Abstract
Glioblastoma (GBM) is a primary malignant tumor characterized by high infiltration and angiogenesis in the brain parenchyma. Glioma stem cells (GSCs), a heterogeneous GBM cell type with the potential for self-renewal and differentiation to tumor cells, are responsible for the high malignancy of GBM. The purpose of the present study was to investigate the roles of significantly differentially expressed genes between GSCs and GBM cells in GBM progression. The gene profiles GSE74304 and GSE124145, containing 10 GSC samples and 12 GBM samples in total, were obtained from the Gene Expression Omnibus (GEO) database. The overlapping differentially expressed genes were identified with GEO2R tools and Venn software online. Subsequently, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis was performed on the 41 upregulated and 142 downregulated differentially expressed genes in GSCs compared with in GBM cells via the DAVID website. Protein-protein interaction and module analyses in Cytoscape with the STRING database revealed 21 hub genes that were downregulated in GSCs compared with in GBM cells. Survival analysis conducted via the GEPIA2 website revealed that low expression levels of the hub genes prolyl 4-hydroxylase subunit α2 (P4HA2), TGF-β induced, integrin subunit α3 and thrombospondin 1 were associated with significantly prolonged survival time in patients with GBM. Further experiments were performed focusing on P4HA2. Reverse transcription-quantitative PCR was used to detect P4HA2 gene expression. In agreement with the bioinformatics analysis, P4HA2 expression was higher in U87 cells than in GSCs. Cell Counting Kit-8, EdU incorporation, cell cycle analysis, wound healing and Transwell assays demonstrated that the cell proliferation and migration increased after P4HA2 overexpression and decreased after P4HA2-knockdown. In conclusion, the present study demonstrated that low P4HA2 expression in GSCs promoted GBM cell proliferation and migration, suggesting that P4HA2 may act as a switch in the transition from GSCs to GBM cells.
Keywords: bioinformatics analysis, glioblastoma, glioma stem cells, microarray, prolyl 4-hydroxylase subunit α2
Introduction
Glioma, the most common primary malignant brain tumor in the central nervous system, is characterized by a high degree of angiogenesis (1). Among gliomas, glioblastoma (GBM) is the most malignant type, with a median survival time of only 12–15 months (2,3). Considerable heterogeneity exists among tumor cells, and the cell types, gene expression patterns and cell proliferation potential can vary (4). One type of tumor cells, known as cancer stem cells, have the characteristics of stem cells, which can proliferate indefinitely, produce tumor cells and drive tumor progression, as well as having important effects on tumor recurrence, metastasis and chemoradiotherapy resistance (5). These cells in GBM are called glioma stem cells (GSCs). GSCs have the potential for self-renewal and multidirectional differentiation, and also promote tumor angiogenesis, trigger immune escape and drive the occurrence and development of GBM (6). These cells are at the root of the continuous cell proliferation, high invasion, infiltration, metastasis and treatment resistance observed in GBM (7). Therefore, therapies targeting GSCs have shown great potential and have been at the forefront of GBM research. At present, although many GSC-associated markers, such as CD133 and A2B5, and the signaling pathways regulating the stemness of GSCs have been revealed (8), the mechanism underlying the continuous conversion of GSCs to GBM cells remains unclear.
The present study selected two gene microarrays from the Gene Expression Omnibus (GEO) database to detect the mRNA expression levels in GSCs and GBM cells and to analyze the overlapping differentially expressed genes (DEGs) between the two types of cells. Subsequently, the genes that may play an essential role in the malignant progression of human GBM were obtained through bioinformatics analysis. Next, several experiments were performed to verify the effects of the selected gene on the proliferation and migration of GBM cells. The current findings may provide new insights for studying the association between GSCs and GBM cells.
Materials and methods
Microarray data information
To analyze the differences between GSCs and GBM cells, gene microarray data were obtained from GSE74304 (9) and GSE124145 (10) in the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/). Both gene profiles were from the GPL570 platform [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array], a typical array for detecting mRNA expression levels. GSE74304 contained data from two patient-derived GSC lines and corresponding GBM cell lines, and GSE124145 contained data from two human GSC lines (X01 and X03), one human GBM tissue and the GBM U251 cell line. Each sample contained two or three replicates. Thus, a total of 10 GSC samples and 12 GBM samples were available for experiments. Profiles filter criteria were as follows: GPL570 platform, publication date between 2015 and 2019, no previous drug treatment and ≥500 DEGs.
Acquisition of DEGs
DEGs were identified using the GEO2R online tools (https://www.ncbi.nlm.nih.gov/geo/geo2r) between two GSC lines and two GBM cell lines in GSE74304, and two GSC lines and GBM tissue or cell lines in GSE124145 with an adjusted P<0.05 and ǀlog (fold-change) FCǀ >2. Overlapping genes were obtained by uploading the DEGs from the two datasets to Venn software online (http://bioinformatics.psb.ugent.be/webtools/Venn/). The DEGs with log FC >0 were considered as upregulated genes and those with log FC <0 were considered as downregulated genes in GSCs compared with in GBM cells.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
GO and KEGG pathway analysis was performed via the DAVID website (https://david.ncifcrf.gov/) to identify significantly enriched molecular functions, cellular components, biological processes and biological pathways [P<0.05; enrichment score = -lg (P-value)]
Protein-protein interaction (PPI) and module analysis
PPI analysis among overlapping DEGs was performed using the STRING database (https://string-db.org/), and network diagrams were constructed via Cytoscape software (v3.6.1; http://www.cytoscape.org/) (maximum number of interactors =0; confidence score ≥0.4). The hub genes were obtained by module analysis with the Molecular Complex Detection (MCODE) plugin in Cytoscape according to the following criteria: Node score cut-off, 0.2; degree cut-off, 2; k-core, 2; MCODE scores, ≥5; and maximum depth, 100.
Survival analysis
To validate the hub genes through survival analysis, the GEPIA2 website (http://gepia2.cancer-pku.cn/#index) was used, an online database for analysis of RNA expression levels, based on the Genotype-Tissue Expression Projects and The Cancer Genome Atlas (11). Kaplan-Meier survival curves were analyzed using the log-rank test. Log-rank P-value, hazard ratio (HR) and HR P-value are displayed in the figures.
