Abstract
Oral squamous cell carcinoma (OSCC) is a multifactorial malignancy, and its high incidence and mortality rate remain a global public health burden. Polymorphisms in the long intergenic noncoding RNA 673 (LINC00673) have been currently connected to the predisposition to various cancer types. The present study attempted to explore the impact of LINC00673 gene polymorphisms on the risk and progression of OSCC. Three LINC00673 single-nucleotide polymorphisms (SNPs), including rs11655237, rs9914618, and rs6501551, were evaluated in 1231 OSCCC cases and 1194 cancer-free controls. We did not observe any significant association of three individual SNPs with the risk of OSCC between the case and control group. However, while assessing the clinicopathological parameters, patients carrying at least one minor allele of rs9914618 (GA and AA; OR, 1.286; 95% CI, 1.008–1.642; p = 0.043) were found to develop lymph node metastasis more often compared to those who are homozygous for the major allele. Further stratification analyses revealed that this genetic correlation with increased risk of lymphatic spread was further fortified in habitual betel quid chewers (OR, 1.534; 95% CI, 1.160–2.028; p = 0.003) or smokers (OR, 1.320; 95% CI, 1.013–1.721; p = 0.040). Moreover, through analyzing the dataset from The Cancer Genome Atlas (TCGA), we found that elevated LINC00673 levels were associated with the development of large tumors in patients with head and neck squamous cell carcinoma and the risk of lymphatic spread in smokers. These data demonstrate a joint effect of LINC00673 rs9914618 with betel nut chewing or smoking on the progression of oral cancer.
Keywords: oral squamous cell carcinoma, long noncoding RNA, single-nucleotide polymorphism, lymph node metastasis
1. Introduction
Oral squamous cell carcinoma (OSCC), representing nearly 90% of oral cancer [1], is a neoplasm of multifactorial nature. Various environmental risks intertwine with inherited parameters in the susceptibility to this malignancy [2]. Well-known external risks of OSCC, including human papillomavirus (HPV) infection [3] and habitual use of cigarettes, alcohol, and betel quid [4], have been reported. Moreover, oral carcinogenesis is found to be mediated by a set of genetic alterations that affect programed cell death, cell division, and DNA repair [5]. Nevertheless, the incidence [1] and mortality [6] of OSCC have not improved remarkably in spite of the advance in surgery and other treatment options [7,8] as well as the recognition of risk factors that cause oral malignancies mentioned above [3,4,5,9]. Considering the high heterogeneity and pathogenicity of OSCC, all these cancer risks appear to be mingled and required to evaluate the disease prognosis.
Current advances in sequencing technologies have led to a paradigm shift in our understanding of the possible functions of the noncoding transcriptome [10], in the majority with the focus on the identification of an expanding class of long noncoding RNAs (lncRNAs). To date, an increasing number of lncRNAs is documented to be causally implicated in a variety of human disorders [11], including cancer. Among them, long intergenic noncoding RNA 673 (LINC00673), also referred to as SLNCR or SLNCR1, was discovered to be differentially expressed in many tumor types and to promote cancer development and progression through distinct molecular mechanisms [12,13,14,15,16,17,18,19,20,21,22]. Specifically, in spite of being noncoding, transcripts of LINC00673 can modulate chromatin dynamics and cancer-causing gene expression by serving as a modular scaffold to interact with lysine-specific demethylase 1 (LSD1) and enhancer of zeste homolog 2 (EZH2) [14,16], recruiting androgen receptor (AR) and brain-specific homeobox protein 3a (Brn3a) to facilitate the expression of metalloproteinase 9 (MMP9) [13,21], stabilizing phosphorylation of dishevelled (Dvl) to activate WNT/β-catenin signaling [15], and acting as a competing endogenous RNA (ceRNA) for miR-150-5p [17], miR-515-5p [20], and miR-1231 [12]. These findings collectively indicate that LINC00673, behaving as a scaffold, decoy, or signal, can influence cancer biology through genomic targeting, transcriptional regulation, epigenetic mechanisms and antisense interference.
In recent years, genome-wide or targeted gene association studies have revealed a connection of LINC00673 gene polymorphisms with the risk of many tumor types, such as cancers of the pancreas [12,23], nerve tissues [24,25], stomach [26], cervix [27], and liver [28]. Yet, the impact of LINC00673 gene polymorphisms on the risk of OSCC remains unclear. Here, we performed a case–control study to investigate the effects of LINC00673 single nucleotide polymorphisms (SNPs) on the predisposition to oral malignancies.
