Abstract
Cotton is one of the most important fiber and oil crops in the world. Chloroplast genomes harbor their own genetic materials and are considered to be highly conserved. Transfer RNAs (tRNAs) act as “bridges” in protein synthesis by carrying amino acids. Currently, the variation and evolutionary characteristics of tRNAs in the cotton chloroplast genome are poorly understood. Here, we analyzed the structural variation and evolution of chloroplast tRNA (cp tRNA) based on eight diploid and two allotetraploid cotton species. We also investigated the nucleotide evolution of chloroplast genomes in cotton species. We found that cp tRNAs in cotton encoded 36 or 37 tRNAs, and 28 or 29 anti-codon types with lengths ranging from 60 to 93 nucleotides. Cotton chloroplast tRNA sequences possessed specific conservation and, in particular, the Ψ-loop contained the conserved U-U-C-X3-U. The cp tRNAs of Gossypium L. contained introns, and cp tRNAIle contained the anti-codon (C-A-U), which was generally the anti-codon of tRNAMet. The transition and transversion analyses showed that cp tRNAs in cotton species were iso-acceptor specific and had undergone unequal rates of evolution. The intergenic region was more variable than coding regions, and non-synonymous mutations have been fixed in cotton cp genomes. On the other hand, phylogeny analyses indicated that cp tRNAs of cotton were derived from several inferred ancestors with greater gene duplications. This study provides new insights into the structural variation and evolution of chloroplast tRNAs in cotton plants. Our findings could contribute to understanding the detailed characteristics and evolutionary variation of the tRNA family.
Keywords: cotton, chloroplast tRNA, evolution, phylogenetic relationship, structural variation
1. Introduction
Chloroplasts (cp) are unique oblate organelles equipped with important photosynthesis functions [1]. Besides the complete photosynthetic components, they have also been reported as active organelles participating in other diverse biomolecule processes including biosynthesis of starch, fatty acids, pigments, amino acids, etc. [2,3,4]. Some previous studies found that chloroplasts originated from cyanobacterial ancestors [5,6]. For most plants, the cp genome is a double-stranded circular unit involving four main regions: two inverted repeats (IRs) separated by a large single copy (LSC) region and a small single copy (SSC) region [7,8]. In addition, the features of non-recombination and maternal inheritance (in most angiosperms) of the cp genome provide the opportunity to employ it as material for evolutionary and genomics research [9,10]. There are many genes, such as protein-coding genes and multiple tRNA genes, present in the cp genome [11]. Relevant studies have shown that most of these genes play a role in the organization of photosynthesis and biochemical reactions [12,13,14]. However, it is worth noting that the genetic and mutational characteristics of cp genes, especially cp tRNA genes in angiosperms, still need research.
Generally, single nucleotide polymorphisms (SNPs) and insertions/deletions are the basis of the differences between most alleles. With the characteristics of high abundance, a fairly low mutation rate, and the adaptability to automatic genotyping, SNPs are employed more frequently than other genetic markers, such as microsatellites [15]. Insertions and deletions (indels) are essential sources of polymorphic markers for high-resolution genetic mapping of traits and association studies based on candidate genes or possibly the whole genome [16]. Furthermore, SNPs and indels are also important in the nucleotide sequence evolution analysis of genomes. The calculation of single nucleotide variation may help estimate and comprehend genetic variations of different genome regions.
According to previous studies, the residue polynucleotide sequences of tRNAs fold back, forming clover leaf-like structures with hydrogen bonds, and then turn into L-shaped tertiary structures [17]. The secondary structure of tRNA contains an acceptor arm, dihydrouridine arm (D-arm), dihydrouridine loop (D-loop), anti-codon arm, anti-codon loop, variable loop, pseudouridine arm (Ψ-arm), and pseudouridine loop (Ψ-loop) [18,19]. With this specific structure, tRNA plays an important role in protein synthesis by carrying amino acids to the ribosome [20,21]. Recently, researchers conducted a series of analyses related to tRNAs on the genomic level. Asymmetric combinations and divided segments in tRNA genes would help to understand the diversity of tRNA molecules [22]. Additionally, studies showed that the heterogeneous tRNA fragments play multiple roles in terms of size, nucleotide composition, biogenesis, and even biological disease [23]. Additionally, wobble modifications were frequently found in tRNAs in diverse species after the discovery of tRNA molecules [24,25,26]. In recent years, studies on the genomic structure and evolution of tRNAs in plants has been attracting researchers’ attention. For example, analyses of detailed molecular aspects in cyanobacterial tRNAs, complete genomic features of tRNAs in Oryza sativa L, and the evolutionary perspective of chloroplast tRNAs (cp tRNAs) in some economic monocots were previously conducted [27,28,29].
Gossypium L., or the cotton plant, is an important economic and oil crop that has been cultivated worldwide. Gossypium is a large genus belonging to the angiosperm family Malvaceae. Fryxell divided the cotton genus into four subgenera, with a total of 51 species, including 46 diploid (2n = 2× = 26) and five tetraploid (2n = 4× = 52) species [30]. The chromosome composition of tetraploid cotton is heterogeneous A and D: Gossypium hirsutum L. (AD1), G. barbadense L. (AD2), G. tomentosum Nuttalex Seemann (AD3), G. mustelinum Miersex Watt (AD4), and G. darwinii Watt (AD5). A total of eight chromosomes of diploid cotton belong to groups A–G and K. Among them, G. hirsutum, G. barbadense, G. arboreum L., and G. herbaceum L. are cultivars, while the others are wild species. Nowadays, the most widely cultivated cotton species are tetraploid G. hirsutum and G. barbadense [31]. At present, with the rapid development of genome sequencing technology, whole-genome sequences of cotton have been released, which provide the foundation for further analysis of cotton genomes [32,33]. The genome-wide landscape of genomic variation of cotton was constructed through SNP distribution density detection, and functional genes that encode proteins involved in regulation of tissue growth, stress responses, and disease resistance were reported [34,35,36,37,38,39]. Moreover, a previous study on variations of repeat sequences and cp evolutionary relationships detected divergence hotspots in plastid genomes and helped to understand phylogenetic relationships among major Gossypium lineages [40]. However, the evolutionary patterns of Gossypium chloroplast tRNAs are still unclear. The study of the genomic and evolutionary characteristics of cotton cp tRNAs seems to be significant.
In this study, we investigated ten globally representative cotton cp genomes, including eight diploid and two allotetraploid species. The aims of our study were: (1) identify the genomic characteristics and diversification of cp tRNAs in cotton; (2) analyze the evolutionary relationship of introns in cp tRNA genes; (3) estimate the evolutionary characteristics of SNPs and the indel mutation rate of the cotton chloroplast genome; and (4) investigate the evolutionary pattern of cp tRNAs in cotton species.
2. Materials and Methods
2.1. Identification of tRNAs
We downloaded the 10 cotton cp genomes (8 diploid and 2 allotetraploid species: Gossypium arboreum L., G. anomalum Wawra and Peyritsch, G. robinsonii (F. Muell.) J. H. Willis, G. klotzschianum Andersson, G. somalense (Gurke) J. B. Hutch., G. longicalyx Hutchinson and Lee, G. bickii Prokhanov, G. populifolium (Benth.) F. Muell., G. hirsutum L., and G. barbadense L.) from the National Center of Biotechnology Information (NCBI) (Table 1). These cotton species are widely distributed near the equator and G. hirsutum, G. barbadense, and G. klotzschianum are mainly distributed in the Americas. Subsequently, tRNA gene sequences were identified and extracted from cp genomes without intergenic regions by the GENEIOUS 8.0.2. program [41].
Table 1.
Gossypium species information in the study.
| Karyotype | Species | Accession Number | Wild/Cultivars |
|---|---|---|---|
| A2 | Gossypium arboreum | NC_016712 | cultivars |
| B1 | G. anomalum | NC_023213 | wild |
| C2 | G. robinsonii | NC_018113 | wild |
| D3-k | G. klotzschianum | NC_033394 | wild |
| E2 | G. somalense | NC_018110 | wild |
| F1 | G. longicalyx | NC_023216 | wild |
| G1 | G. bickii | NC_023214 | wild |
| K2 | G. populifolium | NC_033398 | wild |
| AD1 | G. hirsutum | HQ901196 | cultivars |
| AD2 | G. barbadense | HQ901199 | cultivars |
2.2. Structural Analysis of tRNAs
ARAGORN and tRNAScan-SE software [42,43] were employed to investigate the secondary structure of tRNA sequences of cp genomes. The default parameters of ARAGORN software were set to investigate tRNAs. The parameters of tRNAScan-SE were set as: bacterial for sequence source, default for search mode, formatted (FASTA) for query sequences, and universal for genetic code for tRNA isotype prediction.
