Summary
The Gram‐positive bacterium Bacillus subtilis uses serine not only as a building block for proteins but also as an important precursor in many anabolic reactions. Moreover, a lack of serine results in the initiation of biofilm formation. However, excess serine inhibits the growth of B. subtilis. To unravel the underlying mechanisms, we isolated suppressor mutants that can tolerate toxic serine concentrations by three targeted and non‐targeted genome‐wide screens. All screens as well as genetic complementation in Escherichia coli identified the so far uncharacterized permease YbeC as the major serine transporter of B. subtilis. In addition to YbeC, the threonine transporters BcaP and YbxG make minor contributions to serine uptake. A strain lacking these three transporters was able to tolerate 100 mM serine whereas the wild type strain was already inhibited by 1 mM of the amino acid. The screen for serine‐resistant mutants also identified mutations that result in increased serine degradation and in increased expression of threonine biosynthetic enzymes suggesting that serine toxicity results from interference with threonine biosynthesis.
Introduction
As building block of proteins, amino acids are central to the physiology of any living cell. In addition to their role as substrates in protein biosynthesis, they can be used as carbon and nitrogen sources. Moreover, some amino acids are also required for bacterial cell wall biosynthesis and as osmoprotectants. Accordingly, the acquisition of amino acids is an essential task of all cells. This can be achieved by the direct uptake of amino acids present in the growth medium, by the uptake and intracellular degradation of peptides and by de novo biosynthesis. Many bacteria such as the model organisms Escherichia coli and Bacillus subtilis are capable of synthesizing all amino acids whereas others such as the minimal bacteria of the genus Mycoplasma completely depend on the uptake of amino acids.
While amino acids are essential for the cells, increased concentrations of some amino acids such as glutamate, threonine or serine can be harmful (Lamb and Bott, 1979a; Lamb and Bott, 1979b; Lachowicz et al., 1996; Belitsky and Sonenshein, 1998; Ogawa et al., 1998; Commichau et al., 2008; Belitsky, 2015; Mundhada et al., 2017). Therefore, the homeostasis of the amino acids must be tightly controlled to adjust the intracellular levels of each amino acid to the actual need of the cell. This requires balanced activities of systems for amino acid acquisition and degradation. For the Gram‐positive model bacterium B. subtilis, amino acid metabolism is one of the few functions in core metabolism that have not yet been completely elucidated. This is the case both for the biosynthetic pathways and for amino acid transport.
The genome of B. subtilis encodes 47 known and predicted amino acid transporters (Zhu and Stülke, 2018). For 19 of these transporters, substrates have been identified, and for four additional transporters, tentative substrates can be assigned based on mutant properties (for YbxG; Commichau et al., 2015) and on the assignment to particular regulons (AlsT, YvbW and YvsH; Randazzo et al., 2017; Wels et al., 2008; Rodionov et al., 2003). No functional assignment can so far be made for 11 potential transporters. It is important to note that some proteins that are members of typical amino acid transporter families do actually transport other substrates, such as the recently described potassium transporter KimA (Gundlach et al., 2017). A complete overview on the known and potential amino acid transporters of B. subtilis can be found in Table S1 (see also http://subtiwiki.uni-goettingen.de/v3/category/view/SW%201.2, Zhu and Stülke, 2018). Importantly, no transporters have so far been identified or proposed for alanine, glycine, serine, asparagine, and the aromatic amino acids phenylalanine and tyrosine. The identification of new amino acid transporters is hampered by two peculiarities: For one amino acid, there are often multiple transporters, as has been shown for arginine, proline, or the branched‐chain amino acids (Calogero et al., 1994; Gardan et al., 1995; Sekowska et al., 2001; Zaprasis et al., 2014; Belitsky, 2015). On the other hand, many permeases have a relatively weak substrate specificity, i.e. they are able to transport multiple amino acids, as shown for BcaP or GltT (Belitsky, 2015; Zaprasis et al., 2015; Wicke et al., 2019).
We are interested in the identification of the functions that are required to sustain the life of a minimal cell and in the corresponding set of genes and proteins. In an analysis of the genome of B. subtilis, amino acid transporters were proposed to be kept in a minimal genome rather than biosynthetic genes, since this would require fewer genes (Reuß et al., 2016). However, as indicated above, no transporters have been identified for several amino acids. Accordingly, biosynthetic pathways were included for those amino acids. A minimal organism capable of transporting amino acids but not to produce them is expected to be viable on complex media but would be unable to grow on minimal medium in the absence of added amino acids. As minimal bacterial strains have a huge potential for biotechnological applications (Suárez et al., 2019), the ability to produce amino acids may be important for growth on cheap minimal salts substrates.
Serine is an important amino acid because this molecule is a not only a building block for protein synthesis but also a precursor of nucleotides, phospholipids, redox molecules and other amino acids. In addition, decreased level of intracellular serine can be a signal for initiation of biofilm formation in B. subtilis (Subramanian et al., 2013), suggesting that regulation of serine homeostasis is very important. However, the metabolism of serine is not yet completely understood in B. subtilis. For this amino acid, no transporter has been identified, and the knowledge of biosynthetic pathways has remained limited until recently. Indeed, the serine biosynthesis pathway has been completed just recently in the frame of a genome‐scale deletion study by the identification of the SerB phosphoserine phosphatase that catalyses the last step of the pathway (Koo et al., 2017) (see Fig. 1 for an overview on serine metabolism in B. subtilis). Moreover, the reasons for serine toxicity have remained enigmatic. In E. coli, it has been suggested that serine binds and inactivates the bifunctional enzyme aspartate kinase/homoserine dehydrogenase (ThrA) (Mundhada et al., 2017) thus interfering with threonine biosynthesis. In addition, serine catabolism can give rise to toxic intermediates, the highly reactive and therefore toxic β‐hydroxypyruvate as product of the transamination reaction, and α‐aminoacrylate, which is formed as byproduct of serine deamination (Blatt et al., 1966; Seebeck and Szostak, 2006, see de Lorenzo et al., 2015 for review).
Fig. 1.
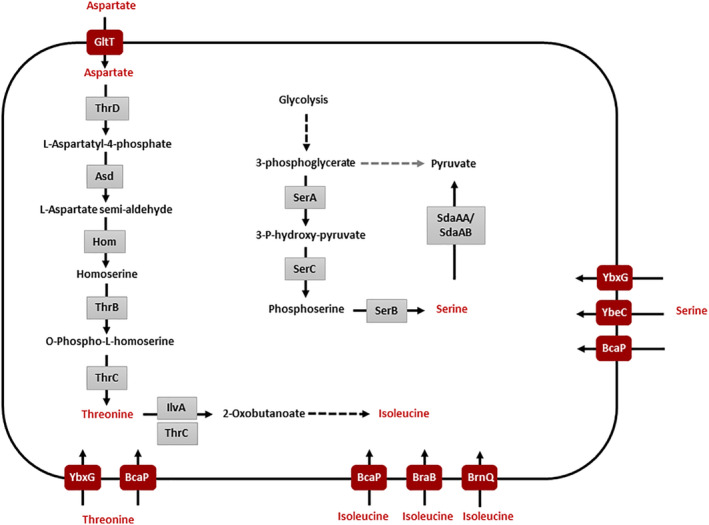
Serine and threonine metabolic pathways in B. subtilis. The model shows the relevant transporters, the biosynthesis of threonine, and its role as a precursor for isoleucine biosynthesis as well as the pathways for serine biosynthesis and degradation. [Color figure can be viewed at wileyonlinelibrary.com]
In this work, we have taken advantage of the serine toxicity phenotype of B. subtilis to devise reverse and forward genetic screens to identify serine transporters. Our analysis identified YbeC as the major serine transporter, and clarified the roles of threonine transporters in uptake of serine. Using a suppressor screen, we also isolated metabolic mutants that circumvent serine toxicity. These mutants exhibit either more efficient serine degradation or overexpression of genes for the threonine and isoleucine biosynthetic pathways suggesting that one or more enzymes in this pathway are inhibited by serine.
