Abstract
Simple Summary
Staphylococcus aureus is present in the microbiota of both humans and some animal species, being recognized as one of the most important opportunistic human pathogens. S. aureus is responsible for causing a variety of infections. Methicillin-resistant S. aureus (MRSA) is particularly important, as it is becoming increasingly prevalent in the population. MRSA has been increasingly reported among wild free-living animals which may impose a public health concern due to its zoonotic potential. To investigate the prevalence and antimicrobial resistance of S. aureus and MRSA in wild synanthropic rodent populations, we conducted this study on 204 rodents captured in port areas in Portugal. The antimicrobial resistance was investigated in all isolates as well as virulence genes and genetic lineages. Thirty-eight S. aureus were isolated. The results showed that six MRSA were detected with particularly interesting mecC-carrying MRSA isolates which had not yet been found in Portugal. A low frequency of antibiotic resistance and virulence genes was observed among the isolates. Nevertheless, a high diversity of clonal lineages was detected among S. aureus some of which are associated with livestock.
Abstract
The frequent carriage of Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA), by wild animals along with its zoonotic potential poses a public health problem. Furthermore, the repeated detection of the mecA gene homologue, mecC, in wildlife raises the question whether these animals may be a reservoir for mecC-MRSA. Thus, we aimed to isolate S. aureus and MRSA from wild rodents living in port areas and to characterize their antimicrobial resistance and genetic lineages. Mouth and rectal swab samples were recovered from 204 wild rodents. The samples were incubated in BHI broth with 6.5% of NaCl and after 24 h at 37 °C the inoculum was seeded onto Baird-Parker agar, Mannitol Salt agar and ORSAB (supplemented with 2 mg/L of oxacillin) plates. Species identification was confirmed by MALDI-TOF MS. The antimicrobial susceptibility testing was performed by the Kirby–Bauer disc diffusion method against 14 antibiotics. The presence of virulence and resistance genes was performed by PCR. The immune evasion cluster (IEC) system was investigated in all S. aureus. All isolates were characterized by MLST, spa- and agr typing. From 204 samples, 38 S. aureus were isolated of which six MRSA were detected. Among the six MRSA isolates, three harbored the mecC gene and the other three, the mecA gene. All mecC-MRSA isolates were ascribed to sequence type (ST) 1945 (which belongs to CC130) and spa-type t1535 whereas the mecA isolates belonged to ST22 and ST36 and spa-types t747 and t018. Twenty-five S. aureus were susceptible to all antibiotics tested. S. aureus isolates were ascribed to 11 MLST and 12 spa-types. S. aureus presents a great diversity of genetic lineages in wild rodents. This is the first report of mecC-MRSA in Portugal.
Keywords: mecC, MRSA, wild rodents, S. aureus
1. Introduction
Staphylococcus aureus is a major opportunistic pathogen that can colonize and infect humans and animals. S. aureus is found as part of the skin and mucous membranes of humans and some animal species. This pathogen is responsible for various types of infections, such as skin and soft tissue infections and toxin-mediated syndromes as well as life-threatening infections such as bacteremia, osteomyelitis and endocarditis [1]. S. aureus has the ability to easily acquire antimicrobial resistance determinants and has an extensive number of virulence factors which are used to establish and maintain infection [2]. S. aureus has been isolated from several animals, including pets and livestock, which are in close contact with humans, and wild animals [3]. Methicillin-resistant Staphylococcus aureus (MRSA) is a major clinical problem in hospitals worldwide [4]. MRSA was initially restricted to the hospital environment causing several types of nosocomial infections and it was named hospital-acquired MRSA (HA-MRSA). Later, MRSA was found in individuals in human communities (community-associated (CA)-MRSA) who have not had previous contact with health facilities [5]. More recently, livestock-associated MRSA (LA-MRSA) has also been widely reported among several species of animals including pigs, poultry and cows [6,7,8]. Moreover, it seems that most mammals can be colonized and/or infected by MRSA since it has also been isolated from pets, such as dogs, cats and horses, and several species of free-living animals [9,10]. The ability of S. aureus to colonize various host species makes it an increasingly recognized zoonotic pathogen. While some clonal complexes (CCs) of S. aureus seem restricted to a certain host, such as ST5 in poultry, other CCs such as CC8, CC22 and CC398 have an extended host spectrum [11]. For instance, initially, S. aureus CC130 was only detected in cattle. More recently, it has been repeatedly found in wild animals and humans and is typically associated with the mecC gene which confers resistance to methicillin [12]. MRSA strains are resistant to almost all beta-lactam antibiotics due to an alteration in the penicillin-binding protein (PBP2a) that is encoded by the mec genes [13]. The mec genes are located in the staphylococcal cassette chromosome mec (SCCmec) which is characterized as a large and potentially transmissible genetic element that not only carries the mec genes but also other antimicrobial resistance genes [14]. SCCmec elements are highly diverse and are currently classified into 14 types [15]. Of all the mec genes, mecA is the predominant variant. However, in 2011, a divergent mecA homologue, mecC, was identified in MRSA strains from human samples in Ireland [16]. Later, two new mec genes were reported, mecB and mecD, which are much less frequent and were both detected in Macrococcus caseolyticus [17,18]. After the first detection of mecC, it has been reported in several countries of all continents and from multiple origins, including humans, animals and the environment [10,19,20,21,22,23,24,25,26]. The origins and reservoirs of the mecC gene in MRSA strains are still unknown. First, mecC was associated with LA-MRSA. However, the continued detection of this gene in wild animals and in the environment indicates that the primary reservoir of the mecC gene may be the natural environment [10]. The mecC gene was first described encoding resistance to methicillin in S. aureus over a decade ago. However, although numerous MRSA studies have been published in recent years in Portugal, mecC has never been detected. Globalized maritime trading routes facilitate the dispersal of synanthropic rodents and their pathogens, with seaports constituting pivotal entry points and potential hotspots of disease. Depending on the extension of the urban matrix, commensal rodents may constitute important vehicles of MRSA not only to humans (directly or indirectly) but also to other resident species with which they may interact. In this study, we isolated methicillin-susceptible S. aureus (MSSA) and MRSA from wild synanthropic rodents captured in two port cities, one continental/highly urbanized and one insular/less urbanized, characterizing all isolates regarding the antimicrobial resistance, virulence and clonal lineages.