2.3. Sequence Alignment
To identify the presence of consensus sequences, sequence alignments were carried out for the intron sequences of the cotton cp tRNAs, Pinus armandii Franch., Marchantia polymorpha L., Raphanus sativus L., Spirogyra maxima (Hassall) Wittrock, Alsophila spinulosa (Wallich ex Hooker) R. M. Tryon Contr. Gray Herb., Zea mays L., Gleocapsa sp. PCC 73106, Nostoc sp. PCC 7107, and Nostoc sp. PCC 7524 using Multalin software, in which default parameters were set [44,45,46].
2.4. Phylogenetic Tree Construction
The phylogenetic tree was constructed using MEGA7.0 software [47,48]. To investigate the evolution of chloroplast tRNAs, a matrix of whole tRNA sequences was created by Clustal Omega software before the phylogenetic tree was constructed. MEGA7 software was employed to turn the matrix file into MEGA file format for tree construction, in which the lowest Bayesian information criterion (BIC) was selected for the model. As a result, the Kimura2 + G + I model was found to have the lowest BIC score, 8980.50. Thus, this model was adopted for the phylogenetic tree construction. The other related parameters were as follows: phylogeny reconstruction for analysis, maximum likelihood model, bootstrap method in phylogeny test, 1000 bootstrap replicates, nucleotide type, γ distributed with invariant sites (G + I) model, 5 discrete γ categories, partial deletion for gaps/missing data treatment, 95% site coverage cutoff, and very strong for branch swap filter.
2.5. Analysis of Disparity Index
A disparity index test of pattern heterogeneity was conducted to check the homogeneity of nucleotide substitutions and find whether all substitutions in nucleotides happened at equal rates, i.e., homogeneity in the process of evolution. The statistical parameters were as follows based on a previous study [29]: disparity index test for substitution pattern homogeneity, in sequence pairs, 10,001 Monte Carlo replications, nucleotide for substitution type, partial deletion of gaps/missing data treatment, and site coverage cutoff 95%.
2.6. Transition/Transversion Analysis
The transition and transversion rates of the tRNAs genes were analyzed according to their isotypes by MEGA 7 software [49]. The parameters were set as follows: substitution pattern estimation (ML) for analysis, automatic (neighbor-joining tree), maximum likelihood statistical method, nucleotide for substitution type, Kimura 2-parameter model, γ distributed (G) site rates, 5 discrete γ categories, partial deletion of gaps/missing data treatment, 95% of site coverage cutoff, and very strong branch swap filter.
2.7. Evolutionary Analysis of Single Nucleotide Polymorphisms
The diversity of single nucleotide polymorphisms of 8 diploid Gossypium cp genomes was calculated based on 3 regions: coding regions, introns, and intergenic spacers. The synonymous (dS) and non-synonymous (dN) substitutions of coding genes were also calculated by DnaSP v5.10 software [50]. The coding regions, introns, and intergenic regions of the studied Gossypium cp genome were extracted through Geneious 8.0.2 software and aligned manually [41].
2.8. Calculation of Mutation Rate
The rate of indel mutation (μ, per site per year) was calculated with the formula:
| μ = m/(nT), | (1) |
where m is the number of sites of observed mutation, n is the total number of sites, and T is the divergence time of Gossypium. The μ value of structural mutations was calculated according to the method of Saitou and Ueda [51]. The T value was obtained through relevant published literature searches in the Fossil works database and the Cenozoic Angiosperm Database [52,53]. Additionally, the mutation rates of protein-coding genes and tRNA genes were calculated.
2.9. Duplication/Loss Analysis of tRNA Genes
To investigate duplication or loss events of the tRNAs, we used the NCBI taxonomy browser to construct the species tree of G. arboreum, G. anomalum, G. robinsonii, G. klotzschianum, G. somalense, G. longicalyx, G. bickii, G. hirsutum, G. barbadense, and G. populifolium. Additionally, the previously constructed phylogenetic tree of the tRNAs was employed as the gene tree. Subsequently, Notung 2.9 software [54,55] was used to reconcile the gene tree and the species tree and obtain the gene duplication and loss nodes.
3. Results
3.1. Basic Characteristics of Cotton Chloroplast tRNAs
The results of detailed genomic analysis showed that G. arboreum, G. anomalum, G. robinsonii, G. klotzschianum, G. somalense, G. hirsutum, G. barbadense, G. longicalyx, and G. populifolium coded 37 tRNAs, respectively, while only G. bickii coded 36 tRNAs (Table 2). The length of the chloroplast tRNAs ranged from 60 nt (tRNAGly in G. arboreum, GCC) to 93 nt (tRNASer, UGA), with an average length of 76 nt (Table S1).
Table 2.
Distribution of tRNA isotypes in cotton chloroplast genome.
| tRNA Isotype | Number of tRNAs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A2 1 | B1 2 | C2 3 | D3-K 4 | E2 5 | F1 6 | AD1 7 | AD2 8 | G1 9 | K2 10 | |
| Alanine | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Glycine | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Proline | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Threonine | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Valine | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Serine | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Arginine | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Leucine | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Phenylalanine | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Asparagine | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Lysine | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Aspartate | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Glutamate | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Histidine | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Glutamine | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Isoleucine | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Methionine | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| Tyrosine | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Cysteine | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Tryptophan | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Selenocysteine | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Suppressor | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 37 | 37 | 37 | 37 | 37 | 37 | 37 | 37 | 36 | 37 |
1Gossypium arboretum; 2G. anomalum; 3G. robinsonii; 4G. klotzschianum; 5G. somalense; 6G. longicalyx; 7G. hirsutum; 8G. barbadense; 9G. bickii; 10G. populifolium.
The genomic analysis results showed that chloroplast tRNA genes of the investigated cotton plants coded 28 or 29 anti-codon types, of which only G. anomalum, G. longicalyx, and G. bickii coded 29 anti-codons. The most common anti-codons observed in cp tRNAs were UGC-tRNAAla, GCC-tRNAGly, GAC-tRNAVal, ACG-tRNAArg, CAA-tRNALeu, GUU-tRNAAsn, CAU-tRNAIle, GAU-tRNAIle, and CAU-tRNAMet. Additionally, each of these genes, except GCC-tRNAGly and CAU-tRNAMet, had two copies (Table S2). GCC (tRNAGly) was found with one copy in G. anomalum, G. longicalyx, and G. bickii but two copies in the other cotton species. Similarly, CAU (tRNAMet) was found with one copy in G. bickii but two copies in the other cotton species. There were 33 tRNAs with various iso-acceptors missing in the cp genome of Gossypium (Table S2). UCC-tRNAGly was observed in G. anomalum, G. longicalyx, and G. bickii. All investigated cotton species were shown to possess at least one anti-codon type for each kind of tRNA. Furthermore, the anti-codon CAU was a typical characteristic of tRNAMet, which harbored only one type of iso-acceptor. Apart from the existence of the anti-codon CAU in tRNAMet, tRNAIle was also observed to encode the anti-codon CAU in the cotton cp genome (Table S2).
All the tRNA gene families were analyzed by multiple sequence alignment, from which the limited conservation and consensus sequences were found in the Ψ-arm and Ψ-loop. The Ψ-arm of tRNAs was observed to contain the G-G consensus sequence, and the Ψ-loop was observed to contain the conserved sequence U-U-C-X3-U (Table 3).
Table 3.
