Abstract
Although composting is effective in deactivating antibiotic substances in manure, the influence of compost fertilization on the occurrence and dissemination of antibiotic resistance in arable soils remains to be controversial. Herein, the abundance and diversity of two sulfonamide resistance genes (sul1 and sul2) in soil fertilized by compost spiked with two concentrations of sulfadiazine (1 and 10 mg kg−1) were studied intensively by qPCR and high throughput sequencing based on a two-month microcosm experiment. The concentration of sulfadiazine decreased rapidly after spiking from 25% at Day 1 to less than 2.7% at Day 60. Relative abundance of both sul1 and sul2 were significantly higher in soil amended with compost than the non-amended control at Day 1 and slightly decreased with incubation time except for sul2 in the S10 treatment. Soil bacterial communities were transiently shifted by compost fertilization regardless of the presence of sulfadiazine. Relative abundance of genera in three hubs positively interlinked with sul1 and sul2 were significantly higher in compost treated soil than the control at Day 1, 7 and 21, but not at Day 60. High throughput sequencing analyses revealed that most detected (>67% in relative abundance) sul1 and sul2 genotypes sharing >99% similarity with those found in gammaproteobacterial pathogens frequently were commonly present in compost and soil. These results indicated that compost fertilization might increase the abundance rather than diversity of sulfadiazine-resistant populations in soil, which may be facilitated by the presence of sulfadiazine.
Keywords: compost, sulfadiazine resistance, high throughput sequencing, sul1, sul2
1. Introduction
A large number of antibiotics are being used as precaution and therapy in industrial high-density animal farms worldwide. In China, the consumption of different antibiotics reached approximately 162,000 tons in 2013 [1]. Sulfonamide, fluoroquinolones, macrolides, β-lactams and tetracycline were the most used in livestock [1,2]. Many antibiotics such as sulfonamides cannot be absorbed by animals and are largely excreted via urine or feces. They are also not or only to a low level degraded during manure storage and may pose a selective pressure on antibiotic resistant bacteria. Ten to ten thousand folds’ elevation of antibiotic-resistance in manure has been reported [3]. Application of manure containing antibiotic-resistant bacteria, residuals antibiotics and potential pathogenic bacteria may facilitate the spread of antibiotic resistance genes (ARGs) into human or animal pathogens, which pose a huge threat to public health [4,5,6,7,8]. A recent study also found that tetracycline and sulfamerazine introduced via manure into soil might be accumulated in Zea mays L. [9]. The presence of antibiotics in manure also induced changes in soil microbial communities and shifted taxa known as human pathogens [10,11]. Several treatments such as elongation of storage, acidification [12], mesophilic digestion [13] and composting [14] have been evaluated for their effects on the mitigation of ARGs or mobile genetic elements in manure. Among these methods, thermophilic aerobic fermentation and thermophilic anaerobic digestion tend to be more effective in deactivating several antibiotics (such as oxytetracycline, macrolide and fluoroquinolone) and ARGs than the corresponding mesophilic treatments [14,15]. However, persistence or elevation of ARGs abundance was still observed in arable soils fertilized by manure or composts [3,16,17]. Relative abundance of two tetracycline resistance genes (tetM and tetK) largely flocculated over growing seasons in a two-year field experiment [18]. Arable soil was regarded as a receptor of manure, and its ARGs, antibiotics and antibiotic residuals served as a reservoir of drug-resistant bacteria, which could be transferred through food chains and other environmental routes [19,20]. Recently, manure-borne microorganisms were suggested to be contributed at a large extent to the elevation of ARGs in manured soil [21]. Several other studies also demonstrated that the majority of microorganisms inhabiting organic fertilizer may not fit for soil environments [22,23], suggesting that the remaining antibiotics in organic fertilizer may be the cause of elevated antibiotic resistance in arable soils.
In the present study, a microcosm experiment with four treatments was conducted to study the effects of compost application and sulfadiazine on soil microbiome and sulfonamides resistant populations. Four treatments included soil (CK), soil amended with compost (S0) and soil amended with compost containing 1 (S1) or 10 (S10) mg kg−1 of sulfadiazine. Dynamic of sulfonamides-resistant populations and total soil microbial population were evaluated by real-time quantitative PCR and Illumina sequencing of sul1, sul2 and 16S rRNA amplicons. This study might provide an in-depth understanding of the shift of sulfadiazine resistome in soil under antibiotic stress or fertilized by compost.
2. Materials and Methods
2.1. Microcosm Experiment
The silt loam soil from a long-term greenhouse experiment was used for the microcosm experiment. All soil was immediately passed through a 2 mm mesh sieve to remove plant debris and stones immediately after sampling in Dec 2014. Then it was stored in an incubator (30 °C) for 10 days for equilibration before the experiment. Four treatments were prepared as follows: 1-kilogram soil (dry weight) was amended with 140 g of compost (S0) or the same amount of compost pre-mixed with sulfadiazine, and soil without compost and sulfadiazine amendment (CK) served as a control. The seeding compost was prepared as follows: 270 g of fresh compost was sprayed with sulfadiazine solutions (50 mL with concentrations of 0, 0.5 and 5 g L−1, respectively) in a closed container, which was shaken vigorously to reach a final concentration of 1 (S1) and 10 (S10) mg kg−1. The amount of compost used was similar to that in the field for spring vegetables in 2014. Five independent replicates for each treatment at each sampling time were included. After mixing, 45 g of soil were redistributed in a 50 mL conical tube, and all tubes were randomly placed and incubated in the dark at 30 °C for 60 days. The moisture of all soil was adjusted to 40%, which was comparable with field conditions by adding sterilized deionized water during the experiment period. A total of 80 soil samples were taken on Day 1, 7, 21 and 60 (5 replicates per treatment × 4 treatments per sampling × 4 samplings).