2. Materials and Methods
2.1. Samples and Bacterial Isolates
From May 2019 to March 2020, mouth and rectal swabs samples were recovered from 204 wild rodents, including nine Mus musculus, 75 Rattus rattus and 120 Rattus norvegicus. Rodents were live trapped with Sherman and Tomahawk traps in port and surrounding areas (up to 10 km) of Lisbon and Ponta Delgada (São Miguel island, Azores), Portugal. Animals were obtained in the framework of the R&D project PTDC/SAU-PUB/29254/2017 and all procedures followed the European directive 2010/63/EU as stated by the Animal Welfare Body ORBEA of the Faculty of Sciences, University of Lisbon (ethics committee statement 4/2018). The location and specific characteristics of the animals are shown in the supplementary material (Table S1). One sample was collected from each animal. The samples were incubated in BHI broth (Oxoid, Basingstoke, Hampshire, England) with 6.5% NaCl for 24 h at 37 °C. The inoculum was seeded onto Baird-Parker agar (Oxoid, Basingstoke, Hampshire, England) supplemted with Egg Yolk Tellurite Emulsion, Mannitol Salt agar (Oxoid, Basingstoke, Hampshire, England) and ORSAB (supplemented with 2 mg/L of oxacillin) Oxoid, Basingstoke, Hampshire, England) plates for S. aureus and MRSA isolation and incubated at 37 °C for 24–48 h. One colony was recovered from each plate. S. aureus species was identified by biochemical tests (Gram staining, DNase and catalase) and confirmed by MALDI-TOF MS (Bruker Daltonics GmbH; Bremen, Germany).
2.2. Antimicrobial Susceptibility Testing
The phenotypic resistance characterization of the isolates was performed by the Kirby–Bauer disk diffusion method against the following 14 antimicrobial agents: cefoxitin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg), erythromycin (15 μg), fusidic acid (10 μg), gentamicin (10 μg), kanamycin (30 μg), linezolid (10 μg), mupirocin (200 μg), penicillin (1 U), tetracycline (30 μg), tobramycin (10 μg), and trimethoprim/sulfamethoxazole (1.25/23.75 μg). The results were evaluated according to the EUCAST 2018 guidelines with the exception of kanamycin which followed the guidelines of CLSI 2017. S. aureus strain ATCC 25923 was used as quality control in the susceptibility assays.
2.3. Antimicrobial Resistance and Virulence Genes
All isolates were screened for the presence antimicrobial resistance genes according to their phenotypic resistance. The presence of mecA and mecC genes was investigated by PCR and sequencing as previously described [27,28]. The following genes were tested: blaZ, blaZ-SCCmecXI, tet(K), tet(M), tet(L), tet(O), erm(A), erm(B), erm(C), erm(T), msr(A/B), mphC, linA, linB, vgaA, vgaB, vgaC, aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa and ant(4′)-Ia [12,29]. The presence of virulence genes encoding Panton–Valentine leucocidin (PVL) (lukF/lukS-PV), alpha- and beta-hemolysins (hla and hlb), exfoliative toxins (eta and etb) and toxic shock syndrome toxin (tst) was determined by PCR as previously described [30]. The immune evasion cluster (IEC) system was studied by PCR [31]. The isolates were screened for the presence of the scn gene, which is a marker of the IEC system, and the presence of chp, sak, sea and sep genes was carried out in scn-positive isolates to determine the IEC group. The presence of the genes encoding for SCCmec were investigated by PCR as previously described [27,28]. Positive and negative controls used in all experiments belonged to the strain collection of University of Trás-os-Montes and Alto Douro.
2.4. Molecular Typing
Multilocus-sequence-typing (MLST) was performed in all isolates and according to Enright et al., 2000 [32]. The sequence type (ST) was obtained by comparing the allelic profile of each isolate to the MLST database. All isolates were typed by spa-typing as previously described [33] and the obtained sequences were analyzed using Ridom® Staph-type software (version 1.5, Ridom GmbH, Würzburg, Germany). All isolates were characterized by agr-typing (I–IV) using specific primers [34].
3. Results
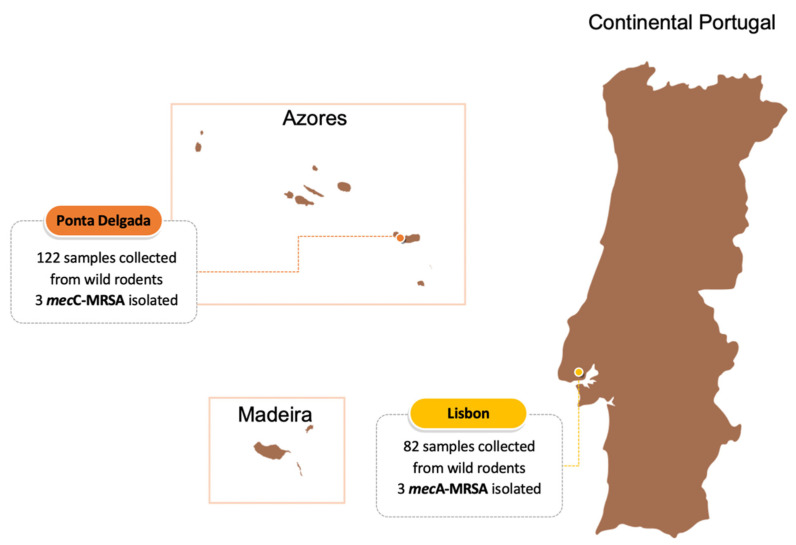
From the 204 rodent samples, 38 (18.6%) S. aureus were isolated from both R. norvegicus and R. rattus, 26 in Lisbon and 12 in Ponta Delgada. From the 38 isolates, six (15.7%) MRSA were identified, three in each study city (Figure 1). Three MRSA were mecA-positive and three were mecC-positive, with the latter only being detected in Ponta Delgada. The characteristics of the isolates are shown in Table 1. S. aureus was isolated from 13 (17.3%) and 25 (20,8%) of the 75 and 120 R. rattus and R. norvegicus, respectively. All mecA-MRSA strains were isolated from R. rattus whereas two mecC-MRSA were isolated from R. rattus and one from R. norvegicus. All mecC-carrying strains were isolated from rodents from S. Miguel island (Azores) and mecA-MRSA isolates were recovered from two different Lisbon locations which are located less than 500 m from hospitals (Table S1). Isolates of mecC-MRSA showed resistance to penicillin and cefoxitin and harbored the blaZ-SCCmecXI. They were all ascribed to spa-types t1535 and ST1945 (which belongs to CC130), to SCCmec type XI and agr type III. The mecC-positive isolates lacked the virulence genes tested for but were positive for scn and sak genes from the IEC system and were, therefore, classified as IEC-type E. Two mecA-positive isolates showed a multidrug-resistant profile, since they were resistant to at least three different classes of antibiotics. All mecA-MRSA were resistant to penicillin, cefoxitin and ciprofloxacin and harbored the blaZ gene. Two strains