Abstract
We synthesized a series of poly(disulfide)s by ring-opening polymerization and demonstrated that the copolymerization of monomer 1 containing diethylenetriamine moieties and monomer 2 containing guanidyl ligands could generate an efficient delivery platform for different forms of CRISPR-Cas9-based genome editors, including plasmid, mRNA, and protein. The excellent delivery performance of designed poly(disulfide)s stems from their delicate molecular structures to interact with genome-editing biomacromolecules, unique delivery pathways to mediate the cellular uptake of CRISPR-Cas9 cargoes, and strong ability to escape the endosome. The degradation of poly(disulfide)s by intracellular glutathione not only promotes the timely release of CRISPR-Cas9 machineries into the cytosol but also minimizes the cytotoxicity that nondegradable polymeric carriers often encounter. These merits collectively account for the excellent ability of poly(disulfide)s to mediate different forms of CRISPR-Cas9 for their efficient genome-editing activities in vitro and in vivo.
Short abstract
Poly(disulfide)s are designed as a universal carrier for the intracellular delivery of different forms of CRISPR/Cas9 machineries, paving a promising avenue for therapeutic genome editing in vivo.
Introduction
The type II clustered, regularly interspaced, short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) are originally part of the adaptive immune system of bacteria and archaea, which have been later repurposed as a ground-breaking tool for genome editing.1,2 In recent years, CRISPR-Cas systems have been demonstrated for their great potentials in a wide range of research areas, including biology, genetics, medicine,3−6 etc. Particularly, CRISPR-Cas9 has been harnessed as biotherapeutics for the treatment of a variety of genetic disorders, such as Duchenne muscular dystrophy7 and Hutchinson–Gilford progeria syndrome,8 largely due to its site-specific feature and precise capability. To be effective, the efficient delivery of CRISPR-Cas9 machineries into cell nuclei is crucial to induce double-strand breaks (DSBs) at the target loci of the genome, which are repaired by either the nonhomologous end-joining (NHEJ) pathway to form insertion/deletion (indel) mutations or the homology-directed repair (HDR) pathway to introduce precise DNA repair templates.9 To date, whereas most therapeutic strategies rely on a virus to transfect plasmids encoding CRISPR-Cas9 elements, viral vectors commonly suffer from inevitable drawbacks including insertional mutagenesis and immunogenicity.10 In addition, some viral vectors, such as the adeno-associated virus, have limited packing capacity and can only accommodate the plasmid up to 4.8 kb.4 As an alternative, nonviral vectors that can deliver CRISPR-Cas9 components into target cells have attracted much attention in recent years, largely owing to their favorable biocompatibility, tailored biophysical properties, and excellent ability to deliver different forms of cargoes.11 A few types of nonviral vectors, such as liposomes,12 gold nanoparticles,13,14 and polymeric carriers,15−20 have demonstrated their ability to deliver CRISPR-Cas9 components. In contrast with viruses, nonviral vectors not only can deliver integrated plasmid encoding Cas9 and single-guide RNA (sgRNA) for genome editing21 but also can be tailored to deliver either Cas9 mRNA/sgRNA complexes22 or Cas9/sgRNA ribonucleoprotein (RNP),23 the delivery forms which are believed to be more clinically relevant. Despite such advances, most nonviral vectors only demonstrated a moderate delivery efficiency, often resulting in lower genome-editing activity in vivo. In addition, under the premise of efficiency, the low cytotoxicity of carriers is also of paramount importance for CRISPR-Cas9-based therapeutic applications because any apparent toxicity may directly or indirectly affect NHEJ or HDR processes, thereby compromising genome-editing activities.24−26
Understanding both fundamental design principles of biomaterials and their various delivery processes is conducive to the development of new nonviral vectors for the delivery of CRISPR-Cas9 machineries. Recently, poly(disulfide)s have been emerging as a fascinating class of functional polymeric materials for a wide range of applications.27−34 Due to their dynamic and reversible chemistry, cell-penetrating poly(disulfide)s (CPDs) synthesized by strain-promoted ring-opening disulfide-exchange polymerization have been explored for their ability to mediate the intracellular delivery of various cargoes, including both small molecules and biomacromolecules.35−39 As compared with other disulfide-containing polycations that are exploited for the delivery of biomacromolecules, this class of poly(disulfide)s presents a few intriguing characteristics. First, racemic lipoic acid offers the strained disulfide that is essential for the ring-opening disulfide-exchange polymerization, leading to the formation of poly(disulfide)s with the reducible linkers in the polymer main chain.38−44 Thus, these poly(disulfide)s are ideally fully biodegradable in the intracellular reductive environment, as opposed to those with the disulfides in the side chain. Second, the disulfide linkers within poly(disulfide)s greatly contribute to thiol-mediated uptake, resulting in the efficient delivery of cargoes into the cytosol.28,29,45 Finally, as lipoic acid is ready for the chemical modification, the monomer can be easily tailored with particular properties. For example, the monomer obtained by conjugating l-arginine with lipoic acid was previously exploited for the polymerization, and the resulting poly(disulfide)s presenting multiple guanidinium cations could mimic cell-penetrating peptides to cross the cellular bilayer membrane.46 These merits greatly motivate us to explore poly(disulfide)s for the delivery of genome-editing biomacromolecules. However, the previous strategies largely rely on the covalent conjugation of delivery cargoes to CPDs,27−31 or exploiting CPDs as an additional component to promote the delivery performance of existing carriers.47 Despite these advances, the direct loading and intracellular delivery of biomacromolecules by CPDs remain to be elusive and have not been demonstrated yet, not to mention their in vivo applications. In addition, as endosomal escape is known to be a limiting step for the intracellular transport of biomacromolecules from the endo/lysosome into the cytosol, it is critical to impart delivery carriers along with the ability to escape endosomes in order to promote cytosolic delivery. In this regard, the rational design is essential to repurpose poly(disulfide)s as a new class of efficient and universal carriers for the delivery of CRISPR-Cas9 machineries in different forms (DNA, mRNA, and RNP), which are of particular interests toward therapeutic genome-editing contexts.
Results and Discussion
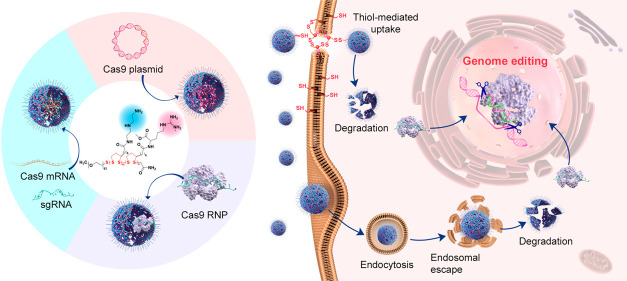
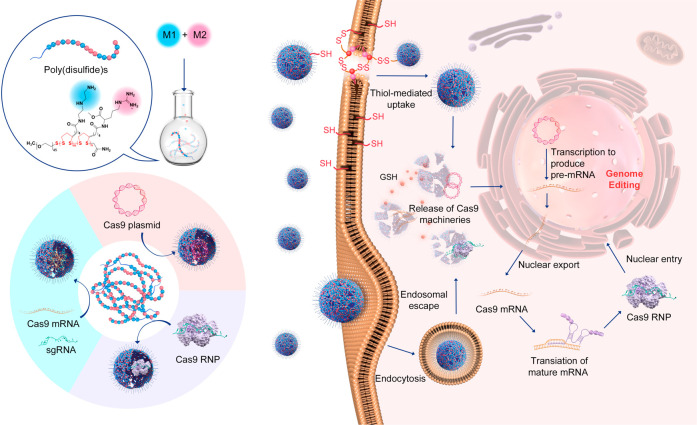
In the current study, we designed a series of poly(disulfide)s and explored their ability to deliver three forms of CRISPR-Cas9 components, namely, Cas9 plasmid (encoding Cas9 protein and sgRNA), Cas9 mRNA and sgRNA (Cas9 mRNA complex), and Cas9 RNP (see Figure 1). To this end, we delicately designed two monomers for the polymerization of poly(disulfide)s. As shown in Figure 2a, whereas monomer 1 (M1) containing cationic diethylenetriamine (DET) moieties may facilitate entrapped biomacromolecules to escape from endo/lysosomes, monomer 2 (M2) containing guanidyl groups is conducive to the transmembrane delivery of CRISPR-Cas9 machineries.48−51 For the complexation of Cas9 RNP, the multivalent display of guanidyl ligands in M2 facilitates the adherence of Cas9 RNP with poly(disulfide)s by forming multiple strong hydrogen bonds and salt bridges between their guanidiniums and oxyanions that exist ubiquitously in proteins,52,53 whereas the primary amines in M1 contribute to the ionic interactions with the carboxyl groups of the Cas9 protein.50−53 In the case of large Cas9 plasmids, both amines and guanidyl ligands contribute to the complexation of CRISPR-Cas9 plasmids, and the polymerization of poly(disulfide)s into the desired molecular weight facilitates the condensation of plasmid DNA. Similarly, the cationic feature of poly(disulfide)s is also important for the binding of Cas9 mRNA and sgRNA. Once polymer/cargo complexes reach the cytoplasm, the disulfide linkers within poly(disulfide)s could be rapidly degraded in the presence of rich intracellular glutathione (1–10 mM) to release CRISPR-Cas9 machineries.25 In the meantime, the intracellular degradation of poly(disulfide)s is also highly desirable to decrease the cytotoxicity by avoiding the accumulation of high-molecular-weight poly(disulfide)s inside the cells. As a result, poly(disulfide)s break up the previous dilemma between efficiency and toxicity for the intracellular delivery of biomacromolecules.54
Figure 1.
