Abstract
Antibiotic resistance is a major health problem worldwide, causing more deaths than diabetes and cancer. The dissemination of vertical and horizontal antibiotic resistance genes has been conducted for a selection of pan-resistant bacteria. Here, we test if the aerobic and anaerobic bacteria from human feces samples in health conditions are carriers of beta-lactamases genes. The samples were cultured in a brain–heart infusion medium and subcultured in blood agar in aerobic and anaerobic conditions for 24 h at 37 °C. The grown colonies were identified by their biochemical profiles. The DNA was extracted and purified by bacterial lysis using thermal shock and were used in the endpoint PCR and next generation sequencing to identify beta-lactamase genes expression (OXA, VIM, SHV, TEM, IMP, ROB, KPC, CMY, DHA, P, CFX, LAP, and BIL). The aerobic bacterias Aeromonas hydrophila, Citrobacter freundii, Proteus mirabilis, Providencia rettgeri, Serratia fonticola, Serratia liquefaciens, Enterobacter aerogenes, Escherichia coli, Klebsiella pneumoniae, Pantoea agglomerans, Enterococcus faecalis, and Enterobacter cloacae, the anaerobic bacteria: Capnocytophaga species, Bacteroides distasonis, Bifidobacterium adolescentis, Bacteroides ovatus, Bacteroides fragilis, Eubacterium species, Eubacterium aerofaciens, Peptostreptococcus anaerobius, Fusobacterium species, Bacteroides species, and Bacteroides vulgatus were isolated and identified. The results showed 49 strains resistant to beta-lactam with the expression of blaSHV (10.2%), blaTEM (100%), blaKPC (10.2%), blaCYM (14.3%), blaP (2%), blaCFX (8.2%), and blaBIL (6.1%). These data support the idea that the human enteric microbiota constitutes an important reservoir of genes for resistance to beta-lactamases and that such genes could be transferred to pathogenic bacteria.
Keywords: microbiome, beta-lactams, beta-lactamases, resistome
1. Introduction
The indiscriminate and unconscious use of antibiotics in the clinic, agriculture, and livestock have been increased the selection pressure in the worldwide microbiome, inducing resistant, multiresistant, and pan-resistant pathogen bacteria selection [1,2,3]. This problem reached a maximal level during the last two years where the antibiotic use has increased drastically due to the pandemic spread of SARS-CoV-2, since more than 70% of COVID-19 patients have received complimentary treatment with antibiotics drugs [4,5].
Bacterial resistance to antibiotics is a situation that has been increasing during the last decades, converting into a wide world public health problem [6,7], that complicates the treatment of the infection while increases the mortality rate in both nosocomial or community-acquired infectious diseases [8,9]. This resistance is reaching a critical point, as the increase in these resistant, multi-resistant, and pan-resistant pathogenic strains causes serious medical complications [10].
The lactam groups of molecules are integrated into four families, i.e., beta-lactam [11], gamma-lactam [12], delta-lactam [13], and epsilon-lactam [14], with a specific action and not necessarily like antibiotics. Specifically, the beta-lactams drugs show antibiotic effects in a wide bacterial variety. They are characterized by a beta-lactam ring presence, which confers the antimicrobial effect by the transpeptidases and carboxypeptidases inactivation (the proteins responsible for the biosynthesis of the cell wall) through their binding to Penicillin Binding Protein (PBPs) receptors [15]. The beta-lactam family is made up of various members, such as penicillins B and G, amoxicillin, ampicillin, cephalosporins (cephalothin, cephalexin, ceftriaxone, and cefepime), carbapenems (imipenem, meropenem, ertapenem), aztreonbactam, monobactam, and some others. The indiscriminate and unconscious use of these antibiotics in daily clinical practice has caused an increase in multiresistant Enterobacteriaceae strains, especially Escherichia coli and Klebsiella pneumoniae, turning this situation into a serious global health problem [16]. During the last decade that there has been a significant increase in the presence of multi-resistant strains worldwide, mainly in the European Union. The European Read for Antimicrobial Resistance Surveillance (EARSNet) has been identified pan-resistant Escherichia coli and Klebsiella pneumoniae strains [17,18]. The bacteria have various resistance mechanisms, mainly associated with plasmid assimilation, which allow cross-resistance against a wide range of antibiotics [19]. The most widely described mechanism of antibiotic resistance that is mediated by plasmid transfer is the enzymatic degradation of the beta-lactam ring through the expression of beta-lactamases [20]. The genes encoding for the beta-lactamases enzymes have been widely spread among the bacterial population, through mobile genetic elements, which confer a plasmid-mediated multi-resistance [21,22]. These multi-resistance bacteria complicate the patient treatments and condition the type of antibiotics that can be administrated for the treatment of common infections [23]. The first-line antibiotic treatment regimens for gastrointestinal and airway infections usually include beta-lactams drugs, such as ampicillin, amoxicillin, or third generation cephalosporins (Ceftriaxone), but the resistance determines the treatment that can be given to patients and decreases the effectiveness significantly of these treatments. The Carbapenems (ertapenem, meropenem, imipenem) have been widely used as an alternative for the treatment of these resistant bacteria, however, in recent years, resistant bacteria strains to these antibiotics have been identified [24]. The main causes associated with this increase in bacterial resistance are the error in the medical prescription, self-medication, incomplete antibiotics treatment, and the use of these drugs by the agricultural and livestock industry for the treatment of crops for human consumption [25].
For these reasons, we test the hypothesis that the Enterobacteriaceae integrating the human gastrointestinal microbiome are carriers of beta-lactamases genes in healthy individuals who have not received antibiotic treatment during their last year of life.
2. Results
2.1. Bacteria Identification
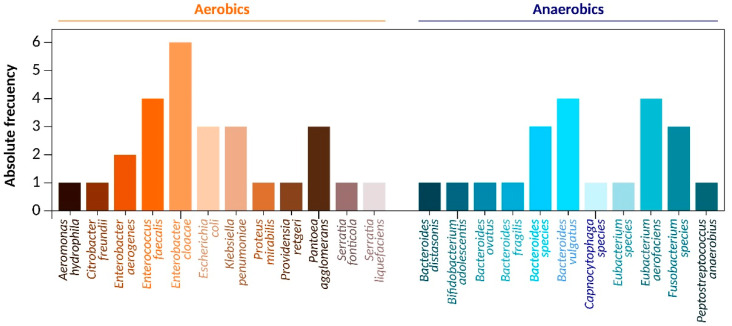
From the sampling of 20 different medicine students in healthy conditions from Mexico City, 49 beta-lactam resistant strains were isolated, founding 27 strands of 12 different aerobic and 22 strand of 11 different anaerobic beta-lactam resistant Enterobacteriaceae species (Figure 1). The distribution of the identified bacteria was as follows. Aerobic: Aeromonas hydrophila 3.7% (n = 1), Citrobacter freundii 3.7% (n = 1), Enterobacter aerogenes 7.4% (n = 2), Enterococcus faecalis 14.8% (n = 4), Enterobacter cloacae 22.2% (n = 6), Escherichia coli 11.1% (n = 3), Klebsiella pneumoniae 11.1%% (n = 3), Proteus mirabilis 3.7% (n = 1), Providencia rettgeri 3.7% (n = 1), Pantoea agglomerans 11.1% (n = 3), Serratia fonticola 3.7% (n = 1), Serratia liquefaciens 3.7% (n = 1), Anaerobic: Bacteroides distasonis 4.5% (n = 1), Bifidobacterium adolescentis 4.5% (n = 1), Bacteroides ovatus 4.5% (n = 1), Bacteroides fragilis 4.5% (n = 1), Bacteroides species 13.6% (n = 3), Bacteroides vulgatus 18.2% (n = 4), Capnocytophaga species 4.5% (n = 1), Eubacterium species 4.5% (n = 1), Eubacterium aerofaciens 18.1% (n = 4), Fusobacterium species 13.6% (n = 3) and Peptostreptococcus anaerobius 9.1% (n = 1) (Figure 1). All these data were confirmed by whole DNA sequencing using NGS (Table 1), showing 98% of concordance with the automatized bacterial identification. All bacteria genomes size sequenced were similar to those reported in the gene bank, observing Δ from 1% to 8% of differences compared with the reported sizes (Table 2).
Figure 1.
Bacterial frequency. The graph shows the frequency of beta-Lactam resistant aerobic and anaerobic Enterobacteriaceae isolated from feces sample of medicine student in healthy conditions.
Table 1.