Results
Overview of the genetic approaches used in this work
All of our screens and selections were based on the fact that addition of serine to minimal medium is toxic for B. subtilis 168, whereas the addition of serine to complex LB medium did not interfere with growth of the bacteria. Moreover, the addition of specific amino acids such as threonine to minimal medium also overcomes serine toxicity (Vandeyar and Zahler, 1986; Lachowicz et al., 1996). These observations suggest that intracellular serine interferes with amino acid metabolism. We might therefore expect that strains would become resistant to serine toxicity either by eliminating the major serine transporter or by altering amino acid metabolism of genes related to serine toxicity. To identify these genes, we used the following approaches:
A targeted screen for transporters
For this purpose, we chose 12 candidate transporters that met two criteria: First, these transporters have been poorly studied in B. subtilis, and second, they are expressed during vegetative growth. These transporters are AapA, AlsT, MtrA, SteT, YbeC, YbgF, YdgF, YecA, YodF and YtnA. Mutants for the corresponding genes (see Table S2) were constructed and analysed for the ability to grow in the presence of 244 μM L‐serine. While all strains were capable of growing on minimal medium in the absence of serine, only the ybeC mutant strain GP1886 was able to grow in the presence of serine, suggesting that YbeC might act as serine transporter.
A suppressor screen aimed at identifying mutants altered in related amino acid metabolism
We selected for loss of serine toxicity in the wild type strain and in a ΔserA mutant that is auxotrophic for serine strain and depends on the uptake of serine for growth. Of eight studied suppressor strains, four were transporter (ybeC) mutants and the remaining strains exhibited genetic lesions related to amino acid metabolism. Interestingly, the ybeC mutation was also found in the serA mutant as a suppressor, indicating that serine can be transported in ybeC mutant.
A screen of the entire B. subtilis deletion library for loss of serine toxicity
In order to make sure that the screens described above were exhaustive, we also made use of the deletion library that encompasses all non‐essential genes of B. subtilis (Koo et al., 2017). In this library, each reading frame is replaced by an antibiotic cassette with a relatively strong outwardly facing promoter, so resistance to serine toxicity could result from gene deletion or from overexpression of downstream genes. To distinguish between these possibilities, we removed the antibiotic cassette and retested the phenotype. If the phenotype was retained following removal of the antibiotic cassette, then the phenotype was caused by gene deletion; if not it was likely due to overexpression of downstream genes. This screen identified both the transporter, YbeC, and genetic lesions related amino acid metabolism (Table 1).
Table 1.
Serine‐resistant mutants identified from the genome‐wide screen.
| Strain a | Genetic context | Serine resistance | Determinant for serine resistance |
|---|---|---|---|
| ΔybeC::erm | ybeC>, <glpQ < glpT | Yes | Deletion of ybeC |
| ΔybeC::lox72 | Yes | ||
| ΔglpQ::erm | ybeC>, <glpQ < glpT | Yes | Inhibition of ybeC expression |
| ΔglpQ::lox72 | No | ||
| ΔyloU::erm | yloU > yloV>, sdaAB > sdaAA > recG> | Yes | Overexpression of sdaAB‐AA |
| ΔyloU::lox72 | No | ||
| ΔyloV::erm | yloU > yloV>, sdaAB > sdaAA > recG> | Yes | Overexpression of sdaAB‐AA |
| ΔyloV::lox72 | No | ||
| ΔthrR::erm | spo0B > obg > thrR > pheA> | Yes | Deletion of thrR |
| ΔthrR::lox72 | Yes | Overexpression of hom‐thrCB | |
| ΔyutH::erm | yutH>, hom > thrC > thrB> | Yes | Overexpression of hom‐thrCB |
| ΔyutH::lox72 | No |
lox72 indicates the scar resulting from looping out of erythromycin‐resistant cassette.
The genes highlighted in bold are directly responsible for the resistance to serine.
Identification of a permease that confers sensitivity to L‐serine
Both our targeted screen of the 13 expressed, uncharacterized transporters and our screen of the B. subtilis deletion library identified only a single putative transporter, YbeC, the loss of which conferred resistance to serine. Supporting the importance of YbeC, 50% of the mutants from the suppressor screen (Selection #2) had mutations targeting ybeC. Additionally, the glpQ mutant (BKE02130), a strain from the whole‐genome screen (Selection #3) that suppressed serine toxicity due to overexpression, likely generates an abundant ybeC antisense RNA (Table 1). The net effect of antisense expression is to decrease ybeC expression, explaining its serine‐resistance phenotype.
To test whether the ybeC mutant is also resistant to higher concentrations of serine, we cultivated the mutant GP1886 at increasing serine concentrations (up to 100 mM), and recorded growth of the bacteria. While the wild type strain was unable to grow at concentrations exceeding 244 μM, the ybeC mutant was able to tolerate as much as 11 mM serine (see Table 2). In addition to serine, the anti‐metabolite serine hydroxamate also inhibits growth of B. subtilis. To test whether loss of YbeC allows growth in the presence of this serine analogue, we cultivated the wild type strain 168 and the ybeC mutant GP1886 in the presence of DL‐serine hydroxamate (1 mg ml−1). As shown in Fig. 2A, the wild type was sensitive to this molecule whereas the ybeC mutant was somewhat more resistant. Thus, loss of YbeC confers resistance to both serine and its toxic analogue serine hydroxamate, suggesting that the protein is a serine transporter.
Table 2.
The resistance of selected B. subtilis mutants towards serine.
| Strain | Relevant genotype | Tolerated serine concentration (mM) a |
|---|---|---|
| 168 | Wild type | <1 |
| GP2786 | ΔybeC | 11 |
| BKE09460 | ΔbcaP | 2 |
| GP2396 | ΔybxG | 1.5 |
| GP2949 | ΔybeC ΔbcaP | 40 |
| GP2951 | ΔybeC ΔybxG | 25 |
| GP2952 | ΔbcaP ΔybxG | 4 |
| GP2950 | ΔybeC ΔbcaP ΔybxG | 100 |
The tolerated serine concentrations were determined by cultivating the strains in liquid C Glc minimal medium in the presence of different serine concentrations. Note that the results obtained with plates and liquid medium can differ slightly.
Fig. 2.
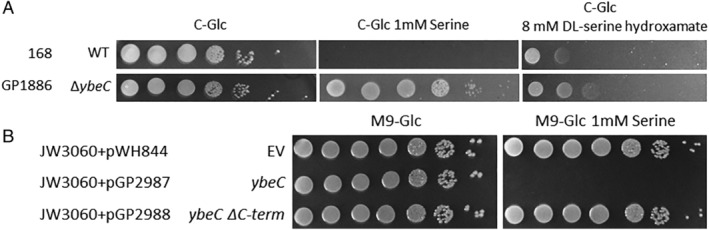
YbeC is a serine transporter.
A. The sensitivity of the wild type strain 168 and the ybeC deletion mutant to serine and the toxic serine analogue DL‐serine hydroxamate. Cells of the wild type 168 and the ybeC deletion mutant were grown in C‐Glc minimal medium to an OD600 of 1.0 and serial dilutions (10‐fold) were prepared. These samples were plated on C‐Glc minimal plates containing no serine, 1 mM serine, or 8 mM DL‐serine hydroxamate. The plates were incubated at 37°C for 48 h.