Schematic illustration of the preparation of poly(disulfide)s, the complexation of genome-editing biomacromolecules (Cas9 plasmid, Cas9 mRNA, and Cas9 ribonucleoprotein) by poly(disulfide)s, and their intracellular delivery processes for genome editing.
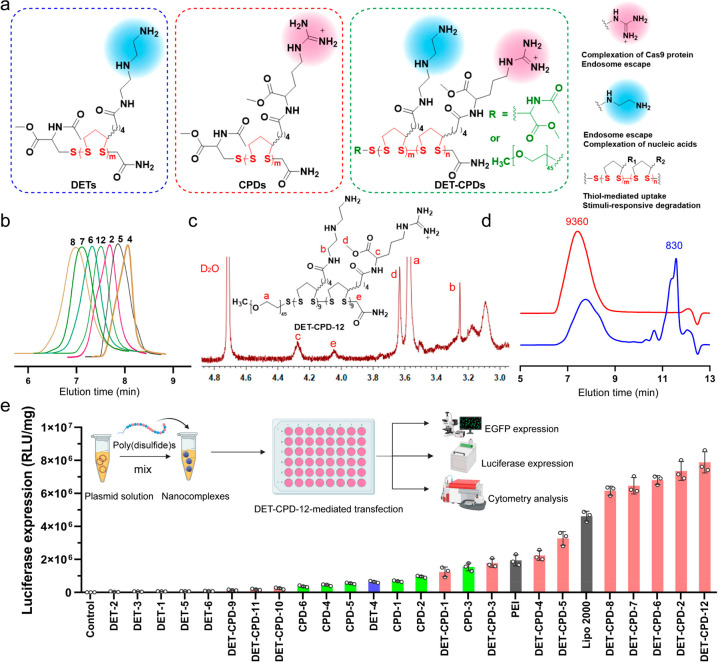
Figure 2.
Design, synthesis, and characterization of poly(disulfide)s. (a) Chemical structure of the DETs, CPDs, and DET-CPDs. (b) Representative GPC profiles of poly(disulfide)s: 4, DET-CPD-4; 5, DET-CPD-5; 2, DET-CPD-2; 6, DET-CPD-6; 7, DET-CPD-7; 8, DET-CPD-8; 12, DET-CPD-12. (c) 1H NMR spectrum of DET-CPD-12. (d) GPC analysis of DET-CPD-12 and the degradation of DET-CPD-12 in the presence of 10 mM GSH. (e) CRISPR-Cas9 plasmid transfected by poly(disulfide)s in 293T cells. Commercially available Lipo 2000 and PEI 25K were used as the positive controls. Data represent mean ± S.D. (n = 3).
To prepare poly(disulfide)s, the strained disulfide monomers were first synthesized by a CDI (1,1′-carbonyldiimidazole)-mediated coupling reaction between the carboxyl group of lipoic acid and the primary amines of l-argininate or diethylenetriamine to obtain M1 or M2, respectively. 1H NMR and 13C NMR spectra confirmed the chemical structure of M1 and M2 (Figures S1 and S2). The copolymerization of M1 and M2 was carried out through the ring-opening polymerization of two monomers using thiolated PEG (polyethylene glycol, MW: 2000 Da) or N-acetyl-l-cysteine methyl ester as the initiator, and the polymerization progress was terminated by adding 2-iodoacetamide. The homopolymerization of either M1 or M2 was also conducted, and all of the polymerization conditions are shown in Table S1. In order to avoid potential gelation and to prevent the cross-linking reactions between reactive thiols in the opened five-membered rings, the polymerization was conducted under a relatively low monomer concentration ([M1] + [M2] = 0.2 M).38,39 All poly(disulfide)s generated from homopolymerization and heteropolymerization were characterized for their molecular weight and distribution (Table S1, Figure 2b, and Figures S3–S5). In general, for both polymerizations, the molecular weight increased with the polymerization time. The resulting poly(disulfide), DET-CPD-12, was characterized by 1H NMR and gel permeation chromatography (GPC). As shown in Figure 2c and Figure S6, the proton signals at δ 4.21–4.35 ppm were associated with methine protons in the vicinity of the amide from M2 moieties (peak c). Similarly, the proton signal at δ 3.25–3.38 ppm was attributed to the methylene protons in the vicinity of the amide from M1 moieties (peak b). The singlet that appeared at δ 4.06 ppm could be ascribed to methylene protons in the terminal of the main chain (peak e). Based on the integration of two peaks, the number of M1 and M2 repeating units per chain was calculated as 9.1 and 9.2, respectively. In the meantime, the number-average molecular weight (Mn) was calculated as ca. 8200 Da based on the integration from the proton peaks of PEG and those of repeating units (peak a). As shown in Figure 2d, DET-CPD-12 displayed a unimodal but rather broad molecular weight distribution in the GPC chromatogram and was eluted at relatively low elution times, indicating its polymeric nature. The peak molecular weight (Mp) was indicated as 9360 Da (PDI = 1.24), which was approximately in agreement with 1H NMR results. After incubating with glutathione (GSH, 10 mM) for 12 h, the peak of DET-CPD-12 eluted out at the same position became much smaller (Figure 2d, Figure S7), whereas a new sharp peak appeared at the longer elution time (11.0–11.5 min), suggesting the degradation of DET-CPD-12 due to the rapid cleavage of disulfides linking five-membered rings in the main chain. These results collectively suggest that poly(disulfide)s with a designed structure were synthesized successfully.
