Abstract
Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) is causing an unprecedented global pandemic demanding the urgent development of therapeutic strategies. Microarray binding experiments, using an extensive heparan sulfate (HS) oligosaccharide library, showed that the receptor binding domain (RBD) of the spike of SARS-CoV-2 can bind HS in a length- and sequence-dependent manner. A hexasaccharide composed of IdoA2S-GlcNS6S repeating units was identified as the minimal binding epitope. Surface plasmon resonance showed the SARS-CoV-2 spike protein binds with a much higher affinity to heparin (KD = 55 nM) compared to the RBD (KD = 1 μM) alone. It was also found that heparin does not interfere in angiotensin-converting enzyme 2 (ACE2) binding or proteolytic processing of the spike. However, exogenous administered heparin or a highly sulfated HS oligosaccharide inhibited RBD binding to cells. Furthermore, an enzymatic removal of HS proteoglycan from physiological relevant tissue resulted in a loss of RBD binding. The data support a model in which HS functions as the point of initial attachment allowing the virus to travel through the glycocalyx by low-affinity high-avidity interactions to reach the cell membrane, where it can engage with ACE2 for cell entry. Microarray binding experiments showed that ACE2 and HS can simultaneously engage with the RBD, and it is likely no dissociation between HS and RBD is required for binding to ACE2. The results highlight the potential of using HS oligosaccharides as a starting material for therapeutic agent development.
Short abstract
Various methods demonstrate that the RBD of SARS-CoV-2 spike can bind to heparan sulfate (HS) in a sequence-dependent manner supporting a model in which HS functions as the initial attachment point.
Introduction
Our response to the severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) pandemic demands an urgent development of therapeutic strategies. An attractive approach is to interfere in the attachment of the virus to the host cell.1 The entry of SARS-CoV-2 into cells is initiated by the binding of the transmembrane spike (S) glycoprotein of the virus to angiotensin-converting enzyme 2 (ACE2) of the host.2 SARS-CoV is closely related to SARS-CoV-2 and employs the same receptor.3 The spike protein of SARS-CoV-2 is comprised of two subunits; S1 is responsible for binding to the host receptor, whereas S2 promotes membrane fusion. The C terminal domain (CTD) of S1 harbors the receptor binding domain (RBD).4 It is known that the spike protein of a number of human coronaviruses can bind to a secondary receptor, or coreceptor, to facilitate cell entry. For example, the Middle East respiratory syndrome (MERS)-CoV employs sialic acid as a coreceptor along with its main receptor DPP4.5 Human CoV-NL63, which also utilizes ACE2 as the receptor, uses heparan sulfate (HS) proteoglycans as a coreceptor.6 It has also been shown that the entry of SARS-CoV pseudotyped virus into Vero E6 and Caco-2 cells can substantially be inhibited by heparin or treatment with heparin lyases, indicating the importance of HS for infectivity.7
There are indications that the SARS-CoV-2 spike also interacts with HS. One early report showed that heparin can induce a conformation change in the RBD of SARS-CoV-2.8 A combined surface plasmon resonance (SPR) and computational study indicated that glycosaminoglycans (GAGs) can bind to the proteolytic cleavage site of the S1 and S2 protein.9,10 Several reports have indicated that heparin or related structures can inhibit the infection process of SARS-CoV-2 in different cell lines.11−14
HS are highly complex O- and N-sulfated polysaccharides that reside as major components on the cell surface and extracellular matrix of all eukaryotic cells.15 Various proteins interact with HS thereby regulating many biological and disease processes, including cell adhesion, proliferation, differentiation, and inflammation. They are also used by many viruses, including herpes simplex virus (HSV), Dengue virus, human immunodeficiency virus (HIV), and various coronaviruses, as receptor or coreceptor.16−18
The biosynthesis of HS is highly regulated, and the length, degree, and pattern of sulfation of HS can differ substantially between different cell types. The so-called HS sulfate code hypothesis is based on the notion that the expression of specific HS epitopes by cells makes it possible to recruit specific HS-binding proteins, thereby controlling a multitude of biological processes.19,20 In support of this hypothesis, several studies have shown that HS binding proteins exhibit preferences for specific HS oligosaccharide motifs.21,22 Therefore, we were compelled to investigate whether the spike of SARS-CoV-2 recognizes specific HS motifs. Such an insight is expected to pave the way to develop inhibitors of viral cell binding and entry.
Previously, we prepared an unprecedented library of structurally well-defined HS oligosaccharides that differ in chain length, backbone composition, and sulfation pattern.23,24 This collection of HS oligosaccharides was used to develop a glycan microarray for the systematic analysis of selectivity of HS-binding proteins. Using this microarray platform in conjugation with detailed binding studies, we found that the RBD domain of SARS-CoV-2 spike can bind HS in a length- and sequence-dependent manner, and the observations support a model in which the RBD confers sequence selectivity, and the affinity of binding is enhanced by additional interactions with other HS binding sites in, for example, the S1/S2 proteolytic cleavage site.9 Identified HS oligosaccharide ligands could inhibit the binding of RBD to cells. Furthermore, tissue-staining studies using biologically relevant tissues indicate that HS proteoglycans (HSPG) are critical for the initial attachment of the virus to cells. It was also found that heparin does not interfere in ACE2 binding or proteolytic processing of the spike. The spike can, however, bind simultaneously with HS and ACE2, and no dissociation between RBD and HS is required before it can engage with ACE2 for cell entry.
Results and Discussion
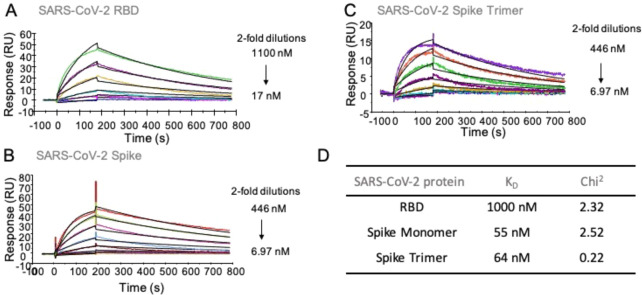
Surface plasmon resonance experiments were performed to probe whether the RBD domain of the SARS-CoV-2 spike protein can bind with heparin. Biotinylated heparin was immobilized on a streptavidin-coated sensor chip, and binding experiments were performed by employing as analytes different concentrations of the RBD, monomeric spike protein, and trimeric spike protein of SARS-CoV-2. The spike glycoprotein of SARS-CoV-2 (S1+S2, extracellular domain, amino acid residues 1–1213) was expressed in insect cells having a C-terminal His-tag.25,26 Recombinant SARS-CoV-2-RBD, containing amino acid residues 319–541, was expressed in HEK293 cells also with a C-terminal His-tag.25,26 The trimer spike, having the furin cleavage site deleted and bearing with two stabilizing mutations, was expressed in HEK293 cells with a C-terminal His-tag.
