Abstract
Simple Summary
Babesia microti, the causative agent of human babesiosis, is an intraerythrocytic protozoan parasite, that circulates among small rodents and ixodid ticks in many countries worldwide. Zoonotic and non-zoonotic B. microti strains have been identified in rodent populations in Europe. Analyzing eight species of small rodents collected from different habitats (meadows, forests and their ecotones) in Lithuania, we checked for the presence of B. microti and found the highest infection prevalence to be in Microtus oeconomus and Microtus agrestis rodents. Of note, this study also detected the first reported cases of Babesia parasites in Micromys minutus mice. In term of habitat, the highest prevalence of Babesia parasites was detected in rodents trapped in meadows. Our results demonstrate that rodents, especially Microtus voles, can play an important role in the circulation of the zoonotic B. microti ‘Jena/Germany’ strain in Lithuania.
Abstract
Babesia microti (Aconoidasida: Piroplasmida) (Franca, 1910) is an emerging tick-borne parasite with rodents serving as the considered reservoir host. However, the distribution of B. microti in Europe is insufficiently characterized. Based on the sample of 1180 rodents from 19 study sites in Lithuania, the objectives of this study were: (1) to investigate the presence of Babesia parasites in eight species of rodents, (2) to determine the prevalence of Babesia parasites in rodents from different habitats, and (3) to characterize the detected Babesia strains using partial sequencing of the 18S rRNR gene. Babesia DNA was detected in 2.8% rodents. The highest prevalence of Babesia was found in Microtus oeconomus (14.5%) and Microtus agrestis (7.1%) followed by Clethrionomys glareolus (2.3%), Apodemus flavicollis (2.2%) and Micromys minutus (1.3%). In M. minutus, Babesia was identified for the first time. The prevalence of Babesia-infected rodents was higher in the meadow (5.67%) than in the ecotone (1.69%) and forest (0.31%) habitats. The sequence analysis of the partial 18S rRNA gene reveals that Babesia isolates derived from rodents were 99–100% identical to human pathogenic B. microti ‘Jena/Germany’ strain.
Keywords: 18S rRNA, Babesia, rodents, voles, mice, Lithuania
1. Introduction
Babesiae are emerging tick-borne protozoan parasites circulating in many countries worldwide in vertebrate hosts and vectors. The Babesia species including Babesia microti, Babesia divergens, B. divergens-like, Babesia venatorum and Babesia duncani are known to cause infection in humans. In Europe, Asia and North America respectively, the main vectors of zoonotic Babesia species are Ixodes ricinus, Ixodes persulcatus and Ixodes scapularis ticks. [1]. B. microti is the main causative agent of human babesiosis, especially in North America [2]. In Europe however, human babesiosis cases are less frequently reported and mostly related to B. divergens, B. divergens-like and B. venatorum [3]. However, a few cases of human babesiosis resulting from B. microti have also been reported in Europe [4,5,6]. To the best of our knowledge, no cases of human babesiosis have been documented in Lithuania.
The common vole (Microtis arvalis), field vole (Microtus agrestis) and root vole (Microtus oeconomus) are microtine rodents that play important roles in the circulation of B. microti in Europe [7,8]. B. microti infection was also detected in yellow-necked mouse (Apodemus flavicollis), striped field mouse (Apodemus agrarius), wood mouse (Apodemus sylvaticus) and bank vole (Clethrionomys glareolus), which are the main hosts for the immature stages of Ixodes ticks [7,9,10,11]. In general, Ixodes trianguliceps (with all three developmental stages feeding on rodents and does not bite humans) is the main vector of B. microti [12,13], while I. ricinus (with larvae and nymphs feeding on rodents) would only serve as a bridge vector of B. microti [9,14,15].
Molecular phylogenetic analysis demonstrated that B. microti consisting of genetically diverse isolates that belong to different clades [16]. B. microti isolates from rodents are subdivided within these clades into the non-zoonotic and zoonotic strains [17,18]. Different strains of B. microti have been reported in rodents in Slovenia, Croatia, Poland, Finland, Germany, Slovakia and France [7,9,12,19,20,21,22]. Various B. microti strains may circulate in rodent community at the same time [7]. However, distributions of B. microti strains in Europe are still insufficiently characterized.
The aims of the present study were: (1) to investigate the presence of Babesia parasites in eight species of Lithuanian rodents, (2) to determine the prevalence of Babesia parasites in rodents from meadows, forests and their ecotones, and (3) to characterize the detected B. microti strains using partial sequencing of 18S rRNR gene.
2. Materials and Methods
2.1. Study Sites
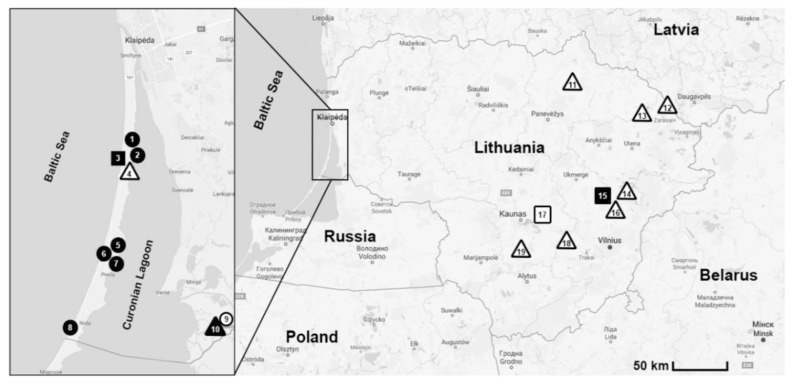
Rodents were trapped in 19 locations of different habitats in western (Curonian Spit; sites 1–8 and Nemunas River Delta; sites 9–10) and eastern (sites 11–19) parts of Lithuania during 2013–2017 (Figure 1). Rodents were captured in meadows, forests and their ecotones. Rodent sampling in the Curonian Spit was conducted in the coastal meadows (sites 1, 2, 5–8), mixed forests (site 4) and meadow-mixed forest ecotone (site 3). In the Nemunas River Delta, the trapping was conducted in two habitats: in a flooded meadow (site 9) and in a spring-flooded black alder stands forest (site 10).