also had resistance to erythromycin and one strain was resistant to aminoglycosides and harbored aph(3′)-IIIa; however, it lacked the aac(6′)-Ie-aph(2″)-Ia gene which confers resistance to gentamicin. Two mecA-MRSA isolates were ascribed to ST22 and spa-type t747 and one isolate was ST36 (CC30) and t018. Three strains were not typeable with respect to the SCCmec types tested and were agr type I. These isolates also harbored the hlb and hld virulence genes. Two MRSA isolates lacked the IEC system genes and one harbored the scn, sak, chp and sea and was classified as IEC-type A. Regarding the MSSA isolates, 26 were susceptible to all antibiotics tested. Among the remaining six MSSA, all were resistant to penicillin and harbored the blaZ gene. All MSSA isolates, except one, carried the virulence gene hld and 22 harbored the hlb gene. All strains were negative for tst, eta and etb genes and lacked the PVL toxin. Four MSSA isolates carried the scn gene and the IEC genes and were further studied. Two MSSA strains were ascribed to IEC-type C, one to type E, one to type A and one strain harbored only the scn gene. Regarding the molecular typing, agr I was detected in 18 isolates, nine isolates were agr III and five were not typeable. The MSSA isolates were distributed in 11 STs, one new ST first described in this study (ST6574) and another 10 STs, including, ST1094 (n = 7), ST130 (n = 7), ST398 (n = 4), ST5926 (n = 3), ST8 (n = 3), ST1245 (n = 2), ST1318, ST1290, ST34 and ST6. Regarding the spa-typing, the MSSA isolates were ascribed to 12 different spa-types, including t516 (n = 7), t843 (n = 7), t1451 (n = 6), t4608 (n = 2), t3256 (n = 2), t1535 (n = 2), t2078, t571, t414, t16615, t131 and t19688 which is first reported in this study.
Figure 1.
Number of samples collected from wild rodents captured in port areas of Lisbon and Ponta Delgada (S. Miguel island, Azores) and distribution of mecA- and mecC-MRSA in each location.
Table 1.
Characteristics of the MRSA and MSSA strains isolated from wild rodents in Portugal.
| Isolate | Host Species | Antimicrobial Resistance | Virulence Factors | Molecular Typing | |||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Genotype | IEC System | Other Genes | ST (CC) | spa | agr | SCCmec | ||
| VS2808 | Rattus rattus | PEN, FOX | mecC, blaZ-SCCmecXI | Type E | - | 1945 (130) | t1535 | III | XI |
| VS2809 | Rattus rattus | PEN, FOX | mecC, blaZ-SCCmecXI | Type E | - | 1945 (130) | t1535 | III | XI |
| VS2810 | Rattus norvegicus | PEN, FOX | mecC, blaZ-SCCmecXI | Type E | - | 1945 (130) | t1535 | III | XI |
| VS2811 | Rattus rattus | PEN, FOX, CIP, ERY | mecA, blaZ | - | hlb, hld | 22 (22) | t747 | I | N.T. |
| VS2812 | Rattus rattus | PEN, FOX, CIP, CN, KAN, ERY, CD | mecA, blaZ, aph(3′)-IIIa, ermA | Type A | hlb, hld | 36 (30) | t018 | I | N.T. |
| VS2813 | Rattus rattus | PEN, FOX, CIP | mecA, blaZ | - | hlb, hld | 22 (22) | t747 | I | N.T. |
| VS2814 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2815 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2816 | Rattus norvegicus | PEN | blaZ | - | hlb, hld | 1094 | t516 | I | - |
| VS2817 | Rattus rattus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2818 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2819 | Rattus rattus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2820 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1094 | t516 | I | - |
| VS2821 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2822 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2823 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2824 | Rattus norvegicus | Susceptible | - | scn | hld | 130 | t843 | III | - |
| VS2825 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 130 | t843 | III | - |
| VS2826 | Rattus rattus | Susceptible | - | - | hlb, hld | 130 | t3256 | III | - |
| VS2827 | Rattus norvegicus | Susceptible | - | - | - | 130 | t3256 | N.T. | - |
| VS2828 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1245 | t843 | III | - |
| VS2829 | Rattus rattus | Susceptible | - | - | hlb, hld | 1245 | t843 | III | - |
| VS2830 | Rattus norvegicus | PEN | blaZ | - | hld | 398 | t1451 | I | - |
| VS2831 | Rattus norvegicus | PEN | blaZ | - | hld | 398 | t1451 | I | - |
| VS2832 | Rattus norvegicus | PEN | blaZ | - | hld | 398 | t1451 | I | - |
| VS2833 | Rattus norvegicus | Susceptible | - | - | hld | 398 | t571 | I | - |
| VS2834 | Rattus rattus | Susceptible | - | Type C | hld | 5926 | t1451 | N.T. | - |
| VS2835 | Rattus norvegicus | Susceptible | - | Type C | hld | 5926 | t1451 | I | - |
| VS2836 | Rattus norvegicus | PEN | blaZ | Type E | hlb, hld | 1318 | t2078 | I | - |
| VS2837 | Rattus norvegicus | Susceptible | - | - | hld | 8 (8) | t4608 | I | - |
| VS2838 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 8 (8) | t19688 | I | - |
| VS2839 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 8 (8) | t4608 | I | - |
| VS2840 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 6574 | t1535 | III | - |
| VS2841 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 6574 | t1535 | N.T. | - |
| VS2842 | Rattus norvegicus | PEN | blaZ | - | hlb, hld | 34 (30) | t414 | N.T. | - |
| VS2843 | Rattus rattus | Susceptible | - | - | hld | 6 (5) | t16615 | I | - |
| VS2844 | Rattus rattus | Susceptible | - | - | hlb, hld | 5926 | t1451 | N.T. | - |
| VS2845 | Rattus norvegicus | Susceptible | - | - | hlb, hld | 1290 (1) | t131 | I | - |
Abbreviations. PEN: penicillin, FOX: cefoxitin, CIP: ciprofloxacin, ERY: erythromycin, CN: gentamicin, KAN: kanamycin, CD: clindamycin, N.T. not typeable.