Sequence alignment and the presence of isotype-specific conserved nucleotide consensus sequences in the cotton chloroplast tRNAs.
| tRNA Isotype | AC-Arm | D-Arm | D-Loop | ANC-Arm | ANC-Loop | Variable Region | Ψ-Arm | Ψ-Loop |
|---|---|---|---|---|---|---|---|---|
| Alanine | GGGGAUA | GCUC | AGUUGGUA | CCGCU | CUUGCAU | AUGUC | AGCGG | UUCGAGU |
| Arginine | GXGXCX2 | Gx3 | AX2GGAUA | ***** | CUXCXAA | GU | GG | UUCGAAU |
| Asparagine | GUCGGGA | GCUC | AGUUGGUA | GUCGG | CUGUUAA | UGGUC | GUAGG | UUCGAAU |
| Aspartate | GGGAUUG | GUUC | AAUUGGUCA | CCGCC | CUGUCAA | AAGCU | GCGGG | UUCGAGC |
| Cysteine | GGCGACA | GCC | GAGCGGUAA | GGGGA | CUGCAAA | UAUUC | CCCAG | UUCAAAU |
| Glutamate | GX2CX3 | GX3 | AGXGGUX1–3 | CX2CX | CUUUCAX | X2GX1–2 | X3GX | UUCXAXU |
| Glutamine | UGGGGCG | GCC | AAGUGGUAA | CGGG | UUUUGGU | CUAUGC | GGAGG | UUCGAAU |
| Glycine | GCGGAUA | GU | CGAAUGGUAAA | UCUCU | UUGCCAA | AGAC | GCGGG | UUCGAUU |
| Histidine | GCGGAUG | GCC | AAGUGGAUCAA | GUGGA | UUGUGAA | CAUGC | GCGGG | UUCAAUU |
| Isoleucine | GCAUCCA | GCU | GAAUGGUUAA | CCCAA | CUCAUAA | AAUUC | GUAGG | UUCAAUU |
| Leucine | GX6 | GXG | AAAUXGX3–4A | X3GX | CUX4A | XGX9–12 | X3GG | UUCXAGU |
| Lysine | GGGUUGC | ACUC | AACGGUA | UCGG | CUUUUAA | CUAGUU | CCGGG | UUCGAGU |
| Methionine | XCX5–6 | X3G | AGUX5–6 | ***** | XUCAUAX | X2GUC | AUXGG | UUCAAAU |
| Phenylalanine | GUCGGGA | GCUC | AGUUGGUA | GAGGA | CUGAAAA | GUGUC | ACCAG | UUCAAAU |
| Proline | AGGGAUG | GCGC | AGCUUGGUA | UUUGU | UUUGGGU | AUGUC | ACGGG | UUCAAAU |
| Serine | GGAGAGA | GCX1–2 | X4GX3–4A | X2GX1–2 | XUXGXAX | X4GX15–19 | GAGGG | UUCGAAU |
| Threonine | XGCCX0–4 | XCUC | AGXGGUA | XCGCX | X3GUAA | X2GUC | AUCGG | UUCX3U |
| Tryptophan | GCGCUCU | GUUC | AGUUCGGUA | UGGGU | CUCCAAA | AUGUC | GUAGG | UUCAAAU |
| Tyrosine | GGGUCGA | CCCG | AGCGGUUAA | ACGGA | CUGUAAA | GGCA | GCUGG | UUCAAAU |
| Valine | AGGGAUA | ACUC | AGCGGUA | UCACC | UUGACGU | AAGUC | AUCAG | UUCGAGC |
Note: The asterisk (*****) shows the absence of conserved nucleotide consensus sequence in the respective region of the chloroplast tRNAs.
Most of the tRNAs possessed a G nucleotide at the first position of the acceptor arm, while tRNAGln, tRNAPro, and tRNAVal were observed to contain a U and an A (Table 3). Except for tRNALys, tRNAMet, tRNAThr, tRNATyr, and tRNAVal, other tRNAs contained a G in the first nucleotide of the D-arm. Additionally, there was usually an A at the last position of the D-loop and ANC-loop. At the final site of the D-arm, except for tRNAArg, tRNAGlu, tRNAIle, tRNALeu, tRNAMet, tRNASer, and tRNATyr, the other investigated tRNAs were found to have a C (Table 3). Additionally, we also observed that all tRNALys, tRNAAla, tRNAGlu, tRNAArg, tRNATyr, and a few tRNALeu had a C-C-A tail in the 3′ end.
3.2. Diversification of tRNA Structure
The tRNA feature of possessing various arms and loops was responsible for protein translation. The results of structure analysis showed that the acceptor arm in cotton chloroplast tRNA contained 6 to 7 nt. Among the 369 investigated tRNA sequences, only nine were found to contain 6 nt, while the remaining 360 tRNAs (97.56%) contained 7 nt (Table S3). In addition, the D-arm was observed to contain 2 to 4 nt, among which 20 contained only 2 nt, and 110 (29.81%) contained 3 nt. The remaining tRNAs (64.50%) were observed to possess 4 nt in the D-arm. Moreover, the D-loop contained 7 to 11 nt. For all the involved tRNAs, 84 contained 7 nt in their D-loops; 69 (18.70%) contained 8; 105 (28.46%) contained 9; 40 (10.84%) contained 10; and the rest (18.98%) contained 11 nt (Table S3).
In all 369 tRNAs, the anti-codon arm had mainly 4 to 5 nt. Among them, 319 (86.45%) had 5 nt, and 50 (13.55%) had 4 nt. In addition, 359 tRNAs (97.29%) contained 7 nt in their anti-codon loops and 10 (only 2.71%) contained 9 nt. This showed that the conservative sequence of anti-codon loops was rather typical (Table 3 and Table S3).
For variable loops, 10 tRNAs (2.71%) contained 2; 30 tRNAs (8.13%) contained 3, 67 tRNAs (18.16%) contained 4; 213 tRNAs (57.72%) contained 5; 39 tRNAs (10.57%) contained 6; and 10 tRNAs (2.71%) contained 8 nt (Table S3). While the Ψ-arms of all the analyzed chloroplast tRNAs were observed to contain 5 nt, most tRNAs (349, 94.58%) had 7 nt in their Ψ-loops, apart from several tRNAArg (Table S3).
3.3. Chloroplast tRNA Contained Introns
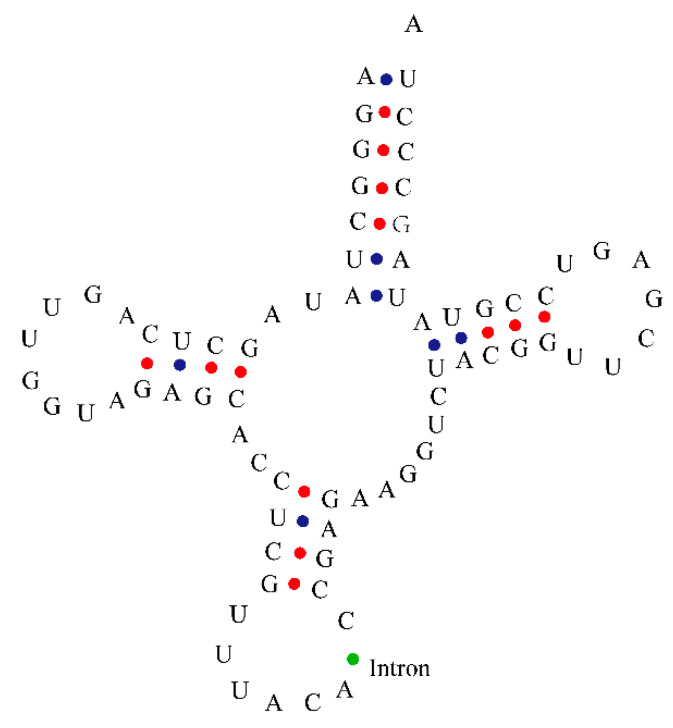
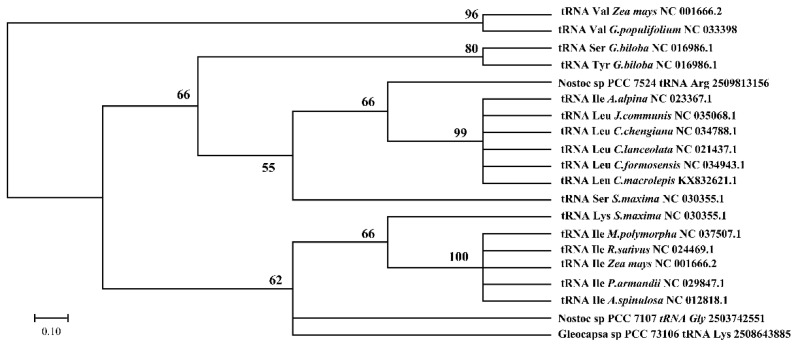
The cotton chloroplast tRNAs had intron annotations according to previous studies. The tRNAVal of G. populifolium was observed to contain an intron in its anti-codon loop region (Figure 1). The introns in bacterial and plant chloroplast tRNAs had conserved G-A-T-T-T and C-T-T-C-A consensus sequences (Figure 2). Phylogenetic analysis showed that chloroplast tRNA introns grouped with the introns in cyanobacteria (Figure 3). Introns contained in the same tRNAs of most plants (tRNAIle and tRNALeu) tended to appear in the same branch. This revealed their close phylogenetic relationship. The introns in chloroplast tRNAVal of G. populifolium and Zea mays L. aggregated to the same branch (Figure 3), showing that the introns of corn and cotton have a close phylogenetic relationship.
Figure 1.
Presence of intron in chloroplast tRNA, found to be located in the tRNA anti-codon loop. The blue dot represents dihydrogen bonds; the red dot represents triple hydrogen bond; and the green dot indicates the position of intron.