Figure 1.
Rodent trapping sites in Lithuania, 2013–2017. ∆/▲—forests; ○/●—meadows; □/■—ecotones; ∆/○/□—rodents negative for Babesia parasites; ▲/●/■—rodents infected with Babesia parasites. Map was created using Open street map data (open source https://www.openstreetmap.org, accessed on 22 April 2020).
Eastern Lithuania was represented by different habitats—mixed forests (sites 11,13, 14, 16,19), mixed forest-meadow ecotones (sites 15, 17), and deciduous forests, one of them in peninsula of Lukštas lake (site 12) and the other on an island in an artificial water body, Elektrėnai Reservoir (site 18) (Table 1).
Table 1.
Prevalence of Babesia parasites in Lithuanian rodents, 2013–2017 (presented as n/N, %) 1.
| No | Habitat | A. flavicollis | A. agrarius | M. musculus | M. minutus | C. glareolus | M. oeconomus | M. agrestis | M. arvalis | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | coastal meadow | 4/59 (6.8) | 1/40 (2.5) | 1/9 | 1/4 | 0/1 | 7/113 (6.2) | |||
| 2 | coastal meadow | 2/2 | 2/2 | |||||||
| 3 | forest-meadow ecotone | 2/192 (1.1) | 0/2 | 0/36 | 1/2 | 0/1 | 3/233 (1.3) | |||
| 4 | mixed forest | 0/29 | 0/29 | |||||||
| 5 | coastal meadow | 0/33 | 0/26 | 1/5 | 0/2 | 1/66 (1.5) | ||||
| 6 | coastal meadow | 2/54 (3.7) | 0/1 | 1/3 | 8/18 (44.5) | 0/1 | 11/77 (14.3) | |||
| 7 | coastal meadow | 3/37 (8.1) | 0/4 | 0/8 | 0/2 | 3/51 (5.9) | ||||
| 8 | coastal meadow | 0/5 | 1/1 | 1/6 | ||||||
| 9 | flooded meadows | 0/52 | 0/3 | 0/19 | 0/40 | 0/12 | 0/126 | |||
| 10 | flooded forest | 0/5 | 0/7 | 0/1 | 1/14 (2.5) | 0/2 | 0/9 | 1/38 (2.6) | ||
| 11 | mixed forest | 0/1 | 0/13 | 0/14 | ||||||
| 12 | deciduous forest | 0/12 | 0/6 | 0/52 | 0/70 | |||||
| 13 | mixed forest | 0/3 | 0/7 | 0/10 | ||||||
| 14 | mixed forest | 0/3 | 0/11 | 0/14 | ||||||
| 15 | forest-meadow ecotone | 0/20 | 4/117 (3.5) | 4/137 (2.9) | ||||||
| 16 | mixed forest | 0/6 | 0/14 | 0/20 | ||||||
| 17 | forest-meadow ecotone | 0/17 | 0/10 | 0/12 | 0/4 | 0/43 | ||||
| 18 | deciduous forest | 0/2 | 0/7 | 0/88 | 0/97 | |||||
| 19 | mixed forest | 0/22 | 0/12 | 0/34 | ||||||
| Total | 11/499 (2.2) | 0/82 | 0/12 | 1/77 (1.3) | 9/396 (2.3) | 10/69 (14.5) | 2/28 (7.1) | 0/17 | 33/1180 (2.8) |
1 n, number of individuals infected; N, number of individuals tested; No, site number.
2.2. Rodent Trapping
Rodents were trapped by using live or snap traps baited with bread immersed in unrefined sunflower oil. One trapping session consist of three days. The traps were checked two times per day [23]. All rodents were identified to species level and gender morphologically and under dissection, with specimens of Microtus voles identified by their teeth [24].