Metagenomic identification of the aerobic and anaerobic beta-lactam resistant bacteria isolated from feces cultures of humans in healthy conditions.
| Bacteria | Total Reads | Classified Reads | Domain | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|---|---|---|
| Aerobic | |||||||||
| Aeromonas hydrophila | 4,502,897 | 4,412,839 (98%) | 4,103,940 (93%) | 3,734,586 (91%) | 3,211,744 (86%) | 2,697,865(84%) | 2,212,249 (82%) | 1,791,922 (81%) | 1,361,861 (76%) |
| Citrobacter freundii | 1,816,430 | 1,798,266 (99%) | 1,744,318 (97%) | 1,657,102 (95%) | 1,491,392 (90%) | 1, 327,339(89%) | 1,101,691 (83%) | 870,336 (79%) | 539,608 (62%) |
| Enterobacter aerogenes | 897,648 | 727,095 (81%) | 567,134 (78%) | 436,693 (77%) | 331,887 (76%) | 248,915 (75%) | 1817,08 (73%) | 130,830 (72%) | 91,581 (70%) |
| Enterococcus faecalis | 3,134,872 | 3,040,826 (97%) | 2,852,734 (91%) | 2,821,385 (90%) | 2,695,990 (86%) | 2,664,641 (85%) | 2,539,246 (81%) | 2,507,898 (80%) | 2,476,549 (79%) |
| Enterobacter cloacae | 945,761 | 898,473 (95%) | 892,798 (94%) | 871,046 (92%) | 866,317 (91%) | 850,239 (89%) | 842,673 (89%) | 836,998 (85%) | 835,107 (88%) |
| Escherichia coli | 2,361,827 | 2,078,408 (88%) | 1,936,698 (82%) | 1,865,843 (79%) | 1,606,042 (68%) | 1,511,569 (64%) | 1,464,333 (62%) | 1,440,714 (61%) | 1,393,478 (59%) |
| Klebsiella pneumoniae | 1,129,675 | 1,039,301 (92%) | 1,016,708 (90%) | 1,005,411 (89%) | 994,114 (88%) | 915,037 (81%) | 881,147 (78%) | 869,850 (77%) | 835,960 (74%) |
| Proteus mirabilis | 816,524 | 808,359 (99%) | 805,909 (98%) | 803,460 (97%) | 767,533 (94%) | 751,202 (92%) | 743,037 (91%) | 738,954 (90%) | 726,706 (89%) |
| Providencia rettgeri | 354,869 | 273,249 (77%) | 266,152 (75%) | 259,054 (73%) | 255,506 (72%) | 237,762 (67%) | 223,567 (63%) | 216,470 (61%) | 191,629 (54%) |
| Pantoea agglomerans | 497,802 | 438,066 (88%) | 433,088 (87%) | 408,198 (82%) | 393,264 (79%) | 388,286 (78%) | 353,439 (71%) | 338,505 (68%) | 333,527 (67%) |
| Serratia fonticola | 1,347,964 | 1,334,484 (99%) | 1,267,086 (94%) | 1,253,607 (93%) | 1,226,647 (91%) | 1,186,208 (88%) | 1,145,769 (85%) | 1,132,290 (84%) | 997,493 (74%) |
| Serratia liquefaciens | 256,789 | 190,024 (74%) | 182,320 (71%) | 179,752 (70%) | 174,617 (68%) | 159,209 (62%) | 156,641 (61%) | 154,073 (60%) | 133,530 (52%) |
| Anaerobic | |||||||||
| Bacteroides distasonis | 845,168 | 752,201 (89%) | 726,844 (86%) | 718,393 (85%) | 693,038 (82%) | 667,683 (79%) | 659,231 (78%) | 625,424 (74%) | 608,521 (72%) |
| Bifidobacterium adolescentis | 748,751 | 718,801 (96%) | 715,806 (95%) | 704,575 (94%) | 700,082 (93%) | 689,600 (92%) | 687,353 (91%) | 682,861 (91%) | 658,901 (88%) |
| Bacteroides ovatus | 965,743 | 905,867 (94%) | 902,004 (94%) | 898,141 (93%) | 893,312 (92%) | 888,484 (91%) | 830,539 (86%) | 743,622 (77%) | 637,390 (66%) |
| Bacteroides fragilis | 986,241 | 976,379 (99%) | 966,516 (98%) | 956,654 (97%) | 951,723 (96.4%) | 946,791 (96.1%) | 927,067 (94.3%) | 917,204 (93%) | 915,232 (92.6%) |
| Bacteroides species | 567,284 | 486,162 (85.7%) | 483,893 (85.3%) | 482,759 (85.1%) | 481,624 (84.9%) | 478,788 (84.4%) | 478,220 (84.3%) | 477,086 (84.1%) | 474,817 (83.7%) |
| Bacteroides vulgatus | 928,712 | 911,995 (98.2%) | 911,066 (98.1%) | 833,055 (89.7%) | 830,269 (89.4%) | 828,411 (89.2%) | 825,625 (88.9%) | 819,124 (88.2%) | 816,338 (87.9%) |
| Capnocytophaga species | 349,817 | 319,733 (91%) | 315,535 (90.2%) | 313,471 (89.6%) | 312,911 (89.4%) | 312,282 (89.2%) | 312,212 (89.1%) | 299,793 (85.7%) | 295,211 (84.3%) |
| Eubacterium species | 567,492 | 542,522 (95.6%) | 541,387 (95.4%) | 540,309 (95.2%) | 539,685 (95.1%) | 537,982 (94.8%) | 536,280 (94.5%) | 535,712 (94.4%) | 534,010 (94.1%) |
| Eubacterium aerofaciens | 814,927 | 620,974 (76.2%) | 616,085 (75.5%) | 604,676 (74.2%) | 599,786 (73.6%) | 595,712 (73.1%) | 594,082 (72.9%) | 581,043 (71.3%) | 576,968 (70.8%) |
| Fusobacterium species | 237,491 | 204,717 (86.2%) | 203,292 (85.6%) | 192,843 (81.2%) | 191,418 (80.6%) | 189,755 (79.9%) | 187,855 (79.1%) | 186,193 (78.4%) | 185,955 (78.3%) |
| Peptostreptococcus anaerobius | 729,358 | 686,691 (94.1%) | 686,399 (94.1%) | 683,044 (93.6%) | 682,314 (93.5%) | 679,251 (93.1%) | 677,282 (92.8%) | 612,442 (83.9%) | 587,498 (80.5%) |
Table 2.
Whole-genome sequencing characteristics of aerobic and anaerobic Enterobacteriaceae rods isolated from human feces in healthy conditions.