Cell culture and transient transfection
The U87 cell line (glioblastoma of unknown origin) was purchased from the American Type Culture Collection (ATCC® HTB14™). U87 cells were maintained in DMEM/F12 (Corning, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and cultured at 37°C with 5% CO2. GSCs were maintained in stem cell medium as previously described (9,12,13). After being cultured at 37°C in a humidified 5% CO2 incubator for 12 h, U87 cells were infected with negative control lentivirus (GV341; empty vector) or prolyl 4-hydroxylase subunit α2 (P4HA2) overexpression lentivirus at a multiplicity of infection of 5 for 12–16 h. The virus was purchased from Shanghai GeneChem Co., Ltd. P4HA2 siRNA-1 (sense, 5′-GAACCAAGUACCAGGCAAUTT-3′ and antisense, 5′-AUUGCCUGGUACUUGGUUCTT-3′), siRNA-2 (sense, 5′-GCAGCAUAUCACAGGGUUATT-3′ and antisense, 5′-UAACCCUGUGAUAUGCUGCTT-3′), siRNA-3 (sense, 5′-GCAAGUGGGUCUCCAAUAATT-3′ and antisense, 5′-UUAUUGGAGACCCACUUGCTT-3′) and scrambled negative control (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) were synthesized by OBiO Technology (Shanghai) Corp., Ltd. After being cultured at 37°C in a humidified 5% CO2 incubator for 12 h, U87 cells were transfected with siRNAs (50 nM) using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol for 12 h at 37°C. Subsequent experiments were performed 48 h after lentivirus infection or 24 h after siRNA transfection.
RNA extraction and reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.), cDNA synthesis was performed with a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol and qPCR was performed with Universal SYBR Green Master Mix (Roche Diagnostics) according to the manufacturer's protocol using the following PCR conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 40 sec, followed by a final elongation step of 10 min at 72°C. P4HA2 expression was normalized to GAPDH expression calculated using the 2-ΔΔCq method (14). The primer sequences were as follows: P4HA2 forward, 5′-GCCAAAGCCCTGATGAGACT-3′ and reverse, 5′-GCTCCATCCACAACACCGTA-3′; and GAPDH forward, 5′-TCATCATCTCTGCCCCCTCT-3′ and reverse, 5′-GTGATGGCATGGACTGTGGT-3′.
EdU incorporation
For EdU incorporation, a Cell-Light EdU Apollo567 In Vitro kit (Guangzhou RiboBio Co., Ltd.) was used for EdU and Hoechst staining according to the manufacturer's instructions. EdU-positive cells were analyzed under a fluorescence microscope (Axio Scope. A1; Zeiss AG) using ×20 magnification.
Cell cycle analysis
U87 cells were harvested 48 h after infection or 24 h after transfection and then fixed with ice-cold 75% ethanol at −20°C for 24 h. After fixation, cells were stained with PI/RNase Staining Buffer (BD Biosciences) and incubated for 30 min at room temperature. Cell cycle analysis was analyzed using the BD FACSCalibur system (BD Biosciences) with ModFit LT v3.3.11 software (Verity Software House, Inc.).
Cell Counting Kit-8 (CCK8) assay
A total of 1,000 U87 cells were plated in 96-well plates and cultured at 37°C with virus infection or siRNA transfection. After culture for 0, 1, 2, 3 and 4 days, 10 µl CCK8 reagent (Dalian Meilun Biology Technology Co., Ltd.) was added into each well and incubated at 37°C for 2 h. The optical density (OD) was examined at a wavelength of 450 nm (OD 450 nm).
Wound healing assay
Infected or transfected cells were seeded in 6-well plates and incubated for 24 h until reaching 90% confluence. The monolayer was scratched with a 1-ml pipette tip and washed with PBS to remove cell debris. Cells were then cultured in DMEM/F12 supplemented with 2% FBS. The wound closure was measured using ImageJ software (v1.52a; National Institutes of Health) in photographs taken using a light microscope (Nikon Corporation) at ×10 magnification at the time of wounding (0 h), and then 24 and 48 h after wounding.
Transwell assay
For migration assays, 3×104 infected or transfected cells in DMEM/F12 were plated in the upper chambers of 24-well Transwell plates (pore size, 8 µm; Corning, Inc.), and the lower chambers were filled with 600 µl DMEM/F12 with 10% FBS. After being cultured at 37°C for 24 h, the non-migrating cells on the upper surface were removed with a cotton swab, and the migrated cells were fixed with 4% formaldehyde at room temperature for 20 min and then stained with 0.2% crystal violet at room temperature for 45 min. The pictures were taken using the AMAFD1000-EVOS™ FL Auto Imaging System (Thermo Fisher Scientific, Inc.) at ×20 magnification.
Statistical analysis
The data were presented as the mean ± SEM. Statistical analysis of the data was performed using GraphPad Prism 8.0 (GraphPad Software, Inc.) on at least three independent samples for each experiment. Two-tailed unpaired Student's t-test was used for comparison between two groups, and two-way ANOVA followed by Sidak's post-hoc test was used for comparing multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs between GSCs and GBM cells
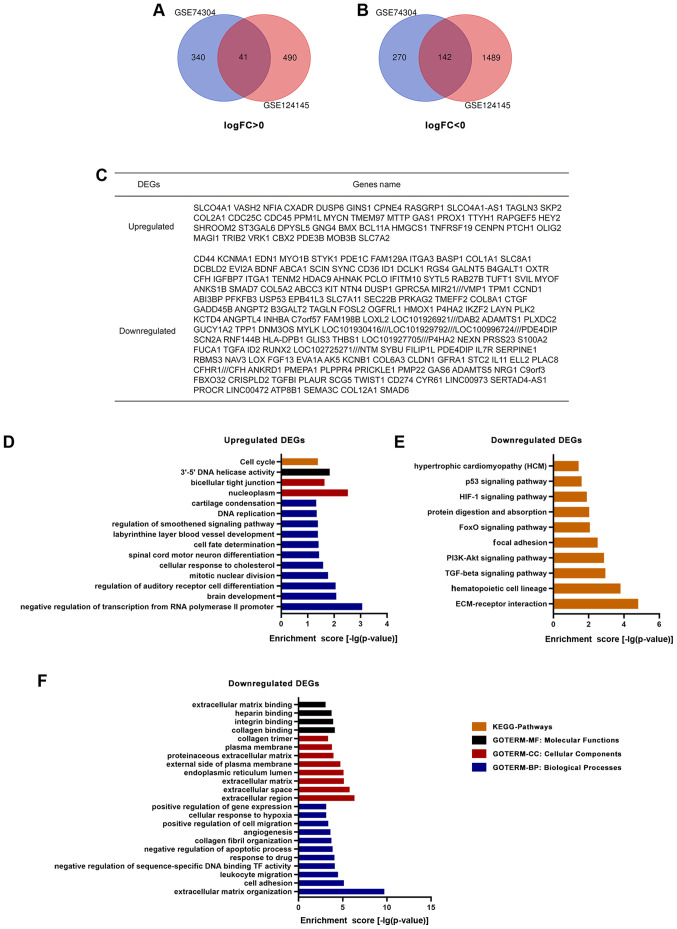
A total of 10 GSC samples and 12 GBM samples were used in the present study. Through GEO2R online tools, 793 DEGs were extracted from GSE74304, with 381 upregulated genes and 412 downregulated genes, and 2,162 DEGs from GSE124145, with 531 upregulated genes and 1,631 downregulated genes in GSCs compared with in GBM cells. Overlapping genes were obtained with Venn software online. A total of 183 DEGs were screened, including 41 upregulated genes (log FC >0) and 142 downregulated genes (log FC <0) in GSCs compared with in GBM cells (Fig. 1A-C).