In order to screen the optimal structure and molecular weight for efficient intracellular delivery of biomacromolecules, 24 poly(disulfide)s which were categorized into 3 series (DETs, CPDs, and DET-CPDs) were synthesized, and their ability to transfect CRISPR-Cas9 plasmids that were cloned with luciferase and enhanced green fluorescence protein (EGFP) tag was evaluated. As shown in Figure 2e, whereas it was hard for poly(disulfide)s of the DET series to transfect the CRISPR-Cas9 plasmid efficiently, we noticed that CPD-3 with a medium degree of polymerization was able to mediate a moderate transfection efficiency, which was close to that mediated by PEI. These results indicated that poly(disulfide)s polymerized from either M1 or M2 were not favorable for intracellular delivery of biomacromolecules, although they showed a good ability to complex the CRISPR-Cas9 plasmid and condense DNA into positively charged nanoparticles (Figures S8–S11). Based on these findings, we further prepared a series of bifunctional poly(disulfide)s (DET-CPDs) by randomly copolymerizing M1 and M2 monomers. By fine-tuning polymerization time, initiator concentration, and monomer ratio (Table S1), we found that the ability of bifunctional poly(disulfide)s to complex with the CRISPR-Cas9 plasmid was greatly improved in comparison with DETs or CPDs, and they formed compact nanocomplexes (Figures S12 and S13). We then carefully studied how polymerization conditions impact the transfection activity of bifunctional poly(disulfide)s (Figure S14 and Table S1). First, we found that the transfection efficiency mediated by bifunctional poly(disulfide)s increased rapidly with the polymerization time, due to the increased molecular weight, but also slightly declined after 1.5 h of polymerization, suggesting that the molecular weight plays a crucial role in their transfection activity. Second, the transfection activity of the copolymer was greatly influenced when the monomer ratio ([M1]/[M2]) changed, though the molecular weight of the resulting copolymers only slightly varied. The monomer ratio at 1:2 (molar ratio) was most favorable for the transfection. Third, the initiator concentration for the random copolymerization also significantly impacted the molecular weight of the resulting bifunctional poly(disulfide)s, thereby affecting the transfection activity. The most efficient bifunctional poly(disulfide) (DET-CPD-2) was obtained when the initiator concentration at 0.03 mM was used for copolymerization. By using thiolated PEG as the initiator, we found that the PEGylation of DET-CPD-2 (DET-CPD-12) was critically conducive to further improve the transfection activity and enhance the water solubility (Figure 2a). These results greatly motivated us to explore whether DET-CPD-12 could serve as the carrier for the intracellular delivery of different formats of genome-editing agents for therapeutic purposes.
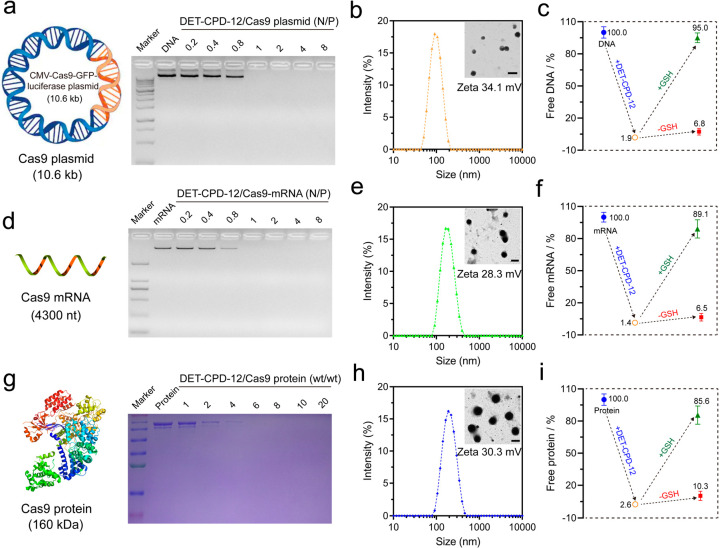
DET-CPD-12 was first examined for its ability to complex with genome-editing biomacromolecules, and the physiochemical properties of the resulting nanocomplexes were evaluated. As shown in Figure 3a, DET-CPD-12 was able to inhibit plasmid DNA from migration when the N/P ratio reached 1, and the average particle size of the resulting DET-CPD-12/DNA complexes was ca. 80 nm at the N/P ratio of 5, as revealed by dynamic light scattering (DLS) analysis (Figure 3b). Likewise, DET-CPD-12 also showed a good ability to inhibit Cas9 mRNA from migration at the N/P ratio of 1 (Figure 3d) but resulted in slightly larger complexes when complexing with Cas9 mRNA at the N/P ratio of 5 (180 nm in average by DLS, Figure 3e). In the case of the Cas9 protein, SDS-PAGE gel electrophoresis was carried out to check the capability of DET-CPD-12 for protein encapsulation. As shown in Figure 3g, protein bands were undetectable when the DET-CPD-12/Cas9 protein weight ratio reached 6, suggesting the complete encapsulation of the protein by DET-CPD-12. All three forms of Cas9 cargoes could be complexed into compact spherical nanoparticles (inserts), and the surface charges of these nanoparticles were positive (Figure 3b,e,h). It is noted that DET-CPD-12/Cas9 RNP complexes (at DET-CPD-12/Cas9 weight ratio of 6) also exhibited a similar size distribution and ζ potential value (Figure S15). In addition, while the encapsulation efficiency and loading capacity for three forms of Cas9 cargoes were generally high, the release of these cargoes in the presence of 10 mM GSH is rapid and efficient (Figure 3c,f,i and Figure S16). Previous studies have indicated that the release of Cas9 plasmid can be manipulated by different means,55−57 such as external light. In the current study, the release of the CRISPR-Cas9 plasmid can be promoted by GSH-triggered polymer degradation in the main chain, which may accelerate the gene expression process of Cas9 and sgRNA. These characteristics collectively indicated that DET-CPD-12 may serve as an efficient vector for the intracellular delivery and the controlled release of genome-editing biomacromolecules.
Figure 3.
Preparation and characterization of DET-CPD-12 polyplexes. Agarose gel electrophoresis assay of DET-CPD-12 polyplexes with Cas9 plasmid (a) and Cas9 mRNA (d) at different N/P ratios. Particle size and ζ potential of DET-CPD-12/Cas9 plasmid complexes (b), DET-CPD-12/Cas9 mRNA complexes (e), and DET-CPD-12/Cas9 protein complexes (h). The inset is the TEM image of the nanoparticles, and all of the scale bars represent 200 nm. Percentage of free genome-editing biomacromolecules after the formation of DET-CPD-12/Cas9 DNA complexes (c), DET-CPD-12/Cas9 mRNA complexes (f), and DET-CPD-12/Cas9 protein complexes (i). The release of Cas9 DNA, mRNA, and protein from their complexes was also evaluated in the presence or absence of 10 mM GSH. All data represent mean ± S.D. (n = 3). (g) SDS-page assay of DET-CPD-12/Cas9 protein complexes at different weight ratios.