4. Discussion
S. aureus colonization and infection in wild animals have only been superficially investigated since most research studies focus on other animal species with economic importance [35]. Nevertheless, to fully understand the process of infection and colonization in humans, an ecological approach is required. In fact, the natural colonization of rodents could have a special interest since laboratory rats and mice are commonly used as an experimental model to study S. aureus infection [36]. However, studies on the prevalence of S. aureus in wild rodents are scarce. In our study, we collected 204 samples of wild rodents living in port and surrounding areas, up to a 10 km radius from the port, both in Lisbon and Ponta Delgada (Azores). These port cities represent different levels of urbanization, with Lisbon portraying a very tight urban matrix in opposition to Ponta Delgada, where, within 2–3 km from the port, a more rural landscape where livestock and farming practices take place. In total, a prevalence of 18.6% of S. aureus colonizing these animals was obtained. Six (2.9%) out of 204 samples (detected both in R. norvegicus and R. rattus) were positive for MRSA, three in Lisbon and three in Ponta Delgada. One study by van de Giessen et al. (2009) reported a prevalence of MSSA and MRSA among rats living on livestock farms of 41.8% and 11.6%, respectively, which was higher than the prevalence of these strains in our study [37]. Raafat et al. (2020) also reported a higher prevalence of S. aureus (25.5%) among free-living wild rats but a lower occurrence of MRSA of 1.38% [11]. Nevertheless, other studies have reported a prevalence of MSSA in wild rodents similar to ours [36,38,39]. Three of the six MRSA harbored the mecC gene and were typed as ST1945-t1535-agrIII-SCCmecXI. ST1945 belongs to CC130 which carries mecC instead of mecA. It has been suggested that there might be a mutual exchange of mecC-MRSA between livestock and wild animals since it was thought that CC130 originated in ruminants [40]. In fact, one of the three MRSA carrying the mecC gene was detected in a R. norvegicus captured in a cattle/dairy farm in the outskirts of Ponta Delgada. ST1945-MRSA-t1535 isolates have only been reported in wild animals (red deer, wild rodents and wild birds) in Spain [38,41,42]. Nevertheless, MRSA spa-type t1535 has been isolated from humans in several European countries, including Germany and Austria [43,44]. ST1945-MRSA has also been isolated from humans in the UK, Spain and France and from animals in Spain, France and Germany but associated with other spa-types [12,45,46,47,48]. ST1945 is usually associated with spa-types t1535 or t843 and always related with mecC-carrying strains [38,41,42]. Studies have shown that mecC-positive ST1945 isolates belong to agr III and usually carry the blaZ-SCCmecXI gene [38,41,42]. All our mecC isolates harbored both the scn and sak genes and they were, consequently, ascribed to IEC type E. Although the presence of IEC genes usually suggests a possible human origin, it has been proposed that IEC-type E might be a conserved trait of ST1945 isolates since several studies conducted in Spain and the UK reported the presence of these genes in mecC-MRSA ST1945 isolates [49]. Furthermore, in most studies reporting mecC-MRSA, the presence of IEC genes was not investigated. Our isolates were susceptible to all antimicrobial agents tested except for ß-lactams and did not present any of the virulence genes tested as reported in other studies [38,41,42]. The blaZ-SCCmecXI is an allotype of the S. aureus blaZ and has 67% amino acid identity [16]. The origin of the mecC gene is unclear; however, mecC-CC130 has been regarded as an animal-adapted lineage of S. aureus which suggests that mecC may have arisen in animals [46]. Therefore, mecC-MRSA strains may impose a zoonotic risk with important public health consequences. Although the presence of MRSA in wild animals is not very common, it seems that MRSA isolated from wildlife are more frequently associated with the mecC gene since it has been isolated from several animal species, including, foxes, deer, hares, hedgehogs, rodents, otters, rabbits, storks, magpies and vultures [10]. However, we also isolated three mecA-positive MRSA in this study. Two mecA isolates were ST22-t747-agrI and one was ST36-t018-agrI. ST22-MRSA-t747 has been reported worldwide, often associated with HA-MRSA. In Portugal, this clone is one of the most frequently found in the nosocomial environment and has been reported associated with several infections [50,51]. The ST22-MRSA-t747 isolate was IEC-type A which may confirm a possible human origin. ST36 belongs to CC30 and it is also a healthcare-associated MRSA clone [52]. MRSA ST36, when associated with SCCmec type II, is known as the epidemic clone EMRSA-16 [52]. However, in our study MRSA ST36 was not typeable regarding the SCCmec. Several studies have reported HA-MRSA strains colonizing wild animals, particularly those in close proximity with human activities [53,54,55]. In our study, mecA-positive strains were isolated in two locations near hospitals which may explain the fact that HA-MRSA strains were identified in wild rodents.