B. Serine transport complementation assay in E. coli. The growth of the E. coli sstT mutant JW3060 harbouring the empty vector (pWH844) was compared with the growth of JW3060 with a plasmid encoding the full‐length YbeC (pGP2987) or YbeC without the C‐terminus (pGP2988) on M9 minimal plates in the presence and absence of serine. The plates were incubated at 37°C for 48 h.
Isolation and initial characterization of mutants that are able to grow in the presence of serine
The targeted analysis of potential amino acid transporters identified YbeC as the only candidate serine transporter. In an attempt to identify more genes involved in serine toxicity, we cultivated the B. subtilis wild type strain 168 in the presence of different concentrations of serine. Moreover, we used the serine auxotrophic serA mutant that depends on the uptake of serine for growth, and the ybeC mutant that already tolerates up to 11 mM of serine (see above). In total, we isolated eight mutants that exhibited increased resistance to serine in five distinct selection experiments. One mutant for each selection was subjected to whole‐genome sequencing to identify the underlying mutations. In one of the mutants (GP2324, isolated from the wild type 168 at 1 mM serine), a ybeC mutation was detected. Thus, we tested the remaining mutants for the presence of mutations in ybeC. Strikingly, four out of the eight mutants had acquired mutations in ybeC. These mutations resulted in the production of truncated and therefore possibly inactive YbeC proteins or in an in‐frame deletion of 236 amino acids (in GP3050). The identification of multiple suppressor mutants affecting YbeC strongly supports the crucial role of YbeC in the resistance to serine.
Of the serine‐resistant strains whose phenotype was not caused by a ybeC mutation, two strains derived from the wild type strain 168 had a duplication of about 16 kb yokD‐thyB chromosomal region. Interestingly, this region contains the ilvA gene encoding threonine dehydratase involved in the biosynthesis of isoleucine from threonine. The remaining three mutants (derived from the serA mutant, and the ybeC mutant at 10 and 17 mM serine, see Table S2 for details) had mutations affecting the repressor for the threonine biosynthetic genes, thrR (Rosenberg et al., 2016), and mutations in the regulatory regions of the sdaAB and hom promoter regions respectively (Table S2). All these genes are involved in serine and threonine metabolism suggesting a close relation between the metabolic pathways for these similar amino acids (see below for further analyses).
YbeC is a serine transporter
All three targeted and unbiased analyses of serine‐resistance mutants identified YbeC as the main player. YbeC is similar to known amino acid transporters, is classified as a member of the amino acid‐polyamine‐organocation superfamily (see Table S1), and the ybeC mutant had the phenotypes expected for a major serine transporter. To test this idea, we made use of an E. coli mutant that lacks the major serine transporter SstT. This strain is less sensitive to growth inhibition by serine (Ogawa et al., 1997; Ogawa et al., 1998). We cloned the ybeC gene into the expression vector pWH844 and used the resulting plasmid pGP2987 to transform the sstT mutant JW3060 (Baba et al., 2006). Indeed, the expression of plasmid‐borne ybeC in E. coli JW3060 restored serine toxicity (Fig. 2B). Taken together, both the genetic characterization and the functional complementation of an E. coli mutant lacking a serine transporter demonstrate that YbeC is indeed a transporter for serine.
The ybeC gene forms a monocistronic transcription unit (Nicolas et al., 2012). To study the activity of the ybeC promoter and its possible regulation by serine, a 257 bp region (222 bp upstream of the ATG translational start codon, and 35 bp of the ybeC coding region) was fused to a promoterless lacZ gene. The resulting strain, GP2965, was cultivated in minimal in the presence and absence of serine as well as in complex (LB) medium. With very similar β‐Galactosidase activities (144 ± 2, 132 ± 8 and 135 ± 10 units mg−1 of protein respectively), this fusion was similarly expressed irrespective of the presence of serine in the medium thus indicating constitutive expression of ybeC.
Serine and threonine share overlapping transporters
Threonine transporters contribute to serine uptake
The identification of viable serA ybeC mutants in the suppressor screen (Screen #2 above) suggested either that the mutant YbeC proteins retained some transport activity or that YbeC is not the only transporter for serine. To resolve this issue, we deleted the ybeC gene in the serA mutant, which is auxotrophic for serine. The resulting double mutant GP2941 depends on serine uptake for viability. Analysis of growth of these mutants demonstrated that both the serA mutant and the serA ybeC double mutant were unable to grow in the absence of serine (C‐glucose medium). In contrast, the strains lacking the ybeC gene were able to grow in minimal medium supplemented with serine (see Fig. 3A). Thus, the ΔybeC mutant is still able to transport serine.
Fig. 3.
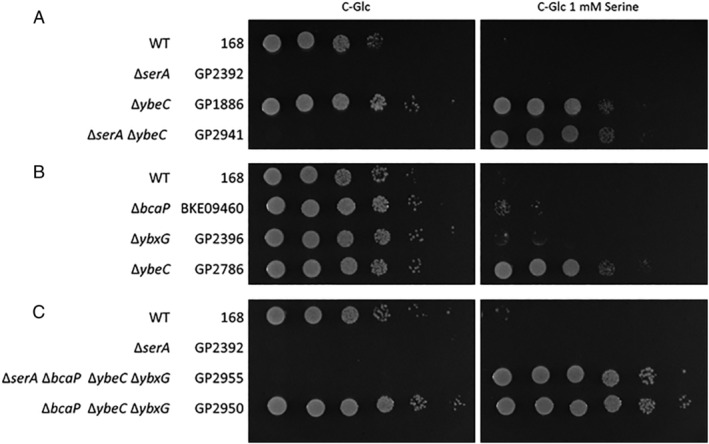
The contribution of threonine transporters to serine uptake. Cells of the indicated strains were grown in C‐Glc minimal medium to an OD600 of 1.0 and serial dilutions (10‐fold) were prepared. These samples were plated on C‐Glc minimal plates containing no serine or 1 mM serine. The plates were incubated at 37°C for 48 h.
A. Combination of the ybeC deletion with the deletion of the serA gene encoding phosphoglycerate dehydrogenase. The growth of the single deletion mutants of ybeC (GP1886) and serA (GP2392) was compared with the growth of the combined deletion strain of ybeC and serA (GP2941).
B. The resistance of the threonine transporter deletion strains to serine. The bcaP (BKE09460) and ybxG deletion strains (GP2396) are compared with the wild type strain 168 and the ybeC deletion strain (GP1886). C. Combination of the serA deletion with the deletion strain if bcaP, ybeC and ybxG. The growth of the wild type strain 168 was compared with GP2392 (serA), GP2955 (serA bcaP ybeC ybxG) and GP2950 (bcaP ybeC ybxG).
Serine and threonine are chemically similar to each other and the E. coli SstT transporter is capable of transporting both serine and threonine (Kim et al., 2002). Therefore, we considered the possibility that threonine transporters might contribute to serine uptake in B. subtilis, and vice versa. Based on the analysis of growth inhibition by threonine and its analogue 4‐hydroxythreonine, the BcaP and YbxG permeases have been identified as tentative threonine transporters in B. subtilis (see Table S1, Belitsky, 2015; Commichau et al., 2015). To test the possible role of these permeases in serine transport, we used single, double and triple mutants lacking ybeC, bcaP and ybxG respectively. The resulting strains were assayed for their resistance towards serine. As shown in Fig. 3B, the single bcaP and ybxG deletions conferred only a weak resistance to growth inhibition by serine, whereas the loss of ybeC resulted in a substantial resistance (see also Table 2). This observation confirms that YbeC is the main transporter for serine in B. subtilis.