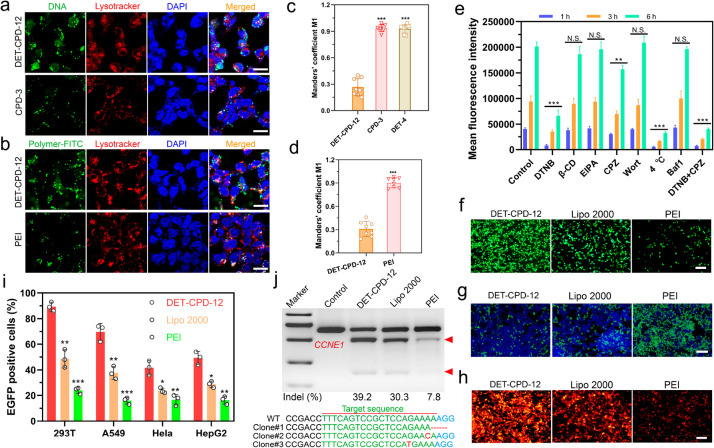
To investigate the mechanism of DET-CPD-12-mediated cellular uptake and endosomal escape, plasmid DNA, lysosome, and DET-CPD-12 were labeled with green or red dye, whereas cell nuclei were stained with blue dye. After 6 h of transfection, green fluorescence from DNA or DET-CPD-12 already spread out from the red fluorescence of the lysosome, suggesting the intracellular release of DNA from the endosome (Figure 4a,b). We also sought to study the endosomal escape by CPD-3 or DET-4 (Figure 4a and Figure S17). To our surprise, endosomal escape by either CPD-3 or DET-4 seems to be limited (Figure 4a,b, Figure S17). To understand how DET-CPD-12 escapes the endosome, we further studied the hemolytic activity of DET-CPD-12 at both endosomal acidic and physiological pH. Whereas a strong hemolytic activity of DET-CPD-12 was observed in the endosomal acidic pH (54%, Figure S18), it dropped to 16% at physiological pH. This suggests that protonation of DET moieties critically contributes to the membrane disruption, and endosomal escape is more likely achieved by protonated amines and guanidyl ligands that collectively lead to the membrane disruption of endocytotic vesicles. These results indicated that the endosomal escape is dominated by a membrane disruption mechanism,48,53 which is attributed to the protonated amines of DET and the penetrating property of guanidyl ligands of CPD. By labeling the delivery vector, we could also clearly observe that a large portion of DET-CPD-12 spread out from LysoTracker, whereas “gold standard” polyethylenimine (PEI) seemed to be less efficient in mediating both cellular uptake and endosomal escape (Figure 4a,b and Figures S19–S21). Manders’ coefficient M1 is close to 1 in the case of PEI-mediated transfection (Figure 4c,d), suggesting poor endosomal escape. In sharp contrast, the Manders’ coefficient M1 in the DET-CPD-12 group is less than 0.3 (Figure 4c,d), which implied more successful endosomal escape by DET-CPD-12. It is noted that the internalization of DET-CPD-12/DNA nanocomplexes by cells was time-dependent, and the majority of nanocomplexes was not trapped by lysosomes at the predefined time points. These facts strongly implied that DET-CPD-12-mediated cytosolic delivery could be promoted by efficient endosomal escape. In order to understand the role of a specific internalization pathway, different inhibitors were supplemented before the transfection to investigate their effects on the internalization of DET-CPD-12/DNA nanocomplexes. It was evident that the addition of DTNB (5,5′-dithiobis-2-nitrobenzoic acid) strongly inhibited the internalization, which implied that DET-CPD-12 primarily followed the thiol-mediated uptake pathway. The addition of chlorpromazine (CPZ), a well-established inhibitor of clathrin-mediated endocytosis, only moderately inhibited the internalization. In contrast, the presence of methyl-β-CD (β-CD), wortmannin (Wort), 5-(N-ethyl-N-isopropyl) amiloride (EIPA), and bafilomycin A1 (Baf1) almost did not inhibit the cellular uptake at different time points and gene expression (Figure 4e and Figure S22). Particularly, the cellular uptake, as indicated by intracellular fluorescence intensity, was clearly blocked when DTNB concentration increased (Figure S23). These results strongly suggest that the cellular uptake of DET-CPD-12/plasmid nanocomplexes primarily follows endocytosis-independent pathways, followed by endocytosis-dependent pathways. For endocytosis-independent pathway, thiol-mediated uptake, which is associated with counterion-mediated binding to the cell surface, takes place first. Subsequently, the formation of transient membrane pores, as proposed previously, can translocate the cargo into the cytosol.58,59 For the endocytosis-dependent pathway, endosomal escape may largely rely on the membrane disruption of endocytotic vesicles, as the presence of bafilomycin A1, a V-type ATPase inhibitor that prevents the acidification of endosomes, did not apparently affect the transfection efficiency. This is in agreement with our early findings where DET-CPD-12 can effectively disrupt the murine erythrocytes in the endosomal acidic pH. In the case of transfecting the smaller CMV-GFP plasmid (3.7 kb), DET-CPD-12 showed similar internalization pathways as transfecting the large CRISPR-Cas9 plasmid (10.6 kb) but clearly differed from PEI-mediated internalization in terms of the thiol-mediated uptake and clathrin-mediated endocytosis (Figure S24). For CPD-3 which only contains guanidyl ligands, whereas the addition of DTNB significantly inhibited the internalization of nanocomplexes, the addition of β-CD almost did not affect its internalization, implying that CPD-3 follows both thiol-mediated translocation and caveolar endocytosis (Figure S25). These results strongly suggested that DET-CPD-12 could successfully deliver CRISPR-Cas9 plasmids into cells primarily following the nonendocytic pathway through disulfide exchange between DET-CPD-12 and exofacial thiols of cell membranes. In the case of the secondary endocytosis-dependent pathway, the DET moieties of DET-CPD-12 also greatly facilitate the endosomal escape by disrupting the membrane of endocytotic vesicles to well avoid the degradation of genome-editing biomacromolecules.60,61
Figure 4.
DET-CPD-12-mediated transfection of the CRISPR-Cas9 plasmid for genome editing in vitro. Cellular uptake and endosomal escape of DET-CPD-12/Cas9 DNA complexes in 293T cells, as evaluated by confocal laser scanning microscopy. Plasmid DNA was labeled by YOYO-1 (a); DET-CPD-12 and PEI by FITC (b); and endosome by red fluorescence dye (LysoTracker) (a, b). (c, d) Quantitative analysis of the colocalization of YOYO-1-labeled plasmid or FITC-labeled carriers with endo/lysosomes labeled with LysoTracker red. Manders’ coefficient M1 denotes the fraction of YOYO-1-labeled plasmid or FITC-labeled carriers in green overlapping with LysoTracker in red. The coefficient is close to 1 if they are highly colocalized (n = 8 images from three independent experiments). (e) Mechanism of cellular uptake of DET-CPD-12/Cas9 DNA complexes in 293T cells by the addition of different inhibitors. Mean fluorescence intensity of cells after YOYO-1 labeled Cas9 plasmid transfection in 293T cells mediated by DET-CPD-12. The cells were incubated with different inhibitors including 5,5′-dithiobis-2-nitrobenzoic acid (DTNB, 2.4 nM), methyl-β-cyclodextrin (β-CD, 50 μM), 5-(N-ethyl-N-isopropyl) amiloride (EIPA, 30 nM), chlorpromazine (CPZ, 30 μM), wortmannin (Wort, 50 nM), and bafilomycin A1 (Baf1, 200 nM) in 2 h, separately. The cells were also incubated at 4 °C (no inhibitor) or in the presence of both DTNB and CPZ before the transfection. The control group was conducted at 37 °C (no inhibitor) treated with DET-CPD-12/YOYO-1 labeled Cas9 plasmid nanoparticles only. The mean fluorescence intensity of the cells was quantified after the transfection for 1, 3, or 6 h, separately, using the flow cytometry. The statistical analysis between the experimental group (1, 3, and 6 h) and the control group (1, 3, and 6 h) was made, separately. (f) GFP expression in 293T after transfecting Cas9 plasmid DNA with the GFP tag. (g) Disruption of EFGP genes after the transfection of the CRISPR-Cas9 plasmid. (h) CRISPR/dCas9-mediated transcriptional activation of mCherry expression after cotransfecting three plasmids (SPH, U6-sgRNA, and CMV-mCherry) in 293T cells. (i) Flow cytometry analysis of GFP-positive cells after Cas9 plasmid transfection in different cell lines. (j) Analysis of indel frequency in CCNE1 locus after the transfection of Cas9 plasmid DNA. The scale bar of parts a and b represents 20 μm. The scale bar of parts f–h represents 200 μm. All quantitative data represent mean ± S.D. (n = 3, one-way ANOVA with a Tukey’s posthoc test: N.S., P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001).