2.3. Molecular Analyses
DNA from rodent spleen was extracted using Genomic DNA Purification Kit (Thermo Fisher Scientific, Vilnius, Lithuania), according to the manufacturer’s protocol. The presence of Babesia pathogens were conducted through the amplification of the 330 bp fragment of the 18S rRNA gene in nested PCR using two primer sets BS1/BS2 and PiroA/PiroC as described by Rar et al. [25,26]. The primary PCR reaction was carried out in a 20 μL final volume containing: 1 × PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs, 10 pmol of each primer, 2 U Taq DNA polymerase (Thermo Fisher Scientific, Lithuania), double-distilled water and 2 μL of DNA template. Reaction was performed according to the conditions: initial denaturation at 94 °C for 3 min, 35 cycles: denaturation at 94 °C for 60 s, annealing at 58 °C for 60 s and extension at 72 °C for 90 s, and final extension step at 72 °C for 3 min. In the second PCR, the reaction mix was similarly prepared as it was in the first step, with exception that instead of the DNA, 1 μL of the PCR product was added. The PCR conditions were: initial denaturation at 94 °C for 3 min, followed by 35 cycles: denaturation at 94 °C for 60 s, annealing at 64 °C for 60 s, and extension at 72 °C for 90 s. The final extending was at 72 °C for 3 min. In each PCR run negative (double-distilled water) and positive (DNA of Babesia positive ticks, infection confirmed by sequencing) controls were used. The PCR products were analyzed by horizontal electrophoresis in 1.5% agarose gel and visualized with ethidium bromide solution (20 ng/μL) using ultra-violet transilluminator UVP GelDoc-It 310 model (Ultra-Violet Products Ltd., Cambridge, UK). The good quality PCR products of Babesia-positive samples were extracted from agarose gel. GenJet PCR purification kit (Thermo Fisher Scientific, Lithuania) was used for purification and, after preparation, samples were sent for direct sequencing by Sanger method to Macrogen Europe company (Amsterdam, The Netherlands).
The partial 18S rRNA sequences were analyzed using MEGA X software package, version 10.0.5. [27] and compared with the sequence data available in NCBI GenBank database using the NCBI BLAST® software (http://blast.ncbi.nlm.nih.gov, accessed on 6 June 2020). A phylogenetic tree was constructed by applying maximum-likelihood (ML) method implemented with Tamura-Nei model. Partial 18S rRNA sequences for representative samples were submitted to GenBank under the accession numbers: MT745579 to MT745583.
2.4. Statistical Analysis
The between-species and between-location differences in the prevalence of Babesia infection were tested. For these differences, we used Fisher’s exact test and the Mantel–Haenszel common odds ratio estimate. Calculations were performed in SPSS software version 22 (IBM SPSS, Chicago, IL, USA), using 95% confidence intervals. We assessed the prevalence of Babesia in all investigated rodent species; calculations were performed in OpenEpi software [28]. 95% CI for prevalence was calculated according to the Wilson method [29]. We tested the significance of differences in the prevalence between species and between habitats. These calculations were performed in WinPepi, ver. 11.39. We used the chi-squared test with Upton’s approximation for small and medium sample sizes. To express the effect size, we used adjusted Cohen’s w [30]. In all tests, p < 0.05 was considered significant.
3. Results
We analyzed 1180 rodent individuals, belonging to eight species, best represented by A. flavicollis and C. glareolus (Table 1). Babesia infected rodents were trapped in nine out of 19 sampling locations (Figure 1). A. flavicollis was the dominant trapped rodent species in Curonian Spit (70.9%; 409/577) with the prevalence of infection ranging in four locations (where the infected rodents were captured) from 1.1% to 8.1%. Babesia infected C. glareolus were found in six of the fifteen sampling locations: with the overall prevalence of infection estimated at 6.5% on the Curonian Spit, 3% in the Nemunas River Delta and 1.3% in the eastern part of the country. Babesia infected M. oeconomus and M. agrestis were found in three and one sampling locations in the Curonian Spit, respectively. One Babesia infected harvest mouse (Micromys minutus) specimen was found in one location in the Curonian Spit (site 1) (Table 1). All trapped house mice (Mus musculus), A. agrarius and M. arvalis were not infected.
3.1. Prevalence of Babesia Parasites in Various Rodent Species
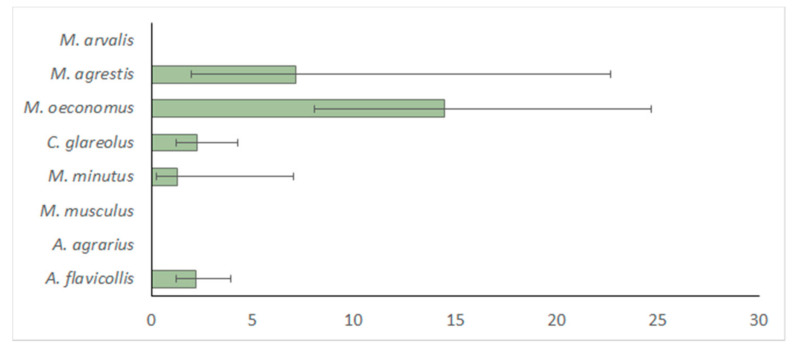
A total of 33 (2.8%, CI = 1.74–4.23%) out of 1180 DNA samples of rodents were positive for Babesia DNA. The species-based differences of prevalence of Babesia parasites were significant (14.5%; OR, 3.6; 95% CI, 1.330–9.625; p < 0.012) and are presented in Figure 2. The highest prevalence of Babesia was found in M. oeconomus and M. agrestis (14.5% vs. 7.1%, χ2 = 0.98, NS; Cohen’s w = 0.101, small effect size). The prevalence in M. oeconomus was significantly higher than that in C. glareolus (χ2 = 23.1, w = 0.221), A. flavicollis (χ2 = 25.7; w = 0.213) and M. minutus (χ2 = 9.0; w = 0.250). All differences are significant at p < 0.001, effect sizes medium.
Figure 2.
Prevalence (in %, bars represent 95% CI) of Babesia in the eight rodent species, irrespective to the habitat.