The double mutants lacking YbeC and one of the threonine transporters exhibited a substantial increase in resistance to serine indicating that both threonine permeases contribute to serine transport (see Table 2). In contrast, the bcaP ybxG double mutant was much more sensitive to serine than the ybeC mutant. This observation supports the conclusion that YbeC is the major serine permease. The analysis of double mutants lacking YbeC and either YbxG or BcaP indicates that the loss of BcaP has a higher contribution to serine resistance as compared with the loss of YbxG (Table 2, compare GP2951 and GP2949). This indicates that BcaP may be more active in serine transport than YbxG. The deletion of the three permease‐encoding genes in the triple mutant GP2950 resulted in an unprecedented resistance to serine up to 100 mM (Table 2). This finding indicates that these three proteins may be responsible for the majority of serine uptake in B. subtilis. If these proteins were the only serine permeases, one would expect that an auxotrophic serA mutant lacking the three permeases would not be viable. However, this mutant (GP2955) was still able to grow on minimal medium in the presence of serine (Fig. 3C). Thus, B. subtilis possesses at least one additional permease that is able to transport serine.
Analysis of threonine transport
Our findings demonstrate that the two previously suggested threonine transporters are also active as minor serine permeases. Next, we asked whether YbeC is also capable of transporting threonine. To address this question, we made use of the observation that threonine is toxic for B. subtilis if added in concentrations exceeding 50 μg ml−1 (Lamb and Bott, 1979a; Lamb and Bott, 1979b). In our experimental setup, threonine (10 mM) inhibits growth of B. subtilis 168. Inactivation of the bcaP gene conferred a growth advantage, the bcaP mutant grew in the presence of threonine as well as the wild type strain in the absence of this amino acid. In contrast, the deletions of ybxG or ybeC had only minor effects (Fig. 4A). This observation is supported by the analysis of the double and triple mutants: While all mutants lacking bcaP showed threonine‐resistant growth, this was not the case for the ybeC ybxG double mutant GP2952 that still expresses BcaP (Fig. 4B). These observations suggest that BcaP is the main threonine transporter in B. subtilis.
Fig. 4.
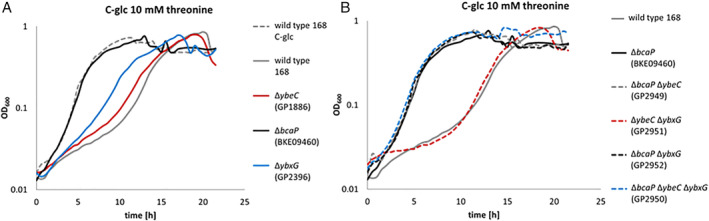
The growth inhibition by threonine.
A The single deletion strains for ybeC (GP1886), bcaP (BKE09460) and ybxG (GP2396) were grown in C‐glc medium with 10 mM threonine in comparison to the wild type strain 168 and the wild type strain 168 in C‐glc medium without threonine.
B The growth of the double deletion mutants bcaP ybeC (GP2949), ybeC ybxG (GP2951) and bcaP ybxG (GP2952) was compared with the growth of the bcaP ybeC ybxG deletion strain and the wild type strain 168 in C‐glc medium with 10 mM threonine. [Color figure can be viewed at wileyonlinelibrary.com]
In order to test the presence of additional threonine transporters, and to get further evidence for the relative roles of BcaP, YbxG and YbeC in threonine uptake, we deleted the thrC gene in the wild type 168 and in relevant transporter mutants. The thrC gene codes for threonine synthase which catalyses the final step in threonine biosynthesis. As expected, the thrC mutant was auxotrophic for threonine (data not shown). The deletion of bcaP alone or in combination with ybeC resulted in improved growth both at 0.04 and 4 mM threonine as compared with the single thrC mutant (see Fig. 5). The combination of the thrC, ybeC and ybxG mutations had no effect as compared with the single thrC deletion supporting the idea that YbeC and YbxG play only very minor roles in threonine uptake. However, the simultaneous deletion of bcaP and ybxG in the thrC mutant resulted in severely reduced growth at 0.04 mM threonine (Fig. 5A). The additional deletion of ybeC had only a minor, if any impact. These findings suggest that BcaP and YbxG act as threonine transporters. Importantly, the thrC mutant lacking BcaP and YbxG (and YbeC) is still able to grow in the presence of threonine suggesting the existence of at least one additional threonine transporter (Fig. 5A and B).
Fig. 5.
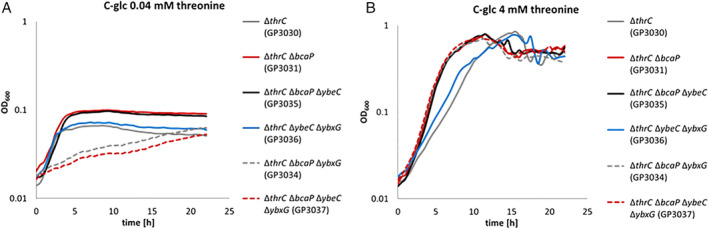
Growth of the auxotrophic strains in combination with transporter deletions in the presence of different amounts of threonine. The growth of the deletion strains GP3031 (thrC bcaP), GP3035 (thrC bcaP ybeC), GP3036 (thrC ybeC ybxG), GP3034 (thrC bcaP ybxG) and GP3037 (thrC bcaP ybeC ybxG) was compared with the thrC deletion mutant (GP3030) in C‐glc medium with 0.04 mM threonine (A) and 4 mM threonine (B). [Color figure can be viewed at wileyonlinelibrary.com]
Taken together, these results indicate that BcaP is the major threonine transporter in B. subtilis, whereas YbxG has a minor threonine permease activity. Our data do not support the annotation of YbeC as a threonine transporter. Moreover, BcaP and YbxG have overlapping activity for both serine and threonine (see Fig. 1).
Putting serine toxicity in its metabolic context
As mentioned above, the addition of serine to minimal medium is toxic, whereas its addition to complex LB medium did not interfere with growth of B. subtilis 168 (data not shown). The inactivation of the ybeC gene to prevent serine uptake or the addition of several individual amino acids such as threonine overcomes serine toxicity (Vandeyar and Zahler, 1986; Lachowicz et al., 1996). These observations suggest that intracellular serine interferes with amino acid metabolism. Mutants from the suppressor screen (Selection #2) and from the genome wide screen (Selection #3) shed light on the origins of serine toxicity.
The role of serine deaminase in overcoming serine toxicity
The sdaAB‐sdaAA operon encodes the two subunits of serine deaminase, which catalyses the degradation of serine to pyruvate and ammonia (Chen et al., 2012). Both the suppressor screen (Selection #2) and the whole‐genome screen (Selection #3) identified overexpression of sdaAB‐sdaAA as relieving serine toxicity. In the suppressor screen, strain GP2971 had a mutation 70 bp upstream of the start of the sdaAB coding sequence suggesting that it might affect expression of the operon. Indeed, a promoter has been identified in the 139 bp intergenic region between the yloV and sdaAB genes (Nicolas et al., 2012). To test this hypothesis, we fused the 166 bp wild type and mutant regions that contain the complete yloV‐sdaAB intergenic region, and thus the sdaAB promoter, to a promoterless lacZ gene, and compared the gene expression driven by these promoters. The strains carrying the lacZ fusions integrated into the amyE gene were cultivated in minimal medium, and their β‐Galactosidase activities were determined. For the wild type promoter, we detected 7.4 ± 2.2 units of β‐Galactosidase per mg of protein. This corresponds to a very weak promoter activity (Schilling et al., 2007). Expression of the lacZ gene from the mutant promoter resulted in 370 ± 48 units of β‐Galactosidase per mg of protein. Thus, the mutation resulted in a 50‐fold increase of promoter activity. A closer inspection of the sequence around the mutation suggests that a TTGCCA sequence had been altered to the perfect −35 sequence, TTGACA. It is tempting to speculate that this perfect −35 region is responsible for the higher expression of the sdaAB‐sdaAA operon and thus for higher intracellular levels of serine deaminase in the mutant. This conclusion is strongly supported by two mutants from the whole‐genome screen, affected in yloV (BKE15840) and yloU (BKE15830) that suppressed serine toxicity due to overexpression of the sdaAB‐sdaAA operon. These strains have their antibiotic cassettes with the strong outwardly facing promoter immediately upstream of sdaAB‐sdaAA, indicating that their overexpression suppresses serine toxicity (Table 1). The increased degradation of serine by serine deaminase is likely to be responsible for the protective action observed upon overexpression of this enzyme.