2. Materials and Methods
2.1. Subjects
This study encompassed 1231 male cases with OSCC and 1194 cancer-free males, with the approval by the institutional review board of Chung Shan Medical University Hospital in Taichung, Taiwan. Subjects, accrued from 2008 to 2020, provided informed written consent at enrollment. Clinical staging and tumor differentiation of OSCC was determined according to the TNM staging system of the American Joint Committee on Cancer (AJCC) [29] while the disease was first diagnosed. Males without self-reported history of cancer of any site and oral precancerous conditions such as oral submucous fibrosis, verrucous hyperplasia, erythroplakia, leukoplakia, etc., were enrolled to the control cohort. Data concerning age, alcohol consumption, betel nut use, and smoking was recorded for every subject. Betel quid chewing and alcohol drinking are defined as excessive use of betel quid (or related products) and alcoholic drinking, respectively. Smoking is determined by recent use of at least one cigarette per day during the latest three months.
2.2. Selection and Genotyping of LINC00673 SNPs
Three common polymorphisms (rs11655237, rs9914618, and rs6501551) from LINC00673 gene explored in this study were chosen based on their functional potential as predicted by using RegulomeDB [30] (RegulomeDB score < 1). Genomic DNA extraction and genotyping were conducted and analyzed as described previously [31].
2.3. Bioinformatics Analysis
The enhancer region bearing rs9914618 in the ENCODE database was visualized by the UCSC genome browser using the hg19 assembly. Prediction of transcription factor binding sites was performed by JASPAR2020 [32].
2.4. Statistical Analysis
Significant variation in demographic data between OSCC cases and non-cancer controls was evaluated by using Mann–Whitney U test and Fisher’s exact test. The association of genotypes with OSCC risk was measured using multiple logistic regression methods and adjusted for potential confounders. The differences of LINC00673 levels in the head and neck squamous cell carcinoma (HNSCC) dataset from The Cancer Genome Atlas (TCGA) were compared by Student’s t-test. Data were calculated with SAS statistical software (Version 9.1, 2005; SAS Institute Inc., Cary, NC, USA). A p-value < 0.05 was considered significant.
3. Results
3.1. Cohort Characteristics
In this investigation, 1231 cases with OSCC and 1194 age-matched non-cancer subjects were recruited to evaluate the impact of LINC00673 SNPs on the development of oral cancer. Only males were enrolled to exclude the potential confounding effect of gender variations on OSCC risk, and cohort characteristics were evaluated (Table 1). Consistent with the findings from others [6,33], significant variations in cigarette smoking, alcohol drinking, and betel quid use were observed between OSCC and control cohort. Among the OSCC cases, lymphatic spread and distal metastasis were observed in 34.7% and 0.9% of patients, respectively.
Table 1.
The distributions of demographical characteristics and clinical parameters in 1195 controls and 1125 cases with OSCC.
| Variable | Controls (N = 1194) | Patients (N = 1231) | p-Value |
|---|---|---|---|
| Age (yrs) | 53.90 ± 10.03 | 55.53 ± 10.85 | |
| <55 | 565 (47.3%) | 574 (46.6%) | p = 0.733 |
| ≥55 | 629 (52.7%) | 657 (53.4%) | |
| Betel quid chewing | |||
| No | 995 (83.3%) | 303 (24.6%) | |
| Yes | 199 (16.7%) | 928 (75.4%) | p < 0.001 * |
| Cigarette smoking | |||
| No | 558 (46.7%) | 193 (15.7%) | |
| Yes | 636 (53.3%) | 1038 (84.3%) | p < 0.001 * |
| Alcohol drinking | |||
| No | 957 (80.2%) | 648 (52.6%) | |
| Yes | 237 (19.8%) | 583 (47.4%) | p < 0.001 * |
| Stage | |||
| I + II | 575 (46.7%) | ||
| III + IV | 656 (53.3%) | ||
| Tumor T status | |||
| T1 + T2 | 605 (49.1%) | ||
| T3 + T4 | 626 (50.9%) | ||
| Lymph node status | |||
| N0 | 804 (65.3%) | ||
| N1 + N2 + N3 | 427 (34.7%) | ||
| Metastasis | |||
| M0 | 1220 (99.1%) | ||
| M1 | 11 (0.9%) | ||
| Cell differentiation | |||
| Well differentiated | 174 (14.1%) | ||
| Moderately or poorly differentiated | 1057 (85.9%) |
Mann–Whitney U test was used between healthy controls and patients with oral cancer. * p-value < 0.05 as statistically significant.
