Abstract
Lumpy skin disease (LSD), an economically significant disease in cattle caused by lumpy skin disease virus (LSDV), is endemic to nearly all of Africa. Since 2012, LSDV has emerged as a significant epizootic pathogen given its rapid spread into new geographical locations outside Africa, including the Middle East, Eastern Europe, and Asia. To assess the genetic diversity of LSDVs in East Africa, we sequenced and analyzed the RPO30 and GPCR genes of LSDV in twenty-two archive samples collected in Ethiopia, Kenya, and Sudan before the appearance of LSD in the Middle East and its incursion into Europe. We compared them to publicly available sequences of LSDVs from the same region and those collected elsewhere. The results showed that the East African field isolates in this study were remarkably similar to each other and to previously sequenced field isolates of LSDV for the RPO30 and GPCR genes. The only exception was LSDV Embu/B338/2011, a field virus collected in Kenya, which displayed mixed features between the LSDV Neethling vaccine and field isolates. LSDV Embu/B338/2011 had the same 12-nucleotide insertion found in LSDV Neethling and KS-1 vaccines. Further analysis of the partial EEV glycoprotein, B22R, RNA helicase, virion core protein, NTPase, and N1R/p28-like protein genes showed that LSDV Embu/B338/2011 differs from previously described LSDV variants carrying the 12-nucleotide insertion in the GPCR gene. These findings highlight the importance of the constant monitoring of genetic variation among LSDV isolates.
Keywords: LSDV, GPCR gene, RPO30 gene, EEV glycoprotein, B22R gene
1. Introduction
Lumpy skin disease (LSD) is a severe contagious disease in cattle, characterized by the appearance of nodules on the skin and enlarged superficial lymph nodes. The causative agent, lumpy skin disease virus (LSDV), belongs to the Capripoxvirus genus of the family Poxviridae [1,2]. LSDV has a double-stranded DNA genome of approximately 151 kb. It is closely related to sheep poxvirus (SPPV) and goat poxvirus (GTPV), two other members of the Capripoxvirus genus. However, the three viruses have several subtle genetic variations, causing differences in the virulence and host range of the capripoxviruses. These differences make LSDV a host-specific pathogen for cattle [3], although LSDV DNA was reported to be found in springbok antelopes in South Africa [4].
As LSD has a considerable economic impact on the cattle industry, the World Organization for Animal Health (OIE) has listed it as a notifiable disease [5]. In East Africa, Kenya was the first country to report LSD in 1957 [6], followed by Sudan in 1971 [7] and Ethiopia and Somalia in 1983 [8]. LSD is endemic to most African countries except Tunisia, Morocco, and Libya. The disease has also spread into most of the Middle East, Eastern Europe, and Asia [2,9,10,11,12,13,14]. East Africa has a large cattle population with important cross-border trade along the boundaries of Kenya, Ethiopia, Somalia, and Sudan. As this region is also a significant livestock export market for North Africa and the Arabian Peninsula, it was suggested that LSDV was re-introduced in Egypt in 2006 via infected cattle imported from the Horn of Africa [15]. This seems to have been the starting point before the disease spread further in the Middle East [16].
Therefore, this work focused on the molecular characterization of LSDV isolates collected in Kenya, Ethiopia, and Sudan before the wave of LSD in the Middle East, Europe, and Asia.
Molecular epidemiological studies of LSDV rely on the analysis of various regions of its genome, such as the GPCR, RPO30, and EEV genes [4,17,18,19]. Before identifying recombinant LSDVs and field-related LSDVs, GPCR and EEV were also suitable to differentiate LSDV vaccines from LSDV field strains.
Although LSD is endemic to East Africa, reports on the molecular characterization of LSDV in the region are inadequate. Previously, the GPCR and RPO30 genes of LSDV field isolates collected between 2008 and 2012 from several areas in Ethiopia were analyzed [18]. Our study further expands this analysis by investigating new outbreaks in Ethiopia and looking at isolates from Kenya and Sudan, where little insight was available on LSDV’s genomic data from post-2000 outbreaks. Because of the large size of the poxvirus genome, multiple-gene or whole-genome analysis is necessary to supplement the available molecular data.
The present study describes the comprehensive molecular characterization of LSDV isolates from East Africa.
2. Materials and Methods
2.1. Samples and DNA Extraction
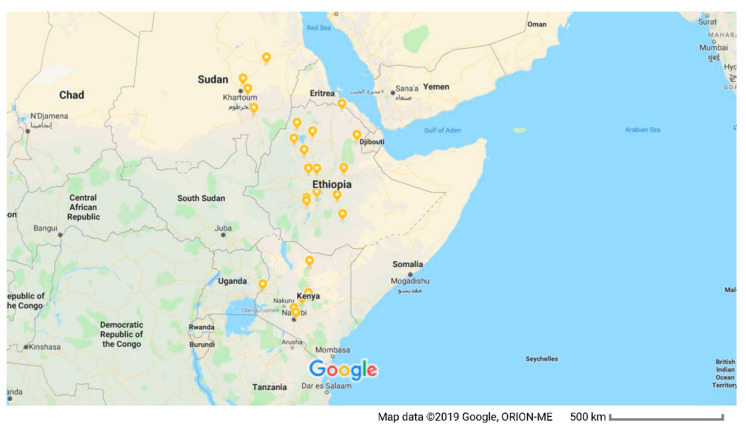
This study included 22 isolates (Table 1) of lumpy skin disease virus collected at various geographical locations in Ethiopia, Kenya, and Sudan (Figure 1). The total DNA was extracted from clinical samples and cell culture supernatants using the AllPrep DNA/RNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA was eluted using 80 μL elution buffer and stored at −20 °C until use.
Table 1.
