Abstract
Corticospinal motor neurons (CSMN) are an indispensable neuron population for the motor neuron circuitry. They are excitatory projection neurons, which collect information from different regions of the brain and transmit it to spinal cord targets, initiating and controlling motor function. CSMN degeneration is pronounced cellular event in motor neurons diseases, such as amyotrophic lateral sclerosis (ALS). Genetic mutations contribute to only about ten percent of ALS. Thus understanding the involvement of other factors, such as epigenetic controls, is immensely valuable. Here, we investigated epigenomic signature of CSMN that become diseased due to misfolded SOD1 toxicity and TDP-43 pathology, by performing quantitative analysis of 5-methylcytosine (5mC) and 5-hydroxymethycytosine (5hmC) expression profiles during end-stage of the disease in hSOD1G93A, and prpTDP-43A315T mice. Our analysis revealed that expression of 5mC was specifically reduced in CSMN of both hSOD1G93A and prpTDP-43A315T mice. However, 5hmC expression was increased in the CSMN that becomes diseased due to misfolded SOD1 and decreased in CSMN that degenerates due to TDP-43 pathology. These results suggest the presence of a distinct difference between different underlying causes. These differential epigenetic events might modulate the expression profiles of select genes, and ultimately contribute to the different paths that lead to CSMN vulnerability in ALS.
Keywords: CSMN, ALS, 5-methylytosine, 5-hydroxymethycytosine, epigenetics, mSOD1, TDP-43
1. Introduction
Corticospinal motor neurons (CSMN) are a unique population of motor neurons that are important for motor function. These neurons collect and integrate inputs from other neurons in the brain, and relay that information to their spinal cord targets, thus maintaining a functional motor neuron circuitry [1]. Degeneration of CSMN results in motor neuron diseases such as hereditary spastic paraplegia (HSP), primary lateral sclerosis (PLS), and amyotrophic lateral sclerosis (ALS) [2]. ALS is a complex and heterogeneous disease that is manifested by progressive degeneration of CSMN, and spinal motor neurons [3]. ALS can be hereditary as well as sporadic, with a major contribution from sporadic factors. CSMN loss is detected in both familial and sporadic forms of ALS [4,5]. Early detection of CSMN loss is beginning to emerge as an early diagnostic marker of ALS [6], and improving CSMN health proved to be beneficial for the health and integrity of motor neuron circuitry [7].
There are about 147 genes mutated in ALS patients that are either causative or associated with ALS [8]. Many of these genes converge on specific cellular defects [8], two of which are prominent in ALS; the mutant SOD1 mediated toxicity and TDP-43 pathology mediated cell death. Among the mouse models generated to understand the disease mechanisms related to SOD1 toxicity and TDP-43 pathology, we find hSOD1G93A and prpTDP-43A315T to be well characterized, and most importantly to faithfully recapitulate ALS pathology at a cellular level [9,10]. Involvement of mSOD1 in ALS pathology is widely studied, and hSOD1G93A mouse shows progressive degeneration of CSMN [11]. Mis-localization and accumulation of TDP-43 protein in the cytoplasm is the most common ALS pathology that is detected in both, sporadic and familial forms of ALS [12]. prpTDP-43A315T mouse model display progressive degeneration of CSMN due to defect in cellular organelles such as mitochondria, ER, and nuclear membrane. Similar cellular defects are also observed in the upper motor neurons of ALS patients with TDP-43 pathology [13]. Interestingly, SOD1 toxicity and TDP-43 pathology have unique and non-overlapping aspects of neurodegeneration. For example, patients with SOD1 mutation did not have TDP-43 pathology in their CNS, even though TDP-43 accumulations are widely observed in a broad spectrum of ALS patients [14]. Similarly, the mouse models also show similar findings, such that ALS mouse models expressing different mutations in SOD1 gene (hSOD1G93A, hSOD1G37R and hSOD1G85R) did not show TDP-43 accumulation [15]. Therefore, investigating epigenetic modification in these two distinct disease models will be immensely valuable in understanding selective vulnerability of CSMN.
Epigenetics is the modulation of gene expression without any changes in the gene sequence. Expression of genes can be modulated via two epigenetic mechanisms: DNA methylation, and DNA hydroxymethylation [16]. Addition of methyl group at 5 carbon position on cytosine by DNA methyltransferase (Dnmt) enzyme converts cytosine into 5-methylcytosine (5mC) by transfer of a methyl group from S-adenosyl-methionine [17]. 5-hydroxymethylcytosine (5hmC) is generated through the oxidation of 5mC by the ten-eleven translocation (TET) family of methylcytosine dioxygenases [18,19]. These epigenetic modifications alter gene expression via differential methylation or hydroxymethylation of promoter regions of a defined population of genes. DNA methylation/hydroxymethylation changes protein–DNA interactions that lead to alterations in chromatin structure and rate of transcription [20]. A growing body of evidence suggests key role of epigenetic mechanisms in ALS pathology [21]. Global epigenome-wide studies of cortex, and spinal cord of sALS patients have shown differential methylation and hydroxymethylation of a select set of genes [22,23]. Recently, TDP-43 pathology was shown to be associated with DNA methylation [24]. Methylation of C9orf72 promoter is reduced while hydroxymethylation is increased during differentiation of ALS patient-derived iPS cells into motor neurons [25]. Additionally, promotor region of C9orf72 have shown increased methylation and decreased transcription in ALS/FTD patients with the pathogenic repeat expansion [26,27,28].