3.2. Habitat-Based Differences
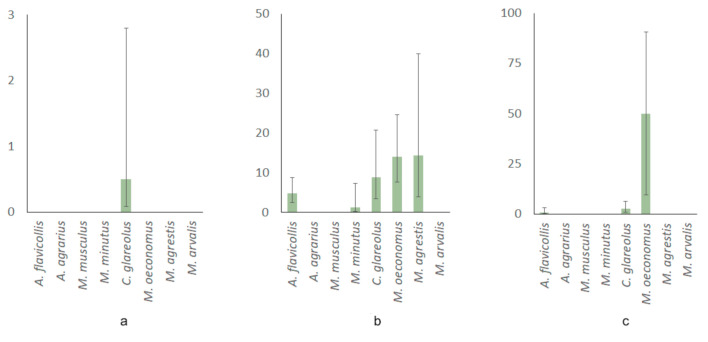
In general, the highest prevalence of Babesia parasites was characteristic to rodents, trapped in meadows (5.67%, CI = 3.87–8.23%), exceeding that in forests (0.31%, CI = 0.05–1.72%), with intermediate prevalence values observed in ecotones (1.69%, CI = 0.82–3.46%). The differences between meadow-forest (χ2 = 16.4, p < 0.001) and meadow-ecotone (χ2 = 9.3, p = 0.002) were highly significant with medium effect size, while that of forest-ecotone (χ2 = 3.3, p = 0.07) had only a trend without effect (w = 0.067).
In the forests, only C. glareolus was infected by Babesia with low prevalence (Figure 3a). In the meadows, minimum observed prevalence of Babesia in M. minutus (Figure 3b) was significantly exceeded by prevalence in M. oeconomus (χ2 = 8.2, p < 0.01; w = 0.245, medium effect size), M. agrestis (χ2 = 5.9, p = 0.015; w = 0.261, medium effect) and C. glareolus (χ2 = 3.9, p < 0.05; w = 0.262, small effect size). Other differences of Babesia prevalence between rodent species in meadows were not significant. In the forest-meadow ecotone, prevalence of Babesia in M. oeconomus was higher than in other species, despite minimum sample size (Figure 3c).
Figure 3.
Habitat-based differences of prevalence (in %, bars represent 95% CI) of Babesia in the eight rodent species: (a) forests, (b) meadows, (c) ecotones.
3.3. Molecular Characterization of Babesia Isolates
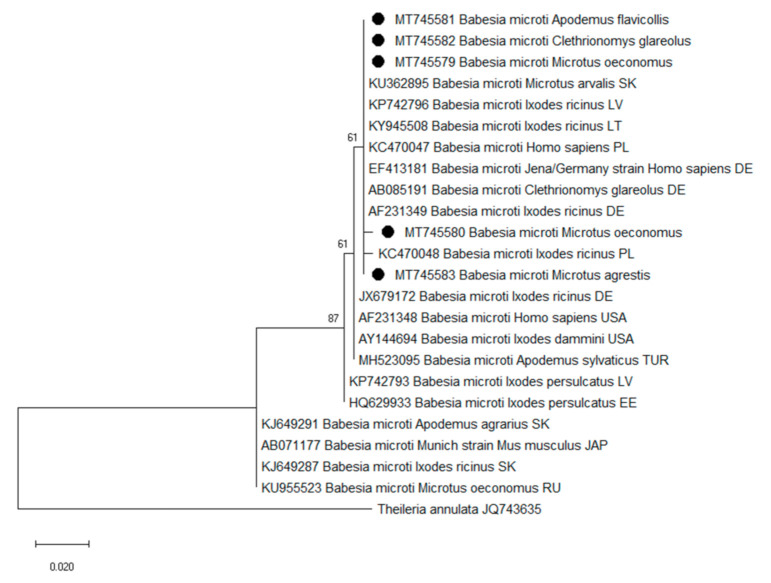
A total of 19 18S rRNA sequences derived from four rodent species A. flavicollis (n = 7), C. glareolus (n = 3), M. oeconomus (n = 7) and M. agrestis (n = 2) were analyzed. The sequence analysis of the partial 18S rRNA gene revealed that Babesia isolates derived from rodents were 99–100% identical to B. microti ‘Jena/Germany’ strain (GenBank: KC470047; EF413181). Two genotypes with one nucleotide difference were detected in M. oeconomus (Figure 4).
Figure 4.
Phylogenetic tree of the partial 18S rRNA gene of Babesia microti inferred by ML method, the Tamura–Nei model and bootstrap analysis of 1000 replicates. Marked with dark circle are samples sequenced in the present study. Sequences MT745579 and MT745581 are representative of six and five other samples obtained in this study (from A. flavicollis and M. oeconomus), respectively. Sequences MT745582 and MT745583 are representative of two and one other samples sequenced in the present study (from C. glareolus and M. agrestis), respectively.