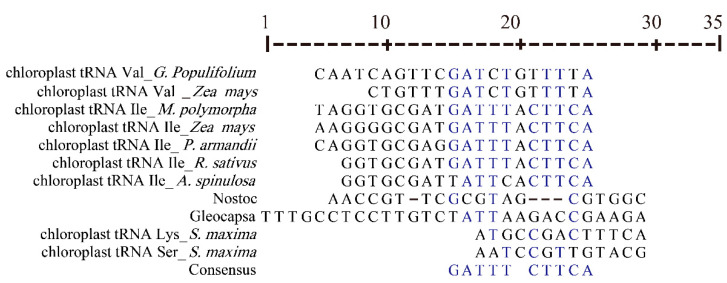
Figure 2.
Multiple sequence alignment of introns in tRNA of plant chloroplast and cyanobacteria. Introns in bacterial and chloroplast tRNAs had conserved G-A-T-T-T and C-T-T-C-A consensus sequences.
Figure 3.
Phylogenetic relationship of introns in chloroplast tRNAs (from angiosperms, gymnosperms, ferns, bryophytes, and algae). Introns of chloroplast tRNA grouped with that of cyanobacteria illustrated a common cyanobacterial origin of introns in chloroplasts. Introns in chloroplast tRNAVal of Gossypium populifolium and Zea mays indicated the same evolutionary ancestors of corn and cotton introns. Introns present in the same tRNA of plants (tRNAIle and tRNALeu) tended to appear in the same branch, showing their close phylogenetic relationship.
3.4. Chloroplast tRNAs with Non-Typical Features
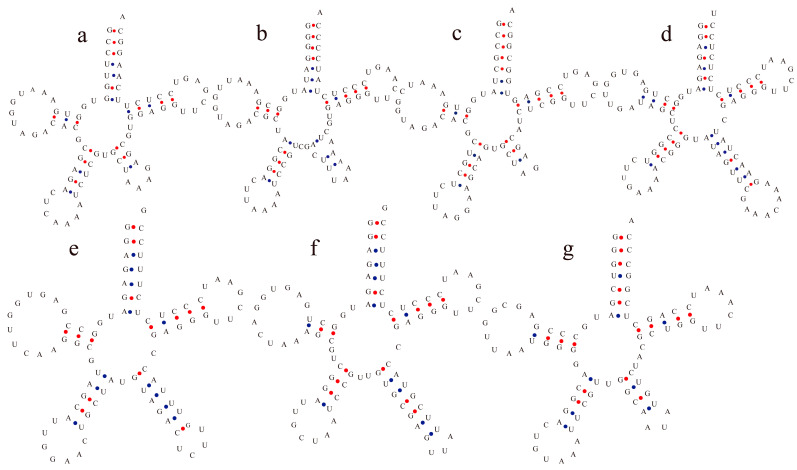
A few unconventional tRNAs were observed in cotton cp genomes. tRNALeu, tRNASer, and tRNATyr were observed to have a loop in the variable region (Figure 4). In these non-typical tRNAs, the anti-codon loop harbored 7 nt with the X-U-X3-A-A consensus sequence, and the stem of the anti-codon loop contained 4 to 5 nt. The variable loop region was observed to have 2 to 8 nt for tRNALeu, tRNASer, and tRNATyr. The stem of the variable region contained 3 to 7 nt pairs. Obviously, tRNASer had mainly 6 or 7 nt pairs (Figure 4). These loop structures in variable regions of tRNAs put forward the question of whether these loops play an important role in the process of protein translation of the chloroplast.
Figure 4.
Structures of the chloroplast tRNAs showed the presence of a loop structure in the variable region: (a–c) tRNALeu; (d–f) tRNASer; (g) tRNATyr. Anti-codon loop had seven nucleotides with the conservative X-U-X3-A-A consensus sequence in these tRNAs. Loop structure of the variable region was observed to contain three to eight nucleotides from tRNALeu, tRNASer, and tRNATyr.
3.5. Cotton Chloroplast tRNAs Were Derived from Several Evolutionary Ancestors
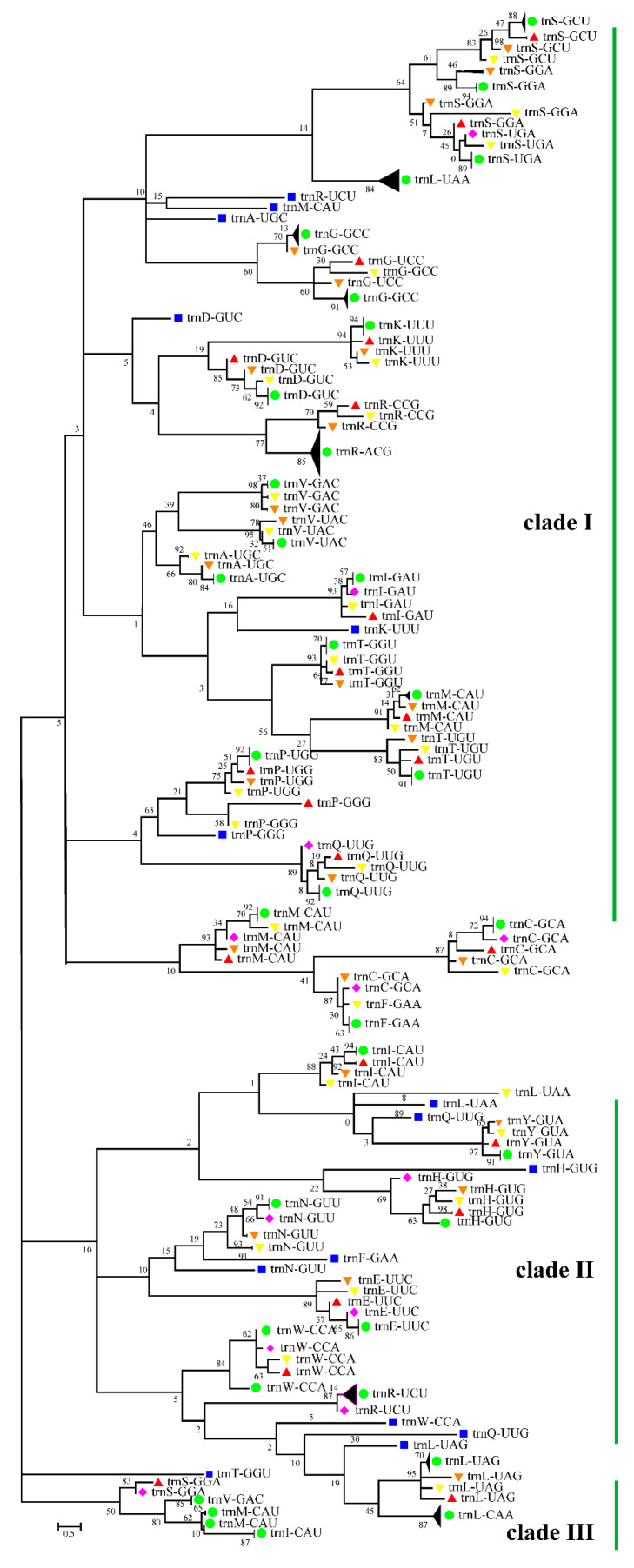
The phylogenetic tree of cotton and other various species’ tRNA sequences from a wide range of taxonomic positions, including algae (Nostoc sp. PCC 7524), bryophytes (Dumortiera hirsuta), ferns (Psilotum nudum), gymnosperms (Pinus taeda), and angiosperms (Arabidopsis lyrata), presented three major clusters (Table 4). In all, there were 87 groups in integrated clade I, 43 groups in integrated clade II, and 7 groups in integrated clade III. In the phylogenetic tree, tRNASer, tRNALeu, tRNAArg, tRNAVal, tRNAIle, tRNAMet, and tRNAGln were polyphyletic, positioned in more than two integrated clades. In integrated clade I, most of tRNAMet was clustered into two polyphyletic sub-clades (containing nine groups), one embedded in the tRNAThr group and another adjacent to tRNACys and tRNAPhe, while tRNAMet of cyanobacteria was independently clustered to its tRNAArg. In integrated clade II, tRNALeu was closely related to cyanobacterial tRNAGln, even with its polyphyly. tRNATrp, except the cyanobacterial one, was clustered into a monophyletic group, while the cyanobacterial one was embedded into the sister clade of the tRNATrp group. tRNAIle was found in all three clades. Interestingly, tRNALeu, tRNAIle, tRNAGln, tRNAPhe, and tRNAArg present in integrated clade I were also found in clade II, and the tRNAs in integrated clade III were also found in clade I (Figure 5).
Table 4.