Figure 1.
Identification and gene enrichment analysis of the DEGs in two datasets from the Gene Expression Omnibus database. Venn diagrams revealed (A) 41 upregulated and (B) 142 downregulated DEGs in glioma stem cells compared with in glioblastoma cells. (C) There were 183 overlapping DEGs between GSE74304 and GSE124145. (D) KEGG pathway and GO analysis of upregulated DEGs. (E) KEGG pathway and (F) GO analysis of downregulated DEGs. DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FC, fold-change.
GO and KEGG pathway analysis
A total of 41 upregulated and 142 downregulated DEGs were analyzed via the DAVID website with the screening criteria of P<0.05 for upregulated genes, and both P<0.05 and enrichment score >3 for downregulated genes. The results of GO analysis revealed that: i) For molecular functions, upregulated DEGs were particularly enriched in the term ‘3′-5′ DNA helicase activity’, and downregulated DEGs were particularly enriched in the terms ‘collagen binding’, ‘integrin binding’, ‘heparin binding’ and ‘extracellular matrix binding’; ii) for cellular components, upregulated DEGs were enriched in the terms ‘nucleoplasm’ and ‘bicellular tight junction’, whereas downregulated DEGs were enriched in the terms ‘extracellular region’, ‘extracellular space’, ‘extracellular matrix’, ‘endoplasmic reticulum lumen’, ‘external side of plasma membrane’, ‘proteinaceous extracellular matrix’, ‘plasma membrane’ and ‘collagen trimer’; iii) for biological processes, upregulated DEGs were significantly enriched in the terms ‘negative regulation of transcription from RNA polymerase II promoter’, ‘brain development’, ‘regulation of auditory receptor cell differentiation’, ‘mitotic nuclear division’, ‘cellular response to cholesterol’, ‘spinal cord motor neuron differentiation’, ‘cell fate determination’, ‘labyrinthine layer blood vessel development’, ‘regulation of smoothened signaling pathway’, ‘DNA replication and cartilage condensation’, and downregulated DEGs were enriched in the terms ‘extracellular matrix organization’, ‘cell adhesion’, ‘leukocyte migration’, ‘negative regulation of sequence-specific DNA binding TF activity’, ‘response to drug’, ‘negative regulation of apoptotic process’, ‘collagen fibril organization’, ‘angiogenesis’, ‘positive regulation of cell migration’, ‘cellular response to hypoxia’ and ‘positive regulation of gene expression’ (Fig. 1D and F). KEGG analysis results demonstrated that upregulated DEGs were particularly enriched in the term ‘cell cycle’, whereas downregulated DEGs were particularly enriched in the terms ‘ECM-receptor interaction’, ‘hematopoietic cell lineage’, ‘TGF-β signaling pathway’, ‘PI3K-Akt signaling pathway’, ‘focal adhesion’, ‘FoxO signaling pathway’, ‘protein digestion and absorption’, ‘HIF-1 signaling pathway’, ‘p53 signaling pathway’ and ‘hypertrophic cardiomyopathy’ (Fig. 1D and E).
PPI and survival analysis
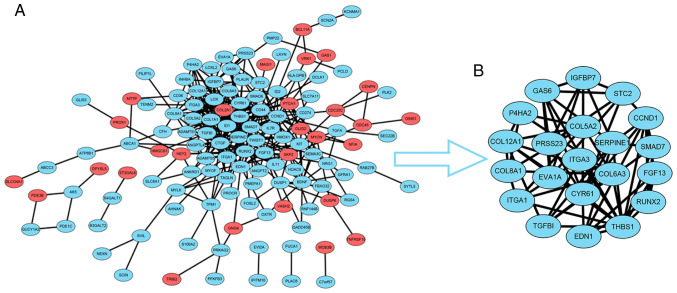
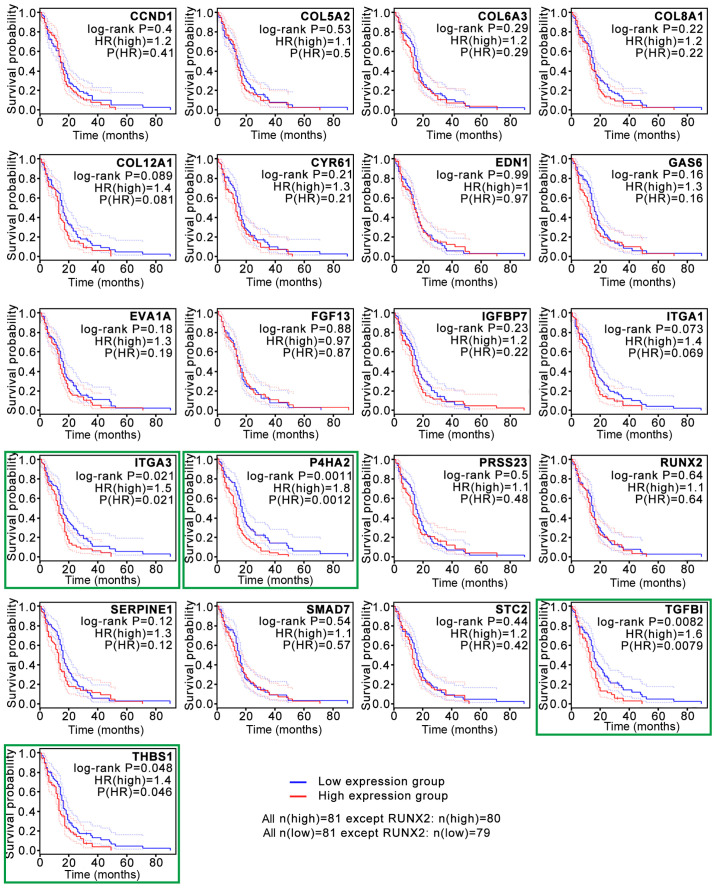
To construct the PPI network, the data for the 183 overlapping DEGs obtained from the STRING database were imported into Cytoscape software. The interaction network contained 41 upregulated genes and 142 downregulated genes (Fig. 2A). For further analysis of the key genes among them, the Cytoscape MCODE plugin was applied, and 21 hub genes were obtained, all of which were downregulated DEGs (Fig. 2B). Subsequently, the association between the expression levels of these genes and survival time in patients with GBM was validated with GEPIA2 analysis (Fig. 3). The survival curves revealed that low expression levels of four hub genes, P4HA2 [log-rank P=0.0011; P(HR)=0.0012], TGF-β induced [TGFBI; log-rank P=0.0082; P(HR)=0.0079], integrin subunit α3 [ITGA3; log-rank P=0.021; P(HR)=0.021] and thrombospondin 1 [THBS1; log-rank P=0.048; P(HR)=0.046], were associated with significantly prolonged survival time in patients with GBM (Fig. 3). The present study then focused on P4HA2 since it had the smallest P-value.
