Abstract
Water deficit is one of the major constraints to crop production and food security worldwide. Some plant growth-promoting rhizobacteria (PGPR) strains are capable of increasing plant drought resistance. Knowledge about the mechanisms underlying bacteria-induced plant drought resistance is important for PGPR applications in agriculture. In this study, we show the drought stress-mitigating effects on tomato plants by the Bacillus megaterium strain TG1-E1, followed by the profiling of plant transcriptomic responses to TG1-E1 and the profiling of bacterial extracellular metabolites. Comparison between the transcriptomes of drought-stressed plants with and without TG1-E1 inoculation revealed bacteria-induced transcriptome reprograming, with highlights on differentially expressed genes belonging to the functional categories including transcription factors, signal transduction, and cell wall biogenesis and organization. Mass spectrometry-based analysis identified over 40 bacterial extracellular metabolites, including several important regulators or osmoprotectant precursors for increasing plant drought resistance. These results demonstrate the importance of plant transcriptional regulation and bacterial metabolites in PGPR-induced plant drought resistance.
Keywords: PGPR, tomato, drought stress, Bacillus megaterium TG1-E1, transcriptome, extracellular metabolites, osmoprotectant
1. Introduction
Drought stress, caused by water deficit, limits the worldwide utilization of arable lands as well as crop productivity [1]. In response to drought stress, plants undergo hyperosmotic signal transduction, leading to transcriptional reprograming for the repair of stress-induced damage, the re-balancing of cellular homeostasis, and the control of growth to adapt to the water deficit condition [2].
Plants live naturally with many microorganisms including plant growth-promoting rhizobacteria (PGPR), which are beneficial soil microorganisms that can stimulate plant growth and/or increase plant resistance to various stress conditions including drought stress [3,4,5]. Each PGPR strain produces a complex array of extracellular metabolites, some of which are responsible for triggering beneficial effects in plants. For instance, many PGPR strains exude the enzyme ACC (1-aminocyclopropane-1-carboxylate) deaminase that reduces plant ethylene levels, as well as siderophores that facilitate root uptake of metal nutrients [3]. Meanwhile, bacterial exopolysaccharides improve soil aggregation and maintain soil moisture in the rhizosphere and thus can help plants survive under water deficit conditions [6].
In response to PGPR, transcriptional reprograming in plant cells plays an important role in transducing the bacterial stimuli to the enhanced plant growth and stress resistance. For instance, Bacillus amyloliquefaciens GB03 enhances Arabidopsis root iron uptake, which is supported by the transcriptional upregulation of the root Fe3+ reductase FRO2 and the Fe2+ transporter IRT1, as well as by the gene induction of FIT1, a transcription activator that controls FRO2 and IRT1 gene expression [7]. Likewise, a group of plant immunity-related genes were repressed by the bacterial volatile compound diacetyl, thereby supporting the function of diacetyl in establishing a beneficial association between PGPR and plants through suppression of immune responses in phosphate-sufficient plants [8].
While PGPR provide potentially powerful tools to improve plant stress resistance in agriculture, understanding the underlying mechanisms is challenging yet crucial for successful applications. In this study, we investigated whether and how Bacillus megaterium TG1-E1, a PGPR strain isolated from a high salinity environment [9], may affect drought stress resistance in tomato plants. The drought stress-alleviating effects of TG1-E1 on two different cultivars are demonstrated, followed by the analyses of plant transcriptome and bacterial extracellular metabolites, which indicate an integrated mechanism mediated through multiple key biological processes in plants as well as bacterial osmoprotectants.
2. Results and Discussion
2.1. Bacillus Megaterium TG1-E1 Increases Drought Resistance in Tomato Seedlings
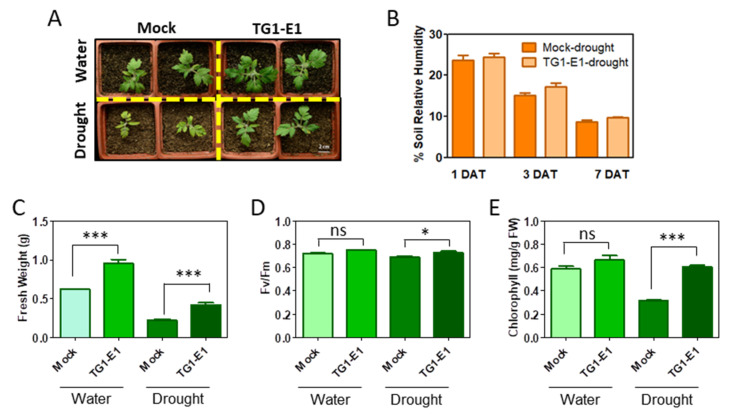
To evaluate the effects of TG1-E1 on tomato drought resistance, we studied two cultivars of tomato, Micro-Tom and Ailsa Craig, with and without bacteria inoculation. At 10 days after the drought treatment started, the drought-treated Micro-Tom plants without TG1-E1 inoculation clearly suffered from water deficit, as evidenced by the arrested growth and leaf yellowing (Figure 1A). In contrast, the drought-treated plants with TG1-E1 inoculation showed more robust growth compared to their non-inoculated counterparts (Figure 1A), demonstrating the capacity of TG1-E1 in enhancing plant drought resistance in tomato. During the period of drought treatment, soil humidity decreased similarly in the soil both with and without TG1-E1 inoculation (Figure 1B), indicating that the increased plant drought resistance was unlikely due to bacteria-dependent water retention in the soil. Under the drought conditions, inoculation with TG1-E1 increased the Micro-Tom plant’s fresh weight by almost 50% (Figure 1C). In addition, plant photosynthesis efficiency and chlorophyll contents were higher in drought-stressed plants with TG1-E1 inoculation than in those without inoculation (Figure 1D,E). Similar results were observed with the Ailsa Craig plants (Figure S1), confirming the drought stress-alleviating effects of TG1-E1 on tomato plants. The plant-beneficial effects were supported by the successful colonization of TG1-E1 to tomato roots under both mock and drought conditions (Figure S2).
Figure 1.
Bacillus megaterium TG1-E1 increases drought resistance in tomato seedlings. (A) TG1-E1 increased drought resistance in tomato (cultivar Micro-Tom) plants. Images were taken at 10 days after the drought treatment (DAT). (B) Relative humidity of the soil where tomato seedlings were grown. (C) Measurements of plant fresh weight. (D) Quantification of photosynthesis efficiency (Fv/Fm). (E) Measurements of chlorophyll contents. Results in panels (C–E) were from plants harvested at 10 DAT. The bar graphs show representative results from three independent experiments. Mean ± SE (n = 6 biological replicates). Asterisks denote significant differences at p < 0.05, Tukey’s multiple comparison test (*, *** and ns denote p < 0.05; p < 0.001 and p > 0.05, respectively).
2.2. Bacillus Megaterium TG1-E1 Induces Transcriptomic Reprograming of in Tomato Plants under Drought Stress
2.2.1. The Overall Functional Categorization of Differentially Expressed Genes (DEGs)
To gain insights into the mechanism for TG1-E1-increased plant drought resistance, we compared the transcriptomes between drought-stressed Micro-Tom with and without TG1-E1 inoculation. While the plants with and without TG1-E1-inoculation showed clearly contrasting phenotypes at 10 days after the drought treatment started (DAT), samples for the transcriptome analysis were harvested at 7 DAT, in order to catch the transcriptional regulation that is responsible for the downstream phenotypic changes. A total of 429 differentially expressed genes (DEGs) (Fold change ≥ 2; p-values ≤ 0.05) were identified, including 250 up-regulated DEGs and 179 down-regulated DEGs (Table S1).