The role of epigenetic modifications, and their contribution to ALS pathogenesis is beginning to emerge. However, CSMN specific epigenetic modifications have not been investigated thus far. This study was designed to investigate whether DNA methylation, and hydroxymethylation levels in CSMN are differentially modulated in the context of ALS. Using hSOD1G93A and prpTDP-43A315T mouse models of ALS, which represent disease mechanisms that occur due to misfolded SOD1 toxicity and TDP-43 pathology, respectively, here, we reveal that CSMN based on the underlying cause of the disease display a distinct pattern of methylation and hydroxymethylation. Epigenetic modifications are an important event in the regulation of gene expression. We find that CSMN that are diseased due to mSOD1 toxicity, and TDP-43 pathology have distinct pattern of DNA methylation/hydroxymethylation status. These results begin to reveal the epigenetic regulation of CSMN vulnerability in ALS.
2. Materials and Methods
2.1. Mice
All animal experiments were performed in compliance with the standards set by National Institutes of Health (NIH) and were approved by the Northwestern University Animal Care and Use committee (NUACUC). The following mouse strains were used in this study: prp-TDP-43A315T (procured from Jackson Laboratory, stock no. 010700), hSOD1G93A (procured from Jackson Laboratory, stock no. 002726), and UCHL1-eGFP (generated by the Ozdinler Lab at Northwestern Targeted Mutagenesis Core Facility, now also available at Jackson Laboratory, stock no. 022476). UCHL1-eGFP is a reporter mice line in which CSMN are specifically labeled with eGFP allowing their precise visualization and investigation [29]. Hemizygous UCHL1-eGFP females were bred to hemizygous prp-TDP-43A315T males to generate prp-TDP-43A315T-UeGFP mice. Similarly, hemizygous UCHL1-eGFP females were bred to hSOD1G93A males to generate hSOD1G93A-UeGFP mice. The analyses were performed at the end stage of hSOD1G93A-UeGFP (P120) and prp-TDP-43A315T-UeGFP (P150) mice, respectively. All the mice used in this study were on C57/BL6 background.
2.2. Tissue Collection and Processing
Mice were deeply anesthetized with intraperitoneal injection of ketamine (90 mg/kg, and xylazine (10 mg/kg; Fort Dodge Animal Health, Fort Dodge, IA, USA) prior to transcardial perfusion with phosphate-buffered saline (PBS) and 4% paraformaldehyde (PFA) in PBS. Intact brain was dissected, post-fixed in 4% PFA overnight and stored for later use in PBS with 0.01% sodium azide at 4 °C. The brain was sectioned at 50 μm using Leica vibratome (Leica VT1000S, Leica Inc., Nussloch, Germany).
2.3. Immunocytochemistry
Before commencing antibody incubation, floating sections were treated with 4N hydrochloric acid for 30 min at room temperature (RT) to denature the DNA and to facilitate the antibody penetration. Sections were washed three times with PBS for 10 min followed by incubation in blocking solution (PBS, 0.05% bovine serum albumin, 2% fetal bovine serum, 1% Triton X-100 and 0.1% saponin) for 30 min at RT prior to addition of primary antibodies: chicken anti-GFP (1:1000, Abcam, Cambridge, MA, USA), mouse anti-5-Methylcytosine (1:500, EpiGentek, Farmingdale, NY, USA), and mouse anti-5-Hydroxymethylcytosine (1:500, EpiGentek, Farmingdale, NY, USA) at 4 °C overnight. Excess antibodies were removed by PBS washes (three washes, 10 min each wash), sections were incubated in appropriate secondary antibodies in blocking solution: anti-chicken Alexa Fluor 488 (1:1000, Thermo Fisher Scientific, Rockford, IL, USA), anti-mouse Alexa Fluor 568 (1:1000, Thermo Fisher Scientific, Rockford, IL, USA) for 2 hrs at RT. Sections were counterstained with DAPI (1:5000). All sections were mounted on slides and coverslipped with Fluoromount G (Electron Microscopy Sciences, Hatfield, PA, USA). All immunostaining were performed at the same time with the same antibody cocktail to reduce experimental variation.