The role of threonine metabolism in serine toxicity
Two different loci related to threonine metabolism were identified in our screens. First, the suppressor screen identified a duplication of the 16 kb yokD‐thyB region containing ilvA as relieving serine toxicity. Second, both the whole‐genome and suppressor screens (Selection #2, 3) identified overexpression of the hom‐thrC‐thrB operon as relieving serine toxicity.
The threonine dehydratase IlvA uses threonine in the initial step of isoleucine biosynthesis. We observed a duplication of the approximately 16 kb yokD‐thyB genomic region encompassing ilvA in two suppressor strains. This observation implies that IlvA may become limiting in the presence of serine or contribute to scavenging excess serine. If IlvA is inhibited by serine, this could be compensated by overexpression of ilvA gene (due to genomic duplication) or by increased synthesis of ThrC with its moonlighting activity as threonine dehydratase (Skarstedt and Greer, 1973; Rosenberg et al., 2016) (see Fig. 1) which resumes isoleucine synthesis. However, this is unlikely since supplementation of isoleucine does not reduce serine toxicity (data not shown). Thus, it is more likely that B. subtilis IlvA may also have serine dehydratase activity, resulting in deamination of serine as has been shown in Salmonella enterica and E. coli (Borchert and Downs, 2018). To test whether IlvA is a major determinant for serine resistance in these suppressor strains, we overexpressed the ilvA gene in the wild type strain 168 using the expression vector pGP2289 (see Fig. 6). Indeed, ilvA overexpression provided resistance to serine. However, the level of resistance of the overexpressing strain was lower than observed for the original suppressor mutation with the genomic duplication (see Discussion).
Fig. 6.
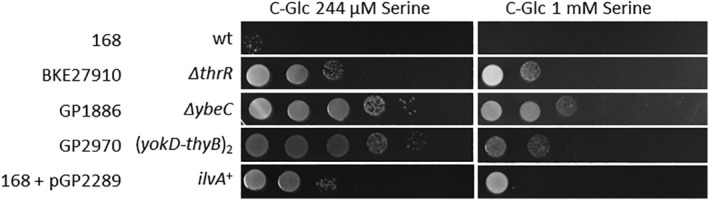
Serine resistance of the thrR deletion mutant and the ilvA overexpression strain. The growth of the wild type strain 168 and the mutant strains BKE27910 (thrR), GP1886 (ybeC), GP2970 (Suppressor with (yokD‐thyB) duplication) and the wild type 168 with the plasmid pGP2289 (ilvA overexpression) was compared on C‐Glc minimal medium plates (10‐fold serial dilution) containing 244 μM or 1 mM serine. The plates were incubated for 48 h at 37°C.
Both the whole‐genome and suppressor screens identified inactivation of the ThrR repressor and overexpression of one of its target operons, hom‐thrC‐thrB as relieving serine toxicity. The suppressor screen (Selection #2) identified a mutation in thrR and a mutation upstream of the hom‐thrC‐thrB operon. The inspection of the mutation in the hom upstream region revealed that this mutation did affect the ThrR binding site (Rosenberg et al., 2016). Moreover, the thrR mutation (deletion of A91) resulted in a frameshift and translation stop after 35 amino acids. This truncation has been observed previously in a different context. It results in an inactive ThrR protein (Rosenberg et al., 2016). This suggests that both the thrR and the hom promoter region mutations result in increased expression of the hom‐thrC‐thrB operon. To test this idea, we tested the activity of the wild type and mutant hom promoters using hom‐lacZ fusions. Strains carrying these fusions were grown in minimal medium and their β‐Galactosidase activities were assayed. For the wild type promoter, we detected 275 ± 35 units of β‐Galactosidase per mg of protein, whereas the mutant promoter resulted in 970 ± 85 units of β‐Galactosidase per mg of protein. These values are similar to those determined previously for the wild type hom promoter and for promoter variants that carry mutations in the ThrR binding site (Rosenberg et al., 2016). Thus, these mutations allow an increased expression of the hom‐thrC‐thrB operon. These findings are supported by the results from the whole‐genome screen (Selection #3): The screen identified a strain with a thrR deletion and as well as overexpression of the hom‐thrC‐thrB operon originating from yutH, which is adjacent to the hom‐thrC‐thrB operon as relieving serine toxicity (Table 1).
Taken together, our results suggest that serine might cause defects in threonine and isoleucine biosynthesis. The defects can be overcome by reducing serine uptake, by degradation of serine, or by an adjustment of threonine and isoleucine metabolism.
Discussion
Metabolite toxicity is one of the least understood areas in the field of microbial metabolism. However, toxic metabolites pose major problems if metabolic pathways are assembled for biotechnological applications or when approaching genome minimization (de Lorenzo et al., 2015; Commichau et al., 2015; Reuß et al., 2016). For B. subtilis, only recently significant effort has been put into the elucidation of resistance mechanisms that allow the bacterium to cope with toxic metabolic intermediates and substrates (Lambrecht et al., 2012; Commichau et al., 2015; Niehaus et al., 2017; Niehaus et al., 2018; Sachla and Helmann, 2019).
In this work, we isolated B. subtilis mutants that are able to grow in minimal medium supplemented with the toxic amino acid serine. Our two laboratories initiated this project independently starting with different aims, the identification of serine transporter and understanding the origin of serine toxicity, but the information obtained from all three strategies was highly similar and complementary. The convergence of the results from the unbiased and the genome‐wide screens strongly suggests that the screens were saturating and that we have elucidated the complete portfolio of possibilities that allows B. subtilis to cope with otherwise toxic serine concentrations.
All three screens identified YbeC as the major serine transporter in B. subtilis. Moreover, this transporter works well in E. coli in which YbeC restores serine sensitivity of a sstT mutant. The common presence of multiple permeases for a single amino acid as well as the weak specificity and promiscuity of amino acid permeases make the identification of these transporters difficult. With YbeC and serine uptake, we had to deal with these challenges: While YbeC is the major transporter for serine, it is not the only one. Our study demonstrates that the BcaP and YbxG transporters that can transport threonine, do also contribute to serine uptake; however, their contribution is rather minor, as can be judged from the analysis of resistance of transporter mutants to serine. Even in the absence of YbeC, YbxG and BcaP, B. subtilis is still able to transport serine from the medium indicating the presence of yet additional serine transporters. The promiscuity of amino acid transporters is important for genome minimization projects (Reuß et al., 2016, Reuß et al., 2017). For example, BcaP alone would be sufficient to transport at least four amino acids. Thus, genes encoding additional transporters for these amino acids (including ybeC) can be deleted as well as the corresponding biosynthetic pathways. It seems that nature has already put this reduction of amino acid acquisition to very few transporters into reality: the highly genome‐reduced Mycoplasma species have lost the ability to produce amino acids and therefore depend completely on their uptake from the medium. Due to the fast evolution of this group of bacteria, it has so far not been possible to identify amino acid transporters based on sequence similarity. However, the independent life of the artificial genome‐reduced organism Mycoplasma mycoides JCVI‐syn3.0 (Hutchison 3rd et al., 2016) indicates that this minimal bacterium possesses a complete set of amino acid transporters.