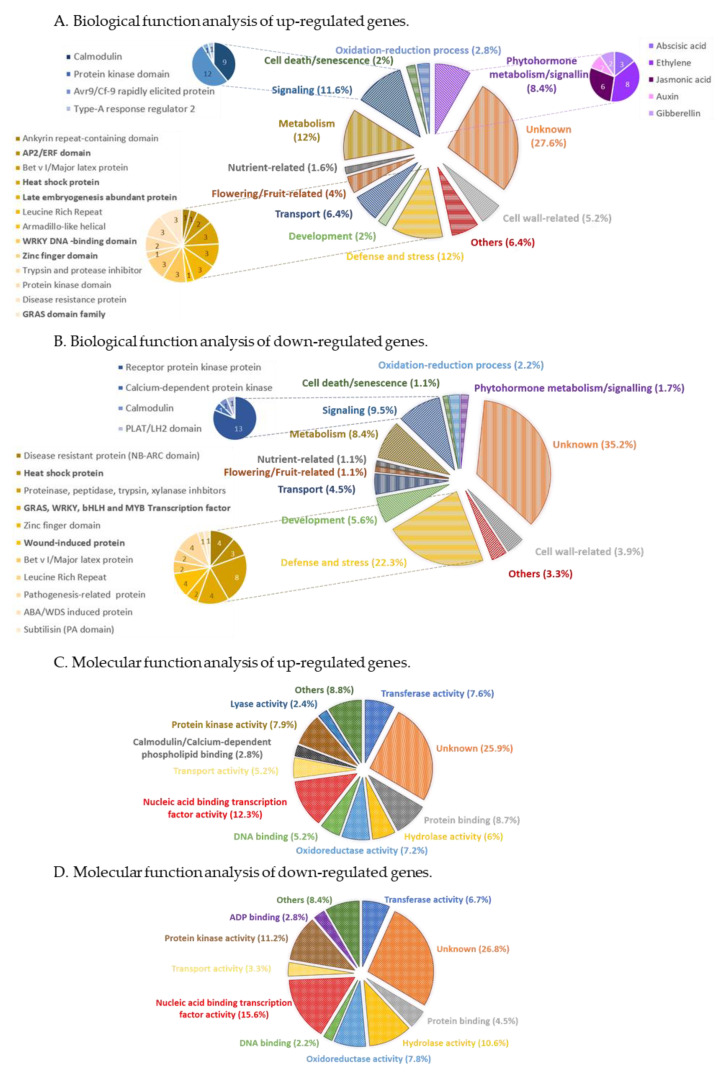
To obtain an overall view of TG1-E1′s impacts on plant cellular functions, the TG1-E1-regualted DEGs were subjected to functional categorization by using AgriGOv2 (Figure 2 and Figure S3). Major biological functional categories of the up-regulated DEGs include unknown (27.6%), metabolism (12%), defense and stress (12%), signaling (11.6%) and phytohormone metabolism and signaling (8.6%) (Figure 2A). Meanwhile, main categories of down-regulated DEGs include unknown (35.2%), defense and stress (22.3%), signaling (9.5%) and metabolism (8.4%) (Figure 2B). Notably, the category of defense and stress accounts for 12% of up-regulated DEGs and 22% of down-regulated DEGs, whereas DEGs in this category are composed of genes encoding proteins with various functions (Figure 1A,B), indicating a complex interaction between plant responses to the biotic stimulus (TG1-E1) and the abiotic stressor (drought). In addition, the category of cell wall biosynthesis and organization was identified in both the up- and down-regulated DEGs (Figure 1A,B), indicating that TG1-E1-triggered plant drought resistance involves alterations in cell wall.
Figure 2.
Functional categorization of tomato DEGs. Differentially expressed genes (DEGs) were identified by the comparison between drought-stressed plants with and without TG1-E1 inoculation. (A,B), Up- and down-regulated genes, respectively, are categorized based on their biological function. (C,D), Up- and down-regulated genes, respectively, are categorized based on their molecular function. Functional categorization was performed by using AgriGOv2. Numbers inside the diagram mean the amount of genes involved in each category.
In order to identify molecular factors that potentially mediate TG1-E1-triggered plant drought resistance, we next performed DEG categorization by molecular function analysis. The results highlighted transcription factors and protein kinases among other categories, because these two categories are both abundant in DEGs which are either up- or down-regulated by TG1-E1 (Figure 2C,D).
2.2.2. DEGs Implicating Regulation Mediated through Transcription Factors
Transcription factors are key regulators of plant responses to abiotic stress conditions including drought [10]. Genes encoding for transcription factors (TFs) accounted for 11% of all identified DEGs (Table 1), indicating that the observed transcriptional regulation by TG1-E1 would lead to a broadened impact on the plant transcriptome later on. Therefore, the enrichment of TFs in the DEGs demonstrates a key role of transcriptional regulation in mediating TG1-E1-triggered plant drought resistance.
Table 1.
Transcription factors that were transcriptionally regulated by TG1-E1 in drought-stressed tomato plants (Micro-Tom). The differentially expressed genes (DEGs) were defined by log2 fold-change values (LogFC) > 1 or < −1 with FDR < 0.01.
| Gene ID | Annotation | Type of Nucleic Acid Binding Transcription Factor Activity | LogFC |
|---|---|---|---|
| Solyc07g061760.2 | Ankyrin repeats | Ankyrin repeat-containing domain | 2.29 |
| Solyc08g014570.3 | PGG domain | Ankyrin repeat-containing domain | 1.06 |
| Solyc06g076050.3 | Ankyrin repeats | Ankyrin repeat-containing domain | 1.04 |
| Solyc05g052570.3 | Ankyrin repeats | Ankyrin repeat-containing domain | 1.11 |
| Solyc05g013540.1 | AP2 domain | AP2/ERF domain | 1.07 |
| Solyc02g090770.1 | AP2 domain | AP2/ERF domain | 2.79 |
| Solyc08g080290.3 | AP2 domain | AP2/ERF domain | 2.58 |
| Solyc08g007830.1 | AP2 domain | AP2/ERF domain | 4.48 |
| Solyc06g035700.1 | AP2 domain | AP2/ERF domain | 3.41 |
| Solyc08g007820.1 | AP2 domain | AP2/ERF domain | 3.02 |
| Solyc10g050970.1 | AP2 domain | AP2/ERF domain | 3.01 |
| Solyc01g108240.3 | AP2 domain | AP2/ERF domain | 4.22 |
| Solyc10g086530.1 | GRAS domain family | GRAS domain | 1.10 |
| Solyc11g012510.2 | GRAS domain family | GRAS domain | 1.28 |
| Solyc12g005340.2 | GRAS domain family | GRAS domain | 1.77 |
| Solyc08g080540.3 | HSF-type DNA-binding | HSF-type DNA binding | 1.64 |
| Solyc02g089200.3 | SRF-type transcription factor | MADS-box | 1.95 |
| Solyc05g015750.3 | SRF-type transcription factor | MADS-box | 1.67 |
| Solyc01g093960.3 | SRF-type transcription factor | MADS-box | 1.04 |
| Solyc06g059970.3 | SRF-type transcription factor | MADS-box | 1.03 |
| Solyc02g071730.3 | SRF-type transcription factor | MADS-box | 1.09 |
| Solyc08g067230.3 | K-box region | MADS-box/K-box | 2.25 |
| Solyc04g081000.3 | K-box region | MADS-box/K-box | 1.42 |
| Solyc05g015840.3 | SBP domain | SBP-box | 1.17 |
| Solyc10g011910.3 | WRKY DNA -binding domain | Zinc finger domain | 1.84 |
| Solyc03g121400.1 | Dof domain, zinc finger | Zinc finger domain | 1.10 |
| Solyc06g053640.1 | Ring finger domain | Zinc finger domain | 1.85 |
| Solyc10g009550.3 | WRKY DNA -binding domain | Zinc finger domain | 1.40 |
| Solyc08g008280.3 | WRKY DNA -binding domain | Zinc finger domain | 1.28 |
| Solyc04g015360.3 | GATA zinc finger | Zinc finger domain | 1.04 |
| Solyc06g075780.2 | C2H2-type zinc finger | Zinc finger domain | 1.49 |
| Solyc05g051200.1 | AP2 domain | AP2/ERF domain | −1.41 |
| Solyc04g007000.1 | AP2 domain | AP2/ERF domain | −1.80 |
| Solyc03g119390.3 | Helix-loop-helix DNA-binding domain | bHLH domain | −1.55 |
| Solyc03g034000.3 | Helix-loop-helix DNA-binding domain | bHLH domain | −1.53 |
| Solyc12g049320.2 | GRAS domain family | GRAS domain | −1.34 |
| Solyc02g077590.1 | Homeobox domain | HD-Zip domain | −1.34 |
| Solyc05g048830.3 | Myb-like DNA-binding domain | Myb domain | −1.05 |
| Solyc05g053330.3 | Myb-like DNA-binding domain | Myb domain | −1.12 |
| Solyc12g089190.1 | Myb-like DNA-binding domain | Myb domain | −1.27 |
| Solyc12g008800.2 | Myb-like DNA-binding domain | Myb domain | −1.42 |
| Solyc04g076220.3 | Domain of unknown function | PPC domain | −1.30 |
| Solyc05g006340.3 | WD domain, G-beta repeat | WD domain | −1.16 |
| Solyc11g006450.2 | Ring finger domain | Zinc finger domain | −1.22 |
| Solyc05g054650.1 | C2H2-type zinc finger | Zinc finger domain | −1.24 |
| Solyc05g051860.3 | zinc-finger of the FCS-type, C2-C2 | Zinc finger domain | −1.33 |
| Solyc05g015850.3 | WRKY DNA -binding domain | Zinc finger domain | −1.48 |
| Solyc06g008020.3 | HIT zinc finger | Zinc finger domain | −1.15 |
| Solyc00g136260.1 | Ring finger domain | Zinc finger domain | −1.97 |
| Solyc12g008830.2 | GATA zinc finger | Zinc finger domain | −1.44 |
| Solyc03g032060.1 | Ring finger domain | Zinc finger domain | −1.11 |
| Solyc12g006230.2 | Ring finger domain | Zinc finger domain | −1.77 |
| Solyc08g067360.3 | WRKY DNA -binding domain | Zinc finger domain | −1.39 |
| Solyc07g045180.3 | B-box zinc finger | Zinc finger domain | −1.23 |
| Solyc08g082680.3 | Ring finger domain | Zinc finger domain | −1.00 |
| Solyc08g067970.3 | zinc-finger of the FCS-type, C2-C2 | Zinc finger domain | −1.11 |
| Solyc06g061240.3 | PLATZ transcription factor | Zinc finger domain | −1.08 |
| Solyc08g065940.3 | Zinc finger C-x8-C-x5-C-x3-H type | Zinc finger domain | −1.08 |
Among the 48 TF DEGs are 22 zinc-finger family members (Table 1), some of which have been reported as responsive to hyperosmotic stress conditions such as drought and high salinity. For example, SlWRKY75 and SlWRKY45, which are known to be repressed by drought stress [11,12], were further down-regulated by TG1-E1 in plants under the drought stress condition; meanwhile, TG1-E1 up-regulated SlWRKY53 that was reported to be highly induced by salt stress [11]. Because TG1-E1 enhances plant resistance to drought stress, the TG1-E1-dependent transcriptional enhancement (either down- or up-regulation) highlights the function of these TFs in mediating plant adaptive responses to drought stress.