4. Discussion
In this study, Babesia DNA was detected in 33 of 1180 (2.8%) spleen tissue samples of five small rodent species. The overall prevalence of Babesia varied among rodent species with the highest prevalence detected in voles M. oeconomus (14.5%) and M. agrestis (7.1%) (Table 1). Babesia infected M. oeconomus and M. agrestis have been found in north-eastern Poland with the 39.5% (30/76) and 17.7% (3/17) prevalence of infection, respectively [8]. These figures are almost three times higher compered than that obtained in this study. A high prevalence of Babesia spp. in M. agrestis has been reported in Austria 30.4% (14/46) [31] and the United Kingdom 27.9% (671/2402) [14], while Šebek [32] reported a much lower 0.5% (1/218) prevalence of infection in this rodent species in the former Czechoslovakia. The low overall prevalence of Babesia in this study was detected in C. glareolus (2.3%). In other European countries, the prevalence of Babesia infection in C. glareolus varied: 39.7% (60/151) reported in Finland [21], 15.9% (60/151) in Slovenia [12], 11.9% (59/495) in north-eastern Poland [33], 6.1% (3/49) in Croatia [19,34], 6% (25/405) in the Netherlands [11], 0.8% (4/498) in Slovakia [7,35], 0.68% (1/147) in France [22], 0.03% (11/396) in Germany [9].
Babesia parasites were found with low prevalence in mice A. flavicollis (2.2%) and M. minutus (1.3%) irrespective of the study sites. In line with our results, a low prevalence of Babesia in A. flavicollis has been reported in Slovakia (1.7%; 12/706; [7,35]) and in Germany (0.01%; 1/178; [9]). However, higher Babesia infection rate detected in A. flavicollis was documented in Croatia 16.9% (11/65) [19,34], in north-eastern Poland 13.1% (8/61) [20] and in Slovenia 11.8% (15/127) [12]. To the best of our knowledge, our study is the first report of Babesia infection in M. minutus.
Although, Babesia parasites were not found in A. agrarius and M. arvalis in this study, it was detected in these rodent species trapped in other locations in Lithuania, with a prevalence of 2.1% and 9.1%, respectively [36].
In the present study, a significantly higher overall prevalence of Babesia among investigated areas was found on the Curonian Spit (4.9%, 28/577; OR, 3.3; 95% CI, 2.346–4.720; p < 0.000) with the highest prevalence of infection (among all locations examined) detected in coastal meadow, site 6 (14.3%, OR, 0.35; 95% CI, 0.173–0.708; p < 0.004). In line with this, five various rodent species have been found positive with Babesia in seven out of eight locations on the Curonian Spit (Table 1). The detected differences in the prevalence of Babesia parasites in the investigated locations might be explained by habitat factors: the highest infection prevalence was detected in the coastal meadows habitat (which on the Curonian Spit most frequent), and additionally by the fact that examined rodents trapped in the Curonian Spit were frequently infested with immature I. ricinus (mostly larvae) [37]. The overall prevalence of infestation with immature I. ricinus varied between rodent hosts and was highest for A. flavicollis (56%). The mean intensity of infestation with I. ricinus was 5.2 per rodent hosts (5.6 in A. flavicollis, 3.3 in M. minutus, 3.0 in M. oeconomus, 3.0 in M. arvalis and 2.0 in C. glareolus) (personal authors data).
Worldwide, seven B. microti strains—‘USA’, ‘Hobetsu’ (‘Otsu’), ‘Nagano’, ‘Kobe’, ‘Jena/Germany’, ‘Munich’ and ‘Baltic’ were identified [18,20,38,39,40]. In Europe, from those, four B. microti strains have been detected: the zoonotic ‘Jena/Germany’ and ‘USA’ strains, and non-zoonotic ‘Munich’ strain reported in Ixodes ticks and rodents [7,8,9,20,22,33] and the ‘Baltic’ strain detected in I. persulcatus collected from Estonia and Latvia [39,41] which pathogenicity for human is not known.
The zoonotic ‘Jena/Germany’ strain has been detected in A. flavicollis, A. agrarius, M. arvalis and C. glareolus from Slovakia [7,35], in M. oeconomus, M. arvalis and M. agrestis from Poland [8], in A. flavicollis, C. glareolus and M. arvalis from Germany [9] and in A. flavicollis and C. glareolus from central Croatia [19], while zoonotic B. microti ‘USA’ strain was detected in microtine rodents from north-eastern Poland [33]. In this study, the B. microti ‘Jena/Germany’ strains were detected in A. flavicollis mice and three voles species—M. oeconomus, M. agrestis and C. glareolus. In previous studies, the zoonotic ‘Jena/Germany’ strains were detected in I. ricinus ticks in Europe, including Baltic countries [39,41,42]. Autochthonous cases of human babesiosis due to the B. microti ‘Jena/Germany’ strain have been reported in Germany [4] and Poland [5]. Worldwide, most of the human babesiosis cases have been related to the zoonotic B. microti ‘USA’ strain. A lower virulence of European B. microti strains compared to those circulating in North America may be the reason of a lack of recognized human cases associated with European B. microti strains, despite human exposure to infectious tick bites in this continent. [43].
The non-zoonotic B. microti ‘Munich’ strain has been found in C. glareolus from Slovakia [7], in A. flavicollis and C. glareolus from central Croatia [19] and in C. glareolus from Finland [21] and France [22]. As a general rule, the B. microti ‘Munich’ strain was not found outside of the I. trianguliceps distribution area (from Great Britain to Baikal). It is thought that I. trianguliceps ticks play an important role for the maintenance of the non-zoonotic B. microti ‘Munich’ strain to mammalian hosts [17]. In Lithuania, I. trianguliceps ticks are present and previously were found on A. flavicollis and C. glareolus rodents [44]. However, in the present study, trapped rodents were infested only with immature I. ricinus.