Representative sensorgrams are shown in Figure 1. Equilibrium dissociation constant (KD) values were determined using a 1:1 Langmuir binding model. Other binding models, including bivalent analyte and two-state binding, provided poorer fits. Detailed fitting data, including residual plots, are shown in Figure S1. The RBD domain binds to heparin with a moderate affinity having a KD value of ∼1 μM (Chi2 = 2.3). The full-length monomeric spike protein showed a much higher binding affinity with a KD value of 55 nM (Chi2 = 2.5). Previously reported computational studies have indicated that the RBD domain may harbor an HS binding site either within or adjacent to the receptor binding domain.9,11 It has also been suggested that another HS-binding site resides in the S1/S2 proteolytic cleavage site of the spike of the S2 domain.9 Thus, the high affinity of the monomeric spike protein may be due to the presence of additional binding sites in the spike protein, which enhances the binding to heparin. The trimeric spike protein displayed a similar binding affinity (KD = 64 nM, Chi2 = 0.22) as the monomer. One of the putative heparin binding sites in the trimeric spike protein, the S1/S2 proteolytic cleavage site, was mutated.25 Thus, a possible increase in avidity due to multivalency may have been offset by a lack of a secondary binding site.
Figure 1.
SPR sensorgrams representing the concentration-dependent kinetic analysis of the binding of immobilized heparin with SARS-CoV-2 related proteins. (A) RBD, (B) spike monomer, (C) spike trimer, (D) summary table of equilibrium dissociation constant (KD) and chi-square (Chi2) goodness-of-fit values. Data were analyzed using Biacore T100 evaluation software, and representative data are shown, which was repeated at least three times.
Intrigued by these results, we examined if the SARS-CoV-2 proteins bind to HS in a sequence-preferred manner. We have developed an HS microarray having well over 100 unique di-, tetra-, hexa-, and octasaccharides differing in backbone composition and sulfation pattern23,24,27 (Figure 2C). The synthetic HS oligosaccharides contain an anomeric aminopentyl linker allowing printing on N-hydroxysuccinimide (NHS)-activated glass slides. The HS oligosaccharides were printed at 100 μM concentration in replicates of six by noncontact piezoelectric printing. The quality of the HS microarray was validated using various well-characterized HS-binding proteins. Subarrays were incubated with different concentrations of SARS-CoV-2 RBD and spike protein in a binding buffer at room temperature for 1 h. After they were washed and dried, the subarrays were exposed to an anti-His antibody labeled with AlexaFluor647 for another hour, washed, and dried, and binding was detected by fluorescent scanning. To analyze the data, the compounds were arranged according to increasing backbone length and, within each group, by increasing numbers of sulfates. Intriguingly, the proteins showed a strong preference for specific HS oligosaccharides (Figure 2A,B). Furthermore, it was found that the RBD, monomeric spike protein, and trimeric spike protein exhibit similar binding patterns (Figure S2). Compounds showing a strong responsiveness (88, 90, 91, and 93) are composed of trisulfated repeating units (IdoA2S-GlcNS6S). On the one hand, the binding is length-dependent, and HS oligosaccharide 93 (IdoA2S-GlcNS6S)4 and 91 (IdoA2S-GlcNS6S)3 having four and three repeating units, respectively, showed the strongest binding. On the other hand, tetrasaccharide 56 (IdoA2S-GlcNS6S)2, which has the same repeating unit structure, gave a very low responsiveness. A similar observation was made for disaccharide 4 (IdoA2S-GlcNS6S).
Figure 2.
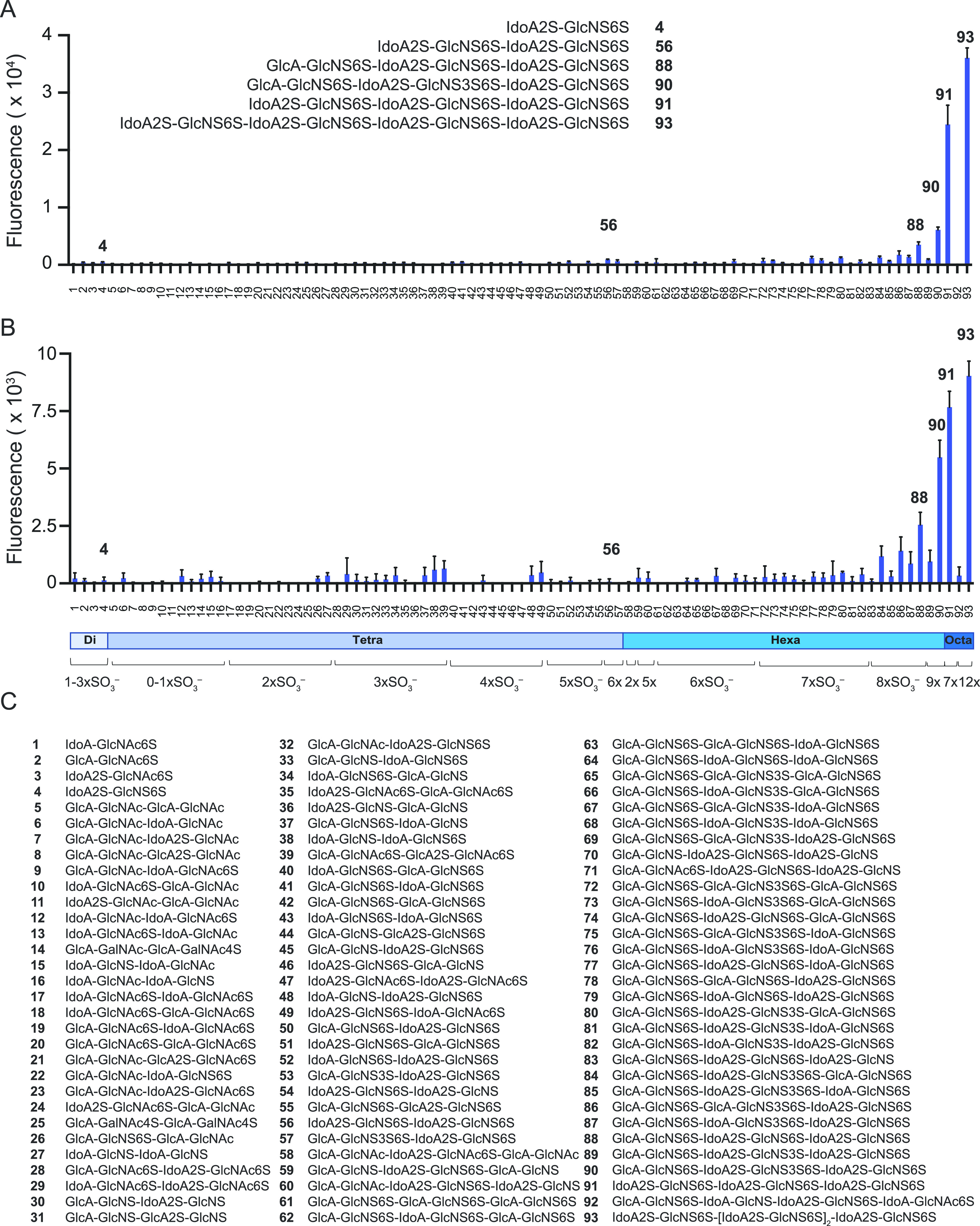
Binding analysis of synthetic HS oligosaccharides to SARS-CoV-2 related proteins by microarray. (A) Spike protein (10 μg/mL); structures of strongest binders are shown as insets. (B) RBD protein (30 μg/mL). (C) Compound numbering and structures. All compounds have a linker at reducing end, R = O(CH2)5NH2. Data are presented as mean ± SD (n = 4). Representative data are shown, which was repeated at least three times.