In addition to the zinc-finger TFs, DEGs encoding for TFs also include 10 APETALA2/ethylene-responsive factor (AP2/ERF) domain family members with eight DEGs being up-regulated, four GRAS family members with 3 DEGs being up-regulated, and seven MADS-box TFs which were all up-regulated by TG1-E1 (Table 1). These results strongly indicate that the amplification of transcriptional regulation is crucial for achieving TG1-E1-triggered plant drought resistance. The GRAS TFs play a significant role in tomato plant growth and development [13], while MADS-box TFs are key regulators of developmental plasticity in different plant species [14]. Therefore, the transcriptional regulation of these TFs provides clues to understanding how TG1-E1-treated plants adjust growth and development to adapt to the water deficit condition.
2.2.3. DEGs Implicating Regulation of Drought-Responsive Signaling
Protein kinases, such as calmodulin-dependent protein kinases (CDPKs), mitogen-activated protein kinases (MAPKs) and receptor protein kinases (RPKs), are important regulatory components of plant signal transduction in response to various biotic and abiotic stress conditions. In the drought-stressed plants, TG1-E1 differentially regulated a total of 38 genes encoding for several types of protein kinases (Table 2). Notably, these DEGs are dominated by genes encoding for RPKs, with 14 and 16 RPKs up-regulated and down-regulated by TG1-E1, respectively (Table 2). The enrichment of RPKs in the kinase DEGs suggests that RPKs are important regulators of TG1-E1-triggered plant drought resistance. Meanwhile, four MAPKs (SlMAPK3, SlMAPKKK14, SlMAPKKK21, SlMAPKKK59), which have been implicated in tomato responses to environmental stress including drought [15], were up-regulated by TG1-E1 (Table 2), suggesting the important roles of these stress-response regulators in mediating TG1-E1-triggered plant drought resistance.
Table 2.
Signaling-related proteins that were transcriptionally regulated by TG1-E1 in drought-stressed tomato plants (Micro-Tom). The differentially expressed genes (DEGs) were defined by log2 fold-change values (LogFC) > 1 or < −1 with FDR < 0.01.
| Gene ID | Annotation | Type of Signaling Protein | LogFC |
|---|---|---|---|
| Solyc11g072930.2 | Carbohydrate-binding protein of the ER | RPK | 1.74 |
| Solyc06g062450.3 | Carbohydrate-binding protein of the ER | RPK | 1.44 |
| Solyc02g094010.2 | Protein kinase domain | RPK | 1.58 |
| Solyc09g014590.3 | Leucine Rich Repeat | RPK | 3.12 |
| Solyc10g006690.3 | Protein tyrosine kinase | RPK | 1.01 |
| Solyc06g007190.3 | Protein phosphatase 2C | ILKAP | 2.04 |
| Solyc12g009550.1 | Leucine rich repeat | RPK | 1.07 |
| Solyc01g005730.3 | Leucine rich repeat | RPK | 1.14 |
| Solyc08g077630.3 | Protein kinase domain | RPK | 1.54 |
| Solyc01g098690.2 | Leucine rich repeat N-terminal domain | RPK | 1.23 |
| Solyc05g053010.1 | Protein kinase domain | RPK | 1.59 |
| Solyc07g064820.1 | Protein kinase domain | MAPK | 1.02 |
| Solyc11g020230.1 | Protein kinase domain | RPK | 1.05 |
| Solyc02g064980.1 | Protein kinase domain | MAPK | 3.15 |
| Solyc02g090970.1 | Protein kinase domain | MAPK | 3.05 |
| Solyc08g077560.3 | Protein tyrosine kinase | RPK | 1.42 |
| Solyc04g074270.3 | Leucine rich repeat | RPK | 1.08 |
| Solyc02g089900.1 | LysM domain | RPK | 1.07 |
| Solyc06g005170.3 | Protein kinase domain | MAPK | 1.65 |
| Solyc09g018280.1 | NAF domain | CDPK | 1.14 |
| Solyc02g092450.3 | E1-E2 ATPase | PM-CA-ATPase | 1.28 |
| Solyc02g064680.3 | E1-E2 ATPase | PM-CA-ATPase | 1.87 |
| Solyc01g099370.3 | C2 domain | CaLB | 1.61 |
| Solyc08g008370.3 | Development and cell death domain | CaLB | 1.66 |
| Solyc03g113980.3 | Calmodulin binding protein-like | CLM | 1.05 |
| Solyc06g053930.3 | EF-hand domain pair | CaM | 1.04 |
| Solyc03g097100.1 | EF-hand domain pair | CLM | 1.10 |
| Solyc11g071740.2 | EF-hand domain pair | CLM | 2.67 |
| Solyc03g118810.1 | EF-hand domain pair | CaM | 1.38 |
| Solyc03g119250.3 | Calmodulin binding protein-like | CLM | 1.99 |
| Solyc06g006020.2 | Leucine Rich Repeat | RPK | −1.31 |
| Solyc12g009510.1 | Leucine rich repeat | RPK | −1.13 |
| Solyc11g017280.2 | Leucine rich repeat | RPK | −1.07 |
| Solyc04g074020.2 | Leucine rich repeat N-terminal domain | RPK | −1.75 |
| Solyc04g009910.3 | Protein kinase domain | CDPK | −1.21 |
| Solyc12g009780.1 | Leucine Rich Repeat | RPK | −1.19 |
| Solyc04g074030.3 | Leucine rich repeat N-terminal domain | RPK | −1.91 |
| Solyc11g011180.2 | Leucine Rich repeat | RPK | −1.23 |
| Solyc05g012430.1 | Leucine rich repeat | RPK | −1.71 |
| Solyc09g090210.3 | Protein tyrosine kinase | RPK | −1.10 |
| Solyc07g006770.2 | TMEM154 protein family | RPK | −1.20 |
| Solyc10g076550.1 | Protein kinase domain | RPK | −1.85 |
| Solyc04g074050.3 | Protein kinase domain | RPK | −1.75 |
| Solyc01g067020.3 | Protein kinase domain | RPK | −1.16 |
| Solyc09g075920.1 | D-mannose binding lectin | RPK | −1.10 |
| Solyc08g016270.2 | Leucine rich repeat | RPK | −1.13 |
| Solyc04g074000.3 | Protein tyrosine kinase | RPK | −1.64 |
| Solyc08g077730.3 | MORN repeat | PIPK | −1.22 |
| Solyc04g009900.3 | Protein kinase domain | PPCK | −1.27 |
| Solyc12g088840.1 | EF-hand domain | CaM | −1.03 |
| Malic acid related DEGs | |||
| Solyc04g080990.2 | Voltage-dependent anion channel | 1.32 | |
| Solyc09g014610.3 | Voltage-dependent anion channel | 1.04 | |
| Solyc03g119640.3 | Aluminum activated malate transporter | 1.30 | |
RPK: Receptor-like kinase protein. ILKAP: Integrin-linked kinase-associated serine/threonine protein. MAPK: Mitogen-activated protein kinase. CDPK: Calcium/calmodulin-dependent protein kinase protein. PM-CA-ATPase: Plasma membrane Ca2+ ATPase protein. CaLB: Calcium-dependent lipid-binding protein. CaM: Calmodulim protein. CLM: Calmodulin-binding protein-like. PIPK: Phosphoinositide kinase-like protein. PPCK: Phosphoenolpyruvate carboxylase kinase.
