtained that the SBF2-AS1 expression was an independent risk factor of LGG (High vs. Low, HR TCGA = 2.35, 95% CI 1.61–3.45, P < 0.001; HR CGGA = 1.56, 95% CI 1.00–2.41, P < 0.05, Table 3). All above results suggested SBF2-AS1 was a reliable prognostic indicator of LGG.
Table 3.
Cox Regression Analysis of SBF2-AS1 Expression and Survival of LGG Patients in the TCGA and CGGA Datasets.
| Univariable Analysis | Multivariable Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables | HR | 95% CI of HR | P | HR | 95% CI of HR | P | |||
| Lower | Upper | Lower | Upper | ||||||
| TCGA | |||||||||
| Age | >40 vs. ≤40 | 2.816 | 1.962 | 4.041 | <0.001 | 1.707 | 0.539 | 5.407 | 0.364 |
| Gender | Male vs. Female | 1.139 | 0.811 | 1.599 | 0.452 | 1.491 | 0.542 | 4.102 | 0.440 |
| Grade | III vs II, | 3.307 | 2.285 | 4.787 | <0.001 | 1.398 | 0.480 | 4.072 | 0.540 |
| IDH mutation status | Wildtype vs. Mutant | 5.532 | 2.065 | 14.820 | 0.001 | 2.891 | 0.909 | 9.193 | 0.072 |
| Signature | High risk vs. low risk | 4.647 | 3.226 | 6.694 | <0.001 | 3.710 | 1.203 | 11.436 | 0.022 |
| CGGA | |||||||||
| Age | >40 vs. ≤40 | 1.188 | 0.892 | 1.581 | 0.239 | 1.186 | 0.882 | 1.596 | 0.258 |
| Gender | Male vs. Female | 1.004 | 0.753 | 1.339 | 0.976 | 1.140 | 0.845 | 1.537 | 0.392 |
| Grade | III vs II | 2.623 | 1.888 | 3.645 | <0.001 | 2.970 | 2.103 | 4.196 | <0.001 |
| IDH mutation status | Wildtype vs. Mutant | 2.245 | 1.642 | 3.069 | <0.001 | 2.043 | 1.439 | 2.902 | <0.001 |
| Signature | High risk vs. low risk | 2.354 | 1.606 | 3.449 | <0.001 | 1.563 | 0.999 | 2.445 | 0.037 |
Prognostic Value of SBF2-AS1 Co-Expressing Network
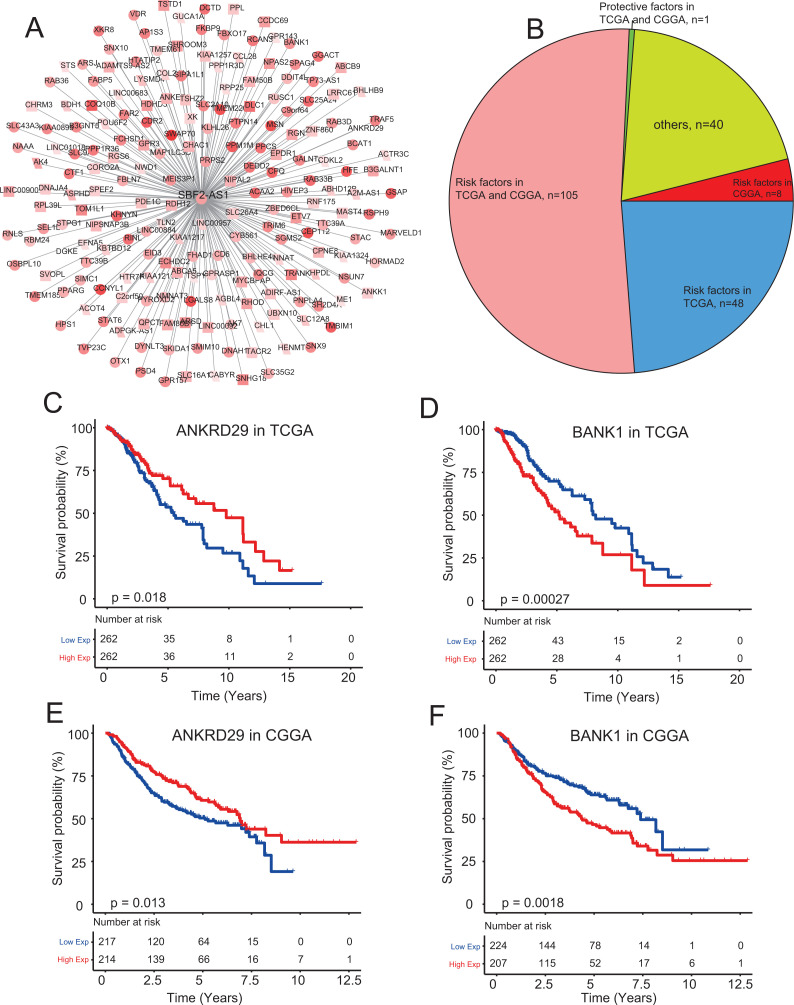