The distribution of types of tRNAs in the three major clusters of the phylogenetic tree.
| Integrated Clades | Types of tRNAs |
|---|---|
| clade I | tRNASer, tRNALeu, tRNAArg, tRNAMet, tRNAAla, tRNAGly, tRNAAsp, tRNALys, tRNAVal, tRNAIle, tRNAThr, tRNAPro, tRNAGln, tRNACys, tRNAPhe |
| clade II | tRNAIle, tRNALeu, tRNAGln, tRNATyr, tRNAHis, tRNAAsn, tRNAPhe, tRNAGlu, tRNATrp, tRNAArg |
| clade III | tRNAThr, tRNASer, tRNAVal, tRNAMet, tRNAIle |
Figure 5.
Phylogenetic tree of chloroplast tRNAs. A, alanine (Ala); R, arginine (Arg); N, asparagine (Asn); D, aspartate (Asp); C, cysteine (Cys); Q, glutamine (Gln); E, glutamate (Glu); G, glycine (Gly); H, histidine (His); I, isoleucine (Ile); L, leucine (Leu); K, lysine (Lys); M, methionine (Met); F, phenylalanine (Phe); P, proline (Pro); S, serine (Ser); T, threonine (Thr); W, tryptophan (Trp); Y, tyrosine (Tyr); V, valine (Val). The green solid circle represents Gossypium; the red solid triangle represents P. taeda; the pink solid diamond represents A. lyrate; the yellow solid triangle represents P. nudum; the orange solid triangle represents D. hirsuta; and the blue solid square represents Nostoc sp. PCC 7524. Phylogenetic analysis showed the polyphyletic origin of chloroplast tRNAs.
3.6. Transition/Transversion of tRNAs
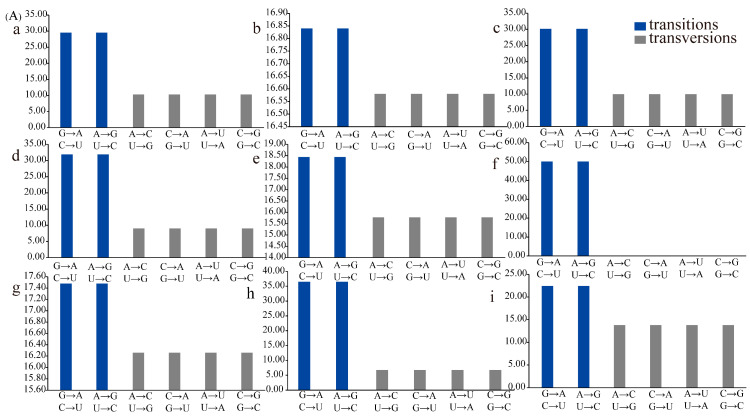
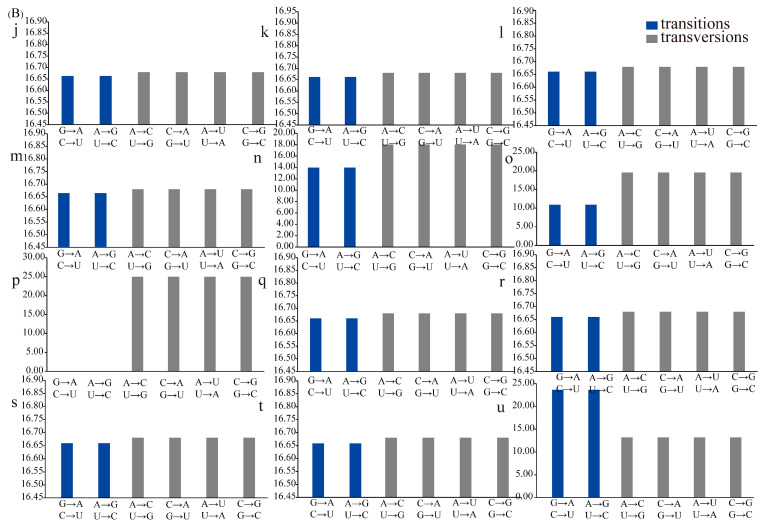
tRNAs have evolved with almost equal transition and transversion rates in spite of the small probability of transition or transversion events in tRNAs. In the present study, we found some intriguing phenomena in the substitution rate of cotton chloroplast tRNAs. A transition rate of 16.66 and transversion rate of 16.68 were found in tRNAAla, tRNAPhe, tRNAAsp, tRNAPro, tRNATyr, tRNAVal, and tRNAIle. This showed that the transversion rate was slightly higher than the transition rate, and these tRNAs evolved with almost the same transition and transversion rates (Figure 6, Table S4). The highest transition rate was 50.00 for tRNATrp and the highest transversion rate was 25.00 for tRNAHis. Correspondingly, the lowest transition rate (0.00) was observed for tRNAHis, and the lowest transversion rate (0.00) for tRNATrp (Figure 6). This indicated that the tRNATrp of the cotton cp genome had experienced a high transition rate without any transversion. Similarly, tRNAHis had experienced a high rate of transversion but no transition. For tRNALys, tRNAArg, tRNAMet, tRNAAsn, tRNACys, tRNASer, tRNAGlu, tRNAGly, and tRNALeu, the transition rate was higher than the transversion rate (Figure 6A). Additionally, the transition rate was two times higher than transversion for tRNAArg, tRNACys, and tRNAGlu, which showed that the evolution of chloroplast tRNAArg, tRNACys, and tRNAGlu tended toward transition rather than transversion. For tRNAGln and tRNAThr, the transversion rate was apparently higher than the transition rate (Figure 6B). This indicated that these tRNA iso-acceptors experienced transversion substitutions more easily than transition. The substitution rates of overall cp tRNAs showed that the average transition rate (23.64) was greater than the transversion rate (13.18) (Figure 6B).
Figure 6.
Rates of transition (blue) and transversion (gray) of chloroplast tRNAs. (A) a–i refer to R, Arg; S, Ser; C, Cys; E, Glu; G, Gly; W, Trp; L, Leu; K, Lys; M, Met. (B) j–u refer to A, Ala; P, Pro; N Asn; D, Asp; T, Thr; Q, Gln; H, His; I, Ile; Y, Tyr; V, Val; F, Phe; overall.
3.7. Evolutionary Characteristics of Single Nucleotide Polymorphisms
The biallelic and parallel mutation SNPs of the eight diploid Gossypium cp genomes were calculated. There were 2709 SNPs in Gossypium cp genomes (Table 5). They were subdivided into coding, intron, and intergenic spacer regions for further analyses. Among the 2709 SNPs, 906 were in coding regions, 299 were in intron regions, and 1504 were in intergenic spacers. The percentage of SNPs to total length was 1.14, 1.39, and 2.90%, respectively. In coding regions, the overall ratio of nonsynonymous mutations to synonymous mutations (dN/dS) was 3.04.
Table 5.
Genomic distribution of biallelic single nucleotide polymorphic loci in the eight chloroplast genomes of diploid Gossypium plants.
| Genome Region | Length (bp) | Value | % |
|---|---|---|---|
| Total substitutions | 162,231 | 2709 | 1.67 |
| Coding regions | 79,244 | 906 | 1.14 |
| Non-synonymous | / | 681.58 | 0.86 |
| Synonymous | / | 224.42 | 0.28 |
| dN/dS | / | 3.04 | / |
| Intron | 21,443 | 299 | 1.39 |
| Intergenic spacer | 51,920 | 1504 | 2.90 |
3.8. Mutation Rate of Chloroplast Genome
The divergence time of Gossypium was speculated to be 9.8 Mya from Theobroma [52,53]. The total length of the eight diploid cotton cp genomes was 156,796 bp. The mutation rate (μ1) of the protein-coding genes and the mutation rate (μ2) of the tRNA genes were calculated using the length of genomes, the number of indel mutations (522 for protein-coding genes and 133 for tRNA genes), and the divergent time. The value of μ1 of 0.34 × 10–9 per site per year for protein-coding genes and the value of μ2 of 0.09 × 10–9 per site per year for tRNA genes were obtained, respectively.
3.9. tRNA Duplication/Loss Events
Besides substitution, duplication and loss events of genes had a vital influence on their evolution. The analysis of duplication or loss events indicated that the investigated cotton chloroplast tRNA genes had experienced 226 duplication and 93 co-duplication events (Figure S1). However, only 63 loss events were observed (Figure S1, Table S5). The results showed that the duplication of genes was greater than the loss of genes for all of the tRNAs.