Calmodulins receive and transduce Ca2+ signals elicited by various stressors. One of the primary responses to drought stress is an increase in the cytosolic Ca2+ concentration and subsequent transduction of Ca2+ signals that promotes appropriate cellular responses in an effort to mitigate potential damages [2,16]. In the drought-stressed tomato, TG1-E1 increased gene expression of seven calmodulins and calmodulin-binding protein-like (CLMs) (Table 2). Notably, the CLM-encoding DEGs include SlCBP60 (Solyc03g119250), which is orthologous to AtSlCBP60g, which positively regulates plant disease resistance and drought resistance in Arabidopsis thaliana [17]. In addition, TG1-E1 also up-regulated gene expression of two calcium-dependent lipid-binding proteins and two plasma-membrane calcium-transporting ATPases (Solyc02g092450; Solyc02g064680) (Table 2), which are key regulators of stress-induced calcium transients in plants [18,19,20]. Together these results indicate that bacterial modulation of plant Ca2+ signaling plays an important role in mediating TG1-E1-triggered drought resistance in tomato.
TG-E1 also induced gene expression of two root C4-dicarboxylate transporter/malic acid transport protein (Solyc04g080990; Solyc09g014610) and an aluminum-activated malate transporter (Solyc03g119640) in drought-stressed tomato (Table 2). As a component of root exudates, malic acid functions in the induction of PGPR chemotaxis and the promotion of biofilm formation for better colonization [21,22]. Therefore, the induction of the malate transporter gene appears to indicate an enhancement in the association between TG1-E1 and the drought-stressed plants.
2.2.4. DEGs Implicating Regulation of Cell Wall Biosynthesis and Organization
Cell wall composition and organization are modified in plants exposed to water deficit [23,24] and bacterial colonization [25]. One of the primary responses of plant cell wall to drought stress is an increase in the biosynthesis of cellulose, xyloglucan and pectin to maintain cell wall integrity and cell turgor pressure, leading to continued cell growth under low water potential [26]. A group of tomato cell wall-related DEGs were identified as regulated by TG1-E1 (Table 3), including two glycosyl hydrolases (Solyc06g073750; Solyc07g006850) involved in xyloglucan metabolism, a cellulose synthase-like protein (Solyc03g097050) and a cellulose synthase-like C1-2 glycosyltransferase (Solyc02g089640) implied in cellulose biosynthesis, and a pectin esterase (Solyc09g075330) involved in pectin metabolism, thereby implying bacteria-triggered regulation of the cell wall structure in response to drought stress.
Table 3.
Cell wall-related proteins that were transcriptionally regulated by TG1-E1 in drought-stressed tomato plants (Micro-Tom). The differentially expressed genes (DEGs) were defined by log2 fold-change values (LogFC) > 1 or < −1 with FDR < 0.01. UDP: Uridine diphosphate; GDP: Guanosine diphosphate; NAD: Nicotinamide adenine dinucleotide.
| Gene ID | Annotation | LogFC |
|---|---|---|
| Solyc06g073750.3 | Glycosyl hydrolase | 1.99 |
| Solyc07g006850.2 | Glycosyl hydrolase | 1.04 |
| Solyc03g115380.2 | UDP-glucose/GDP-mannose dehydrogenase | 1.78 |
| Solyc02g094010.2 | Protein kinase domain | 1.58 |
| Solyc03g110890.1 | Polysaccharide biosynthesis related protein | 1.13 |
| Solyc09g075330.3 | Pectinesterase | 1.08 |
| Solyc09g011860.3 | GDP-fucose protein O-fucosyltransferase | 1.69 |
| Solyc12g010540.1 | GDP-mannose 4,6 dehydratase | 1.07 |
| Solyc04g005040.1 | Putative peptidoglycan binding domain | 1.37 |
| Solyc02g089640.3 | Glycosyltransferase-like | 1.03 |
| Solyc03g123620.4 | Plant invertase/pectin methylesterase inhibitor | 1.35 |
| Solyc03g097050.3 | RING/Ubox-like zinc-binding domain | 1.42 |
| Solyc06g074670.3 | NAD dependent epimerase/dehydratase | 1.31 |
| Solyc04g072280.3 | Multicopper oxidase | −2.95 |
| Solyc03g026360.1 | LysM domain | −1.23 |
| Solyc11g044910.2 | Glycosyl hydrolase domain | −1.66 |
In drought-stressed tomato plants, TG1-E1 up-regulated gene expression of several glycosyltransferases (Table 3), which may function in regulating plant cell wall polysaccharides [27]. Particularly, gene up-regulation was observed for a UDP-glucose/GDP-mannose dehydrogenase (Solyc03g115380), a GDP-fucose protein O-fucosyltransferase (Solyc09g011860), orthologous to At2g03280, and a GDP-mannose 4,6 dehydratase (Solyc12g010540), which are involved in pectin metabolism [28,29], a potential glucuronoxylan 4-O-methyltransferase-like protein (Solyc03g110890), orthologous to At4g24910, implied in xylan modifications [30], and a NAD-dependent epimerase/dehydratase (Solyc06g074670) that catalyzes the conversion of UDP-D-glucuronate to UDP-D-xylose, providing nucleotide sugars for cell-wall polymers. In addition, a glycosyl hydrolase (Solyc11g044910), required for pectic arabinan modification, was down-regulated by TG1-E1. Plant polysaccharides, including arabinogalactan, pectin, and xylan, can act as environmental cues that trigger biofilm formation in certain PGPR strains and consequently support bacterial root colonization [31]. Thus, it is intriguing whether these glycosyltransferases might be involved in TG1-E1 colonization to tomato roots.
We also found a putative matrix metalloproteinase (Solyc04g005040) that is potentially involved in the degradation and remodeling of the extracellular matrix, a cell surface continuum beyond the cell wall, which plays a vital role in cell adhesion, cell-cell communication, cell wall modification, and protection against stresses [32]. Altogether, these cell wall-related DEGs indicate that TG1-E1 might trigger modifications in plant cell wall structure and composition, which potentially contributed to TG1-E1 colonization and/or plant adaption to the altered cell turgor pressure.
3. Hyperosmotic Stress Induces TG1-E1 Production of Extracellular Metabolites Including Potential Osmoprotectants
To identify the bacterial factors that potentially caused the increased plant drought resistance, we collected TG1-E1 extracellular metabolites under normal and hyperosmotic stressed conditions (PEG30). Subsequently, analysis by gas chromatography coupled with mass spectrometry (GC-MS) identified 49 metabolites as being significantly altered (Fold change ≥ 1.9; p-values ≤ 0.05) in TG1-E1 by the hyperosmotic stress (Table 4).
Table 4.