2.2. Sulfadiazine Quantitation
Concentrations of sulfadiazine in soil and compost were analyzed according to the previous method [24]. Fresh soil and compost samples were lyophilized by a vacuum freeze drier, then sieved through a 2-mm mesh and stored in the dark at room temperature until analysis. Samples were extracted by adding multiple extracting agents (e.g., EDTA-Mcllvaine buffer, methanol and acetone) and ultrasound-assisted. The standard solutions of sulfadiazine were made using a 10-fold series dilution of stock solution (10 mg mL−1) for five gradients and stored at 4 °C. The analysis was performed using an Agilent HPLC-MS system (Waters, Milford, MA, USA). Peak areas of sulfadiazine for each sample and standard solutions were recorded for further calculations.
2.3. TC-DNA Extraction, Real Time qPCR of sul1, sul2 and 16S rRNA Gene
Total community DNA was extracted from 0.5 g of soil or compost using FastDNA spin Kit for soil (MP, Biomedicals, Santa Ana, Carlsbad, CA, USA). Quantification of 16S rRNA genes and sulfadiazine resistance genes sul1 and sul2 were performed according to previous studies [25,26,27]. For sul1 and sul2, a 50 μL reaction volume contained 2.5U Taq DNA polymerase and 25 μL buffer; both were made by TaKaRa (Bao-TaKaRa company, Dalian, China), 0.5 μL of each primer (10 μM), 0.5 μL probe (10 μM), 2.5 μL BSA (0.5%) and 5 μL template. Thermocycles for sul1 were 5 min at 94 °C and 40 cycles consisting of 15 s at 95 °C, 1 min at 60 °C, while for sul2 were 5 min at 94 °C and 40 cycles consisting of 15 s at 95 °C, 15 s at 53 °C, 1 min at 60 °C. The sequence of all primers and Taqman probes of sul1 and sul2 were given in Table 1. Real-time qPCR reactions were performed in an iQ-5 real-time PCR detection system (Bio-Rad, Hercules, California, USA). R2 values were more than 0.99, and the amplification efficiencies ranged between 84% and 98%. Gene copy numbers of sul1 and sul2 were adjusted to 16S rRNA for further analysis. One-way ANOVA in conjunction with Tukey’s honest significant difference (HSD) test (p < 0.05) was used to compare different treatment and sampling times.
Table 1.
Probes and primers used for the real-time qPCR.
| Genes | Sequences of Probes | Sequences of Forward Primer (5′-3′) |
Sequences of Reverse Primer (5′-3′) |
References |
|---|---|---|---|---|
| 16S rRNA | CTTGTACACACCGCCCGTC | CGGTGAATACGTTCYCGG | GGWTACCTTGTTACGACTT | [25] |
| sul1 | CAGCGAGCCTTGCGGCGG | CCGTTGGCCTTCCTGTAAAG | TTGCCGATCGCGTGAAGT | [26] |
| sul2 | CGGTGCTTCTGTCTGTTTCGCGC | CGGCTGCGCTTCGATT | CGCGCGCAGAAAGGATT | [27] |
2.4. High Throughput Sequencing Analysis of Bacterial sul1, sul2 and 16S rRNA Gene Amplicon
Fragments of sul1 and sul2 were amplified with barcode-fused primers used in the qPCR analyses. Due to the length of sul1 or sul2 amplicon being shorter than 250 bp (the read length of illumine sequencing), an in-silicon PCR was performed to extract the proper fragment from each read. Then, the reads were assigned to each sample based on barcode sequences, and the primer regions were trimmed. A standalone BLASTP analysis was used to identify the translation frames, and only those sequences where the deduced ammonia acid sequence has no stop codon were included for further analysis. The sul1 or sul2 sequences were assigned to different genotypes based on the deduced ammonia acid sequence using usearch software [28]. The amplification, purification, sequencing and analysis of the 16S rRNA gene were performed according to previous descriptions [29,30,31,32,33,34,35]. All sequences have been submitted to GenBank (SRP126466).
Beta diversity of microbial community was compared using or PCoA based on the Bray–Curtis distance. Chao1 was calculated by 100 times of re-sampling an equal number of sequences from each sample using R-add-on vegan packages to attenuate the biases caused by different read numbers [34]. The relative abundance of bacterial taxa, sul1 and sul2 genotype were calculated by dividing the read number for each taxon or genotype with the total read number for each sample. Co-occurrence network analysis was performed based on the Spearman correlation (cor > 0.6, p < 0.001). The network was analyzed with the software, Gephi [36]. Microbial hubs that were significantly different on the relative abundance were identified by a generalized linear model for binominal data using the R add-on package “multicomp” [37]. All statistical analyses and plots were performed with the software R 3.1.2 (http://www.r-project.org/, downloaded in 2020), and these tools mentioned above have been implemented into a galaxy instance (www.freebioinfo.org, processed data from January to March of 2021).