Most of the MSSA isolates from wild rodents had a very low frequency of antimicrobial resistance. In fact, the great majority of the isolates were susceptible to all tested antibiotics. These results are in accordance with other studies conducted in wild animals [41,56]. This low prevalence of antimicrobial resistance determinants in wildlife may be explained by the fact that these animals do not have direct contact with antibiotics and live in the absence of selective pressure [38,56,57]. Nevertheless, other studies have shown that wild animals with no apparent contact with antibiotics carried antimicrobial resistant strains [58,59]. Therefore, wildlife may be considered a sentinel of antimicrobial resistance, environmental pollution and, in consequence, the prevalence of resistance in wild reservoirs will depend on the geographical area where they are living. The most frequently detected resistance in MSSA was to penicillin which was found in six out of 32 isolates. The molecular typing revealed a high diversity of genetic lineages among the MSSA isolates. Eleven STs and 12 spa-types were detected. ST1094 and ST130 were the predominant STs found in our study. ST1094 is a singleton and was only found in strains ascribed to spa-type t516 and agr type I. All ST1094-MSSA-t516 isolates were susceptible to all antibiotics tested with the exception of one penicillin-resistant isolate. ST1094 have been previously reported in wild rodents in Boston, associated with low resistance rates, but all strains were typed as t933 [60]. ST1094 has also been found in Asia in human samples in Myanmar (associated with t516) and in ready-to-eat food in China [61,62]. Five out of seven MSSA ST130 isolates were typed as t843 and two isolates were typed as t3256. CC130 was firstly associated with MSSA but more recently this CC has been continuedly reported as mecC-MRSA. CC130 is known to be a livestock-associated lineage particularly common in small ruminants such as domestic sheep and goats [63]. Nevertheless, MSSA CC130-t843 has been isolated from wild animals, including wild rodents and boars [36,37,64]. As for CC130-t3256, this strain has been isolated from humans, bovine mastitis and wild animals, but always in MRSA strains harboring the mecC gene [44,65,66]. Four MSSA isolates were ST398 (CC398) of which three were typed as t1451 and one as t571 which seem to be the spa-types commonly associated with MSSA CC398 [67]. None of the isolates belonged to the spa-type t011 which is the most frequent spa-type in CC398 strains [56]. CC398 strains are broadly disseminated across Europe and the rest of the world. Additionally, both CC398 MRSA and MSSA do not seem to have host specificity as they have been isolated from livestock, particularly pigs, but also from humans and wild animals [54,56,68]. In our study, CC398 isolates did not exhibited the antimicrobial resistance patterns, predominantly resistance to tetracycline, often observed in MRSA CC398 and commonly associated with livestock [68]. The spa-type t1451 was detected in ST398 strains and also in ST5926 isolates. Interestingly, one ST5926 isolate was not typeable regarding the agr and one was agr I. Furthermore, both ST5926 isolates were positive for scn and chp genes, being ascribed to IEC-type C, which points to a possible human origin. One isolate ascribed to ST1318 and spa-type t2078 was also positive for IEC genes, namely, scn and sak (IEC-type E). ST6574 was described in this study for the first time, and it was found in two isolates which were spa-type t1535. t1535 has been reported in wild animals associated with mecC-MRSA as in our study. The spa-type t19688 was also firstly reported in this study and was associated with CC8. Antimicrobial resistance is an important public health problem in both human and veterinary medicine. Epidemiological and surveillance data on new lineages and antimicrobial resistance of S. aureus and MRSA will be useful in devising an effective antimicrobial stewardship program in hospitals and also in choosing treatment strategies [69].
5. Conclusions
Wild synanthropic rodents are the first mecC-positive detected hosts in Portugal. So far, mecC-MRSA was only detected in the Azores, but despite the narrow geographic scope of this study, it was found in both R. norvegicus and R. rattus, in a livestock farm and a forest area, respectively. Furthermore, wild rodents seem to be a natural host of S. aureus strains, including MRSA, detected both in continental in insular settings, which can represent a dangerous vector for those strains with zoonotic potential. Besides, rats are considered a common urban pest species, especially in port areas, and their associated pathogens may spread to other animals or humans. Also, the detection of mecA-MRSA in R. rattus in the Lisbon port is a clear indication of the potential passive worldwide dispersion of these rodents and their zoonotic pathogens, including MRSA, through maritime transport.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ani11061537/s1, Table S1: location and specific characteristics of wild rodents captured in port areas of Lisbon and Azores island.
Author Contributions
Conceptualization, V.S. and P.P.; methodology, V.S., M.C. and P.P.; investigation, V.S.; visualization, V.M., E.F. and L.R.; resources, S.I.G., S.B.B. and M.T.T.-J.; data curation, V.S.; writing—original draft preparation, V.S.; writing—review and editing, V.S., S.I.G. and P.P.; supervision, M.C., J.L.C., G.I. and P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the R&D Project CAREBIO2—Comparative assessment of antimicrobial resistance in environmental biofilms through proteomics—towards innovative theranostic biomarkers, with reference NORTE-01-0145-FEDER-030101; PTDC/SAU-INF/30101/2017, financed by the European Regional Development Fund (ERDF) through the Northern Regional Operational Program (NORTE 2020) and the Foundation for Science and Technology (FCT); and PTDC/SAU-PUB/29254/2017 funded by FCT. This work was also supported by the Associate Laboratory for Green Chemistry-LAQV which is financed by national funds from FCT/MCTES (UID/QUI/50006/2019) and by the Associate Laboratory CESAM (UIDP/50017/2020+UIDB/50017/2020), financed by FCT/MCTES through national funds. Vanessa Silva is grateful to FCT (Fundação para a Ciência e a Tecnologia) for financial support through the PhD grant SFRH/BD/137947/2018. Sofia I. Gabriel was funded by national funds (OE), through FCT, in the scope of the framework contract foreseen in the numbers 4, 5 and 6 of article 23, Decree-Law 57/2016 of 29 August, changed by Law 57/2017, of 19 July.
Institutional Review Board Statement
Animals were obtained in the framework of the R&D project PTDC/SAU-PUB/29254/2017 and all procedures followed the European directive 2010/63/EU as stated by the Animal Welfare Body ORBEA of the Faculty of Sciences, University of Lisbon (ethics committee statement 4/2018 approved on 12/12/2018).