3.2. Association of LINC00673 Gene Polymorphism with the Progression of OSCC
To examine the possible impact of LINC00673 gene variants on OSCC progression, three SNPs from LINC00673 gene (rs11655237, rs9914618, and rs6501551) were genotyped in this investigation. The distributions of genotype frequencies for each SNP in our cohort were evaluated (Table 2). No deviation (p > 0.05) from Hardy–Weinberg equilibrium in both case and control cohorts was detected for all three SNPs. We failed to individually observe any significant correlation of these LINC00673 variants with the occurrence of OSCC between the case and control group. Moreover, we investigated the correlations of polymorphic genotypes of LINC00673 with clinicopathological characteristics of OSCC patients. We found that patients who carry at least one minor allele of rs9914618 (GA and AA; OR, 1.286; 95% CI, 1.008–1.642; p = 0.043) were more prone to develop lymph node metastasis as compared with those homologous for the major allele (Table 3). In our stratified analysis, this genetic correlation with increased risk of lymphatic spread was further fortified in patients who were habitual betel quid chewers (OR, 1.534; 95% CI, 1.160–2.028; p = 0.003) or smokers (OR, 1.320; 95% CI, 1.013–1.721; p = 0.040) (Table 4). These data demonstrate a joint effect of LINC00673 SNPs and environmental triggers on the progression of oral cancer.
Table 2.
Odds ratio (OR) and 95% confidence interval (CI) of OSCC associated with LINC00673 genotypic frequencies.
| Variable | Controls (N = 1194) n (%) | Patients (N = 1231) n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs11655237 | ||||
| CC | 772 (64.7%) | 784 (63.7%) | 1.000 | 1.000 |
| CT | 371 (31.1%) | 398 (32.3%) | 1.056 (0.889–1.256) | 1.034 (0.834–1.282) |
| TT | 51 (4.2%) | 49 (4.0%) | 0.946 (0.631–1.418) | 0.675 (0.405–1.125) |
| CT + TT | 422 (35.3%) | 447 (36.3%) | 1.043 (0.883–1.231) | 0.986 (0.802–1.213) |
| rs9914618 | ||||
| GG | 770 (64.5%) | 799 (64.9%) | 1.000 (reference) | 1.000 (reference) |
| GA | 370 (31.0%) | 380 (30.9%) | 0.990 (0.832–1.178) | 0.954 (0.768–1.184) |
| AA | 54 (4.5%) | 52 (4.2%) | 0.928 (0.626–1.375) | 0.652 (0.398–1.068) |
| GA + AA | 424 (35.5%) | 432 (35.1%) | 0.982 (0.831–1.160) | 0.911 (0.740–1.121) |
| rs6501551 | ||||
| AA | 890 (74.5%) | 919 (74.7%) | 1.000 (reference) | 1.000 (reference) |
| AG | 279 (23.4%) | 294 (23.9%) | 1.021 (0.846–1.232) | 1.039 (0.822–1.312) |
| GG | 25 (2.1%) | 18 (1.4%) | 0.697 (0.378–1.287) | 0.689 (0.323–1.471) |
| AG + GG | 304 (25.5%) | 312 (25.3%) | 0.994 (0.828–1.193) | 0.911 (0.740–1.121) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models. The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for betel nut chewing, alcohol and tobacco consumption.
Table 3.
Clinical statuses and LINC00673 rs9914618 genotype frequencies in oral cancer.
| Variable | LINC00673 (rs9914618) | |||
|---|---|---|---|---|
| GG (%) (n = 799) | GA + AA (%) (n = 432) | OR (95% CI) | p-Value | |
| Clinical Stage | ||||
| Stage I/II | 373 (46.7%) | 202 (46.8%) | 1.00 | p = 0.980 |
| Stage III/IV | 426 (53.3%) | 230 (53.2%) | 0.997 (0.788–1.261) | |
| Tumor size | ||||
| T1 + T2 | 393 (49.2%) | 212 (49.1%) | 1.00 | p = 0.970 |
| T3 + T4 | 406 (50.8%) | 220 (50.9%) | 1.005 (0.795–1.270) | |
| Lymph node metastasis | ||||
| No | 538 (67.3%) | 266 (61.6%) | 1.00 | p = 0.043 |
| Yes | 261 (32.7%) | 166 (38.4%) | 1.286 (1.008–1.642) | |
| Distant metastasis | ||||
| No | 794 (99.4%) | 426 (98.6%) | 1.00 | p = 0.175 |
| Yes | 5 (0.6%) | 6 (1.4%) | 2.237 (0.679–7.371) | |
| Cell differentiation | ||||
| well | 111 (13.9%) | 63 (14.6%) | 1.00 | p = 0.740 |
| Moderate/poor | 688 (86.1%) | 369 (85.4%) | 0.945 (0.677–1.320) | |
Table 4.