Extracellular metabolites released from B. megaterium TG1-E1. The ratios show metabolite levels from TG1-E1 with hyperosmotic stress (by PEG30) compared to the non-stressed condition. N = 3 biological replicates. All ratios have p < 0.05, Student’s t-test. MeOX: methoximated-derived compound.
| Compound | Mock | PEG30 | LogFC |
|---|---|---|---|
| Piruvate | 0.04 | 0.14 | 1.72 |
| beta-Aminoisobutiric acid | 3.99 | 11.99 | 1.59 |
| acid α-ketocaproate | 0.35 | 2.88 | 3.06 |
| Urea | 20.53 | 55.66 | 1.44 |
| Ethanolamine | 2.11 | 15.58 | 2.88 |
| Leucine | 2666.87 | 6457.33 | 1.28 |
| Succinate | 51.39 | 130.40 | 1.34 |
| Glycerate | 6.04 | 16.87 | 1.48 |
| Fumarate | 4.64 | 9.08 | 0.97 |
| Serine | 7.56 | 34.32 | 2.18 |
| Threonine | 8.33 | 50.54 | 2.60 |
| beta-Alanine | 1.19 | 17.05 | 3.84 |
| Glutamine | 1.51 | 6.59 | 2.13 |
| Malate | 2.20 | 5.55 | 1.33 |
| Cytosine | 0.05 | 0.17 | 1.72 |
| L-Aspartate | 48.05 | 160.37 | 1.74 |
| Ornithin-Citrullin-Arginine | 1.09 | 4.55 | 2.06 |
| Xylose MeOX1 | 0.12 | 1.55 | 3.65 |
| Ribose | 0.22 | 0.64 | 1.54 |
| cis-Aconitate | 0.18 | 0.56 | 1.65 |
| 1-Methyl-L-histidine | 0.11 | 0.49 | 2.19 |
| Glycerate-3-P | 0.40 | 0.97 | 1.28 |
| Citrullin-Ornithin-Arginine | 6.14 | 78.11 | 3.67 |
| Citrate | 2.13 | 6.83 | 1.68 |
| Isocitrate | 0.52 | 1.45 | 1.47 |
| Arginine-NH3 | 8.62 | 16.92 | 0.97 |
| Pinitol | 0.10 | 0.28 | 1.50 |
| Fructose MEOX1 | 0.09 | 0.30 | 1.78 |
| Fructose MEOX2 | 0.04 | 0.12 | 1.52 |
| Galactose MeOX1 | 0.11 | 0.38 | 1.84 |
| Glucose MEOX1 | 0.46 | 1.46 | 1.67 |
| Glucose MEOX2 | 0.04 | 0.17 | 1.99 |
| Glucuronic acid MEOX1 | 0.03 | 0.13 | 2.04 |
| Gluconate | 0.23 | 0.59 | 1.35 |
| myo-Inositol | 7.21 | 24.71 | 1.78 |
| Spermidine | 0.45 | 3.41 | 2.93 |
| L-Cystathionine | 0.71 | 3.80 | 2.42 |
| Tryptophan | 112.05 | 222.43 | 0.99 |
| Fructose-6-P | 0.04 | 0.09 | 1.18 |
| Gluconate-6-P | 0.03 | 0.13 | 2.17 |
| Sucrose | 0.59 | 2.00 | 1.75 |
| Trehalose | 0.33 | 0.66 | 0.98 |
| Lysine | 4737.93 | 12276.77 | 1.37 |
| Histidine | 193.44 | 393.00 | 1.02 |
| Maleic acid | 9.10 | 2.61 | −1.80 |
| Uracil | 51.66 | 13.95 | −1.89 |
| Thymine | 6.94 | 0.16 | −5.46 |
| 4-Aminobutyrate (GABA) | 20.49 | 3.28 | −2.64 |
3.1. Sugars
TG1-E1 under hyperosmotic stress released higher levels of components of the glycolytic pathway, including glucose-6-P, fructose-6-P, glycerate-3-P, pyruvate and trehalose (Table 4). The glycolytic pathway have been related to increased levels of trehalose as an osmoprotectant molecule against drought in plants [33,34] and bacteria [35,36]. In this sense, we found an increase in trehalose upon PEG treatment, indicating that TG1-E1 might enhance plant drought tolerance by this osmoprotectant sugar.
Under the hyperosmotic stress condition, TG1-E1 also exuded higher levels of xylose, galactose, sucrose, and arabinose (Table 4). These sugars may function as a carbon reservoir, which protects microorganisms from fluctuations in water potential by enhancing water retention and regulating the diffusion of carbon sources [37,38]. Although it is unclear whether the drought-stressed plants would take in these bacteria-derived sugars, these sugars have been identified in other Bacillus strains as bacterial metabolites with a role in improving plant drought resistance [39]. In cells under dehydration stress, the hydroxyl group of cellular sugar alcohols can substitute the hydroxyl group of water to maintain the hydrophilic interactions with the membrane lipids and proteins, thereby helping the maintenance of membrane structural integrity [40].
3.2. Amino Acids
TG1-E1 under hyperosmotic stress accumulated a higher level of extracellular arginine, together with several other amino acids (Table 4). Arginine can be used as the substrate for producing polyamines, including spermidine, spermine and their diamine precursor putrescine, which are the major polyamines involved in drought resistance [41,42,43]. Inoculation of A. thaliana with Pseudomonas putida GAP-P45 caused significant fluctuations in the expression of most polyamine biosynthetic genes and cellular levels of polyamines, including putrescine and spermidine, which positively correlated with the water stress tolerant phenotype of A. thaliana in response to P. putida GAP-P45 inoculation [44,45]. In our RNAseq results, TG1-E1 significantly up-regulated the expression of several genes involved in polyamine production, including Arginine decarboxylase 1 (SlADC1; Solyc10g054440), which is a rate-limiting enzyme for the biosynthesis of putrescine and other polyamines [46,47], as well as Adenosylmethionine decarboxylase 2 (SlSAMDC2; Solyc02g089610) and S-adenosyl-l-methionine decarboxylase (Solyc02g089615) which are involved in spermidine and spermine biosynthesis [48,49] (Table 5). In contrast, the expression of genes involved in arginine biosynthesis was not affected by TG1-E1. Therefore, it is possible that TG1-E1-produced arginine may be utilized by the plants for polyamine production, thereby contributing to the bacteria-induced plant drought resistance. In addition, TG1-E1 under hyperosmotic stress exuded more spermidine than under the non-stressed condition, indicating that the plants might also directly take up bacterial polyamine to increase drought resistance.
Table 5.
Transcriptional regulation of gene potentially regulated by TG1-E1 extracellular metabolites under hyperosmotic stress. The differentially expressed genes (DEGs) were defined by log2 fold-change values (LogFC) > 1 or < −1 with FDR < 0.01.
| Gene ID | Annotation | LogFC |
|---|---|---|
| Solyc10g054440.2 | Pyridoxal-dependent decarboxylase | 1.46 |
| Solyc02g089610.2 | Adenosylmethionine decarboxylase | 1.39 |
| Solyc02g089615.1 | S-adenosyl-l-methionine decarboxylase leader peptide | 1.31 |
| Solyc08g007820.1 | Dehydration-responsive element-binding protein 1E-like | 3.02 |
| Solyc03g080090.3 | No apical meristem (NAM) protein | 1.08 |
| Solyc07g008300.2 | Ring hydroxylating alpha subunit | 2.28 |
| Solyc08g068600.3 | Pyridoxal-dependent decarboxylase conserved domain | 1.20 |
TG1-E1 under hyperosmotic stress accumulated a higher level of extracellular glutamine (Table 4). Meanwhile, TG1-E1 up-regulated gene expression of Dehydration-responsive element-binding protein 1E-like (DREB1a-like; Solyc08g007820) and Nam-like protein 1 (Solyc03g080090) in drought-stressed tomato. These two tomato genes are orthologous to rice OsDREB1A and OsNAC5, respectively, which are inducible by exogenous glutamine, and play positive roles in rice plant drought resistance [50,51,52]. Thus, it appears that the gene induction of tomato DREB1a-like and Nam-like protein 1 by bacterial glutamine also contributes to TG1-E1-triggered plant drought resistance. Consistent with a positive role of glutamine in plant drought resistance, pepper plants inoculated with Microbacterium sp. 3J1 accumulated showed increased levels of glutamine as a major change in metabolites for balancing the hyperosmotic stress [53].
3.3. Precursors of the Osmoprotectant Glycine Betaine
The level of ethanolamine in TG1-E1 exudates was increased by hyperosmotic stress (Table 4). Ethanolamine is a precursor of glycine betaine and proline which are two important osmoprotectants in plants [54]. In fact, exogenous application of ethanolamine stimulates glycine betaine biosynthesis in Nicotiana rustica, resulting in improved plant resistance to salt stress [55]. TG1-E1 up-regulated gene expression of Solyc07g008300 (Table 5), which is a choline monooxygenase functioning in the first step of glycine betaine biosynthesis [56]. TG1-E1 up-regulated gene expression of Solyc08g068600 (Table 5), a putative serine decarboxylase that is involved in ethanolamine metabolism. Meanwhile, the level of TG1-E1 exuded serine was increased by hyperosmotic stress (Table 4). Altogether, these results indicate that TG1-E1-produced ethanolamine may be utilized by the plants for glycine betaine production, thereby contributing to the bacteria-induced plant drought resistance.