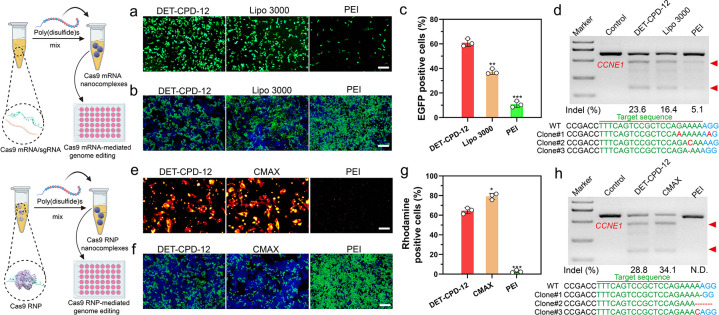
Next, we further evaluated the transfection efficiency and genome-editing activity of CRISPR-Cas9 machineries of different forms. First, we found that DET-CPD-12 exhibited a strong ability to transfect the plasmid in different cell lines at the optimal N/P ratio (N/P = 5) and is more efficient over Lipofectamine 2000 (Lipo 2000, a commercial transfection agent) and PEI of 25 kDa (a gold-standard nonviral transfection agent) (Figure 4f,i and Figures S26 and S27). In contrast to the transfection of the large CRISPR-Cas9 plasmid, DET-CPD-12 showed comparable transfection activities when transfecting the CMV-GFP plasmid, as compared with Lipo 2000 or PEI (Figure S28). These results corroborated that the transfection mediated by DET-CPD-12 was independent of plasmid size, which is in sharp contrast with frequently used nonviral transfection agents.24 It is noted that the transfection efficiency slightly dropped under the serum concentration of 10% in the cell culture and significantly decreased when the serum concentration reached 30%, indicating the moderate degree ability of DET-CPD-12 to resist serum (Figure S29). We also found that DET-CPD-12-mediated transfection of the CRISPR-Cas9 plasmid targeting EGFP gene could successfully disrupt GFP expression in 293T-EGFP cells (Figure 4g and Figure S30d), and such a delivery system also works well for dCas9-mediated transcriptional activation of exogenous gene expression (mCherry expression) when cotransfecting three plasmids (SPH, U6-sgRNA, and CMV-mCherry) in 293T cells (Figure 4h and Figure S31). In the meantime, the T7E1 assay indicated that DET-CPD-12-mediated genome editing at the CCNE1 locus in 293T cells caused a frequency of indels (insertions and deletions) up to 39.2%, which was higher than Lipo 2000- or PEI-mediated genome editing (Figure 4j). The mutation frequency at the target genome was also confirmed by deep sequencing (Figure S32). In the meantime, we evaluated the transfection efficiency of the Cas9 plasmid and the resulting indel frequency at the target genome (CCNE1) in three types of liver cells, including AML-12 cells (hepatocytes), mouse hepatic stellate cells (HSCs, nonparenchymal cells), and Hepa1–6 cells (liver cancer cells). As shown in Figure S33, DET-CPD-12 could mediate efficient transfection in all three types of cells and caused the substantial indel frequency at CCNE1. Furthermore, the transfection of Cas9-GFP or GFP mRNA by DET-CPD-12 also resulted in strong GFP expression in 293T cells, and 60% EGFP-positive cells (Cas9-GFP mRNA) were detected by flow cytometry (Figure 5a,c, and Figure S34). We also found that DET-CPD-12-mediated transfection of the Cas9-mRNA targeting EGFP gene was capable of disrupting GFP expression in 293T-EGFP cells (Figure 5b and Figure S30e), which was more superior to Lipo 3000- or PEI-mediated transfection. The excellent performance for the transfection of Cas9 mRNA by DET-CPD-12 also turned into efficient genome editing at CCNE1, resulting in the indel frequency of 23.6% (Figure 5d). Lastly, we also found that the intracellular delivery of rhodamine-labeled Cas9 was efficient, which resulted in strong GFP disruption by complexing sgRNA targeting EGFP (Figure 5e–g and Figure S30f). Though the intracellular delivery of Cas9 RNP by DET-CPD-12 was slightly less efficient in comparison with the commercial transfection agent (Lipofectamine CRISPRMAX, CMAX), it still induced an indel frequency up to 28.8% in the CCNE1 locus (Figure 5h). It should be noted that DET-CPD-12 loading with different forms of CRISPR-Cas9 cargoes did not exhibit any apparent cytotoxicity up to a dose of 64 μg/mL (Figures S35 and S36). Whereas the cell viability only slightly decreased with time, the treatment of cells with d,l-buthionine sulfoximine (BSO), a specific γ-glutamylcysteine synthetase inhibitor that limits the biosynthesis of intracellular GSH, resulted in the evident decrease of cell viability with the increased incubation time (Figures S37 and S38). The above information suggested that the disulfide bonds linking the monomers critically contributed to the low cytotoxicity nature of DET-CPD-12. Collectively, these results demonstrated that DET-CPD-12 was efficient and safe in mediating CRISPR-Cas-based genome editing at the target genomic locus.
Figure 5.
DET-CPD-12-mediated transfection of Cas9 mRNA or Cas9 RNP for genome editing in vitro. (a) GFP expression in 293T after transfecting Cas9 mRNA with the GFP tag. (c) Flow cytometry analysis of GFP-positive cells after Cas9 mRNA transfection in 293T cells was quantified. Disruption of EFGP genes after the transfection of Cas9 mRNA/sgRNA (b) or Cas9 RNP (f). Intracellular delivery of rhodamine-labeled Cas9 protein (e) (scale bar, 50 μm) and quantitative analysis of rhodamine-positive 293T cells by flow cytometry analysis (g). Analysis of indel frequency in the CCNE1 locus after the transfection of Cas9 mRNA/sgRNA (d), or Cas9 RNP (h) by DET-CPD-12. The scale bar of parts a, b, and f represents 200 μm. All quantitative data represent mean ± S.D. (n = 3, one-way ANOVA with a Tukey’s posthoc test: N.S., P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001).
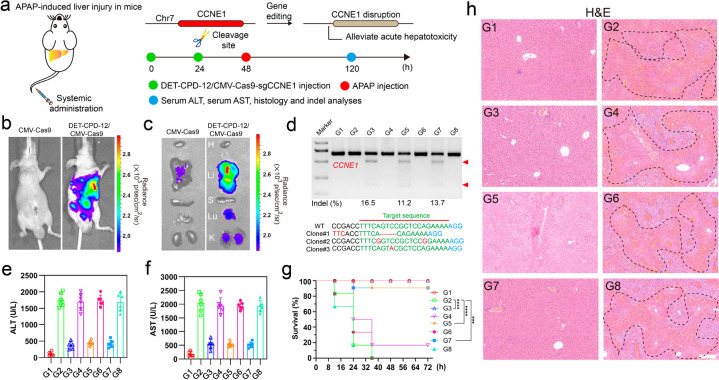
Finally, we explored the therapeutic potential through the delivery of genome-editing biomacromolecules by DET-CPD-12 in vivo. To this end, we first investigated the in vivo distribution of DET-CPD-12/DNA nanocomplexes 48 h after systemic administration (Figure 6a). For different forms of fluorescence-labeled CRISPR-Cas9 machineries (including plasmid, mRNA, and RNP), the fluorescence signals were clearly observed to accumulate primarily in the liver, followed by the lung and kidney (Figures S39–S41). As reflected by luciferase signals, the strong expression of the CRISPR-Cas9 plasmid was observed in the liver, followed by moderate or weak expression in the lung and kidneys (Figure 6b,c). Nevertheless, the tail-vein injection of the naked CRISPR-Cas9 plasmid generated poor luciferase expression (Figure 6b,c). Therefore, we are curious about whether the systemic administration of genome-editing agents would possibly treat liver-associated diseases. Acute liver failure (ALF), also known as fulminant hepatic failure, is defined by coagulopathy and hepatic encephalopathy, which is usually induced by hepatitis viruses or drugs. ALF can cause serious complications and significant liver dysfunctions, a complicated process involving hepatic parenchymal and nonparenchymal cells.62,63 As previous studies have shown that the delivery of siRNA targeting CCNE1 prevents the liver from liver fibrogenesis,64 we therefore designed sgRNA targeting CCNE1 and delivered different forms of CRISPR-Cas9 in vivo to investigate their therapeutic efficacies for the prevention of fulminant hepatic failure (FHF).64,65 The mice from acetaminophen (APAP)-induced FHF were first treated with DET-CPD-12 loaded with the CRISPR-Cas9 plasmid targeting CCNE1, which induced the indel frequency of 16.5%. Similarly, the delivery of CRIPSR/Cas9 mRNA and Cas9 RNP resulted in the indel frequency of 11.2% and 13.7% (Figure 6d), respectively. The mutation frequency was also confirmed by deep-sequencing analysis (Figure S42). Blood biochemistry suggested that these treatments significantly decreased AST (aspartate transaminase) and ALT (alanine aminotransferase) levels and effectively extended the survival time of the mice with FHF (Figure 6e–g). In the meantime, reduced hyperemia and the necrotic area in the liver were clearly observed for those mice received with CRISPR-Cas9 treatments, in sharp contrast with ones treated with CRISPR-Cas9 with scramble sgRNA (Figure 6h and Figures S43). Hematological indicators were also assessed 5 days after the systemic administration of these genome-editing agents, and we found that the administration of DET-CPD-12 at the therapy-relevant dose of 100 μg/mL merely induced any toxicity in the liver and kidney during the observational period (Figure S44). The above results were further confirmed by H&E staining of lung and kidney sections, where no serious side effects were observed (Figure S45). These results demonstrated that the bifunctional poly(disulfide)s we have developed are effective for the in vivo delivery of CRISPR-Cas9 biomacromolecules for therapeutic genome editing.
Figure 6.