Clinical statuses and LINC00673 rs9914618 genotype frequencies in oral cancer among 928 betel quid chewing and 1038 smoking.
| Variable | LINC00673 (rs9914618) | |||||
|---|---|---|---|---|---|---|
| Betel Quid Chewing (n = 928) | Smoking (n = 1038) | |||||
| GG (%) (n = 590) | GA + AA (%) (n = 338) | p-Value | GG (%) (n = 672) | GA + AA (%) (n = 366) | p-Value | |
| Clinical Stage | ||||||
| Stage I/II | 288 (48.8%) | 148 (43.8%) | p = 0.140 | 318 (47.3%) | 165 (45.1%) | p = 0.489 |
| Stage III/IV | 302 (51.2%) | 190 (56.2%) | 354 (52.7%) | 201 (54.9%) | ||
| Tumor size | ||||||
| ≤T2 | 300 (50.8%) | 163 (48.2%) | p = 0.442 | 345 (51.3%) | 180 (49.2%) | p = 0.506 |
| >T2 | 290 (49.2%) | 175 (51.8%) | 327 (48.7%) | 186 (50.8%) | ||
| Lymph node metastasis | ||||||
| No | 410 (69.5%) | 202 (59.8%) | p = 0.003 *,a | 454 (67.6%) | 224 (61.2%) | p = 0.040 *,b |
| Yes | 180 (30.5%) | 136 (40.2%) | 218 (32.4%) | 142 (38.8%) | ||
| Distant metastasis | ||||||
| No | 586 (99.3%) | 334 (98.8%) | p = 0.423 | 668 (99.4%) | 360 (98.4%) | p = 0.100 |
| Yes | 4 (0.7%) | 4 (1.2%) | 4 (0.6%) | 6 (1.6%) | ||
| Cell differentiation | ||||||
| well | 94 (15.9%) | 47 (13.9%) | p = 0.408 | 97 (14.4%) | 59 (16.1%) | p = 0.468 |
| Moderate/poor | 496 (84.1%) | 291 (86.1%) | 575 (85.6%) | 307 (83.9%) | ||
* p < 0.05; a OR (95% CI):1.534 (1.160–2.028); b OR (95% CI):1.320 (1.013–1.721).
3.3. Clinical and Functional Insights of LINC00673 into OSCC
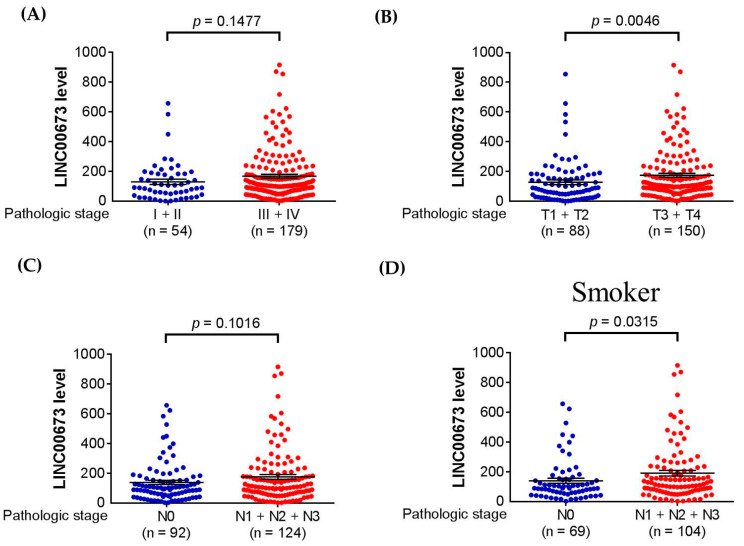
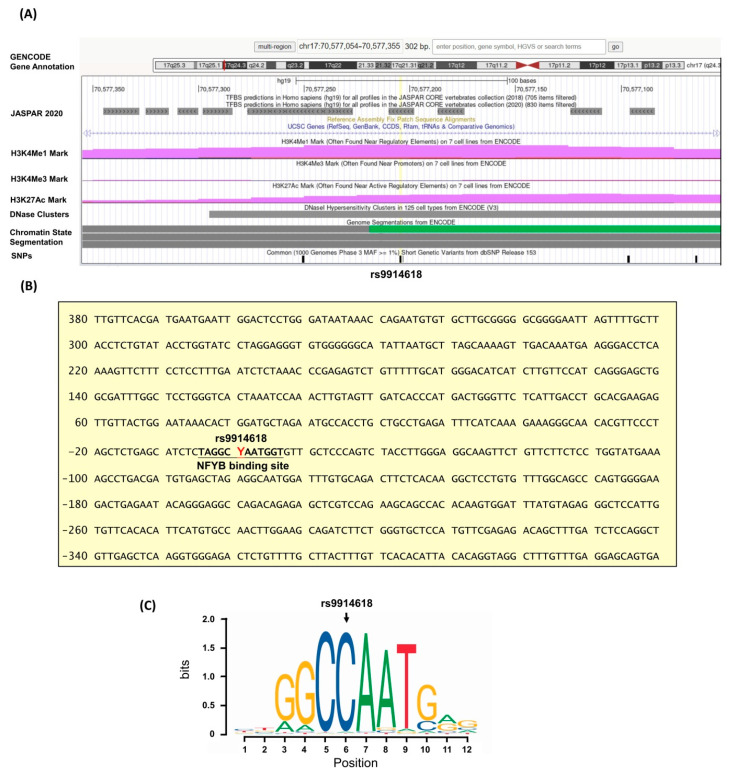
As a genetic association of LINC00673 with oral cancer was noted, additional analyses using public datasets were performed to gain clinical relevance of this gene. We found that higher LINC00673 expression levels were seen in large tumors in cases with HNSCC in TCGA dataset (Figure 1). Moreover, in HNSCC patients who were habitual smokers, increased LINC00673 expression levels were associated with lymphatic spread. These data support genetic associations detected in our study and suggest that habitual exposure to environmental risks in combination with altered expression levels of LINC00673 may affect OSCC progression. Since LINC00673 SNP, rs9914618, was found to be associated with OSCC progression, we then performed a pilot assessment of the tentative functional relevance of this SNP. We found that rs9914618 is lying on the first intron of the LINC00673 gene, within an active enhancer region in which multiple epigenetic actions (predominantly H3K4me1) occurred [34] (Figure 2). A