5. Conclusions
Our findings suggest that rodents, especially Microtus voles, play an important role in the circulation of the zoonotic B. microti ‘Jena/Germany’ strain in Lithuania. The highest prevalence of Babesia parasites was detected in rodents trapped in coastal meadows. This study also detected Babesia infection in M. minutus, the first recorded infection in this species to the best of the authors’ knowledge.
Acknowledgments
The authors are grateful to Vaclovas Gedminas for collection of rodents on the Curonian Spit.
Author Contributions
Conceptualization, A.P., J.R. and D.M.-B.; methodology, L.B., J.R. and D.M.-B.; software, L.B., J.R. and D.M.-B.; formal analysis, L.B., J.R. and D.M.-B.; investigation, J.R. and D.M.-B.; writing—original draft preparation, A.P., J.R., L.B. and D.M.-B.; writing—review and editing, A.P., J.R., L.B. and D.M.-B.; funding acquisition, A.P. and J.R. All authors have read and agreed to the published version of the manuscript.
Funding
Part of this research is funded by the European Social Fund under the Nr. 09.3.3-LMT-K-712-09-0270 “Development of Competences of Scientists, other Researchers and Students through Practical Research Activities” measure.
Institutional Review Board Statement
Rodent sampling was conducted with permission from the Environmental Protection Agency (EPA) and approved by the Ministry of Environment of the Republic of Lithuania, licenses (No. 1 (10 April 2013), No. 15 (31 March 2014), No. 22 (10 April 2015), No. 12 (30 March 2016) and No. 13 (22 March 2017)) in accordance with Lithuanian (the Republic of Lithuania Law on Welfare and Protection of Animals No. XI-2271) and European legislation (Directive 2010/63/EU) on the protection of animals.
Data Availability Statement
Partial 18S rRNA sequences for representative samples were submitted to GenBank under the accession numbers: MT745579 to MT745583.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Young K.M., Corrin T., Wilhelm B., Uhland C., Greig J., Mascarenhas M., Waddell L.A. Zoonotic Babesia: A scoping review of the global evidence. PLoS ONE. 2019;14:e0226781. doi: 10.1371/journal.pone.0226781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westblade L.F., Simon M.S., Mathison B.A., Kirkman L.A. Babesia microti: From mice to ticks to an increasing number of highly susceptible humans. J. Clin. Microbiol. 2017;55:2903–2912. doi: 10.1128/JCM.00504-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildebrandt A., Gray J.S., Hunfeld K.P. Human Babesiosis in Europe: What clinicians need to know. Infection. 2013;41:1057–1072. doi: 10.1007/s15010-013-0526-8. [DOI] [PubMed] [Google Scholar]
- 4.Hildebrandt A., Hunfeld K.P., Baier M., Krumbholz A., Sachse S., Lorenzen T., Kiehntopf M., Fricke H.-J., Straube E. First confirmed autochthonous case of human Babesia microti infection in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 2007;26:595–601. doi: 10.1007/s10096-007-0333-1. [DOI] [PubMed] [Google Scholar]
- 5.Welc-Falęciak R., Pawełczyk A., Radkowski M., Pancewicz S.A., Zajkowska J., Siński E. First report of two asymptomatic cases of human infection with Babesia microti (Franca, 1910) in Poland. Ann. Agric. Environ. Med. 2015;22:51–54. doi: 10.5604/12321966.1141394. [DOI] [PubMed] [Google Scholar]
- 6.Moniuszko-Malinowska A., Swiecicka I., Dunaj J., Zajkowska J., Czupryna P., Zambrowski G., Czupryna P., Garkowski A. Infection with Babesia microti in humans with non-specific symptoms in North East Poland. Infect. Dis. 2016;48:537–543. doi: 10.3109/23744235.2016.1164339. [DOI] [PubMed] [Google Scholar]
- 7.Blaňarová L., Stanko M., Miklisová D., Víchová B., Mošanský L., Kraljik J., Bona M., Derdáková M. Presence of Candidatus Neoehrlichia mikurensis and Babesia microti in rodents and two tick species (Ixodes ricinus and Ixodes trianguliceps) in Slovakia. Ticks Tick Borne Dis. 2016;7:319–326. doi: 10.1016/j.ttbdis.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Tołkacz K., Bednarska M., Alsarraf M., Dwużnik D., Grzybek M., Welc-Falęciak R., Behnke J.M., Bajer A. Prevalence, genetic identity and vertical transmission of Babesia microti in three naturally infected species of vole, Microtus spp. (Cricetidae) Parasites Vectors. 2017;10:66. doi: 10.1186/s13071-017-2007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obiegala A., Pfeffer M., Pfister K., Karnath C., Silaghi C. Molecular examinations of Babesia microti in rodents and rodent-attached ticks from urban and sylvatic habitats in Germany. Ticks Tick Borne Dis. 2015;6:445–449. doi: 10.1016/j.ttbdis.