3. Results
3.1. Concentration of Sulfadiazine
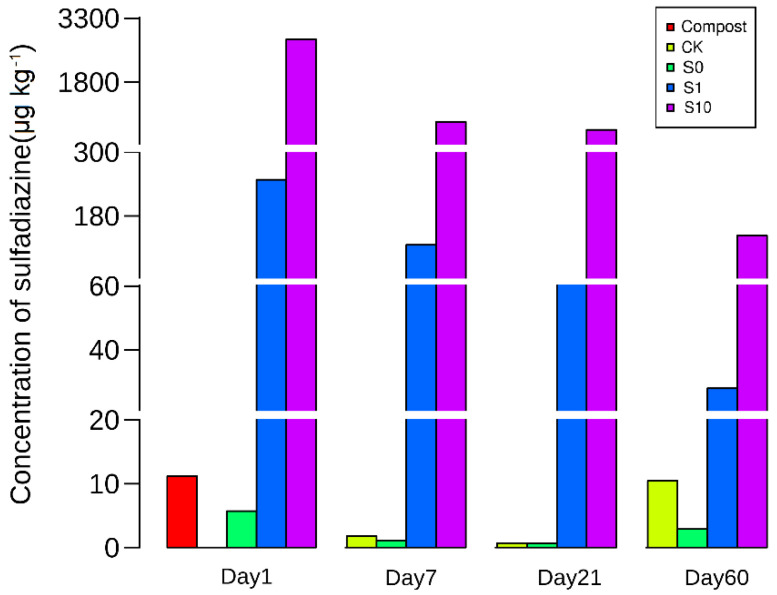
A small amount of sulfadiazine was also detected from soil (1.8 μg kg−1) and compost (11.2 μg kg−1) (Figure 1). The concentration of sulfadiazine decreased rapidly after spiking (Figure 1). On Day 1, the concentrations of sulfadiazine were only 251 and 2596 µg kg−1 in S1 and S10 soils, respectively, accounting for ca 25% of the concentration spiked (Figure 1). The concentrations rapidly decreased to 121 and 782 µg kg−1 at Day7 and 51 and 607 µg kg−1 at Day21 (Figure 1). The concentrations of sulfadiazine were only 27.1 and 140.2 µg kg−1 in S1 and S10 soils at Day 60, respectively, accounting for 2.7% and 1.4% of the concentration spiked (Figure 1).
Figure 1.
The concentration of sulfadiazine in compost and soils in different treatments. CK: soil; S0: soil amended with compost; S1: soil amended with compost and 1 mg kg−1 of sulfadiazine; S10: soil amended with compost and 10 mg kg−1 of sulfadiazine.
3.2. Abundance of Bacterial sul1 and sul2 Genes
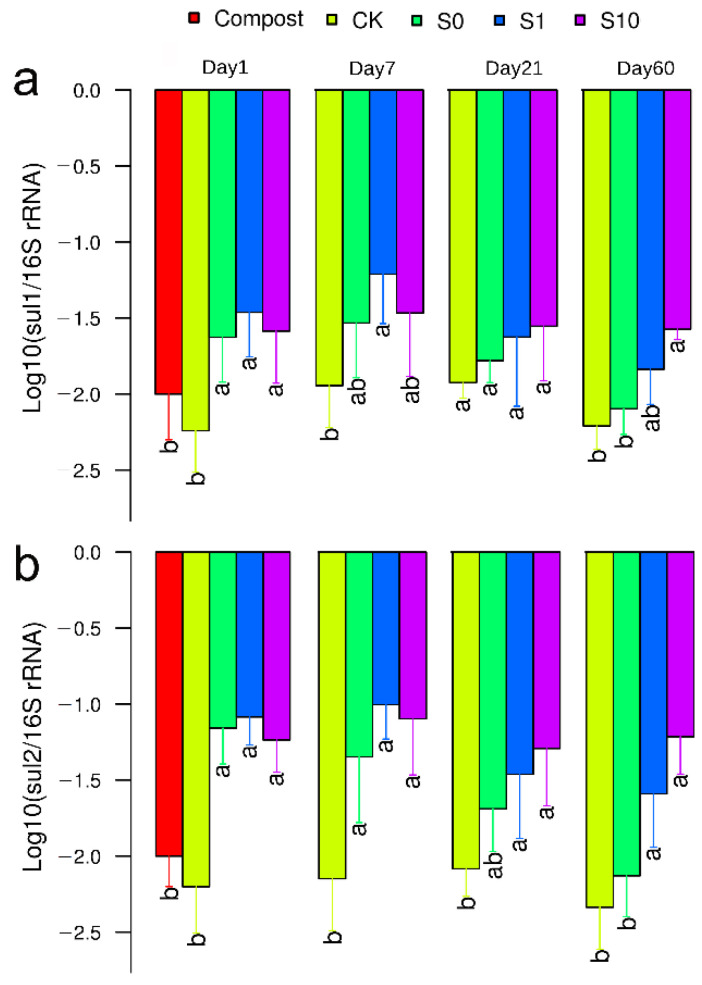
The copy numbers of 16S rRNA genes in different soils were comparable among all treatments over two months (Supplementary Materials, Figure S1). The relative abundance of sul2 was significantly higher in those soils amended with compost than the non-amended control at Day 1 and 7 (Figure 2b). Interestingly, a decrease in sul2 with incubation time was only observed for S0 and S1 but not for S10 (Figure 2b), suggesting that a higher concentration of sulfadiazine in soil may facilitate the persistence of resistant bacteria in soils. The relative abundance of the sul1 gene tended to be lower in soils amended with compost than the non-amended control (Figure 2a). Again, a slight decrease in sul1 with the incubation time was also observed for S0 and S1, but not for S10 (Figure 2a).
Figure 2.
The relative abundance of sul1 (a) and sul2 (b) in different treatments. CK: soil; S0: soil amended with compost; S1: soil amended with compost and 1 mg kg−1 of sulfadiazine; S10: soil amended with compost and 10 mg kg−1 of sulfadiazine. The different letters above the columns in the same sampling indicate significant differences (p < 0.05) between treatments.