further motif search of this enhancer region identified the position of rs9914618 within a putative CCAAT box, a unique pattern of nucleotides present in a high number of the promoter and enhancer regions in eukaryotic genes for the binding with specific transcription factors (e.g., nuclear transcription factor Y, NF-Y) [35,36], suggesting that this intronic variant may be a functional SNP in OSCC progression.
Figure 1.
LINC00673 expression levels are associated with clinicopathological parameters in HNSCC. Correlations of increased LINC00673 expression with the clinical staging (A), tumor size (B), and lymph node metastasis (C) of HNSCC from The Cancer Genome Atlas (TCGA) database. (D) In HNSCC patients who were habitual smokers, increased LINC00673 expression levels were associated with lymphatic spread. p values are calculated with Student’s t-test.
Figure 2.
Intron structure of LINC00673 (NR_137201.2) and the features of rs9914618. (A) The intron spanning the chromosome position chr17:70,577,054 to 70,577,355 (reference genome GRCh37.p13) is shown, and the SNPs of LNC00673 are indicated by the black bars. rs9914628 is centered and marked with reference SNP ID number. The H3K4Me1, H3K4Me3, and H3K27Ac tracks exhibit the enrichment of the mono-methylation of lysine 4, trimethylation of lysine 4, and acetylation of lysine 27 of the H3 histone protein, as determined in ENCODE project. DNase clusters track indicates DNase hypersensitivity regions. Chromatin State Segmentation tracks display chromatin state segmentations by integrating ChIPseq data with a hidden Markov model for H1-hESC embryonic stem cells. The chromatin state region predicted for transcribed regions is highlighted in green. (B) Sequence of the intron 1 region. The putative NFYB binding motif (MA0502.2) is shown in bold fonts. Y indicates the position of rs9914628 and denotes C or T. (C) Motif logo of NYFB consensus sequences.
4. Discussion
Accumulative evidence has manifested that OSCC progression is a multi-step process orchestrated by both genetic and environmental factors. Here, we reported that LINC00673 SNP, rs9914618, mediated the invasive potential of OSCC but did not confer the susceptibility to oral malignancies. When comparing only patients who were habitual betel quid or cigarette users, the association of rs9914618 with lymph node metastasis in oral cancer was further strengthened. Our results, for the first time, reveal an interactive effect of rs9914618 with betel nut use or smoking on OSCC progression.