As mentioned above, serine can inhibit bacterial metabolism both by the interference of the amino acid with other enzymatic reactions, and by the high reactivity toxicity of intrinsic products of serine catabolism (de Lorenzo et al., 2015). One might therefore expect the isolation of different classes of suppressor mutants depending on the ability of the strain to synthesize serine or not. However, all our screens suggest that the amino acid serine rather than derived metabolites is the major obstacle for the metabolic network of B. subtilis. The toxicity of serine can not only be mitigated by the loss of the major serine transporter, YbeC. In addition, our screens also identified other ways to cope with increased serine concentrations, i.e. (i) the rapid conversion of serine to other metabolites, mostly pyruvate and (ii) the overexpression of genes involved in the synthesis of threonine (the hom‐thrC‐thrB operon). The serine deaminase complex SdaAA‐AB converts serine to pyruvate and ammonia, thus detoxifying excess serine as well as allowing cells to use serine as carbon and nitrogen source. It is therefore not surprising that removal of this enzyme activity was attempted to increase the yield of serine production in E. coli (Li et al., 2012). On the other hand, it was reported that serine deaminase deficiency in E. coli resulted in abnormal cell division even in lysogeny broth medium (Zhang and Newman, 2008). Serine deaminase can produce toxic α‐aminoacrylate, and overexpression of the enzyme might theoretically result in the accumulation of this intermediate. However, α‐aminoacrylate can be disposed of by the activity of the B. subtilis YabJ protein (Lambrecht et al., 2012). Importantly, the YabJ protein is much more abundant than the serine deaminase (1660 vs. 90 molecules per cell during growth in minimal medium) (Muntel et al., 2014). The high abundance of YabJ as well as the fact that we did not isolate any suppressor mutants with changed metabolism of serine degradation products suggest that the accumulation of toxic degradation products plays only a minor role in serine toxicity in B. subtilis.
There are a couple of ways in which increased expression of the hom‐thrC‐thrB operon could suppress serine toxicity. First, increased levels of threonine biosynthetic enzymes may produce more threonine. As the addition of threonine to serine‐containing minimal medium can partially overcome serine toxicity, it is likely that serine addition deprives the cell of threonine, which can be overcome either by increased threonine synthesis or by external supplementation. Second, L‐serine toxicity in E. coli works by inhibiting both the aspartate kinase and homoserine dehydrogenase activities of the fused enzyme ThrA (Costrejean and Truffa‐Bachi, 1977), and may function analogously in B. subtilis. Consistent with this idea, we found that supplementation of homoserine restored the growth of wild type B. subtilis in the presence of serine (data not shown). Biochemical analysis with purified B. subtilis homoserine dehydrogenase would provide clear evidence for this hypothesis. We attempted to purify B. subtilis homoserine dehydrogenase from hom overexpressing E. coli strain but failed to get active enzyme. It is tempting to speculate that the increased expression of ilvA upon the duplication the yokD‐thyB genomic region is the major determinant for serine‐resistant phenotype in this suppressor mutant since we observed that overexpression of ilvA phenocopied it, even though only partially (Fig. 6). One explanation for the incomplete effect of IlvA overexpression is that the enzyme not only suppresses serine toxicity but also is itself toxic to cell due to the accumulation of toxic levels of 2‐oxobutanoate or 2‐aminoacrylate (Borchert and Downs, 2018). Strikingly, the ilvA gene is present in two copies in the suppressor strain whereas it is present on multiple plasmid copies and expressed from a strong constitutive promoter in the artificial overexpression system. This may be too much of a good thing!
This study provides novel insights into important aspects of serine metabolism in B. subtilis and into its integration into the amino acid acquisition network. This network consists not only of biosynthetic enzymes with overlapping activities but also of the transporters that are often promiscuous and transport multiple amino acids. Our work provides a starting point for further analysis of the complex and interlocking set of proteins that carry out amino acid transport in B. subtilis.
Methods
Bacterial strains and growth conditions
All B. subtilis strains used in this work are derived from the laboratory wild type strain 168. They are listed in Table S2. Bacillus subtilis was grown in LB (Lysogeny broth) medium, SP (sporulation) medium and in C minimal medium containing glucose and ammonium as basic sources of carbon and nitrogen respectively (Commichau et al., 2008). Minimal medium was supplemented with auxotrophic requirements (at 50 mg L−1) and amino acids as indicated. Plates were prepared by the addition of 17 g Bacto agar/l (Difco) to the liquid medium. Escherichia coli DH5α and JW3060 (Sambrook et al., 1989; Baba et al., 2006) were used for cloning and complementation experiments respectively. JW3060 was grown in M9 minimal medium (Sambrook et al., 1989) with glucose (1% w/v) as the carbon source, but lacking casamino acids. Serine was added as indicated. For the determination of the tolerated serine concentrations, bacteria were grown in C glucose minimal medium to an OD600 of 1.0 and plated on C‐Glc plates containing a wide range of serine concentrations (1–100 mM). The growth was compared after incubation of the plates at 37°C for 48 h.
DNA manipulation and genome sequencing
Plasmid DNA extraction from E. coli were performed using standard procedures (Sambrook et al., 1989). Restriction enzymes, T4 DNA ligase and DNA polymerases were used as recommended by the manufacturers. Fusion DNA polymerase (Biozym, Germany) was used for the polymerase chain reaction as recommended by the manufacturer. DNA fragments were purified using the Qiaquick PCR Purification kit (Qiagen, Germany). DNA sequences were determined using the dideoxy chain termination method (Sambrook et al., 1989). All plasmid inserts derived from PCR products were verified by DNA sequencing. Chromosomal DNA of B. subtilis was isolated as described (Commichau et al., 2008). To identify the mutations in the suppressor mutant strains GP2324, GP2969, GP2970, GP2971 and GP2972 (see Table S2), the genomic DNA was subjected to whole‐genome sequencing (Reuß et al., 2019). Briefly, the reads were mapped on the reference genome of B. subtilis 168 (GenBank accession number: NC_000964) (Barbe et al., 2009). Mapping of the reads was performed using the Geneious software package (Biomatters, New Zealand) (Kearse et al., 2012). Single nucleotide polymorphisms were considered as significant when the total coverage depth exceeded 25 reads with a variant frequency of ≥90%. All identified mutations were verified by PCR amplification and Sanger sequencing.
Transformation and phenotypic analysis
Standard procedures were used to transform E. coli (Sambrook et al., 1989) and transformants were selected on LB plates containing ampicillin (100 μg ml−1). Bacillus subtilis was transformed with plasmid or chromosomal DNA according to the two‐step protocol described previously (Kunst and Rapoport, 1995). Transformants were selected on SP plates containing chloramphenicol (Cm 5 μg ml−1), kanamycin (Km 5 μg ml−1), spectinomycin (Spc 150 μg ml−1), or erythromycin plus lincomycin (Em 25 μg ml−1 and Lin 25 μg ml−1).
In B. subtilis, amylase activity was detected after growth on plates containing nutrient broth (7.5 g L−1), 17 g Bacto agar/L (Difco) and 5 g hydrolyzed starch/L (Connaught). Starch degradation was detected by sublimating iodine onto the plates.
Quantitative studies of lacZ expression in B. subtilis were performed as follows: cells were grown in LB medium or in C glucose medium supplemented with serine as indicated. Cells were harvested at OD600 of 0.6 to 0.8. β‐Galactosidase specific activities were determined with cell extracts obtained by lysozyme treatment as described previously (Kunst and Rapoport, 1995). One unit of β‐Galactosidase is defined as the amount of enzyme, which produces 1 nmol of o‐nitrophenol min−1 at 28° C.