| Bacteria | CDS | Number of Sequence Contigs | Genome Size (bp) | ||
|---|---|---|---|---|---|
| Assembled | Reported | Difference | |||
| Aerobic | |||||
| Aeromonas hydrophila | 4,428 | 1,124 | 5,124,487 | 4,911,246 | 213,241 |
| Citrobacter freundii | 5,064 | 789 | 5,343,952 | 5,297,052 | 46,900 |
| Enterobacter aerogenes | 4,545 | 1,484 | 5,578,724 | 5,280,350 | 298,374 |
| Enterococcus faecalis | 2,969 | 951 | 3,111,017 | 3,038,914 | 72,103 |
| Enterobacter cloacae | 4,545 | 1,484 | 4,982,176 | 4,772,910 | 209,266 |
| Escherichia coli | 5,704 | 1,561 | 5,689,156 | 5,615,389 | 73,767 |
| Klebsiella pneumoniae | 5,071 | 583 | 5,479,173 | 5,315,120 | 164,053 |
| Proteus mirabilis | 3,772 | 584 | 4,183,869 | 4,209,445 | 25,576 |
| Providencia rettgeri | 4,497 | 638 | 4,954,326 | 4,780,676 | 173,650 |
| Pantoea agglomerans | 4,255 | 738 | 4,090,220 | 4,047,712 | 42,508 |
| Serratia fonticola | 5,945 | 916 | 6,483,043 | 6,000,511 | 482,532 |
| Serratia liquefaciens | 4,936 | 495 | 5,706,987 | 5,395,544 | 311,443 |
| Anaerobic | |||||
| Bacteroides distasonis | 3,896 | 603 | 4,952,323 | 4,812,038 | 140,285 |
| Bifidobacterium adolescentis | 1,742 | 418 | 2,173,720 | 2,089,645 | 84,075 |
| Bacteroides ovatus | 4,803 | 756 | 6,425,267 | 6,472,489 | 47,222 |
| Bacteroides fragilis | 4,100 | 1,025 | 5,188,967 | 5,474,541 | 285,574 |
| Bacteroides species | 2,401 | 904 | 2,658,624 | 2,628,345 | 30,279 |
| Bacteroides vulgatus | 5,055 | 406 | 5,311,454 | 5,681,290 | 369,836 |
| Capnocytophaga species | 2,208 | 502 | 2,614,527 | 2,837,214 | 222,687 |
| Eubacterium species | 2,197 | 505 | 2,628,803 | 2,450,450 | 178,353 |
| Eubacterium aerofaciens | 1,887 | 1,548 | 2,128,754 | 2,264,854 | 136,100 |
| Fusobacterium species | 2,113 | 738 | 2,159,799 | 2,185,897 | 26,098 |
| Peptostreptococcus anaerobius | 1,793 | 847 | 1,989,753 | 2,106,123 | 116,370 |
2.2. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility test of beta lactam resistant bacteria showed, that the 88.9% (n = 24) showed resistance to amoxicillin/clavulanic acid, ampicillin, and vancomycin, followed by resistance to cefixime with 81.5% (n = 22). Resistance towards cefuroxime and towards cephalothin occurred in both cases in 70.4% (n = 19) of the isolated aerobic bacteria, followed by resistance towards cefazolin and cefaclor, both with 66.7% (n = 18). 63% (n = 17) of the aerobic bacteria isolates showed resistance to piperacillin, followed by resistance to doxycycline with 51.9% (n = 14) resistance. The lowest resistance was observed towards piperacillin/tazobactam 18.5% (n = 5), ceftizoxime 14.8% (n = 4) and towards meropenem and imipenem, both with 11.1% (n = 3), While 95.5% (n = 21) of the isolated anaerobic bacteria showed resistance towards vancomycin followed by resistance towards cefixime with 81.8% (n = 18) and towards piperacillin, cephalothin, cefaclor and cefuroxime, each with 72.7% (n = 18). 68.2% (n = 15) of this population was resistant to cefazolin, followed by resistance towards doxycycline 63.6% (n = 14) and towards Amoxicillin/Clavulanic acid and Ampicillin, both with 54.5% (n = 12) and towards Ceftizoxime 41.5% (n = 10). The lowest resistance was observed towards Piperacillin/Tazobactam 27.3% (n = 6) and to Meropenem and Imipenem, each with 4.5% (n = 1), and non-beta-lactam resistance is showed in Table 3.
Table 3.
Whole antibiograms characteristics of beta-lactam resistant bacteria isolated from feces cultures of humans in healthy conditions.
| Antibiotic Families | Drugs | Aerobics | Anaerobic | ||
|---|---|---|---|---|---|
| Frequency | % | Frequency | % | ||
| Beta-Lactam | Amoxicillin/Clavulanic acid (AmC-30) | 24 | 88.9 | 12 | 54.5 |
| Piperacillin (PIP-100) | 17 | 63.0 | 16 | 72.7 | |
| Piperacillin/Tazobactam (TZP-110) | 5 | 18.5 | 6 | 27.3 | |
| Doxycycline (D-30) | 14 | 51.9 | 14 | 63.6 | |
| Ampicillin 10(AM-10) | 24 | 88.9 | 12 | 54.5 | |
| Cephalothin 1° (CF-30) | 19 | 70.4 | 16 | 72.7 | |
| Cefazolin 1° (CZ-30) | 18 | 66.7 | 15 | 68.2 | |
| Cefaclor 2° (CEC-30) | 18 | 66.7 | 16 | 72.7 | |
| Cefuroxime 2° (CXM-30) | 19 | 70.4 | 16 | 72.7 | |
| Cefixime 3° (CFM-5) | 22 | 81.5 | 18 | 81.8 | |
| Ceftizoxime 3a (ZOX-30) | 4 | 14.8 | 10 | 45.5 | |
| Meropenem (MEM-10) | 3 | 11.1 | 1 | 4.5 | |
| Imipenem (IPM-10) | 3 | 11.1 | 1 | 4.5 | |
2.3. Beta-Lactamases Gene Family Identification
The endpoint PCR results showed that 100 % of the obtained bacterial strain carries at least a gene of Beta-Lactamases, identified the gene families transcribed and the phenotype responsible for Beta-Lactam resistance: 49 strains were positive to blaTEM (100%), five positive strains for the blaSHV (10.2%), five positive strains for blaKPC (10.2%), seven positive strains for blaCYM (14.3%), one positive strain for blaP (2%), four positive strain for blaCFX (8.2%), and three positive strains for blaBIL (6.1%) gene families (Table 4). No members of gene families blaOXA, blaVIM, blaIMP, blaROB, blaCTX, blaDHA, and blaLAP were found in all obtained strains from feces of human and healthy conditions. These data were also confirmed by whole-genome sequencing of isolated strains (Table 3 and Table 4).
Table 4.
Frequency of beta-lactamase gene families identified by endpoint PCR in roads cultured from gut microbiota in healthy conditions.
| Gene Family | Positive Strains (n) | % | Gene Family | Positive Strains (n) | % |
|---|---|---|---|---|---|
| blaTEM | 49 | 100.0 | blaBIL | 3 | 6.1 |
| blaSHV | 5 | 10.2 | blaP | 1 | 2.0 |
| blaKPC | 5 | 10.2 | blaCFX | 4 | 8.2 |
| blaCYM | 7 | 14.3 |
The isolated strains from human feces in healthy conditions showed a restricted pattern of resistance to beta-lactam drugs. The individual analysis showed that of the blaTEM was present in all (100% n = 49) of isolated strains, from these the 77.5% (n = 38) was carrier only for this gene family, the other 25.5% of isolated strains showed a mixed genotype for multiple gene families encoding beta-lactamase and specific for each strain, conformed from two (blaTEM + blaBIL, blaTEM + blaCYM, or blaTEM + blaP) to five (blaTEM + blaSHV + blaKPC + blaCYMX + blaDHA + blaBIL) gene families of beta-lactamases (Table 5).
Table 5.
Beta-Lactamases (bla) gene families carried by the aerobic and anaerobic Enterobacteria of humans in healthy conditions.
| Aerobic Bacteria | Beta-Lactamases Families | Anaerobic Bacteria | Beta-Lactamases Families | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TEM | SHV | KPC | CYM | BIL | CFX | TEM | CYM | BIL | P | ||
| Aeromonas hydrophila | + | + | + | Capnocytophaga species | + | ||||||
| Citrobacter freundii | + | Bacteroides distasonis | + | + | |||||||
| Proteus mirabilis | + | + | + | + | Bifidobacterium adolescenti | + | |||||
| Providencia rettgeri | + | Bacteroides ovatus | + | ||||||||
| Serratia fonticola | + | + | Bacteroides fragilis | + | + | ||||||
| Serratia liquefaciens | + | Eubacterium species | + | ||||||||
| Enterobacter aerogenes | + | + | Eubacterium aerofaciens | + | |||||||
| Escherichia coli | + | + | + | + | Peptostreptocuccus anaerobius | + | |||||
| Klebsiella pneumoniae | + | + | + | + | + | Fusobacterium species | + | + | |||
| Pantoea agglomerans | + | + | Bacteroides vulgatus | + | + | ||||||
| Enterococcus faecalis | + | + | Bacteroides species | + | |||||||
| Enterobacter cloacae | + | + | + | + | + | ||||||
(+) Positive for the beta-lactamase gene family.
2.4. Beta-Lactamases Genotype
The NGS sequencing showed seven members of the TEM beta-lactamases gene family including blaTEM-1, blaTEM-2, blaTEM-3, blaTEM-10, blaTEM-12, blaTEM-24, and blaTEM-52 distributed in the 49 blaTEM positive strains, two members of KPC beta-lactamases gene family (blaKPC-2 and blaKPC-3), only one member of each one of CYM (blaCMY-2), BIL (blaBIL-1), CFX (blaCTX-M-15), and P (blaP) beta-lactamases gene families were found (Table 4).