LSDV isolates from East Africa included in this study and previous isolates.
| No. | Strain Name | Host | Origin | Sample Type | Vaccination History | Year | Accession Number | |
|---|---|---|---|---|---|---|---|---|
| RPO30 | GPCR | |||||||
| 1 | * Marsabit/B291/2007 | Cattle | Kenya | Skin lesion | Non-vaccinated | 2007 | MK302092 | MK302070 |
| 2 | * Embu/B338/2011 | Cattle | Kenya | Skin lesion | Vaccinated | 2011 | MK302093 | MK302071 |
| 3 | * Bungoma/B624/2010 | Cattle | Kenya | Skin lesion | Non-vaccinated | 2010 | MK302094 | MK302072 |
| 4 | * Arsi/B1/2011 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2011 | MK302095 | MK302073 |
| 5 | * Asella/B2/2011 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2011 | MK302096 | MK302074 |
| 6 | * Ziway/B3/2011 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2011 | MK302097 | MK302075 |
| 7 | * Adama/B4/2011 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2011 | MK302098 | MK302076 |
| 8 | * Guder/B5/2008 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2008 | MK302099 | MK302077 |
| 9 | * Sodo/B22/2010 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2010 | MK302100 | MK302078 |
| 10 | * Humbo/B23/2010 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2010 | MK302101 | MK302079 |
| 11 | * Sodo/B24/2010 | Cattle | Ethiopia | Skin lesion | Non-vaccinated | 2010 | MK302102 | MK302080 |
| 12 | * Sundus/1970 | Cattle | Sudan | Skin lesion | Non-vaccinated | 1971 | MK302103 | MK302081 |
| 13 | * Sudan North/2008 | Cattle | Sudan | Skin lesion | Non-vaccinated | 2008 | MK302104 | MK302082 |
| 14 | * Sinnar/2006 | Cattle | Sudan | Skin lesion | Non-vaccinated | 2006 | MK302105 | MK302083 |
| 15 | * Toke/B6/2008 | Cattle | Ethiopia | Swab samples | Non-vaccinated | 2008 | MK302106 | MK302084 |
| 16 | * Gindo/B7/2008 | Cattle | Ethiopia | Swab samples | Non-vaccinated | 2008 | MK302107 | MK302085 |
| 17 | * Ambo/B8/2008 | Cattle | Ethiopia | Swab samples | Non-vaccinated | 2008 | MK302108 | MK302086 |
| 18 | * Holeta/B9/2008 | Cattle | Ethiopia | Swab samples | Non-vaccinated | 2008 | MK302109 | MK302087 |
| 19 | * Ginchi/B10/2008 | Cattle | Ethiopia | Swab samples | Non-vaccinated | 2008 | MK302110 | MK302088 |
| 20 | * Chilimo/B11/2008 | Cattle | Ethiopia | Swab samples | Non-vaccinated | 2008 | MK302111 | MK302089 |
| 21 | * Galesa/B12/2008 | Cattle | Ethiopia | Swab samples | Non-vaccinated | 2008 | MK302112 | MK302090 |
| 22 | * Massalamia/P04/1971 | Cattle | Sudan | Cell culture | Non-vaccinated | 1971 | MK302113 | MK302091 |
| 23 | Sudan/99-Atbara | Cattle | Sudan | 1999 | GU119944 | FJ869367 | ||
| 24 | Sudan/06-Obied | Cattle | Sudan | 2006 | GU119938 | FJ869369 | ||
| 25 | KSGP 0240 | Sheep | Kenya | 1976 | KX683219 | KX683219 | ||
| 26 | KS-1 | Sheep | Kenya | 1976 | KJ818290 | KJ818283 | ||
| 27 | NI-2490 | Cattle | Kenya | 1958 | AF325528 | AF325528 | ||
| 28 | Kenya | Cattle | Kenya | 1950 | MN072619 | MN072619 | ||
| 29 | Adama/B01/2011 | Cattle | Ethiopia | 2011 | KP663667 | KP663690 | ||
| 30 | Andassa/B04/2012 | Cattle | Ethiopia | 2012 | KP663671 | KP663694 | ||
| 31 | Kadjima/B01/2009 | Cattle | Ethiopia | 2009 | KP663681 | KP663704 | ||
| 32 | Mojo/B01/2011 | Cattle | Ethiopia | 2011 | KP663683 | KP663706 | ||
| 33 | Wenji/B01/2011 | Cattle | Ethiopia | 2011 | KP663687 | KP663710 | ||
* LSDV sequenced for this study.
Figure 1.
Map of East Africa showing Sudan, Kenya, and Ethiopia, and the approximate geographical origins (highlighted in orange) of LSDV isolates included in this study.
2.2. Amplification of RPO30, GPCR, EEV Glycoprotein, and B22R Genes
For all 22 samples, RPO30 and GPCR were amplified as previously described [18]. A pair of primers, EEVGly F-5′-ATGGGAATAGTATCTGTTGTATACG-3′ and EEVGly R-5′-CGAACCCCTATTTACTTGAGAA-3′ [11], were used for the amplification of fragments containing the partial EEV glycoprotein (encoded by ORF LSDV126) and hypothetical protein LSDV 127 gene.
The PCR reaction was performed in a reaction volume of 20 μL containing 500 nM of each of the forward and reverse primers, 0.2 mM of dNTPs, 1× buffer (Qiagen), 2.5 U of Taq DNA polymerase (Qiagen), and 2 μL of template DNA. The thermal cycler (Bio-Rad, USA) parameters were: initial denaturation at 95 °C for 4 min, followed by 35 cycles at 95 °C for 40 s, 55 °C for 30 s, and 72 °C for 1 min, and a final extension step at 72 °C for 7 min. The partial B22R gene was amplified as described by [20].
The amplified PCR amplicons were separated by electrophoresis on a 1.5% agarose gel at 100 V for 60 min and visualized using a Gel Documentation System (Bio-Rad, Hercules, CA, USA).