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Krawczyk A.I., van Duijvendijk G.L.A., Swart A., Heylen D., Jaarsma R.I., Jacobs F.H.H., Fonville M., Sprong H., Takken W. Effect of rodent density on tick and tick-borne pathogen populations: Consequences for infectious disease risk. Parasites Vectors. 2020;13:34. doi: 10.1186/s13071-020-3902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azagi T., Jaarsma R.I., Docters van Leeuwen A., Fonville M., Maas M., Franssen F.F.J., Kik M., Rijks J.M., Montizaan M.G., Groenevelt M., et al. Circulation of Babesia Species and Their Exposure to Humans through Ixodes ricinus. Pathogens. 2021;10:386. doi: 10.3390/pathogens10040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duh D., Petrovec M., Trilar T., Avsic-Zupanc T. The molecular evidence of Babesia microti infection in small mammals collected in Slovenia. Parasitology. 2003;126:113–117. doi: 10.1017/S0031182002002743. [DOI] [PubMed] [Google Scholar]
- 13.Nefedova V.V., Korenberg E.I., Kovalevskii Y.V., Samokhvalov M.V., Gorelova N.B. The role of Ixodes trianguliceps tick larvae in circulation of Babesia microti in the Middle Urals. Entomol. Rev. 2013;93:258–266. doi: 10.1134/S0013873813020152. [DOI] [Google Scholar]
- 14.Bown K.J., Lambin X., Telford G.R., Ogden N., Telfer S., Woldehiwet Z., Birtles R. Relative importance of Ixodes ricinus and Ixodes trianguliceps as vectors for Anaplasma phagocytophilum and Babesia microti in field vole (Microtus agrestis) populations. Appl. Environ. Microbiol. 2008;74:7118–7125. doi: 10.1128/AEM.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cayol C., Jääskeläinen A., Koskela E., Kyröläinen S., Mappes T., Siukkola A., Kallio E.R. Sympatric Ixodes-tick species: Pattern of distribution and pathogen transmission within wild rodent populations. Sci. Rep. 2018;8:16660. doi: 10.1038/s41598-018-35031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baneth G., Florin-Christensen M., Cardoso L., Schnittger L. Reclassification of Theileria annae as Babesia vulpes sp. nov. Parasites Vectors. 2015;8:207. doi: 10.1186/s13071-015-0830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rar V., Yakimenko V., Makenov M., Tikunov A., Epikhina T., Tancev A., Bobrova O., Tikunova N. High prevalence of Babesia microti ‘Munich’ type in small mammals from an Ixodes persulcatus/Ixodes trianguliceps sympatric area in the Omsk region, Russia. Parasitol. Res. 2016;115:3619–3629. doi: 10.1007/s00436-016-5128-9. [DOI] [PubMed] [Google Scholar]
- 18.Usluca S., Celebi B., Karasartova D., Gureser A.S., Matur F., Oktem M.A., Sozen M., Karatas A., Babur C., Mumcuoglu K.Y., et al. Molecular Survey of Babesia microti (Aconoidasida: Piroplasmida) in Wild Rodents in Turkey. J. Med. Entomol. 2019;56:1605–1609. doi: 10.1093/jme/tjz084. [DOI] [PubMed] [Google Scholar]
- 19.Beck R., Vojta L., Ćurković S., Mrljak V., Margaletić J., Habrun B. Molecular survey of Babesia microti in wild rodents in central Croatia. Vector Borne Zoonotic Dis. 2011;11:81–83. doi: 10.1089/vbz.2009.0260. [DOI] [PubMed] [Google Scholar]
- 20.Welc-Falęciak R., Bajer A., Paziewska-Harris A., Baumann-Popczyk A., Siński E. Diversity of Babesia in Ixodes ricinus ticks in Poland. Adv. Med. Sci. 2012;57:364–369. doi: 10.2478/v10039-012-0023-9. [DOI] [PubMed] [Google Scholar]
- 21.Kallio E.R., Begon M., Birtles R.J., Bown K.J., Koskela E., Mappes T., Watts P.C. First report of Anaplasma phagocytophilum and Babesia microti in rodents in Finland. Vector Borne Zoonotic Dis. 2014;14:389–393. doi: 10.1089/vbz.2013.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jouglin M., Perez G., Butet A., Malandrin L., Bastian S. Low prevalence of zoonotic Babesia in small mammals and Ixodes ricinus in Brittany, France. Vet. Parasitol. 2017;238:58–60. doi: 10.1016/j.vetpar.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Balčiauskas L. Methods of Investigation of Terrestrial Ecosystems. Part I. Animal Surveys. Vilnius University; Vilnius, Lithuania: 2004. p. 183. [Google Scholar]
- 24.Prūsaitė J., Mažeikytė R., Pauža D., Paužienė N., Baleišis R., Juškaitis R., Mickus A., Grušas A., Skeiveris R., Bluzma P.M. Fauna of Lithuania. Mammals. Mokslas; Vilnius, Lithuania: 1988. p. 295. [Google Scholar]
- 25.Rar V.A., Fomenko N.V., Dobrotvorsky A.K., Livanova N.N., Rudakova S.A., Fedorov E.G., Astanin V.B., Morozova O.V. Tick-borne pathogen detection, western Siberia, Russia. Emerg. Infect. Dis. 2005;11:1708. doi: 10.3201/eid1111.041195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rar V.A., Epikhina T.I., Livanova N.N., Panov V.V. Genetic diversity of Babesia in Ixodes persulcatus and small mammals from North Ural and West Siberia, Russia. Parasitology. 2011;138:175–182. doi: 10.1017/S0031182010001162. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown L.D., Cat T.T., DasGupta A. Interval Estimation for a Proportion. Stat. Sci. 2001;16:101–133. doi: 10.1214/ss/1009213286. [DOI] [Google Scholar]