3.3. Bacterial Community Composition
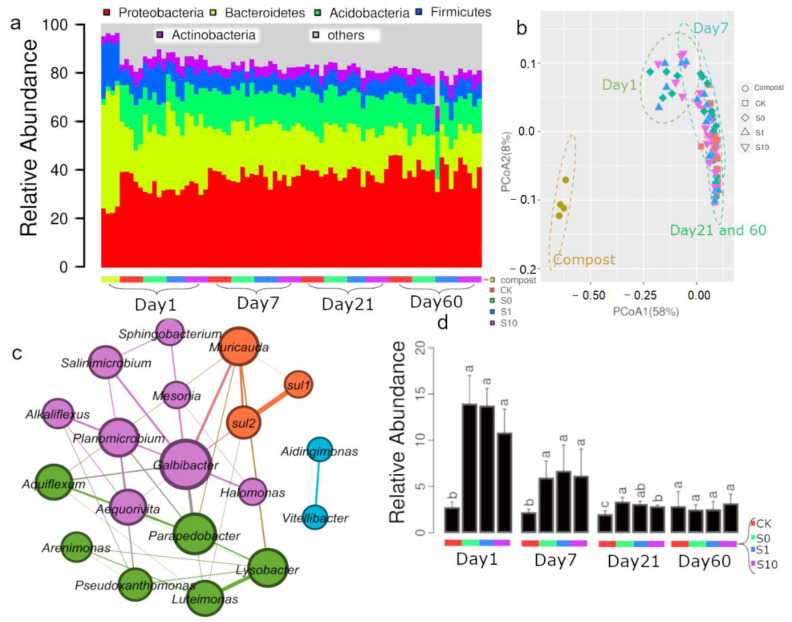
In contrast to sul1 or sul2, bacterial communities were dramatically different between compost and soil. Bacteroidetes (52.4%), Proteobacteria (21.5%), Firmicutes (17.7%), Actinobacteria (3.0%) and Acidobacteria (2.2%) were most detected phyla from compost (Figure 3a). Relative abundance of Bacteroidetes and Firmicutes was significantly higher in compost than soil, in contrast to Proteobacteria, Acidobacteria, Chloreflexi, Planctomycetes and Nitrospirae (Figure 3a). Interestingly, the relative abundance of Firmicutes in soils fertilized by compost decreased rapidly with incubation time and was comparable to the non-fertilized soil at Day 7 (Figure 3a). While the relative abundances of Bacteroidetes were comparable between compost fertilized and non-fertilized soil at Day 60 (Figure 3a). Principal coordinate analysis (PCoA) confirmed that the bacterial community was largely shaped by compost fertilization and incubation time (Figure 3b). While the similarity in community composition between compost fertilized and non-fertilized soil increased with incubation time (Figure 3b), suggesting that soil bacterial communities were resilient to the perturbation by compost fertilization.
Figure 3.
The relative abundance of dominant compost bacteria (a), PCoA (principal coordinate analysis) of bacterial community (b), the co-occurrence network between sul1, sul2 and bacterial taxa (c) and the relative abundance of the three interlinked hubs in different treatments (d). CK: soil; S0: soil amended with compost; S1: soil amended with compost and 1 mg kg−1 of sulfadiazine; S10: soil amended with compost and 10 mg kg−1 of sulfadiazine.
Co-occurrence network analysis was applied to study the correlation between sul1 or sul2 and bacterial taxa. The majority of these positively correlated genera were affiliated with Bacteroidetes (Aequorivita, Alkaliflexus, Aquiflexum, Galbibacter, Mesonia, Muricauda, Parapedobacter, Salinimicrobium, Sphingobacterium and Vitellibacter) and Proteobacteria (Aidingimonas, Arenimonas, Halomonas, Luteimonas, Lysobacter and Pseudoxanthomonas) (Figure 3c). Sul1, sul2 and these positively correlated genera formed four hubs, and three hubs were positively interlinked (Figure 3c). Both sul1 and sul2 were positively correlated with the genera Muricauda (Figure 3c). Additionally, Galbibacter and Lysobacter were also significantly correlated with sul2 (Figure 3c). Relative abundances of three interlinked hubs were significantly higher in compost treated soil than the control at Day 1, 7 and 21 (Figure 3d).
3.4. Diversity of sul1 and sul2 Genes
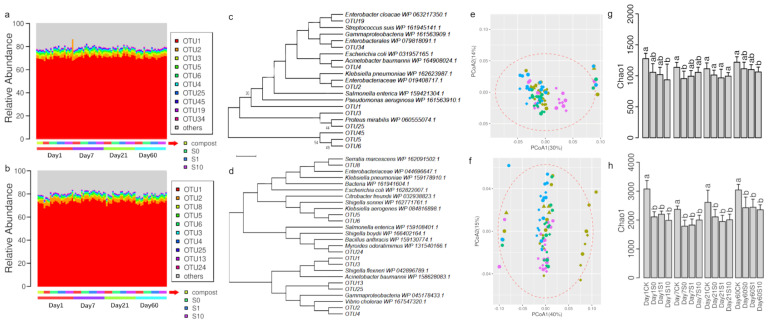
Both sul1 and sul2 gene fragments were subjected to Illumina Hiseq 2500 analyses. Totally 6,475,646 and 5,236,210 sequences were acquired for sul1 and sul2, respectively. The detected diversities of sul1 and sul2 genes were extremely high with 64,148 and 59,461 unique putative amino acid sequences, respectively. Interestingly, the most detected genotype accounted for 67.8–74.5% for sul1 and 68.0–77.3% for sul2 (Figure 4a,b). BLASTP analysis revealed that the deduced amino acid sequence of most detected sul1 (OTU1) shared >99% similarity with those genes encoded within genomes of Salmonella enterica, Klebsiella pneumonia or Escherichia coli (Figure 4c). The most detected sul2 genotype (OTU1) was similar (>99%) to those bacteria carried by Shingella boydii, Acinetobacter Baumannii (Figure 4d). Other most detected genotypes were also similar (>97% similarity) to sul1 or sul2 genes carried by those Gamma proteobacteria (Figure 4c,d). The composition of sul1 or sul2 genes was highly similar between compost or soils (>78% similarity) (data not shown). Alpha-diversities of sul1 tended to be higher in control soil than those compost fertilized soils (Figure 4e). However, the significant differences were only detected between CK and S10 at Day 1 and 60 or between CK and S0 at Day 7 (Figure 4e). The alpha-diversity of sul2 was significantly higher in CK than compost fertilized soil at all samplings (Figure 4f), suggesting slight effects of composting fertilization on alpha-diversity of sul1 and sul2. No effect of sulfadiazine spiking on the alpha-diversity of sul1 and sul2 was detected (Figure 4e,f).