Data Availability Statement
The data presented in this study are available in Supplementary Material Table S1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Papadopoulos P., Papadopoulos T., Angelidis A.S., Boukouvala E., Zdragas A., Papa A., Hadjichristodoulou C., Sergelidis D. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 2018;69:43–50. doi: 10.1016/j.fm.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian D., Harper L., Shopsin B., Torres V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017;75 doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monecke S., Gavier-Widén D., Hotzel H., Peters M., Guenther S., Lazaris A., Loncaric I., Müller E., Reissig A., Ruppelt-Lorz A., et al. Diversity of Staphylococcus aureus Isolates in European Wildlife. PLoS ONE. 2016;11:e0168433. doi: 10.1371/journal.pone.0168433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao J., Chen L., Wang J., Wang W., Chen D., Li L., Li B., Deng Y., Xu Z. Current methodologies on genotyping for nosocomial pathogen methicillin-resistant Staphylococcus aureus (MRSA) Microb. Pathog. 2017;107:17–28. doi: 10.1016/j.micpath.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Khan A., Wilson B., Gould I.M. Current and future treatment options for community-associated MRSA infection. Expert Opin. Pharmacother. 2018;19:457–470. doi: 10.1080/14656566.2018.1442826. [DOI] [PubMed] [Google Scholar]
- 6.Sieber R.N., Larsen A.R., Urth T.R., Iversen S., Møller C.H., Skov R.L., Larsen J., Stegger M. Genome investigations show host adaptation and transmission of LA-MRSA CC398 from pigs into Danish healthcare institutions. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-55086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernier-Lachance J., Arsenault J., Usongo V., Parent É., Labrie J., Jacques M., Malouin F., Archambault M. Prevalence and characteristics of Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec, Canada. PLoS ONE. 2020;15:e0227183. doi: 10.1371/journal.pone.0227183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadlec K., Entorf M., Peters T. Occurrence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus in quarter milk samples from dairy cows in Germany. Front. Microbiol. 2019;10:1295. doi: 10.3389/fmicb.2019.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loncaric I., Lepuschitz S., Ruppitsch W., Trstan A., Andreadis T., Bouchlis N., Marbach H., Schauer B., Szostak M.P., Feßler A.T., et al. Increased genetic diversity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from companion animals. Vet. Microbiol. 2019;235:118–126. doi: 10.1016/j.vetmic.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Silva V., Capelo J.L., Igrejas G., Poeta P. Molecular Epidemiology of Staphylococcus aureus Lineages in Wild Animals in Europe: A Review. Antibiotics. 2020;9:122. doi: 10.3390/antibiotics9030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raafat D., Mrochen D.M., Al’Sholui F., Heuser E., Ryll R., Pritchett-Corning K.R., Jacob J., Walther B., Matuschka F.-R., Richter D., et al. Molecular Epidemiology of Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus in Wild, Captive and Laboratory Rats: Effect of Habitat on the Nasal S. aureus Population. Toxins. 2020;12:80. doi: 10.3390/toxins12020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Álvarez L., Holden M.T.G., Lindsay H., Webb C.R., Brown D.F.J., Curran M.D., Walpole E., Brooks K., Pickard D.J., Teale C., et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet. Infect. Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ba X., Harrison E.M., Edwards G.F., Holden M.T.G., Larsen A.R., Petersen A., Skov R.L., Peacock S.J., Parkhill J., Paterson G.K. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J. Antimicrob. Chemother. 2014;69:594–597. doi: 10.1093/jac/dkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker K. Methicillin-Resistant Staphylococci and Macrococci at the Interface of Human and Animal Health. Toxins. 2021;13:61. doi: 10.3390/toxins13010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urushibara N., Aung M.S., Kawaguchiya M., Kobayashi N. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan. J. Antimicrob. Chemother. 2020;75:46–50. doi: 10.1093/jac/dkz406. [DOI] [PubMed] [Google Scholar]
- 16.Shore A.C., Deasy E.C., Slickers P., Brennan G., O’Connell B., Monecke S., Ehricht R., Coleman D.C. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2011;55:3765–3773. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker K., van Alen S., Idelevich E.A., Schleimer N., Seggewiß J., Mellmann A., Kaspar U., Peters G. Plasmid-Encoded Transferable mecB-Mediated Methicillin Resistance in Staphylococcus aureus. Emerg. Infect. Dis. 2018;24:242–248. doi: 10.3201/eid2402.171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakhundi S., Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018;31:e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan A.A., Ali A., Tharmalingam N., Mylonakis E., Zahra R. First report of mecC gene in clinical methicillin resistant S. aureus (MRSA) from tertiary care hospital Islamabad, Pakistan. J. Infect. Public Health. 2020;13:1501–1507. doi: 10.1016/j.jiph.2020.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Dweba C.C., Zishiri O.T., El Zowalaty M.E. Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus. Pathogens. 2019;8:79. doi: 10.3390/pathogens8020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aklilu E., Chia H.Y. First mecC and mecA Positive Livestock-Associated Methicillin Resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from Dairy Cattle in Malaysia. Microorganisms. 2020;8:147. doi: 10.3390/microorganisms8020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bietrix J., Kolenda C., Sapin A., Haenni M., Madec J.-Y., Bes M., Dupieux C., Tasse J., Laurent F. Persistence and Diffusion of mecC-Positive CC130 MRSA Isolates in Dairy Farms in Meurthe-et-Moselle County (France) Front. Microbiol. 2019;10:47. doi: 10.3389/fmicb.2019.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincze S., Stamm I., Kopp P.A., Hermes J., Adlhoch C., Semmler T., Wieler L.H., Lübke-Becker A., Walther B. Alarming Proportions of Methicillin-Resistant Staphylococcus aureus (MRSA) in Wound Samples from Companion Animals, Germany 2010–2012. PLoS ONE. 2014;9:e85656. doi: 10.1371/journal.pone.0085656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worthing K.A., Coombs G.W., Pang S., Abraham S., Saputra S., Trott D.J., Jordan D., Wong H.S., Abraham R.J., Norris J.M. Isolation of mecC MRSA in Australia. J. Antimicrob. Chemother. 2016;71:2348–2349. doi: 10.1093/jac/dkw138. [DOI] [PubMed] [Google Scholar]