2.3. Sequencing and Phylogenetic Analysis
The positive PCR products were purified using the Wizard SV Gel and PCR clean-up system kit (Promega, Dane County, WI, USA) according to the manufacturer’s instructions. The purified PCR products were sequenced at LGC Genomics (Berlin, Germany). The sequence data were assembled using Vector NTI 11.5 software (Invitrogen, Carlsbad, CA, USA). The generated sequences were submitted to GenBank under accession numbers MK302070 to MK302091 and MK302092 to MK302113 for the RPO30 and GPCR genes, respectively, and MN161848 for the partial EEV glycoprotein of LSDV Embu/B338/2011.
Nucleotide sequences were aligned using the MUSCLE algorithm and the codon option implemented in MEGA software version 7.0.26 [21]. The complete RPO30 and GPCR gene sequences of 11 additional LSDVs from East Africa (Table 1), 17 LSDVs from other regions, 7 GTPVs, and 4 SPPVs retrieved from GenBank were included for comparative analyses.
Bayesian phylogenetic inference was performed with BEAST v1.8.4 [22] using the HKY+G nucleotide substitution and a UPGMA starting tree option [22]. The Markov chain Monte Carlo method was run with BEAST for 10,000,000 generations, with a sample taken every 10,000 generations. The maximum clade credibility (MCC) was produced using TreeAnnotator after discarding the 10% burn-in determined using the TRACER program. The tree was visualized with the associated meta-data using the ggtree package in R [23]. A section of the alignment of the GPCR, between nucleotide positions 80 and 120, was visualized together with the GPCR tree [23].
2.4. Targeted Next-Generation Sequencing of Selected Variable Sites in the LSDV Genome
To further examine the differences between LSDV Embu and previously described LSDVs, and to rule out the possibility of a mixture between vaccine and field LSDV strains, we analyzed 7 selected hotspots using next-generation sequencing. These hotspots contain sites with nucleotide differences between the LSDV Neethling vaccine and field viruses or sites with nucleotide differences between the historical LSDVs from Kenya and vaccine-related field strains [24].
A PCR reaction was performed in a reaction volume of 20 μL containing 500 nM of each of the forward and reverse primers (Supplementary Table S1), 0.2 mM of dNTPs, 1× buffer (Qiagen), 2.5 U of Taq DNA polymerase (Qiagen), and 2 μL of template DNA. The thermal cycler (Bio-Rad, USA) parameters were: initial denaturation at 95 °C for 4 min, followed by 35 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, and a final extension step at 72 °C for 7 min. The positive PCRs were checked on a 1.5% (w/v) agarose gel. The amplicons were pooled and further purified using a 1.8X Agencourt AMPure XP kit (Beckman Coulter, Brea, CA, USA), and their concentrations were estimated using nanodrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Approximately 50–100 ng of the purified amplicons were enzymatically fragmented to 200 bp length using Ion Shear Plus reagents (Thermo Fisher Scientific, Waltham, MA, USA).
The sequencing library was prepared using an Ion Xpress™ Plus Fragment Library Kit (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s protocol and size-selected using Pippin Prep (Sage Science, Inc., Beverly, MA, USA). The libraries were clonally amplified and enriched using the Ion OneTouch system with the Ion 540™ Kit- OT2 reagents and Ion One Touch ES (enrichment system) as per the manufacturer’s instructions. Template-enriched ISPs were loaded onto the Ion540 chip and were sequenced with 500 flows to generate 200 bp reads on the Ion Torrent S5 sequencer (Thermo Fisher Scientific). The raw sequences were cleaned to remove low-quality and short reads using fastq-mcf v1.04.676 (ea-utils), and their quality was assessed with FastQC (v. 011.5). After mapping the cleaned raw reads against the reference sequence (LSDV NI-2490, NC_003027) using bowtie (v0.7.17), SAMtools (v1.11) was used to generate Mpileup files, and variant calling was performed using BCFtools (v1.9). The sorted reads were displayed within the Integrated Genome Viewer (IGV, v2.8.0) browser [25] to visualize possible mix populations at base mismatch sites (known to vary between the LSDV Neethling-related viruses and common LSDV field isolates). The consensus sequence for each targeted fragment was extracted using the IGV browser. Finally, the consensus sequences were compared to the corresponding fragments in publicly available complete genome sequences of LSDVs by multiple sequence alignments using BioEdit (v7.2.5).
3. Results
3.1. PCR Amplification and Sequencing
For each of the 22 samples, we successfully amplified and sequenced two fragments for the RPO30 gene (554 bp and 520 bp) and three for the GPCR gene (617 bp, 603 bp, and 684 bp) and assembled them to produce the complete RPO30 and GPCR gene sequences. We amplified and sequenced the partial EEV glycoprotein (931–958 bp) and the B22R (250 bp) genes of LSDV Embu/B338/2011.
