Abstract
We adopted a bioinformatics-based technique to identify strain-specific markers, which were then used to quantify the abundances of three distinct B. longum sup. longum strains in fecal samples of humans and mice. A pangenome analysis of 205 B. longum sup. longum genomes revealed the accumulation of considerable strain-specific genes within this species; specifically, 28.7% of the total identified genes were strain-specific. We identified 32, 14, and 49 genes specific to B. longum sup. longum RG4-1, B. longum sup. longum M1-20-R01-3, and B. longum sup. longum FGSZY6M4, respectively. After performing an in silico validation of these strain-specific markers using a nucleotide BLAST against both the B. longum sup. longum genome database and an NR/NT database, RG4-1_01874 (1331 bp), M1-20-R01-3_00324 (1745 bp), and FGSZY6M4_01477 (1691 bp) were chosen as target genes for strain-specific quantification. The specificities of the qPCR primers were validated against 47 non-target microorganisms and fecal baseline microbiota to ensure that they produced no PCR amplification products. The performance of the qPCR primer-based analysis was further assessed using fecal samples. After oral administration, the target B. longum strains appeared to efficiently colonize both the human and mouse guts, with average population levels of >108 CFU/g feces. The bioinformatics pipeline proposed here can be applied to the quantification of various bacterial species.
Keywords: strain-specific qualification, probiotics, B. longum sup. longum, bioinformatics, Roary, gut colonization
1. Introduction
Intestinal commensals play an important role in host health via being involved in various aspects of host physiology, such as tissue development, metabolism, and immunomodulation [1,2]. Many of these organisms are believed to be beneficial to the host. Bifidobacterium is a genus of bacterial species that colonizes the gut early in life [3] and is considered beneficial to host health [4]. The abundances of various Bifidobacterium species in the gut vary widely among individuals according to differences in dietary patterns [5,6], age groups [7], and physiological statuses [8]. Among these species, B. longum stands out as a member of the core human microbiome [9] and the most dominant species within the Bifidobacterium genus in the gut, regardless of the host age [7]. B. longum is distributed broadly across subjects of various ages [10], and is among the limited number of bacterial species that can colonize the gut over years [11]. Therefore, B. longum is an excellent example of host–microbe co-evolution, and is considered to be among the most potent probiotic species that are likely to engraft and persist in the gut after oral ingestion [12].
Compared with probiotic strains that merely transit through the gut, those probiotic strains that are able to successfully reside in the gut, would interact closely with the gut immune system, mucosa, epithelial cells, and native microbial communities, thereby possibly harboring better probiotic effects. However, as a consequence of the difficulty of strain-level detection, there is clearly a knowledge gap regarding gut colonization mechanisms of probiotics [13]. Current generally used approaches to detect strain colonization include plate counting and species-level PCR [14,15,16]. However, these methods are not accurate, considering the natural occurrence of phylogenetically related species with the ingested probiotic strains in the indigenous microbiota. Therefore, detection and quantitation of probiotics at the strain level are critically important for accessing gut colonization by various strains, and further understanding their functionality and related mechanistic insight.
Multiple approaches have been developed to measure the presence and abundance of specific probiotic strains in the gastrointestinal tract. Initially, methods based on selective culture medium and colony identification (e.g., bacterial morphology, biochemical analysis, pulsed field gel electrophoresis (PFGE), 16S ribosomal DNA (rDNA) PCR, internally transcribed spacer (ITS)-PCR, random amplified polymorphic DNA (RAPD)-PCR, and monoclonal antibodies) were commonly used [14,17,18,19,20,21]. However, these methods are time consuming, laborious, and often inaccurate. Fluorescence [22] or antibiotic labeling [23], and group-specific fluorescence in situ hybridization (FISH) [24] are also ineffective, because of the recurrent loss of plasmids with these tags by strains during gut transition, the low detection sensitivity of fluorescence signals, and safety considerations regarding the application of these approaches in human subjects. Species-specific PCR assays that target 16S rDNA variable regions or 16S-23S ITS rDNA sequences have also been used to directly determine ingested probiotic strains in fecal samples [25,26]. However, this approach cannot distinguish the target strain from phylogenetically related species present in the baseline microbiota. Recently, with the accumulation of sequenced bacterial genomes, strain-specific gene markers have been identified at unprecedented speeds, impelling us to use these unique markers to detect and quantify strains using molecular methods.
Strain-specific detection depends on the identification of DNA regions unique to specific strains. Before the era of large-scale genomic sequencing, selected RAPD electrophoresis bands, specific DNA fragments from suppression subtractive hybridization (SSH), or known sequences related to specific traits (e.g., Lactobacillus rhamnosus GG (LGG) harbors a pili structure, but LC705 does not) were used to design strain-specific primers for qualification of some probiotic strains in the gut/fecal samples, including LGG [27], B. bifidum OLB6378 [28], L. gasseri K7 [29], B. breve Yakult [30], and L. reuteri DSM 16350 [31]. However, this strain specificity remained within narrow confidence intervals because the identification of the strain-specific DNA regions was based on a limited number of bacterial strains. Additionally, these methods usually required pure cultures of various bacterial strains for laborious electrophoretic analyses. Fortunately, recent emerged bioinformatics strategies based on the sequenced bacterial genomes provide an alternative to find nearly “true” strain-specific DNA sequences. Theoretically, some bioinformatics pipelines (Pan-Seq [32], PGAT [33], PGAP [34], and Roary [35]) can be used to search bacterial strain-specific DNA segments, which can subsequently be used as templates for strain-specific primers. However, no previous study has used these bioinformatics tools to identify strain-specific sequences and achieve strain-level bacterial detection.
In this study, we selected B. longum sup. longum as an example due to the fact that it has been reported to be the most potent probiotic species with long-term gut colonization potential, and used genomics analyses to identify DNA sequences specific to three B. longum sup. longum strains isolated from the fecal samples of three Chinese subjects. First, we used a Roary-based pangenome analysis to identify unique gene markers that were present only in a single strains of B. longum sup. longum but absent from all the other strains of this species. Next, we validated this strain specificity in the context of other microbes and the baseline microbiota, and then targeted these unique sequences to design strain-specific primers. Finally, we applied these strain-specific primers and quantitative PCR (qPCR) to quantify the colonized biomasses of these three B. longum sup. longum strains in the feces of humans and mice after ingestion.
2. Materials and Methods
2.1. Bacterial Strains, Culture Conditions and Genomic DNA Extraction
As shown in Table 1, 48 bacterial strains were used in this study. The genomic DNA of each bacterial strain was extracted using rapid bacterial genomic DNA isolation kit (Sangon Biotech Co., Ltd., Shanghai, China).
Table 1.
Bacterial strains used for primer validation via electrophoresis a.
| Species | Accession Number | Culture Conditions |
|---|---|---|
| Bifidobacterium | deMan Rogosa Sharpe (MRS) broth supplemented with 0.1% L-cysteine HCl at 37 °C | |
| B. longum | RG4-1 b, FGSZY6M4 b, M1-20-R01-3 b, 274 b, FSHHK13M1 b, FSDLZ57M1 b, NaTon 49-4 b, FJSWXJ11M1 b, HUB 36-17 b, 28-10 b, ZCC7 b | |
| B. breve | DSM 20213 c | |
| B. bifidum | DSM 20456 c | |
| B. pseudocatenulatum | FQHXN5M4 b | |
| B. pseudolongum | 56M2 b | |
| B. animalis | BB12d | |
| B. adolescentis | L2-32 e | |
| Lactobacillus | MRS broth at 37 °C | |
| L. salivarius | DSM 20555 c | |
| L. gasseri | DSM 20243 c | |
| L. casei | DSM 20011 c | |
| L. acidophilus | DSM 20079 c | |
| L. plantarum | DSM 20174 c | |
| L. reuteri | DSM 20016 c | |
| L. rhamnosus | LMS2-1 e | |
| Non-lactic acid bacteria (LAB) | ||
| Escherichia coli | CMCC 44102 f | Luria-Bertani (LB) broth at 37 °C |
| Akkermansia muciniphila | FJLHD50M21 b | Brain Heart Infusion (BHI) broth at 37 °C |
| Faecalibacterium prausnitzii | DSM 17677 c | BHI broth containing 3.7% BHI powder supplemented with 0.5% yeast extract, 0.0005% hemin, 0.0005% vitamin K and 0.2% L-cysteine HCl at 37 °C |
| Enterococcus faecalis | CCFM596 b | BHI broth at 37 °C |
| Bacteroides fragilis | ATCC 25285/NCTC 9343 g | BHI broth supplemented with 0.1% L-cysteine HCl, 0.001% hemin and 0.0002% vitamin K at 37 °C |
| Bacteroides thetaiotaomicron | FNMHLBE9-K-7 b | |
| Bacteroides eggerthii | FSDTA-HCK-B-9 b | |
| Bacteroides cellulosilyticus | FSDTA-ELI-BHI-5 b | |
| Bacteroides nordii | FNMHLBE13K2 b | |
| Bacteroides stercoris | FJSWX62K34 b | |
| Bacteroides uniformis | FJSWX62K43 b | |
| Bacteroides caccae | FFJLY22K5 b | |
| Parabacteroides distasonis | FSDTA-HCM-XY-12 b | |
| Bacteroides dorei | FJSWX61E4 b | |
| Bacteroides faecis | FTJS2E2 b | |
| Bacteroides intestinalis | FBJ60K5 b | |
| Bacteroides vulgatus | FSDLZ51K1 b | |
| Bacteroides finegoldii | FNMHLBE11E1 b | |
| Bacteroides ovatus | FBJ10-K-10 b | |
| Bacteroides clarus | F-FJ-LY 22-K-22 b | |
| Bacteroides salyersiae | FSDTA-ELI-BHI-9 b | |
| Bacteroides xylanisolvens | FSDTAHCMXY17 b | |
| Parabacteroides merdae | FSDTAELIBHI4 b | |
| Clostridium butyricum | FJSCZD1G10 b | Reinforced Clostridial Medium (RCM) at 37 °C |
a Anaerobes (Bifidobacterium, Akkermansia muciniphila, Faecalibacterium prausnitzii, Bacteroides strains and Clostridium butyricum) were maintained in anaerobic chamber (80% N2, 10% H2, 10% CO2) during cultivation. b These strains were retrieved from Culture Collection of Food Microorganisms, Jiangnan university. c These strains were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). d The strain was isolated from the commercial probiotic product. e The strains were kindly provided by Biodefense and Emerging Infections Research Resources Repository (BEI Resources). f The strain was purchased from National Center for Medical Culture Collections (CMCC). g The strain was purchased from American Type Culture Collection (ATCC).
2.2. Bacterial Genome Sequencing and Retrieval of Publicly Available Genomes
Three B. longum strains (RG4-1, FGSZY6M4, and M1-20-R01-3) were isolated from fecal samples of three Chinese individuals, and under genome sequencing by Illumina HiSeq 2000. Briefly, a paired-end sequencing library (average insert size of 350 bp and maximum read length of 150 bp) was built according to manufacturers’ instructions (Illumina Inc., San Diego, CA, USA). On average, 3 GB paired-end raw reads were generated for each sample. After removing adaptors and low-quality reads, the resulting clean reads were assembled using SOAPdenovo v2.04 Software for short-read de novo assembler, BGI HK Research Institute: Hong Kong, China, 2012 [36], as described previously [37]. A total of 202 publicly available B. longum genomes were downloaded from National Center for Biotechnology Information (NCBI) database (Table 2). In total, 205 B. longum assemblies were finally used in this study.
Table 2.