Figure 4.
The relative abundance of most detected genotype for sul1 (a) and sul2 (b) gene, BLASTP analysis of sul1 (c) and sul2 (d) gene, PCoA (principal coordinate analysis) of sul1 (e) and sul2 (f) gene composition, Chao1 index of sul1 (g) and sul2 (h) in different treatments. CK: soil; S0: soil amended with compost; S1: soil amended with compost and 1 mg kg−1 of sulfadiazine; S10: soil amended with compost and 10 mg kg−1 of sulfadiazine.
4. Discussion
Antibiotic resistance genes were widespread in environmental bacteria. Several antibiotic resistance genes were ubiquitous in environments, and some of which were believed to be pristine from antibiotic contamination [38]. For example, a Paenibacillus bacterium isolated from an underground cave that is believed to be isolated from the surface for over 4 Myr is resistant to most clinically used antibiotics [39]. A large-scale survey also revealed that the relative abundance of sulfonamide resistance genes ranged from 10−6 to 10−2 gene copies per 16S rRNA gene copies in the arable soils of China [40]. The functional metagenomic analysis also revealed that diverse sulfonamides resistance genes were also detected from different soil microbial communities, indicating that sulfonamides resistance was ubiquitously present in several soil environments [41]. In vitro studies have long demonstrated that the spreading of antibiotic resistance among bacteria could be strengthened under the selection pressure of antibiotics [42,43]. However, bacteria carrying antibiotic resistance genes may need more energy to replicate their ARGs during reproduction, which might be a disadvantage if there were no selective pressure from antibiotics [44]. Thus, the fate of antibiotic resistance genes in environmental samples remains to be elusive.
4.1. Diversities of sul1 or sul2 Were Extremely High but Only Few Common Dominant Genotypes were Prevalent in Soil or Compost
Herein, we employed Illumina sequencing to analyze PCR amplicons of sul1 and sul2 genes, and the acquired diversities of both sul1 and sul2 genes were extremely high in soil and compost samples, suggesting that both soil and compost are reservoir rich in diverse sulfonamide resistance genes. These findings indicated that resistance to antibiotics might evolve rapidly in Bacteria [45], which may exchange with one another antibiotic resistance gene by horizontal gene transferring mechanisms [46,47] or mutate its own genes to become resistant [48]. Several environmental stressors such as starvation, antimicrobials may drive the evolution of antibiotic resistance [49,50,51] or contribute to their maintenance in environments [14,47]. Although sequencing errors may cause artificial diversity [52], we analyzed the dataset in a stringent manner by checking primer region, translation frame and deduced amino acids. In contrast, it is still possible that novel sul1 or sul2 genes may not be undetected due to that the spectrum of genes that can be amplified has been defined by the primer sequences. Recently, researchers detected novel sulfonamide resistance genes which shared relatively low similarity with known entities in reference database via metagenomics analysis, highlighting a requirement of extensive study on environmental resistomes [41].
Despite the immense diversities of sul1 or sul2, the community compositions were highly similar between compost and soil, of which distinct bacterial communities were detected. It is likely due to that the most detected genotypes of sul1 or sul2 (accounting for more than 67%) were commonly present in both compost and soil, and their proportions were not affected by compost fertilization, sulfadiazine spiking or incubation. These genotypes were highly similar to those hosted by species such as Salmonella enterica, Klebsiella pneumonia, Escherichia coli Shingella boydii and Acinetobacter baumannii, which were known as pathogenic bacteria [53,54,55,56]. It is also worth noting that all these species except for Klebsiella pneumonia (relative abundance <0.03%) were not detected from both soil and compost by the 16S rRNA gene analysis. Since both sul1 and sul2 genes were reported to be present on plasmids [57,58], which could spread into indigenous soil microorganisms [46,47]. In consideration of the relative abundance of sul1 and sul2 by qPCR, these data suggested the dominant sul1 or sul2 genotypes were possibly present in a wide spectrum of different taxonomic groups. However, the spectrum of their hosts and which mechanisms drive the dominance of these genotypes in different environmental bacteria remains to be explored.
4.2. Compost Fertilization Elevated the Abundance of sul1 and sul2 in Soils, and the Co-Introduced Sulfadiazine may Facilitate the Persistence of Such Resistance
Quantitative PCR analysis revealed that sul1 and sul2 genes were significantly higher in the compost treated soils than the control shortly after the fertilization. This result indicated that compost amendment possibly stimulated the growth of bacteria carrying sul1 or sul2 genes. In general, these findings agree with other studies that ARGs in soil were frequently elevated after amendment with an organic fertilizer such as manure or compost [3,18,21,46]. That transient enrichments of sulfonamide or other antibiotic resistance with different manure applications without known selective pressure were also observed in other studies [21,59]. However, repeated application of manure containing antibiotics could cause an increase in antibiotic resistance in soil [46]. Herein, the decrease in sul1 or sul2 with incubation was slower in S10 than in other treatments. Thus, the presence of sulfadiazine may facilitate the persistence of sul1 or sul2 in soils. In the other aspect, sulfadiazine might be not a long-living persistent selective pressure for sul1 or sul2 as its concentration decreased dramatically with incubation time (Figure 1). Despite sulfonamide antibiotics were adsorbed by soil or compost [60,61], the sorption coefficients were very low, indicating that these substances were highly mobile [62].