The mixed gene expression pattern was present some aerobic of Enterobacteriaceae species: Aeromonas hydrophila (blaTEM-1 + blaSHV-12 + blaBIL-1), Enterobacter aerogenes (blaTEM-1 + blaKPC-2), Enterococcus faecalis (blaTEM-1 + blaSHV-1), Enterobacter cloacae (blaTEM-12 + blaSHV-1), Escherichia coli (blaTEM-52 + blaSHV-12 + blaCMY-2 + blaCTX-M-15), Klebsiella pneumoniae (blaTEM-10 + blaSHV-12 + blaKPC-3 + blaCMY-2 + blaCTX-M-15), Proteus mirabilis (blaTEM-1 + blaKPC-2 + blaCMY-2 + blaCTX-M-15), Pantoea agglomerans (blaTEM-1 + blaCMY-2), Serratia fonticola (blaTEM-1 + blaKPC-2), while the anaerobic bacteria that express mixed genotype were: Bacteroides distasonis (blaTEM-1 + blaBIL-1), Bacteroides fragilis (blaTEM-2 + blaCMY-2), Bacteroides vulgatus (blaTEM-1 + blaCMY-2), and Fusobacterium species (blaTEM-12 + blaP). The NGS analyses for all the bacterial strains tested were negative to blaOXA, blaVIM, blaIMP, blaROB, blaCTX, blaDHA, and blaLAP (Table 6).
Table 6.
Mixed Genotype of the beta-lactam antibiotic resistome identified by next-generation sequencing in the aerobic and anaerobic Enterobacteria from feces of humans in healthy conditions.
| Bacteria | Phenotype of Beta-Lactamases | ||||||
|---|---|---|---|---|---|---|---|
| TEM | SHV | KPC | CYM | BIL | CFX | P | |
| Aerobic | |||||||
| Aeromonas hydrophila | blaTEM-1 | blaSHV-12 | blaBIL-1 | ||||
| Citrobacter freundii | blaTEM-2 | ||||||
| Enterobacter aerogenes | blaTEM-1 | blaKPC-2 | |||||
| Enterococcus faecalis | blaTEM-1 | blaSHV-1 | |||||
| Enterobacter cloacae | blaTEM-12 | blaSHV-1 | |||||
| Escherichia coli | blaTEM-52 | blaSHV-12 | blaCMY-2 | blaCTX-M-15 | |||
| Klebsiella pneumoniae | blaTEM-10 | blaSHV-12 | blaKPC-3 | blaCMY-2 | blaCTX-M-15 | ||
| Proteus mirabilis | blaTEM-1 | blaKPC-2 | blaCMY-2 | blaCTX-M-15 | |||
| Providencia rettgeri | blaTEM-24 | ||||||
| Pantoea agglomerans | blaTEM-1 | blaCMY-2 | |||||
| Serratia fonticola | blaTEM-1 | blaKPC-2 | |||||
| Serratia liquefaciens | blaTEM-12 | ||||||
| Anaerobic | |||||||
| Bacteroides distasonis | blaTEM-1 | blaBIL-1 | |||||
| Bifidobacterium adolescentis | blaTEM-1 | ||||||
| Bacteroides ovatus | blaTEM-1 | ||||||
| Bacteroides fragilis | blaTEM-2 | blaCMY-2 | |||||
| Bacteroides species | blaTEM-1 | ||||||
| Bacteroides vulgatus | blaTEM-1 | blaCMY-2 | |||||
| Capnocytophaga species | blaTEM-1 | ||||||
| Eubacterium species | blaTEM-1 | ||||||
| Eubacterium aerofaciens | blaTEM-1 | ||||||
| Fusobacterium species | blaTEM-12 | blaP-1 | |||||
| Peptostreptococcus anaerobius | blaTEM-1 | ||||||
3. Discussion
The wide use of antibiotics in the clinic, agriculture, and livestock induce a high selection pressure in the worldwide microbiome allowing the resistant, multi-resistant, and pan-resistant bacteria selection and colonization of the gut of human and some other animals including insects and seafoodn [1,2,3]. In the same way, contamination affects vegetables for human consumption [26,27,28,29,30,31,32,33,34,35,36,37]. Even though many of these bacteria do not produce pathology, they are capable of transferring these antibiotic resistances mechanisms to human gut microbiota members and pathogenic bacteria, making it a reservoir for antibiotic resistance genes [37]. Following this idea, our research group characterized the beta-Lactam resistome carried by the aerobic and anaerobic Enterobacteriaceae from healthy medical students of Mexico City.
3.1. Enterobacteriaceae Characterization
The data analysis demonstrates the presence of multiple members of aerobic and anaerobic beta-lactam resistant Enterobacteriaceae, the identified bacteria distribution was as follows. Aerobic: Aeromonas hydrophila, Citrobacter freundii, Proteus mirabilis, Providencia rettgeri, Serratia fonticola, and Serratia liquefaciens. Each one represents 3.7% of all beta-lactam resistant aerobic Enterobacteriaceae, the Enterobacter aerogenes represent 7.4%, Enterococcus faecalis was present in 14.8%, Enterobacter cloacae were in the 22.2% of all isolated strands, the more abundant Enterobacteriaceae were Escherichia coli, Klebsiella pneumoniae in and Pantoea agglomerans were found in the 11.1% each one (Figure 1, Table 1 and Table 2). The founded species from human feces are in concordance with previous results were reported the repertoire and variations of human gut microbiota, they characterized 113 different bacteria, including Gram-positive bacteria (Bifidobacterium, Eubacterium, Peptostreptococcus, Ruminococcus, Lactobacillus, and Clostridium genera) and Gram-negative bacteria (Bacteroides, Fusobacteria genera) [38]. All our bacterial culture data were corroborated by NGS sequencing show 98% of concordance in the final bacterial identification. These data are supported by previously published works. Our NGS data are in concordance with other NGS studies that found 71% of all bacterial community was conformed by Firmicutes, 9% was Actinobacteria, while Bacteroidetes, Proteobacteria, and Cyanobacteria represent a relatively low abundance ranging from 3% to 7%, the bacterial families found were: Lactobacillacea, Ruminococcacea, Lancnospiraceacea, Clostridiaceacea. Streptococceacea, Streptophyta, Staphylococceacea, Verrucomicrobiaceace, and Enterobacteriaceae among others [39]. However, the composition of the intestinal microbiota varies depending on the type of diet [40]. The obtained results from feces samples reveal the potentially pathogenic enterobacteria presence in the human digestive tract in healthy conditions, their presence as the gastrointestinal resident microbiome, and not necessarily associated with an active gut infection. To determine the potential of these batteries to cause infection, it would be necessary to study other factors, such as virulence and pathogenesis genes expressed by each bacterium, bacterial load, and subjects’ immunological status.
3.2. Beta-Lactam Gene Families
Due to the isolated Enterobacteriaceae being isolated in presence of the beta-Lactam drug in the culture media (Table 2), all strains were expected to be carriers at least one member of the different gene families that code for beta-Lactamases. The endpoint PCR identified the gene families transcribed and the phenotype responsible for Beta-Lactam resistance: 100% of the isolated strain were positive to blaTEM, 10.2% positive for the blaSHV, 10.2% positive to blaKPC, 10.2% positive for blaCYM (14.3%), 2% positive to blaP, 8.2% positive to blaCFX and 6.1 positive blaBIL gene family (Table 3 and Table 4), no strand positives to members of blaOXA, blaVIM, blaIMP, blaROB, blaCTX, blaDHA, and blaLAP gene families were found (Table 3 and Table 4). These results accord with previous works, finding that Enterobacteriaceae expressed a member of blaCTX, blaSHV, and blaTEM genes families [41], while around the world the main reported beta-lactamase families in the multidrug-resistant Enterobacteriaceae are: blaKPC, blaTEM, blaOKP, blaOXA, blaSHV, blaVIM and blaNDM [42], in agreement with our results.