3.4. Pinitol
An increase in the level of pinitol, a polyol that has been described as an osmoprotectant in several plant species [57,58,59], was also observed in TG1-E1 extracellular metabolites with the hyperosmotic stress treatment (Table 4). Synthesis of D-ononitol, a transient intermediate of D-pinitol, increased in leaves under drought stress conditions in Arabidopsis thaliana; subsequently, the D-ononitol was transported to the roots to synthesize D-pinitol, which then acts as osmoprotectant [58]. Therefore, pinitol produced by TG1-E1 may directly protect tomato roots from drought stress.
Together, these results potentially provide a mechanism for TG1-E1-induced plant drought resistance. It should be noted that in a real system, i.e., plants with root-colonizing bacteria, the dynamic interaction between the roots and the bacteria would likely result in more complex profiles of bacteria extracellular metabolites.
In this study, the plants were transferred from sterile growth medium to a steam-sterilized (20 min at 120 °C) soil mixture of peat and vermiculite (1:1, v/v). The plants were then grown in a clean growth chamber. For performing the bacteria inoculation, the bacteria was centrifuged from its original culture solution and then resuspended in sterile 0.45% NaCl solution. Although these actions could not completely prevent unwanted growth of other bacteria, they helped minimize possible involvements of other bacteria. Sometimes the inoculant may fail to colonize and consequently fail to trigger beneficial effects. However, it is unlikely that inoculation of one bacteria strain under these conditions would result in dominant growth of another strain, which then happens to cause the same beneficial effects to the plants. Nonetheless, our study of the soil-grown plants cannot completely exclude the possibility that certain environmental factors other than the inoculant may contribute to the plant-beneficial effects.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum) seeds of two different cultivars, Micro-Tom and Ailsa Craig, were surface-sterilized by soaking in 5% v/v sodium hypochlorite for 5 min. After washing by sterile distilled water, the seeds were planted on 1/2-strength Murashige and Skoog medium (0.5 MS) with 0.7% (w/v) agar and 1.5% (w/v) sucrose. The plates were then placed in the dark for 4 d at 25 °C. Seedlings were grown under sterile conditions with 200 µmol photons/m2/s light, 25 ± 2 °C, at a 16 h light/8 h dark cycle. Ten days after germination, tomato seedlings were transferred into a steam-sterilized (20 min at 120 °C) soil mixture of peat and vermiculite (1:1, v/v). Plant growth and treatments took place in a growth chamber (16 h day at 25 °C + 8 h night at 22 °C).
4.2. Bacterial Culture and Inoculation
To make the inoculum, Bacillus megaterium TG1-E1 was cultured in liquid Luria–Bertani (LB) medium at 37 °C and 180 rpm overnight. Bacterial growth was estimated by measuring the optical density at 600 nm using a spectrophotometer. The bacterial suspension was centrifuged (5000 rpm, 20 min) and resuspended in 0.45% (w/v) NaCl sterile solution. Inoculation with B. megaterium TG1-E1 was carried out 2 days after tomato seedlings were planting in 200-mL pots. Each seedling was grown in a separate pot and was inoculated with 50 mL bacterial suspension (108–109 CFU/mL) in 0.45% NaCl sterile solution, and non-inoculated controls were watered with 0.45% NaCl sterile solution.
4.3. Monitoring of Plant Growth
After 10 days of bacterial inoculation, the drought treatment was started by stopping the watering. Fresh weight (FW), relative water content (RWC), maximum chlorophyll fluorescence (Fv/Fm) and chlorophyll content of tomato seedlings were measured 10 days after the drought treatment started. The RWC of the leaves was calculated as RWC = (FW − DW) × (TW − DW) − 1. The soil relative humidity was measured by a WET DELTA-T device (DELTA-T DEVICES LTD, Cambridge, England) at 1, 3 and 7 days after the drought treatment started, following the protocol provided by the manufacturer.
4.4. Bacteria Root Colonization
Root colonization test was carried out based on Vílchez et al. [60]. Briefly, TG1-E1-inoculated and control tomato seedlings were harvest seven days after disruption of watering. Roots were separated from the aerial portions and were surface-sterilized by immersion in ethanol (75%; vol/vol) for 1 min, then washed for four times using sterile double-distilled water. Separated roots were then grinded using pistils in 1.5 mL tubes. A 0.45% NaCl solution was used to prepare serial dilutions for a drop-by-drop seeding on LB agar plates. After 24 h culturing at 37 °C, bacteria colonies were counted and expressed as c.f.u. mg−1 root dry weight.
4.5. Quantification of Chlorophyll Contents and Photosynthesis Efficiency
Two grams of fresh tomato leaves were immersed in 10 mL of 96% ethanol for 24 h at room temperature in the dark and then centrifuged at 5000 rpm for 10 min at room temperature. The chlorophyll contents of the supernatants were measured by reading optical density (OD) using a Microplate Reader Thermo Varioskan Flash 13. The wave length for optical density measurements were 665 (OD665) and 649 (OD649) nm. The concentrations of total chlorophyll, chlorophyll a, and chlorophyll b were calculated by the following equations:
Chlorophyll a concentration (Ca) = 13.95 × OD665 − 6.88 × OD649.
Chlorophyll b concentration (Cb) = 24.96 × OD649 − 7.32 × OD665.
Total chlorophyll concentration (CT) = Ca + Cb.
Content of chlorophyll = (CT × V)/W, where V is the volume of supernatant and W is the weight of fresh tissue.
The ratio of variable fluorescence/maximum fluorescence (Fv/Fm), which indicates the potential quantum yield of PSII photochemistry, was determined with a Fluorpen FP 100 (Photon Systems Instruments, Brno, Czech Republic) following the protocol provided by Photon Systems Instruments.
4.6. Bacterial Extracellular Metabolite Collection and GC-MS Analysis
Extraction of bacterial exudates under regular and hyperosmotic stress (by supplying 30% wt/vol of polyethyleneglycol; PEG-30; 24 h) conditions, were prepared as in [53]. In brief, a 50 mL aliquot of a LB-cultured strain was filtered by a 22 µm mesh, frozen at −80 °C and freeze-dried for further processing. Then, 20 mg per sample were treated with 80% methanol and 1 μM of ribitol was added as an internal control. After being derivatized by using 100 μL methoxylamine hydrochloride in pyridine and treated with 100 μL MSTFA, samples were ready to be run in a TraceGC (up to 1 μL per sample), coupled to a PolarisQ ion trap mass spectrometer (Thermo Finnigan, Dreieich, Germany). Compounds in the sample were separated on a 30 m × 0.25 mm Equity-5 column with 0.25 μm coating of 5% diphenyl 95% dimethylsiloxane (Supelco, Bellefonte, California, CA, USA). Finally, compounds were identified by comparison with purified standards, the NIST 2005 database (NIST, Gaithersburg, MD, USA) and the Golm Metabolome Database. Identities were confirmed by matching mass spectral data and chromatographic retention time. Peak area quantification was prepared with Xcalibur 1.4 software (Thermo Finnigan, Dreieich, Germany) and normalized with ribitol standard and dry mass of the sample. Six replicates per condition were carried out in this analysis.
4.7. RNAseq Analysis
Tomato Micro-Tom cultivar was grown and treated as described in the “Plant growth conditions” section. TG1-E1-inoculated and control tomato seedlings were harvested 7 days after disruption of watering. RNA was extracted by using Plant RNeasy Kit (Qiagen, Hilden, Germany), and the integrity was monitored using the RNA Nano 6000 Assay of the Agilent Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA). RNA purity and concentration were checked using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
RNAseq was performed at the Core Facility for Genomics, Shanghai Center for Plant Stress Biology, China. Three biological replicates of each treatment were generated. Total RNA (1 µg) from each sample was used for library preparation with NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, E7420L), following the manufacturer’s instructions. The prepared libraries were assessed for quality by using NGS High-Sensitivity Kit on a Fragment Analyzer (AATI) and for quantity by using Qubit 2.0 Fluorometer (Thermo Fisher Scientific).