Figure 2.
PPI network analysis of the overlapping DEGs. (A) PPI network was obtained from the STRING database. Red indicates upregulated DEGs, while blue indicates downregulated DEGs. (B) Module analysis performed by the Cytoscape MCODE plugin revealed 21 hub genes all downregulated in glioma stem cells. PPI, protein-protein interaction; DEGs, differentially expressed genes.
Figure 3.
Survival analysis based on the overlapping differentially expressed genes. Survival analysis conducted on the GEPIA2 website revealed that low expression levels of 4 hub genes, P4HA2, TGFBI, ITGA3 and THBS1 (green boxes), were associated with significantly prolonged survival time in patients with glioblastoma. P4HA2, prolyl 4-hydroxylase subunit α2; TGFBI, TGF-β induced; ITGA3, integrin subunit α3; THBS1, thrombospondin 1; HR, hazard ratio.
P4HA2 promotes the proliferation of GBM cells
GSCs were generated from the U87 cell line, a human malignant glioblastoma cell line, as previously described (9,12,13). RT-qPCR was used to detect P4HA2 expression. In agreement with the bioinformatics analysis results, P4HA2 expression was significantly higher in U87 cells than in GSCs (Fig. 4A). Subsequently, U87 cells were infected with empty vector (Ctrl) or P4HA2 vector (P4HA2) expression lentiviruses. The RT-qPCR results revealed that P4HA2 mRNA expression was significantly increased after P4HA2 overexpression (Fig. 4B). To explore the effects on GBM cell proliferation, CCK8, EdU incorporation and cell cycle analyses were performed. As shown in Fig. 4C, the overexpression of P4HA2, compared with the control, induced significant increases in cell proliferation from day 1. The EdU-positive cells also significantly increased after P4HA2 viral transfection (Fig. 4D and E). Cell cycle analysis by flow cytometry aims to calculate the proportion of cells at different stages of mitosis based on the DNA content, so as to evaluate the proliferative ability of cells. Therefore, this method was used to further confirm the effect of P4HA2 on U87 cell proliferation. According to the amount of DNA, the whole cell cycle can be divided into G0/G1, S and G2/M phases (15). DNA in the G0/G1 phase is diploid, while it is tetraploid in the G2/M phase and in between in S phase (16,17). The ratio of cells in S phase and G2/M phase is usually used as an index to assess the cell proliferation state (15,17). As shown in Fig. 4F-H, P4HA2 overexpression promoted the entry of cells into S phase and significantly increased the proportion of cells in S+G2 phase, thus explaining the robust proliferation of U87 cells. Overall, the present results demonstrated that P4HA2 increased the proliferation of GBM cells.
Figure 4.
Overexpression of P4HA2 promotes the proliferation of glioblastoma cells. (A) Reverse transcription-quantitative PCR assay revealed that the mRNA expression levels of P4HA2 were lower in GSCs than in U87 cells. (B) Infection with the P4HA2 overexpression lentivirus in U87 cells significantly increased P4HA2 mRNA expression. (C) CCK8 assay performed on day 0, 1, 2, 3 and 4 after infection revealed that P4HA2 promoted U87 cell proliferation. (D and E) EdU-positive cells increased after P4HA2 overexpression. Scale bar, 50 µm. (F and G) Cell cycle analysis indicated that the percentage of P4HA2-overexpressing cells at S+G2 phase was increased compared with in the control group. (H) Statistical analysis of flow cytometry plots. Data were presented as the mean ± SEM. **P<0.01 and ***P<0.001 vs. U87 or Ctrl. Ctrl, control; OD, optical density; CCK8, Cell Counting Kit-8; GSCs, glioma stem cells; P4HA2, prolyl 4-hydroxylase subunit α2.
P4HA2 promotes the migration of GBM cells
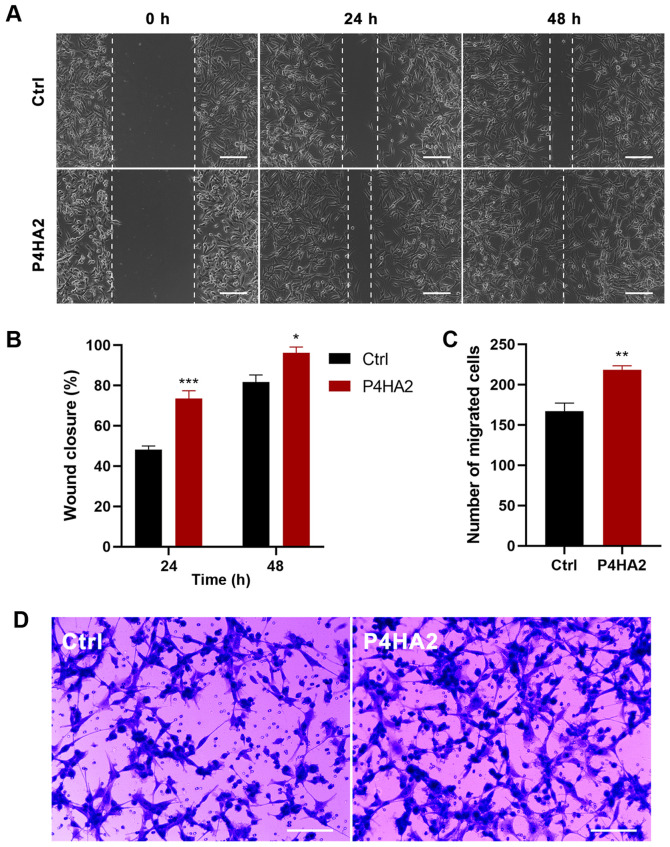
Since metastasis is the main malignant characteristic of GBM, wound healing and Transwell assays were conducted to investigate the effect of P4HA2 on U87 cell migration. In the wound healing assays, the scratch width narrowed significantly in the P4HA2 group from 24 h after transfection, thereby indicating that cells with P4HA2 overexpression exhibited increased migratory ability (Fig. 5A and B). In agreement with this finding, Transwell assays also demonstrated that the overexpression of P4HA2 promoted the migration of U87 cells compared with cells in the control group (Fig. 5C and D).