Publicly available B. longum genomes used in this study.
| Genome | Strain | BioSample | Size (Mb) | GC% | Scaffolds | CDS |
|---|---|---|---|---|---|---|
| GCA_001576955.1_ASM157695v1 | 121.2 | SAMN04497913 | 1.87256 | 60.3 | 234 | 1453 |
| GCA_002331305.1_ASM233130v1 | UBA2088 | SAMN06457477 | 1.87849 | 59.2 | 227 | 0 |
| GCF_900157195.1_Bifido_02_v1 | Bifido_02 | SAMEA51816418 | 2.33429 | 60.1 | 97 | 1860 |
| GCF_900157165.1_Bifido_12_v1 | Bifido_12 | SAMEA51823918 | 2.07288 | 60.5 | 681 | 1743 |
| GCF_900157155.1_Bifido_06_v1 | Bifido_06 | SAMEA51819418 | 2.42182 | 60 | 48 | 1978 |
| GCF_900157145.1_Bifido_03_v1 | Bifido_03 | SAMEA51817168 | 2.41363 | 60.1 | 82 | 1962 |
| GCF_900157115.1_Bifido_05_v1 | Bifido_05 | SAMEA51818668 | 2.33474 | 59.9 | 88 | 1906 |
| GCF_900157095.1_Bifido_01_v1 | Bifido_01 | SAMEA51815668 | 2.33463 | 59.9 | 39 | 1907 |
| GCF_900157055.1_Bifido_09_v1 | Bifido_09 | SAMEA51821668 | 2.66124 | 59.9 | 68 | 2225 |
| GCF_900104835.1_IMG-taxon_2634166334_annotated_assembly | DSM 20219 | SAMN04489748 | 2.44902 | 60.3 | 6 | 1942 |
| GCF_004334865.1_ASM433486v1 | MCC10119 | SAMN06368669 | 2.48689 | 60.1 | 45 | 2023 |
| GCF_004334855.1_ASM433485v1 | MCC10122 | SAMN06368672 | 2.46043 | 60.1 | 49 | 1978 |
| GCF_004334815.1_ASM433481v1 | MCC10123 | SAMN06368673 | 2.5065 | 59.7 | 49 | 2060 |
| GCF_004334795.1_ASM433479v1 | MCC10125 | SAMN06368675 | 2.44774 | 60.2 | 43 | 1979 |
| GCF_004334785.1_ASM433478v1 | MCC10128 | SAMN06368678 | 2.50961 | 59.9 | 57 | 2089 |
| GCF_004334775.1_ASM433477v1 | MCC10129 | SAMN06368679 | 2.27353 | 60.1 | 18 | 1812 |
| GCF_004334745.1_ASM433474v1 | MCC10117 | SAMN06368667 | 2.30167 | 59.9 | 35 | 1807 |
| GCF_004334715.1_ASM433471v1 | MCC10120 | SAMN06368670 | 2.48377 | 60.2 | 62 | 2014 |
| GCF_004334705.1_ASM433470v1 | MCC10118 | SAMN06368668 | 2.34975 | 59.9 | 36 | 1897 |
| GCF_004334695.1_ASM433469v1 | MCC10121 | SAMN06368671 | 2.38447 | 60 | 26 | 1916 |
| GCF_004334645.1_ASM433464v1 | MCC10124 | SAMN06368674 | 2.45702 | 60.1 | 47 | 1985 |
| GCF_004334635.1_ASM433463v1 | MCC10126 | SAMN06368676 | 2.55307 | 59.8 | 68 | 2055 |
| GCF_004334625.1_ASM433462v1 | MCC10130 | SAMN06368680 | 2.36857 | 60 | 59 | 1908 |
| GCF_004334615.1_ASM433461v1 | MCC10127 | SAMN06368677 | 2.35673 | 60.1 | 41 | 1874 |
| GCF_004334555.1_ASM433455v1 | MCC10212 | SAMN06368681 | 2.36547 | 59.9 | 28 | 1904 |
| GCF_004334545.1_ASM433454v1 | MCC10002 | SAMN06368569 | 2.63451 | 60 | 59 | 2202 |
| GCF_004334535.1_ASM433453v1 | MCC10006 | SAMN06368572 | 2.45014 | 60.4 | 83 | 1991 |
| GCF_004334515.1_ASM433451v1 | MCC10009 | SAMN06368575 | 2.5237 | 60.1 | 60 | 2035 |
| GCF_004334485.1_ASM433448v1 | MCC10011 | SAMN06368577 | 2.39746 | 59.9 | 34 | 1931 |
| GCF_004334465.1_ASM433446v1 | MCC10016 | SAMN06368581 | 2.37118 | 60 | 66 | 1893 |
| GCF_004334445.1_ASM433444v1 | MCC10027 | SAMN06368589 | 2.50583 | 59.9 | 59 | 2042 |
| GCF_004334435.1_ASM433443v1 | MCC10019 | SAMN06368584 | 2.30453 | 60.1 | 41 | 1837 |
| GCF_004334425.1_ASM433442v1 | MCC10028 | SAMN06368590 | 2.42541 | 60.2 | 44 | 1983 |
| GCF_004334365.1_ASM433436v1 | MCC10038 | SAMN06368598 | 2.36701 | 59.9 | 38 | 1968 |
| GCF_004334355.1_ASM433435v1 | MCC10030 | SAMN06368592 | 2.51844 | 60.1 | 64 | 2023 |
| GCF_004334345.1_ASM433434v1 | MCC10040 | SAMN06368600 | 2.44996 | 60.2 | 45 | 1996 |
| GCF_004334335.1_ASM433433v1 | MCC10039 | SAMN06368599 | 2.38905 | 60 | 34 | 1924 |
| GCF_004334325.1_ASM433432v1 | MCC10047 | SAMN06368607 | 2.36451 | 59.9 | 64 | 1861 |
| GCF_004334285.1_ASM433428v1 | MCC10051 | SAMN06368610 | 2.30363 | 60.1 | 42 | 1830 |
| GCF_004334255.1_ASM433425v1 | MCC10057 | SAMN06368616 | 2.18862 | 59.9 | 73 | 1725 |
| GCF_004334245.1_ASM433424v1 | MCC10054 | SAMN06368613 | 2.29564 | 60 | 61 | 1842 |
| GCF_004334235.1_ASM433423v1 | MCC10059 | SAMN06368618 | 2.43718 | 60 | 47 | 2005 |
| GCF_004334215.1_ASM433421v1 | MCC10058 | SAMN06368617 | 2.32956 | 60.2 | 65 | 1886 |
| GCF_004334205.1_ASM433420v1 | MCC10072 | SAMN06368628 | 2.23388 | 60 | 23 | 1742 |
| GCF_004334165.1_ASM433416v1 | MCC10074 | SAMN06368630 | 2.41001 | 59.8 | 34 | 1965 |
| GCF_004334155.1_ASM433415v1 | MCC10077 | SAMN06368633 | 2.41162 | 60 | 40 | 1957 |
| GCF_004334145.1_ASM433414v1 | MCC10083 | SAMN06368638 | 2.47557 | 60.2 | 64 | 1953 |
| GCF_004334105.1_ASM433410v1 | MCC10085 | SAMN06368640 | 2.3663 | 60 | 64 | 1908 |
| GCF_004334075.1_ASM433407v1 | MCC10003 | SAMN06368570 | 2.52677 | 60.1 | 69 | 2047 |
| GCF_004334065.1_ASM433406v1 | MCC10004 | SAMN06368571 | 2.55173 | 60 | 52 | 2028 |
| GCF_004334045.1_ASM433404v1 | MCC10007 | SAMN06368573 | 2.48463 | 60.2 | 59 | 2024 |
| GCF_004334035.1_ASM433403v1 | MCC10008 | SAMN06368574 | 2.54367 | 60 | 80 | 2119 |
| GCF_004334005.1_ASM433400v1 | MCC10010 | SAMN06368576 | 2.45245 | 60.4 | 93 | 1984 |
| GCF_004333995.1_ASM433399v1 | MCC10012 | SAMN06368578 | 2.47987 | 59.6 | 67 | 1977 |
| GCF_004333975.1_ASM433397v1 | MCC10014 | SAMN06368579 | 2.46621 | 60.2 | 59 | 2007 |
| GCF_004333935.1_ASM433393v1 | MCC10017 | SAMN06368582 | 2.44883 | 59.7 | 60 | 1972 |
| GCF_004333925.1_ASM433392v1 | MCC10015 | SAMN06368580 | 2.63029 | 59.9 | 83 | 2177 |
| GCF_004333905.1_ASM433390v1 | MCC10018 | SAMN06368583 | 2.33041 | 60 | 32 | 1891 |
| GCF_004333895.1_ASM433389v1 | MCC10022 | SAMN06368586 | 2.41904 | 59.7 | 55 | 1950 |
| GCF_004333875.1_ASM433387v1 | MCC10021 | SAMN06368585 | 2.23575 | 60 | 58 | 1774 |
| GCF_004333855.1_ASM433385v1 | MCC10023 | SAMN06368587 | 2.30816 | 60.3 | 41 | 1853 |
| GCF_004333845.1_ASM433384v1 | MCC10025 | SAMN06368588 | 2.40884 | 60.1 | 56 | 1912 |
| GCF_004333795.1_ASM433379v1 | MCC10029 | SAMN06368591 | 2.3461 | 59.9 | 55 | 1877 |
| GCF_004333785.1_ASM433378v1 | MCC10031 | SAMN06368593 | 2.40115 | 60.1 | 41 | 1916 |
| GCF_004333775.1_ASM433377v1 | MCC10033 | SAMN06368594 | 2.39204 | 60 | 63 | 1929 |
| GCF_004333765.1_ASM433376v1 | MCC10034 | SAMN06368595 | 2.25363 | 60 | 70 | 1771 |
| GCF_004333735.1_ASM433373v1 | MCC10036 | SAMN06368597 | 2.24998 | 59.9 | 22 | 1811 |
| GCF_004333715.1_ASM433371v1 | MCC10035 | SAMN06368596 | 2.4575 | 59.8 | 52 | 2012 |
| GCF_004333695.1_ASM433369v1 | MCC10041 | SAMN06368601 | 2.37064 | 60.1 | 47 | 1906 |
| GCF_004333675.1_ASM433367v1 | MCC10042 | SAMN06368602 | 2.32056 | 60.1 | 53 | 1867 |
| GCF_004333645.1_ASM433364v1 | MCC10044 | SAMN06368604 | 2.51281 | 60.3 | 50 | 2055 |
| GCF_004333635.1_ASM433363v1 | MCC10043 | SAMN06368603 | 2.62386 | 59.5 | 62 | 2115 |
| GCF_004333625.1_ASM433362v1 | MCC10045 | SAMN06368605 | 2.43778 | 60.3 | 61 | 1973 |
| GCF_004333575.1_ASM433357v1 | MCC10046 | SAMN06368606 | 2.28641 | 59.9 | 71 | 1761 |
| GCF_004333565.1_ASM433356v1 | MCC10048 | SAMN06368608 | 2.45602 | 59.8 | 71 | 1949 |
| GCF_004333555.1_ASM433355v1 | MCC10050 | SAMN06368609 | 2.28037 | 59.8 | 34 | 1782 |
| GCF_004333535.1_ASM433353v1 | MCC10052 | SAMN06368611 | 2.42874 | 60.1 | 65 | 1956 |
| GCF_004333515.1_ASM433351v1 | MCC10053 | SAMN06368612 | 2.42582 | 60.3 | 51 | 1942 |
| GCF_004333475.1_ASM433347v1 | MCC10056 | SAMN06368615 | 2.29975 | 60 | 79 | 1837 |
| GCF_004333465.1_ASM433346v1 | MCC10060 | SAMN06368619 | 2.31311 | 60.2 | 59 | 1831 |
| GCF_004333455.1_ASM433345v1 | MCC10055 | SAMN06368614 | 2.48603 | 60.1 | 72 | 2019 |
| GCF_004333445.1_ASM433344v1 | MCC10062 | SAMN06368620 | 2.32592 | 59.8 | 59 | 1837 |
| GCF_004333425.1_ASM433342v1 | MCC10064 | SAMN06368621 | 2.26578 | 60 | 40 | 1790 |
| GCF_004333385.1_ASM433338v1 | MCC10066 | SAMN06368622 | 2.29383 | 59.8 | 53 | 1852 |
| GCF_004333375.1_ASM433337v1 | MCC10067 | SAMN06368623 | 2.39437 | 59.6 | 55 | 1910 |
| GCF_004333365.1_ASM433336v1 | MCC10068 | SAMN06368624 | 2.40787 | 59.7 | 58 | 1894 |
| GCF_004333335.1_ASM433333v1 | MCC10069 | SAMN06368625 | 2.36718 | 59.9 | 57 | 1893 |
| GCF_004333325.1_ASM433332v1 | MCC10070 | SAMN06368626 | 2.50822 | 59.6 | 48 | 2038 |
| GCF_004333305.1_ASM433330v1 | MCC10071 | SAMN06368627 | 2.28772 | 60 | 49 | 1793 |
| GCF_004333275.1_ASM433327v1 | MCC10073 | SAMN06368629 | 2.2843 | 59.7 | 42 | 1829 |
| GCF_004333265.1_ASM433326v1 | MCC10075 | SAMN06368631 | 2.38497 | 60.1 | 53 | 1947 |
| GCF_004333235.1_ASM433323v1 | MCC10076 | SAMN06368632 | 2.56355 | 60.2 | 50 | 2152 |
| GCF_004333215.1_ASM433321v1 | MCC10079 | SAMN06368635 | 2.38272 | 59.9 | 77 | 1936 |
| GCF_004333205.1_ASM433320v1 | MCC10078 | SAMN06368634 | 2.27198 | 59.8 | 58 | 1766 |
| GCF_004333175.1_ASM433317v1 | MCC10080 | SAMN06368636 | 2.52893 | 60.2 | 59 | 2014 |
| GCF_004333165.1_ASM433316v1 | MCC10081 | SAMN06368637 | 2.32346 | 59.9 | 56 | 1911 |
| GCF_004333125.1_ASM433312v1 | MCC10084 | SAMN06368639 | 2.26435 | 60.1 | 46 | 1788 |
| GCF_004333115.1_ASM433311v1 | MCC10086 | SAMN06368641 | 2.28382 | 59.8 | 48 | 1783 |