3.2. Analysis of the RPO30 Gene
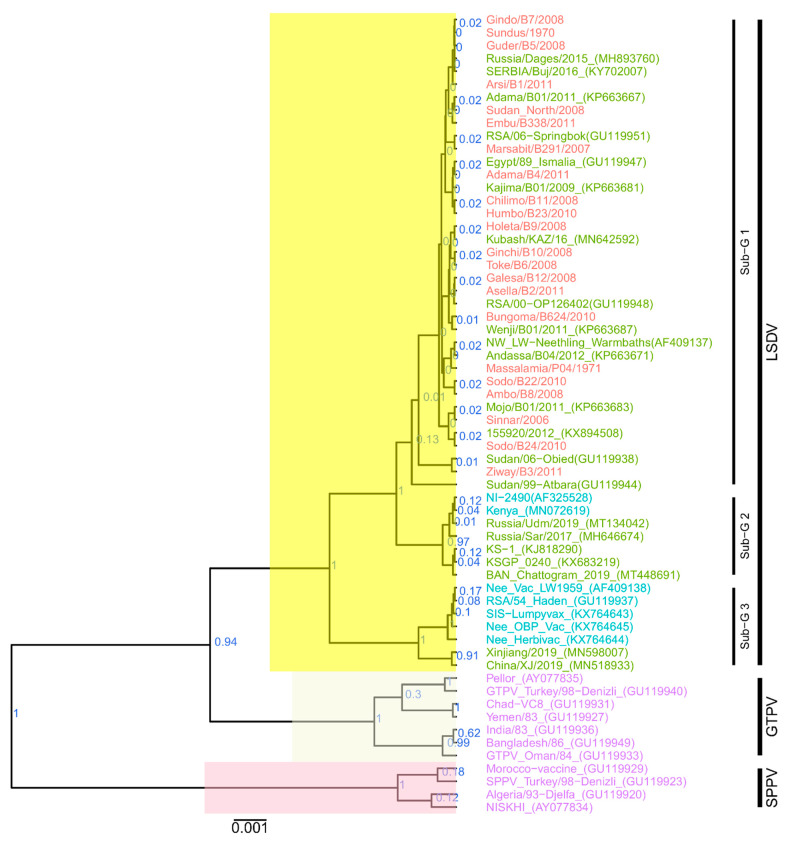
The phylogenetic tree based on the RPO30 gene showed that all 22 isolates in this study belong to LSDV. The phylogenetic reconstructions produced three subgroups for LSDVs with substantial posterior probabilities (Figure 2). Subgroup 1 included all the isolates in this study and common LSDV field isolates from East Africa, South Africa, North Africa, West Africa, and Europe. Subgroup 2 included mainly isolates from Kenya, such as LSDV NI-2490 (AF325528) and LSDV KSGP_O240 (KX683219); LSDV from Bangladesh; and two recombinant LSDV field isolates from Russia, LSDV Russia/Udmurtiya/2019 (MT134042) and LSDV Russia/Saratov/2017 (MH646674). Subgroup 3 included LSDV Neethling-derived vaccines and RSA/54 Haden (FJ869376), both from South Africa and recovered from outbreaks that occurred before 1960, and the LSDV field isolates from China (Figure 2).
Figure 2.
Maximum clade credibility (MCC) tree based on the complete RPO30 gene sequences of capripoxviruses. The posterior probabilities are plotted as respective node labels. The sequences of this study are highlighted in red, and reference sequences are represented with their accession numbers (blue for LSDVs collected before 1960 and green for those collected after 1960). SPPVs and GTPVs are shown in purple.
Multiple sequence alignments of the RPO30 gene showed 100% identity among the East African isolates of this study, at both the nucleotide and amino acid levels, except for LSDV Bungoma/B624/2010. This isolate, collected in Kenya, presented a non-synonymous mutation (A/G, position 338), leading to an E to G amino acid change.
The comparison of the newly sequenced RPO30 genes with publicly available RPO30 sequences from East Africa showed high similarity between the sequences. LSDV KSGP_O240 (KX683219) and similar vaccines as well as LSDV NI-2490 (AF325528) and LSDV Kenya (MN072619), two field isolates from Kenya. Each presented a non-synonymous mutation, T/C (position 292), leading to an S to P change in their amino acid sequences. LSDV Sudan/06-Obied (GU119938) and LSDV Sudan/99-Atbara (GU119944), two previously sequenced isolates from Sudan, each presented one non-synonymous mutation, C/A (position 41) and A/G (position 305), respectively, leading to T to N and D to G amino acid changes, respectively.
3.3. Analysis of the GPCR Gene
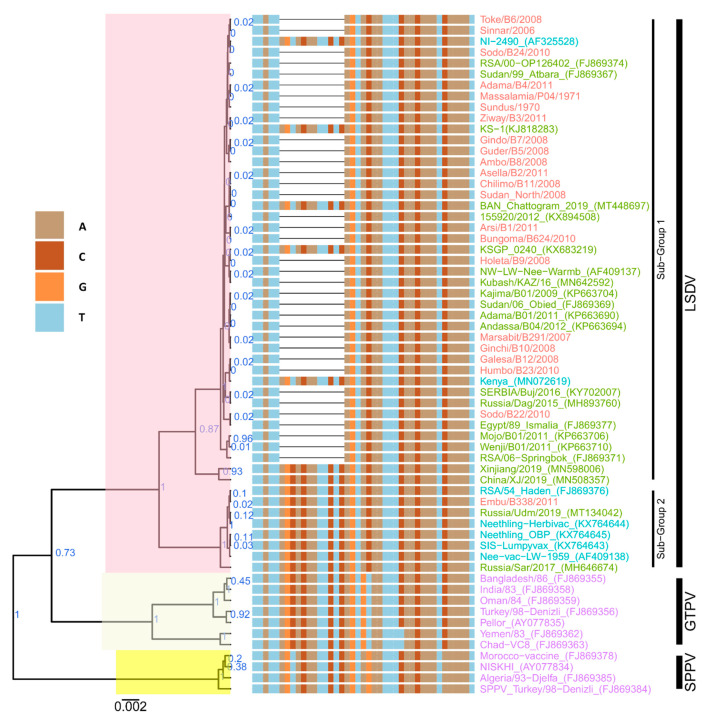
As observed for the RPO30 gene, the phylogenetic tree of the GPCR genes also confirmed all 22 isolates in this study to be LSDVs. In contrast to the RPO30 tree, the GPCR tree produced only two subgroups with strong posterior probability values: 21 out of 22 LSDVs in this study clustered with subgroup 1, along with previously sequenced LSDV isolates from East Africa and other African countries; common LSDV field isolates from Europe; LSDVs from Bangladesh and China; and LSDV KSGP_O240 (KX683219), LSDV NI-2490 (AF325528), and LSDV Kenya (MN072619). Subgroup 2 contained the isolate LSDV Embu/B338/2011 from Kenya, together with LSDV Neethling vaccine LW-1959 (AF409138) and similar vaccines, LSDV RSA/54 Haden (FJ869376) from South Africa, and two recombinant LSDVs from Russia (Figure 3).
Figure 3.
