Abstract
Trypanosoma rangeli is a generalist hemoflagellate that infects mammals and is transmitted by triatomines around Latin America. Due to its high genetic diversity, it can be classified into two to five lineages. In Brazil, its distribution outside the Amazon region is virtually unknown, and knowledge on the ecology of its lineages and on host species diversity requires further investigation. Here, we analyzed 57 T. rangeli samples obtained from hemocultures and blood clots of 1392 mammals captured in different Brazilian biomes. The samples were subjected to small subunit (SSU) rDNA amplification and sequencing to confirm T. rangeli infection. Phylogenetic inferences and haplotype networks were reconstructed to classify T. rangeli lineages and to infer the genetic diversity of the samples. The results obtained in our study highlighted both the mammalian host range and distribution of T. rangeli in Brazil: infection was observed in five new species (Procyon cancrivorous, Priodontes maximum, Alouatta belzebul, Sapajus libidinosus, and Trinomys dimidiatus), and transmission was observed in the Caatinga biome. The coati (Nasua nasua) and capuchin monkey (S. libidinosus) are the key hosts of T. rangeli. We identified all four T. rangeli lineages previously reported in Brazil (A, B, D, and E) and possibly two new genotypes.
Keywords: Trypanosoma rangeli, mammals, parasite ecology, lineages, Brazilian biomes
1. Introduction
Trypanosoma rangeli (Kinetoplastea: Tripanosomatidae) was first described by Tejera in 1920 [1], infecting the triatomine species Rhodnius prolixus (Hemiptera: Reduviidae). This protozoan infects different mammal species, including humans in Latin America and the Caribbean [2,3,4,5,6,7]. Infection is usually considered non-pathogenic to mammals [2,8], but T. rangeli induces low-level and long-lasting parasitemia that can last for years [9,10]. Unlike mammals, the parasite is pathogenic to triatomine species of the genus Rhodnius [8,11,12]. Trypanosoma rangeli and T. cruzi usually share mammalian hosts, and their distributions in nature often overlap in simple or mixed infections in mammals and triatomines [4,13,14,15,16]. However, there are marked differences in the transmission routes, life cycles, and host-parasite interactions of these two species. To date, T. rangeli metacyclic forms have been found in the salivary glands of only Rhodnius species, and parasite transmission by the inoculative route is associated with this triatomine genus [8,17,18,19].
Trypanosoma rangeli genetic diversity has been demonstrated by nuclear and mitochondrial markers [4,5,20,21,22,23]. Variability in the kDNA minicircles, as demonstrated by the presence or absence of the KP1 minicircle, separated T. rangeli into two groups, called KP1 (+) and KP1 (–) [7,23,24,25,26]. The use of spliced leader rRNA, ITS rDNA, SSU rDNA, and other nuclear sequences showed that T. rangeli is composed of two main lineages: one containing lineages A, C, D, and E and the other formed by the phylogenetically basal B lineage [6,27,28,29,30,31,32]. In Brazil, T. rangeli is mainly described in mammals and triatomines from the Amazon region; moreover, there are also sporadic reports of its occurrence in the Pantanal, Atlantic Forest, Cerrado, and Caatinga biomes, where it infects different mammal species of the orders Chiroptera, Carnivora, Didelphimorphia, Rodentia, Pilosa, Primates, and Cingulata [5,7,28,33,34,35,36,37,38,39,40]. Lineages A and B were described as having the greatest diversity of mammalian hosts and vectors. Lineage A was reported in bats of Central Brazil and the Amazon [6,39] and in primates and marsupials from the Amazon region [5,29]. Lineage B was originally reported in the Amazon, infecting non-human primates, triatomines, sloths, anteater, and humans [27,28,29], and recently in the Atlantic Forest infecting a bat species [37]. Rhodnius robustus was found harboring lineage A in the Amazon biome [28]. Lineage C is not reported in Brazil but in countries such as Colombia, Panamá, Costa Rica, and El Salvador [27,28]. Lineage D was only reported in the Atlantic Forest, first in the rodent species Echimys dasythrix and then in a bat species (Carollia perspicillata) [36,37,39]. Lineage E was described only in bats from Central Brazil and R. pictipes from the Amazon region [6].
In this context, even though T. rangeli is one of the most studied trypanosomatids, previous studies demonstrated only a small piece of what occurs in nature. We started from the hypothesis that T. rangeli is a parasite with wide dispersion in nature and that is transmitted among animals that use all forest strata, and that is probably transmitted by other triatomines besides Rhodnius. Our study included samples from a much larger number of mammal species from a wide geographical range and identified new hosts, and extended the known geographical distribution of T. rangeli and its lineages. Since Brazil is a country of continental proportions, we were able to survey several types of landscapes. We characterized through the small subunit (SSU) rDNA barcoding 57 samples of T. rangeli deposited in our DNA collection (http://coltryp.fiocruz.br/) (accessed on 31 March 2021). These T. rangeli samples were obtained from hemocultures and blood clots of free-ranging wild animals and domestic dogs in five different biomes over a period of 12 years. This is the first time that such a large number of T. rangeli samples were evaluated in Brazilian biomes, and the data generated will improve the understanding of this parasite’s ecology.