Therapeutic effects of genome editing in vivo. Schematic illustration of DET-CPD-12 for in vivo delivery of CMV-Cas9-sgCCNE1 for the treatment of APAP-induced hepatic injury. (a) APAP was administered through intraperitoneal injection, and DET-CPD-12/CMV-Cas9-sgCCNE1 complexes were administered through the tail vein; hematological and histological analysis was carried out 5 days after the first treatment. In vivo luciferase expression of the Cas9 plasmid with the luciferase tag in the whole mice (b) or in the dislodged organs from treated mice (c): H, heart; Li, liver; S, spleen; Lu, lung; K, kidney. (d) Frequency of indel mutation detected by the T7E1 assay from liver tissue and representative Sanger sequencing results of T-A cloning from liver tissue after treatments (Clone 1, DET-CPD-12/CMV-Cas9-sgCCNE1; Clone 2, DET-CPD-12/Cas9 mRNA-sgCCNE1; Clone 3, DET-CPD-12/Cas9 RNP-sgCCNE1). The serum ALT (e) and AST (f) levels after different treatments. (g) Survival rates after different treatments (n = 6). Statistical significance was calculated by log-rank test (***P < 0.001, ****P < 0.0001). (h) H&E staining of the liver sections from the mice after the specified treatments. The regions within the dotted lines denote the accumulation of blood cells. Scale bar, 200 μm. For parts d–h, the code denotes the following: G1, mice without APAP treatment treated with PBS; G2, APAP-treated mice treated with PBS; G3, APAP-treated mice treated with the DET-CPD-12/Cas9-sgCCNE1 plasmid; G4, APAP-treated mice treated with the DET-CPD-12/Cas9-sgMock plasmid; G5, APAP-treated mice treated with DET-CPD-12/Cas9-sgCCNE1 mRNA; G6, APAP-treated mice treated with DET-CPD-12/Cas9-sgMock mRNA; G7, APAP-treated mice treated with DET-CPD-12/RNP-sgCCNE1; G8, APAP-treated mice treated with DET-CPD-12/RNP-sgMock. Error bars represent the standard error (mean ± S.D., n = 6).
In conclusion, we have developed a series of poly(disulfide)s and demonstrated that the bifunctional poly(disulfide)s with DET and CPD blocks exhibited great potentials for the efficient delivery of different forms of genome-editing biomacromolecules in vitro, and in vivo delivery of CRISPR-Cas9 targeting CCNE1 by DET-CPD-12 could successfully protect mice for FHF. The excellent delivery performance of DET-CPD-12 stems from their delicate molecular structure to interact with these biomacromolecules, the efficient intracellular delivery through the thiol-mediated uptake pathway, and strong endosomal escape capability by membrane disruption. Because the disulfide linkers within the main chain of poly(disulfide)s are readily cleavable under the intracellular glutathione level, the degradation of DET-CPD-12 not only triggers the timely release of CRISPR-Cas9 machineries but also well minimizes the cytotoxicity induced by the intracellular accumulation of the polymer. These merits collectively account for the excellent capability of DET-CPD-12 to mediate different forms to CRISPR-Cas9 for the efficient genome-editing activities at different levels. In the future, further functionalization of DET-CPD-12 is still highly desirable to promote the delivery precision, such as organ or tissue specificity in vivo. Collectively, the current study not only successfully develops a delivery platform for the efficient intracellular transport of CRISPR-Cas9 machineries but also offers useful insights for the rational design of polymers for the therapeutic delivery of genome-editing agents.
Acknowledgments
This work was supported by National Natural Science Foundation of China (82073779), Natural Science Foundation of Zhejiang Province (Distinguished Young Scholar Program, LR21H300002), Fundamental Research Funds for the Zhejiang Provincial Universities (2021XZZX036), National Key Research and Development Program of China (2018YFA0901800), and Leading Talent of “Ten Thousand Plan”—National High-Level Talents Special Support Plan.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01648.
Methods for the synthesis of M1 and M2, the polymerization of poly(disulfide)s, sgRNA design, cell culture and transfection, cell viability, characterization, gel retardation assay, Cas9 release, cellular uptake, T7E1 assay, off-target analysis, mouse model of fulminant hepatic failure, hemolysis assay, and statistical analysis approaches; NMR data, GPC profiles, agarose gel electrophoresis assay, hydrodynamic size and ζ potential, luciferase expression, release data, CLSM images, hemolytic activity, mechanism of cellular uptake, transfection data, EGFP gene disruption, deep-sequencing analysis, cell viability, in vivo fluorescence images, necrotic areas and hematological evaluation, and H&E staining; polymerization conditions, primer sequences for sgRNA and PCR amplification, sequences of off-target sites and PCR amplification, and off-target site analysis(PDF)
Author Contributions
∥ J.G. and T.W. contributed equally to this work. Y.P. conceived the project and designed experiment. J.G. and B.L. synthesized and characterized the materials. J.G., T.W. and Q.P. performed the biological experiments, J.G. and T.W. analyzed the data. H.X. constructed the plasmids and mRNA. Y.P. supervised the project and wrote manuscript.
The authors declare no competing financial interest.
This paper was published ASAP on May 27, 2021 with an error in Figure 5. The corrected version was reposted on June 7, 2021.
Supplementary Material
References
- Cong L.; Ran F.; Cox D.; Lin S.; Barretto R.; Habib N.; Hsu P. D.; Wu X.; Wu W.; Marraffini L. A.; Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Q.; Joung J. K. Defining and improving the genome wide specificities of CRISPR–Cas9 nucleases. Nat. Rev. Genet. 2016, 17, 300–312. 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Zhao W.; Luo X.; Zhang X.; Li C.; Zeng C.; Dong Y. Engineering CRISPR–Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat. Biomed. Eng. 2017, 1, 1–10. 10.1038/s41551-017-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan T.; Niu D.; Wu C.; Xu F. J.; Church G. M.; Ping Y. Material solutions for delivery of CRISPR/Cas-based genome editing tools: Current status and future outlook. Mater. Today 2019, 26, 40–46. 10.1016/j.mattod.2018.12.003. [DOI] [Google Scholar]
- Wan T.; Ping Y. Delivery of genome-editing biomacromolecules for treatment of lung genetic disorders. Adv. Drug Delivery Rev. 2021, 168, 196–216. 10.1016/j.addr.2020.05.002. [DOI] [PubMed] [Google Scholar]