We constructed the co-expression network of SBF2-AS1 based on TCGA (n = 524) and CGGA (n = 431) datasets by Pearson test and obtained 202 genes that co-expressed with SBF2-AS1 in both datasets (Pearson coefficient > 0.4/<− 0.4, P < 0.001, Figure 4A). KM survival analysis identified 106 genes which were associated with the prognosis of LGG in the 2 sets of data sets (log Rank P < 0.05). Among them, ANKRD29 was a protective factor (Cox coefficient < 0, P < 0.05) and other 105 genes were risk factors for LGG (Cox coefficient > 0, P < 0.05, Figure 4B). SBF2-AS1 co-expressed with 105 genes related to poor prognosis, suggesting that SBF2-AS1 may interact with these genes and promote tumor progression. To display the correlation of these prognostic genes with survival, KM survival curves of the protective gene (ANKRD29) and one randomly selected risk gene (BANK1) from the above 105 risk genes were showed (Figure 4C-F).
Figure 4.
Prognostic value of SBF2-AS1 co-expressing network (A) Constructing the SBF2-AS1 co-expressing network based on TCGA and CGGA datasets by Pearson test (Pearson coefficient > 0.4/<− 0.4, P < 0.001). Distribution statistics of the 202 genes associated with LGG prognosis by Kaplan–Meier and Cox regression analysis (B). Kaplan–Meier curves of ANKRD29 and BANK1 in TCGA (C and D) and CGGA datasets (E and F).
Function Analysis of SBF2-AS1 by GSEA
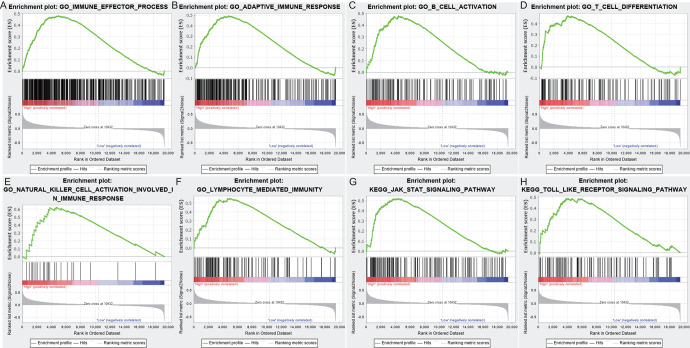
Based on the SBF2-AS1 expression phenotype label in 2 datasets, we performed GSEA analysis to explore significantly different pathways between high- and low-expressed SBF2-AS1 groups in TCGA and CGGA datasets, respectively. We obtained 126 GO terms and 6 KEGG pathways that were significantly enriched in both datasets (NOM P value < 0.05, Supplementary Table S2). Then we found the above 126 GO terms and 6 KEGG pathways were mainly clustered in immune-related GO terms and KEGG pathways, such as adaptive immune response, T cell differentiation, B cell activation, immune effector process, lymphocyte mediated immunity, natural killer cell activation, JAK STAT signaling pathway, and Toll like receptor signaling pathway (Figure 5).
Figure 5.