For data analysis, SolexaQA 2.0 [61] and cutadapt v1.10 [62] were used to remove low quality regions and adapter sequences in the raw reads, and clean reads with length longer than 25 bp and phred score greater than 17 were mapped to the Tair10 reference genome using TopHat 2.0.10 [63] with default parameters. The reads that were mapped to each annotated gene were counted by HTseq-count (version 0.9.1) [64]. Using edgeR [65], the raw counts of each gene were normalized, and differentially expressed genes were identified using fold change >2 and false discovery rate < 0.01 as significance cutoffs. Three biological replicates of each condition were used to perform transcriptome analyses.
4.8. Statistical Analysis
Statistical analyses were performed with Prism software (https://www.graphpad.com/scientific-software/prism/, Accessed 4 April 2020). Significant difference between treatments was based on p-values ≤ 0.05.
5. Conclusions
PGPR are promising tools for increasing crop drought resistance. In this study, we show that B. megaterium TG1-E1 increases drought resistance in different tomato cultivars. Our transcriptome analysis reveals several key mediators of TG1-E1-induced transcriptional regulation in tomato plants, including transcription factors, stress signaling components and regulators, and putative regulators of cell wall organization. In addition, our analysis of TG1-E1 extracellular metabolites identifies some important compounds, including sugars, amino acid, ethanolamine, and pinitol, which are potentially regulators (or precursors of regulators) of TG1-E1-triggered plant drought resistance. These findings not only contribute to our understanding of PGPR-triggered drought resistance in tomato plants, but also provide important clues for future elucidation of the underlying molecular mechanisms.
Acknowledgments
Research in H.Z. Lab has been supported by the Chinese Academy of Sciences (CAS).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/metabo11060369/s1, Figure S1: Bacillus megaterium TG1-E1 alleviate drought stress and maintain growth in Ailsa-Craig tomato plants; Figure S2: Root colonization by TG1-E1 in tomato seedlings exposed to drought. Figure S3: Distribution of enriched Gene Ontology (GO). Supplementary Table S1: Differentially expressed genes (DEGs) regulated by TG1-E1 in drought stressed tomato plants (Micro-Tom) compared to un-inoculated plants. Excel files: Supplementary table Metabolites and Supplementary table RNAseq.
Author Contributions
H.Z. (Huiming Zhang) designed the project with the input of R.J.L.M. R.J.L.M., J.I.V. and S.Z. to-gether performed/participated in all experiments or data analysis. H.Z. (Hailing Zi) and R.L. performed R.N.A. seq raw data analysis. K.N. performed the G.C.-M.S. analysis. R.K. and D.H. participated in the experiments. H.Z. (Huiming Zhang) and R.J.L.M. wrote the manuscript with input from A.K.H. and J.I.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Chinese Academy of Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bailey-Serres J., Parker J.E., Ainsworth E.A., Oldroyd G.E.D., Schroeder J.I. Genetic strategies for improving crop yields. Nature. 2019;575:109–118. doi: 10.1038/s41586-019-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glick B.R. Overview of plant growth-promoting bacteria. In: Patten B.R.N., Holguin G., Penrose D.M., editors. Biochemical and Genetic Mechanisms Used by Plant Growth Promoting Bacteria. Imperial College Press; London, UK: 1999. pp. 1–13. [Google Scholar]
- 4.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 5.Liu X.M., Zhang H. The effects of bacterial volatile emissions on plant abiotic stress resistance. Front. Plant. Sci. 2015;6:774. doi: 10.3389/fpls.2015.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amellal N., Burtin G., Bartoli F., Heulin T. Colonization of wheat rhizosphere by EPS producing Pantoea agglomerans and its effect on soil aggregation. Appl. Environ. Microbiol. 1998;64:3740–3747. doi: 10.1128/AEM.64.10.3740-3747.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H., Sun Y., Xie X., Kim M.S., Dowd S.E., Paré P.W. A soil iron via deficiency-inducible mechanisms. Plant J. 2009;58:568–577. doi: 10.1111/j.1365-313X.2009.03803.x. [DOI] [PubMed] [Google Scholar]
- 8.Morcillo R.J.L., Singh S.K., He D., An G., Vílchez J.I., Tang K., Yuan F., Sun Y., Shao C., Zhang S., et al. Rhizobacterium-derived diacetyl modulates plant immunity in a phosphate-dependent manner. EMBO J. 2019;39:e102602. doi: 10.15252/embj.2019102602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vílchez J.I., Tang Q., Kaushal R., Wang W., Lv S., He D., Chu Z., Zhang H., Liu R., Zhang H. Complete Genome Sequence of Bacillus megaterium Strain TG1-E1, a Plant Drought Resistance-Enhancing Bacterium. Microbiol. Resour. Announc. 2018;7:e00842-18. doi: 10.1128/MRA.00842-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and resistance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 11.Huang S., Gao Y., Liu J., Peng X., Niu X., Fei Z., Cao S., Liu Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012;287:495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- 12.Karkute S.G., Gujjar R.S., Rai A., Akhtar M., Singh M., Singh B. Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene. 2018;13:8–17. doi: 10.1016/j.plgene.2017.11.002. [DOI] [Google Scholar]
- 13.Niu Y., Zhao T., Xu X., Li J. Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum) PeerJ. 2017;8:e3955. doi: 10.7717/peerj.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castelán-Muñoz N., Herrera J., Cajero-Sánchez W., Arrizubieta M., Trejo C., García-Ponce B., Sánchez M., Álvarez-Buylla E.R., Garay-Arroyo A. MADS-Box Genes Are Key Components of Genetic Regulatory Networks Involved in Abiotic Stress and Plastic Developmental Responses in Plants. Front. Plant Sci. 2019;10:853. doi: 10.3389/fpls.2019.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J., Wang J., Pan C., Guan X., Wang Y., Liu S., He Y., Chen J., Chen L., Lu G. Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS ONE. 2014;18:e103032. doi: 10.1371/journal.pone.0103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong L., Schumaker K.S., Zhu J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan D., Li R., Zou B., Zhang X., Cong J., Wang R., Xia Y., Li G. Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought resistance in Arabidopsis. Plant Cell Rep. 2012;31:1269–1281. doi: 10.1007/s00299-012-1247-7. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Prat E., Narashimhan M.L., Binzel M.L., Botella M.A., Chen Z., Valpuesta V., Bressan R.A., Hasegawa P.M. Induction of aputative Ca2+-ATPase mRNA in NaCl adapted cells. Plant Physiol. 1992;100:1471–1478. doi: 10.1104/pp.100.3.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wimmers L.E., Ewing N.N., Bennett A.B. Higher plant Ca2+ATPase: Primary structure and regulation of mRNA abundance by salt. Proc. Natl. Acad. Sci. USA. 1992;89:9205–9209. doi: 10.1073/pnas.89.19.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huda K.M., Banu M.S., Garg B., Tula S., Tuteja R., Tuteja N. OsACA6, a P-type IIB Ca²⁺ ATPase promotes salinity and drought stress resistance in tobacco by ROS scavenging and enhancing the expression of stress-responsive genes. Plant J. 2013;76:997–1015. doi: 10.1111/tpj.12352. [DOI] [PubMed] [Google Scholar]
- 21.Rudrappa T., Czymmek K.J., Paré P.W., Bais H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rekha K., Baskar B., Srinath S., Usha B. Plant-growth-promoting rhizobacteria Bacillus subtilis RR4 isolated from rice rhizosphere induces malic acid biosynthesis in rice roots. Can. J. Microbiol. 2018;64:20–27. doi: 10.1139/cjm-2017-0409. [DOI] [PubMed] [Google Scholar]
- 23.Tenhaken R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015;7:771. doi: 10.3389/fpls.2014.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Gall H., Philippe F., Domon J.M., Gillet F., Pelloux J., Rayon C. Cell Wall Metabolism in Response to Abiotic Stress. Plants. 2015;16:112. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakshmanan V., Castaneda R., Rudrappa T., Bais H.P. Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta. 2013;238:657–668. doi: 10.1007/s00425-013-1920-2. [DOI] [PubMed] [Google Scholar]
- 26.Piro G., Leucci M.R., Waldron K., Dalessandro G. Exposure to water stress causes changes in the biosynthesis of cell wall polysaccharides in roots of wheat cultivars varying in drought resistance. Plant Sci. 2003;165:559–569. doi: 10.1016/S0168-9452(03)00215-2. [DOI] [Google Scholar]