| GCF_004333105.1_ASM433310v1 | MCC10087 | SAMN06368642 | 2.30319 | 60 | 49 | 1819 |
| GCF_004333065.1_ASM433306v1 | MCC10089 | SAMN06368643 | 2.32481 | 60.3 | 41 | 1853 |
| GCF_004333045.1_ASM433304v1 | MCC10096 | SAMN06368650 | 2.57129 | 59.7 | 39 | 2099 |
| GCF_004333035.1_ASM433303v1 | MCC10090 | SAMN06368644 | 2.34731 | 59.8 | 49 | 1883 |
| GCF_004333015.1_ASM433301v1 | MCC10091 | SAMN06368645 | 2.39421 | 60 | 64 | 1931 |
| GCF_004333005.1_ASM433300v1 | MCC10103 | SAMN06368657 | 2.39468 | 59.9 | 16 | 1933 |
| GCF_004332965.1_ASM433296v1 | MCC10100 | SAMN06368654 | 2.518 | 60.1 | 56 | 2037 |
| GCF_004332945.1_ASM433294v1 | MCC10116 | SAMN06368666 | 2.62898 | 60 | 47 | 2167 |
| GCF_004332935.1_ASM433293v1 | MCC10112 | SAMN06368662 | 2.27757 | 60 | 58 | 1804 |
| GCF_004332925.1_ASM433292v1 | MCC10092 | SAMN06368646 | 2.23483 | 59.9 | 89 | 1742 |
| GCF_004332895.1_ASM433289v1 | MCC10094 | SAMN06368648 | 2.50962 | 59.9 | 39 | 2078 |
| GCF_004332865.1_ASM433286v1 | MCC10093 | SAMN06368647 | 2.45894 | 60.2 | 57 | 2000 |
| GCF_004332855.1_ASM433285v1 | MCC10095 | SAMN06368649 | 2.35682 | 60.3 | 95 | 1915 |
| GCF_004332835.1_ASM433283v1 | MCC10098 | SAMN06368652 | 2.32667 | 59.9 | 58 | 1815 |
| GCF_004332825.1_ASM433282v1 | MCC10097 | SAMN06368651 | 2.28035 | 60 | 52 | 1805 |
| GCF_004332765.1_ASM433276v1 | MCC10107 | SAMN06368659 | 2.38475 | 59.8 | 45 | 1931 |
| GCF_004332755.1_ASM433275v1 | MCC10102 | SAMN06368656 | 2.53875 | 60.1 | 56 | 2077 |
| GCF_004332745.1_ASM433274v1 | MCC10099 | SAMN06368653 | 2.34007 | 60.1 | 66 | 1870 |
| GCF_004332735.1_ASM433273v1 | MCC10106 | SAMN06368658 | 2.41503 | 60.1 | 74 | 1947 |
| GCF_004332725.1_ASM433272v1 | MCC10101 | SAMN06368655 | 2.41806 | 60.1 | 65 | 1953 |
| GCF_004332665.1_ASM433266v1 | MCC10108 | SAMN06368660 | 2.41875 | 60.3 | 73 | 1972 |
| GCF_004332655.1_ASM433265v1 | MCC10111 | SAMN06368661 | 2.43826 | 60 | 49 | 2007 |
| GCF_004332645.1_ASM433264v1 | MCC10115 | SAMN06368665 | 2.43372 | 60.3 | 54 | 2017 |
| GCF_004332635.1_ASM433263v1 | MCC10113 | SAMN06368663 | 2.46411 | 60 | 62 | 2021 |
| GCF_004332625.1_ASM433262v1 | MCC10114 | SAMN06368664 | 2.45459 | 59.8 | 58 | 2005 |
| GCF_002900845.1_ASM290084v1 | CECT 7210 | SAMEA3158508 | 2.4677 | 59.9 | 1 | 2009 |
| GCF_002861445.1_ASM286144v1 | UMB0788 | SAMN08193649 | 2.45493 | 60.2 | 33 | 2051 |
| GCF_002833285.1_ASM283328v1 | APC1466 | SAMN07958351 | 2.41998 | 59.8 | 51 | 1967 |
| GCF_002833265.1_ASM283326v1 | APC1476 | SAMN07958355 | 2.53254 | 60 | 48 | 2094 |
| GCF_002833255.1_ASM283325v1 | DPC6320 | SAMN07958364 | 2.33037 | 59.9 | 25 | 1807 |
| GCF_002833215.1_ASM283321v1 | DPC6323 | SAMN07958366 | 2.39696 | 60.2 | 52 | 1911 |
| GCF_002833205.1_ASM283320v1 | APC1462 | SAMN07958348 | 2.41778 | 60.3 | 27 | 1953 |
| GCF_002833185.1_ASM283318v1 | APC1464 | SAMN07958349 | 2.34652 | 60.1 | 31 | 1873 |
| GCF_002833175.1_ASM283317v1 | APC1465 | SAMN07958350 | 2.45221 | 59.7 | 57 | 1976 |
| GCF_002833135.1_ASM283313v1 | APC1468 | SAMN07958352 | 2.39516 | 60.2 | 45 | 1966 |
| GCF_002833125.1_ASM283312v1 | APC1473 | SAMN07958354 | 2.31707 | 59.8 | 39 | 1817 |
| GCF_002833115.1_ASM283311v1 | APC1472 | SAMN07958353 | 2.36404 | 60.2 | 50 | 1863 |
| GCF_002833075.1_ASM283307v1 | APC1477 | SAMN07958356 | 2.22881 | 59.8 | 24 | 1726 |
| GCF_002833065.1_ASM283306v1 | APC1480 | SAMN07958358 | 2.47775 | 59.9 | 27 | 2022 |
| GCF_002833055.1_ASM283305v1 | APC1478 | SAMN07958357 | 2.22335 | 59.8 | 21 | 1729 |
| GCF_002833035.1_ASM283303v1 | APC1482 | SAMN07958359 | 2.33744 | 60.2 | 72 | 1858 |
| GCF_002833015.1_ASM283301v1 | DPC6316 | SAMN07958362 | 2.39397 | 60.4 | 32 | 1912 |
| GCF_002832995.1_ASM283299v1 | DPC6321 | SAMN07958365 | 2.38236 | 59.9 | 28 | 1894 |
| GCF_002832985.1_ASM283298v1 | APC1503 | SAMN07958360 | 2.5627 | 59.7 | 39 | 2103 |
| GCF_002832955.1_ASM283295v1 | APC1504 | SAMN07958361 | 2.31029 | 60.2 | 51 | 1860 |
| GCF_002832945.1_ASM283294v1 | DPC6317 | SAMN07958363 | 2.44863 | 60.2 | 20 | 1918 |
| GCF_002276185.1_ASM227618v1 | Indica | SAMN07503177 | 2.37423 | 60 | 43 | 1948 |
| GCF_002076095.1_Bbif1886B | 1886B | SAMN06621706 | 2.47375 | 60.2 | 47 | 2083 |
| GCF_002076015.1_Bbif1890B | 1890B | SAMN06621710 | 2.34167 | 59.9 | 109 | 1846 |
| GCF_002075875.1_Bbif1898B | 1898B | SAMN06621716 | 2.47439 | 59.9 | 41 | 1998 |
| GCF_001940535.1_BlonW11v1 | W11 | SAMN06109230 | 2.32998 | 59.9 | 22 | 1857 |
| GCF_001892965.1_ASM189296v1 | 296B | SAMN05916052 | 2.25318 | 59.9 | 40 | 1685 |
| GCF_001725985.1_ASM172598v1 | AH1206 | SAMN04576213 | 2.42129 | 60.2 | 1 | 1967 |
| GCF_001719085.1_ASM171908v1 | 35624 | SAMN04254466 | 2.26406 | 60 | 1 | 1773 |
| GCF_001686245.1_ASM168624v1 | LO-K29b | SAMD00047623 | 2.37271 | 60.1 | 97 | 1866 |
| GCF_001686225.1_ASM168622v1 | LO-K29a | SAMD00047622 | 2.44918 | 60 | 85 | 1874 |
| GCF_001686205.1_ASM168620v1 | LO-C29 | SAMD00047621 | 2.48387 | 60 | 49 | 1927 |
| GCF_001686185.1_ASM168618v1 | LO-21 | SAMD00047620 | 2.65603 | 60.1 | 71 | 2034 |
| GCF_001686165.1_ASM168616v1 | LO-10 | SAMD00047619 | 2.54024 | 60.3 | 80 | 1988 |
| GCF_001686145.1_ASM168614v1 | LO-06 | SAMD00047618 | 2.43747 | 60 | 77 | 1926 |
| GCF_001595465.1_ASM159546v1 | 379 | SAMN04155602 | 2.38762 | 60.2 | 24 | 1921 |
| GCF_001546275.1_ASM154627v1 | CMW7750 | SAMN03842222 | 2.37208 | 60 | 39 | 1894 |
| GCF_001516925.1_ASM151692v1 | MC-42 | SAMN04263942 | 2.28825 | 59.8 | 29 | 1792 |
| GCF_001447975.1_ASM144797v1 | 7 | SAMN04129533 | 2.23558 | 60 | 36 | 1766 |
| GCF_001447955.1_ASM144795v1 | 9 | SAMN04129541 | 2.23377 | 60 | 31 | 1765 |
| GCF_001446275.1_ASM144627v1 | CCUG30698 | SAMN03785819 | 2.458 | 60.2 | 1 | 1956 |
| GCF_001446255.1_ASM144625v1 | NCIMB8809 | SAMN03785818 | 2.34099 | 60.1 | 1 | 1807 |
| GCF_001293145.1_ASM129314v1 | BG7 | SAMN03271682 | 2.45576 | 60.0068 | 2 | 1926 |
| GCF_001275745.1_assBLOI2 | BLOI2 | SAMN03775040 | 2.41759 | 60 | 72 | 1937 |
| GCF_001051015.2_ASM105101v2 | CECT 7210 | SAMEA3158508 | 2.4677 | 59.9 | 1 | 1992 |
| GCF_001050555.1_ASM105055v1 | CECT 7347 | SAMEA3146249 | 2.32722 | 60 | 128 | 1868 |
| GCF_000829295.1_ASM82929v1 | 105-A | SAMD00019943 | 2.29014 | 60.1 | 1 | 1772 |
| GCF_000786175.1_ASM78617v1 | VMKB44 | SAMN03105207 | 2.50193 | 60.3 | 34 | 2080 |
| GCF_000772485.1_ASM77248v1 | GT15 | SAMN03093230 | 2.33752 | 60 | 1 | 1815 |
| GCF_000741245.1_Biflon_sub.lon | LMG 13197 | SAMN02673437 | 2.3847 | 60.3 | 8 | 1803 |
| GCF_000730135.1_ASM73013v1 | EK13 | SAMN02862997 | 2.47453 | 60 | 39 | 2043 |
| GCF_000730105.1_ASM73010v1 | 1-5B | SAMN02862991 | 2.36751 | 60.1 | 25 | 1902 |
| GCF_000730055.1_ASM73005v1 | 7-1B | SAMN02862992 | 2.40709 | 59.8 | 34 | 1904 |
| GCF_000730045.1_ASM73004v1 | 72B | SAMN02862994 | 2.37445 | 60.3 | 30 | 1950 |
| GCF_000730035.1_ASM73003v1 | 17-1B | SAMN02862993 | 2.4672 | 60.2 | 20 | 1962 |
| GCF_000730025.1_ASM73002v1 | EK5 | SAMN02862996 | 2.23129 | 59.7 | 28 | 1780 |
| GCF_000497735.1_BLONGv1.0 | E18 | SAMN02471972 | 2.37297 | 60 | 1 | 1912 |
| GCF_000478525.1_blongD2957 | D2957 | SAMN02472064 | 2.33023 | 60.4 | 13 | 1812 |
| GCF_000261265.1_Blongum44Bv1.0 | 44B | SAMN00829148 | 2.55922 | 59.7 | 62 | 2109 |
| GCF_000261245.1_Blongum16Bv1.0 | 1-6B | SAMN00829154 | 2.68677 | 59.6 | 171 | 2215 |
| GCF_000261225.1_Blongum35Bv1.0 | 35B | SAMN00829158 | 2.51443 | 60.1 | 131 | 1967 |
| GCF_000261205.1_Blongum22Bv1.0 | 2-2B | SAMN00829155 | 2.6257 | 59.7 | 141 | 2089 |
| GCF_000219455.1_ASM21945v1 | KACC 91563 | SAMN02603656 | 2.39576 | 59.8115 | 3 | 1856 |
| GCF_000210755.1_ASM21075v1 | F8 | SAMEA3138379 | 2.38499 | 59.9 | 1 | 1884 |
| GCF_000196575.1_ASM19657v1 | 157F | SAMD00060953 | 2.40883 | 60.111 | 3 | 1923 |
| GCF_000196555.1_ASM19655v1 | JCM 1217 | SAMD00060951 | 2.38516 | 60.3 | 1 | 1870 |
| GCF_000185665.1_ASM18566v1 | 12_1_47BFAA | SAMN02463822 | 2.40599 | 60.1 | 61 | 1981 |
| GCF_000166895.2_ASM16689v2 | DJO10A | SAMN02441414 | 2.37528 | 59.9 | 120 | 1792 |
| GCF_000166315.1_ASM16631v1 | BBMN68 | SAMN02603469 | 2.26594 | 59.9 | 1 | 1740 |
| GCF_000155415.1_ASM15541v1 | CCUG 52486 | SAMN02463677 | 2.48085 | 60 | 22 | 2034 |
| GCF_000008945.1_ASM894v1 | DJO10A | SAMN02603512 | 2.38953 | 60.1182 | 3 | 1932 |
| GCF_000007525.1_ASM752v1 | NCC2705 | SAMN02603675 | 2.26027 | 60.1075 | 2 | 1773 |
| GCF_000003135.1_ASM313v1 | ATCC 55813 | SAMN00001475 | 2.39636 | 60.1 | 114 | 1901 |
| GCF_003094635.1_ASM309463v1 | DS9_3 | SAMN06464100 | 2.39717 | 59.9 | 13 | 1955 |
| GCF_003094855.1_ASM309485v1 | DS15_3 | SAMN06464097 | 2.39818 | 59.9 | 21 | 1956 |
| GCF_003094935.1_ASM309493v1 | DS18_3 | SAMN06464098 | 2.44826 | 59.7 | 174 | 2002 |
| GCF_003094955.1_ASM309495v1 | DS1_3 | SAMN06464096 | 2.41728 | 60 | 170 | 1958 |