Figure 5.
Overexpression of P4HA2 promotes the migration of glioblastoma cells. (A and B) Wound healing assay performed at 24 and 48 h after infection showed that the percentage of the wound closure in the P4HA2 overexpression group was higher than in the control group. Scale bar, 200 µm. (C and D) Transwell assay demonstrated that the number of migrated cells increased after P4HA2 overexpression. Scale bar, 200 µm. Data were presented as the mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001 vs. Ctrl. Ctrl, control; P4HA2, prolyl 4-hydroxylase subunit α2.
Knockdown of P4HA2 inhibits GBM cell proliferation and migration
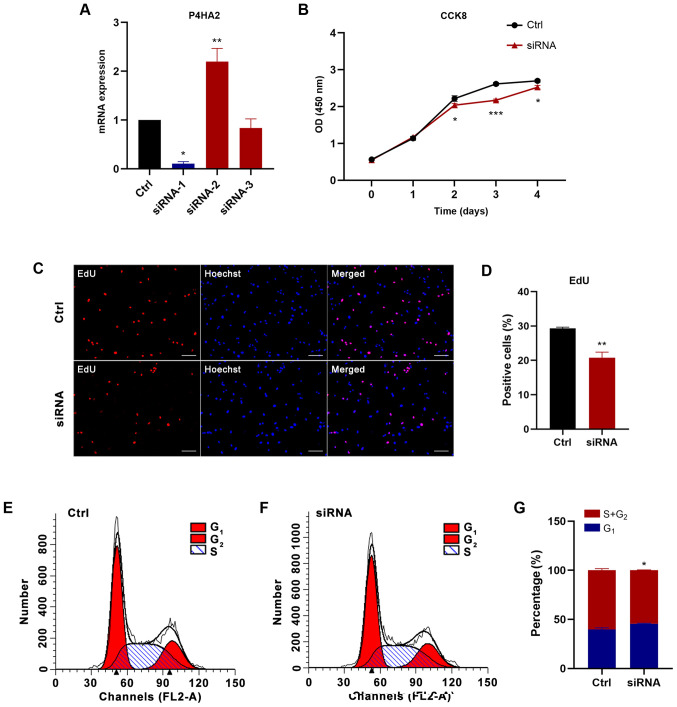
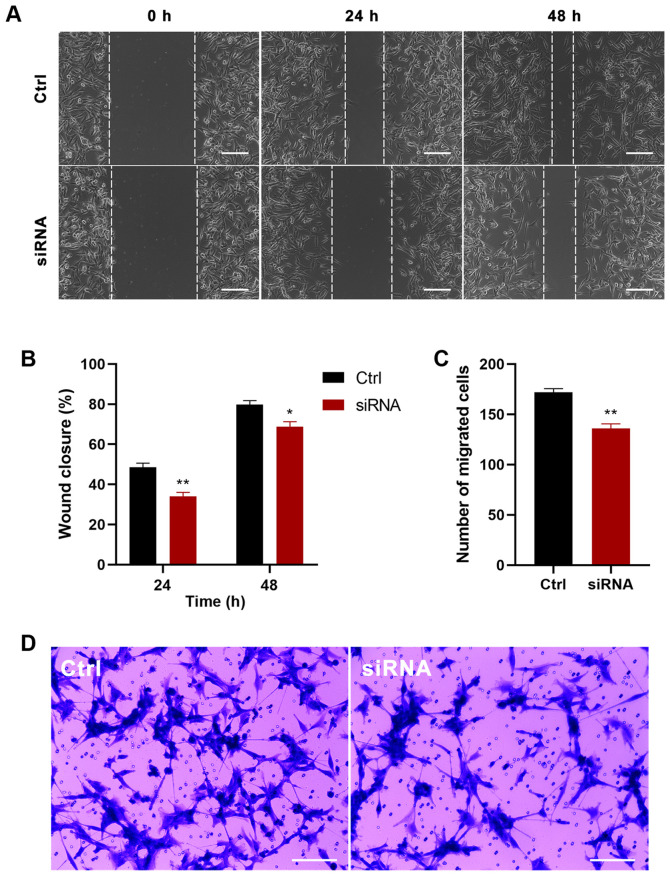
To further determine the function of P4HA2 in GBM, P4HA2 was knocked down in U87 cells with siRNA transfection. First, three siRNAs were designed targeting P4HA2; siRNA-1 (hereafter called siRNA) was selected for further experiments, since it exhibited the highest knockdown efficiency, as verified by RT-qPCR (Fig. 6A). To investigate the influence of P4HA2-knockdown on GBM, CCK8, EdU incorporation and cell cycle analyses were performed to examine cell proliferation, and wound healing and Transwell assays were performed to examine cell migration. As shown in Fig. 6B, P4HA2-knockdown induced significant decreases in cell proliferation from day 2, compared with the control group. The EdU-positive cells also significantly decreased after siRNA transfection (Fig. 6C and D). The results of cell cycle analysis by flow cytometry indicated that P4HA2-knockdown promoted G1 phase arrest and delayed entry into S phase (Fig. 6E-G). Moreover, both the wound healing (Fig. 7A and B) and Transwell assays (Fig. 7C and D) revealed that P4HA2-knockdown significantly suppressed the migratory ability of U87 cells. Thus, P2HA2 overexpression promoted the proliferation and migration of GBM cells, while P4HA2-knockdown inhibited these processes.
Figure 6.
P4HA2-knockdown inhibits the proliferation of glioblastoma cells. (A) Reverse transcription-quantitative PCR assay revealed that siRNA-1 exhibited the highest P4HA2-knockdown efficiency. (B) CCK8 assay performed on day 0, 1, 2, 3 and 4 after transfection showed that P4HA2-knockdown suppressed U87 cell proliferation. (C and D) EdU-positive cells were decreased after siRNA transfection. Scale bar, 50 µm. (E and F) Cell cycle analysis indicated that the percentage of P4HA2-knockdown cells at S+G2 phase was decreased compared with cells in the control group. (G) Statistical analysis of flow cytometry plots. Data were presented as the mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001 vs. Ctrl. Ctrl, control; OD, optical density; CCK8, Cell Counting Kit-8; P4HA2, prolyl 4-hydroxylase subunit α2; siRNA, small interfering RNA.
Figure 7.
P4HA2-knockdown inhibits the migration of glioblastoma cells. (A and B) Wound healing assay performed at 24 and 48 h after siRNA transfection showed that the percentage of the wound closure in the P4HA2-knockdown group was lower than in that in the control group. Scale bar, 200 µm. (C and D) Transwell assay demonstrated that the number of migrated cells decreased after P4HA2-knockdown. Scale bar, 200 µm. Data were presented as the mean ± SEM. *P<0.05 and **P<0.01 vs. Ctrl. Ctrl, control; P4HA2, prolyl 4-hydroxylasesubunit α2; siRNA, small interfering RNA.