4.3. Resilience of Soil Bacterial Community to the Perturbation of Compost
Similar to other organic fertilizers, compost application not only introduced a complex of nutrients or carbons but also exogenous microbial communities into the soil. The bacterial compositions in compost dramatically differed from those in soil. Those soil bacterial communities shifted by compost rapidly become to be similar to the control, indicating that bacteria in compost may diminish after entering into the soil (Figure 4a). A previous study demonstrated that indigenous soil microorganisms inhibited the invasion and establishment of exogenous soil microorganisms from manure [21]. Co-occurrence network analysis also suggested that both sul1 or sul2 genes were positively correlated with several genera, which were also enriched after compost fertilization (Figure 3c). Taken together, these results suggested that compost fertilization may trigger the growth of indigenous soil microorganisms carrying sulfonamide resistance.
5. Conclusions
Compost fertilization triggered a transient increase in sulfonamides resistant bacteria in soil, and the presence of sulfadiazine might facilitate the persistence of resistance populations. These findings highlight the importance of deactivating antibiotics or other selective pressure on mitigating ARGs spreading in agricultural systems. The dominant genotype of sul1 and sul2 might be widely distributed in different bacteria inhabiting in soil and compost and have evolved into huge genetic diversity.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/10/6/699/s1, Figure S1: The abundance of bacteria in different treatments.
Author Contributions
Conceptualization, G.D. and J.L.; methodology, G.D.; software, G.D.; validation, H.H., M.B., Y.C., Y.G., H.Y. and M.W.; formal Analysis, G.D.; data curation, G.D.; writing—original draft preparation, G.D.; writing—review and editing, H.H., T.X., Q.C., Y.W. and J.L.; visualization, G.D.; supervision, G.D. and J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program (grant numbers: 2019YFD1002000, 2016YFD0800602, 2016YFD0800601); the National Nature Science Foundation of China [grant numbers:32071552].
Data Availability Statement
The data presented in this study are available in GenBank (SRP126466) or freebioinfo.org.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang Q.Q., Ying G.G., Pan C.G., Liu Y.S., Zhao J.L. Comprehensive evaluation of antibiotics emission and fate in the river basins of china: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 2015;49:6772–6782. doi: 10.1021/acs.est.5b00729. [DOI] [PubMed] [Google Scholar]
- 2.Yao L.H., Li Y., Li Z.Q., Shen D.S., Feng H.J., Zhou H.H., Wang M.Z. Prevalence of fluoroquinolone, macrolide and sulfonamide-related resistance genes in landfills from East China, mainly driven by MGEs. Ecotoxicol. Environ. Saf. 2019;190:110131. doi: 10.1016/j.ecoenv.2019.110131. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y.G., Johnson T.A., Su J.Q., Qiao M., Guo G.X., Stedtfeld R.D., Hashsham S.A., Tiedje J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsberg K.J., Reyes A., Wang B., Selleck E.M., Sommer M.O.A., Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y.W., Cheng D.M., Xue J.M., Weaver L., Wakelin S.A., Feng Y., Li Z.J. Changes in microbial community structure during pig manure composting and its relationship to the fate of antibiotics and antibiotic resistance genes. J. Hazard. Mater. 2020;389:122082. doi: 10.1016/j.jhazmat.2020.122082. [DOI] [PubMed] [Google Scholar]
- 6.Pruden A., Pei R.T., Storteboom H., Carlson K. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006;40:7445–7450. doi: 10.1021/es060413l. [DOI] [PubMed] [Google Scholar]
- 7.Wichmann F., Udikovickolic N., Andrew S.M., Handelsman J. Diverse antibiotic resistance genes in dairy cow manure. mBio. 2014;5:e01017-13. doi: 10.1128/mBio.01017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y.G., Gillings M., Simonet P., Stekel D., Banwart S., Penuelas J. Microbial mass movements. Science. 2017;357:1099–1100. doi: 10.1126/science.aao3007. [DOI] [PubMed] [Google Scholar]
- 9.Mullen R.A., Hurst J.J., Naas K.M., Sassoubre L.M., Aga D.S. Assessing uptake of antimicrobials by Zea mays L. and prevalence of antimicrobial resistance genes in manure-fertilized soil. Sci. Total Environ. 2018;646:409–415. doi: 10.1016/j.scitotenv.2018.07.199. [DOI] [PubMed] [Google Scholar]
- 10.Ding G.C., Radl V., Schloterhai B., Jechalke S., Heuer H., Smalla K., Schloter M. Dynamics of soil bacterial communities in response to repeated application of manure containing sulfadiazine. PLoS ONE. 2014;9:e92958. doi: 10.1371/journal.pone.0092958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urra J., Alkorta I., Lanzén A., Mijangos I., Garbisu C. The application of fresh and composted horse and chicken manure affects soil quality, microbial composition and antibiotic resistance. Appl. Soil Ecol. 2019;135:73–84. doi: 10.1016/j.apsoil.2018.11.005. [DOI] [Google Scholar]
- 12.Lin H., Sun W.C., Yu Q.G., Ma J.W. Acidic conditions enhance the removal of sulfonamide antibiotics and antibiotic resistance determinants in swine manure. Environ. Pollut. 2020;263:114439. doi: 10.1016/j.envpol.2020.114439. [DOI] [PubMed] [Google Scholar]
- 13.Wolters B., Ding G.C., Kreuzig R., Smalla K. Full-scale mesophilic biogas plants using manure as c-source: Bacterial community shifts along the process cause changes in the abundance of resistance genes and mobile genetic elements. FEMS Microbiol. Ecol. 2016;92:fiv163. doi: 10.1093/femsec/fiv163. [DOI] [PubMed] [Google Scholar]