The structure–binding data show that perturbations in the backbone or sulfation pattern led to substantial reductions or loss in binding. The importance of the IdoA2S residue is highlighted by comparing hexasaccharides 91 with 88 in which a single IdoA2S in the distal disaccharide repeating unit is replaced with GlcA. This modification leads to a substantial reduction in responsiveness. Further replacements of IdoA2S with GlcA in compound 88 completely abolish binding, as evident for compounds 78, 74, and 61. The structure–activity data also showed that the 2-O-sulfates are crucial, and binding was lost when such functionalities were not present (88 vs 79, 77, and 64). Lack of one or more 6-O-sulfates also resulted in substantial reductions in binding (88 vs 83 and 70). The array contains a series of hexasaccharide motifs modified by 3-O-sulfate that have been observed in HS.27 The presence of a 3-O-sulfate somewhat enhanced binding (86 vs 78, 87 vs 79, and 90 vs 88). The fact that the removal of only one sulfate from a hexasaccharide such as 90 or 91 resulted in substantial reductions or loss in responsiveness indicates that the sulfates engage in specific interactions with the protein. The importance of electrostatic interactions, can, however, not be excluded and may also contribute to binding. Although SARS-CoV-2 spike and RBD showed similar selectivities, the binding of the spike appeared stronger, and much higher fluorescent readings were observed at the same protein concentration (Figure S2).
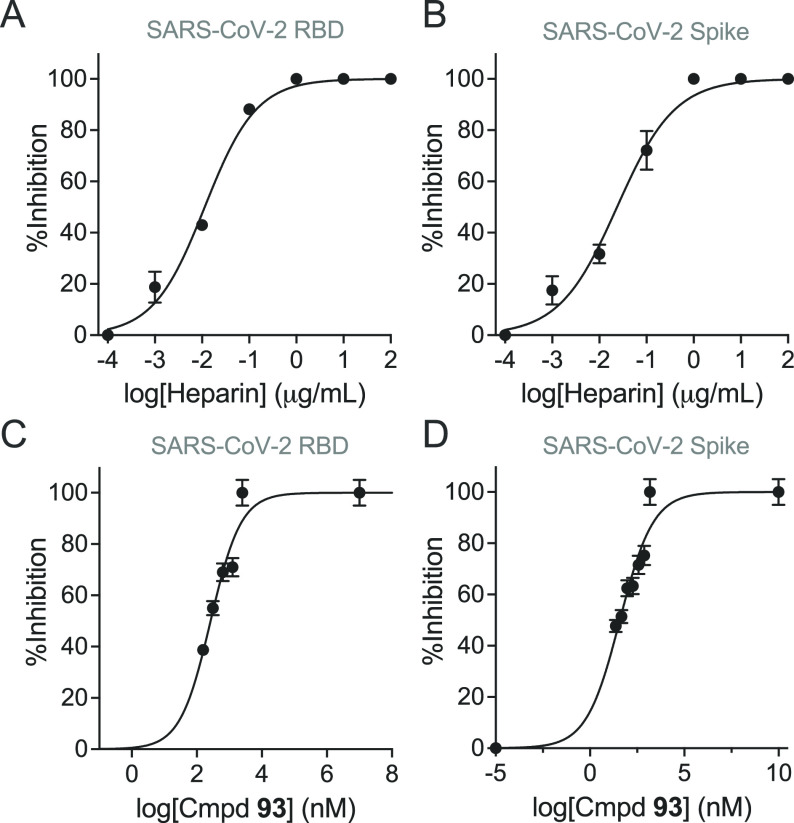
Next, we examined whether octasaccharide 93 can interfere in the interaction of the spike or RBD with immobilized heparin. Thus, the spike protein (150 nM) or RBD (2.4 μM) were premixed with different concentrations of compound 93 and then used as analytes in SPR experiments employing a sensor chip modified by heparin. A concentration-dependent reduction in SPR response units (RUs) was observed. IC50 values (50% reduction in RUs) were determined by a nonlinear fitting of log(inhibitor) versus response using a variable slope (Figure 3). The resulting values for spike and RBD are 38 nM (0.1 μg/mL) and 264 nM (0.73 μg/mL), respectively. Similar inhibition studies with unfractionated heparin (UFH) gave IC50 values for spike of 0.02 μg/mL and RBD of 0.01 μg/mL. Thus, on the basis of weight, UFH is a substantially better inhibitor compared to octasaccharide 93, indicating that increasingly long saccharides provide better binders. Computational studies have indicated multiple HS binding sites in the spike, and thus it is conceivable that polymeric compounds can make additional interactions.
Figure 3.