3.3. Beta-Lactam Phenotypes
The sequencing data shows that 83.7% of all resistant Enterobacteriaceae was a carrier of blaTEM gene family including blaTEM-1, blaTEM-2, blaTEM-10, blaTEM-12, blaTEM-24, and blaTEM-52 members, like a unique or multiple beta-lactam resistance mechanisms, for the blaSHV and blaKPC gene family we found only two members of each family including blaSHV-1, blaSHV-12, blaKPC-2 and blaKPC-3, while for families blaCMY, blaBIL, blaCTX, and blaP we only find a member of each one of them including blaCMY-2, blaBIL-1, blaCTX-M-15, and blaP-1, all of these beta-lactamases genes were found in combination with a member of the blaTEM gene family. In specific, we found that blaTEM-1 a non-xtended-spectrum beta-lactamase (ESBL) was dominant in unique beta-lactamase gene expression, while the 16.3% of all resistant Enterobacteriaceae was carrier of mixed phenotype with an ESBL, in specific in Aeromonas hydrophila (blaTEM-1 + blaSHV-12 + blaBIL-1), Enterobacter aerogenes (blaTEM-1 + blaKPC-2), Enterococcus faecalis (blaTEM-1 + blaSHV-1), Enterobacter cloacae (blaTEM-12 + blaSHV-1), Escherichia coli (blaTEM-52 + blaSHV-12 + blaCMY-2 + blaCTX-M-15), Klebsiella pneumoniae (blaTEM-10 + blaSHV-12 + blaKPC-3 + blaCMY-2 + blaCTX-M-15), Proteus mirabilis (blaTEM-1 + blaKPC-2 + blaCMY-2 + blaCTX-M-15), Pantoea agglomerans (blaTEM-1 + blaCMY-2), Serratia fonticola (blaTEM-1 + blaKPC-2), while the anaerobic bacteria that express mixed genotype were: Bacteroides distasonis (blaTEM-1 + blaBIL-1), Bacteroides fragilis (blaTEM-2 + blaCMY-2), Bacteroides vulgatus (blaTEM-1 + blaCMY-2) and Fusobacterium species (blaTEM-12 + blaP), in the NGS analysis, all bacterial strains tested were negative to blaOXA, blaVIM, blaIMP, blaROB, blaDHA and blaLAP (Table 5). These results are in agreement with a Korean report were they found that the blaTEM-1 is the most prevalent beta-lactamase expressed by the stool Enterobacteriaceae of healthy human [43]. In the same way, the Swiss study conducted with Enterobacteriaceae isolated from healthy humans found that the blaCTX-M-14 and blaCTX-M-15 are the most ESBL prevalent and are generally expressed as unique genes. Here, too, it is reported that blaTEM-1 + blaCTX-M1, blaTEM-1 + blaCTX-M-14 and blaTEM-1 + blaCTX-M-14 phenotypes are the most prevalent [44], in agreement with our results, which found that blaCTX-M-15 is always expressed in mixed genotype.
The bacterial isolates obtained were identified with simple or complex beta-lactam resistome. This is especially relevant since the resistance gene can be transmitted from among a wide range of Enterobacteriaceae through plasmid and mobile genetic elements, conferring antibiotic resistance to pathogenic bacteria that are causing gastrointestinal and systemic infection, complicating their treatment. These findings are especially important since the complex resistome expressed by the Enterobacteriaceae were integrated by one member of blaTEM gene family, including blaTEM-1, blaTEM-2, blaTEM-10, blaTEM-12, blaTEM-24, and blaTEM-52, unique or in combination one or more member of blaSHV, blaKPC, blaCMY, blaBIL, blaCTX, and blaP gen families, including blaSHV-1, blaSHV-12, blaKPC-2, blaKPC-3, blaCMY-2, blaBIL-1, blaCTX-M-15, or blaP-1 (Table 3 and Table 4), which confer resistance to ampicillin, penicillins including oxacillin, extended-spectrum cephalosporins (cefotaxime, ceftazidime), cephamycin, carbapenems, and monobactams characteristics that limit the therapeutic option of infections caused by bacterial carriers of these genes.
The data analysis showed multiple members of aerobic and anaerobic Enterobacteriaceae that show multi-resistance molecular mechanisms to beta-lactams drugs, mediated by unique resistance genes or conforming a beta-lactam resistome, in asymptomatic individuals. This can be widely explained by mechanisms such as antibiotics self-medication, abuse, and error in their medical prescription, as well as lack of adherence to antibiotic treatment, including via the consumption of contaminated water [45,46,47] or foods, mainly meats [48,49], fruits [50], and vegetables [26,51]. Moreover, the agriculture [52] and livestock industries [53,54] indiscriminately use the beta-lactam drugs to maximize production [55,56].
4. Materials and Methods
4.1. Bioinformatic Analysis and Primer Design
The bioinformatic analysis and primer design were previously described and reported by our research group [26]. Briefly, we obtained the beta-Lactamases integrons and DNA sequences from the GenBank of NCBI, all reported sequences of beta-lactamases we found were used to design primer for the PCR. The DNA sequence alignments were made with ClustalW v.2 software (http://www.clustal.org/clustal2/ accessed on 10 April 2021) [27,28,29], to obtain the phylogenetic trees by using FigTree V1.4.0 software (http://tree.bio.ed.ac.uk/software/figtree/ accessed 10 April 2021) [30]. The conserved sequences found in the alignment were used to design a set of specific and degenerate primers (PerPrimer v1.1.21 Software, http://perlprimer.sourceforge.net accessed 10 April 2021 [31] under astringent criteria, length (18–25 bp), Tm (60–62 °C), GC (40–60 %), ∆T° (1 °C), Amplicons (83–230 bp) [26].
4.2. Sampling
Twenty volunteers (10 men and 10 women) were selected for this study. All of them were medicine students from Mexico City enrolled at the Anahuac University, aged between 20 and 22 years, with no history of beta-lactam antibiotics treatment in the last year or have been suffering diarrhea during that period. Donors were provided with the sample collection material, which consisted of plastic containers, gloves, a cardboard paper sheet, and wooden spatulas; all the material was sterile. The donors were asked to deposit a portion of their stool on the cardboard sheet and from this, they collect with the wooden spatula, an approximate volume of 10 cubic cm, and place it in the sterile container. The time elapsed from when the sample was taken to its beginning of processing in the laboratory was not more than 2 h, this in order not to alter, as far as possible, the bacterial viability. Then, five g of each human feces samples were weighed and cultured in BHI (brain and heart infusion broth) (Becton Dickinson, Franklin Lakes, NJ) incubated at 37 °C for 24 h, in both aerobic and anaerobic conditions.
4.3. Bacterial Isolation
For the anaerobic conditions, the BHI tubes containing samples were immediately placed in the BD BBL ™ GasPak ™ anaerobic jar, one envelope of BD BBL ™ CO2 gas generators and another of BD BBL ™ GasPak ™ anaerobic indicator were placed on them. Later they were incubated at 37 °C during 24 h. For the aerobic conditions, the tubes with BHI containing the samples were directly incubated at 37 °C for 24 h. After is incubation, 10 μL from the BHI were taken, subcultured in MacConkey agar (Becton Dickinson, Franklin Lakes, NJ, USA), and incubated at 37 °C for 24 h. The obtained colonies were subcultured in blood agar and incubated at 37 °C for 24 h, in both aerobic or anaerobic conditions.
4.4. Biochemical Bacterial Identification
The automatized bacterial identification was made according to previously reported by our research group. Briefly, bacterial suspensions were made, by depositing two or three medium-sized colonies (2 to 3 mm) in BBL™ Crystal™ Inoculum Broth (Becton Dickinson, Franklin Lakes, NJ, USA). Said inoculum was adjusted to a Mac Farland 1.0 scale (Expected CFU/mL 3.0 × 108). The inoculum was deposited in BBL™ Crystal™ Enteric/Nonfermenter or Anaerobe ID Kit plates, incubated at 37 °C for 18 h, without CO2 and 40–60% humidity. Finally, the plates were read by the BBL™ Crystal™ AutoReader (Becton Dickinson, Franklin Lakes, NJ, USA) and the results were analyzed with the BBL™ Crystal™ MIND v.5.05 Software (Becton Dickinson, Franklin Lakes, NJ, USA) [26].
4.5. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility testing was done following a previously reported method [26]. Briefly, the pure colonies obtained from blood agar were resuspended in bacterial suspensions, and were made depositing two or three medium-sized colonies (2 to 3 mm) in BBL Crystal Inoculum Broth (Becton Dickinson; Franklin Lakes, NJ, USA). The obtained inoculum was adjusted while using a Mac Farland 0.5 reading (Expected CFU/mL 1.5 × 108), and cultured in Müeller Hinton 150 × 15 mm2 media BD BBL (Becton Dickinson Franklin Lakes, NJ, USA). The antibiotic discs were applied with the Sensi-Disc Designer Dispenser System. The antibiotics panel was conformed by ampicillin (10 µg), ampicillin/sulbactam (10/10 µg), mezlocillin (75 µg), carbenicillin (100 µg), piperacillin/tazobactam (100/10 µg), cefazolin (30 µg), cefaclor (30 µg), cefepime (30 µg), cefoperazone (75 µg), and cefotetan (30 µg) from Becton Dickinson (Franklin Lakes, NJ, USA), Müeller Hinton medium were incubated at 37 °C for 24 h in both aerobic and anaerobic conditions. In anaerobic conditions, the media were placed in an anaerobic jar (BD BBL™ GasPak™), with a C02 gas generators envelope (BD BBL™) and another envelope of BD BBL™ GasPak™ anaerobic indicator placed on them.