2.4. Imaging
Nikon Eclipse TE2000-E (Nikon Inc., Melville, NY, USA) and Zeiss 880 confocal microscope (Carl Zeiss microscopy, Jena, Germany) were used to acquire low-and high-magnification images, respectively. Z-stacks were processed using ImageJ (National Institutes of Health, Bethesda, MD, USA, http://imagej.nih.gov/ij accessed on 16 April 2021) to generate maximum intensity projections.
2.5. Quantification and Data Analysis
CSMN from WT-UeGFP, hSOD1G93A-UeGFP, and prp-TDP-43A315T-UeGFP mice (n = 3) expressing 5mC, and 5hmC were counted using maximum projection images acquired from 50-μm thick sections. An equivalent area of the motor cortex (1.55 mm × 1.55 mm area corresponding to the 10× objective field size) was defined in three serial sections (~600 μm apart) per mouse. Neurons with pyramidal morphology and a visible apical dendrite were counted in each area, with experimenter blind to age and genotype of mice.
For intensity measurement, a maximum intensity projection image was opened using ImageJ (NIH), and CSMN were traced using free hand tool. Intensity of signals was measured in the defines area, and integrated density was recorded. A similar area was drawn in the background where there were no signals present. The integrated density of background was deducted from the integrated density of signal to measure the intensity of expression of 5mC, and 5hmC.
2.6. Statistical Analyses
GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analyses. At least n = 3 mice and n = 3 brain sections for each mouse were used for each genotype and group. D’Agostino and Pearson normality tests were performed on all data sets. Student’s t test was used to determine statistical differences between experimental groups depending on the genotype, experimental conditions, and the disease group. Data are shown as mean ± SEM of at least three replicates and are representative of three independent experiments unless otherwise stated and statistically significant differences were taken at p < 0.05, and p values are reported in the text.
3. Results
3.1. CSMN Diseased Due to mSOD1 Toxicity and TDP-43 Pathology Displayed Reduced Methylation
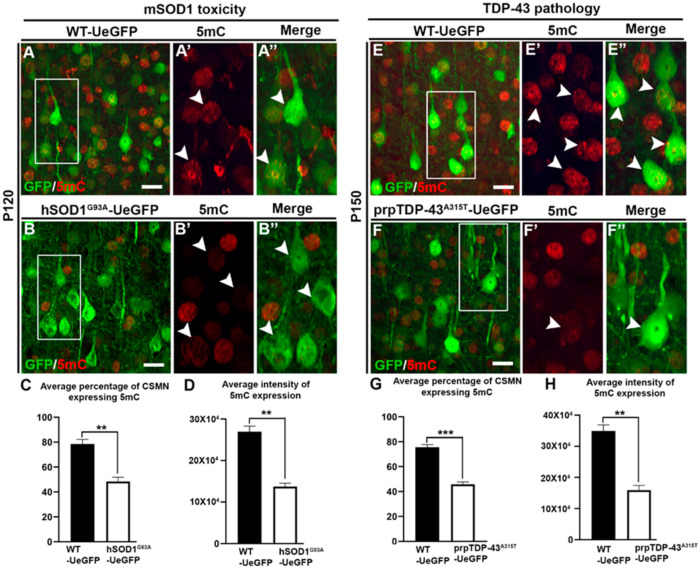
Methylation status of CSMN was analyzed to investigate whether vulnerable CSMN have increased or decreased methylation levels. We used expression of 5-methylcytosine to determine the methylation levels in CSMN of hSOD1G93A and prp-TDP-43A315T mouse models of ALS, because progressive CSMN degeneration is well characterized in these two mouse models that represent two different underlying causes of the disease [11,13]. Analysis of CSMN expressing 5mC showed that most CSMN failed to express 5mc in hSOD1G93A mouse (WT-UeGFP: 78.5 ± 3.7%; hSOD1G93A-UeGFP: 48.4 ± 3.41%, n = 3, p < 0.003, Figure 1A–C), and even the CSMN which continued to express 5mC had less fluorescence intensity (WT-UeGFP: 269,717 ± 13,116; hSOD1G93A-UeGFP: 137,023 ± 8441, n = 3, p < 0.001, Figure 1D). CSMN which become diseased due to TDP-43 pathology also showed less methylation (WT-UeGFP: 75.6 ± 2%; prp-TDP-43A315T-UeGFP: 45.8 ± 1.9%, n = 3, p < 0.0005, Figure 1E–G), and decreased levels of fluorescent intensity for 5mC expression (WT-UeGFP: 349,130 ± 19,675, prp-TDP-43A315T-UeGFP: 159,238 ± 15,042, n = 3, p < 0.001, Figure 1H).
Figure 1.