Discussion
GBM, a primary malignant tumor of the central nervous system, is characterized by high infiltration and angiogenesis in the brain parenchyma (1). Its 5-year survival rate is <10%, and it has high recurrence and metastasis rates (18). The treatment of GBM remains a major challenge, owing to the poor efficacy of current therapeutics, including surgery, radiotherapy and drugs (19). Therefore, new therapeutic targets for GBM are urgently needed. Increasing studies have indicated that the main reason for the high malignancy of GBM involves GSCs, a heterogeneous cell type existing among GBM cells, which have the potential for self-renewal and differentiation into tumor cells (4–6). GSCs are involved in the immune escape of tumor cells, which is the most important reason underlying GBM recurrence and metastasis, as well as resistance to chemoradiotherapy (6,7). Therefore, exploring the GSC-specific surface markers and conversion mechanisms from GSCs to GBM cells has become a research hotspot in recent years.
To analyze the differences in gene expression levels between GSCs and GBM cells, mRNA microarray data were obtained from accession codes GSE74304 and GSE124145 from the GPL570 platform in the GEO database. A total of 183 DEGs were identified, of which 41 were upregulated and 142 were downregulated in GSCs compared with in GBM cells. PPI analysis of these genes revealed 21 key genes, denoted hub genes. Notably, these 21 hub genes were all downregulated in GSCs. GO and KEGG pathway analysis indicated that the downregulated genes were likely to be involved in numerous biological processes, such as construction of the extracellular matrix and collagen binding. Subsequent survival analysis of the 21 hub genes revealed that low expression levels of four hub genes, P4HA2, TGFBI, ITGA3 and THBS1, were significantly associated with longer survival time, with P4HA2 exhibiting the most significant differential expression.
The P4HA2 gene encodes prolyl 4-hydroxylase (P4H) subunit α2, one of several different types of α subunits of P4H, an enzyme composed of two identical α subunits and two β subunits (20,21). P4H catalyzes proline hydroxylation to 4-hydroxyproline, an important post-translational modification that frequently occurs in the Gly-3Hyp-4Hyp sequence of collagen, and is as a crucial factor in the structural stability of collagen (20,22). Collagen is an important component of the extracellular matrix, which can regulate cell proliferation, migration and invasion (23). P4HA2 is essential for the proper and stable three-dimensional folding of newly synthesized procollagen chains, and it is involved in several biological processes, such as collagen maturation, epithelial-mesenchymal transition, abnormal construction of the extracellular matrix in tumor progression and amyloid-β peptide deposition in Alzheimer's disease (24–28). P4HA2 is a risk gene for high myopia, owing to structural and quantitative alterations of collagen, which cause myopic axial elongation of the eyeball (29–32). Additionally, high P4HA2 expression is associated with the poor prognosis of some malignant tumors, such as hepatocellular carcinoma (27,33), gastric cancer (34), lung cancer (35), breast cancer (36–38), pancreatic cancer (39), cervical cancer (26,40), oral squamous cell carcinoma (41), melanoma (23) and B-cell lymphoma (42). A previous study has suggested that P53, a tumor suppressor gene, activates P4HA2 expression, thereby exerting anti-angiogenic effects and inhibiting tumor growth (43). By contrast, another study has indicated that although P53 can upregulate the mRNA expression levels of P4HA2 in GBM, it does not lead to an increase in the expression levels of P4HA2 protein or anti-angiogenic endostatin (44). Therefore, the role of P4HA2 in GBM is unclear. Given its effects on numerous diseases, P4HA2 may also play a vital role in the occurrence and development of GBM.
As aforementioned, GSCs promote the malignant progression of GBM (5,6). The present study originally sought to analyze the differences in gene expression levels between GSCs and GBM cells to identify genes with high expression in GSCs that significantly affected the prognosis of GBM for further research. However, through bioinformatics analysis, it was revealed that all the hub genes were downregulated in GSCs, and although the expression levels of the gene with the most statistically significant difference, P4HA2, were lower in GSCs than in GBM cells, survival curve analysis indicated that lower expression levels of P4HA2 corresponded to longer survival times. Subsequently, CCK8, EdU incorporation, cell cycle, wound healing and Transwell analyses were performed, demonstrating that P4HA2 promoted the proliferation and migration of U87 cells, and the effect of promoting migration was more pronounced than that of promoting proliferation, which was consistent with a previous study (23). However, there are some limitations in the present study. First, although as many samples as possible were searched on the GPL570 platform, the sample size was not large enough. Thus, future studies should attempt to directly analyze the clinical tissue samples of patients with GBM in order to obtain more definite conclusions. Additionally, the mechanism by which P4HA2 may promote GBM progression is unclear, which is undoubtedly an obstacle for clinical application. It is well known that GSCs promote the progression of GBM, and P4HA2 has a cancer-promoting effect. Therefore, low P4HA2 expression in GSCs may be responsible for longer survival times in patients with GBM. In addition, the present study hypothesized that P4HA2 may possibly act as a switch mediating the transition from GSCs to GBM. According to this hypothesis, in GSCs, when P4HA2 expression is low, the switch is off, but after a stimulus increases P4HA2 expression, P4HA2 may induce GSCs to transform into GBM cells. P4HA2, together with the changed extracellular matrix, may then promote the continuous proliferation, migration and invasion of GBM cells. Further investigation will be necessary to verify this hypothesis in future studies. There have been similar studies comparing GSCs and GBM samples to find specific biomarkers (45–48), but few studies have focused on the switch that transforms GSCs into GBM cells (49). The current findings may provide a new therapeutic target for blocking the conversion of GSCs to GBM cells and preventing GSCs from promoting the malignant progression of GBM.
In conclusion, the present study identified that P4HA2 expression in U87 cells was higher than in GSCs, and P4HA2 promoted U87 cell proliferation and migration via both bioinformatics analysis and experimental verification. The current results may provide new insights for further studies on the transition from GSCs to GBM cells.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant no. 31171038), Jiangsu Natural Science Foundation (grant no. BK2011385), Jiangsu ‘333’ program funding (grant no. BRA2016450), Application Research Project of Nantong City (grant no. MS12017015-3) and the Training Program of Innovation and Entrepreneurship for Undergraduates of Xinglin College (grant no. 201813993007Y).