4.6. DNA Extraction
4.6.1. Crude Extract
Five mL of Luria-Bertani broth (LB) were inoculated with an isolated bacterial colony and incubated overnight at 37 °C with continuous shaking (200 rpm), the bacterial culture was centrifuged at 4000 rpm for five minutes at room temperature, bacterial the pellet was resuspended on 1 mL of sterile free-RNAse and free-DNAse deionized water, the bacterial suspension was heated at 94 °C for 10 min followed by ice shock. Finally, the sample of lysed bacterial was kept at –80 °C until use [26].
4.6.2. Genomic DNA Extraction
The DNA extraction was done following a previously reported method [26]. Briefly, 500 µL of bacterial culture was obtained from a tested single bacterial colony for genomic DNA isolation by RTP pathogen kit (Invitek, Germany) following the manufacturer’s instruction. The eluted DNA solution was quantified by absorbance and its integrity was verify by 2% agarose gel electrophoresis. The sample was kept at –20 °C until use.
4.6.3. Plasmid DNA Extraction
The plasmid DNA extraction was done following a previously reported method [26], by using plasmid DNA isolation by using PureLink HiPure Plasmid DNA Purification Kit (Invitrogen, USA) following the manufacturer’s instructions. Plasmid DNA solution was quantified by digital spectrophotometry by using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and its integrity was verified by electrophoresis in 2 % agarose gel, the sample was kept at –20 °C until use.
4.7. Endpoint PCR
The endpoint PCR to identify the β-Lactamases gene families was made by using previously reported specific and degenerated primes (Table 7) [26]: Amplification was made in 25 μL of the reaction mixture containing 2.5 μL of 10x PCR buffer (100 mM TRIS•HCI, 15 mM MgCl2, and 500 mM KCl, pH 8.3), 200 nM each dNTP, 10 μM each primer, 1 U Taq DNA polymerase (Invitrogen; Carlsbad, CA, USA), and 2 μL of crude extract or 10 ng of DNA (genomic or plasmidic). The PCR conditions were 94 °C for 5 min, then 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and finally 72 °C for 10 min. The PCR products were analyzed by electrophoresis in a 2% Agarose gel prestained with ethidium bromide and the image was digitized in a GelLogistic 3000 photodocumenter.
Table 7.
Primers sequences and amplicon size for beta-lactamase gene family [26].
| Gene Family | Primer Name | Primer Sequence (5’ to 3’) | Tm (°C) | Position | Amplicon (bp) |
|---|---|---|---|---|---|
| blaOXA | BlaOXA-FW | GGTTTCGGTAATGCTGAAATTGG | 61.18 | 214–236 | 114 |
| BlaOXA-RW | GCTGTGTATGTGCTAATTGGGA | 61.19 | 327–306 | ||
| blaVIM | BlaVIM-FW | CGACAGTCARCGAAATTCC | 61.39 | 105–123 | 133 |
| BlaVIM-RW | CAATGGTCTSATTGTCCGTG | 61.34 | 238–219 | ||
| blaSHV | BlaSHV-FW1 | CGTAGGCATGATAGAAATGGATC | 61.04 | 133–155 | 106 |
| BlaSHV-RW1 | CGCAGAGCACTACTTTAAAGG | 61.33 | 239–218 | ||
| BlaSHV-FW2 | GCCTCATTCAGTTCCGTTTC | 61.62 | 399–418 | 141 | |
| BlaSHV-RW2 | CCATTACCATGAGCGATAACAG | 61.22 | 540–518 | ||
| blaTEM | BlaTEM-FW | GCCAACTTACTTCTGACAACG | 61.80 | 1699–1719 | 213 |
| BlaTEM-RW | CGTTTGGAATGGCTTCATTC | 60.13 | 1912–1892 | ||
| blaIMP | BlaIMP-FW1 | GGAATAGARTGGCTTAAYTCTCG | 60.92 | 319–332 | 183 |
| BlaIMP-RW1 | CYASTASGTTATCTKGAGTGTG | 62.45 | 502–480 | ||
| BlaIMP-FW2 | GGTGGAATAGARTGGCTTAAYTC | 61.11 | 316–339 | 192 | |
| BlaIMP-RW2 | CCAAACCACTACGTTATCTKGAG | 61.29 | 508–485 | ||
| blaROB | BlaROB-FW | CCAACATCGTGGAAAGTGTAG | 61.27 | 718–739 | 126 |
| BlaROB-RW | GTAAATTGCGTACTCATGATTGC | 60.90 | 844–821 | ||
| blaKPC | BlaKPC-FW | GCTAAACTCGAACAGGACTTTG | 61.79 | 100–121 | 117 |
| BlaKPC-RW | CTTGAATGAGCTGCACAGTG | 61.90 | 216–197 | ||
| blaCTX | BlaCTX-FW1 | GATACCGCAGATAATACGCAG | 60.79 | 161–181 | 116 |
| BlaCTX-RW1 | CGTTTTGCGTTTCACTCTG | 60.28 | 276–258 | ||
| BlaCTX-FW2 | GCTGATTCTGGTCACTTACTTC | 61.02 | 789–810 | 83 | |
| BlaCTX-RW2 | CGCCGACGCTAATACATC | 60.69 | 855–872 | ||
| BlaCTX-FW3 | CTGCTTAACTACAATCCSATTGC | 62.17 | 314–336 | 226 | |
| BlaCTX-RW3 | GGAATGGCGGTATTKAGC | 60.86 | 539–522 | ||
| blaCMY | BlaCMY-FW1 | GTTTGAGCTAGGATCGGTTAG | 60.25 | 337–357 | 123 |
| BlaCMY-RW1 | CTGTTTGCCTGTCAGTTCTG | 61.48 | 460–441 | ||
| BlaCMY-FW2 | GAACGAAGGCTACGTAGCT | 61.71 | 213–231 | 160 | |
| BlaCMY-RW2 | CTGAAACGTGATTCGATCATCA | 61.08 | 372–351 | ||
| blaDHA | BlaDHA-FW1 | GCATATTGATCTGCATATCTCCAC | 61.60 | 399–422 | 200 |
| BlaDHA-RW1 | GCTGCTGTAACTGTTCTGC | 61.62 | 598–580 | ||
| BlaDHA-FW2 | GCGGATCTGCTGAATTTCTATC | 61.54 | 464–485 | 147 | |
| BlaDHA-RW2 | GCAGTCAGCAACTGCTCATAC | 61.05 | 610–591 | ||
| BlaDHA-FW3 | GTAAGATTCCGCATCAAGCTG | 61.74 | 430–450 | 117 | |
| BlaDHA-RW3 | GGGTTATCTCACACCTTTATTACTG | 61.08 | 546–522 | ||
| blaP | BlaP-FW | GGAGAATATTGGGATTACAATGGC | 61.74 | 271–294 | 204 |
| BlaP-RW | CGCATCATCGAGTGTGATTG | 61.80 | 474–455 | ||
| blaCFX | BlaCFX-FW | CCAGTCATATCATTGACAGTGAG | 60.86 | 437–459 | 177 |
| BlaCFX-RW | GACATTTCCTCTTCCGTATAAGC | 61.16 | 613–591 | ||
| blaLAP | BlaLAP-FW | AGGGCTTGAACAACTTGAAC | 61.07 | 249–268 | 126 |
| BlaLAP-RW | GTAATGGCAGCATTGCATAAC | 60.59 | 374–354 | ||
| blaBIL | BlaBIL-FW | GCCGATATCGTTAATCGCAC | 61.65 | 100–119 | 128 |
| BlaBIL-RW | GTTATTGGCGATATCGGCTTTA | 60.98 | 227–206 |
4.8. Whole and Plasmid DNA Sequencing
The DNA sequencing was performed following a previously reported method by our research group [26], the indexed libraries that were prepared using a standard Illumina Nextera XT DNA Sample Preparation Kit (FC-131-1096) for small genomes and were sequenced on the MiSeq platform (Illumina; San Diego, CA, USA). Adapters and barcodes were trimmed by the default setting in the Illumina experiment manager, generating 300-bp paired-end reads. The quality of the unprocessed reads was assessed using FastQC High Throughput Sequence QC Report v:0.11.5 (Babraham Bioinformatics, Babraham Institute; Cambridge, UK) [32]. A minimum Q score of more than 30 for at least 85% of all reads was attained. All reads were mapped using BWA-MEM aligner version 0.7.7-r441 (Wellome trust, Sanger Institute, Hinxton, UK) [33] and SAMtools version 1.3.1 (Wellome trust, Sanger Institute, Hinxton, UK). The NOVO genome assembly was done using the SPAdes Genome Assembler software version 3.11 (CAB, St. Petersburg State University, St. Petersburg, Russia) [34]. The metagenomic analysis for the taxonomic classification of bacteria was done by using the software Kraken taxonomic sequence classification system Version 0.10.5-beta (CCB, Johns Hopkins University, Baltimore, MD, USA) [35]. The beta-lactamase genes were identified by the comparative analysis while using the Basic Local Alignment Search Tool (BLAST, NCBI-NIH, Bethesda, MD, USA) [36].