- 14.Qian X., Sun W., Gu J., Wang X.J., Sun J.J., Yin Y.N., Duan M.L. Variable effects of oxytetracycline on antibiotic resistance gene abundance and the bacterial community during aerobic composting of cow manure. J. Hazard. Mater. 2016;315:61–69. doi: 10.1016/j.jhazmat.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Liu H., Pu C.J., Yu X.L., Sun Y., Chen J.H. Removal of tetracyclines, sulfonamides, and quinolones by industrial-scale composting and anaerobic digestion processes. Environ. Sci. Pollut. Res. 2018;25:35835–35844. doi: 10.1007/s11356-018-1487-3. [DOI] [PubMed] [Google Scholar]
- 16.Lin H., Chapman S.J., Freitag T.E., Kyle C., Ma J.W., Yang Y.Y., Zhang Z.L. Fate of tetracycline and sulfonamide resistance genes in a grassland soil amended with different organic fertilizers. Ecotoxicol. Environ. Saf. 2019;170:39–46. doi: 10.1016/j.ecoenv.2018.11.059. [DOI] [PubMed] [Google Scholar]
- 17.Wang N., Yang X.H., Jiao S.J., Zhang J., Ye B.P., Gao S.X. Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS ONE. 2014;9:e112626. doi: 10.1371/journal.pone.0112626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M., Han H., Zheng X.N., Bai M.H., Xu T., Ding G.C., Li J. Dynamics of oxytetracycline and resistance genes in soil under long-term intensive compost fertilization in Northern China. Environ. Sci. Pollut. Res. 2019;26:21381–21393. doi: 10.1007/s11356-019-05173-3. [DOI] [PubMed] [Google Scholar]
- 19.Guerra B., Junker E., Helmuth R. Incidence of the recently described sulfonamide resistance gene sul3 among german salmonella enterica strains isolated from livestock and food. Antimicrob. Agents Chemother. 2004;48:2712–2715. doi: 10.1128/AAC.48.7.2712-2715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y.J., Hu H.W., Chen Q.L., Singh B.K., Yan H., Chen D., He J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019;130:104912. doi: 10.1016/j.envint.2019.104912. [DOI] [PubMed] [Google Scholar]
- 21.Chen Q.L., An X.L., Li H., Zhu Y.G., Su J.Q., Cui L. Do manure-borne or indigenous soil microorganisms influence the spread of antibiotic resistance genes in manured soil? Soil Biol. Biochem. 2017;114:229–237. doi: 10.1016/j.soilbio.2017.07.022. [DOI] [Google Scholar]
- 22.Tien Y.C., Li B., Zhang T., Scott A., Murray R., Sabourin L., Marti R., Topp E. Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci. Total Environ. 2017;581–582:32–39. doi: 10.1016/j.scitotenv.2016.12.138. [DOI] [PubMed] [Google Scholar]
- 23.Liu W.B., Ling N., Guo J.J., Ruan Y., Wang M., Shen Q.R., Guo S.W. Dynamics of the antibiotic resistome in agricultural soils amended with different sources of animal manures over three consecutive years. J. Hazard. Mater. 2021;401:123399. doi: 10.1016/j.jhazmat.2020.123399. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y., Wei C.J., Zhang W.J., Liu Y.W., Li Z.J., Hu H.Y., Xue J.M., Davis M. A simple and economic method for simultaneous determination of 11 antibiotics in manure by solid-phase extraction and high-performance liquid chromatography. J. Soils Sediments. 2016;16:2242–2251. doi: 10.1007/s11368-016-1414-5. [DOI] [Google Scholar]
- 25.Suzuki M.T., Taylor L.T., DeLong E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 2000;66:4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heuer H., Smalla K. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 2007;9:657–666. doi: 10.1111/j.1462-2920.2006.01185.x. [DOI] [PubMed] [Google Scholar]
- 27.Heuer H., Focks A., Lamshöft M., Smalla K., Matthies M., Spiteller M. Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol. Biochem. 2008;40:1892–1900. doi: 10.1016/j.soilbio.2008.03.014. [DOI] [Google Scholar]
- 28.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 29.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H.X., Ding X.Y., Chen C., Zheng X.N., Han H., Li C.N., Gong J.Y., Xu T., Li Q.X., Ding G.C., et al. Enrichment of phosphate solubilizing bacteria during late developmental stages of eggplant (Solanum melongena L.) FEMS Microbiol. Ecol. 2019;95:fiz023. doi: 10.1093/femsec/fiz023. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding G.C., Heuer H., Smalla K. Dynamics of bacterial communities in two unpolluted soils after spiking with phenanthrene: Soil type specific and common responders. Front. Microbiol. 2012;3:290. doi: 10.3389/fmicb.2012.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding G.C., Bai M.H., Han H., Li H.X., Ding X.Y., Yang H.F., Xu T., Li J. Microbial taxonomic, nitrogen cycling, and phosphorus recycling community composition during long-term organic greenhouse farming. FEMS Microbiol. Ecol. 2019;95:fiz042. doi: 10.1093/femsec/fiz042. [DOI] [PubMed] [Google Scholar]
- 36.Bastian M., Heymann S., Jacomy M. Gephi: An open source software for exploring and manipulating networks; Proceedings of the Third International Conference on Weblogs and Social Media; San Jose, CA, USA. 17–20 May 2009; pp. 1–2. [Google Scholar]
- 37.Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom. J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z.Y., Chen Q.W., Zhang J.Y., Guan T.S., Chen Y.C., Shi W.Q. Critical roles of cyanobacteria as reservoir and source for antibiotic resistance genes. Environ. Int. 2020;144:106034. doi: 10.1016/j.envint.2020.106034. [DOI] [PubMed] [Google Scholar]