CSMN that are vulnerable in ALS due to different underlying causes display reduced levels of 5-methylcytosine (5mC). (A) Representative image of layer V of motor cortex from WT-UeGFP mouse at P120 showing GFP labelled CSMN and 5mC expression. Box enlarged (A′,A″, arrow heads). (B) Representative image of layer V of motor cortex from hSOD1G93A-UeGFP mouse at P120 showing GFP labelled CSMN and 5mC expression. Box enlarged (B′,B″, arrow heads). (C) Quantitative analysis of average percentage of CSMN expressing 5mC in hSOD1G93A-UeGFP mouse at P120. (D) Quantitative analysis of average intensity of 5mC expression in CSMN of hSOD1G93A-UeGFP mouse at P120. (E) Representative image of layer V of motor cortex from WT-UeGFP mouse at P150 showing GFP labelled CSMN and 5mC expression. Box enlarged (E′,E″, arrow heads). (F) Representative image of layer V of motor cortex from prp-TDP-43A315T-UeGFP mouse at P150 showing GFP labelled CSMN and 5mC expression. Box enlarged (F′,F″, arrow heads). (G) Quantitative analysis of average percentage of CSMN expressing 5mC in prp-TDP-43A315T-UeGFP mouse at P150. (H) Quantitative analysis of average intensity of 5mC expression in CSMN of prp-TDP-43A315T-UeGFP mouse at P150. ** p < 0.001, *** p < 0.0005; Scale bar: 20 µm.
3.2. CSMN Show Increased Hydroxymethylation Due to mSOD1 Toxicity, but Decreased Hydroxymethylation Due to TDP-43 Pathology
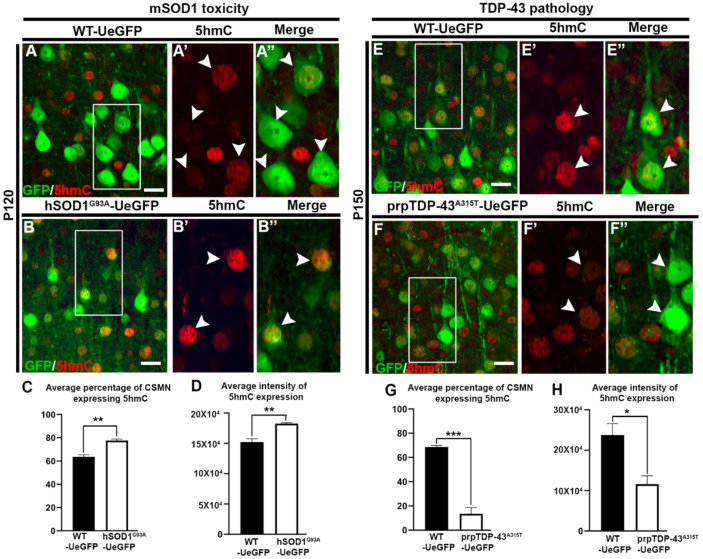
Hydroxymethylation levels of CSMN were analyzed in hSOD1G93A and prp-TDP-43A315T mouse models of ALS during end-stage of the disease. Analysis of 5-hydroxymethylcytosine expression was used to assess hydroxymethylation levels in diseased CSMN. CSMN of hSOD1G93A mice showed significantly increased expression of 5hmC when compared to CSMN of healthy controls (WT-UeGFP: 63.5 ± 1.7%; hSOD1G93A-UeGFP: 77.6 ± 1.2%, n = 3, p < 0.002, Figure 2A–C). Interestingly, the intensity of 5hmC expression was also increased in diseased CSMN (WT-UeGFP: 152,016 ± 5466; hSOD1G93A-UeGFP: 182,636 ± 1484, n = 3, p < 0.005, Figure 2D). In striking contrast to CSMN that become diseased due to mSOD1 toxicity, CSMN of prp-TDP-43A315T mouse displayed reduced hydroxymethylation (WT-UeGFP: 68.6 ± 1.3%; prp-TDP-43A315T-UeGFP: 19.3 ± 5.1%, n = 3, p < 0.0005, Figure 2E–E″,G). Measurement of intensity of expression also confirmed reduction in 5hmC expression in CSMN of prp-TDP-43A315T mouse (WT-UeGFP: 237,614 ± 28,228, prpTDP-43A315T-UeGFP: 115,764 ± 20,764, n = 3, p < 0.025, Figure 2H).
Figure 2.