Functional prediction of SBF2-AS1 by GSEA. (A) Immune effector process, (B) Adaptive immune response, (C) B cell activation, (D) T cell differentiation (E) Natural killer cell activation involved ln immune response (F) Lymphocyte mediated immunity, (G) JAK-STAT signaling pathway, (H) Toll like receptor signaling pathway.
Discussion
Patient survival is largely affected by the ability of the tumor to migrate to surrounding normal tissues. 26 LGG is highly invasive and heterogeneous, so the prognosis of patients is large and difficult to assess. Over the past decades, the abnormal gene expression (including mRNA, microRNA and lncRNA), gene mutations and epigenetic modifications in LGG have been the research hot-spots of glioma prognostic biomarker. Recently, researchers find that lncRNAs have the advantages of stable existence and can be detected in blood, urine or other body fluids, and have good clinical application value.
Here, after analyzing the expression of SBF2-AS1 in 955 patients with LGG from TCGA and CGGA database, we found that SBF2-AS1 could predict LGG overall survival and be an independent risk factor of overall survival in the 2 large queues of LGG.LGG patients with high expression of SBF2-AS1 were proved to have worse prognosis than patients with low expression. Additionally, the expression of SBF2-AS1 was associated with IDH mutation status, which was regarded as ne of the prognostic indicators of glioma. We also found that patients with IDH mutation had a good prognosis and high expression of SBF2-AS1, and vice versa, confirming the argument that SBF2-AS1 was an indicator of poor prognosis. GSEA analysis found SBF2-AS1 mainly enriched in immune-related GO terms and KEGG pathways, such as adaptive immune response, T cell differentiation, B cell activation, immune effector process, lymphocyte mediated immunity, natural killer cell activation, JAK STAT signaling pathway, and Toll like receptor signaling pathway, suggesting that SBF2-AS1 could participate in tumor progression by regulating immunity.
LncRNA SBF2-AS1 has been reported to serve as an oncogene in a variety of cancers such as lung cancer, 27,28 gastric cancer, 29 colorectal cancer 30 hepatocellular carcinoma, 15 breast cancer, 16 acute myeloid leukemia, 31 and clear cell renal cell carcinoma. 32 It has been reported that SBF2-AS1 is highly expressed in the tissues of above tumors, and downregulated SBF2-AS1 could inhibit tumor cells proliferation and promote apoptosis. Moreover, SBF2-AS1 has been demonstrated to be associated with poor prognosis in non-small cell lung cancer 33 and hepatocellular carcinoma, 15 indicating its prognostic value in cancer. In glioblastoma, Hai Yu et al found SBF2-AS1 was highly expressed and played an important role in the GBM angiogenesis through NFAT5/SBF2-AS1/miR-338-3p/EGFL7 pathway. 34 Zhuoran Zhang et al discovered that SBF2-AS1 was mainly expressed in the cytoplasm of GBM cells and could secrete into serum by exosomes, and identified another function of the oncogenic SBF2-AS1, mediating TMZ resistance. 35 Fangkun Luan et al found that SBF2-AS1 was an independent unfavorable prognostic factor in glioma. 36 However, there are few reports on the specific function of SBF2-AS1 in LGG patients and its prognostic significance. This article found that SBF2-AS1 had a prognostic value in LGG, which was consistent with the reported results of SBF2-AS1 in other cancers. Furthermore, we found that SBF2-AS1 was related to immunity in functional analysis, suggesting that it may be a target for LGG immunotherapy. In the later research, we need explore the the carcinogenic mechanism of lncRNA SBF2-AS1 in LGG through more biological experiments such as cell proliferation assay, colony formation assay, transwell cell migration and invasion assay. From the perspective of bioinformatics analysis, we plan to explore the specific mechanism of SBF2-AS1 in LGG through constructing a network composed of lncRNA, genes and miRNA. 37
Overall, we identified that lncRNA SBF2-AS1 could be a prognostic risk indicator of LGG and the SBF2-AS1 expression was associated with unfavorable survival. SBF2-AS1 has potential to become a good prognostic biomarker.