Maximum clade credibility (MCC) tree based on the complete GPCR gene sequences of capripoxviruses, plotted together with multiple sequence alignment. Only the portion of the alignment between positions 80 and 120 is shown. The posterior probabilities are plotted as respective node labels. Study sequences are highlighted in red and reference sequences are represented with their accession numbers (blue for LSDVs collected before 1960 and green for those collected after 1960). SPPVs and GTPVs are shown in purple.
A close inspection of the multiple sequence alignments of GPCR nucleotide sequences showed that the GPCR genes of the isolates in this study were almost 100% similar to each other at both the nucleotide and amino acid levels, except for LSDV Embu/B338/2011. Expanding the comparison to previously sequenced LSDV isolates from East Africa, we observed that the GPCR gene sequences shared 100% identity, except for isolates LSDV Wenji/B01/2011 (KP663710) and LSDV Mojo/B02/2011 (KP663707) from Ethiopia, as well as LSDV KSGP_O240 (KX683219) and LSDV NI-2490 (AF325528) and LSDV Kenya (MN072619), two historical field isolates from Kenya.
LSDV Wenji/B01/2011 differed from other East African field LSDVs with a T/C mutation (position 908 in the GPCR gene of common LSDV field isolates), leading to an F to P change in the amino acid sequence. There was also one synonymous nucleotide change each in LSDV Wenji/B01/2011 (KP663710) and LSDV Mojo/B02/2011 (KP663707). LSDV Embu/B338/2011 contained an insertion of 12 nucleotides (Figure 3), corresponding to amino acids I, L, S, and T, and was 100% identical to LSDV Neethling vaccine LW-1959 (AF409138) in the GPCR gene. The 12-nucleotide insertion was also present in LSDV KSGP_O240 (KX683219), two historical field isolates from Kenya (LSDV NI-2490 (AF325528) and LSDV Kenya (MN072619)), LSDVs from Bangladesh, LSDV RSA/54 Haden (GU119937) from South Africa, recombinant LSDVs from Russia (LSDV Russia/Udmurtiya/2019, LSDV Russia/Saratov/2017, and LSDV Dergachevskyi), and recent LSDV isolates from China (Figure 3). LSDV KSGP_O240 (KX683219) and the historical Kenyan isolates (LSDV Kenya and LSDV NI-2490) differed from LSDV Embu/B338/2011 and LSDV Neethling vaccine by 23 additional nucleotides, leading to four further amino acid changes (S/N, M/I, L/I, M/T).
3.4. Analysis of the EEV Glycoprotein and the B22R Genes of LSDV Embu/B338/2011
As the LSDV Embu/B338/2011 showed significant similarity to LSDV vaccines on the GPCR gene and LSDV field isolates on the RPO30 gene, we sequenced the EEV glycoprotein gene. We compared the results to the publicly available sequences for further elucidation. The EEV glycoprotein gene sequence of LSDV Embu/B338/2011 contained an insertion of 27 nucleotides (9 amino acids), as previously reported in LSDV field isolates; LSDV KSGP-0240-derived vaccines; LSDVs from Bangladesh; and two historical LSDVs, LSDV NI2490 (1958) and LSDV Kenya (1950), both from Kenya. This fragment was absent in LSDV Neethling vaccine-like viruses, LSDVs from China, and the recombinant LSDVs described in Russia (Figure 4).
Figure 4.
Multiple sequence alignments of the partial EEV glycoprotein nucleotide sequences of LSDV Embu/B338/2011 (highlighted in red, with a star after the name) with sequences of 22 additional LSDVs retrieved from GenBank. A unique sequence signature of 27 nucleotides only in LSDV Neethling-like viruses is highlighted in the box. Note also the A to G change highlighted the red box. Identical nucleotides are indicated with dots.
This result confirmed that LSDV Embu/B338/2011 differed from the LSDV Neethling vaccine. We also sequenced the B22R gene of LSDV Embu/B338/2011 and compared it with publicly available LSDV sequences. The results showed that part of the B22R gene of LSDV Embu/B338/2011 was identical to that of field isolates and contained four mutations (A/G, A/G, C/T, A/G) compared with LSDV Neethling vaccine and LSDV Russia/Saratov/2017, a recombinant-like virus from Russia (Figure 5).
Figure 5.
Multiple sequence alignments of the partial B22R gene sequences of LSDV Embu/B338/2011 (highlighted in red, with a star after the name) and 23 additional LSDVs retrieved from GenBank. The variable sites appear as letters, and identical nucleotides appear as dots.
3.5. Analysis of Additional Variable Sites in the LSDV Genome by Targeted Next-Generation Sequencing
The comparative analysis of the partial sequences of the RNA helicase gene (LSDV049), virion core protein p4b gene (LSDV094), the NTPase (LSDV083) gene, and the N1R/p28-like protein gene (LSDV140), derived from the mapping of the NGS reads, revealed additional differences between LSDV Embu/B338/2011 and previously described LSDVs (Supplementary Table S2). There were three synonymous changes on the RNA helicase gene (C/T, A/G, and A/G at positions corresponding to 44,018, 44,168, and 44,202, respectively, in NC_003027) and another single synonymous mutation on the NTPase gene (A/T at the position corresponding to 75,218 in NC_003027) between LSDV Embu/B338/2011 and the two recombinant LSDVs from Russia (LSDV Saratov/2017 and LSDV Russia/Udm/2019). Similarly, we also found noticeable differences between LSDV Embu/B338/2011 and the South African vaccine-related field LSDVs. Hence, two synonymous nucleotide changes occurred on the NTPase gene (A/G and G/A at positions corresponding to 75,253 and 75,325, respectively, in NC_003027) and five occurred on the N1R/p28-like protein gene (A/G, G/A, G/T, A/T, and G/A, at positions corresponding to 133,035, 133,056, 133,108, 133,126, and 133,137, respectively, in NC_003027).
The visualization of the mapped and sorted reads for selected hotspots (Supplementary Figure S1) showed only one virus variant at each variable site. This indicates that only one viral population was present in the sample; therefore, LSDV Embu/B338/2011 is a new LSDV variant, not a mixture of two viruses.