| GCF_003094975.1_ASM309497v1 | DS7_3 | SAMN06464099 | 2.23729 | 60 | 17 | 1765 |
| GCF_003094995.1_ASM309499v1 | DS32_3 | SAMN08949007 | 2.23593 | 60.1 | 28 | 1761 |
2.3. Single Nucleotide Polymorphism (SNP) Calling and Phylogeny Reconstruction
The SNPs were recalled for 205 B. longum genomes by mapping the assemblies against the reference genome (B. longum NCC 2705) using MUMmer [38], as previously described [37], and only bi-allelic SNPs in the core genome were included in following analysis. The sequences of concatenated SNPs were used to construct phylogenetic tree (neighbor-joining method) using TreeBest (http://treesoft.sourceforge.net/treebest.shtml, accessed on 23 September 2020). It should be mentioned that before conducting the above phylogenetic analysis, we confirmed that the used 205 B. longum assemblies belonged to B. longum sup. longum by building a phylogenetic tree based on core genome bi-SNPs with the genomes of B. longum subsp. suillum, B. longum subsp. infantis, and B. longum subsp. suis as the outgroup.
2.4. Identification of Strain-Specific Markers
B. longum genomes were re-annotated using Prokka [39], and the obtained protein sequences were used to perform pangenome analysis via Roary (with a minimum BLASTP percentage identity of 90%) [35]. The genes that were only present in a single newly sequenced strain (RG4-1, FGSZY6M4 or M1-20-R01-3) and absent from all the other 204 strains were preliminarily identified. Considering the above gene presence/absence analysis was conducted at the protein level, we further validated the specificity of these strain-specific genes in nucleotide level. A nucleotide database containing 205 B. longum genomes was constructed using the makeblastdb command, and the above strain-specific gene sequences were analyzed through BLASTN against this database [40]. The DNA sequences that were only present in the target strain were retained. After the validation for intra-species specificity, we then tested the specificity of the DNA sequences under the background of all the representative microbes via website-based nucleotide BLAST against NR/NT database of NCBI database. The DNA sequences showing no hit in the database were finally selected.
2.5. Design of Strain-Specific Primers and Validation of Their Specificity via Electrophoresis
Based on the selected strain-specific DNA sequence for each B. longum strain, we designed corresponding qPCR primer pairs using Primer Premier5.0. The specificity of each primer pair was checked by Primer-blast in NCBI database. We conducted three titers of electrophoresis analysis to evaluate the primer specificity. For the intra-species specificity, another ten B. longum strains, in addition to each target strain (RG4-1, FGSZY6M4 or M1-20-R01-3), were used. For the intra-genus specificity within Bifidobacterium, six other Bifidobacterium species were selected. For the specificity against other gut bacteria, 32 representative members of intestinal microbes were adopted. Genome DNA was extracted for each strain, and amplification was performed using the designed strain-specific primers. Bio-Rad T100 Thermal Cycler was used for PCR amplifications. Each reaction mixture (50 µL) consisted of 2 µL bacterial genomic DNA, 25 µL 2× Taq Plus MasterMix, 2 µL forward primer (0.4 µmol in the final mixture), 2 µL reverse primer (0.4 µmol in the final mixture), and 19 µL ddH2O. The PCR conditions were as follows: 94 °C for 2 min, followed by 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s conducting 35 cycles, and then 72 °C for 2 min. The electrophoresis analysis was conducted to separate PCR products at 120 V using a 1.5% agarose gel.
2.6. Strain-Specific qPCR Designs and Standard Curves for Absolute Quantification
To evaluate the specificity of these strain-specific primer pairs against the background of complex fecal bacterial communities, we collected fecal samples from 30 humans and 15 mice. These samples were all the bassline samples (i.e., before B. longum administration) in the following described animal experiment and human trial. The fecal DNA was extracted using FastDNA Spin Kit for Soil (Catalog number: 116570200, MP Biomedicals, Santa Ana, CA, USA) according to the manufacturers’ instructions. The PCR program was optimized to ensure no positive amplification for these baseline samples. Positive control with DNA of each target B. longum strain and negative control using water instead of genomic DNA were included in all PCR runs. The PCR system (20 μL) consisted of 2 μL genomic DNA (template), 10 μL 2× supermix (BIO-RAD), 2 µL forward primer (0.4 µmol in the final mixture), 2 µL reverse primer (0.4 µmol in the final mixture), and 4 µL ddH2O. The following qPCR program yielded the most selective amplification: an initial denaturation step at 95 °C for 2 min; 30 cycles of denaturation at 95 °C for 5 s and annealing/extension at 65 °C for 30 s; a melt curve analysis between 65 °C and 95 °C in 0.5 °C increments at 2–5 s/step; and polymerase activation and DNA denaturation at 95 °C for 5 min.
For absolute quantification of target B. longum strains, standard curves were prepared. A pure culture-based standard series of each target B. longum strain was obtained using DNA extracted from a tenfold dilution series of each B. longum strain in MRS (16 h). The exact bacterial cell numbers of the first serial decimal dilution were determined using plate counting. Cycle threshold values (CT) were plotted versus equivalent log cell numbers. The amplification efficiency of each design was determined by the slope of the standard curves: E (%) = (10−1/slope − 1) × 100.
2.7. Quantification of Ingested B. longum Strains in the Fecal Samples
Each B. longum-specific qPCR system was used to access the colonized biomass of corresponding target strain in the gut of mice after strain administration. For animal experiments, 5-week-old male Balb/c mice used in this study were purchased from the Shanghai Laboratory Animal Center (Shanghai, China). Animal care and study protocols were approved by the Ethics Committee of Jiangnan University, China (JN. No20181130b1200130[261]). All of the applicable institutional and national guidelines for the care and use of animals were followed. All mice were kept in the mouse facility of the Laboratory Animal Center of the Department of Food Science and Technology, Jiangnan University, Wuxi, China, on a 12-h light/dark cycle in a temperature-(22 °C ± 1 °C) and humidity-controlled (55% ± 10%) room. Mice were assigned to three experiment groups (n = 5): the RG4-1 group, the FGSZY6M4 group and the M1-20-R01-3 group. The experimental period was 14 days, including a 7-day accommodation period, and followed by a 7-day B. longum intervention period. MRS broth-cultured B. longum strains, after being resuspended in sterile saline, were prepared each day, and routinely subjected to plate counting to ensure a gavage dose of 108–109 CFU/d for each mouse. Fecal samples were collected at day 7 and day 14.
For the human trial, the 30 volunteers were students at Jiangnan University who ranged in age from 20 to 30 years. No use of antibiotics was reported by the subjects within the 1 month before or during the study, and probiotic foods were not allowed during the trial. The experimental period included a 2-week baseline period (without any treatment) and a 2-week probiotic intervention period. The subjects were randomly assigned to three groups (n = 10 for each group), in which each subject in each B. longum intervention group (RG4-1, FGSZY6M4 or M1-20-R01-3) was administrated 109–1010 viable cells/d of the corresponding B. longum strain. Fecal samples were collected at day 14 (±3 days) of the baseline period, and day 14 (±3 days) of the treatment period. The colonized biomass of the three B. longum strains in fecal samples was determined using the respective qPCR primers. All volunteers provided informed consent. The Ethics Committee of Jiangnan University (Wuxi, China) provided ethical clearance for this human trial in accordance with the Declaration of Helsinki.
The colonized biomass of each B. longum strains was confirmed by selective culture medium with colony typing with the designed strain-specific primers. In brief, fecal samples were used to isolate bifidobacteria by cultivation on deMan Rogosa Sharpe (MRS) agar supplemented with 50 mg/L mupirocin and 0.1% L-cysteine HCl. After incubation at 37 °C for 48 h in an anaerobic chamber (80% N2, 10% H2, 10% CO2), colonies were counted, picked, and then subjected to conventional PCR using the strain-specific primers.