CSMN that are diseased owing to mSOD1 toxicity, and TDP-43 pathology display differential expression of 5-hydroxymethylcytosine (5hmC). (A) Representative image of layer V of motor cortex from WT-UeGFP mouse at P120 showing GFP-labelled CSMN and 5hmC expression. Box enlarged (A′,A″, arrow heads). (B) Representative image of layer V of motor cortex from hSOD1G93A-UeGFP mouse at P120 showing GFP-labelled CSMN and 5hmC expression. Box enlarged (B′,B″, arrow heads). (C) Quantitative analysis of average percentage of CSMN expressing 5hmC in hSOD1G93A-UeGFP mouse at P120. (D) Quantitative analysis of average intensity of 5hmC expression in CSMN of hSOD1G93A-UeGFP mouse at P120. (E) Representative image of layer V of motor cortex from WT-UeGFP mouse at P150 showing GFP-labelled CSMN and 5hmC expression. Box enlarged (E′,E″, arrow heads). (F) Representative image of layer V of motor cortex from prp-TDP-43A315T-UeGFP mouse at P150 showing GFP-labelled CSMN and 5hmC expression. Box enlarged (F′,F″, arrow heads). (G) Quantitative analysis of average percentage of CSMN expressing 5hmC in prp-TDP-43A315T-UeGFP mouse at P150. (H) Quantitative analysis of average intensity of 5hmC expression in CSMN of prp-TDP-43A315T-UeGFP mouse at P150. * p < 0.025, ** p < 0.001, *** p < 0.0005; Scale bar: 20 µm.
Our analyses of 5mC and 5hmC expression were restricted to eGFP expressing CSMN in both hSOD1G93A-UeGFP and prp-TDP-43A315T–UeGFP mice. 5mC and 5hmC expressions were also observed in other cells adjacent to CSMN. However, the identity of these cells could not be ascertained since we did not perform 5mC and 5hmC co-immunostaining with GFAP (astrocyte), Iba1 (microglia), and NeuN/MAP2 (neuron).
4. Discussion
Epigenetic modification of DNA is emerging as a key contributory factor in ALS pathology [30]. In sporadic ALS patients, 5mC expression is increased in the motor cortical neurons, and the elevated 5mC levels are a potential contributor to apoptosis of these neurons [30]. These modifications result in differential DNA methylation that leads to modulation of gene expression in ALS [31]. Whole methylation profiling followed by transcriptomics have identified global gene expression changes in spinal cord and blood cell of ALS patients [23,32]. These gene expression alterations are implicated in various pathways such as immune response and cellular transport [33,34,35].
However, majority of these studies investigated the global changes in DNA methylation of either blood or brain and/or spinal cord tissues of ALS patients. Motor neurons degenerate selectively and progressively in ALS. Thus, investigating motor neuron specific DNA methylation, and hydroxymethylation would reveal cell-type-specific vulnerability that might have been masked in global epigenome profiles. Here, we report that CSMN that are diseased due to mSOD1 toxicity and TDP-43 pathology undergo reduction in methylation. In ALS patients, upper as well as lower motor neurons display accumulation of DNA damage response that leads to lower methylation levels in the promoter region of specific genes such as Ogg1, Apex1, Pnkp, and Aptx in ALS [36]. This phenomenon is also observed in iPS motor neurons derived from familial ALS patients [36]. Recently, reduced methylation levels were observed in mitochondrial DNA regulatory region (D-loop) of ALS-associated genes such as C9orf72, SOD1, FUS, and TDP-43 [37]. Differential methylation was reported in RAD9B and C8orf46, CCNF, DPP6, RAMP3, and CCS genes in monozygotic twins and triplets discordant for amyotrophic lateral sclerosis [38]. C9orf72 has been shown to have increased methylation and decreased transcription in ALS/FTD patients with the pathogenic repeat expansion [26,27,39]. Therefore, convincing evidence suggesting a key role of differential DNA methylation in ALS, is beginning to emerge.
We found that CSMN of hSOD1G93A mouse showed higher levels of 5hmC. Hydroxymethylation is critical for postnatal neurodevelopment and aging [40,41]. The presence of 5hmC has generally been associated with increased gene expression [42]. 5hmC-mediated epigenetic regulation is implicated in the development of human cerebellum [43]. In human brains, the promoter region coding for genes involved in ion transport, neuronal development, and cell death has increased 5hmC [42]. Emerging evidence suggests that dysregulation of hydroxymethylation contributes to neurodegenerative diseases including Alzheimer’s, Parkinson’s, Huntington’s disease, and cerebrovascular diseases [44,45]. Selective loss of 5hmC in the cortex and hippocampus of AD patients and 3xTg mouse model promotes AD pathology [46]. Increased levels of hydroxymethylation were observed in hippocampal dentate gyrus, CA3 and CA1–2 regions of aging mice, and caloric restriction helped lower hmC levels [47]. Our results suggest that diseased CSMN acquire hydroxymethylation. Overall, this study reveals that approximately 60% degenerating CSMN lose 5mC expression both in hSOD1G93A and prp-TDP-43A315T mouse. Intriguingly, about 80% degenerating CSMN express 5hmC in hSOD1G93A mouse but about 80% CSMN in prp-TDP-43A315T lose 5hmC expression. CSMN expressing mutated form of hTDP-43 showed reduced levels of 5hmC. Discrepancy in 5hmC expression between hSOD1G93A and prp-TDP-43A315T could be attributed to dysregulation of TDP-43. In a recent study, it was found that neurons that show aberrant localization of TDP-43 into the cytoplasm showed lower levels of 5hmC [24]. Thus, our results suggest that TDP-43 pathology modulates DNA hydroxymethylation specifically in CSMN, and differential expression of 5mC and 5hmC could also be attributed to different underlying causes of CSMN vulnerability [48].