Construction of deletion mutants
Deletion of amino acid transporter and biosynthetic genes was achieved by transformation with PCR products constructed using appropriate oligonucleotides to amplify DNA fragments flanking the target genes and intervening antibiotic resistance cassettes (Guérout‐Fleury et al., 1995) as described previously (Wach, 1996).
Whole‐genome growth phenotype screen
The screen was carried out as described previously (Koo et al., 2017) with modifications that optimized screening for serine toxicity. Plates for screening were allowed to dry for 2 days. The BKE (ErmR) library was arrayed in 384‐well plates using a Biomek FX liquid handling robot (Beckman Coulter) and stored as glycerol stock. To screen the whole BKE library, cells were pinned from glycerol stocks onto rectangular LB agar plates in 384‐format using a Singer Rotor robot, then four 384‐format plates were combined and pinned to 1536‐format. For each screen, exponentially growing cells in 1536‐format were then pinned to glucose minimal agar plates (growth control) and glucose minimal plates supplemented with three different concentrations of L‐serine (0.38, 0.75 and 1.5 mM). Then, plates were incubated at 37°C in a humidified incubator for about 24–44 h. Plates were imaged using a Powershot G10 camera (Canon) and serine‐resistant mutants were identified by their position in the plates. Each mutant was confirmed by sequencing of their barcodes.
Plasmids
Plasmid pAC5 (Martin‐Verstraete et al., 1992) was used to construct translational fusions of the ybeC, sdaAB and hom control regions with the lacZ gene. For this purpose, the regions upstream of these genes were amplified using appropriate oligonucleotides. The PCR products were digested with EcoRI and BamHI PCR and cloned into pAC5 linearized with the same enzymes. The resulting plasmids were pGP2287 (ybeC), pGP2295 (sdaAB), pGP2294 (sdaAB*) and pGP2296 (hom*).
For the expression of YbeC in E. coli, we constructed plasmid pGP2987. For this purpose, the ybeC gene was amplified using chromosomal DNA of B. subtilis as a template. The PCR product was digested with BamHI and SalI and cloned into the expression vector pWH844 (Schirmer et al., 1997).
For the expression of the threonine dehydratase IlvA in B. subtilis, plasmid pGP2289 was constructed by cloning a DNA fragment covering the ilvA gene between the BamHI and SalI restriction sites of the overexpression vector pBQ200 (Martin‐Verstraete et al., 1994).
Supporting information
Table S1 Known and potential amino acid transporters in B. subtilis
Table S2. Bacillus subtilis strains used in this study.
Acknowledgements
We are grateful to Christina Herzberg for the help with some drop dilution assays and to Fabian M. Commichau for helpful discussions. This work was supported by the EU Horizons 2020 program (Rafts4Biotech, 720776 to J.S.), the Deutsche Forschungsgemeinschaft via priority program SPP1879 (to J.S.) and the NIH (R35 GM118061 to C.A.G.).
References
- Baba, T. , Ara, T. , Hasegawa, M. , Takai, Y. , Okumura, Y. , et al. (2006) Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbe, V. , Cruveiller, S. , Kunst, F. , Lenoble, P. , Meurice, G. , Sekowska, A. , et al. (2009) From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155: 1758–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky, B.R. (2015) Role of branched‐chain amino acid transport in Bacillus subtilis CodY activity. J Bacteriol 197: 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky, B.R. , and Sonenshein, A.L. (1998) Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180: 6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, L. , Dorer, F.E. , and Sallach, H.J. (1966) Occurrence of hydroxypyruvate‐L‐glutamate transaminase in Escherichia coli and its separation from hydroxypyruvate‐phosphate‐L‐glutamate transaminase. J Bacteriol 92: 678–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert, A.J. , and Downs, D.M. (2018) Analyses of variants of the Ser/Thr dehydratase IlvA provide insight into 2‐aminoacrylate metabolism in Salmonella enterica . J Biol Chem 293: 19240–19249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calogero, S. , Gardan, R. , Glaser, P. , Schweizer, J. , Rapoport, G. , and Débarbouillé, M. (1994) RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol 176: 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Xu, X.L. , and Grant, G.A. (2012) Allosteric activation and contrasting properties of L‐serine dehydratase types 1 and 2. Biochemistry 51: 5320–5328. [DOI] [PubMed] [Google Scholar]
- Commichau, F.M. , Alzinger, A. , Sande, R. , Bretzel, W. , Reuß, D.R. , Dormeyer, M. , et al. (2015) Engineering Bacillus subtilis for the conversion of the antimetabolite 4‐hydroxy‐L‐threonine to pyridoxine. Metab Eng 29: 196–207. [DOI] [PubMed] [Google Scholar]
- Commichau, F.M. , Gunka, K. , Landmann, J.J. , and Stülke, J. (2008) Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations in the system. J Bacteriol 190: 3557–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costrejean, J.M. , and Truffa‐Bachi, P. (1977) Threonine‐sensitive homoserine dehydrogenase and aspartokinase activities of Escherichia coli K12. Kinetic and spectroscopic effects upon binding of serine and threonine. J Biol Chem 252: 5332–5336. [PubMed] [Google Scholar]
- De Lorenzo, V. , Sekowska, A. , and Danchin, A. (2015) Chemical reactivity drives spatiotemporal organisation of bacterial metabolism. FEMS Microbiol Rev 39: 96–119. [DOI] [PubMed] [Google Scholar]
- Gardan, R. , Rapoport, G. , and Débarbouillé, M. (1995) Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis . J Mol Biol 249: 843–856. [DOI] [PubMed] [Google Scholar]
- Guérout‐Fleury, A.M. , Shazand, K. , Frandsen, N. , and Stragier, P. (1995) Antibiotic resistance cassettes for Bacillus subtilis . Gene 167: 335–336. [DOI] [PubMed] [Google Scholar]
- Gundlach, J. , Herzberg, C. , Kaever, V. , Gunka, K. , Hoffmann, T. , Weiß, M. , et al. (2017) Control of potassium homeostasis is an essential function of the second messenger cyclic di‐AMP in Bacillus subtilis . Sci Signal 10: eaal3011. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.A., 3rd , Chuang, R.Y. , Noskov, V.N. , Assad‐Garcia, N. , Deerinck, T.J. , et al. (2016) Design and synthesis of a minimal bacterial genome. Science 351: aad6253. [DOI] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , et al. (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.M. , Ogawa, W. , Tamai, E. , Kuroda, T. , Mizushima, T. , and Tsuchiya, T. (2002) Purification, reconstitution, and characterization of Na(+)/serine symporter, SstT, of Escherichia coli . J Biochem 132: 71–76. [DOI] [PubMed] [Google Scholar]
- Koo, B.M. , Kritikos, G. , Farelli, J.D. , Todor, H. , Tong, K. , Kimsey, H. , et al. (2017) Construction and analysis of two genome‐scale libraries for Bacillus subtilis . Cell Syst 4: 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst, F. , and Rapoport, G. (1995) Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis . J Bacteriol 177: 2403–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowicz, T.M. , Morzejko, E. , Panek, E. , and Piątkowski, J. (1996) Inhibitory action of serine on growth of bacteria of the genus Bacillus on mineral synthetic media. Folia Microbiol 41: 21–25. [Google Scholar]
- Lamb, D.H. , and Bott, K.F. (1979a) Threonine inhibition of growth of Bacillus subtilis: positive selection for isoleucine auxotrophy. J Gen Microbiol 111: 433–435. [DOI] [PubMed] [Google Scholar]