- Sun W. J.; Ji W.; Hall J. M.; Hu Q. Y.; Wang C.; Beisel C. L.; Gu Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem., Int. Ed. 2015, 54, 12029–12033. 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Li H.; Min Y. L.; Ortiz E. S.; Hang J.; Mlreault A. A.; Shelton J. M.; Kim J.; Mammen P. P.; Duby R. B.; Olson E. N. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci. Adv. 2020, 6, eaay6812. 10.1126/sciadv.aay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Fernandez O.; Osorio F. G.; Quesada V.; Rodriguez F.; Basso S.; Maeso D.; Rolas L.; Barkaway A.; Nourshargh S.; Folgueras A. R.; Frejie J. P.; Otin C. L. Development of a CRISPR/Cas9-based therapy for Hutchinson–Gilford progeria syndrome. Nat. Med. 2019, 25, 423–426. 10.1038/s41591-018-0338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.; Kim J. S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Chew W. L.; Tabebordbar M.; Cheng J. K.; Mali P.; Wu E. Y.; Zhu K.; Wagers A. J.; Church G. M. A multifunctional AAV–CRISPR–Cas9 and its host response. Nat. Methods 2016, 13, 868–874. 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. X.; Li M.; Lee C. M.; Chakraborty S.; Kim H. W.; Bao G.; Leong K. W. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev. 2017, 117, 9874–9906. 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- Cheng Q.; Wei T.; Farbiak L.; Johnson L. T.; Dilliard S. A.; Siegwart D. J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas9 gene editing. Nat. Nanotechnol. 2020, 15, 313–320. 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.; Lee K.; Panda S.; Rojas R. G.; Chong A.; Bugay V.; Park H. M.; Brenner R.; Murthy N.; Lee H. Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2, 497–507. 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Chen Y.; Xin H.; Wan T.; Ping Y. Near-Infrared Optogenetic Engineering of Photothermal NanoCRISPR for Programmable Genome Editing. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 2395–2405. 10.1073/pnas.1912220117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Wan T.; Wang H.; Zhang S.; Ping Y.; Cheng Y. A boronic acid–rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019, 5, eaaw8922. 10.1126/sciadv.aaw8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Wang J.; Hu Q.; Zhou X.; Khademhosseini A.; Gu Z. CRISPR-Cas12a delivery by DNA-mediated bioresponsive editing for cholesterol regulation. Sci. Adv. 2020, 6, eaba2983. 10.1126/sciadv.aba2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y.; Wilson D. R.; Choi J.; Varanasi M.; Sanders K.; Karlsson J.; Lim M.; Green J. J. Carboxylated branched poly (β-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019, 5, eaay3255. 10.1126/sciadv.aay3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y.; Varanasi M.; Mendes S.; Yamagata H. M.; Wilson D. R.; Green J. J. Poly (Beta-Amino Ester) Nanoparticles Enable Nonviral Delivery of CRISPR-Cas9 Plasmids for Gene Knockout and Gene Deletion. Mol. Ther.--Nucleic Acids 2020, 20, 661–672. 10.1016/j.omtn.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Zhao W.; Nguyen G. N.; Zhang C.; Zeng C.; Yan J.; Du S.; Hou X.; Li W.; Jiang J.; Deng B.; Mccomb D. W.; Dorkin R.; Shah A.; Barrera L.; Gregoire F.; Singh M.; Chen D.; Sabatino D. E.; Dong Y. Functionalized lipid-like nanoparticles for in vivo mRNA delivery and base editing Functionalized lipid-like delivery and base editing. Sci. Adv. 2020, 6, eabc2315. 10.1126/sciadv.abc2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzmann J. A.; Ho D.; Evans C. W.; Lam J. P.; Bloj B. G.; Mohamed A. E.; Omara M. L.; Ford E.; Tan D. K.; Lister R.; Blancafort P.; Norret M.; Lyer K. S. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem. Sci. 2017, 8, 2923–2930. 10.1039/C7SC00097A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K.; Jeong E.; Lee J.; Jung M.; shin E.; Kim Y. H.; Lee K.; Jung I.; Kim D.; Kim S.; Kim J. S. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.; Ashwaniumar N.; Robinson E.; Xia Y.; Mihai C.; Grifith J. P.; Hou S.; Esposito A. A.; Ketova T.; Welsher K.; Joyal J. L.; Almarsson O.; Sahay G. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020, 11, 983. 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T.; Cheng Q.; Min Y. L.; Olson E. N.; Sigwart D. J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 2020, 11, 3232. 10.1038/s41467-020-17029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Hu S.; Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmus V.; Woodward M.; Lin L.; Murthy N.; Stayton P.; Hoffman A. A new pH-responsive and glutathione-reactive, endosomal membrane-disruptive polymeric carrier for intracellular delivery of biomolecular drugs. J. Controlled Release 2003, 93, 105–120. 10.1016/j.jconrel.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Sun W.; Gu Z. Tailoring non-viral delivery vehicles for transporting genome-editing tools. Sci. China. Mater. 2017, 60, 511–515. 10.1007/s40843-016-5154-4. [DOI] [Google Scholar]
- Fu J.; Yu C.; Li L.; Yao S. Q. Intracellular Delivery of Functional Proteins and Native Drugs by Cell-Penetrating Poly(disulfide)s. J. Am. Chem. Soc. 2015, 137, 12153–12160. 10.1021/jacs.5b08130. [DOI] [PubMed] [Google Scholar]
- Gasparini G.; Matile S. Protein delivery with cell-penetrating poly(disulfide)s. Chem. Commun. 2015, 51, 17160–17162. 10.1039/C5CC07460F. [DOI] [PubMed] [Google Scholar]
- Yu C.; Qian L.; Ge J.; Fu J.; Yuan P.; Yao S. L.; Yao S. Q. Cell-Penetrating Poly(disulfide) Assisted Intracellular Delivery of Mesoporous Silica Nanoparticles for Inhibition of miR-21 Function and Detection of Subsequent Therapeutic Effects. Angew. Chem., Int. Ed. 2016, 55, 9272–9276. 10.1002/anie.201602188. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Wang Y.; Lian J.; Shen Q.; Wang C.; Ma B.; Zhang Y.; Xu T.; Li J.; Shao Y.; Xu F.; Zhu J. J. Engineering the Surface of Smart Nanocarriers Using a pH-/Thermal-/GSH-Responsive Polymer Zipper for Precise Tumor Targeting Therapy In Vivo. Adv. Mater. 2017, 29, 1702311. 10.1002/adma.201702311. [DOI] [PubMed] [Google Scholar]
- Derivery E.; Bartolami E.; Matile S.; Gaitan M. G. Efficient Delivery of Quantum Dots into the Cytosol of Cells Using Cell-Penetrating Poly(disulfide)s. J. Am. Chem. Soc. 2017, 139, 10172–10175. 10.1021/jacs.7b02952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abegg D.; Gasparini G.; Hoch D. G.; Shuster A.; Bartolami E.; Matile S.; Adibekian A. Strained Cyclic Disulfides Enable Cellular Uptake by Reacting with the Transferrin Receptor. J. Am. Chem. Soc. 2017, 139, 231–238. 10.1021/jacs.6b09643. [DOI] [PubMed] [Google Scholar]
- Shu Z.; Tanaka I.; Ota A.; Fushihara D.; Abe N.; Kawaguchi S.; Nakamoto K.; Tomoike F.; Tada S.; Ito Y.; Kimura Y.; Abe H. Disulfide-Unit Conjugation Enables Ultrafast Cytosolic Internalization of Antisense DNA and siRNA. Angew. Chem., Int. Ed. 2019, 58, 6611–6615. 10.1002/anie.201900993. [DOI] [PubMed] [Google Scholar]
- Yuan P.; Zhang H.; Qian L.; Mao X.; Du S.; Yu C.; Peng B.; Yao S. Q. Intracellular Delivery of Functional Native Antibodies under Hypoxic Conditions by Using a Biodegradable Silica Nanoquencher. Angew. Chem., Int. Ed. 2017, 56, 12481–12485. 10.1002/anie.201705578. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Waymouth R. M. 1,2-Dithiolane-Derived Dynamic, Covalent Materials: Cooperative Self-Assembly and Reversible Cross-Linking. J. Am. Chem. Soc. 2017, 139, 3822–3833. 10.1021/jacs.7b00039. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Sun L.; Wang L.; Liu Y.; Li J.; Li J.; Li J.; Yang H. Self-Assembled and Size-controllable Oligonucleotides Nanospheres for Effective Antisense Gene Delivery via Endocytosis-independent Pathway. Angew. Chem., Int. Ed. 2019, 58, 5236–5240. 10.1002/anie.201813665. [DOI] [PubMed] [Google Scholar]
- Mosquera J.; Garcia I.; Marzan L. M. Cellular Uptake of Nanoparticles versus Small Molecular: A Matter of Size. Acc. Chem. Res. 2018, 51, 2305–2313. 10.1021/acs.accounts.8b00292. [DOI] [PubMed] [Google Scholar]