5. Conclusions
In summary, this study sheds light on differential DNA methylation and DNA hydroxymethylation specific to CSMN that are vulnerable to degeneration due to different underlying causes in ALS. Further investigation into genes associated with differential methylome, and hydroxymethylome would lead to understanding the basis of selective vulnerability of CSMN in ALS and other upper motor neuron diseases.
Acknowledgments
We thank ‘A Long Swim’ for Ellen McConnell Blakeman Fellowship (M.G.). The authors thank P Hande Ozdinler for critically reading the manuscript, and for her valuable suggestions. We thank Jolanta Topczewska, Ann and Robert H Lurie Children’s Hospital of Chicago, for helping with confocal microscopy. We thank the Spastic Paraplegia Foundation for covering the publication cost.
Author Contributions
Conceptualization, M.G.; methodology, M.G. and T.O.; formal analysis, M.G. and T.O.; writing—original draft preparation, M.G. All authors have read and agreed to the published version of the manuscript.
Funding
Ellen McConnell Blakeman Fellowship (M.G.).
Institutional Review Board Statement
Animals used in these studies were housed under National Institutes of Health guidelines and all experiments were conducted in accordance compliance with the standards set by National Institutes of Health and were approved by the Northwestern University Animal Care and Use committee (IS00009980).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lemon R.N. Descending pathways in motor control. Annu. Rev. Neurosci. 2008;31:195–218. doi: 10.1146/annurev.neuro.31.060407.125547. [DOI] [PubMed] [Google Scholar]
- 2.Fink J.K. Progressive spastic paraparesis: Hereditary spastic paraplegia and its relation to primary and amyotrophic lateral sclerosis. Semin. Neurol. 2001;21:199–207. doi: 10.1055/s-2001-15265. [DOI] [PubMed] [Google Scholar]
- 3.Eisen A. The Dying Forward Hypothesis of ALS: Tracing Its History. Brain Sci. 2021;11:300. doi: 10.3390/brainsci11030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burg T., Bichara C., Scekic-Zahirovic J., Fischer M., Stuart-Lopez G., Brunet A., Lefebvre F., Cordero-Erausquin M., Rouaux C. Absence of Subcerebral Projection Neurons Is Beneficial in a Mouse Model of Amyotrophic Lateral Sclerosis. Ann. Neurol. 2020;88:688–702. doi: 10.1002/ana.25833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marques C., Burg T., Scekic-Zahirovic J., Fischer M., Rouaux C. Upper and Lower Motor Neuron Degenerations Are Somatotopically Related and Temporally Ordered in the Sod1 Mouse Model of Amyotrophic Lateral Sclerosis. Brain Sci. 2021;11:369. doi: 10.3390/brainsci11030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geevasinga N., Menon P., Ozdinler P.H., Kiernan M.C., Vucic S. Pathophysiological and diagnostic implications of cortical dysfunction in ALS. Nat. Rev. Neurol. 2016;12:651–661. doi: 10.1038/nrneurol.2016.140. [DOI] [PubMed] [Google Scholar]
- 7.Thomsen G.M., Gowing G., Latter J., Chen M., Vit J.P., Staggenborg K., Avalos P., Alkaslasi M., Ferraiuolo L., Likhite S., et al. Delayed disease onset and extended survival in the SOD1G93A rat model of amyotrophic lateral sclerosis after suppression of mutant SOD1 in the motor cortex. J. Neurosci. 2014;34:15587–15600. doi: 10.1523/JNEUROSCI.2037-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dervishi I., Gozutok O., Murnan K., Gautam M., Heller D., Bigio E., Ozdinler P.H. Protein-protein interactions reveal key canonical pathways, upstream regulators, interactome domains, and novel targets in ALS. Sci. Rep. 2018;8:14732. doi: 10.1038/s41598-018-32902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X., et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 10.Wegorzewska I., Bell S., Cairns N.J., Miller T.M., Baloh R.H. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozdinler P.H., Benn S., Yamamoto T.H., Guzel M., Brown R.H., Jr., Macklis J.D. Corticospinal motor neurons and related subcerebral projection neurons undergo early and specific neurodegeneration in hSOD1G (9)(3)A transgenic ALS mice. J. Neurosci. 2011;31:4166–4177. doi: 10.1523/JNEUROSCI.4184-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautam M., Jara J.H., Kocak N., Rylaarsdam L.E., Kim K.D., Bigio E.H., Hande Ozdinler P. Mitochondria, ER, and nuclear membrane defects reveal early mechanisms for upper motor neuron vulnerability with respect to TDP-43 pathology. Acta Neuropathol. 2019;137:47–69. doi: 10.1007/s00401-018-1934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenzie I.R., Bigio E.H., Ince P.G., Geser F., Neumann M., Cairns N.J., Kwong L.K., Forman M.S., Ravits J., Stewart H., et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann. Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 15.Robertson J., Sanelli T., Xiao S., Yang W., Horne P., Hammond R., Pioro E.P., Strong M.J. Lack of TDP-43 abnormalities in mutant SOD1 transgenic mice shows disparity with ALS. Neurosci. Lett. 2007;420:128–132. doi: 10.1016/j.neulet.2007.03.066. [DOI] [PubMed] [Google Scholar]