4. Discussion
In this study, we analyzed LSDV isolates collected in East Africa before the appearance of LSD in the Middle East and its incursion into Europe. The East African LSDV field isolates covered in this study (newly sequenced and recovered from public databases) were collected between 1950 and 2012. Overall, except for LSDV Embu/B338/2011, the East African field isolates in this study were highly similar to each other and to the previously sequenced East African field isolates of LSDV for the RPO30 and the GPCR genes. Likewise, the East African isolates were also highly similar to LSDV field isolates encountered elsewhere, including the Middle Eastern and European isolates. As the LSDV genome is stable, limited variability is likely to have occurred following the escape of the virus from Africa to the Middle East and then to Europe.
The only exceptions were LSDV Embu/B338/2011, collected in 2011 in Kenya, which differed from other field isolates by an insertion of 12 nucleotides in its GPCR gene; this was previously described for several historical isolates, namely LSDV Neethling vaccine LW-1959 (AF409138), LSDV RSA/54 Haden (FJ869376), LSDV NI-2490 (AF325528) [3], LSDV Kenya (MN072619), LSDVs from Bangladesh [11], recent LSDV from China, and recombinant field LSDVs from Russia [26,27,28].
In contrast to our findings on the GPCR gene, LSDV Embu/B338/2011 was identical to LSDV field isolates and LSDV KSGP_O240 (KX683219) for the full RP30 gene, the partial EEV glycoprotein gene, and the partial B22R gene. Altogether, the analysis of these four genes showed that LSDV Embu/B338/2011 differs from the two commonly used LSDV vaccines in East Africa: the LSDV KSGP_O240 (KX683219)-derived vaccines and the LSDV Neethling vaccine.
Based on the full GPCR and RPO30 genes and the partial EEV glycoprotein and B22R genes, LSDV Embu/B338/2011 appears to be a mix between an LSDV field isolate and the LSDV Neethling vaccine. In a previous study, based on a comparative analysis of detailed physical maps of the genomes of four Capripoxvirus isolates, the authors suggested that the isolate Yemen goat-1, with mixed features of the Kenya cattle-1 Capripoxvirus field strain and the Iraq goat-1 vaccine strain, was a recombinant virus [29]. More recently, some reports highlighted the presence of recombinant field LSDVs in Russia [26,28]. Analyzing the full genome of LSDV Russia/Saratov/2017 collected at the Russian border with Kazakhstan, the authors suggested that this virus was a recombinant escape of one of the LSDV vaccines derived from the Neethling vaccine strain and a field isolate [26]. Interestingly, the comparison of the EEV glycoprotein and the B22R genes of LSDV Embu/B338/2011 to that of the Russian recombinant-like isolates LSDV Russia/Saratov/2017 suggests that they are different. While LSDV RUSSIA/Saratov/2017 resembled LSDV Neethling vaccine on the EEV glycoprotein and the B22R genes, LSDV Embu/B338/2011 was more related to field isolates on the same genes. This was further confirmed by the presence of additional variable sites on the RNA helicase and the NTPase genes of the sequences derived from the targeted NGS. Our results also highlighted some differences between LSDV Embu/B338/2011 and the recently sequenced vaccine-related field viruses from South Africa [24].
The epidemiological data could support the hypothesis that LSDV Embu/B338/2011 arose through recombination; however, because the isolate was from a previously vaccinated herd, it is also possible that such an isolate was circulating before the vaccination. For instance, LSDV NI-2490 and LSDV Kenya, two historical field isolates collected in Kenya in 1950 and 1958, respectively, also contain a 12-nucleotide insertion in the GPCR gene [4,30].
Several reports have suggested that the occurrences of LSDV in vaccinated flocks were because of vaccination failures or the potential residual virulence of vaccines such as LSDV KS1 [18,31,32,33]. However, our data show that LSDV KSGP O240 and LSDV Embu/B338/2011 are genetically different.
It is also interesting to note that LSDV isolates carrying the 12-nucleotide insertion in their GPCR gene were previously rare. For instance, LSDV Kenya occurred in 1950, followed by LSDV RSA/54 in 1954, LSDV NI-2490 in 1958, LSDV Neethling vaccine LW-1959 in 1958 and 1959, LSDV KS-1 in 1976, and the LSDV Embu/B338/2011 of this study in 2011. However, following the detection of recombinant LSDV RUSSIA/Saratov/2017 in 2017 and LSDV Russia/Udmurtiya/2019, all the new outbreaks characterized so far in Asia involved LSDV field isolates carrying the 12-nucleotide insertion in their GPCR gene. Additionally, the sequencing of archive samples showed that most vaccine-related field viruses from South Africa carry this 12-nucleotide insertion [24].
5. Conclusions
Our data suggest that LSDV genomes were remarkably stable over decades in East Africa; however, LSDV variants such as those with this 12-nucleotide insertion in their GPCR gene were circulating without being readily noticed.
These findings on LSDV Embu/B338/2011 highlight the importance of constant monitoring of LSDV genetics in the field. They also show the importance of analyzing multiple genes while addressing the differentiation between vaccines and field isolates.
Acknowledgments
The authors would like to thank Jenna E. Achenbach (Battelle, VA, USA) for the help rendered the assessment of the manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9061142/s1, Figure S1: Visualization of targeted next-generation sequencing reads for four selected genes in LSDV Embu/B338/2011. The reads were mapped against the LSDV NI-2490 (NC_003027), sorted, and displayed in the IGV browser. The blue arrows indicate the positions of the variable sites in comparison to the reference. Note the absence of subpopulation (only one color per variable site), Table S1: Primers used for targeted sequencing by NGS. The primer sequences, the amplicon size and their positions in LSDV NI-2490 (NC_003027) and the targeted genes are provided, Table S2: Nucleotides polymorphism identified between representative LSDVs for vaccine strains (AF409138 and KX683219), field strains (KY829023 and NC_003027), recombinant viruses (MH646674 and MT134042), and vaccine-related field viruses (MN636841). The polymorphic nucleotides in four targeted genes and their position in LSDV NI-2490 (NC_003027) are shown. Nucleotide changes in LSDV Embu/B338/2011 compared to recombinant viruses (MH646674 and MT134042) are highlighted in yellow. The differences to the South African vaccine-related field virus are highlighted in green.