SPR-based competition assays on heparin-immobilized surface. (A) RBD protein in the presence of UFH. (B) Spike protein in the presence of UFH. (C) RBD protein in the presence of HS-octasaccharide (93). (D) Spike protein in the presence of 93. Concentrations of RBD and spike proteins were 5 μM and 150 nM, respectively. The IC50 values were calculated using dose–response equations [nonlinear regression, log(inhibitor) vs response-variable slope (four parameters)] built in Prism software 9 (GraphPad Software, Inc.). Experiments were performed (in duplicate) three times at the minimum.
To further determine the possible role of HS in the infection process, we examined the binding affinities of spike proteins to ACE2 and compared these with binding affinities for heparin. Biotinylated ACE2 was immobilized on a streptavidin-coated sensor chip, and binding experiments were performed with different concentrations of SARS-CoV-2 derived proteins. Representative sensorgrams for the RBD domain, monomeric spike protein, and trimeric spike protein are shown in Figure S3. KD values of 3.6, 24.5, and 0.7 nM were determined using a 1:1 Langmuir binding model, respectively, which are in agreement with reported data.28 It shows convincingly that the RBD domain has a much higher affinity for ACE2 compared to that of heparin.
A number of reports have indicated that heparin and related compounds can block an infection of cells by SARS-CoV-2. Therefore, we were compelled to investigate the molecular mechanisms by which heparin blocks a viral entry.2,10,12 It is possible that the antiviral properties of heparin are due to binding to the RBD domain thereby blocking the interaction with ACE2. In this respect, a computational study indicated that ACE2 and HS bind to the same region of the RBD.9 Another study also indicated overlapping binding sites and suggested that heparin binding can induce a conformational change and inhibit ACE2 binding.12 A docking study located, however, the HS binding site adjacent to the ACE2-binding site and inferred a model in which RBD, HS, and ACE2 can form a ternary complex.11 Alternatively, heparin may interfere in the proteolytic processing of the spike protein thereby preventing membrane fusion. In this respect, the spike of SARS-CoV-2 contains a unique furin cleavage site, which is not present in other human CoVs, and has been proposed to contribute to high infectivity,29 because the cleavage of the spike protein is a prerequisite for membrane fusion. Modeling studies have indicated that the furin cleavage site may harbor a binding site for HS.9 Finally, HSPG may function as an initial attachment factor, and the addition of exogenous heparin may interfere in this process.
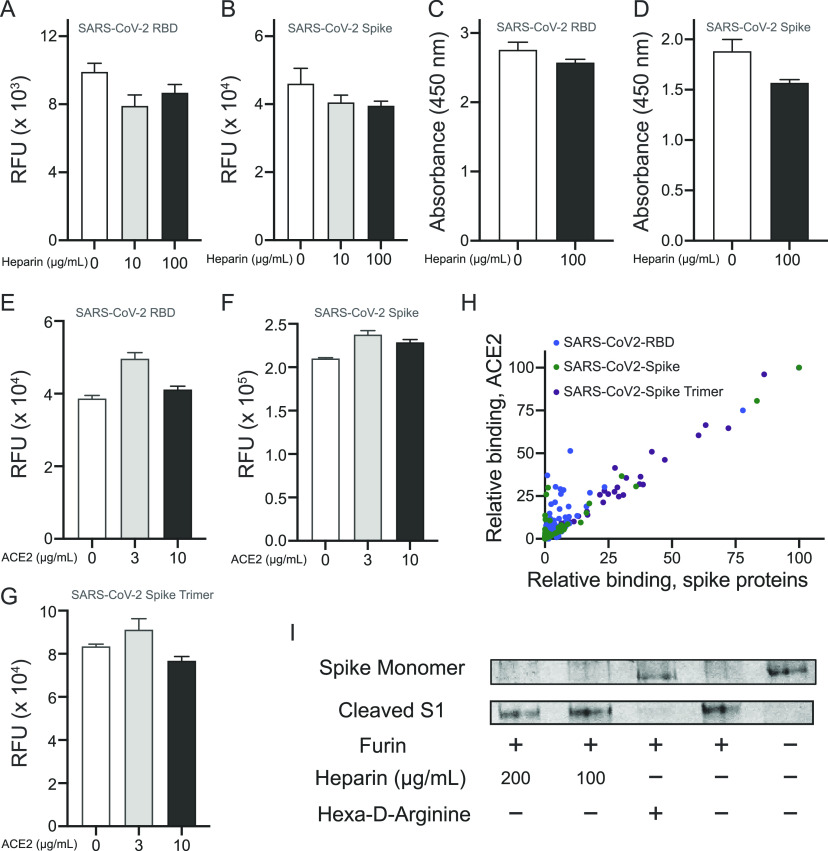
To examine whether heparin can interfere in the binding of the spike to ACE2, we performed microarray experiments in which biotinylated Fc-tagged ACE2 (50 μg/mL) was printed onto streptavidin-coated microarray slides. The printing quality was confirmed by using a goat antihuman-Fc antibody conjugated with AlexaFluoro647 (Figure S4A). Next, His-tagged RBD and monomeric spike protein were premixed with different concentrations of heparin, and binding of the proteins to immobilized ACE2 was evaluated by an anti-His antibody. A commercially available spike protein inhibitor was used as a positive control. Although the known inhibitor efficiently blocked RBD and spike binding (Figure S4B,C), only a small reduction in binding was observed in the presence of 10 and 100 μg/mL heparin (Figure 4A,B). In addition, we immobilized the RBD and monomeric spike proteins on enzyme-linked immunosorbent assay (ELISA) plates and assayed the binding of ACE2 to the spike proteins in the presence or absence of heparin (Figure 4C,D). Soluble human ACE2 was used as a positive control, which as expected exhibited a potent inhibition. At 100 μg/mL heparin, no inhibition of binding was observed for either RBD or monomeric spike protein. These results indicate that heparin does not substantially interfere in the interaction of spike with ACE2. It was also examined whether binding to ACE2 can affect the ability of the spike to bind with HS oligosaccharides. Thus, His-tagged RBD, monomeric spike protein, and trimeric spike protein were premixed with 3 and 10 μg/mL of ACE2, and the mixtures were incubated on the HS microarray. The binding of the spike proteins was detected by anti-His antibody (Figure 4E–G). The results indicate that the presence of ACE2 does not substantially interfere in the spike protein’s ability to bind with HS.
Figure 4.