Supplemental Material
Supplemental Material, sj-xls-1-tct-10.1177_15330338211011966 for Prognostic Value of Immune-Related lncRNA SBF2-AS1 in Diffuse Lower-Grade Glioma by Qiang Zhang, Xiao-Jun Liu, Yang Li, Xiao-Wei Ying and Lu Chen in Technology in Cancer Research & Treatment
Supplemental Material, sj-xls-2-tct-10.1177_15330338211011966 for Prognostic Value of Immune-Related lncRNA SBF2-AS1 in Diffuse Lower-Grade Glioma by Qiang Zhang, Xiao-Jun Liu, Yang Li, Xiao-Wei Ying and Lu Chen in Technology in Cancer Research & Treatment
Footnotes
Authors’ Note: Publicly available datasets can be found here,
Author Contributions: Qiang Zhang, Xiao-Jun Liu, Yang Li: data collection, data analysis, interpretation and drafting; Xiao-Wei Ying, Lu Chen: study design, study supervision and final approval of the manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Xiao-Wei Ying  https://orcid.org/0000-0003-1308-340X
https://orcid.org/0000-0003-1308-340X
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Cancer Genome Atlas Research Network,Brat DJ, Verhaak RG, et al. Comprehensive, Integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14(Suppl 5):v1–v49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Batsios G, Viswanath P, Subramani E, et al. PI3K/mTOR inhibition of IDH1 mutant glioma leads to reduced 2HG production that is associated with increased survival. Sci Rep. 2019;9(1):10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang C, Yu R, Li Z, et al. Comprehensive analysis of genes based on chr1p/19q co-deletion reveals a robust 4-gene prognostic signature for lower grade glioma. Cancer Manag Res. 2019;11:4971–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179(5):1033–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ye J, Zhu J, Chen H, et al. A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int J Cancer, 2020;146(1):248–261. [DOI] [PubMed] [Google Scholar]
- 8. Yang C, Wang L, Sun J, et al. Identification of long non-coding RNA HERC2P2 as a tumor suppressor in glioma. Carcinogenesis. 2019;40(8):956–964. [DOI] [PubMed] [Google Scholar]
- 9. Xiao Y, Zhu Z, Li J, et al. Expression and prognostic value of long non-coding RNA H19 in glioma via integrated bioinformatics analyses. Aging (Albany NY). 2020;12(4):3407–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salavaty A, Motlagh FM, Barabadi M, et al. Potential role of RAB6C-AS1 long noncoding RNA in different cancers. J Cell Physiol. 2018;234(1):891–903. [DOI] [PubMed] [Google Scholar]
- 11. Wang Y, Xin S, Zhang K, et al. Low GAS5 levels as a predictor of poor survival in patients with lower-grade gliomas. J Oncol. 2019;2019:1785042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu Z, Wang G, Zhang C, et al. LncRNA SBF2-AS1 affects the radiosensitivity of non-small cell lung cancer via modulating microRNA-302a/MBNL3 axis. Cell Cycle. 2020;19(3):300–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao F, Feng J, Yao H, Li Y, Xi J, Yang J. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782. [DOI] [PubMed] [Google Scholar]
- 14. Chen R, Xia W, Wang X, et al. Upregulated long non-coding RNA SBF2-AS1 promotes proliferation in esophageal squamous cell carcinoma. Oncol Lett. 2018;15(4):5071–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang YT, Li BP, Zhang B, et al. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22(19):6333–6341. [DOI] [PubMed] [Google Scholar]
- 16. Xia W, Liu Y, Cheng T, Xu T, Dong M, Hu X. Down-regulated lncRNA SBF2-AS1 inhibits tumorigenesis and progression of breast cancer by sponging microRNA-143 and repressing RRS1. J Exp Clin Cancer Res. 2020;39(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song LR, Weng JC, Huo XL, et al. , Identification and validation of a 21-mRNA prognostic signature in diffuse lower-grade gliomas. J Neurooncol. 2020;146(1):207–217. [DOI] [PubMed] [Google Scholar]