- Lamb, D.H. , and Bott, K.F. (1979b) Inhibition of Bacillus subtilis growth and sporulation by threonine. J Bacteriol 137: 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht, J.A. , Flynn, J.M. , and Downs, D.M. (2012) Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′‐phosphate (PLP)‐dependent enzymes. J Biol Chem 287: 3454–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Chen, G.K. , Tong, X.W. , Zhang, H.T. , Liu, X.G. , Liu, Y.H. , and Lu, F.P. (2012) Construction of Escherichia coli strains producing L‐serine from glucose. Biotechnol Lett 34: 1525–1530. [DOI] [PubMed] [Google Scholar]
- Martin‐Verstraete, I. , Débarbouillé, M. , Klier, A. , and Rapoport, G. (1992) Mutagenesis of the Bacillus subtilis "‐12,‐24" promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol 226: 85–99. [DOI] [PubMed] [Google Scholar]
- Martin‐Verstraete, I. , Débarbouillé, M. , Klier, A. , and Rapoport, G. (1994) Interactions of wild‐type and truncated LevR of Bacillus subtilis with the upstream activating sequence of the levanase operon. J Mol Biol 241: 178–192. [DOI] [PubMed] [Google Scholar]
- Mundhada, H. , Seoane, J.M. , Schneider, K. , Koza, A. , Christensen, H.B. , Klein, T. , et al. (2017) Increased production of L‐serine in Escherichia coli through adaptive laboratory evolution. Metab Eng 39: 141–150. [DOI] [PubMed] [Google Scholar]
- Muntel, J. , Fromion, V. , Goelzer, A. , Maaß, S. , Mäder, U. , et al. (2014) Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis by data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MS(E)). Mol Cell Proteomics 13: 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, P. , Mäder, U. , Dervyn, E. , Rochat, T. , Leduc, A. , et al. (2012) Condition‐dependent transcriptome reveals high‐level regulatory architecture in Bacillus subtilis . Science 335: 1103–1106. [DOI] [PubMed] [Google Scholar]
- Niehaus, T.D. , Elbadawi, M. , de Crécy‐Lagard, V. , Fiehn, O. , and Hanson, A.D. (2017) Discovery of a widespread prokaryotic 5‐oxoprolinase that was hiding in plain sight. J Biol Chem 292: 16360–16367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus, T.D. , Folz, J. , McCarty, D.R. , Cooper, A.J.L. , Amador, D.M. , Fiehn, O. , and Hanson, A.D. (2018) Identification of a metabolic disposal route for the oncometabolite S‐(2‐succino)cysteine in Bacillus subtilis . J Biol Chem 293: 8255–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, W. , Kayahara, T. , Tsuda, M. , Mizushima, T. , and Tsuchiya, T. (1997) Isolation and characterization of an Escherichia coli mutant lacking the major serine transporter, and cloning of a serine transporter gene. J Biochem 122: 1241–1245. [DOI] [PubMed] [Google Scholar]
- Ogawa, W. , Kim, Y.M. , Mizushima, T. , and Tsuchiya, T. (1998) Cloning and expression of the gene for the Na+‐coupled serine transporter from Escherichia coli and characteristics of the transporter. J Bacteriol 180: 6749–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, P. , Aucouturier, A. , Delumeau, O. , and Auger, S. (2017) Revisiting the in vivo GlnR‐binding sites at the genome scale in Bacillus subtilis . BMC Res Notes 10: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuß, D.R. , Altenbuchner, J. , Mäder, U. , Rath, H. , Ischebeck, T. , Sappa, P.K. , et al. (2017) Large‐scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res 27: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuß, D.R. , Commichau, F.M. , Gundlach, J. , Zhu, B. , and Stülke, J. (2016) The blueprint of a minimal cell: MiniBacillus . Microbiol Mol Biol Rev 80: 955–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuß, D.R. , Faßhauer, P. , Mroch, P.J. , Ul‐Haq, I. , Koo, B.M. , Pöhlein, A. , et al. (2019) Topoisomerase IV can functionally replace all type 1A topoisomerases in Bacillus subtilis . Nucleic Acids Res 47: 5231–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov, D.A. , Vitreschak, A.G. , Mironov, A.A. , and Gelfand, M.S. (2003) Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res 31: 6748–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, J. , Müller, P. , Lentes, S. , Thiele, M.J. , Zeigler, D.R. , Tödter, D. , et al. (2016) ThrR, a DNA‐binding transcription factor involved in controlling threonine biosynthesis in Bacillus subtilis . Mol Microbiol 101: 879–893. [DOI] [PubMed] [Google Scholar]
- Sachla, A.J. , and Helmann, J.D. (2019) A bacterial checkpoint protein for ribosome assembly moonlights as an essential metabolite‐proofreading enzyme. Nat Commun 10: 1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schilling, O. , Frick, O. , Herzberg, C. , Ehrenreich, A. , Heinzle, E. , Wittmann, C. , and Stülke, J. (2007) Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Appl Environ Microbiol 73: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, F. , Ehrt, S. , and Hillen, W. (1997) Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J Bacteriol 179: 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebeck, F.P. , and Szostak, J.W. (2006) Ribosomal synthesis of dehydroalanine‐containing peptides. J Am Chem Soc 128: 7150–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska, A. , Robin, S. , Daudin, J.J. , Hénaut, A. , and Danchin, A. (2001) Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis . Genome Biol 2: RESEARCH0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstedt, M.T. , and Greer, S.B. (1973) Threonine synthetase of Bacillus subtilis . J Biol Chem 248: 1032–1044. [PubMed] [Google Scholar]
- Suárez, R.A. , Stülke, J. , and van Dijl, J.M. (2019) Less is more: towards a genome‐reduced Bacillus cell factory for “difficult proteins”. ACS Synth Biol 8: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A.R. , DeLoughery, A. , Bradshaw, N. , Chen, Y. , O'Shea, E. , et al. (2013) A serine sensor for multicellularity in a bacterium. eLife 2: 01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeyar, M.A. , and Zahler, S.A. (1986) Chromosomal insertions of Tn917 in Bacillus subtilis . J Bacteriol 167: 530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A. (1996) PCR‐synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae . Yeast 12: 259–265. [DOI] [PubMed] [Google Scholar]
- Wels, M. , Groot Kormelink, T. , Kleerebezem, M. , Siezen, R.J. , and Francke, C. (2008) An in silico analysis of T‐box regulated genes and T‐box evolution in prokaryotes, with emphasis on prediction of substrate specificity of transporters. BMC Genomics 9: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke, D. , Schulz, L.M. , Lentes, S. , Scholz, P. , Poehlein, A. , Gibhardt, J. , et al. (2019) Identification of the first glyphosate transporter by genomic adaptation. Environ Microbiol 21: 1287–1305. [DOI] [PubMed] [Google Scholar]
- Zaprasis, A. , Bleisteiner, M. , Kerres, A. , Hoffmann, T. , and Bremer, E. (2015) Uptake of amino acids and their metabolic conversion into the compatible solute proline confers osmoprotection to Bacillus subtilis . Appl Environ Microbiol 81: 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaprasis, A. , Hoffmann, T. , Stannek, L. , Gunka, K. , Commichau, F.M. , and Bremer, E. (2014) The γ‐aminobutyrate permease GabP serves as the third proline transporter of Bacillus subtilis . J Bacteriol 196: 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , and Newman, E. (2008) Deficiency in L‐serine deaminase results in abnormal growth and cell division of Escherichia coli K‐12. Mol Microbiol 69: 870–881. [DOI] [PubMed] [Google Scholar]
- Zhu, B. , and Stülke, J. (2018) SubtiWiki in 2018: from genes and proteins to functional network annotation of the model organism Bacillus subtilis . Nucleic Acids Res 46: D743–D748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Known and potential amino acid transporters in B. subtilis
Table S2. Bacillus subtilis strains used in this study.