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 31171038), Jiangsu Natural Science Foundation (grant no. BK2011385), Jiangsu ‘333’ program funding (grant no. BRA2016450), Application Research Project of Nantong City (grant no. MS12017015-3) and the Training Program of Innovation and Entrepreneurship for Undergraduates of Xinglin College (grant no. 201813993007Y).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
XHZ conceived and designed the study. YW collected and analyzed the data, performed the experiments and drafted the paper. XRZ, JW and RJ helped with experiments and preparing figures. LZ and JQ helped with the data analysis and manuscript revising. MT and GJ analyzed the data and revised the final paper. XHZ and YW confirm the authenticity of the data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Camelo-Piragua S, Kesari S. Further understanding of the pathology of glioma: Implications for the clinic. Expert Rev Neurother. 2016;16:1055–1065. doi: 10.1080/14737175.2016.1194755. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Davis ME. Glioblastoma: Overview of disease and treatment. Clin J Oncol Nurs. 2016;20(Suppl 5):S2–S8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Sousa E, Melo F, Vermeulen L, Fessler E, Medema JP. Cancer heterogeneity - a multifaceted view. EMBO Rep. 2013;14:686–695. doi: 10.1038/embor.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida GJ, Saya H. Therapeutic strategies targeting cancer stem cells. Cancer Sci. 2016;107:5–11. doi: 10.1111/cas.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan MA, Canman CE. Replication stress: An Achilles heel of glioma cancer stem-like cells. Cancer Res. 2018;78:6713–6716. doi: 10.1158/0008-5472.CAN-18-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134:505–512. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouchi R, Okabe S, Migita T, Nakano I, Seimiya H. Senescence from glioma stem cell differentiation promotes tumor growth. Biochem Biophys Res Commun. 2016;470:275–281. doi: 10.1016/j.bbrc.2016.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakamoto D, Takagi T, Fujita M, Omura S, Yoshida Y, Iida T, Yoshimura S. Basic gene expression characteristics of glioma stem cells and human glioblastoma. Anticancer Res. 2019;39:597–607. doi: 10.21873/anticanres.13153. [DOI] [PubMed] [Google Scholar]
- 11.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakano I, Joshi K, Visnyei K, Hu B, Watanabe M, Lam D, Wexler E, Saigusa K, Nakamura Y, Laks DR, et al. Siomycin A targets brain tumor stem cells partially through a MELK-mediated pathway. Neuro-oncol. 2011;13:622–634. doi: 10.1093/neuonc/nor023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano I, Masterman-Smith M, Saigusa K, Paucar AA, Horvath S, Shoemaker L, Watanabe M, Negro A, Bajpai R, Howes A, et al. Maternal embryonic leucine zipper kinase is a key regulator of the proliferation of malignant brain tumors, including brain tumor stem cells. J Neurosci Res. 2008;86:48–60. doi: 10.1002/jnr.21471. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Xu L, Wang Y, Liu J, Zhu W, Mao S. Morphological adaptation of sheep's rumen epithelium to high-grain diet entails alteration in the expression of genes involved in cell cycle regulation, cell proliferation and apoptosis. J Anim Sci Biotechnol. 2018;9:32. doi: 10.1186/s40104-018-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Y, El Agha E, Turcatel G, Chen H, Chiu J, Warburton D, Bellusci S, Qian BP, Menke DB, Shi W. Mesenchymal adenomatous polyposis coli plays critical and diverse roles in regulating lung development. BMC Biol. 2015;13:42. doi: 10.1186/s12915-015-0153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Hahn H, Odze RD, Goyal RK. Metaplastic esophageal columnar epithelium without goblet cells shows DNA content abnormalities similar to goblet cell-containing epithelium. Am J Gastroenterol. 2009;104:816–824. doi: 10.1038/ajg.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wesseling P, Capper D. WHO 2016 Classification of gliomas. Neuropathol Appl Neurobiol. 2018;44:139–150. doi: 10.1111/nan.12432. [DOI] [PubMed] [Google Scholar]
- 19.Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatzimichael E, Lo Nigro C, Lattanzio L, Syed N, Shah R, Dasoula A, Janczar K, Vivenza D, Monteverde M, Merlano M, et al. The collagen prolyl hydroxylases are novel transcriptionally silenced genes in lymphoma. Br J Cancer. 2012;107:1423–1432. doi: 10.1038/bjc.2012.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aro E, Salo AM, Khatri R, Finnilä M, Miinalainen I, Sormunen R, Pakkanen O, Holster T, Soininen R, Prein C, et al. Severe extracellular matrix abnormalities and chondrodysplasia in mice lacking collagen prolyl 4-hydroxylase isoenzyme II in combination with a reduced amount of isoenzyme I. J Biol Chem. 2015;290:16964–16978. doi: 10.1074/jbc.M115.662635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martini D, Giannaccini M, Guadagni V, Marracci S, Giudetti G, Andreazzoli M. Comparative analysis of p4ha1 and p4ha2 expression during Xenopus laevis development. Int J Dev Biol. 2019;63:311–316. doi: 10.1387/ijdb.190067ma. [DOI] [PubMed] [Google Scholar]
- 23.Atkinson A, Renziehausen A, Wang H, Lo Nigro C, Lattanzio L, Merlano M, Rao B, Weir L, Evans A, Matin R, et al. Collagen prolyl hydroxylases are bifunctional growth regulators in melanoma. J Invest Dermatol. 2019;139:1118–1126. doi: 10.1016/j.jid.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nokelainen M, Nissi R, Kukkola L, Helaakoski T, Myllyharju J. Characterization of the human and mouse genes for the alpha subunit of type II prolyl 4-hydroxylase. Identification of a previously unknown alternatively spliced exon and its expression in various tissues. Eur J Biochem. 2001;268:5300–5309. doi: 10.1046/j.0014-2956.2001.02464.x. [DOI] [PubMed] [Google Scholar]