Consistent with our finding that LINC00673 gene polymorphism was associated with lymphatic spread in oral cancer, the oncogenic role of LINC00673 in promoting cancer invasion and metastasis through diverse molecular mechanisms has been reported [13,14,15,16,17]. By acting as a scaffold molecule to interact with LSD1, a chromatin modifier that demethylates histone H3 lysine 4 and non-histone targets [37,38], LINC00673 has been shown to exhibit pro-metastatic properties in gastric cancer [16]. It was also demonstrated that LSD1 functions as a key epigenetic regulator in oral cancer metastasis [39]. Other than LSD1, LINC00673 facilitated the interaction between DDX3 (DEAD box RNA helicase) and CK1ε (casein kinase 1ε) and thus the phosphorylation of dishevelled, leading to activation of WNT/β-catenin signaling and aggressiveness of lung adenocarcinoma [15]. Our results and findings from others imply that polymorphic transcripts of LINC00673 may exert differential affinities with distinct binding partners to manage cancer invasiveness and metastasis. In addition to protein molecules, LINC00673 could sponge miR-150-5p and modulate epithelial-mesenchymal transition in lung cancer [17]. In a recent study aiming to discover specific plasma microRNAs as OSCC biomarkers, miR-150-5p was identified to be useful to monitor malignant progression in oral cancer [40]. These data support the genetic association of LINC00673 with lymphatic spread observed in our study and reveal a multifaceted role of LINC00673 in cancer progression via [27].
LINC00673 SNP, rs11655237, has been extensively reported for its association with the occurrence, progression, and prognosis of multiple tumor types [41]. This genetic variation (G > A) creates a miR-1231–binding site, which diminishes the effect of LINC00673 and, thus, may confer susceptibility to cancer [12]. In addition to the changes in functionality, the polymorphic allele of rs11655237 was proposed to cause reduced expression of LINC00673 [27]. Of note, unlike several previous reports linking rs11655237 to cancer risk [12,24,25,26,27,28], we failed to detect a genetic association of rs11655237 with the predisposition to OSCC. This discrepancy may be, in part, accounted for by a relative weak effect of specific SNPs or influence from other potential etiological confounders.
In the present study, we reported that LINC00673 gene polymorphism, rs9914618, was linked to the invasive potential of OSCC. Our exploratory evaluation of functional relevance demonstrated that rs9914618 is located within a CCAAT box, a putative binding motif of NF-Y (nuclear transcription factor Y) [36] and C/EBPs (CCAAT/enhancer binding proteins) [35]. NF-Y has been emerging as a key transcriptional regulator for many genes overexpressed in several cancer types [42], and a tumor suppressive role for C/EBPs has previously been demonstrated in HNSCC [43]. These findings suggest that rs9914618 polymorphisms may render a distinct expression profile of cancer-related genes mainly through impaired interactions with NF-Y or C/EBPs, thereby mediating OSCC progression.
Our data revealed an influence of LINC00673 SNPs on the lymph node metastasis of oral cancer; yet, extra work is required to address several limitations in the study. One is that the effects of LINC00673 gene polymorphisms on the risk of developing OSCC may be underestimated because of a lack of enumerative definition for betel quid consumption, alcohol use, and smoking. Another weakness is that the molecular mechanism underlying the promotive role of rs9914618 in oral cancer metastasis remains an open question. Whether the genetic polymorphism (G > A) controls its expression or alters the binding to its interacting proteins or microRNAs, thereby affecting OSCC biology, requires further investigations. Additionally, RNA is unavailable in situ hybridization analysis of LINC00673 on neoplasm lesion and/or lymph node metastasis lesion in OSCC. Moreover, the findings reported in the present investigation might be unable to be applied to other ethnic groups unless replication experiments are conducted.
5. Conclusions
In conclusion, our results demonstrate an association of LINC00673 rs9914618 with the metastatic potential in oral cancer. Elevated LINC00673 expression levels contribute to an inclination to develop large tumors in HNSCC patients and lymph node metastasis in habitual smokers. These findings reveal a novel genetic relationship between LINC00673 variants and OSCC progression.
Acknowledgments
We thank Tissue Bank at Chang Gung Memorial Hospital, Keelung for sample preparation. This study was supported by a research grant from Chang Gung Memorial Hospital (BMRPE97) to SCS.