- 19. Zhang JH, Hou R, Pan Y, et al. A five-microRNA signature for individualized prognosis evaluation and radiotherapy guidance in patients with diffuse lower-grade glioma. J Cell Mol Med. 2020;24(13):7504–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC cancer genomics browser. Sci Rep. 2013;3(1):2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu H, Mu Q, Bao Z, et al. Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell. 2018;175(6):1665–1678. e18. [DOI] [PubMed] [Google Scholar]
- 22. Jiang T, Mao Y, Ma W, et al. , CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375(2):263–273. [DOI] [PubMed] [Google Scholar]
- 23. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. [DOI] [PubMed] [Google Scholar]
- 24. Hothorn T, Zeileis A. Generalized maximally selected statistics. Biometrics. 2008;64(4):1263–1269. [DOI] [PubMed] [Google Scholar]
- 25. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang WG, Sanders AJ, Katoh M, et al. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol. 2015;35 (Suppl):S244–S275. [DOI] [PubMed] [Google Scholar]
- 27. Wang A, Wang J, E2F1-induced overexpression of long noncoding RNA SBF2-AF1 promotes non-small-cell lung cancer metastasis through regulating miR-362-3p/GRB2 Axis. DNA Cell Biol. 2020;39(7):1290–1298. [DOI] [PubMed] [Google Scholar]
- 28. Chen R, Xia W, Wang S, et al. Long noncoding RNA SBF2-AS1 is critical for tumorigenesis of early-stage lung adenocarcinoma. Mol Ther Nucleic Acids. 2019;16:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang C, Yue C, Liang C, et al. The long non-coding RNA SBF2-AS1 exerts oncogenic functions in gastric cancer by targeting the miR-302b-3p/E2F transcription factor 3 Axis. Onco Targets Ther. 2019;12:8879–8893. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Chen G, Gu Y, Han P, Li Z, Zhao JL, Gao MZ. Long noncoding RNA SBF2-AS1 promotes colorectal cancer proliferation and invasion by inhibiting miR-619-5p activity and facilitating HDAC3 expression. J Cell Physiol. 2019;234(10):18688–18696. [DOI] [PubMed] [Google Scholar]
- 31. Tian YJ, Wang YH, Xiao AJ, et al. Long noncoding RNA SBF2-AS1 act as a ceRNA to modulate cell proliferation via binding with miR-188-5p in acute myeloid leukemia. Artif Cells Nanomed Biotechnol. 2019;47(1):1730–1737. [DOI] [PubMed] [Google Scholar]
- 32. Yang X, Zhang Y, Fan H. Downregulation of SBF2-AS1 functions as a tumor suppressor in clear cell renal cell carcinoma by inhibiting miR-338-3p-targeted ETS1. Cancer Gene Ther. 2020:1–5. [DOI] [PubMed] [Google Scholar]
- 33. Zhao QS, Fan H, Li L, et al. Over-expression of lncRNA SBF2-AS1 is associated with advanced tumor progression and poor prognosis in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2016;20(14):3031–3034. [PubMed] [Google Scholar]
- 34. Yu H, Zheng J, Liu X, et al. Transcription factor NFAT5 promotes glioblastoma cell-driven angiogenesis via SBF2-AS1/miR-338-3p-mediated EGFL7 expression change. Front Mol Neurosci. 2017;10:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer Res. 2019;38(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luan F, Chen W, Chen M, et al. An autophagy-related long non-coding RNA signature for glioma. FEBS Open Bio. 2019;9(4):653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bendahou MA, Ibrahimi A, Boutarbouch M. Bioinformatics analysis of differentially expressed genes and miRNAs in low-grade gliomas. Cancer Inform. 2020;19:1176935120969692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-xls-1-tct-10.1177_15330338211011966 for Prognostic Value of Immune-Related lncRNA SBF2-AS1 in Diffuse Lower-Grade Glioma by Qiang Zhang, Xiao-Jun Liu, Yang Li, Xiao-Wei Ying and Lu Chen in Technology in Cancer Research & Treatment
Supplemental Material, sj-xls-2-tct-10.1177_15330338211011966 for Prognostic Value of Immune-Related lncRNA SBF2-AS1 in Diffuse Lower-Grade Glioma by Qiang Zhang, Xiao-Jun Liu, Yang Li, Xiao-Wei Ying and Lu Chen in Technology in Cancer Research & Treatment