- 25.Porrero M.C., Harrison E., Fernández-Garayzábal J.F., Paterson G.K., Díez-Guerrier A., Holmes M.A., Domínguez L. Detection of mecC-Methicillin-resistant Staphylococcus aureus isolates in river water: A potential role for water in the environmental dissemination. Environ. Microbiol. Rep. 2014;6:705–708. doi: 10.1111/1758-2229.12191. [DOI] [PubMed] [Google Scholar]
- 26.Maria de Fatima N.F., Penna B., Pereira R.F.A., Geraldo R.B., Folly E., Castro H.C., Aguiar-Alves F. First report of meticillin-resistant Staphylococcus aureus harboring mecC gene in milk samples from cows with mastitis in southeastern Brazil. Brazilian J. Microbiol. 2020;51:2175–2179. doi: 10.1007/s42770-020-00385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuny C., Layer F., Strommenger B., Witte W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS ONE. 2011;6:e24360. doi: 10.1371/journal.pone.0024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K., Sparling J., Chow B.L., Elsayed S., Hussain Z., Church D.L., Gregson D.B., Louie T., Conly J.M. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2004;42:4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva V., Almeida F., Silva A., Correia S., Carvalho J.A., Castro A.P., Ferreira E., Manageiro V., Caniça M., Igrejas G., et al. First report of linezolid-resistant cfr-positive methicillin-resistant Staphylococcus aureus in humans in Portugal. J. Glob. Antimicrob. Resist. 2019;17:323–325. doi: 10.1016/j.jgar.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Jarraud S., Mougel C., Thioulouse J., Lina G., Meugnier H., Forey F., Etienne J., Vandenesch F., Nesme X. Relationships between Staphylococcus aureus Genetic Background, Virulence Factors, agr Groups (Alleles), and Human Disease. Infect. Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Wamel W.J.B., Rooijakkers S.H.M., Ruyken M., van Kessel K.P.M., van Strijp J.A.G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006;188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmsen D., Claus H.H.H.H., Witte W., Rothgänger J., Claus H.H.H.H., Turnwald D., Vogel U. Typing of Methicillin-Resistant Staphylococcus aureus in a University Hospital Setting by Using Novel Software for spa Repeat Determination and Database Management. J. Clin. Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shopsin B., Mathema B., Alcabes P., Said-Salim B., Lina G., Matsuka A., Martinez J., Kreiswirth B.N. Prevalence of agr Specificity Groups among Staphylococcus aureus Strains Colonizing Children and Their Guardians. J. Clin. Microbiol. 2003;41:456–459. doi: 10.1128/JCM.41.1.456-459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haag A.F., Fitzgerald J.R., Penadés J.R. Staphylococcus aureus in Animals. Gram-Positive Pathog. 2019:731–746. doi: 10.1128/9781683670131.ch46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mrochen D.M., Schulz D., Fischer S., Jeske K., El Gohary H., Reil D., Imholt C., Trübe P., Suchomel J., Tricaud E., et al. Wild rodents and shrews are natural hosts of Staphylococcus aureus. Int. J. Med. Microbiol. 2018;308:590–597. doi: 10.1016/j.ijmm.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 37.van de Giessen A.W., van Santen-Verheuvel M.G., Hengeveld P.D., Bosch T., Broens E.M., Reusken C.B.E.M. Occurrence of methicillin-resistant Staphylococcus aureus in rats living on pig farms. Prev. Vet. Med. 2009;91:270–273. doi: 10.1016/j.prevetmed.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 38.Gómez P., González-Barrio D., Benito D., García J.T., Viñuela J., Zarazaga M., Ruiz-Fons F., Torres C. Detection of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. J. Antimicrob. Chemother. 2014;69:2061–2064. doi: 10.1093/jac/dku100. [DOI] [PubMed] [Google Scholar]
- 39.Kmeť V., Čuvalová A., Stanko M. Small mammals as sentinels of antimicrobial-resistant staphylococci. Folia Microbiol. (Praha) 2018;63:665–668. doi: 10.1007/s12223-018-0594-3. [DOI] [PubMed] [Google Scholar]
- 40.Loncaric I., Kübber-Heiss A., Posautz A., Stalder G.L., Hoffmann D., Rosengarten R., Walzer C. mecC-and mecA-positive meticillin-resistant Staphylococcus aureus (MRSA) isolated from livestock sharing habitat with wildlife previously tested positive for mecC-positive MRSA. Vet. Dermatol. 2014;25:147–148. doi: 10.1111/vde.12116. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Ripa L., Gómez P., Alonso C.A., Camacho M.C., de la Puente J., Fernández-Fernández R., Ramiro Y., Quevedo M.A., Blanco J.M., Zarazaga M., et al. Detection of MRSA of Lineages CC130-mecC and CC398-mecA and Staphylococcus delphini-lnu(A) in Magpies and Cinereous Vultures in Spain. Microb. Ecol. 2019;78:409–415. doi: 10.1007/s00248-019-01328-4. [DOI] [PubMed] [Google Scholar]
- 42.Gómez P., Lozano C., González-Barrio D., Zarazaga M., Ruiz-Fons F., Torres C. High prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in a semi-extensive red deer (Cervus elaphus hispanicus) farm in Southern Spain. Vet. Microbiol. 2015;177:326–331. doi: 10.1016/j.vetmic.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 43.Kriegeskorte A., Ballhausen B., Idelevich E.A., Köck R., Friedrich A.W., Karch H., Peters G., Becker K. Human MRSA isolates with novel genetic homolog, Germany. Emerg. Infect. Dis. 2012;18:1016–1018. doi: 10.3201/eid1806.110910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerschner H., Harrison E.M., Hartl R., Holmes M.A., Apfalter P. First report of mecC MRSA in human samples from Austria: Molecular characteristics and clinical data. New Microbes New Infect. 2015;3:4–9. doi: 10.1016/j.nmni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walther B., Wieler L.H., Vincze S., Antão E.-M., Brandenburg A., Stamm I., Kopp P.A., Kohn B., Semmler T., Lübke-Becker A. MRSA variant in companion animals. Emerg. Infect. Dis. 2012;18:2017–2020. doi: 10.3201/eid1812.120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paterson G.K., Harrison E.M., Holmes M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22:42–47. doi: 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García-Garrote F., Cercenado E., Marín M., Bal M., Trincado P., Corredoira J., Ballesteros C., Pita J., Alonso P., Vindel A. Methicillin-resistant Staphylococcus aureus carrying the mecC gene: Emergence in Spain and report of a fatal case of bacteraemia. J. Antimicrob. Chemother. 2014;69:45–50. doi: 10.1093/jac/dkt327. [DOI] [PubMed] [Google Scholar]