4. Discussion
The nucleotide composition of tRNA is closely related to its senior structure, which is responsible for the translation process. In many species, the tRNA family is conserved in evolution [56,57]. As one of the major gene components of semi-autonomous chloroplast, tRNAs were shown to have several basic conserved genomic features. tRNALeu and tRNASer were observed to have 80 nt or more. These two tRNA isotypes were also found to harbor more than 83 nt in Adoxaceae plants, which shows that the gene sequences of tRNASer and tRNALeu are longer than other tRNAs [58]. In addition, some tRNA iso-acceptors were not observed in the cp genome of Gossypium, which was similar to that of the species in Gramineae [28]. Additionally, this is perhaps related to the codon usage and wobble base pairing in the genomic constitution of plants [59]. Furthermore, selenocysteine and suppressor tRNAs were found lacking in the cotton cp genome, which were present in Oryza sativa, Sachharum officinarum L., Sorghum bicolor (L.) Moench, Triticum aestivum L., and Zea mays. This may be related to the biotoxin accumulation ability of these two tRNAs [29]. The differences between the quantity (36 or 37) and anti-codon types (28 or 29) of tRNAs in diploid and allotetraploid cotton species were not significant. This may be related to the stability and conservation of tRNA genes and the long cultivation of cotton species, which reduced the level of interspecific diversity [60,61].
The tRNA family was conserved in its genomic composition. In this study, most of the involved tRNAs were observed to harbor the conservative sequence U-U-C-X3-U at their Ψ-loop (Table 3). Additionally, in Adoxaceae, U-U-C-A conservative nucleotides at its Ψ-loop region were found [58]. This showed that the short consensus sequences might be important components of the molecular recognizer in the Ψ-loop, and are associated with the recognition of ribosomal RNA during the translation process [62]. In our study, we found that the acceptor arm of cotton chloroplast tRNA contained 6 to 7 nt; the D-arm contained 2 to 4 nt; the D-loop contained 7 to 11 nt; the anti-codon loops contained 7 and 9 nt; the Ψ-arms contained 5 nt; and the Ψ-loops had 7 nt. Additionally, in gymnosperm chloroplast genomes, the acceptor arm of tRNAs harbors 6 bp to 7 bp; the D-arm has 3 bp or 4 bp; and the D-loop contains 7 nt to 11 nt; the anti-codon loop contains 7 nt; the Ψ-arm contains 5 bp; and the Ψ-loop has 7 nt [63]. Our results and previous findings together suggest that chloroplast RNAs are significantly conserved, though a few tRNAs contain rare secondary structures [64,65]. In the present study, some tRNALeu, tRNASer, and tRNATyr were found in the variable region including a loop structure. The presence of this unconventional structure in the variable region exhibited the structural variation existing in tRNAs and this might be relevant to the maintenance of tRNA structures and the interaction with the D-arm and Ψ-arm [66].
Introns are previously reported in archaeal and eukaryotic genomes that break the continuity of numerous eukaryotic genes [67]. Here, the intron was observed in chloroplast tRNAs of G. populifolium. This is consistent with previous reports in other organisms [68,69]. In general, the group I intron has a few hundred nucleotides because of its built-in ribozyme unit [70]. tRNAs contain sequences of less than 100 polynucleotides that fold into a clover-type secondary structure [71]. Thus, the intron we observed in this study might be a part of intron I that was contained within the cotton chloroplast genome.
Phylogenetic analysis revealed that the introns in chloroplast tRNAVal of Gossypium populifolium and Zea mays aggregated to the same branch (Figure 3), suggesting that the introns of corn and cotton have a close phylogenetic relationship. Additionally, chloroplast tRNAs with introns were grouped with cyanobacteria, providing supportive evidence for a common cyanobacterial lineage source of cp tRNAs [58].
In addition to the anti-codon CAU of tRNAMet, tRNAIle was observed to code the anti-codon CAU in the cotton chloroplast genome. This may be associated with the modification of tRNA [72]. Three types of tRNAs—tRNAfMet, tRNAMet, and tRNAkIle, with anti-codon CAU—were found in a bacterial genome, and tRNAkIle was able to identify the codon of isoleucine instead of methionine after anti-codon modification [73]. For the tRNAIle CAU observed in the genomes of plants, the first position in the anti-codon of these tRNAs has a lysidine-like nucleotide. On the other hand, a methionine-discerning anti-codon CAT was found in the genes. After the modification of the C residue of the CAT anti-codon, the mature tRNA shows isoleucine-identifying activity rather than methionine-identifying activity [74,75].
The presence of three obvious clusters and diverse groupings, and the appearance of tRNA groups from different plants with a wide range of taxonomic positions in integrated clade I and II, clade I and III, and clade I, II, and III of the phylogenetic tree, indicate frequent duplication and divergence during their evolution. In addition, tRNASer, tRNALeu, tRNAArg, tRNAVal, tRNAIle, tRNAMet, and tRNAGln were found in more than two integrated clades. This indicates their multiple evolutionary origins. Diverse groupings and the overlapping of tRNA groups from different plants suggest that the tRNAs have several inferred ancestors, including tRNAMet, tRNAIle, etc., in the evolutionary history [63]. The interlaced emergence of tRNA groups (tRNAMet, tRNAThr, tRNAArg, tRNATrp, tRNAVal, and tRNASer) also suggests that evolutionary relationships among these tRNA types are relatively close.
Just as the cases with the most genetic variations, the transition rate was greater than the transversion rate of Gossypium cp tRNAs at the overall level. However, tRNATrp and tRNAHis showed different substitutive choices, which may be caused by various factors, such as their neighbor bases and the efficiency of the repair system of DNA strands [76]. The percentage of SNPs in coding regions, intron regions, and intergenic spacers compared to the total cotton cp genomes was 1.14, 1.39, and 2.90%, respectively, which implies that the density of SNPs was different in the cotton cp genome and the intergenic spacer of Gossypium was more variable than intron regions. This may be associated with the richness of A/T and G/C repetitive units in certain regions of the cp genome. It also implies that the evolutionary mutation potential of different positions in the genome is unbalanced [77]. In coding regions, the overall ratio of non-synonymous to synonymous mutations (dN/dS) was 3.04, showing that non-synonymous mutations had been fixed in the cotton cp genome. In the involved diploid Gossypium cp genomes, the mutation rate (μ1), 0.34 × 10–9 per site per year of protein-coding genes, was significantly higher than the mutation rate (μ2), 0.09 × 10–9 per site per year of tRNA genes (more than three times than that of μ2), which also implies that compared with protein-coding genes, tRNA genes and structures have considerable evolutionary stability [78]. In addition, it should be mentioned that the genomic data employed in our study still have limitations. Perhaps the addition of more diverse gene sequences would provide more interesting results.
In concert with multiple factors of tRNA evolution, most important gene functions have evolved from gene duplication events [79,80]. In this study, there were about five times as many duplication events as loss events. Many studies have also shown that frequent duplication greatly promotes evolution and functional diversification in gene families [81,82]. This may provide further insights to confirm that the evolution of cotton tRNAs derived from polyphyletic evolutionary ancestors.
5. Conclusions
Cotton chloroplast tRNAs encode 28 or 29 anti-codon types, and 36 or 37 anti-codon-specific tRNAs with lengths ranging from 60 (tRNAGly) to 93 nucleotides (tRNASer). Thirty-three anti-codon types including AGC-tRNAAla are absent in the cp genome of Gossypium. The CAU anti-codon is encoded in both tRNAMet and tRNAIle. The acceptor arm of cotton chloroplast tRNA contains 6 to 7 nt; the D-arm contains 2 to 4 nt; the D-loop contains 7 to 11 nt; the anti-codon loop contains 7 and 9 nt; the Ψ-arms contains 5 nt; and the Ψ-loop has 7 nt. The Ψ-arm of cotton chloroplast tRNAs contains the G-G consensus sequence, and the Ψ-loop contains the conserved U-U-C-X3-U motifs. Additionally, cotton chloroplast tRNAs were found to contain introns and a few tRNALeu, tRNASer, and tRNATyr were observed to have a loop in the variable region. Furthermore, phylogenetic analysis suggests that tRNAs possibly have several inferred ancestors, including tRNAMet, tRNAIle, etc., in the evolutionary history. On the other hand, the average transition rate of all involved cp tRNAs was greater than their transversion rate. The density of SNPs was unbalanced and the intergenic spacer of Gossypium was more variable than intron regions. Gene duplication events (226 duplication and 93 co-duplication) have occurred more frequently than gene loss events (63) in cotton chloroplast tRNAs. These results provide helpful insights into the detailed characteristics and evolutionary variation of the tRNA family.