- 27.Lairson L.L., Henrissat B., Davies G.J., Withers S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 28.Ding X., Li J., Pan Y., Zhang Y., Ni L., Wang Y., Zhang X. Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int. J. Mol. Sci. 2018;19:1583. doi: 10.3390/ijms19061583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takenaka Y., Kato K., Ogawa-Ohnishi M., Tsuruhama K., Kajiura H., Yagyu K., Takeda A., Takeda Y., Kunieda T., Hara-Nishimura I., et al. Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat. Plants. 2018;4:669–676. doi: 10.1038/s41477-018-0217-7. [DOI] [PubMed] [Google Scholar]
- 30.Lee C., Teng Q., Zhong R., Yuan Y., Haghighat M., Ye Z.H. Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-o-methylation of glucuronic acid on xylan. Plant Cell Physiol. 2012;53:1934–1949. doi: 10.1093/pcp/pcs138. [DOI] [PubMed] [Google Scholar]
- 31.Beauregard P.B., Chai Y., Vlamakis H., Losick R., Kolter R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl Acad. Sci. USA. 2013;110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das P.K., Biswas R., Anjum N., Das A.K., Maiti M.K. Rice matrix metalloproteinase OsMMP1 plays pleiotropic roles in plant development and symplastic-apoplastic transport by modulating cellulose and callose depositions. Sci. Rep. 2018;8:2783. doi: 10.1038/s41598-018-20070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vílchez J.I., García-Fontana C., Román-Naranjo D., González-López J., Manzanera M. Plant drought resistance enhancement by trehalose production of desiccation-tolerant microorganisms. Front. Microbiol. 2016;7:1577. doi: 10.3389/fmicb.2016.01577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iordachescu M., Imai R. Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 2008;50:1223–1229. doi: 10.1111/j.1744-7909.2008.00736.x. [DOI] [PubMed] [Google Scholar]
- 35.Jain N.K., Roy I. Effect of trehalose on protein structure. Protein Sci. 2009;18:24–36. doi: 10.1002/pro.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H., Liu G.L., Chi Z., Hu Z., Chi Z.M. Genetics of trehalose biosynthesis in desert-derived Aureobasidium melanogenum and role of trehalose in the adaptation of the yeast to extreme environments. Curr. Genet. 2018;64:479–491. doi: 10.1007/s00294-017-0762-z. [DOI] [PubMed] [Google Scholar]
- 37.Chenu C., Roberson E.B. Diffusion of glucose in microbial extracellular polysaccharide as affected by water potential. Soil Biol. Biochem. 1996;28:877–884. doi: 10.1016/0038-0717(96)00070-3. [DOI] [Google Scholar]
- 38.Sandhya V., Ali S.Z. The production of exopolysaccharide by Pseudomonas putida GAP-P45 under various abiotic stress conditions and its role in soil aggregation. Microbiology. 2015;84:512–519. doi: 10.1134/S0026261715040153. [DOI] [Google Scholar]
- 39.Vardharajula V., Ali S.Z., Grover M., Reddy G., Bandi V. Drought-tolerant plant growth promoting Bacillus spp.: Effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact. 2011;6:1–14. doi: 10.1080/17429145.2010.535178. [DOI] [Google Scholar]
- 40.ElSayed A.I., Rafudeen M.S., Golldack D. Physiological aspects of raffinose family oligosaccharides in plants: Protection against abiotic stress. Plant Biol. 2014;16:1–8. doi: 10.1111/plb.12053. [DOI] [PubMed] [Google Scholar]
- 41.Alcázar R., Marco F., Cuevas J.C., Patron M., Ferrando A., Carrasco P., Tiburcio A.F., Altabella T. Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 2006;28:1867–1876. doi: 10.1007/s10529-006-9179-3. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi T., Kakehi J.I. Polyamines: Ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 2010;105:1–6. doi: 10.1093/aob/mcp259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi H., Chan Z. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J. Integr. Plant Biol. 2014;56:114–121. doi: 10.1111/jipb.12128. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh D., Sen S., Mohapatra S. Modulation of proline metabolic gene expression in Arabidopsis thaliana under water-stressed conditions by a drought-mitigating Pseudomonas putida strain. Ann. Microbiol. 2017;67:655–668. doi: 10.1007/s13213-017-1294-y. [DOI] [Google Scholar]
- 45.Sen S., Ghosh D., Mohapatra S. Modulation of polyamine biosynthesis in Arabidopsis thaliana by a drought mitigating Pseudomonas putida strain. Plant Physiol. Biochem. 2018;129:180–188. doi: 10.1016/j.plaphy.2018.05.034. [DOI] [PubMed] [Google Scholar]
- 46.Rastogi R., Dulson J., Rothstein S.J. Cloning of tomato (Lycopersicon esculenfum Mill.) Arginine decarboxylase gene and its expression during fruit ripening. Plant Physiol. 1993;103:829–834. doi: 10.1104/pp.103.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urano K., Yoshiba Y., Nanjo T., Igarashi Y., Seki M., Sekiguchi F., Yamaguchi-Shinozaki K., Shinozaki K. Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant. Cell Environ. 2003;26:1917–1926. doi: 10.1046/j.1365-3040.2003.01108.x. [DOI] [Google Scholar]
- 48.Kakkar R.K., Sawhney V.K. Polyamine research in plants—A changing perspective. Physiol. Plant. 2002;116:281–288. doi: 10.1034/j.1399-3054.2002.1160302.x. [DOI] [Google Scholar]
- 49.Sinha R., Rajam M.V. RNAi silencing of three homologues of S-adenosylmethionine decarboxylase gene in tapetal tissue of tomato results in male sterility. Plant Mol. Biol. 2013;82:169–180. doi: 10.1007/s11103-013-0051-2. [DOI] [PubMed] [Google Scholar]
- 50.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeong J.S., Kim Y.S., Redillas M.C., Jang G., Jung H., Bang S.W., Choi Y.D., Ha S.H., Reuzeau C., Kim J.K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought resistance and increased grain yield in the field. Plant Biotechnol. J. 2013;11:101–114. doi: 10.1111/pbi.12011. [DOI] [PubMed] [Google Scholar]
- 52.Kan C.C., Chung T.Y., Juo Y.A., Hsieh M.H. Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genom. 2015;16:731. doi: 10.1186/s12864-015-1892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vílchez J.I., Niehaus K., Dowling D.N., González-López J., Manzanera M. Protection of pepper plants from drought by Microbacterium sp. 3J1 by modulation of the plant’s glutamine and a-ketoglutarate content: A comparative metabolomics approach. Front. Microbiol. 2018;9:284. doi: 10.3389/fmicb.2018.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Igamberdiev A.U., Kleczkowski L.A. The glycerate and phosphorylated pathways of serine synthesis in plants: The branches of plant glycolysis linking carbon and nitrogen metabolism. Front. Plant Sci. 2018;9:318. doi: 10.3389/fpls.2018.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajaeian S., Ehsanpour A.A., Javadi M., Shojaee B. Ethanolamine induced modification in glycine betaine and proline metabolism in Nicotiana rustica under salt stress. Biol. Plant. 2017;61:797–800. doi: 10.1007/s10535-017-0704-0. [DOI] [Google Scholar]
- 56.Bhuiyan N.H., Hamada A., Yamada N., Rai V., Hibino T., Takabe T. Regulation of betaine synthesis by precursor supply and choline monooxygenase expression in Amaranthus tricolor. J. Exp. Bot. 2007;58:4203–4212. doi: 10.1093/jxb/erm278. [DOI] [PubMed] [Google Scholar]
- 57.Streeter J.G., Lohnes D.G., Fioritto R.J. Patterns of pinitol accumulation in soybean plants and relationships to drought resistance. Plant Cell Environ. 2001;24:429–438. doi: 10.1046/j.1365-3040.2001.00690.x. [DOI] [Google Scholar]
- 58.Ahn C.H., Hossain M.D., Lee E., Kanth B.K., Park P.B. Increased salt and drought resistance by D-pinitol production in transgenic Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018;504:315–320. doi: 10.1016/j.bbrc.2018.08.183. [DOI] [PubMed] [Google Scholar]
- 59.Gargallo-Garriga A., Preece C., Sardans J., Oravec M., Urban O., Peñuelas J. Root exudate metabolomes change under drought and show limited capacity for recovery. Sci. Rep. 2018;8:12696. doi: 10.1038/s41598-018-30150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vílchez J.I., Yang Y., Yi D., Zhang H. Measurements of root colonized bacteria species. Bio-Protoc. 2021;11:e3976. doi: 10.21769/BioProtoc.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox M.P., Peterson D.A., Biggs P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 63.Trapnell C., Pachter L., Salzberg S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or Supplementary Material.