- 16.Guibert S., Weber M. Functions of DNA methylation and hydroxymethylation in mammalian development. Curr. Top. Dev. Biol. 2013;104:47–83. doi: 10.1016/B978-0-12-416027-9.00002-4. [DOI] [PubMed] [Google Scholar]
- 17.Cheng X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 1995;24:293–318. doi: 10.1146/annurev.bb.24.060195.001453. [DOI] [PubMed] [Google Scholar]
- 18.Guo J.U., Ma D.K., Mo H., Ball M.P., Jang M.H., Bonaguidi M.A., Balazer J.A., Eaves H.L., Xie B., Ford E., et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto H., Liu Y., Upadhyay A.K., Chang Y., Howerton S.B., Vertino P.M., Zhang X., Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 21.Paez-Colasante X., Figueroa-Romero C., Sakowski S.A., Goutman S.A., Feldman E.L. Amyotrophic lateral sclerosis: Mechanisms and therapeutics in the epigenomic era. Nat. Rev. Neurol. 2015;11:266–279. doi: 10.1038/nrneurol.2015.57. [DOI] [PubMed] [Google Scholar]
- 22.Morahan J.M., Yu B., Trent R.J., Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2009;10:418–429. doi: 10.3109/17482960802635397. [DOI] [PubMed] [Google Scholar]
- 23.Figueroa-Romero C., Hur J., Bender D.E., Delaney C.E., Cataldo M.D., Smith A.L., Yung R., Ruden D.M., Callaghan B.C., Feldman E.L. Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PLoS ONE. 2012;7:e52672. doi: 10.1371/journal.pone.0052672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Appleby-Mallinder C., Schaber E., Kirby J., Shaw P.J., Cooper-Knock J., Heath P.R., Highley J.R. TDP43 proteinopathy is associated with aberrant DNA methylation in human amyotrophic lateral sclerosis. Neuropathol. Appl. Neurobiol. 2021;47:61–72. doi: 10.1111/nan.12625. [DOI] [PubMed] [Google Scholar]
- 25.Esanov R., Belle K.C., van Blitterswijk M., Belzil V.V., Rademakers R., Dickson D.W., Petrucelli L., Boylan K.B., Dykxhoorn D.M., Wuu J., et al. C9orf72 promoter hypermethylation is reduced while hydroxymethylation is acquired during reprogramming of ALS patient cells. Exp. Neurol. 2016;277:171–177. doi: 10.1016/j.expneurol.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belzil V.V., Bauer P.O., Gendron T.F., Murray M.E., Dickson D., Petrucelli L. Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res. 2014;1584:15–21. doi: 10.1016/j.brainres.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belzil V.V., Bauer P.O., Prudencio M., Gendron T.F., Stetler C.T., Yan I.K., Pregent L., Daughrity L., Baker M.C., Rademakers R., et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi Z., Zinman L., Moreno D., Schymick J., Liang Y., Sato C., Zheng Y., Ghani M., Dib S., Keith J., et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am. J. Hum. Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasvoina M.V., Genc B., Jara J.H., Sheets P.L., Quinlan K.A., Milosevic A., Shepherd G.M.G., Heckman C.J., Özdinler P.H. eGFP Expression under UCHL1 promoter genetically labels corticospinal motor neurons and a subpopulation of degeneration-resistant spinal motor neurons in an ALS mouse model. J. Neurosci. 2013;33:7890–7904. doi: 10.1523/JNEUROSCI.2787-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chestnut B.A., Chang Q., Price A., Lesuisse C., Wong M., Martin L.J. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andres-Benito P., Moreno J., Aso E., Povedano M., Ferrer I. Amyotrophic lateral sclerosis, gene deregulation in the anterior horn of the spinal cord and frontal cortex area 8: Implications in frontotemporal lobar degeneration. Aging. 2017;9:823–851. doi: 10.18632/aging.101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremolizzo L., Messina P., Conti E., Sala G., Cecchi M., Airoldi L., Pastorelli R., Pupillo E., Bandettini Di Poggio M., Filosto M., et al. Whole-blood global DNA methylation is increased in amyotrophic lateral sclerosis independently of age of onset. Amyotroph. Lateral Scler. Front. Degener. 2014;15:98–105. doi: 10.3109/21678421.2013.851247. [DOI] [PubMed] [Google Scholar]