Acknowledgments
We thank Qiu-Yi Zhong for her kindly help for the manuscript improvements.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12060822/s1. Figure S1: Duplication and loss events of the chloroplast tRNAs. Blue: duplication events; gray: loss events; D: duplication node; cD: conditional duplication node. Analysis indicates that the chloroplast tRNAs underwent frequent duplication events during their evolution with subsequent diversification, Table S1: Information registration of the analyzed tRNA genes, Table S2: Distribution of anti-codons in cotton chloroplast genome, Table S3: Nucleotide composition in acceptor arm (AA), D-arm (DA), D-loop (DL), anti-codon arm (ACA), anti-codon loop (ACL), variable loop (VL), pseudouridine arm (ΨA), and pseudouridine loop (ΨL) of chloroplast tRNA, Table S4: Results of transition and transversion rates of chloroplast tRNAs, Table S5: Loss events of the chloroplast tRNAs.
Author Contributions
Conceptualization, Z.-H.L. and T.-T.Z.; methodology, T.-T.Z., Y.Y., and X.-Y.S.; software, X.-L.Z.; validation, X.-Y.G., J.-J.Z., and X.-Y.S.; formal analysis, K.-H.Z.; investigation, C.-B.Z.; resources, W.L.; data curation, D.-G.Y. and X.-F.M.; writing—review and editing, Z.-H.L., Y.Y., and T.-T.Z.; supervision, Z.-H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the National Key R&D Program for Crop Breeding (2016YFD0100306), the National Natural Science Foundation of China (No. 31401431), Shaanxi Science and Technology Innovation Team (2019TD-012), Fourth National Survey of Traditional Chinese Medicine Resources (2018-43 and 2019-39), and the Open Foundation of Key Laboratory of Resource Biology and Biotechnology in Western China (Ministry of Education) (grant nos. ZSK2017007 and ZSK2019008).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arnon D.I. The chloroplast as a complete photosynthetic unit. Science. 1955;122:9–16. doi: 10.1126/science.122.3157.9. [DOI] [PubMed] [Google Scholar]
- 2.Roughan P.G., Holland R., Slack C.R. On the control of long-chain-fatty acid synthesis in isolated intact spinach (Spinacia oleracea) chloroplasts. Biochem. J. 1979;184:193–202. doi: 10.1042/bj1840193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuhaus H.E., Emes M.J. Nonphotosyntheticmetabolism inplastids. Annu. Rev. Plant Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- 4.Spetea C., Hundal T., Lundin B., Heddad M., Adamska I., Andersson B. Multiple evidence for nucleotide metabolism in the chloroplast thylakoid lumen. Proc. Natl. Acad. Sci. USA. 2004;101:1409–1414. doi: 10.1073/pnas.0308164100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holsinger K.E., Soltis P.S., Soltis D.E., Doyle J.J. Molecular systematics of plants. Syst. Bot. 1993;18:539. doi: 10.2307/2419426. [DOI] [Google Scholar]
- 6.Gross J., Meurer J., Bhattacharya D. Evidence of a chimeric genome in the cyanobacterial ancestor of plastids. BMC Evol. Biol. 2008;8:117. doi: 10.1186/1471-2148-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C.-P., Wu C.-S., Huang Y.-Y., Chaw S.-M. The complete chloroplast genome of ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol. Evol. 2012;4:374–381. doi: 10.1093/gbe/evs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen P.A.T., Kim J.S., Kim J.-H. The complete chloroplast genome of colchicine plants (Colchicum autumnale L. and Gloriosa superba L.) and its application for identifying the genus. Planta. 2015;242:223–237. doi: 10.1007/s00425-015-2303-7. [DOI] [PubMed] [Google Scholar]
- 9.Ruf S., Karcher D., Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc. Natl. Acad. Sci. USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo L.-Y., Tang T.-Y., Li F.-W., Su H.-J., Chiou W.-L., Huang Y.-M., Wang C.-N. Organelle genome inheritance in Deparia Ferns (Athyriaceae, Aspleniineae, Polypodiales) Front. Plant Sci. 2018;9:486. doi: 10.3389/fpls.2018.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R.-J., Cheng C.-L., Chang C.-C., Wu C.-L., Su T.-M., Chaw S.-M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008;8:36. doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fish L.E., Jagendorf A.T. High rates of potein synthesis by isolated chloroplasts. Plant Physiol. 1982;70:1107–1114. doi: 10.1104/pp.70.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinozaki K., Hayashida N., Sugiura M. Nicotiana chloroplast genes for components of the photosynthetic apparatus. Photosynth. Res. 1988;18:7–31. doi: 10.1007/BF00042978. [DOI] [PubMed] [Google Scholar]
- 14.Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2014;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 15.Brumfield R.T., Beerli P., Nickerson D.A., Edwards S.V. Single nucleotide polymorphisms (SNPs) as markers in phylogeography. Trends Ecol. Evol. 2003;18:249–256. doi: 10.1016/S0169-5347(03)00018-1. [DOI] [Google Scholar]
- 16.Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002;5:94–100. doi: 10.1016/S1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- 17.Holley R.W., Apgar J., Everett G.A., Madison J.T., Marquisee M., Merrill S.H., Penswick J.R., Zamir A. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 18.Nicoghosian K., Bigras M., Sankoff D., Cedergren R. Archetypical features in tRNA families. J. Mol. Evol. 1987;26:341–346. doi: 10.1007/BF02101153. [DOI] [PubMed] [Google Scholar]
- 19.Giegé R., Puglisi J.D., Florentz C. tRNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol. 1993;45:129–206. doi: 10.1016/S0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- 20.De Pouplana L.R., Dedon P.C. More than an adaptor molecule: The emerging role of tRNA in cell signaling and disease. FEBS Lett. 2014;588:4267. doi: 10.1016/j.febslet.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blee E., Joyard J. Envelope membranes from spinach chloroplasts are a site of metabolism of fatty acid hydroperoxides. Plant Physiol. 1996;110:445–454. doi: 10.1104/pp.110.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanai A. Disrupted tRNA genes and tRNA fragments: A perspective on tRNA gene evolution. Life. 2015;5:321–331. doi: 10.3390/life5010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson P., Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashdan M.A., Dudock B.S. The gene for a spinach chloroplast isoleucine tRNA has a methionine anticodon. J. Biol. Chem. 1982;257:11191–11194. doi: 10.1016/S0021-9258(18)33737-2. [DOI] [PubMed] [Google Scholar]
- 25.Yasukawa T., Kirino Y., Ishii N., Holt I., Jacobs H.T., Makifuchi T., Fukuhara N., Ohta S., Suzuki T., Watanabe K. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 2005;579:2948–2952. doi: 10.1016/j.febslet.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 26.Morscher R.J., Ducker G.S., Li S.H.-J., Mayer J.A., Gitai Z., Sperl W., Rabinowitz J.D. Mitochondrial translation requires folate-dependent tRNA methylation. Nat. Cell Biol. 2018;554:128–132. doi: 10.1038/nature25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohanta T.K., Syed A.S., Ameen F., Bae H. Novel genomic and evolutionary perspective of cyanobacterial tRNAs. Front. Genet. 2017;8:200. doi: 10.3389/fgene.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohanta T.K., Bae H. Analyses of genomic tRNA reveal presence of novel tRNAs in Oryza sativa. Front. Genet. 2017;8:90. doi: 10.3389/fgene.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanta T.K., Khan A.L., Hashem A., Allah E.F.A., Yadav D., Al-Harrasi A. Genomic and evolutionary aspects of chloroplast tRNA in monocot plants. BMC Plant Biol. 2019;19:39. doi: 10.1186/s12870-018-1625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fryxell P.A. A classification of Gossypium L. (Malvaceae) Taxon. 1969;18:585–591. doi: 10.2307/1218405. [DOI] [Google Scholar]
- 31.Beasley J.O. The origin of American Tetraploid Gossypium species. Am. Nat. 1940;74:285–286. doi: 10.1086/280895. [DOI] [Google Scholar]
- 32.Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R.J., Ma Z., Shang H., Ma X., Wu J., et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015;33:524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 33.Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., Zhang J., A Saski C., E Scheffler B., Stelly D., et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 34.Sun R., Wang K., Guo T., Jones D.C., Cobb J., Zhang B., Wang Q. Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct. Integr. Genom. 2015;15:481–493. doi: 10.1007/s10142-015-0437-0. [DOI] [PubMed] [Google Scholar]
- 35.Yuan D., Tang Z., Wang M., Gao W., Tu L., Jin X., Chen L., He Y., Zhang L., Zhu L., et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 2016;5:17662. doi: 10.1038/srep17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Q., Jones D.C., Li W., Xie F., Ma J., Sun R., Wang Q., Zhu S., Zhang B. Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci. Rep. 2016;6:22980. doi: 10.1038/srep22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L.-F., Li Y., Chen Y., Li X.-B. Improved drought and salt tolerance of Arabidopsis thaliana by ectopic expression of a cotton (Gossypium hirsutum) CBF gene. Plant Cell Tissue Organ Cult. 2016;124:583–598. doi: 10.1007/s11240-015-0917-x. [DOI] [Google Scholar]