Interplay of interactions of HS and ACE2 with spike/RBD. (A) Influence of heparin on the binding of His-tagged RBD or (B) His-tagged spike monomer to biotinylated human ACE2 immobilized on streptavidin-coated microarray slides. Detection of RBD and spike was accomplished using an anti-His antibody labeled with AlexaFluor 647. (C) Influence of heparin on the binding of biotinylated human ACE2 to RBD and (D) to immobilized spike monomer immobilized to high surface microtiter plates. Binding was detected by treatment with streptavidin-horseradish peroxidase (HRP) followed by an addition of a colorimetric HRP substrate. (E). Effect of ACE2 on binding of RBD, (F) spike monomer, and (G) spike trimer to the HS microarray. Binding intensity corresponding to compound 93 was used. (H) Binding of ACE2-spike protein complex to HS oligosaccharides on the microarray, by detecting spike proteins (x-axis) and ACE2 (y-axis). Each spot represents an individual compound on the HS array. (I) SDS-PAGE analysis of furin-mediated cleavage of spike monomer in the presence and absence of heparin or a known furin inhibitor (hexa-d-arginine).
Next, we explored if spike, ACE2, and HS can form a ternary complex. Thus, biotinylated ACE2 was premixed with His-tagged RBD, monomeric spike, or trimeric spike, and then exposed to the HS oligosaccharide microarray. After the proteins were incubated and washed, detection of the individual proteins was performed using either an anti-His antibody or streptavidin labeled with AlexaFluoro647. While biotinylated ACE2 alone did not exhibit any binding to the HS oligosaccharides, in a complex with a spike it could readily be detected, and a good correlation was observed between the intensity of the spike and ACE2 binding (Figure 4H), indicating that a ternary complex had been formed. To support this mode of binding, His-tagged spike proteins and biotinylated ACE2 were premixed to form a complex followed by the addition of an anti-His antibody labeled with AlexaFluoro647 and streptavidin-Cy3. The mixture was exposed to the heparan sulfate microarray, and after the mixture was washed, a scan was performed at 635 and 532 nm to detect spike and ACE2, respectively (Figure S6) The two sets of signals overlaid very well further supporting the formation of a ternary complex. The observations are in agreement with the ability of ACE2 to bind to spike protein immobilized on heparin-bovine serum albumin (BSA).11
To investigate whether the binding of heparin can hinder the cleavage of a spike by furin, monomeric spike was exposed to furin in the presence of different concentrations of heparin, and protein cleavage was examined by sodium dodecyl sulfate poly(acrylamide) gel electrophoresis (SDS-PAGE). The spike was readily cleaved by furin even in the presence of a high concentration of heparin (400 μg/mL), while 50 μg/mL of a known furin inhibitor completely abolished the cleavage (Figure 4I).
To confirm the inhibitory effects of GAGs on binding SARS-CoV-2 to cells, trimeric RBD was pretreated with UFH, non-anticoagulant heparin (NACH), which is a chemically modified heparin devoid of anticoagulant activity,30 and HS oligosaccharides 91 and 93 (Figure 5A, Figure S7, and Figure S8 provide the date for various treatments at different concentrations). Hyaluronic acid (HA) was used as a control. Both UFH and NACH inhibited trimeric RBD binding up to a concentration of 10 μg/mL, whereas HA failed to elicit any inhibition even at a very high concentration (250 μg/mL). When applied at a 10-fold higher concentration based on weight, the synthetic oligosaccharides afforded a similar inhibition as that of UFH. This observation is in agreement with the SPR-based inhibition studies that demonstrated that heparin is a more potent inhibitor than octasaccharide 93 (Figure 3). A preincubation of all GAGs at 250 μg/mL did not result in an inhibition of the ACE2 antibody (Figure 5B).
Figure 5.

Binding of SARS-CoV-2 RBD pretreated with GAGs to Vero-E6 cells. (A) (top to bottom) Untreated RBD, HA (250 μg/mL), UFH (10 μg/mL), NACH (10 μg/mL), and octasaccharide 93 (100 μg/mL), complete dilution series are shown in Figure S7 and Figure S8. (B) ACE2 antibody pretreated with GAGs.
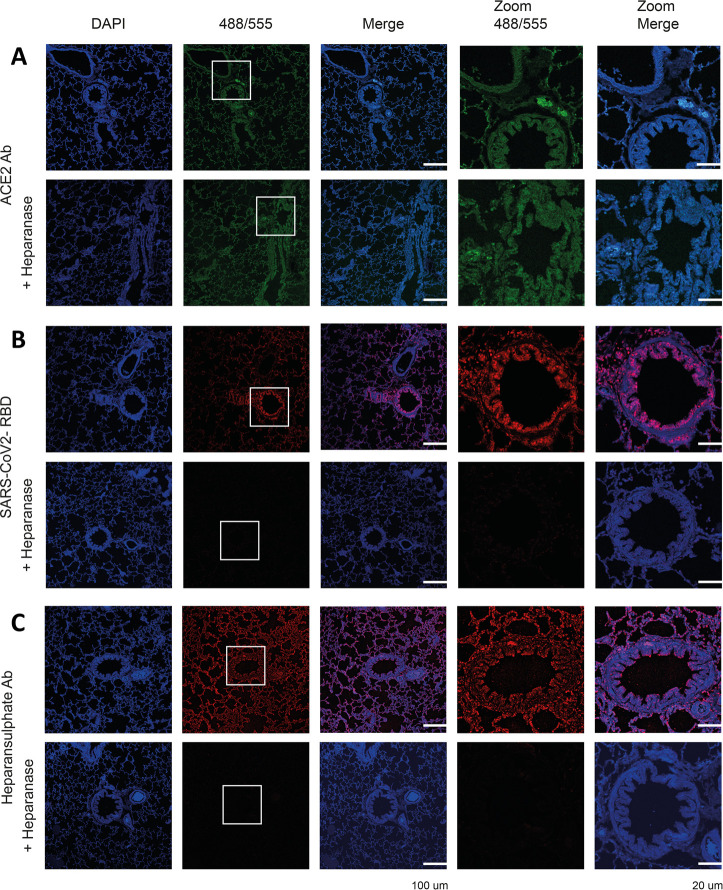
It is possible that heparin interferes in the initial attachment of the virus to the glycocalyx thereby preventing infection. Therefore, we examined the importance of HS for the binding of trimeric RBD to relevant tissues.31 Ferrets are a susceptible animal model for SARS-CoV-2,32,33 and closely related minks are easily infected on farms.34 Formalin-fixed, paraffin-embedded lung tissue slides resemble the complex membrane structures to which spike proteins need to bind before it can engage with ACE2 for cell entry. The expression of ACE2 was assessed using an ACE2 antibody allowing us to compare the binding with the SARS-CoV-2 RBD protein and binding localization and dependency on HS. The ACE2 antibody (Figure 6A) and the RBD trimer bound efficiently to the ferret lung tissues (Figure 6B). We also examined a commonly used heparan sulfate antibody (10E4), which bound efficiently to ferret lung tissue, indicating the omnipresence of HS. On the one hand, after an overnight exposure to heparanase (HPSE), the ACE2 antibody staining was mostly unaffected, indicating HSPG-independent binding. On the other hand, the SARS-CoV-2 RBD trimer was not able to engage with the ferret lung tissue slide after the HPSE treatment. No staining was observed with 10E4, indicating all HS had been removed (Figure 6C). These results indicate HS is required for an initial cell attachment before the spike engages with ACE2.