Author Contributions
Conceptualization, S.-C.S., C.-W.L., M.-J.H. and S.-F.Y.; formal analysis, P.-C.J., L.-C.C., Y.-F.L. and S.-F.Y.; resources, C.-Y.C.; writing—original draft preparation, S.-C.S., C.-W.L., M.-J.H. and S.-F.Y.; writing—review and editing, S.-C.S., C.-W.L., M.-J.H. and S.-F.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Chung Shan Medical University Hospital (CSMUH No: CS15125).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Krishna Rao S.V., Mejia G., Roberts-Thomson K., Logan R. Epidemiology of oral cancer in Asia in the past decade—An update (2000–2012) Asian Pac. J. Cancer Prev. 2013;14:5567–5577. doi: 10.7314/apjcp.2013.14.10.5567. [DOI] [PubMed] [Google Scholar]
- 3.Gupta K., Metgud R. Evidences Suggesting Involvement of Viruses in Oral Squamous Cell Carcinoma. Pathol. Res. Int. 2013;2013:642496. doi: 10.1155/2013/642496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scully C., Bagan J. Oral squamous cell carcinoma overview. Oral Oncol. 2009;45:301–308. doi: 10.1016/j.oraloncology.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Scully C., Field J.K., Tanzawa H. Genetic aberrations in oral or head and neck squamous cell carcinoma (SCCHN): 1. Carcinogen metabolism, DNA repair and cell cycle control. Oral Oncol. 2000;36:256–263. doi: 10.1016/S1368-8375(00)00007-5. [DOI] [PubMed] [Google Scholar]
- 6.Zini A., Czerninski R., Sgan-Cohen H.D. Oral cancer over four decades: Epidemiology, trends, histology, and survival by anatomical sites. J. Oral Pathol. Med. 2010;39:299–305. doi: 10.1111/j.1600-0714.2009.00845.x. [DOI] [PubMed] [Google Scholar]
- 7.Gougis P., Moreau-Bachelard C., Kamal M., Gan H.K., Borcoman E., Torossian N., Bieche I., Le Tourneau C. Clinical Development of Molecular Targeted Therapy in Head and Neck Squamous Cell Carcinoma. JNCI Cancer Spectr. 2019;3:pkz055. doi: 10.1093/jncics/pkz055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanna R., Dalvi S., Benedicenti S., Amaroli A., Salagean T., Pop I.D., Todea D., Bordea I.R. Photobiomodulation Therapy in Oral Mucositis and Potentially Malignant Oral Lesions: A Therapy Towards the Future. Cancers. 2020;12:1949. doi: 10.3390/cancers12071949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scully C., Field J.K., Tanzawa H. Genetic aberrations in oral or head and neck squamous cell carcinoma 2: Chromosomal aberrations. Oral Oncol. 2000;36:311–327. doi: 10.1016/S1368-8375(00)00021-X. [DOI] [PubMed] [Google Scholar]
- 10.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng J., Huang X., Tan W., Yu D., Du Z., Chang J., Wei L., Han Y., Wang C., Che X., et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet. 2016;48:747–757. doi: 10.1038/ng.3568. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt K., Joyce C.E., Buquicchio F., Brown A., Ritz J., Distel R.J., Yoon C.H., Novina C.D. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016;15:2025–2037. doi: 10.1016/j.celrep.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi X., Ma C., Zhu Q., Yuan D., Sun M., Gu X., Wu G., Lv T., Song Y. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7:25558–25575. doi: 10.18632/oncotarget.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan H., Zhu T., Wu S., Liu S., Liu B., Wu J., Cai J., Zhu X., Zhang X., Zeng M., et al. Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/beta-catenin signaling. Proc. Natl. Acad. Sci. USA. 2019;116:14019–14028. doi: 10.1073/pnas.1900997116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang M., Hou J., Wang Y., Xie M., Wei C., Nie F., Wang Z., Sun M. Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with LSD1 and EZH2 in Gastric Cancer. Mol. Ther. 2017;25:1014–1026. doi: 10.1016/j.ymthe.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Lu W., Zhang H., Niu Y., Wu Y., Sun W., Li H., Kong J., Ding K., Shen H.M., Wu H., et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150–5p. Mol. Cancer. 2017;16:118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W., Xu Q., Kuang D., Wang Z., Lu Q., Lin Q., Wu H., Chen L. Long noncoding RNA SLNCR1 regulates nonsmall cell lung cancer migration, invasion and stemness through interactions with secretory phospholipase A2. Mol. Med. Rep. 2019;20:2591–2596. doi: 10.3892/mmr.2019.10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia E., Bhandari A., Shen Y., Zhou X., Wang O. lncRNA LINC00673 induces proliferation, metastasis and epithelial-mesenchymal transition in thyroid carcinoma via Kruppel-like factor 2. Int. J. Oncol. 2018;53:1927–1938. doi: 10.3892/ijo.2018.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao K., Ning S., Wan L., Wu H., Wang Q., Zhang X., Xu S., Pang D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:418. doi: 10.1186/s13046-019-1421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt K., Carroll J.S., Yee E., Thomas D.D., Wert-Lamas L., Neier S.C., Sheynkman G., Ritz J., Novina C.D. The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21. Cell Rep. 2019;27:2493–2507.e4. doi: 10.1016/j.celrep.2019.04.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng L.M., Zhao D.W., Li S.J., Huang J. Association of the upregulation of LncRNA00673 with poor prognosis for colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:687–694. doi: 10.26355/eurrev_201802_14294. [DOI] [PubMed] [Google Scholar]