- 39.Pawlowski A.C., Wang W., Koteva K., Barton H.A., McArthur A.G., Wright G.D. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 2016;7:13803. doi: 10.1038/ncomms13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y.T., Niu L.L., Zhu S.Y., Lu H.J., Liu W.P. Occurrence, abundance, and distribution of sulfonamide and tetracycline resistance genes in agricultural soils across china. Sci. Total Environ. 2017;599–600:1977–1983. doi: 10.1016/j.scitotenv.2017.05.152. [DOI] [PubMed] [Google Scholar]
- 41.Willms I.M., Kamran A., Aßmann N.F., Krone D., Bolz S.H., Fiedler F., Nacke H. Discovery of novel antibiotic resistance determinants in forest and grassland soil metagenomes. Front. Microbiol. 2019;10:460. doi: 10.3389/fmicb.2019.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson C.M., Grossman A.D. Integrative and conjugative elements (ICEs): What they do and how they work. Annu. Rev. Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao R.X., Yu K., Zhang J.Y., Zhang G.J., Huang J., Ma L.P., Deng C.F., Li X.Y., Li B. Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches. Water Res. 2020;186:116318. doi: 10.1016/j.watres.2020.116318. [DOI] [PubMed] [Google Scholar]
- 44.Wei H.W., Ma J.Y., Su Y.L., Xie B. Effect of nutritional energy regulation on the fate of antibiotic resistance genes during composting of sewage sludge. Bioresour. Technol. 2019;297:122513. doi: 10.1016/j.biortech.2019.122513. [DOI] [PubMed] [Google Scholar]
- 45.Hall J.P.J., Harrison E. Bacterial evolution: Resistance is a numbers game. Nat. Microbiol. 2016;1:16235. doi: 10.1038/nmicrobiol.2016.235. [DOI] [PubMed] [Google Scholar]
- 46.Heuer H., Solehati Q., Zimmerling U., Kleineidam K., Schloter M., Muller T., Focks A., Thielebruhn S., Smalla K. Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl. Environ. Microbiol. 2011;77:2527–2530. doi: 10.1128/AEM.02577-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jechalke S., Heuer H., Siemens J., Amelung W., Smalla K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014;22:536–545. doi: 10.1016/j.tim.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Knapp C.W., Engemann C.A., Hanson M.L., Keen P.L., Hall K.J., Graham D.W. Indirect evidence of transposon-mediated selection of antibiotic resistance genes in aquatic systems at low-level oxytetracycline exposures. Environ. Sci. Technol. 2008;42:5348–5353. doi: 10.1021/es703199g. [DOI] [PubMed] [Google Scholar]
- 49.Cirz R.T., Chin J.K., Andes D.R., de Crécy Lagard V., Craig W.A., Romesberg F.E. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:1024–1033. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohanski M.A., DePristo M.A., Collins J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fitzgerald D.M., Rosenberg S.M. What is mutation? A chapter in the series: How microbes ‘jeopardize’ the modern synthesis. PLoS Genet. 2019;15:e1007995. doi: 10.1371/journal.pgen.1007995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunin V., Engelbrektson A., Ochman H., Hugenholtz P. Wrinkles in the rare biosphere: Pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 53.Hyma K.E., Lacher D.W., Nelson A.M., Bumbaugh A.C., Whittam T.S. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 2005;187:619–628. doi: 10.1128/JB.187.2.619-628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhle V., Abrahams G.L., Hensel M. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic. 2010;7:716–730. doi: 10.1111/j.1600-0854.2006.00422.x. [DOI] [PubMed] [Google Scholar]
- 55.Peleg A.Y., Harald S., Paterson D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pomakova D.K., Hsiao C.B., Beanan J.M., Olson R., Macdonald U., Keynan Y., Russo T.A. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: An emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 57.Hamidian M., Ambrose S.J., Hall R.M. A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid. 2016;87–88:43–50. doi: 10.1016/j.plasmid.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Oliveira C.S., Moura A., Henriques I., Brown C.J., Rogers L.M., Top E.M., Correia A. Comparative genomics of IncP-1ε plasmids from water environments reveals diverse and unique accessory genetic elements. Plasmid. 2013;70:412–419. doi: 10.1016/j.plasmid.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Fahrenfeld N., Knowlton K., Krometis L.A., Hession W.C., Xia K., Lipscomb E., Libuit K., Green B.L., Pruden A. Effect of manure application on abundance of antibiotic resistance genes and their attenuation rates in soil: Field-scale mass balance approach. Environ. Sci. Technol. 2014;48:2643–2650. doi: 10.1021/es404988k. [DOI] [PubMed] [Google Scholar]
- 60.Ho Y.B., Zakaria M.P., Latif P.A., Saari N. Degradation of veterinary antibiotics and hormone during broiler manure composting. Bioresour. Technol. 2013;131:476–484. doi: 10.1016/j.biortech.2012.12.194. [DOI] [PubMed] [Google Scholar]
- 61.Yang J.F., Ying G.G., Yang L.H., Zhao J.L., Liu F., Tao R., Yu Z.Q., Peng P.A. Degradation behavior of sulfadiazine in soils under different conditions. J. Environ. Sci. Health Part B. 2009;44:241–248. doi: 10.1080/03601230902728245. [DOI] [PubMed] [Google Scholar]
- 62.Boxall A.B.A., Blackwell P., Cavallo R., Kay P., Tolls J. The sorption and transport of a sulphonamide antibiotic in soil systems. Toxicol. Lett. 2002;131:19–28. doi: 10.1016/S0378-4274(02)00063-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in GenBank (SRP126466) or freebioinfo.org.