- Bang E. K.; Gasparini G.; Moinard G.; Roux A.; Sakai N.; Matile S. Substrate-Initiated Synthesis of Cell-Penetrating Poly(disulfide)s. J. Am. Chem. Soc. 2013, 135, 2088–2091. 10.1021/ja311961k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini G.; Bang E. K.; Moinard G.; Tulumello D. V.; Ward S.; Kelley S. O.; Roux A.; Sakai N.; Matile S. Cellular Uptake of Substrate-Initiated Cell-Penetrating Poly(disulfide)s. J. Am. Chem. Soc. 2014, 136, 6069–6074. 10.1021/ja501581b. [DOI] [PubMed] [Google Scholar]
- Gasparini G.; Sargsyan G.; Bang E. K.; Sakai N.; Matile S. Ring Tension Applied to Thiol-Mediated Cellular Uptake. Angew. Chem., Int. Ed. 2015, 54, 7328–7331. 10.1002/anie.201502358. [DOI] [PubMed] [Google Scholar]
- Zong L.; Bartolami E.; Abegg D.; Adibekian A.; Sakai N.; Matile S. Epidithiodiketopiperazines: Strain-Promoted Thiol-Mediated Cellular Uptake at the Highest Tension. ACS Cent. Sci. 2017, 3, 449–453. 10.1021/acscentsci.7b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulisauer L.; Kathriner N.; Prenrecaj M.; Gauthier M. A.; Leroux J.-C. Tracking the Bioreduction of Disulfide-Containing Cationic Dendrimers. Angew. Chem., Int. Ed. 2012, 51, 12454–12458. 10.1002/anie.201207070. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Johnston A. P.; Dodds S. J.; Kamphuis M. J.; Ferguson C.; Parton R. G.; Nice E. C.; Heath J. K.; Caruso F. Uptake and Intracellular Fate of Disulfide-Bonded Polymer Hydrogel Capsules for Doxorubicin Delivery to Colorectal Cancer Cells. ACS Nano 2010, 4, 2928–2936. 10.1021/nn100173h. [DOI] [PubMed] [Google Scholar]
- Shimoni O.; Postma A.; Yan Y.; Scott A. M.; Heath J. K.; Nice E. C.; Zelikin A.; Caruso F. Macromolecule Functionalization of Disulfide-Bonded Polymer Hydrogel Capsules and Cancer Cell Targeting. ACS Nano 2012, 6, 1463–1472. 10.1021/nn204319b. [DOI] [PubMed] [Google Scholar]
- Yuan P.; Zhang H.; Qian L.; Mao X.; Du S.; Yu C.; Peng B.; Yao S. Q. Intracellular Delivery of Functional Native Antibodies under Hypoxic Conditions by Using a Biodegradable Silica Nanoquencher. Angew. Chem., Int. Ed. 2017, 56, 12481–12485. 10.1002/anie.201705578. [DOI] [PubMed] [Google Scholar]
- Chuard N.; Gasparini G.; Moreau D.; Lorcher S.; Palivan C.; Meier W.; Sakai N.; Matile S. Strain-Promoted Thiol-Mediated Cellular Uptake of Giant Substrates: Liposomes and Polymersomes. Angew. Chem., Int. Ed. 2017, 56, 2947–2950. 10.1002/anie.201611772. [DOI] [PubMed] [Google Scholar]
- Qin X.; Yu C.; Wei J.; Li L.; Zhang C.; Wu Q.; Liu J.; Yao S. Q.; Huang W. Rational Design of Nanocarriers for Intracellular Protein Delivery. Adv. Mater. 2019, 31, 1902791. 10.1002/adma.201902791. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Oba M.; Nakanishi M.; Fukushima S.; Yamasaki Y.; Koyama H.; Nishiyama N.; Kataoka K. Polyplexes from Poly(aspartamide) Bearing 1,2-Diaminoethane Side Chains Induce pH-Selective, Endosomal Membrane Destabilization with Amplified Transfection and Negligible Cytotoxicity. J. Am. Chem. Soc. 2008, 130, 16287–16294. 10.1021/ja804561g. [DOI] [PubMed] [Google Scholar]
- Sun M.; Wang K.; Oupicky D. Advances in Stimulus-Responsive Polymeric Materials for Systemic Delivery of Nucleic Acids. Adv. Healthcare Mater. 2018, 7, 1701070. 10.1002/adhm.201701070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Shen W.; Ling J.; Yan Y.; Hu J.; Cheng Y. The Fluorination Effect of Fluoroamphiphiles in Cytosolic Protein Delivery. Nat. Commun. 2018, 9, 1377. 10.1038/s41467-018-03779-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan T.; Chen Y.; Pan Q.; Xu X.; Kang Y.; Gao X.; Huang F.; Wu C.; Ping Y. Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J. Controlled Release 2020, 322, 236–247. 10.1016/j.jconrel.2020.03.015. [DOI] [PubMed] [Google Scholar]
- Chang H.; Lv J.; Gao X.; Wang X.; Wang H.; Chen H.; Hu X.; Li L.; Cheng Y. Rational Design of a Polymer with Robust Efficacy for Intracellular Protein and Peptide Delivery. Nano Lett. 2017, 17, 1678–1684. 10.1021/acs.nanolett.6b04955. [DOI] [PubMed] [Google Scholar]
- Okuro K.; Kinbara K.; Tsumoto K.; Ishii N.; Aida T. Molecular Glues Carrying Multiple Guanidinium Ion Pendants via an Oligoether Spacer: Stabilization of Microtubules against Depolymerization. J. Am. Chem. Soc. 2009, 131, 1626–1627. 10.1021/ja800491v. [DOI] [PubMed] [Google Scholar]
- Yang J.; Zhang Q.; Chang H.; Cheng Y. Surface-Engineered Dendrimers in Gene Delivery. Chem. Rev. 2015, 115, 5274–5300. 10.1021/cr500542t. [DOI] [PubMed] [Google Scholar]
- Lyu Y.; Cui D.; Sun H.; Miao Y.; Duan H.; Pu K. Dendronized Semiconducting Polymer as Photothermal Nanocarrier for Remote Activation of Gene Expression. Angew. Chem., Int. Ed. 2017, 56, 9155–9159. 10.1002/anie.201705543. [DOI] [PubMed] [Google Scholar]
- Lyu Y.; He S.; Li J.; Jiang Y.; Sun H.; Miao Y.; Pu K. A Photolabile Semiconducting Polymer Nanotransducer for Near-Infrared Regulation of CRISPR/Cas9 Gene Editing. Angew. Chem., Int. Ed. 2019, 58, 18197–18201. 10.1002/anie.201909264. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Kuang G.; He S.; Liu S.; Lu H.; Li X.; Zhou D.; Huang Y. Chain-shattering Pt(IV)-backboned polymeric nanoplatform for efficient CRISPR/Cas9 gene editing to enhance synergistic cancer therapy. Nano Res. 2021, 14, 601–610. 10.1007/s12274-020-3066-4. [DOI] [Google Scholar]
- Derakhshankhah H.; Jafari S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomed. Pharmacother. 2018, 108, 1090–1096. 10.1016/j.biopha.2018.09.097. [DOI] [PubMed] [Google Scholar]
- Du S.; Liew S. S.; Li L.; Yao S. Q. Bypassing Endocytosis: Direct Cytosolic Delivery of Proteins. J. Am. Chem. Soc. 2018, 140, 15986–15996. 10.1021/jacs.8b06584. [DOI] [PubMed] [Google Scholar]
- Itaka K.; Ishii T.; Hasegawa Y.; Kataoka K. Biodegradable polyamino acid-based polycations as safe and effective gene carrier minimizing cumulative toxicity. Biomaterials 2010, 31, 3707–3714. 10.1016/j.biomaterials.2009.11.072. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Nishiyama N.; Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem. Soc. Rev. 2012, 41, 2562–2574. 10.1039/C1CS15258K. [DOI] [PubMed] [Google Scholar]
- Stickel F.; Kessebohm K.; Weimann R.; Seitz H. K. Review of liver injury associated with dietary supplements. Liver Int. 2011, 31, 595–605. 10.1111/j.1478-3231.2010.02439.x. [DOI] [PubMed] [Google Scholar]
- Iredale J. P. Hepatic Stellate Cell Behavior during Resolution of Liver Injury. Semin. Liver Dis. 2001, 21, 427–436. 10.1055/s-2001-17557. [DOI] [PubMed] [Google Scholar]
- Bangen J. M.; Hammerish L.; Sonntag R.; Baues M.; Haas U.; Lamertz D.; Longerish T.; Lammers T.; Tacke F.; Trautwein C.; Liedtke C. Targeting CCl4-Induced Liver Fibrosis by RNA Interference–Mediated Inhibition of Cyclin E1 in Mice. Hepatology 2017, 66, 1242–1257. 10.1002/hep.29275. [DOI] [PubMed] [Google Scholar]
- Chen D.; Ni H. M.; Wang L.; Ma X.; Yu J.; Ding W. X.; Zhang L. PUMA induction mediates acetaminophen-induced necrosis and liver injury. Hepatology 2019, 69, 2164–2179. 10.1002/hep.30422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.