- 23.Childs E.J., Mocci E., Campa D., Bracci P.M., Gallinger S., Goggins M., Li D., Neale R.E., Olson S.H., Scelo G., et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet. 2015;47:911–916. doi: 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Zhuo Z.J., Zhou H., Liu J., Liu Z., Zhang J., Cheng J., Li S., Zhou H., Zhou R., et al. Additional data support the role of LINC00673 rs11655237 C>T in the development of neuroblastoma. Aging. 2019;11:2369–2377. doi: 10.18632/aging.101920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Chang Y., Jia W., Zhang J., Zhang R., Zhu J., Yang T., Xia H., Zou Y., He J. LINC00673 rs11655237 C>T confers neuroblastoma susceptibility in Chinese population. Biosci. Rep. 2018;38 doi: 10.1042/BSR20171667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao K., Zhang R., Li T., Xiong Z. Functional variants of lncRNA LINC00673 and gastric cancer susceptibility: A case-control study in a Chinese population. Cancer Manag. Res. 2019;11:3861–3868. doi: 10.2147/CMAR.S187011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Wang Y., Luo T. LINC00673 rs11655237 Polymorphism Is Associated with Increased Risk of Cervical Cancer in a Chinese Population. Cancer Control. 2018;25:1073274818803942. doi: 10.1177/1073274818803942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang T., Li J., Wen Y., Tan T., Yang J., Pan J., Hu C., Yao Y., Zhang J., Xin Y., et al. LINC00673 rs11655237 C>T Polymorphism Impacts Hepatoblastoma Susceptibility in Chinese Children. Front. Genet. 2019;10:506. doi: 10.3389/fgene.2019.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edge S.B., Compton C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 30.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su S.C., Hsieh M.J., Lin C.W., Chuang C.Y., Liu Y.F., Yeh C.M., Yang S.F. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J. Dent. Res. 2018;97:717–724. doi: 10.1177/0022034517749451. [DOI] [PubMed] [Google Scholar]
- 32.Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranasic D., et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jadhav K.B., Gupta N. Clinicopathological Prognostic Implicators of Oral Squamous Cell Carcinoma: Need to Understand and Revise. N. Am. J. Med. Sci. 2013;5:671–679. doi: 10.4103/1947-2714.123239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hon G.C., Hawkins R.D., Ren B. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 2009;18:R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramji D.P., Foka P. CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/bj20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/S0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 37.Huang J., Sengupta R., Espejo A.B., Lee M.G., Dorsey J.A., Richter M., Opravil S., Shiekhattar R., Bedford M.T., Jenuwein T., et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Hevi S., Kurash J.K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 39.Alsaqer S.F., Tashkandi M.M., Kartha V.K., Yang Y.T., Alkheriji Y., Salama A., Varelas X., Kukuruzinska M., Monti S., Bais M.V. Inhibition of LSD1 epigenetically attenuates oral cancer growth and metastasis. Oncotarget. 2017;8:73372–73386. doi: 10.18632/oncotarget.19637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Y.A., Weng S.L., Yang S.F., Chou C.H., Huang W.C., Tu S.J., Chang T.H., Huang C.N., Jong Y.J., Huang H.D. A Three-MicroRNA Signature as a Potential Biomarker for the Early Detection of Oral Cancer. Int. J. Mol. Sci. 2018;19:758. doi: 10.3390/ijms19030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li N., Cui Z., Huang D., Gao M., Li S., Song M., Wang Y., Tong L., Yin Z. Association of LINC00673 rs11655237 polymorphism with cancer susceptibility: A meta-analysis based on 23,478 subjects. Genomics. 2020;112:4148–4154. doi: 10.1016/j.ygeno.2020.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Dolfini D., Mantovani R. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y? Cell Death Differ. 2013;20:676–685. doi: 10.1038/cdd.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett K.L., Hackanson B., Smith L.T., Morrison C.D., Lang J.C., Schuller D.E., Weber F., Eng C., Plass C. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in head and neck squamous cell carcinoma. Cancer Res. 2007;67:4657–4664. doi: 10.1158/0008-5472.CAN-06-4793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.