Author Contributions
Conceptualization, C.E.L. and T.R.C.; methodology, T.R.C., Y.L., C.E.L., R.G., and A.L.; software, Y.L. and C.E.L.; formal analysis, T.R.C., C.E.L., and T.B.K.S.; investigation, T.R.C., J.K.L., M.S. (Mesfin Sahle), Y.A.E., and T.I.B.A.; resources, A.L., M.S. (Melaku Sombo), J.K.L., M.S. (Mesfin Sahle), Y.A.E., and T.I.B.A.; data curation, M.S. (Melaku Sombo), Y.A.E., and T.I.B.A.; writing—original draft preparation, T.R.C., C.E.L., F.J.B., and R.G.; writing—review and editing, F.J.B., A.D., G.C., R.G., A.L., M.S. (Melaku Sombo), J.K.L., T.B.K.S., M.S. (Mesfin Sahle), Y.A.E., and T.I.B.A.; visualization, Y.L. and C.E.L.; supervision, A.D., G.C., R.G., and C.E.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the VETLAB network initiative of the Joint FAO/IAEA Division, funded through the African Renaissance and International Cooperation Fund of South Africa and the Peaceful Uses Initiatives (PUI) funded by Japan and the United States of America.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ (accessed on 25 May 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Buller R.M., Arif B.M., Black D.N., Dumbell K.R., Esposito J.J., Lefkowitz E.J., McFadden G., Moss B., Mercer A.A., Moyer R.W., et al. Family poxviridae. In: Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, CA, USA: 2005. pp. 117–133. [DOI] [Google Scholar]
- 2.Babiuk S., Bowden T.R., Boyle D.B., Wallace D.B., Kitching R.P. Capripoxviruses: An Emerging Worldwide Threat to Sheep, Goats and Cattle. Transbound. Emerg. Dis. 2008;55:263–272. doi: 10.1111/j.1865-1682.2008.01043.x. [DOI] [PubMed] [Google Scholar]
- 3.Tulman E.R., Afonso C.L., Lu Z., Zsak L., Kutish G.F., Rock D.L. Genome of Lumpy Skin Disease Virus. J. Virol. 2001;75:7122–7130. doi: 10.1128/JVI.75.15.7122-7130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Goff C., Lamien C.E., Fakhfakh E., Chadeyras A., Aba-Adulugba E., Libeau G., Tuppurainen E., Wallace D.B., Adam T., Silber R., et al. Capripoxvirus G-protein-coupled chemokine receptor: A host-range gene suitable for virus animal origin discrimination. J. Gen. Virol. 2009;90:1967–1977. doi: 10.1099/vir.0.010686-0. [DOI] [PubMed] [Google Scholar]
- 5.OIE . Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. OIE; Paris, France: 2017. Lumpy skin disease. [Google Scholar]
- 6.Davies F.G. Characteristics of a virus causing a pox disease in sheep and goats in Kenya, with observations on the epidemiology and control. J. Hyg. (Lond) 1976;76:163–171. doi: 10.1017/S0022172400055066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali B.B.H. Investigation of the First Outbreaks of Lumpy Skin Disease in the Sudan. Br. Vet. J. 1977;133:184–189. doi: 10.1016/S0007-1935(17)34140-4. [DOI] [PubMed] [Google Scholar]
- 8.Mebratu C.Y. Observation on the outbreak of lumpy skin disease in Ethiopia. Rev. Elev. Med. Vet. Pays Trop. 1984;37:395–399. [PubMed] [Google Scholar]
- 9.Tuppurainen E.S.M., Oura C.A.L. Review: Lumpy Skin Disease: An Emerging Threat to Europe, the Middle East and Asia. Transbound. Emerg. Dis. 2012;59:40–48. doi: 10.1111/j.1865-1682.2011.01242.x. [DOI] [PubMed] [Google Scholar]
- 10.Mercier A., Arsevska E., Bournez L., Bronner A., Calavas D., Cauchard J., Falala S., Caufour P., Tisseuil C., Lefrançois T., et al. Spread rate of lumpy skin disease in the Balkans, 2015–2016. Transbound. Emerg. Dis. 2018;65:240–243. doi: 10.1111/tbed.12624. [DOI] [PubMed] [Google Scholar]
- 11.Badhy S.C., Chowdhury M.G.A., Settypalli T.B.K., Cattoli G., Lamien C.E., Fakir M.A.U., Akter S., Osmani M.G., Talukdar F., Begum N., et al. Molecular characterization of lumpy skin disease virus (LSDV) emerged in Bangladesh reveals unique genetic features compared to contemporary field strains. BMC Vet. Res. 2021;17:1–11. doi: 10.1186/s12917-021-02751-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu G., Xie J., Luo J., Shao R., Jia K., Li S. Lumpy skin disease outbreaks in China, since 3 August 2019. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13898. [DOI] [PubMed] [Google Scholar]
- 13.Sudhakar S.B., Mishra N., Kalaiyarasu S., Jhade S.K., Hemadri D., Sood R., Bal G.C., Nayak M.K., Pradhan S.K., Singh V.P. Lumpy skin disease (LSD) outbreaks in cattle in Odisha state, India in August 2019: Epidemiological features and molecular studies. Transbound. Emerg. Dis. 2020;67:2408–2422. doi: 10.1111/tbed.13579. [DOI] [PubMed] [Google Scholar]
- 14.Tran H.T.T., Truong A.D., Dang A.K., Ly D.V., Nguyen C.T., Chu N.T., Hoang T.V., Nguyen H.T., Nguyen V.T., Dang H.V. Lumpy skin disease outbreaks in vietnam, 2020. Transbound. Emerg. Dis. 2021 doi: 10.1111/tbed.14022. [DOI] [PubMed] [Google Scholar]
- 15.El-Kholy A.A., Soliman H.M.T., Abdelrahman K.A. Polymerase chain reaction for rapid diagnosis of a recent lumpy skin disease virus incursion to Egypt. Arab J. Biotechnol. 2008;11:293–302. [Google Scholar]