Figure 6.
Binding of ACE2 antibody, SARS-CoV-2 RBD, and heparan sulfate antibody to ferret lung serial tissue slides. (A) ACE2 antibody staining without and after HPSE treatment. (B) SARS-CoV-2 RBD staining without and after HPSE treatment. (C) Heparan sulfate antibody (10E4) staining without and after HPSE treatment. HPSE treatment was achieved by an overnight incubation of the tissues with HPSE (0.2 μg/mL) at 37 °C.
Discussion and Conclusions
The glycan microarray and SPR results indicate that the spike of SARS-CoV-2 can bind HS in a length- and sequence-dependent manner, and a hexasaccharide composed of IdoA2S-GlcNS6S repeating units was identified as the minimal epitope. The data support a model in which the RBD of the spike confers sequence specificity and an additional HS binding site in the S1/S2 proteolytic cleavage site9 may enhance the avidity of binding probably by nonspecific interactions. In a BioRxiv preprint, we presented, for the first time, experimental support for such a model, and subsequent papers9,11,12 have confirmed the RBD of SARS-CoV-2 harbors an HS binding site. Although IdoA2S-GlcNS6S sequons are abundantly present in heparin, it is a minor component of HS.35 Interestingly, it has been reported that the expression of a (GlcNS6S-IdoA2S)3 motif is highly regulated and plays a crucial role in cell behavior and disease including endothelial cell activation.36 Severe thrombosis in COVID-19 patients is associated with endothelial dysfunction,37 and a connection may exist between SARS-CoV-2’s ability to bind to HS and thrombotic disorder. It is also possible that HS is a determinant of the cell and tissue tropism.
The microarray studies demonstrated that the removal of only one sulfate from the highly sulfated hexasaccharide (GlcNS6S-IdoA2S)3 resulted in a substantial reduction or complete loss of responsiveness, indicating that the sulfates engage in specific interactions with the protein. The importance of electrostatic interactions can, however, not be ruled out, and structural studies are required to uncover the molecular basis of HS binding.
A number of reports have shown that heparin and related products can block infection by a pseudotyped virus or an authentic SARS-CoV-2 virus.9,11,12,14 We explored the possibility that the binding of heparin blocks the RBD from interacting with ACE2. However, in two experimental formats such properties were not observed. We found that the affinity of the RBD for heparin is much lower than that for ACE2, providing a rationale for the inability of heparin to inhibit the binding between RBD or spike with ACE2. In addition, microarray binding experiments, in which RBD or spike and ACE can individually be detected, indicated that ACE2 and HS can simultaneously engage with the viral proteins.
We employed physiological relevant tissues to explore the importance of HS for SARS-CoV-2 adhesion and demonstrated that an HPSE treatment greatly reduces RBD binding but not that of ACE2. In addition, heparin and the highly sulfated octasaccharide 93 could inhibit the attachment of RBD to Vero E6 cells. The data support a model in which HS functions as an initial host attachment factor that facilitates SARS-CoV-2 infection. It has also been proposed that a viral attachment and infection involves an HS-dependent enhancement of binding to ACE2.9,11 Furthermore, electron microscopy graphs of spike protein gave some indications that heparin can enhance the open conformation of the RBD that is important for ACE2 binding. Although RBD and spike have a high affinity for ACE2 in the absence of heparin, the removal of HS from cells or physiological relevant tissues abolished binding of RBD and spike protein. This indicates HS is critical for the initial attachment to cells. In addition, exogenous administered heparin or a highly sulfated HS oligosaccharide can inhibit spike binding to cells that can be infected by SARS-CoV-2. Thus, it is likely that, after an initial attachment to the cell surface HSPG, the virus travels through the glycocalyx by low-affinity high-avidity interactions to reach the cell membrane, where it engages with ACE2 for cell entry. The importance of a ternary complex formation between viral spike protein, HSPG, and ACE2 for infectivity remains to be determined.
The current clinical guidelines call for the use of unfractionated heparin or low molecular weight heparin (LMWH) for the treatment of all COVID-19 patients for systemic clotting in the absence of contradictions.38,39 A heparin treatment may have additional benefits and may compete with the binding of the spike protein to cell surface HS thereby preventing infectivity. Our data suggest that noncoagulating heparin or HS preparations can be developed that reduce cell binding and infectivity without a risk of causing bleeding. In this respect, an administration of heparin requires great care, because its anticoagulant activity can result in excessive bleeding. Antithrombin III (AT-III), which confers anticoagulant activity, binds the specific pentasaccharide GlcNAc(6S)-GlcA-GlcNS(3S)(6S)-IdoA2S-GlcNS(6S) embedded in HS or heparin. Removal of the sulfate at C-3 of N-sulfoglucosamine (GlcNS3S) of the pentasaccharide results in a 105-fold reduction in the binding affinity.40 Importantly, such a functionality is not present in the identified HS ligand of the SARS-CoV-2 spike, and therefore compounds can be developed that can inhibit cell binding but do not interact with AT-III. As a result, such preparations can be used at higher doses without causing adverse side effects. Our data also show that multivalent interactions of the spike with HS results in a high avidity of the binding. This observation provides opportunities to develop glyco-polymers modified by HS oligosaccharides as inhibitors of SARS-CoV-2 cell binding to prevent or treat COVID-19.
Acknowledgments
This research was supported by the National Institutes of Health (P41GM103390 and R01HL151617 to G.-J.B.). R.P.dV is a recipient of an ERC Starting Grant from the European Commission (802780) and a Beijerinck Premium of the Royal Dutch Academy of Sciences. We thank S. Herfst (Department of Viroscience, Erasmus Medical Center) for the ferret tissues and G. Wright (Addgene) for providing HPSE-bio-His (Plasmid No. 53407). Plasmids for expression of SARS-CoV-2 spike and RBD proteins were provided by Dr. F. Krammer (Icahn School of Medicine at Mount Sinai, produced under NIAID CEIRS contract HHSN272201400008C). The production of recombinant proteins was supported by NIAID Centers of Excellence for Influenza Research and Surveillance contract HHSN272201400004C to S.M.T.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00010.