3. Results
3.1. Genomic Diversity of B. longum
We reconstructed a phylogenetic tree based on three newly sequenced and 202 publicly available B. longum strain genomes (Table 2 and Table 3, and Figure 1A), and calculated the pair-wise genetic distances and accessory gene numbers to reveal the intra-species genomic diversity (Figure 1B–D). The parameters for the newly sequenced genomes are shown in Table 3. As shown in Figure 1A, the three B. longum strains (RG4-1, M1-20-R01-3, and FGSZY6M4) isolated from the fecal samples of three Chinese individuals are closely clustered in the phylogenetic tree. This is unsurprising, because the majority of publicly available strains were isolated from geographically distant areas (i.e., other countries or continents). Nevertheless, the three strains exhibited obvious genetic distances indicative of their distinct genotypes.
Table 3.
Parameters for self-sequenced genomes.
| Scaffold Number | Length | Gap | Average Length | N50 | N90 | GC Content (%) | |
|---|---|---|---|---|---|---|---|
| FGSZY6M4 | 52 | 2,321,455 | 3574 | 44,643.37 | 202,550 | 83,023 | 59.72 |
| RG4-1 | 51 | 2,601,515 | 3420 | 51,010.1 | 224,480 | 60,103 | 60.21 |
| M1-20-R01-3 | 46 | 2,237,922 | 3258 | 48,650.48 | 232,217 | 63,500 | 60.02 |
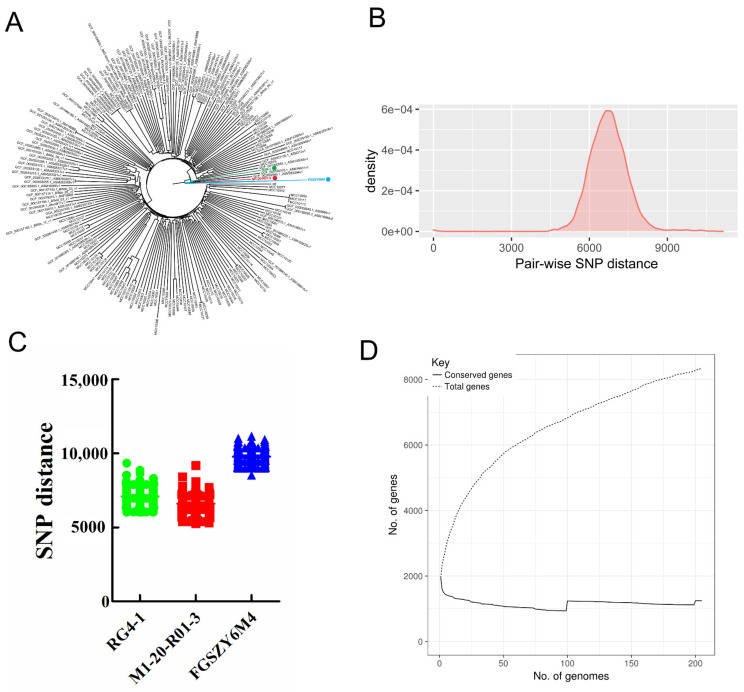
Figure 1.
Phylogenetic relationship and genomic diversity of B. longum. (A) Phylogenetic tree (neighbor-joining method) of 205 B. longum strains. The three target strains used for strain-specific detection are colored. (B) Distribution of pair-wise SNP distances between 205 strains. (C) Pair-wise SNP distances between each of the target strains and all the other strains in the dataset. (D) Pangenome curve depicting the number of total genes detected versus the number of conserved genes as the number of included genomes increases.
We also determined the number of variable SNPs in the core genomes of 205 B. longum strains (Figure 1B). The results indicated that the average SNP distance across the 205 strains was 6691. The most phylogenetically related strains exhibited an SNP distance of 0, while the most phylogenetically distant strains exhibited an SNP distance of 11,145. For each of the three novel B. longum strains, the minimum genetic distances between the respective strain and the other 204 strains in the dataset were 6000 (RG4-1), 5257 (M1-20-R01-3), and 8514 (FGSZY6M4). These data further confirm the genetic differences between each of these three B. longum strains and the other strains (Figure 1C). As shown in Figure 1D, the pangenome analysis indicated that accessory genes accounted for 85.1% of the total genes, and that new genes were accumulated frequently as new strains were added to the analysis.
Overall, B. longum showed a high level of intra-species genomic diversity in terms of the pair-wise SNP distances and the total numbers of accessory genes. The target strains selected for strain-specific detection were phylogenetically distinct and exhibited considerable genetic dissimilarity with the other strains in this dataset, thus implying the possibility of finding appropriate strain-specific markers.
3.2. In Silico Identification and Validation of Strain-Specific Gene Markers
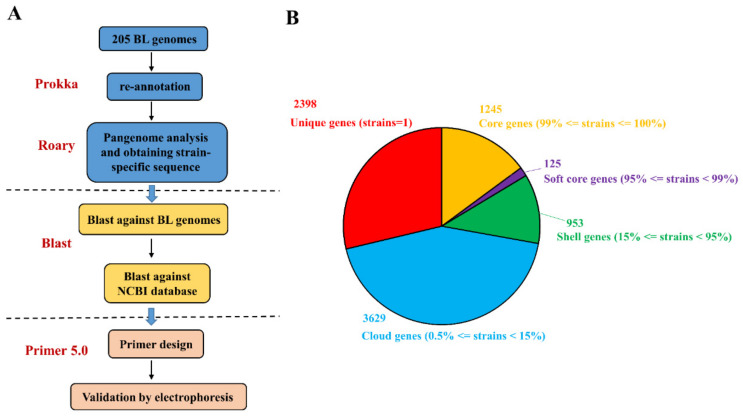
Considering the relatively high intra-species genetic similarity relative to inter-species and inter-genus similarities, we initially searched for strain-specific genes within the B. longum genomes. The pipeline used to construct a strain-specific detection tool is shown in Figure 2A. The publicly available B. longum genomes used in this study were derived from different projects, and the formats of their annotation files were not uniform, which prevented a pangenome analysis. Accordingly, we re-annotated these publicly available genomes and the three target B. longum strain genomes using Prokka, and then conducted a gene presence/absence analysis based on the annotated protein sequences. We defined unique genes (present in only one strain within the dataset of 205 strains), core genes (present in ≥99% of strains), soft core genes (present in ≥95% to <99%), shell genes (present in ≥15% to <95%), and cloud genes (present in ≥0.5% to <15%). As shown in Figure 2B, this analysis preliminarily identified 2398 strain-specific genes across the 205 B. longum strains, which accounted for 28.7% of the total genes. Notably, 32, 14, and 49 strain-specific genes were identified, respectively, for B. longum RG4-1 (Table 4), B. longum M1-20-R01-3 (Table 5), and B. longum FGSZY6M4 (Table 6).
Figure 2.
The analysis pipeline for strain-specific primer design corresponding to the three B. longum strains (A) and the pangenome readout (B).
Table 4.
Strain-specific genes identified by Roary for B. longum RG4-1.
| Gene | Non-Unique Gene Name | Annotation | Avg Group Size Nuc | Gene Tag |
|---|---|---|---|---|
| group_8150 | hypothetical protein | 227 | RG4-1_00079 | |
| group_8151 | hypothetical protein | 296 | RG4-1_00112 | |
| group_8152 | hypothetical protein | 257 | RG4-1_00222 | |
| group_8153 | xerC_1 | Tyrosine recombinase XerC | 1286 | RG4-1_00224 |
| group_8154 | Helix-turn-helix domain protein | 185 | RG4-1_00225 | |
| group_8155 | Helix-turn-helix domain protein | 395 | RG4-1_00226 | |
| group_8156 | Helix-turn-helix domain protein | 341 | RG4-1_00227 | |
| group_8157 | site-specific tyrosine recombinase XerC | 1349 | RG4-1_00228 | |
| group_8158 | hypothetical protein | 209 | RG4-1_00545 | |
| group_8160 | hypothetical protein | 236 | RG4-1_00568 | |
| group_8161 | hypothetical protein | 272 | RG4-1_01044 | |
| group_8162 | mdeA_1 | Methionine gamma-lyase | 1277 | RG4-1_01045 |
| group_8163 | Phage-related minor tail protein | 3317 | RG4-1_01165 | |
| group_8164 | Phage tail protein | 788 | RG4-1_01166 | |
| group_8165 | hypothetical protein | 1115 | RG4-1_01167 | |
| group_8166 | smc_4 | Chromosome partition protein Smc | 1124 | RG4-1_01168 |
| group_8167 | hypothetical protein | 359 | RG4-1_01169 | |
| group_8168 | hypothetical protein | 1871 | RG4-1_01170 | |
| group_8169 | hypothetical protein | 185 | RG4-1_01171 | |
| group_8170 | acm | Lysozyme M1 precursor | 1280 | RG4-1_01174 |
| yoaD | Putative 2-hydroxyacid dehydrogenase YoaD | 968 | RG4-1_01873 | |
| group_8172 | araN_4 | putative arabinose-binding protein precursor | 1331 | RG4-1_01874 |
| ycjP_2 | Inner membrane ABC transporter permease protein YcjP | 830 | RG4-1_01875 | |
| group_8174 | ycjO_1 | Inner membrane ABC transporter permease protein YcjO | 899 | RG4-1_01876 |
| group_8175 | hypothetical protein | 161 | RG4-1_01877 | |
| group_8176 | nanE | Putative N-acetylmannosamine-6-phosphate 2-epimerase | 689 | RG4-1_01878 |
| group_8177 | nanA | N-acetylneuraminate lyase | 917 | RG4-1_01879 |
| bglK | Beta-glucoside kinase | 914 | RG4-1_01880 | |
| rpiR | HTH-type transcriptional regulator RpiR | 905 | RG4-1_01881 | |
| group_8180 | Chitinase class I | 551 | RG4-1_02208 | |
| group_8181 | Thaumatin family protein | 284 | RG4-1_02209 | |
| group_8182 | hypothetical protein | 197 | RG4-1_02210 |
Table 5.
Strain-specific genes identified by Roary for B. longum M1-20-R01-3.
| Gene | Non-Unique Gene Name | Annotation | Avg Group Size Nuc | Gene Tag |
|---|---|---|---|---|
| group_6841 | hypothetical protein | 194 | M1-20-R01-3_00305 | |
| group_6842 | hypothetical protein | 998 | M1-20-R01-3_00310 | |
| group_6843 | hypothetical protein | 221 | M1-20-R01-3_00311 | |
| group_6844 | hypothetical protein | 233 | M1-20-R01-3_00316 | |
| group_6845 | hypothetical protein | 257 | M1-20-R01-3_00318 | |
| group_6846 | hypothetical protein | 623 | M1-20-R01-3_00319 | |
| group_6847 | hypothetical protein | 587 | M1-20-R01-3_00320 | |
| group_6848 | hypothetical protein | 1745 | M1-20-R01-3_00324 | |
| group_6849 | hypothetical protein | 236 | M1-20-R01-3_00325 | |
| group_6850 | hypothetical protein | 581 | M1-20-R01-3_00326 | |
| group_6851 | hypothetical protein | 191 | M1-20-R01-3_00327 | |
| group_6852 | YcfA-like protein | 224 | M1-20-R01-3_00328 | |
| group_6853 | hypothetical protein | 413 | M1-20-R01-3_00329 | |
| group_6854 | hypothetical protein | 617 | M1-20-R01-3_00562 |
Table 6.