- 29.Abramson J.H. WINPEPI Updated: Computer Programs for Epidemiologists, and their Teaching Potential. Epidemiol. Perspect. Innov. 2011;8:1. doi: 10.1186/1742-5573-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas J.R., Salazar W., Landers D.M. What is Missing in p <0.05? Effect Size. Res. Q. Exerc. Sport. 1991;62:344–348. doi: 10.1080/02701367.1991.10608733. [DOI] [PubMed] [Google Scholar]
- 31.Šebek Z., Sixl W., Stünzner D., Valová M., Hubálek Z., Troger H. Zur Kenntnis der Blutparasiten wildlebender Kleinsäuger in der Steiermark und im Burgenland [Blood parasites of small wild mammals in Steiermark and Burgenland] Folia Parasitol. 1980;27:295–301. (In German) [PubMed] [Google Scholar]
- 32.Šebek Z. Blutparasiten der wildlebenden Kleinsäuger in der Tschechoslowakei [Blood parasites of small wild mammals in Czechoslovakia] Folia Parasitol. 1975;22:11–20. (In German) [PubMed] [Google Scholar]
- 33.Welc-Falęciak R., Bajer A., Behnke J.M., Siński E. Effects of host diversity and the community composition of hard ticks (Ixodidae) on Babesia microti infection. Int. J. Med. Microbiol. 2008;298:235–242. doi: 10.1016/j.ijmm.2007.12.002. [DOI] [Google Scholar]
- 34.Tadin A., Turk N., Korva M., Margaletić J., Beck R., Vucelja M., Habuš J., Svoboda P., Županc T.A., Henttonen H., et al. Multiple co-infections of rodents with hantaviruses, Leptospira, and Babesia in Croatia. Vector Borne Zoonotic Dis. 2012;12:388–392. doi: 10.1089/vbz.2011.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamšíková Z., Kazimírová M., Haruštiaková D., Mahríková L., Slovák M., Berthová L., Kocianová E., Schnittger L. Babesia spp. in ticks and wildlife in different habitat types of Slovakia. Parasites Vectors. 2016;9:292. doi: 10.1186/s13071-016-1560-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baltrūnaitė L., Kitrytė N., Križanauskienė A. Blood parasites (Babesia, Hepatozoon and Trypanosoma) of rodents, Lithuania: Part I. Molecular and traditional microscopy approach. Parasitol. Res. 2020;119:687–694. doi: 10.1007/s00436-019-06577-3. [DOI] [PubMed] [Google Scholar]
- 37.Radzijevskaja J., Kaminskienė E., Lipatova I., Mardosaitė-Busaitienė D., Balčiauskas L., Stanko M., Paulauskas A. Prevalence and diversity of Rickettsia species in ectoparasites collected from small rodents in Lithuania. Parasites Vectors. 2018;11:375. doi: 10.1186/s13071-018-2947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito-Ito A., Kawai A., Ohmori S., Nagano-Fujii M. Continuous in vivo culture and indirect fluorescent antibody test for zoonotic protozoa of Babesia microti. Parasitol. Int. 2016;65:526–531. doi: 10.1016/j.parint.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Capligina V., Berzina I., Bormane A., Salmane I., Vilks K., Kazarina A., Bandere D., Baumanis V., Ranka R. Prevalence and phylogenetic analysis of Babesia spp. in Ixodes ricinus and Ixodes persulcatus ticks in Latvia. Exp. Appl. Acarol. 2016;68:325–336. doi: 10.1007/s10493-015-9978-0. [DOI] [PubMed] [Google Scholar]
- 40.Wei C.Y., Wang X.M., Wang Z.S., Wang Z.H., Guan Z.Z., Zhang L.H., Dou X.F., Wang H. High prevalence of Babesia microti in small mammals in Beijing. Infect. Dis. Poverty. 2020;9:1–11. doi: 10.1186/s40249-020-00775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katargina O., Geller J., Vasilenko V., Kuznetsova T., Järvekülg L., Vene S., Lundkvist Å., Golovljova I. Detection and characterization of Babesia species in Ixodes ticks in Estonia. Vector-Borne Zoonotic Dis. 2011;11:923–928. doi: 10.1089/vbz.2010.0199. [DOI] [PubMed] [Google Scholar]
- 42.Radzijevskaja J., Mardosaitė-Busaitienė D., Aleksandravičienė A., Paulauskas A. Investigation of Babesia spp. in sympatric populations of Dermacentor reticulatus and Ixodes ricinus ticks in Lithuania and Latvia. Ticks Tick Borne Dis. 2018;9:270–274. doi: 10.1016/j.ttbdis.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Rudolf I., Golovchenko M., Sikutová S., Rudenko N. Babesia microti (Piroplasmida: Babesiidae) in nymphal Ixodes ricinus (Acari: Ixodidae) in the Czech Republic. Folia Parasitol. 2005;52:274. doi: 10.14411/fp.2005.036. [DOI] [PubMed] [Google Scholar]
- 44.Paulauskas A., Radzijevskaja J., Turčinavičienė J., Ambrasienė D., Galdikaitė E. Data on some Ixodid tick species (Acari, Ixodidae) in the Baltic countries. New Rare Lith. Insect Species. 2010;22:43–51. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Partial 18S rRNA sequences for representative samples were submitted to GenBank under the accession numbers: MT745579 to MT745583.