- 38.Salih H., Gong W., He S., Sun G., Sun J., Du X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 2016;17:1–12. doi: 10.1186/s12863-016-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Q., Wang G., Zhang X., Zhang X., Qiao P., Long L., Yuan Y., Cai Y. Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci. Rep. 2017;7:42418. doi: 10.1038/srep42418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y., Liu F., Yang D.-G., Li W., Zhou X.-J., Pei X.-Y., Liu Y.-G., He K.-L., Zhang W.-S., Ren Z.-Y., et al. Comparative chloroplast genomics of Gossypium species: Insights into repeat sequence variations and phylogeny. Front. Plant Sci. 2018;9:376. doi: 10.3389/fpls.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowe T.M., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:0955–0964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe T.M., Chan P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell C. MultAlin–multiple sequence alignment. Bioinformatics. 1993;9:614. doi: 10.1093/bioinformatics/9.5.614. [DOI] [Google Scholar]
- 45.Mohanta T.K., Arora P.K., Mohanta N., Parida P., Bae H. Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genom. 2015;16:1–20. doi: 10.1186/s12864-015-1244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohanta T.K., Mohanta N., Parida P., Panda S.K., Ponpandian L.N., Bae H. Genome-wide identification of mitogen-activated protein kinase gene family across fungal lineage shows presence of novel and diverse activation loop motifs. PLoS ONE. 2016;11:e0149861. doi: 10.1371/journal.pone.0149861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S., Nei M., Dudley J., Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- 50.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 51.Saitou N., Ueda S. Evolutionary rates of insertion and deletion in noncoding nucleotide sequences of primates. Mol. Biol. Evol. 1994;11:504–512. doi: 10.1016/0303-7207(94)90253-4. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z., Feng K., Grover C.E., Li P., Liu F., Wang Y., Xu Q., Shang M., Zhou Z., Cai X., et al. Chloroplast DNA structural variation, phylogeny, and age of divergence among Diploid Cotton species. PLoS ONE. 2016;11:e0157183. doi: 10.1371/journal.pone.0157183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grover C.E., Gallagher J.P., Jareczek J.J., Page J.T., Udall J.A., Gore M., Wendel J.F. Re-evaluating the phylogeny of allopolyploid Gossypium L. Mol. Phylogenet. Evol. 2015;92:45–52. doi: 10.1016/j.ympev.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 54.Chen K., Durand D., Farach-Colton M. NOTUNG: A program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 2000;7:429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- 55.Vernot B., Stolzer M., Goldman A., Durand D. Reconciliation with non-binary species trees. J. Comput. Biol. 2008;15:981–1006. doi: 10.1089/cmb.2008.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumazawa Y., Nishida M. Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics. J. Mol. Evol. 1993;37:380–398. doi: 10.1007/BF00178868. [DOI] [PubMed] [Google Scholar]
- 57.Shepotinovskaya I., Uhlenbeck O.C. tRNA residues evolved to promote translational accuracy. RNA. 2013;19:510–516. doi: 10.1261/rna.036038.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong Q., Fu X., Zhang T., Zhou T., Yue M., Liu J., Li Z. Phylogeny and evolution of chloroplast tRNAs in Adoxaceae. Ecol. Evol. 2021;11:1294–1309. doi: 10.1002/ece3.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong K.-L., Wong J.-F. Anticodon and wobble evolution. Gene. 2004;333:169–177. doi: 10.1016/j.gene.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 60.Iqbal M.J., Aziz N., Saeed N.A., Zafar Y., Malik K.A. Genetic diversity evaluation of some elite cotton varieties by RAPD analysis. Theor. Appl. Genet. 1997;94:139–144. doi: 10.1007/s001220050392. [DOI] [PubMed] [Google Scholar]
- 61.Kumar P., Anaya J., Mudunuri S.B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:1–14. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Charette M.W.G.M. Pseudouridine in RNA: What, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 63.Zhang T.-T., Hou Y.-K., Yang T., Zhang S.-Y., Yue M., Liu J., Li Z. Evolutionary analysis of chloroplast tRNA of Gymnosperm revealed the novel structural variation and evolutionary aspect. PeerJ. 2020;8:e10312. doi: 10.7717/peerj.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nardi F., Carapelli A., Fanciulli P.P., Dallai R., Frati F. The complete mitochondrial DNA sequence of the basal Hexapod Tetrodontophora bielanensis: Evidence for Heteroplasmy and tRNA translocations. Mol. Biol. Evol. 2001;18:1293–1304. doi: 10.1093/oxfordjournals.molbev.a003914. [DOI] [PubMed] [Google Scholar]
- 65.Jühling T., Duchardt-Ferner E., Bonin S., Wöhnert J., Pütz J., Florentz C., Betat H., Sauter C., Mörl M. Small but large enough: Structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax. Nucleic Acids Res. 2018;46:9170–9180. doi: 10.1093/nar/gky593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W., Chen X., Wolin S.L., Xiong Y. Structural basis for tRNA mimicry by a bacterial Y RNA. Structure. 2018;26:1635–1644. doi: 10.1016/j.str.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshihisa T. Handling tRNA introns, archaeal way and eukaryotic way. Front. Genet. 2014;5:213. doi: 10.3389/fgene.2014.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawach O., Voß C., Wolff J., Hadfi K., Maier U.-G., Zauner S. Unique tRNA introns of an enslaved algal cell. Mol. Biol. Evol. 2005;22:1694–1701. doi: 10.1093/molbev/msi161. [DOI] [PubMed] [Google Scholar]
- 69.Hoser S.M., Hoffmann A., Meindl A., Gamper M., Fallmann J., Bernhart S.H., Müller L., Ploner M., Misslinger M., Kremser L., et al. Intronic tRNAs of mitochondrial origin regulate constitutive and alternative splicing. Genome Biol. 2020;21:1–35. doi: 10.1186/s13059-020-02199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A. Crystal structure of a self-splicing group I intron with both exons. Nat. Cell Biol. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 71.Wilusz J.E. Controlling translation via modulation of tRNA levels. Wiley Interdiscip. Rev. RNA. 2015;6:453–470. doi: 10.1002/wrna.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Köhrer C., Mandal D., Gaston K.W., Grosjean H., Limbach P.A., Rajbhandary U.L. Life without tRNAIle-lysidine synthetase: Translation of the isoleucine codon AUA in Bacillus subtilis lacking the canonical tRNA2Ile. Nucleic Acids Res. 2013;42:1904–1915. doi: 10.1093/nar/gkt1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sonawane K.D., Tewari R. Conformational Preferences of Hypermodified Nucleoside Lysidine (k2C) Occurring at “Wobble” position in Anticodon Loop of tRNAIle. Nucleosides Nucleotides Nucleic Acids. 2008;27:1158–1174. doi: 10.1080/15257770802341475. [DOI] [PubMed] [Google Scholar]
- 74.Weber F., Dietrich A., Weil J.-H., Maréchal-Drouard L. A potato mitochondrial isoleucine tRNA is coded for by a mitochondrial gene possessing a methionine anticodon. Nucleic Acids Res. 1990;18:5027–5030. doi: 10.1093/nar/18.17.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soma A., Ikeuchi Y., Kanemasa S., Kobayashi K., Ogasawara N., Ote T., Kato J.-I., Watanabe K., Sekine Y., Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/S1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 76.Shrestha B., Weng M.-L., Theriot E.C., Gilbert L.E., Ruhlman T.A., Krosnick S.E., Jansen R.K. Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol. Phylogenet. Evol. 2019;138:53–64. doi: 10.1016/j.ympev.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 77.Zhao Z., Fu Y.-X., Hewett-Emmett D., Boerwinkle E. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene. 2003;312:207–213. doi: 10.1016/S0378-1119(03)00670-X. [DOI] [PubMed] [Google Scholar]
- 78.Yarham J.W., McFarland R., Taylor R.W., Elson J.L. A proposed consensus panel of organisms for determining evolutionary conservation of mt-tRNA point mutations. Mitochondrion. 2012;12:533–538. doi: 10.1016/j.mito.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jelesko J.G., Harper R., Furuya M., Gruissem W. Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1999;96:10302–10307. doi: 10.1073/pnas.96.18.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao H., Jiang N., Schaffner E., Stockinger E.J., Van Der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 81.Rensing S.A. Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 2014;17:43–48. doi: 10.1016/j.pbi.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Panchy N., Lehti-Shiu M., Shiu S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.