- 16.Alkhamis M.A., VanderWaal K. Spatial and temporal epidemiology of lumpy skin disease in the Middle East, 2012–2015. Front. Vet. Sci. 2016;3:19. doi: 10.3389/fvets.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamien C.E., Le Goff C., Silber R., Wallace D.B., Gulyaz V., Tuppurainen E., Madani H., Caufour P., Adam T., Harrak M.E., et al. Use of the Capripoxvirus homologue of Vaccinia virus 30 kDa RNA polymerase subunit (RPO30) gene as a novel diagnostic and genotyping target: Development of a classical PCR method to differentiate Goat poxvirus from Sheep poxvirus. Vet. Microbiol. 2011;149:30–39. doi: 10.1016/j.vetmic.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 18.Gelaye E., Belay A., Ayelet G., Jenberie S., Yami M., Loitsch A., Tuppurainen E., Grabherr R., Diallo A., Lamien C.E. Capripox disease in Ethiopia: Genetic differences between field isolates and vaccine strain, and implications for vaccination failure. Antivir. Res. 2015;119:28–35. doi: 10.1016/j.antiviral.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Menasherow S., Erster O., Rubinstein-Giuni M., Kovtunenko A., Eyngor E., Gelman B., Khinich E., Stram Y. A high-resolution melting (HRM) assay for the differentiation between Israeli field and Neethling vaccine lumpy skin disease viruses. J. Virol. Methods. 2016;232:12–15. doi: 10.1016/j.jviromet.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Chibssa T.R., Settypalli T.B.K., Berguido F.J., Grabherr R., Loitsch A., Tuppurainen E., Nwankpa N., Tounkara K., Madani H., Omani A., et al. An HRM Assay to Differentiate Sheeppox Virus Vaccine Strains from Sheeppox Virus Field Isolates and other Capripoxvirus Species. Sci. Rep. 2019;9:6646. doi: 10.1038/s41598-019-43158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0. molecular biology and evolution. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012 doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.Y. Ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017 doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 24.Van Schalkwyk A., Kara P., Ebersohn K., Mather A., Annandale C.H., Venter E.H., Wallace D.B. Potential link of single nucleotide polymorphisms to virulence of vaccine-associated field strains of lumpy skin disease virus in South Africa. Transbound. Emerg. Dis. 2020;67:2946–2960. doi: 10.1111/tbed.13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sprygin A., Babin Y., Pestova Y., Kononova S., Wallace D.B., Van Schalkwyk A., Byadovskaya O., Diev V., Lozovoy D., Kononov A. Analysis and insights into recombination signals in lumpy skin disease virus recovered in the field. PLoS ONE. 2018;13:e0207480. doi: 10.1371/journal.pone.0207480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kononov A., Byadovskaya O., Kononova S., Yashin R., Zinyakov N., Mischenko V., Perevozchikova N., Sprygin A. Detection of vaccine-like strains of lumpy skin disease virus in outbreaks in Russia in 2017. Arch. Virol. 2019;164:1575–1585. doi: 10.1007/s00705-019-04229-6. [DOI] [PubMed] [Google Scholar]
- 28.Sprygin A., Pestova Y., Bjadovskaya O., Prutnikov P., Zinyakov N., Kononova S., Ruchnova O., Lozovoy D., Chvala I., Kononov A. Evidence of recombination of vaccine strains of lumpy skin disease virus with field strains, causing disease. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0232584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gershon P.D., Kitching R.P., Hammond J.M., Black D.N. Poxvirus genetic recombination during natural virus transmission. J. Gen. Virol. 1989;70:485–489. doi: 10.1099/0022-1317-70-2-485. [DOI] [PubMed] [Google Scholar]
- 30.Agianniotaki E.I., Tasioudi K.E., Chaintoutis S.C., Iliadou P., Mangana-Vougiouka O., Kirtzalidou A., Alexandropoulos T., Sachpatzidis A., Plevraki E., Dovas C.I., et al. Lumpy skin disease outbreaks in Greece during 2015–16, implementation of emergency immunization and genetic differentiation between field isolates and vaccine virus strains. Vet. Microbiol. 2017;201:78–84. doi: 10.1016/j.vetmic.2016.12.037. [DOI] [PubMed] [Google Scholar]
- 31.Yeruham I., Perl S., Nyska A., Abraham A., Davidson M., Haymovitch M., Zamir O., Grinstein H. Adverse reactions in cattle to a capripox vaccine. Vet. Rec. 1994;135:330–332. doi: 10.1136/vr.135.14.330. [DOI] [PubMed] [Google Scholar]
- 32.Tuppurainen E.S.M., Pearson C.R., Bachanek-bankowska K., Knowles N.J., Amareen S., Frost L., Henstock M.R., Lamien C.E., Diallo A., Mertens P.P.C. Characterization of sheep pox virus vaccine for cattle against lumpy skin disease virus. Antivir. Res. 2014;109:1–6. doi: 10.1016/j.antiviral.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abutarbush S.M., Hananeh W.M., Ramadan W., Al Sheyab O.M., Alnajjar A.R., Al Zoubi I.G., Knowles N.J., Bachanek-Bankowska K., Tuppurainen E.S.M. Adverse Reactions to Field Vaccination Against Lumpy Skin Disease in Jordan. Transbound. Emerg. Dis. 2016;63:e213–e219. doi: 10.1111/tbed.12257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ (accessed on 25 May 2021).