Strain-specific genes identified by Roary for B. longum FGSZY6M4.
| Gene | Non-Unique Gene Name | Annotation | Avg Group Size Nuc | Gene Tag |
|---|---|---|---|---|
| group_3844 | hypothetical protein | 203 | FGSZY6M4_00001 | |
| group_3845 | pepD_1 | Dipeptidase | 1637 | FGSZY6M4_00002 |
| group_3846 | epsH | Putative glycosyltransferase EpsH | 1034 | FGSZY6M4_00003 |
| group_3847 | transcriptional regulator BetI | 821 | FGSZY6M4_00004 | |
| group_3850 | N-acetylmuramoyl-L-alanine amidase | 878 | FGSZY6M4_00052 | |
| group_3869 | hypothetical protein | 419 | FGSZY6M4_00335 | |
| group_3870 | putative ABC transporter ATP-binding protein/MT1014 | 434 | FGSZY6M4_00336 | |
| zur_2 | Zinc uptake regulation protein | 500 | FGSZY6M4_00339 | |
| group_3880 | hypothetical protein | 578 | FGSZY6M4_00378 | |
| group_3899 | hypothetical protein | 518 | FGSZY6M4_00692 | |
| yhcR_2 | Endonuclease YhcR precursor | 3566 | FGSZY6M4_01406 | |
| group_3960 | hypothetical protein | 332 | FGSZY6M4_01466 | |
| group_3961 | hypothetical protein | 692 | FGSZY6M4_01467 | |
| group_3962 | YcaO-like family protein | 1610 | FGSZY6M4_01468 | |
| group_3963 | ABC-2 type transporter | 731 | FGSZY6M4_01469 | |
| group_3964 | yxlF | putative ABC transporter ATP-binding protein YxlF | 908 | FGSZY6M4_01470 |
| group_3965 | hypothetical protein | 188 | FGSZY6M4_01471 | |
| group_3966 | hypothetical protein | 1079 | FGSZY6M4_01472 | |
| group_3967 | hypothetical protein | 1061 | FGSZY6M4_01473 | |
| group_3968 | hypothetical protein | 2594 | FGSZY6M4_01474 | |
| group_3969 | Nitroreductase family protein | 1532 | FGSZY6M4_01475 | |
| group_3970 | YcaO-like family protein | 1607 | FGSZY6M4_01476 | |
| group_3971 | hypothetical protein | 1691 | FGSZY6M4_01477 | |
| group_3972 | hypothetical protein | 176 | FGSZY6M4_01478 | |
| group_4002 | hypothetical protein | 254 | FGSZY6M4_01825 | |
| group_4004 | hypothetical protein | 308 | FGSZY6M4_01832 | |
| group_4005 | hypothetical protein | 227 | FGSZY6M4_01836 | |
| group_4008 | hypothetical protein | 365 | FGSZY6M4_01863 | |
| group_4009 | hypothetical protein | 359 | FGSZY6M4_01864 | |
| group_4010 | hypothetical protein | 197 | FGSZY6M4_01866 | |
| group_4011 | hypothetical protein | 455 | FGSZY6M4_01867 | |
| group_4012 | whiB1_2 | Transcriptional regulator WhiB1 | 245 | FGSZY6M4_01868 |
| group_4013 | hypothetical protein | 329 | FGSZY6M4_01869 | |
| group_4014 | hypothetical protein | 170 | FGSZY6M4_01871 | |
| group_4036 | hypothetical protein | 4835 | FGSZY6M4_01918 | |
| group_4037 | hypothetical protein | 566 | FGSZY6M4_01919 | |
| group_4038 | hypothetical protein | 440 | FGSZY6M4_01920 | |
| group_4039 | hypothetical protein | 176 | FGSZY6M4_01921 | |
| group_4040 | hypothetical protein | 527 | FGSZY6M4_01922 | |
| group_4041 | hypothetical protein | 368 | FGSZY6M4_01923 | |
| group_4042 | hypothetical protein | 566 | FGSZY6M4_01939 | |
| group_4048 | hypothetical protein | 338 | FGSZY6M4_01977 | |
| group_4049 | hypothetical protein | 419 | FGSZY6M4_01981 | |
| group_4050 | hypothetical protein | 1190 | FGSZY6M4_01982 | |
| bvgA | Virulence factors putative positive transcription regulator BvgA | 668 | FGSZY6M4_01983 | |
| group_4052 | enterobactin exporter EntS | 1262 | FGSZY6M4_01984 | |
| group_4053 | hypothetical protein | 197 | FGSZY6M4_01985 | |
| aacA-aphD | Bifunctional AAC/APH | 1340 | FGSZY6M4_01986 | |
| group_4055 | Zein seed storage protein | 755 | FGSZY6M4_01993 |
Next, we explored whether the specificities of the identified strain-specific genes would be maintained against other microbial taxa. Rather than using amino acid sequences in Roary analysis, we tested the specificities of the preliminary strain-specific genes at the nucleotide level using an in-house nucleotide BLAST against a B. longum genome database to further ensure the strain-specificity of the identified genes. We also used a website-based nucleotide BLAST tool against the NR/NT database (NCBI) to determine which sequences could not be identified in any other representative microbes. Finally, this screening process identified RG4-1_01874 (1331 bp), M1-20-R01-3_00324 (1745 bp), and FGSZY6M4_01477 (1691 bp), which were selected as the target DNA sequences for strain-specific quantification.
3.3. Strain-Specific qPCR Designs and Electrophoretic Validation
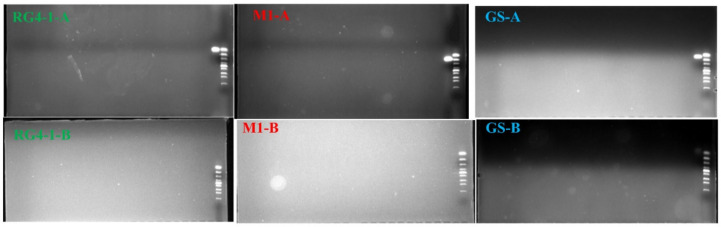
Next, qPCR primer pairs were designed for each of the three B. longum strains (RG4-1, M1-20-R01-3, and FGSZY6M4) based on their respective strain-specific DNA sequences (Table 7), with predicted product sizes of 115, 199, and 144 bp, respectively. Using electrophoresis, the specificities of the three qPCR probe sets were validated against genomic DNA derived from various microbial strains (Table 1), including 10 other B. longum strains, 6 other Bifidobacterium species, and 32 representative members of the intestinal microbiome. The results indicated that PCR amplification based on the strain-specific primers was only successful for each of the respective target strains, and no amplification of genomic material from non-target microorganisms was observed (Figure 3). Multiple qPCR primer pairs were designed for each target strain, and the primers with the best performance at this stage were selected and are reported here.
Table 7.
Strain-specific primers for three B. longum strains.
| Strain | Primer Sequence (5′-3′) | Primer Length (bp) | Primer Score | Product Length (bp) |
|---|---|---|---|---|
| RG4-1 | F: ACCATCTGGGTGGAGAAAGTG | 21 | 100 | 115 |
| R: TGGCGGAAATGAACTCGTAAT | 21 | 100 | ||
| M1-20-R01-3 | F: GATGGCACCAGCACAGG | 17 | 100 | 199 |
| R: GGAGCACGGCGACTATG | 17 | 100 | ||
| FGSZY6M4 | F: TCCCGAATCCGACTATGA | 18 | 100 | 144 |
| R: TCGCTGCCAACTACTAAAA | 19 | 100 |
Figure 3.
Electrophoresis results of PCR products generated using each strain-specific qPCR primer pair against DNA from the respective target B. longum strains and non-target microorganisms. Each gel includes 25 lanes (including a lane for the gene ruler). Order of microorganisms were as follows: for RG4-1-A (from right to left), gene ruler, B. longum RG4-1, B. longum FGSZY6M4, B. longum M1-20-R01-3, B. longum 274, B. longum FSHHK13M1, B. longum FSDLZ57M1, B. longum NaTon 49-4, B. longum FJSWXJ11M1, B. longum HUB 36-17, B. longum 28-10, B. longum ZCC7, Bifidobacterium breve DSM 20213, Bifidobacterium bifidum DSM 20456, Bifidobacterium pseudocatenulatum FQHXN5M4, Bifidobacterium pseudolongum 56M2, Bifidobacterium animalis BB12, Bifidobacterium adolescentis L2-32, Lactobacillus salivarius DSM 20555, Lactobacillus gasseri DSM 20243, Lactobacillus casei DSM 20011, Lactobacillus acidophilus DSM 20079, Lactobacillus plantarum DSM 20174, Lactobacillus reuteri DSM 20016, and Lactobacillus rhamnosus LMS2-1; for M1-A (from right to left), gene ruler, B. longum M1-20-R01-3, B. longum RG4-1, B. longum FGSZY6M4, and the order of following strains was the same as that of RG4-1-A; for GS-A (from right to left), gene ruler, B. longum FGSZY6M4, B. longum M1-20-R01-3, B. longum RG4-1, and the order of following strains was the same as that of RG4-1-A; for RG4-1-B, M1-B and GS-B (from right to left), Escherichia coli CMCC44102, Akkermansia muciniphila FJLHD50M21, Faecalibacterium prausnitzii ATCC 27768, Enterococcus faecalis CCFM596, Bacteroides fragilis NCTC9343, Bacteroides thetaiotaomicron FNMHLBE9-K-7, Bacteroides eggerthii FSDTA-HCK-B-9, Bacteroides cellulosilyticus FSDTA-ELI-BHI-5, Bacteroides nordii FNMHLBE13K2, Bacteroides stercoris FJSWX62K34, Bacteroides uniformis FJSWX62K43, Bacteroides caccae FFJLY22K5, Parabacteroides distasonis FSDTA-HCM-XY-12, Bacteroides dorei FJSWX61E4, Bacteroides faecis FTJS2E2, Bacteroides intestinalis FBJ60K5, Bacteroides vulgatus FSDLZ51K1, Bacteroides finegoldii FNMHLBE11E1, Bacteroides ovatus FBJ10-K-10, Bacteroides clarus F-FJ-LY 22-K-22, Bacteroides salyersiae FSDTA-ELI-BHI-9, Bacteroides xylanisolvens FSDTAHCMXY17, Parabacteroides merdae FSDTAELIBHI4 and Clostridium butyricum FJSCZD1G10.
3.4. Specificities, Standard Curves, and Amplification Efficiencies of qPCR Assays
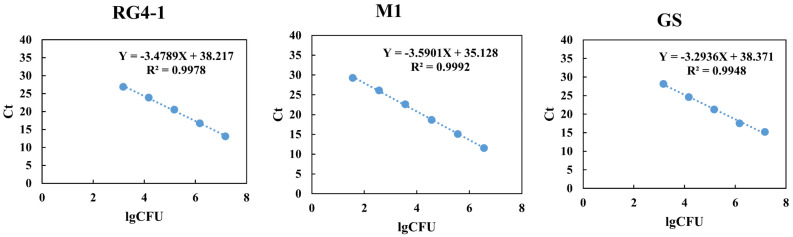
Primer specificity was evaluated by qPCR against a complex microbial community present in baseline fecal samples that had not been enriched for the target strain (i.e., pre-treatment). The DNA of each target B. longum strain was used as a positive control in the qPCR runs. Under optimized conditions, no amplification was observed for the baseline samples, whereas the positive controls exhibited good amplification. As shown in Figure 4, standard curves corresponding to RG4-1, M1-20-R01-3, and FGSZY6M4 exhibited good linearity over a 4-log range (103–107 CFU/qPCR system, R2 > 0.99), a 5-log range (101–106 CFU/qPCR system, R2 > 0.99) and a 4-log range (103–107 CFU/qPCR system, R2 > 0.99), respectively. The equations of the regression curves for RG4-1, M1-20-R01-3, and FGSZY6M4 were as follows: Ct = −3.4789lgCFU + 38.217, Ct = −3.5901lgCFU + 35.128, and Ct = −3.2936lgCFU + 38.371; and the corresponding amplification efficiencies were 93.8%, 90.0% and 101.2%, respectively. Collectively, the three qPCR primer pairs exhibited specificity in the context of the complex bacterial communities in fecal samples from both humans and mice, the standard curves used for absolute quantification were of good linearity, and the amplification efficiencies were qualified, suggesting that these primers can be used to detect the abundances of the target B. longum strains in fecal samples.
Figure 4.
qPCR standard curves for the three B. longum strains.