- 33.Ebbert M.T.W., Ross C.A., Pregent L.J., Lank R.J., Zhang C., Katzman R.B., Jansen-West K., Song Y., da Rocha E.L., Palmucci C., et al. Conserved DNA methylation combined with differential frontal cortex and cerebellar expression distinguishes C9orf72-associated and sporadic ALS, and implicates SERPINA1 in disease. Acta Neuropathol. 2017;134:715–728. doi: 10.1007/s00401-017-1760-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Rheenen W., Diekstra F.P., Harschnitz O., Westeneng H.J., van Eijk K.R., Saris C.G.J., Groen E.J.N., van Es M.A., Blauw H.M., van Vught P.W.J., et al. Whole blood transcriptome analysis in amyotrophic lateral sclerosis: A biomarker study. PLoS ONE. 2018;13:e0198874. doi: 10.1371/journal.pone.0198874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao W., Beers D.R., Hooten K.G., Sieglaff D.H., Zhang A., Kalyana-Sundaram S., Traini C.M., Halsey W.S., Hughes A.M., Sathe G.M., et al. Characterization of Gene Expression Phenotype in Amyotrophic Lateral Sclerosis Monocytes. JAMA Neurol. 2017;74:677–685. doi: 10.1001/jamaneurol.2017.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim B.W., Jeong Y.E., Wong M., Martin L.J. DNA damage accumulates and responses are engaged in human ALS brain and spinal motor neurons and DNA repair is activatable in iPSC-derived motor neurons with SOD1 mutations. Acta Neuropathol. Commun. 2020;8:7. doi: 10.1186/s40478-019-0874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoccoro A., Smith A.R., Mosca L., Marocchi A., Gerardi F., Lunetta C., Cereda C., Gagliardi S., Lunnon K., Migliore L., et al. Reduced mitochondrial D-loop methylation levels in sporadic amyotrophic lateral sclerosis. Clin. Epigenet. 2020;12:137. doi: 10.1186/s13148-020-00933-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarr I.S., McCann E.P., Benyamin B., Peters T.J., Twine N.A., Zhang K.Y., Zhao Q., Zhang Z.H., Rowe D.B., Nicholson G.A., et al. Monozygotic twins and triplets discordant for amyotrophic lateral sclerosis display differential methylation and gene expression. Sci. Rep. 2019;9:8254. doi: 10.1038/s41598-019-44765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xi Z., Rainero I., Rubino E., Pinessi L., Bruni A.C., Maletta R.G., Nacmias B., Sorbi S., Galimberti D., Surace E.I., et al. Hypermethylation of the CpG-island near the C9orf72 G (4)C(2)-repeat expansion in FTLD patients. Hum. Mol. Genet. 2014;23:5630–5637. doi: 10.1093/hmg/ddu279. [DOI] [PubMed] [Google Scholar]
- 40.Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I., et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szwagierczak A., Bultmann S., Schmidt C.S., Spada F., Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin S.G., Wu X., Li A.X., Pfeifer G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang T., Pan Q., Lin L., Szulwach K.E., Song C.X., He C., Wu H., Warren S.T., Jin P., Duan R., et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Iglesias O., Carrera I., Carril J.C., Fernandez-Novoa L., Cacabelos N., Cacabelos R. DNA Methylation in Neurodegenerative and Cerebrovascular Disorders. Int. J. Mol. Sci. 2020;21:2220. doi: 10.3390/ijms21062220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherwani S.I., Khan H.A. Role of 5-hydroxymethylcytosine in neurodegeneration. Gene. 2015;570:17–24. doi: 10.1016/j.gene.2015.06.052. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Zhang Z., Li L., Xu K., Ma Z., Chow H.M., Herrup K., Li J. Selective loss of 5hmC promotes neurodegeneration in the mouse model of Alzheimer’s disease. FASEB J. 2020;34:16364–16382. doi: 10.1096/fj.202001271R. [DOI] [PubMed] [Google Scholar]
- 47.Chouliaras L., van den Hove D.L., Kenis G., Keitel S., Hof P.R., van Os J., Steinbusch H.W., Schmitz C., Rutten B.P. Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr. Alzheimer Res. 2012;9:536–544. doi: 10.2174/156720512800618035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin L.J., Chang Q. DNA Damage Response and Repair, DNA Methylation, and Cell Death in Human Neurons and Experimental Animal Neurons Are Different. J. Neuropathol. Exp. Neurol. 2018;77:636–655. doi: 10.1093/jnen/nly040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are contained within the article.