Materials, methods, additional data, and figures including microarray printing and screening, preparation of biotinylated heparin, surface plasma resonance experiments, competition experiments using immobilized ACE2 on streptavidin-coated slides, ability of heparin to inhibit spike protein RBD or spike protein monomer by ELISA, furin cleavage assay, tissue binding experiments, additional references (PDF)
Author Contributions
⊥ (L.L., P.C., and X.L.) These authors contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Dimitrov D. S. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004, 2 (2), 109–122. 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Park Y. J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181 (2), 281–292. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Li W.; Farzan M.; Harrison S. C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309 (5742), 1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Monteil V.; Kwon H.; Prado P.; Hagelkruys A.; Wimmer R. A.; Stahl M.; Leopoldi A.; Garreta E.; Hurtado Del Pozo C.; Prosper F.; Romero J. P.; Wirnsberger G.; Zhang H.; Slutsky A. S.; Conder R.; Montserrat N.; Mirazimi A.; Penninger J. M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020, 181 (4), 905–913. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Hulswit R. J. G.; Widjaja I.; Raj V. S.; McBride R.; Peng W.; Widagdo W.; Tortorici M. A.; van Dieren B.; Lang Y.; van Lent J. W. M.; Paulson J. C.; de Haan C. A. M.; de Groot R. J.; van Kuppeveld F. J. M.; Haagmans B. L.; Bosch B. J. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (40), E8508–E8517. 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A.; Zarebski M.; Nowak P.; Stozek K.; Potempa J.; Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014, 88 (22), 13221–13230. 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J.; Yang N.; Deng J.; Liu K.; Yang P.; Zhang G.; Jiang C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One 2011, 6 (8), e23710 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West C.; Su D.; Elli S.; Li Y.; Guimond S.; Miller G.; Turnbull J.; Yates E.; Guerrini M.; Fernig D.; Lima M.; Skidmore M.. The 2019 coronavirus (SARS-CoV-2) surface protein (spike) S1 receptor binding domain undergoes conformational change upon heparin binding. bioRxiv 2020, 10.1101/2020.02.29.971093. [DOI] [Google Scholar]
- Kim S. Y.; Jin W.; Sood A.; Montgomery D. W.; Grant O. C.; Fuster M. M.; Fu L.; Dordick J. S.; Woods R. J.; Zhang F.; Linhardt R. J. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Res. 2020, 181, 104873. 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.; Bidon M.; Jaimes J. A.; Whittaker G. R.; Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020, 178, 104792. 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T. M.; Sandoval D. R.; Spliid C. B.; Pihl J.; Perrett H. R.; Painter C. D.; Narayanan A.; Majowicz S. A.; Kwong E. M.; McVicar R. N.; Thacker B. E.; Glass C. A.; Yang Z.; Torres J. L.; Golden G. J.; Bartels P. L.; Porell R. N.; Garretson A. F.; Laubach L.; Feldman J.; Yin X.; Pu Y.; Hauser B. M.; Caradonna T. M.; Kellman B. P.; Martino C.; Gordts P.; Chanda S. K.; Schmidt A. G.; Godula K.; Leibel S. L.; Jose J.; Corbett K. D.; Ward A. B.; Carlin A. F.; Esko J. D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 2020, 183 (4), 1043–1057. 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West C. J.; Su D.; Pagani I.; Rudd T. R.; Elli S.; Gandhi N. S.; Guimond S. E.; Miller G. J.; Meneghetti M. C. Z.; Nader H. B.; Li Y.; Nunes Q. M.; Procter P.; Mancini N.; Clementi M.; Bisio A.; Forsyth N. R.; Ferro V.; Turnbull J. E.; Guerrini M.; Fernig D. G.; Vicenzi E.; Yates E. A.; Lima M. A.; Skidmore M. A. Heparin inhibits cellular invasion by SARS-CoV-2: Structural dependence of the interaction of the spike S1 receptor-binding domain with heparin. Thromb. Haemostasis 2020, 120 (12), 1700–1715. 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L. J.; Urwin L.; Nicklin M. J. H.; James D. C.; Green L. R.; Monk P. N.. ACE2-independent interaction of SARS-CoV-2 spike protein to human epithelial cells can be inhibited by unfractionated heparin. bioRxiv 2020, 10.1101/2020.05.21.107870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond S. E.; Mycroft-West C. J.; Gandhi N. S.; Tree J. A.; Buttigieg K. R.; Coombes N.; Nystrom K.; Said J.; Setoh Y. X.; Amarilla A.; Modhiran N.; Julian Sng D. J.; Chhabra M.; Watterson D.; Young P. R.; Khromykh A. A.; Lima M. A.; Fernig D. G.; Su D.; Yates E. A.; Hammond E.; Dredge K.; Carroll M. W.; Trybala E.; Bergstrom T.; Ferro V.; Skidmore M. A.; Turnbull J. E.. Synthetic heparan sulfate mimetic Pixatimod (PG545) potently inhibits SARS-CoV-2 by disrupting the spike-ACE2 interaction. bioRxiv 2021, 10.1101/2020.06.24.169334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. R.; Schuksz M.; Esko J. D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446 (7139), 1030–1037. 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Cagno V.; Tseligka E. D.; Jones S. T.; Tapparel C. Heparan sulfate proteoglycans and viral attachment: True receptors or adaptation bias?. Viruses 2019, 11 (7), 596. 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C. A.; Haijema B. J.; Schellen P.; Wichgers Schreur P.; te Lintelo E.; Vennema H.; Rottier P. J. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 2008, 82 (12), 6078–6083. 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C. A.; Li Z.; te Lintelo E.; Bosch B. J.; Haijema B. J.; Rottier P. J. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 2005, 79 (22), 14451–14456. 10.1128/JVI.79.22.14451-14456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S.; Lamanna W. C.; Esko J. D. Heparan sulfate proteoglycans. Cold Spring Harbor Perspect. Biol. 2011, 3 (7), a004952 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D.; Esko J. D. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 2014, 83, 129–157. 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi E.; Joo E. J.; Dordick J. S.; Linhardt R. J. Glycosaminoglycans in infectious disease. Biol. Rev. Camb. Philos. Soc. 2013, 88 (4), 928–943. 10.1111/brv.12034. [DOI] [PubMed] [Google Scholar]