5. Conclusions
The human gut bacteriome is an important reservoir and mediator for the environmental beta-lactam resistome. The cumulative beta-lactam resistome in the Enterobacteriaceae is indicative of the indiscriminate and irrational use of beta-lactam drugs in practically all economical worldwide activities, especially in Mexico City. The gut bacteriome express beta-lactam resistome in healthy conditions is integrated by multiple beta-lactamases gen families, such as blaTEM, blaSHV, blaKPC, blaCYM, blaBIL, blaCFX, blaCYM, blaBIL, and blaP, which altogether confer resistant, multi-resistant, and pan-resistant characteristics against beta-lactam antibiotics.
Acknowledgments
The authors thankfully acknowledge to María Goti Rodríguez (Principal of Hospital Regional “Primero de Octubre”) and Jose Damian Carrillo Ruiz director of the Faculty of Health Sciences, Antonio Ibarra Coordinator of CICSA, and José Pozón Director of Academic Vice-rectory Research, all of them for their support in the development of part of this research by the Universidad Anáhuac México.
Author Contributions
Conceptualization, R.V.-L. and J.A.G.-B.; methodology, D.A.Á.-H., J.A.G.-B.; software, M.L.-L.; validation, S.S.-G., C.P.-L., and J.A.G.-B.; formal analysis, R.V.-L., T.G.-G., and J.A.A.-A.; investigation, J.A.G.-B.; resources, J.A.G.-B.; data curation, R.V.-L., E.G.-C., and S.S.-G.; writing original draft preparation, R.V.-L.; writing review and editing, J.A.G.-B.; project administration, R.V.-L., and J.A.G.-B.; funding acquisition, J.A.G.-B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Council for Science and Technology (CONACYT) Mexico through the Sectoral Fund for Research in Health and Social Security (FOSISS), grant MOD.ORD./2012/SSA/IMSS/ISSSTE-CONACYT/000000000181384 “and it was also supported by CICSA of Faculty of Health Sciences of the Universidad Anáhuac México Project Number 201917.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Facultad de Ciencias de la Salud Universidad Anáhuac Mexico Campus Norte and of the “1 October” Regional Hospital, ISSSTE, approval code E015-591 (13 January 2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kardas P., Devine S., Golembesky A., Roberts C. A systematic review and meta-analysis of misuse of antibiotic therapies in the community. Int. J. Antimicrob. Agents. 2005;26:106–113. doi: 10.1016/j.ijantimicag.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 2.Kadima M.G. Master’s Thesis. University of KwaZulu-Natal; Scottsville, Pietermaritzburg, South Africa: Jan, 2019. Concurrent Use of Traditional Medicine and Prescribed Antibiotics and/or Antiviral Medicines and Their Effects on Antimicrobial Resistance and Treatment Failure in Ilembe District, Kwazulu-Natal Province, South Africa. [Google Scholar]
- 3.Huttner A., Harbarth S., Carlet J., Cosgrove S., Goossens H., Holmes A., Jarlier V., Voss A., Pittet D. Antimicrobial resistance: A global view from the 2013 World Healthcare-Associated Infections Forum. Antimicrob. Resist. Infect. Control. 2013;2:31. doi: 10.1186/2047-2994-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucien M.A.B., Canarie M.F., Kilgore P.E., Jean-Denis G., Fénélon N., Pierre M., Cerpa M., Joseph G.A., Maki G., Zervos M.J., et al. Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings. Int. J. Infect. Dis. 2021;104:250–254. doi: 10.1016/j.ijid.2020.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z., Guo J., Jiang Y., Shao Y. High concentration and high dose of disinfectants and antibiotics used during the COVID-19 pandemic threaten human health. Environ. Sci. Eur. 2021;33:1–14. doi: 10.1186/s12302-021-00456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K., Bradford P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020;33:e00047-19. doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera-Tapia J.A. Antibiotic resistance, public health problem. An. Med. Asoc. Med. Hosp. ABC. 2003;48:42–47. [Google Scholar]
- 8.Kang C.-I., Kim S.-H., Park W.B., Lee K.-D., Kim H.-B., Kim E.-C., Oh M.-D., Choe K.W. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: Risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob. Agents Chemother. 2005;49:760–766. doi: 10.1128/AAC.49.2.760-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zilberberg M.D., Shorr A.F., Micek S.T., Vazquez-Guillamet C., Kollef M.H. Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in Gram-negative severe sepsis and septic shock: A retrospective cohort study. Crit. Care. 2014;18:596. doi: 10.1186/s13054-014-0596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor A., Abate B.A., Deák P., Fodor L., Gyenge E., Klein M.G., Koncz Z., Muvevi J., Ötvös L., Székely G., et al. Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review. Pathogens. 2020;9:522. doi: 10.3390/pathogens9070522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong K.-F., Schneper L., Mathee K. Beta-lactam antibiotics: From antibiosis to resistance and bacteriology. APMIS. 2010;118:1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caruano J., Muccioli G.G., Robiette R. Biologically active γ-lactams: Synthesis and natural sources. Org. Biomol. Chem. 2016;14:10134–10156. doi: 10.1039/C6OB01349J. [DOI] [PubMed] [Google Scholar]
- 13.Lee E., Lim D.Y., Lee K.A. Synthesis of A δ-Lactam Analogue of Penem Carboxylate. Bull. Korean Chem. Soc. 1990;11:361–362. [Google Scholar]
- 14.Dutton F.E., Lee B.H. Epsilon-lactam analogs of the anthelmintic cyclodepsipeptide PF1022A. Tetrahedron Lett. 1998;39:5313–5316. doi: 10.1016/S0040-4039(98)01075-2. [DOI] [Google Scholar]
- 15.Heesemann J. Resistenzmechanismen gegen Betalaktamantibiotika. Infection. 1993;21:S4–S9. doi: 10.1007/BF01710336. [DOI] [PubMed] [Google Scholar]
- 16.Mathers A.J., Peirano G., Pitout J.D.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coque T.M., Baquero F., Cantón R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance. 2008;13:19044. doi: 10.2807/ese.13.47.19044-en. [DOI] [PubMed] [Google Scholar]
- 18.Vatopoulos A. High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece—A review of the current evidence. Eurosurveillance. 2008;13:7–8. doi: 10.2807/ese.13.04.08023-en. [DOI] [PubMed] [Google Scholar]
- 19.Brolund A., Sandegren L. Characterization of ESBL disseminating plasmids. Infect. Dis. 2015;48:18–25. doi: 10.3109/23744235.2015.1062536. [DOI] [PubMed] [Google Scholar]
- 20.De Angelis G., Del Giacomo P., Posteraro B., Sanguinetti M., Tumbarello M. Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in enterobacteriaceae. Int. J. Mol. Sci. 2020;21:5090. doi: 10.3390/ijms21145090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultsz C., Geerlings S. Plasmid-mediated resistance in enterobacteriaceae: Changing landscape and implications for therapy. Drugs. 2012;72:1–16. doi: 10.2165/11597960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 22.El Salabi A., Walsh T.R., Chouchani C. Extended spectrum β-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gram-negative bacteria. Crit. Rev. Microbiol. 2013;39:113–122. doi: 10.3109/1040841X.2012.691870. [DOI] [PubMed] [Google Scholar]
- 23.Perez F., El Chakhtoura N.G., Papp-Wallace K.M., Wilson B.M., Bonomo R.A. Treatment options for infections caused by carbapenem-resistant Enterobacteriaceae: Can we apply “precision medicine” to antimicrobial chemotherapy? Expert Opin. Pharm. 2016;17:761–781. doi: 10.1517/14656566.2016.1145658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Codjoe F.S., Donkor E.S. Carbapenem Resistance: A Review. Med. Sci. 2017;6:1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills M.C., Lee J. The threat of carbapenem-resistant bacteria in the environment: Evidence of widespread contamination of reservoirs at a global scale. Environ. Pollut. 2019;255:113143. doi: 10.1016/j.envpol.2019.113143. [DOI] [PubMed] [Google Scholar]
- 26.Vázquez-López R., Solano-Gálvez S., León-Chávez B.A., Thompson-Bonilla M.R., Guerrero-González T., Gómez-Conde E., Martínez-Fong D., González-Barrios J.A. Characterization of gene families encoding beta-lactamases of gram-negative rods isolated from ready-to-eat vegetables in Mexico City. High. Throughput. 2018;7:36. doi: 10.3390/ht7040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauretti L., Riccio M.L., Mazzariol A., Cornaglia G., Amicosante G., Fontana R., Rossolini G.M. Cloning and characterization of bla(VIM), a new integron-borne metallo- β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999;43:1584–1590. doi: 10.1128/AAC.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de M. Rangel W., de Oliveira Longatti S.M., Ferreira P.A.A., Bonaldi D.S., Guimarães A.A., Thijs S., Weyens N., Vangronsveld J., Moreira F.M.S. Leguminosae native nodulating bacteria from a gold mine As-contaminated soil: Multi-resistance to trace elements, and possible role in plant growth and mineral nutrition. Int. J. Phytoremediation. 2017;19:925–936. doi: 10.1080/15226514.2017.1303812. [DOI] [PubMed] [Google Scholar]
- 29.Sievert D.M., Ricks P., Edwards J.R., Schneider A., Patel J., Srinivasan A., Kallen A., Limbago B., Fridkin S. National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 30.Centers for Disease Control and Prevention Vital Signs: Carbapenem-Resistant Enterobacteriaceae. [(accessed on 12 April 2021)]; Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6209a3.htm.