- 25.Shi R, Gao S, Smith AH, Li H, Shao M, Shangguan J, Zhang J, Xu J, Ye J, Graham LM, et al. Superoxide-induced Type I collagen secretion depends on prolyl 4-hydroxylases. Biochem Biophys Res Commun. 2020;529:1011–1017. doi: 10.1016/j.bbrc.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Han Q, Li J, Jia Y, Zhang R, Shi H. P4HA2 contributes to cervical cancer progression via inducing epithelial-mesenchymal transition. J Cancer. 2020;11:2788–2799. doi: 10.7150/jca.38401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, Fu X, Jin T, Zhang L, Liu B, Wu Y, Xu F, Wang X, Ye K, Zhang W, et al. Aspirin targets P4HA2 through inhibiting NF-κB and LMCD1-AS1/let-7g to inhibit tumour growth and collagen deposition in hepatocellular carcinoma. EBioMedicine. 2019;45:168–180. doi: 10.1016/j.ebiom.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Saar V, Leung KL, Chen L, Wong G. Human amyloid β peptide and tau co-expression impairs behavior and causes specific gene expression changes in Caenorhabditis elegans. Neurobiol Dis. 2018;109:88–101. doi: 10.1016/j.nbd.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Napolitano F, Di Iorio V, Testa F, Tirozzi A, Reccia MG, Lombardi L, Farina O, Simonelli F, Gianfrancesco F, Di Iorio G, et al. Autosomal-dominant myopia associated to a novel P4HA2 missense variant and defective collagen hydroxylation. Clin Genet. 2018;93:982–991. doi: 10.1111/cge.13217. [DOI] [PubMed] [Google Scholar]
- 30.Guo H, Tong P, Liu Y, Xia L, Wang T, Tian Q, Li Y, Hu Y, Zheng Y, Jin X, et al. Mutations of P4HA2 encoding prolyl 4-hydroxylase 2 are associated with nonsyndromic high myopia. Genet Med. 2015;17:300–306. doi: 10.1038/gim.2015.28. [DOI] [PubMed] [Google Scholar]
- 31.Napolitano F, Di Iorio V, Di Iorio G, Melone MA, Gianfrancesco F, Simonelli F, Esposito T, Testa F, Sampaolo S. Early posterior vitreous detachment is associated with LAMA5 dominant mutation. Ophthalmic Genet. 2019;40:39–42. doi: 10.1080/13816810.2018.1558261. [DOI] [PubMed] [Google Scholar]
- 32.Cai XB, Zheng YH, Chen DF, Zhou FY, Xia LQ, Wen XR, Yuan YM, Han F, Piao SY, Zhuang W, et al. Expanding the phenotypic and genotypic landscape of nonsyndromic high myopia: A cross-sectional study in 731 chinese patients. Invest Ophthalmol Vis Sci. 2019;60:4052–4062. doi: 10.1167/iovs.19-27921. [DOI] [PubMed] [Google Scholar]
- 33.Feng GX, Li J, Yang Z, Zhang SQ, Liu YX, Zhang WY, Ye LH, Zhang XD. Hepatitis B virus X protein promotes the development of liver fibrosis and hepatoma through downregulation of miR-30e targeting P4HA2 mRNA. Oncogene. 2017;36:6895–6905. doi: 10.1038/onc.2017.291. [DOI] [PubMed] [Google Scholar]
- 34.Zhang T, Piao HY, Guo S, Zhao Y, Wang Y, Zheng ZC, Zhang J. LncRNA PCGEM1 enhances metastasis and gastric cancer invasion through targeting of miR-129-5p to regulate P4HA2 expression. Exp Mol Pathol. 2020;116:104487. doi: 10.1016/j.yexmp.2020.104487. [DOI] [PubMed] [Google Scholar]
- 35.Pankova D, Jiang Y, Chatzifrangkeskou M, Vendrell I, Buzzelli J, Ryan A, Brown C, O'Neill E. RASSF1A controls tissue stiffness and cancer stem-like cells in lung adenocarcinoma. EMBO J. 2019;38:e100532. doi: 10.15252/embj.2018100532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toss MS, Miligy IM, Gorringe KL, AlKawaz A, Khout H, Ellis IO, Green AR, Rakha EA. Prolyl-4-hydroxylase α subunit 2 (P4HA2) expression is a predictor of poor outcome in breast ductal carcinoma in situ (DCIS) Br J Cancer. 2018;119:1518–1526. doi: 10.1038/s41416-018-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong G, Deng L, Zhu J, Rychahou PG, Xu R. Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer. 2014;14:1. doi: 10.1186/1471-2407-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Hu D, Ansari D, Pawłowski K, Zhou Q, Sasor A, Welinder C, Kristl T, Bauden M, Rezeli M, Jiang Y, et al. Proteomic analyses identify prognostic biomarkers for pancreatic ductal adenocarcinoma. Oncotarget. 2018;9:9789–9807. doi: 10.18632/oncotarget.23929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q, Wang Q, Zhang Q, Zhang J, Zhang J. Collagen prolyl 4-hydroxylase 2 predicts worse prognosis and promotes glycolysis in cervical cancer. Am J Transl Res. 2019;11:6938–6951. [PMC free article] [PubMed] [Google Scholar]
- 41.Chang KP, Yu JS, Chien KY, Lee CW, Liang Y, Liao CT, Yen TC, Lee LY, Huang LL, Liu SC, et al. Identification of PRDX4 and P4HA2 as metastasis-associated proteins in oral cavity squamous cell carcinoma by comparative tissue proteomics of microdissected specimens using iTRAQ technology. J Proteome Res. 2011;10:4935–4947. doi: 10.1021/pr200311p. [DOI] [PubMed] [Google Scholar]
- 42.Jiang W, Zhou X, Li Z, Liu K, Wang W, Tan R, Cong X, Shan J, Zhan Y, Cui Z, et al. Prolyl 4-hydroxylase 2 promotes B-cell lymphoma progression via hydroxylation of Carabin. Blood. 2018;131:1325–1336. doi: 10.1182/blood-2017-07-794875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teodoro JG, Parker AE, Zhu X, Green MR. p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science. 2006;313:968–971. doi: 10.1126/science.1126391. [DOI] [PubMed] [Google Scholar]
- 44.Berger B, Capper D, Lemke D, Pfenning PN, Platten M, Weller M, von Deimling A, Wick W, Weiler M. Defective p53 antiangiogenic signaling in glioblastoma. Neuro-oncol. 2010;12:894–907. doi: 10.1093/neuonc/noq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trépant AL, Bouchart C, Rorive S, Sauvage S, Decaestecker C, Demetter P, Salmon I. Identification of OLIG2 as the most specific glioblastoma stem cell marker starting from comparative analysis of data from similar DNA chip microarray platforms. Tumour Biol. 2015;36:1943–1953. doi: 10.1007/s13277-014-2800-5. [DOI] [PubMed] [Google Scholar]
- 46.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradshaw A, Wickremsekera A, Tan ST, Peng L, Davis PF, Itinteang T. Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front Surg. 2016;3:21. doi: 10.3389/fsurg.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, Guryanova OA, Zhou W, Liu C, Huang Z, Fang X, Wang X, Chen C, Wu Q, He Z, et al. Ibrutinib inactivates BMX-STAT3 in glioma stem cells to impair malignant growth and radioresistance. Sci Transl Med. 2018;10:10. doi: 10.1126/scitranslmed.aah6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Wang J, Marzese DM, Wang X, Yang Z, Li C, Zhang H, Zhang J, Chen CC, Kelly DF, et al. B7H3 regulates differentiation and serves as a potential biomarker and theranostic target for human glioblastoma. Lab Invest. 2019;99:1117–1129. doi: 10.1038/s41374-019-0238-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/).