3.5. Quantification of Ingested B. longum Strains in Mouse and Human Fecal Samples
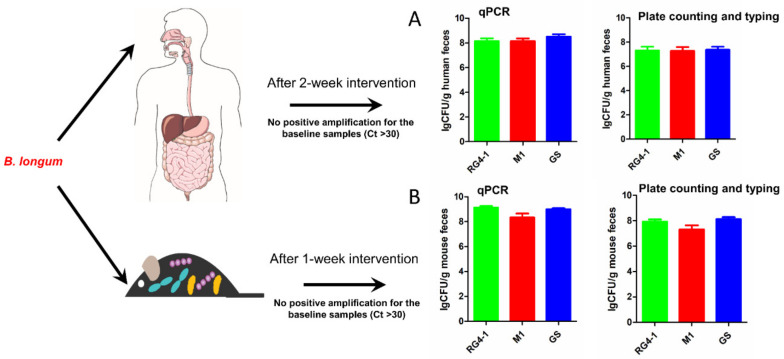
We used the strain-specific primers to detect the abundances of three target B. longum strains in the feces of mice and human volunteers who had consumed these B. longum strains (Figure 5). For the animal experiments, the colonized biomasses of the three B. longum strains in samples collected 1 week after oral administration exceeded 108 CFU/g feces, with average abundances of 1.54 × 109 CFU/g feces for RG4-1, 4.60 × 108 CFU/g feces for M1-20-R01-3, and 1.06 × 109 CFU/g feces for FGSZY6M4. For the human trial, the average population levels of each strain reached >108 CFU/g feces, with average abundances of 4.00 × 108 CFU/g feces for RG4-1, 3.78 × 108 CFU/g feces for M1-20-R01-3, and 7.18 × 108 CFU/g feces for FGSZY6M4. These results indicate that orally ingested B. longum strains can be detected at considerable levels in the feces of both mice and humans during a period of intervention, suggesting short-term engraftment of the ingested bacteria in the gut. The colonized abundances of the three B. longum strains obtained from selective culture medium combined with colony typing using our specific qPCR primers further confirmed the conclusion of short-term engraftment by those strains, despite the bacterial numbers detected by this culture-dependent method were generally 10-fold lower (Figure 5).
Figure 5.
Colonized biomasses of the target B. longum strains in fecal samples from humans and mice. Panel (A), the results collected from the human trail; Panel (B), the results collected from the animal experiments.
4. Discussion
It has become increasingly clear that the beneficial effects of probiotic bacteria on the host are strain-specific, and the viability of probiotic strains in the gut after ingestion is believed to be an essential factor for them to demonstrate health-promoting functions. Therefore, determining the presence and colonized biomasses of specific probiotic strains in the gut is a key step toward the informed use of probiotics for therapeutic ends. Compared with traditional microbiological methods (e.g., selective media with colony identification), PCR-based molecular methods, which can distinguish the target strain from the baseline microbiota, are the most popular because of their superior sensitivity and specificity [13]. Here, we adopted a pangenome analysis-based approach to identify strain-specific DNA sequences and designed qPCR primers based on these unique markers, which enabled us to detect B. longum strains in fecal samples at a strain-level resolution. In addition to determining the abundances of probiotic bacteria at the strain level, this strategy advantageously involves the identification of strain-specific sequences based on a large set of bacterial genomes. The resulting strain-specificity was more “true” than those identified in an RAPD analysis based on a restricted number of pure cultured bacteria.
This is the first known example of using a bioinformatics method to search for unique gene markers and design strain-specific molecular tools to detect and quantify individual bacterial strains. Previous studies have used the abovementioned RAPD methods to identify strain-specific sequences against a background of a limited number of strains of the same species. Daranas et al. screened a unique marker of L. plantarum PM411 against seven other L. plantarum strains in an assay based on seven random RAPD primers [41]. In another study, B. bifidum BF-1 was detected by targeting a strain-specific sequence in an analysis involving 27 RAPD primers and 30 B. bifidum strains [42]. In this study, we identified 32, 14, and 49 strain-specific genes corresponding to B. longum RG4-1 (Table 5), B. longum M1-20-R01-3 (Table 6), and B. longum FGSZY6M4, respectively (Table 7), in the context of 205 B. longum genomes. Obviously, our pangenome-based approach could identify a larger number of strain-specific fragments within a wider confidence interval, because it uses whole genome sequences rather than random PCR products, and a large set of bacterial genomes rather than a limited number of available pure cultures. In addition, we also observed a high frequency of strain-specific sequences among B. longum isolates. Specifically, we detected 2398 strain-specific genes among 205 B. longum strains, which accounted for 28.7% of the total genes, and 178 of these strains harbored unique gene markers. Our data suggest the existence of considerable intra-species genomic diversity within B. longum in terms of the accessory gene number. This finding allowed us to construct strain-specific primers to identify and quantify most of these strains based on single gene markers. For those strains without unique genes, future approaches may use two or more gene sequences as unique combination amplification targets.
The strain-specific sequences identified in this study were first confirmed in silico, by electrophoresis and by the absence of qPCR amplification signals in baseline microbiota of the tested fecal samples derived from humans and mice. In previous studies, strain-specific sequences identified using RAPD methods often showed homology with sequences from other microbes and thus were not truly strain-specific. A previous BLAST analysis of a B. bifidum OLB6378-specific RAPD fragment showed a high similarity (98%) to the parB gene sequences in publicly available B. bifidum genomes [28]. The strain-specific RAPD band identified by Karjalainen et al., and used for the strain-level detection of Propionibacterium freudenreichii (P. freudenreichii). JS was found to encode a 103-bp region with 91% identity to P. freudenreichii ssp. shermanii CIRM-BIA1 [43]. Similarly, the unique RAPD band for B. breve 99 was shown to contain regions homologous to those detected in strains of B. longum, B. adolescentis, B. dentium Bd1, and B. animalis ssp. lactis [43]. In this study, we validated the strain-specific sequences identified preliminarily via nucleotide BLAST against a self-constructed database of the 205 B. longum genomes and further checked the specificities of the sequences against all available gene information from representative microbes via searching the NR/NT NCBI database. Our selected strain-specific fragments (RG4-1_01874, M1-20-R01-3_00324, and FGSZY6M4_01477) passed the nucleotide BLAST against the B. longum genome database and generated no hits in the NR/NT database. Therefore, the strain-specific sequences identified in this study were highly specific against a background of various microbes and can probably be applied in a broader taxonomic context. We further validated the specificities of the newly designed qPCR primers via conventional PCR against 10 other B. longum strains, 6 other Bifidobacterium species, and 32 representative members of the intestinal microbiome. We additionally confirmed the specificities of the primers by the absence of qPCR amplification products from fecal samples derived from both humans and mice that were free from the target B. longum strains.
The bioinformatics pipeline proposed in this study can be expanded to search for strain-specific markers and achieve strain-level qualifications of various bacteria, including probiotic species. A large number of genomes corresponding to probiotic Lactobacillus and Bifidobacterium species have been sequenced and made publicly available, including 544 L. plantarum, 198 L. paracasei, 112 B. breve, and 176 L. salivarius genomes in the NCBI database. Previous genomic analyses of these species have demonstrated open pangenomes, high intra-species genomic diversity, and high new gene discovery rates for L. salivarius [44,45] and L. casei [46] as the number of included genomes increases. Therefore, the pipeline constructed here can be directly translated for use in other probiotic species, provided that sufficient genomic data and abundant strain-specific genes are available. For this study, we selected Roary because of the simple command line and relatively higher running speed compared with other available tools. Other pangenome analysis tools, such as Pan-Seq [32], PGAT [33], and PGAP [34], are also suitable choices for constructing strain-specific primers.
Our qPCR assays further indicated that the three target B. longum strains could colonize the guts of both humans and mice at high levels of abundance (>108 CFU/g feces for both humans and mice) during an intervention period (1 week for mice and 2 weeks for humans), and the colonized biomasses were validated using selective culture and colony-typing methods with the strain-specific primers. Absolute qualification was achieved using highly linear standard curves (R2 > 0.99) and amplification efficiency was qualified (>90.0%), both of which are comparable to previous studies [41]. In line with our results, a previous study demonstrated that at 2 weeks post-ingestion, the abundances of B. longum AH1206 ranged from 107 to 1010 cells/g feces in human subjects [47]. Selective culture methods combined with strain typing using our strain-specific primers confirmed the colonization of these target B. longum strains. However, the bacterial numbers generated using this culture-based method were nearly 10-fold lower than those determined using qPCR. We attribute this underestimation to an inherent defect of culture-based methods, as previous studies have reported that both the frequencies and numbers of bacterial cells detected in feces by culture methods were substantially lower than the corresponding values obtained using qPCR [42,48]. In addition, PCR with our strain-specific primers enabled us to identify colonies of the three B. longum strains on selective agar efficiently and accurately.
5. Conclusions
In conclusion, benefiting from the large number of sequenced genomes of probiotic species, we took the most potent probiotic gut colonizer, B. longum, as an example, and proposed a precedent in which a pangenome analysis-based approach can be used to identify unique gene markers for a given bacterial strain, and targeted these markers to achieve strain-level qualification. The qPCR primers designed in this study were able to successfully detect and quantify the colonized biomasses of the given B. longum strains in fecal samples from humans and mice. Therefore, we demonstrated the ability of these efficient in silico analyses to replace existing time- and labor-intensive RAPD methods. Furthermore, by including as many bacterial genomes as possible, the annotated unique sequences are highly specific and can be applied in a broader taxonomic context involving a more complex microbial ecology. The pipeline constructed herein can also be adapted to identify strain-specific markers and design strain-level qPCR primers for other probiotic species.
Author Contributions
Conceptualization, Y.X., C.W., J.Z., H.Z., W.C. and Q.Z.; Data curation, Y.X.; Formal analysis, Y.X.; Funding acquisition, W.C. and Q.Z.; Investigation, Y.X. and Q.Z.; Methodology, Y.X.; Project administration, J.Z., H.Z., W.C. and Q.Z.; Resources, J.Z., H.Z., W.C. and Q.Z.; Software, Y.X.; Supervision, J.Z., H.Z., W.C. and Q.Z.; Validation, Y.X. and Q.Z.; Visualization, Y.X. and C.W.; Writing—original draft, Y.X. and J.Z.; Writing—review & editing, Y.X., C.W., J.Z., H.Z., W.C. and Q.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China Program [No. 31820103010 and No. 31871773]; Projects of Innovation and Development Pillar Program for Key Industries in Southern Xinjiang of Xinjiang Production and Construction Corps [2018DB002]; National Key Research and Development Project [No. 2018YFC1604206]; National First-Class Discipline Program of Food Science and Technology [JUFSTR20180102]; the BBSRC Newton Fund Joint Centre Award; and Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.
Institutional Review Board Statement
Animal care and study protocols were approved by the Ethics Committee of Jiangnan University, China (JN. No20181130b1200130[261]). All of the applicable institutional and national guidelines for the care and use of animals were followed. For the human trial, the Ethics Committee of Jiangnan University (Wuxi, China) provided ethical clearance for this human trial according to the Helsinki Declaration.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data in this study are available from the authors upon request.