- Garcia B.; Merayo-Lloves J.; Martin C.; Alcalde I.; Quiros L. M.; Vazquez F. Surface proteoglycans as mediators in bacterial pathogens infections. Front. Microbiol. 2016, 7, 220. 10.3389/fmicb.2016.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arungundram S.; Al-Mafraji K.; Asong J.; Leach F. E. III; Amster I. J.; Venot A.; Turnbull J. E.; Boons G. J. Modular synthesis of heparan sulfate oligosaccharides for structure-activity relationship studies. J. Am. Chem. Soc. 2009, 131 (47), 17394–17405. 10.1021/ja907358k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C.; Venot A.; Li X.; Lu W.; Xiao W.; Wilkes J. L.; Salanga C. L.; Handel T. M.; Wang L.; Wolfert M. A.; Boons G. J. Heparan sulfate microarray reveals that heparan sulfate-protein binding exhibits different ligand requirements. J. Am. Chem. Soc. 2017, 139 (28), 9534–9543. 10.1021/jacs.7b01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F.; Stadlbauer D.; Strohmeier S.; Nguyen T. H. O.; Chromikova V.; McMahon M.; Jiang K.; Arunkumar G. A.; Jurczyszak D.; Polanco J.; Bermudez-Gonzalez M.; Kleiner G.; Aydillo T.; Miorin L.; Fierer D. S.; Lugo L. A.; Kojic E. M.; Stoever J.; Liu S. T. H.; Cunningham-Rundles C.; Felgner P. L.; Moran T.; Garcia-Sastre A.; Caplivski D.; Cheng A. C.; Kedzierska K.; Vapalahti O.; Hepojoki J. M.; Simon V.; Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26 (7), 1033–1036. 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D.; Amanat F.; Chromikova V.; Jiang K.; Strohmeier S.; Arunkumar G. A.; Tan J.; Bhavsar D.; Capuano C.; Kirkpatrick E.; Meade P.; Brito R. N.; Teo C.; McMahon M.; Simon V.; Krammer F. SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020, 57 (1), e100 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra P.; Joshi A.; Wu J.; Lu W.; Yadavalli T.; Wolfert M. A.; Shukla D.; Zaia J.; Boons G. J. 3-O-Sulfation of heparan sulfate modulates protein binding and lyase degradation. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (3), e2012935118 10.1073/pnas.2012935118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H.; Geng Q.; Auerbach A.; Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581 (7807), 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S.; Lan Q.; Su S.; Wang X.; Xu W.; Liu Z.; Zhu Y.; Wang Q.; Lu L.; Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020, 5 (1), 92. 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T.; Butler M.; Sikkander S. A.; Toida T.; Linhardt R. J. Further evidence that periodate cleavage of heparin occurs primarily through the antithrombin binding site. Carbohydr. Res. 2002, 337 (21–23), 2239–2243. 10.1016/S0008-6215(02)00229-X. [DOI] [PubMed] [Google Scholar]
- Bouwman K. M.; Tomris I.; Turner H. L.; van der Woude R.; Shamorkina T. M.; Bosman G. P.; Rockx B.; Herfst S.; Snijder J.; Haagmans B. L.; Ward A. B.; Boons G. J.; de Vries R. P. Multimerization- and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins. PLoS Pathog. 2021, 17 (2), e1009282 10.1371/journal.ppat.1009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. I.; Kim S. G.; Kim S. M.; Kim E. H.; Park S. J.; Yu K. M.; Chang J. H.; Kim E. J.; Lee S.; Casel M. A. B.; Um J.; Song M. S.; Jeong H. W.; Lai V. D.; Kim Y.; Chin B. S.; Park J. S.; Chung K. H.; Foo S. S.; Poo H.; Mo I. P.; Lee O. J.; Webby R. J.; Jung J. U.; Choi Y. K. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020, 27 (5), 704–709. 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M.; Kok A.; de Meulder D.; Bestebroer T. M.; Lamers M. M.; Okba N. M. A.; Fentener van Vlissingen M.; Rockx B.; Haagmans B. L.; Koopmans M. P. G.; Fouchier R. A. M.; Herfst S. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020, 11 (1), 3496. 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N.; Molenaar R. J.; Vreman S.; Harders F.; Oude Munnink B. B.; Hakze-Van der Honing R. W.; Gerhards N.; Tolsma P.; Bouwstra R.; Sikkema R. S.; Tacken M. G.; de Rooij M. M.; Weesendorp E.; Engelsma M. Y.; Bruschke C. J.; Smit L. A.; Koopmans M.; van der Poel W. H.; Stegeman A. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25 (23), 2001005. 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein D. L. Heparin and heparan sulfate: structure and function. Nat. Prod. Rep. 2002, 19 (3), 312–331. 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- Smits N. C.; Kurup S.; Rops A. L.; ten Dam G. B.; Massuger L. F.; Hafmans T.; Turnbull J. E.; Spillmann D.; Li J. P.; Kennel S. J.; Wall J. S.; Shworak N. W.; Dekhuijzen P. N.; van der Vlag J.; van Kuppevelt T. H. The heparan sulfate motif (GlcNS6S-IdoA2S)3, common in heparin, has a strict topography and is involved in cell behavior and disease. J. Biol. Chem. 2010, 285 (52), 41143–41151. 10.1074/jbc.M110.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardu C.; Gambardella J.; Morelli M. B.; Wang X.; Marfella R.; Santulli G. Hypertension, thrombosis, kidney failure, and diabetes: Is covid-19 an endothelial disease? A comprehensive evaluation of clinical and basic evidence. J. Clin. Med. 2020, 9 (5), 1417. 10.3390/jcm9051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N.; Bai H.; Chen X.; Gong J.; Li D.; Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemostasis 2020, 18 (5), 1094–1099. 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thachil J.; Tang N.; Gando S.; Falanga A.; Cattaneo M.; Levi M.; Clark C.; Iba T. ISTH interim guidance on recognition and management of coagulopathy in COVID-19. J. Thromb. Haemostasis 2020, 18 (5), 1023–1026. 10.1111/jth.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker B. E.; Xu D.; Lawrence R.; Esko J. D. Heparan sulfate 3-O-sulfation: a rare modification in search of a function. Matrix Biol. 2014, 35, 60–72. 10.1016/j.matbio.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.