- 48.Laurent F., Chardon H., Haenni M., Bes M., Reverdy M.-E., Madec J.-Y., Lagier E., Vandenesch F., Tristan A. MRSA harboring mecA variant gene mecC, France. Emerg. Infect. Dis. 2012;18:1465. doi: 10.3201/eid1809.111920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison E.M., Coll F., Toleman M.S., Blane B., Brown N.M., Török M.E., Parkhill J., Peacock S.J. Genomic surveillance reveals low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus in the East of England. Sci. Rep. 2017;7:1–7. doi: 10.1038/s41598-017-07662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva V., Almeida F., Carvalho J.A., Castro A.P., Ferreira E., Manageiro V., Tejedor-Junco M.T., Caniça M., Igrejas G., Poeta P. Emergence of community-acquired methicillin-resistant Staphylococcus aureus EMRSA-15 clone as the predominant cause of diabetic foot ulcer infections in Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:179–186. doi: 10.1007/s10096-019-03709-6. [DOI] [PubMed] [Google Scholar]
- 51.Silva V., Hermenegildo S., Ferreira C., Manaia C.M., Capita R., Alonso-Calleja C., Carvalho I., Pereira J.E., Maltez L., Capelo J.L. Genetic Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Human Bloodstream Infections: Detection of MLSB Resistance. Antibiotics. 2020;9:375. doi: 10.3390/antibiotics9070375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ledda A., Price J.R., Cole K., Llewelyn M.J., Kearns A.M., Crook D.W., Paul J., Didelot X. Re-emergence of methicillin susceptibility in a resistant lineage of Staphylococcus aureus. J. Antimicrob. Chemother. 2017;72:1285–1288. doi: 10.1093/jac/dkw570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts M.C., Joshi P.R., Monecke S., Ehricht R., Müller E., Gawlik D., Paudel S., Acharya M., Bhattarai S., Pokharel S., et al. MRSA Strains in Nepalese Rhesus Macaques (Macaca mulatta) and Their Environment. Front. Microbiol. 2019;10:2505. doi: 10.3389/fmicb.2019.02505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gómez P., Lozano C., Camacho M.C., Lima-Barbero J.-F., Hernández J.-M., Zarazaga M., Höfle Ú., Torres C. Detection of MRSA ST3061-t843-mecC and ST398-t011-mecA in white stork nestlings exposed to human residues. J. Antimicrob. Chemother. 2015;71:53–57. doi: 10.1093/jac/dkv314. [DOI] [PubMed] [Google Scholar]
- 55.Kraushaar B., Fetsch A. First description of PVL-positive methicillin-resistant Staphylococcus aureus (MRSA) in wild boar meat. Int. J. Food Microbiol. 2014;186:68–73. doi: 10.1016/j.ijfoodmicro.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 56.Porrero M.C., Mentaberre G., Sánchez S., Fernández-Llario P., Casas-Díaz E., Mateos A., Vidal D., Lavín S., Fernández-Garayzábal J.-F., Domínguez L. Carriage of Staphylococcus aureus by Free-Living Wild Animals in Spain. Appl. Environ. Microbiol. 2014;80:4865–4870. doi: 10.1128/AEM.00647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seinige D., Von Altrock A., Kehrenberg C. Genetic diversity and antibiotic susceptibility of Staphylococcus aureus isolates from wild boars. Comp. Immunol. Microbiol. Infect. Dis. 2017;54:7–12. doi: 10.1016/j.cimid.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Darwich L., Vidal A., Seminati C., Albamonte A., Casado A., López F., Molina-López R.A., Migura-Garcia L. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE. 2019;14:e0210686. doi: 10.1371/journal.pone.0210686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silva V., Pereira J.E., Maltez L., Ferreira E., Manageiro V., Caniça M., Capelo J.L., Igrejas G., Poeta P. Diversity of methicillin-resistant staphylococci among wild Lepus granatensis: First detection of mecA-MRSA in hares. FEMS Microbiol. Ecol. 2020;96 doi: 10.1093/femsec/fiz204. [DOI] [PubMed] [Google Scholar]
- 60.Gerbig G.R. Ph.D. Thesis. Kent State University Honors College; Kent, OH, USA: 2020. Characterization and Whole-Genome Sequencing of Staphylococcus aureus Collected from Boston Rats. [Google Scholar]
- 61.Yang X., Yu S., Wu Q., Zhang J., Wu S., Rong D. Multilocus Sequence Typing and Virulence-Associated Gene Profile Analysis of Staphylococcus aureus Isolates From Retail Ready-to-Eat Food in China. Front. Microbiol. 2018;9:197. doi: 10.3389/fmicb.2018.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aung M.S., San T., Urushibara N., San N., Oo W.M., Soe P.E., Kyaw Y., Ko P.M., Thu P.P., Hlaing M.S., et al. Molecular Characterization of Methicillin-Susceptible and -Resistant Staphylococcus aureus Harboring Panton-Valentine Leukocidin-Encoding Bacteriophages in a Tertiary Care Hospital in Myanmar. Microb. Drug Resist. 2019;26:360–367. doi: 10.1089/mdr.2019.0208. [DOI] [PubMed] [Google Scholar]
- 63.Eriksson J., Espinosa-Gongora C., Stamphøj I., Larsen A.R., Guardabassi L. Carriage frequency, diversity and methicillin resistance of Staphylococcus aureus in Danish small ruminants. Vet. Microbiol. 2013;163:110–115. doi: 10.1016/j.vetmic.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 64.Mama O.M., Ruiz-Ripa L., Fernández-Fernández R., González-Barrio D., Ruiz-Fons J.F., Torres C. High frequency of coagulase-positive staphylococci carriage in healthy wild boar with detection of MRSA of lineage ST398-t011. FEMS Microbiol. Lett. 2019;366:fny292. doi: 10.1093/femsle/fny292. [DOI] [PubMed] [Google Scholar]
- 65.Loncaric I., Kübber-Heiss A., Posautz A., Stalder G.L., Hoffmann D., Rosengarten R., Walzer C. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother. 2013;68:2222–2225. doi: 10.1093/jac/dkt186. [DOI] [PubMed] [Google Scholar]
- 66.Gindonis V., Taponen S., Myllyniemi A.-L., Pyörälä S., Nykäsenoja S., Salmenlinna S., Lindholm L., Rantala M. Occurrence and characterization of methicillin-resistant staphylococci from bovine mastitis milk samples in Finland. Acta Vet. Scand. 2013;55:61. doi: 10.1186/1751-0147-55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mama O.M., Aspiroz C., Ruiz-Ripa L., Ceballos S., Iñiguez-Barrio M., Cercenado E., Azcona J.M., López-Cerero L., Seral C., López-Calleja A.I., et al. Prevalence and Genetic Characteristics of Staphylococcus aureus CC398 Isolates From Invasive Infections in Spanish Hospitals, Focusing on the Livestock-Independent CC398-MSSA Clade. Front. Microbiol. 2021;12:623108. doi: 10.3389/fmicb.2021.623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porrero M.C., Mentaberre G., Sánchez S., Fernández-Llario P., Gómez-Barrero S., Navarro-Gonzalez N., Serrano E., Casas-Díaz E., Marco I., Fernández-Garayzabal J.F., et al. Methicillin resistant Staphylococcus aureus (MRSA) carriage in different free-living wild animal species in Spain. Vet. J. 2013;198:127–130. doi: 10.1016/j.tvjl.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 69.Akova M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence. 2016;7:252–266. doi: 10.1080/21505594.2016.1159366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Material Table S1.