Conflicts of Interest
There are no conflict of interest to declare.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kamada N., Chen G.Y., Inohara N., Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nat. Cell Biol. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korpela K., de Vos W.M. Early life colonization of the human gut: Microbes matter everywhere. Curr. Opin. Microbiol. 2018;44:70–78. doi: 10.1016/j.mib.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo-Cantabrana C., Delgado S., Ruiz L., Ruas-Madiedo P., Sánchez B., Margolles A. Bifidobacteria and Their Health-Promoting Effects. Tuberc. Nontuberculous Mycobact. Infect. 2018:73–98. doi: 10.1128/9781555819705.ch3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krumbeck J.A., Maldonado-Gomez M.X., Martínez I., Frese S.A., Burkey T.E., Rasineni K., Ramer-Tait A.E., Harris E.N., Hutkins R.W., Walter J. In Vivo Selection To Identify Bacterial Strains with Enhanced Ecological Performance in Synbiotic Applications. Appl. Environ. Microbiol. 2015;81:2455–2465. doi: 10.1128/AEM.03903-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G., Zhang C., Wu H., Wang R., Shen J., Wang L., Zhao Y., Pang X., Zhang X., Zhao L., et al. Genomic Microdiversity of Bifidobacterium pseudocatenulatum Underlying Differential Strain-Level Responses to Dietary Carbohydrate Intervention. mBio. 2017;8:e02348-16. doi: 10.1128/mBio.02348-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arboleya S., Watkins C., Stanton C., Ross R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevers D., Kugathasan S., Denson L.A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S.J., Yassour M., et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schloissnig S., Arumugam M., Sunagawa S., Mitreva M., Tap J., Zhu A., Waller A., Mende D.R., Kultima J.R., Martin J., et al. Genomic variation landscape of the human gut microbiome. Nat. Cell Biol. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odamaki T., Bottacini F., Kato K., Mitsuyama E., Yoshida K., Horigome A., Xiao J.-Z., Van Sinderen D. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-017-18391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faith J.J., Guruge J.L., Charbonneau M., Subramanian S., Seedorf H., Goodman A.L., Clemente J.C., Knight R., Heath A.C., Leibel R.L., et al. The Long-Term Stability of the Human Gut Microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter J., Maldonado-Gómez M.X., Martínez I. To engraft or not to engraft: An ecological framework for gut microbiome modulation with live microbes. Curr. Opin. Biotechnol. 2018;49:129–139. doi: 10.1016/j.copbio.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao Y., Zhao J., Zhang H., Zhai Q., Chen W. Mining Lactobacillus and Bifidobacterium for organisms with long-term gut colonization potential. Clin. Nutr. 2020;39:1315–1323. doi: 10.1016/j.clnu.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Panigrahi P., Parida S., Pradhan L., Mohapatra S.S., Misra P.R., Johnson J.A., Chaudhry R., Taylor S., Hansen N.I., Gewolb I.H. Long-term Colonization of a Lactobacillus plantarum Synbiotic Preparation in the Neonatal Gut. J. Pediatr. Gastroenterol. Nutr. 2008;47:45–53. doi: 10.1097/MPG.0b013e31815a5f2c. [DOI] [PubMed] [Google Scholar]
- 15.Spanhaak S., Havenaar R., Schaafsma G. The effect of consumption of milk fermented by Lactobacillus casei strain Shirota on the intestinal microflora and immune parameters in humans. Eur. J. Clin. Nutr. 1998;52:899–907. doi: 10.1038/sj.ejcn.1600663. [DOI] [PubMed] [Google Scholar]
- 16.Toscano M., De Grandi R., Miniello V.L., Mattina R., Drago L. Ability of Lactobacillus kefiri LKF01 (DSM32079) to colonize the intestinal environment and modify the gut microbiota composition of healthy individuals. Dig. Liver Dis. 2017;49:261–267. doi: 10.1016/j.dld.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen C.N., Nielsen V.R., Hayford A.E., Møller P.L., Michaelsen K.F., Pærregaard A., Sandström B., Tvede M., Jakobsen M. Screening of Probiotic Activities of Forty-Seven Strains of Lactobacillus spp. by In Vitro Techniques and Evaluation of the Colonization Ability of Five Selected Strains in Humans. Appl. Environ. Microbiol. 1999;65:4949–4956. doi: 10.1128/AEM.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara S., Seto Y., Kimura A., Hashiba H. Intestinal transit of an orally administered streptomycin-rifampicin-resistant variant ofBifidobacterium longumSBT2928: Its long-term survival and effect on the intestinal microflora and metabolism. J. Appl. Microbiol. 2001;90:43–52. doi: 10.1046/j.1365-2672.2001.01205.x. [DOI] [PubMed] [Google Scholar]
- 19.Frese S.A., Hutton A.A., Contreras L.N., Shaw C.A., Palumbo M., Casaburi G., Xu G., Davis J.C.C., Lebrilla C.B., Henrick B.M., et al. Persistence of Supplemented Bifidobacterium longum subsp. infantis EVC001 in Breastfed Infants. mSphere. 2017;2:e00501-17. doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxelin M., Pessi T., Salminen S. Fecal recovery following oral administration of Lactobacillus Strain GG (ATCC 53103) in gelatine capsules to healthy volunteers. Int. J. Food Microbiol. 1995;25:199–203. doi: 10.1016/0168-1605(94)00091-J. [DOI] [PubMed] [Google Scholar]
- 21.Yuki N., Watanabe K., Mike A., Tagami Y., Tanaka R., Ohwaki M., Morotomi M. Survival of a probiotic, Lactobacillus casei strain Shirota, in the gastrointestinal tract: Selective isolation from faeces and identification using monoclonal antibodies. Int. J. Food Microbiol. 1999;48:51–57. doi: 10.1016/S0168-1605(99)00029-X. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y.K., Ho P.S., Low C.S., Arvilommi H., Salminen S. Permanent Colonization by Lactobacillus casei Is Hindered by the Low Rate of Cell Division in Mouse Gut. Appl. Environ. Microbiol. 2004;70:670–674. doi: 10.1128/AEM.70.2.670-674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denou E., Pridmore R.D., Berger B., Panoff J.-M., Arigoni F., Brüssow H. Identification of Genes Associated with the Long-Gut-Persistence Phenotype of the Probiotic Lactobacillus johnsonii Strain NCC533 Using a Combination of Genomics and Transcriptome Analysis. J. Bacteriol. 2008;190:3161–3168. doi: 10.1128/JB.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valeur N., Engel P., Carbajal N., Connolly E., Ladefoged K. Colonization and Immunomodulation by Lactobacillus reuteri ATCC 55730 in the Human Gastrointestinal Tract. Appl. Environ. Microbiol. 2004;70:1176–1181. doi: 10.1128/AEM.70.2.1176-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobin J.M., Garland S.M., Jacobs S.E., Pirotta M., Tabrizi S.N. Rapid assay to assess colonization patterns following in-vivo probiotic ingestion. BMC Res. Notes. 2013;6:252. doi: 10.1186/1756-0500-6-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massi M., Vitali B., Federici F., Matteuzzi D., Brigidi P. Identification method based on PCR combined with automated ribotyping for tracking probiotic Lactobacillus strains colonizing the human gut and vagina. J. Appl. Microbiol. 2004;96:777–786. doi: 10.1111/j.1365-2672.2004.02228.x. [DOI] [PubMed] [Google Scholar]
- 27.Kankainen M., Paulin L., Tynkkynen S., von Ossowski I., Reunanen J., Partanen P., Satokari R., Vesterlund S., Hendrickx A.P.A., Lebeer S., et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toshimitsu T., Nakamura M., Ikegami S., Terahara M., Itou H. Strain-Specific Identification ofBifidobacterium bifidumOLB6378 by PCR. Biosci. Biotechnol. Biochem. 2013;77:572–576. doi: 10.1271/bbb.120803. [DOI] [PubMed] [Google Scholar]
- 29.Treven P., Turkova K., Trmčić A., Obermajer T., Rogelj I., Matijašić B.B. Detection and quantification of probiotic strain Lactobacillus gasseri K7 in faecal samples by targeting bacteriocin genes. Folia Microbiol. 2013;58:623–630. doi: 10.1007/s12223-013-0252-8. [DOI] [PubMed] [Google Scholar]
- 30.Fujimoto J., Tanigawa K., Kudo Y., Makino H., Watanabe K. Identification and quantification of viable Bifidobacterium breve strain Yakult in human faeces by using strain-specific primers and propidium monoazide. J. Appl. Microbiol. 2010;110:209–217. doi: 10.1111/j.1365-2672.2010.04873.x. [DOI] [PubMed] [Google Scholar]
- 31.Sattler V.A., Mohnl M., Klose V. Development of a Strain-Specific Real-Time PCR Assay for Enumeration of a Probiotic Lactobacillus reuteri in Chicken Feed and Intestine. PLoS ONE. 2014;9:e90208. doi: 10.1371/journal.pone.0090208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laing C., Buchanan C., Taboada E.N., Zhang Y., Kropinski A., Villegas A., Thomas J.E., Gannon V.P.J. Pan-genome sequence analysis using Panseq: An online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinform. 2010;11:461. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brittnacher M.J., Fong C., Hayden H.S., Jacobs M.A., Radey M., Rohmer L. PGAT: A multistrain analysis resource for microbial genomes. Bioinformatics. 2011;27:2429–2430. doi: 10.1093/bioinformatics/btr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y., Wu J., Yang J., Sun S., Xiao J., Yu J. PGAP: Pan-genomes analysis pipeline. Bioinformatics. 2011;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. Erratum: SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience. 2015;4:30. doi: 10.1186/s13742-015-0069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui Y., Yang X., Didelot X., Guo C., Li D., Yan Y., Zhang Y., Yuan Y., Yang H., Wang J., et al. Epidemic Clones, Oceanic Gene Pools, and Eco-LD in the Free Living Marine Pathogen Vibrio parahaemolyticus. Mol. Biol. Evol. 2015;32:1396–1410. doi: 10.1093/molbev/msv009. [DOI] [PubMed] [Google Scholar]
- 38.Delcher A.L., Salzberg S.L., Phillippy A.M. Using MUMmer to Identify Similar Regions in Large Sequence Sets. Curr. Protoc. Bioinform. 2003;10:10.3.1–10.3.18. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 39.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 40.Madden T. The NCBI Handbook. 2nd ed. National Center for Biotechnology Information (US); Bethesda, MD, USA: 2013. [Google Scholar]
- 41.Daranas N., Bonaterra A., Francés J., Cabrefiga J., Montesinos E., Badosa E. Monitoring Viable Cells of the Biological Control AgentLactobacillus plantarumPM411 in Aerial Plant Surfaces by Means of a Strain-Specific Viability Quantitative PCR Method. Appl. Environ. Microbiol. 2018;84:84. doi: 10.1128/AEM.00107-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimoto J., Watanabe K. Quantitative Detection of Viable Bifidobacterium bifidum BF-1 Cells in Human Feces by Using Propidium Monoazide and Strain-Specific Primers. Appl. Environ. Microbiol. 2013;79:2182–2188. doi: 10.1128/AEM.03294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karjalainen H., Ahlroos T., Myllyluoma E., Tynkkynen S. Real-time PCR assays for strain-specific quantification of probiotic strains in human faecal samples. Int. Dairy J. 2012;27:58–64. doi: 10.1016/j.idairyj.2012.07.006. [DOI] [Google Scholar]
- 44.Zhai Q., Shen X., Cen S., Zhang C., Tian F., Zhao J., Zhang H., Xue Y., Chen W. Screening of Lactobacillus salivarius strains from the feces of Chinese populations and the evaluation of their effects against intestinal inflammation in mice. Food Funct. 2020;11:221–235. doi: 10.1039/C9FO02116G. [DOI] [PubMed] [Google Scholar]
- 45.Harris H.M.B., Bourin M.J.B., Claesson M.J., O’Toole P.W. Phylogenomics and comparative genomics of Lactobacillus salivarius, a mammalian gut commensal. Microb. Genom. 2017;3:e000115. doi: 10.1099/mgen.0.000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuyts S., Wittouck S., De Boeck I., Allonsius C.N., Pasolli E., Segata N., Lebeer S. Large-Scale Phylogenomics of the Lactobacillus casei Group Highlights Taxonomic Inconsistencies and Reveals Novel Clade-Associated Features. mSystems. 2017;2:e00061-17. doi: 10.1128/mSystems.00061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maldonado-Gómez M.X., Martínez I., Bottacini F., O’Callaghan A., Ventura M., van Sinderen D., Hillmann B., Vangay P., Knights D., Hutkins R.W., et al. Stable Engraftment of Bifidobacterium longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Sato N., Seo G., Benno Y. Development of Strain-Specific PCR Primers for Quantitative Detection of Bacillus mesentericus Strain TO-A in Human Feces. Biol. Pharm. Bull. 2014;37:123–129. doi: 10.1248/bpb.b13-00641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data in this study are available from the authors upon request.