- 31.Ye Q., Wu Q., Zhang S., Zhang J., Yang G., Wang J., Xue L., Chen M. Characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from retail food in China. Front. Microbiol. 2018;9:1709. doi: 10.3389/fmicb.2018.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babraham Bioinformatics—FastQC Babraham Bioinformatics—FastQC A Quality Control tool for High Throughput Sequence Data. [(accessed on 12 April 2021)]; Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 33.Burrows-Wheeler Aligner Burrows-Wheeler Aligner / [Bio-bwa-help] Citing bwa-mem. [(accessed on 12 April 2021)]; Available online: https://sourceforge.net/p/bio-bwa/mailman/message/30894287/
- 34.CodonCode Corporation Sequence Assembly and Alignment with CodonCode Aligner. [(accessed on 12 April 2021)]; Available online: https://www.codoncode.com/aligner/
- 35.System K.T.S.C. Kraken Taxonomic Sequence Classification System. [(accessed on 12 April 2021)]; Available online: https://ccb.jhu.edu/software/kraken/
- 36.BLAST BLAST: Basic Local Alignment Search Tool. [(accessed on 12 April 2021)]; Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 37.Hu Y., Yang X., Qin J., Lu N., Cheng G., Wu N., Pan Y., Li J., Zhu L., Wang X., et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013;4:2151. doi: 10.1038/ncomms3151. [DOI] [PubMed] [Google Scholar]
- 38.Lagier J.C., Million M., Hugon P., Armougom F., Raoult D. Human gut microbiota: Repertoire and variations. Front. Cell. Infect. Microbiol. 2012;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parma L., Candela M., Soverini M., Turroni S., Consolandi C., Brigidi P., Mandrioli L., Sirri R., Fontanillas R., Gatta P.P., et al. Next-generation sequencing characterization of the gut bacterial community of gilthead sea bream (Sparus aurata, L.) fed low fishmeal based diets with increasing soybean meal levels. Anim. Feed Sci. Technol. 2016;222:204–216. doi: 10.1016/j.anifeedsci.2016.10.022. [DOI] [Google Scholar]
- 40.Tidjani Alou M., Lagier J.-C., Raoult D. Diet influence on the gut microbiota and dysbiosis related to nutritional disorders. Hum. Microbiome J. 2016;1:3–11. doi: 10.1016/j.humic.2016.09.001. [DOI] [Google Scholar]
- 41.Korzeniewska E., Harnisz M. Beta-lactamase-producing Enterobacteriaceae in hospital effluents. J. Environ. Manag. 2013;123:1–7. doi: 10.1016/j.jenvman.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 42.Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 2010;13:558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Joo E.-J., Kim S.J., Baek M., Choi Y., Seo J., Yeom J.-S., Ko K.S. Fecal carriage of antimicrobial-resistant enterobacteriaceae in healthy Korean adults. J. Microbiol. Biotechnol. 2018;28:1178–1184. doi: 10.4014/jmb.1801.12060. [DOI] [PubMed] [Google Scholar]
- 44.Geser N., Stephan R., Korczak B.M., Beutin L., Hächler H. Molecular identification of extended-spectrum-β-lactamase genes from Enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob. Agents Chemother. 2012;56:1609–1612. doi: 10.1128/AAC.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cacace D., Fatta-Kassinos D., Manaia C.M., Cytryn E., Kreuzinger N., Rizzo L., Karaolia P., Schwartz T., Alexander J., Merlin C., et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019;162:320–330. doi: 10.1016/j.watres.2019.06.039. [DOI] [PubMed] [Google Scholar]
- 46.Adesoji A.T., Ogunjobi A.A. Detection of Extended Spectrum Beta-Lactamases Resistance Genes among Bacteria Isolated from Selected Drinking Water Distribution Channels in Southwestern Nigeria. Biomed. Res. Int. 2016;2016:7149295. doi: 10.1155/2016/7149295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delgado-Gardea M.C.E., Tamez-Guerra P., Gomez-Flores R., de la Serna F.J.Z.-D., Eroza-de la Vega G., Nevárez-Moorillón G.V., Pérez-Recoder M.C., Sánchez-Ramírez B., González-Horta M.D.C., Infante-Ramírez R. Multidrug-resistant bacteria isolated from surface water in Bassaseachic Falls National Park, Mexico. Int. J. Environ. Res. Public Health. 2016;13:597. doi: 10.3390/ijerph13060597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trott D. β-lactam Resistance in Gram-negative Pathogens Isolated from Animals. Curr. Pharm. Des. 2012;19:239–249. doi: 10.2174/1381612811306020239. [DOI] [PubMed] [Google Scholar]
- 49.Chen C.-M., Ke S.-C., Li C.-R., Wu Y.-C., Chen T.-H., Lai C.-H., Wu X.-X., Wu L.-T. High Diversity of Antimicrobial Resistance Genes, Class 1 Integrons, and Genotypes of Multidrug-Resistant Escherichia coli in Beef Carcasses. Microb. Drug Resist. 2017;23:915–924. doi: 10.1089/mdr.2016.0223. [DOI] [PubMed] [Google Scholar]
- 50.Heaton J.C., Jones K. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: A review. J. Appl. Microbiol. 2008;104:613–626. doi: 10.1111/j.1365-2672.2007.03587.x. [DOI] [PubMed] [Google Scholar]
- 51.Gudda F.O., Waigi M.G., Odinga E.S., Yang B., Carter L., Gao Y. Antibiotic-contaminated wastewater irrigated vegetables pose resistance selection risks to the gut microbiome. Environ. Pollut. 2020;264:114752. doi: 10.1016/j.envpol.2020.114752. [DOI] [PubMed] [Google Scholar]
- 52.Iwu C.D., Korsten L., Okoh A.I. The incidence of antibiotic resistance within and beyond the agricultural ecosystem: A concern for public health. Microbiologyopen. 2020;9:e1035. doi: 10.1002/mbo3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tekiner İ.H., Özpınar H. Occurrence and characteristics of extended spectrum beta-lactamases-producing enterobacteriaceae from foods of animal origin. Braz. J. Microbiol. 2016;47:444–451. doi: 10.1016/j.bjm.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mesa R.J., Blanc V., Blanch A.R., Cortés P., González J.J., Lavilla S., Miró E., Muniesa M., Saco M., Tórtola M.T., et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage) J. Antimicrob. Chemother. 2006;58:211–215. doi: 10.1093/jac/dkl211. [DOI] [PubMed] [Google Scholar]
- 55.Ruppé E., Andremont A., Armand-Lefèvre L. Digestive tract colonization by multidrug-resistant Enterobacteriaceae in travellers: An update. Travel Med. Infect. Dis. 2018;21:28–35. doi: 10.1016/j.tmaid.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Armand-Lefèvre L., Andremont A., Ruppé E. Travel and acquisition of multidrug-resistant Enterobacteriaceae. Med. Mal. Infect. 2018;48:431–441. doi: 10.1016/j.medmal.2018.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available from the corresponding author.