2. Results
From a total of 1392 free-ranging wild mammals examined between 2005 and 2017, Trypanosoma rangeli was confirmed in 57 DNA samples (4.1%) (Table 1). We have demonstrated that T. rangeli is a widely dispersed multi-host trypanosomatid since it was found infecting 15 mammal species of six orders dispersed in five different Brazilian biomes (Table 1). Trypanosoma rangeli infection was observed for the first time in the raccoon species Procyon cancrivorus (n = 1), in a giant armadillo, Priodontes maximus (n = 1), in a howler monkey, Alouatta belzebul (n = 1), in capuchin monkeys, Sapajus libidinosus (n = 15), and in the rodent Trinomys dimidiatus (n = 1). Blood samples of the giant armadillo (C752, C776, and C792 samples) collected at different periods (June, October, and December 2017) were all positive for T. rangeli (Supplementary Materials Table S1). The Pantanal biome had the largest number of T. rangeli occurrences in mammals, followed by the Cerrado-Amazon transition area and Atlantic Forest (Table 1).
Table 1.
Trypanosoma rangeli infection was demonstrated in different mammalian species in Brazilian biomes between 2005 and 2017.
| Order | Species | T. rangeli Infection/Total Examined | Infection Rate (%) | Biome | T. rangeli Lineage |
|---|---|---|---|---|---|
| Carnivora | Nasua nasua | 22/189 | 11.6 | Pantanal | A and B |
| Canis familiaris | 4/119 | 3.4 | Amazon Forest (n = 2) | A and E | |
| Atlantic Forest (n = 2 *) | E | ||||
| Procyon cancrivorus | 1/17 | 5.9 | Pantanal | E | |
| Cingulata | Priodontes maximus | 3 **/10 | 30 | Pantanal | E |
| Chiroptera | Carollia perspicillata | 3/279 | 1.1 | Amazon Forest (n = 1) | A |
| Atlantic Forest (n = 2) | B and D | ||||
| Didelphimorphia | Didelphis albiventris | 1/303 | 0.3 | Caatinga | A |
| Didelphis aurita | 1/271 | 0.4 | Atlantic Forest | D | |
| Didelphis marsupialis | 1/58 | 1.7 | Amazon Forest | A | |
| Philander opossum | 1/59 | 1.7 | Amazon Forest | E | |
| Primates | Alouatta belzebul | 1/6 | 16.7 | Amazon-Cerrado transition area | B |
| Alouatta caraya | 1/5 | 20 | Amazon-Cerrado transition area | B | |
| Sapajus libidinosus | 15/46 | 32.6 | Amazon-Cerrado transition area | A and B | |
| Saguinus bicolor bicolor | 1/24 | 4.2 | Amazon | B | |
| Rodentia | Coendou prehensilis | 1/5 | 20 | Amazon-Cerrado transition area | A |
| Trinomys dimidiatus | 1/1 | 100 | Atlantic Forest | D | |
| Total | 57/1392 | 4.1% | - | - |
* The two samples from the Atlantic Forest were characterized from blood clot samples. ** The three T. rangeli infections from P. maximus are from the same animal collected at different time points.
Nasua nasua (coatis) and capuchin monkeys had the highest number of individuals infected with T. rangeli (22/189 (11.6%) and 15/46 (32.6%), respectively) (Table 1). According to both t-tests, T. rangeli infection was statistically significant for the mammals analyzed (Tables S2 and S3).
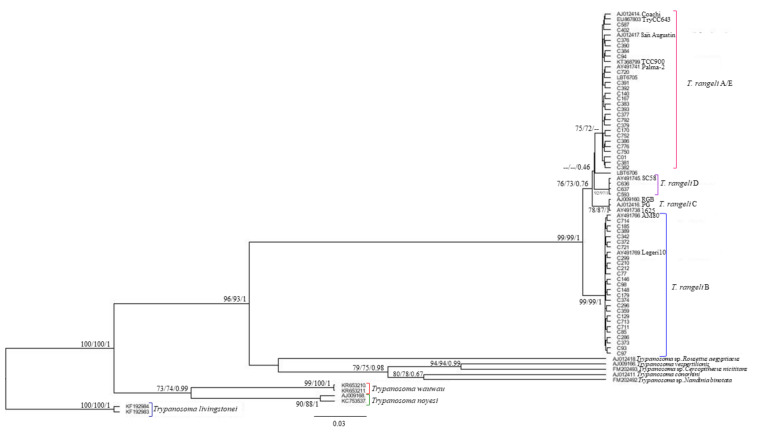
Fifty-four T. rangeli sequence samples were subjected to phylogenetic reconstruction by maximum likelihood (ML) and Bayesian inference (BI) (Figure 1). The sequences presented a base pair (bp) range from 556 to 912 nucleotides, and the final alignment presented 554 bp. Samples C375 (lineage A), C710, and RM2028 (lineage B) were not included in the phylogenetic reconstruction because they presented sequences with fewer than 554 nucleotides (441, 415, and 394 bp, respectively); thus, their lineage classification was performed solely based on BLAST screening. Both the ML and BI phylogenetic tree reconstructions grouped T. rangeli sequences into four lineage groups instead of five (Figure 1), as the A and E lineages were grouped together in the same clade, with some branch support, as observed in the ultra-bootstrap and SH-aLRT values (>70). The other three clusters were formed by T. rangeli lineages B, C, and D, all with high ultra-bootstrap, SH-aLRT (>70), and posterior probability values (PP = 1.0) (Figure 1). The lineage B cluster was the most distant from the other lineages (99 and 1.0 for the ultra-bootstrap, SH-aLRT, and posterior probability values, respectively). Although the lineage C cluster was separated from the other lineages with some branch support (>70), the separation between clusters A/E, LBT6706, and lineage D was not supported in either the ML or BI phylogenetic reconstructions (PP < 0.46).
Figure 1.
SSU rDNA reconstruction and phylogenetic trees based on 554 bp fragment alignment. Maximum likelihood ultrafast bootstrapping, SH-aLRT values, and Bayesian posterior probabilities are given near the nodes. Four T. rangeli lineage groups are shown: T. rangeli A/E; T. rangeli D; T. rangeli C; and T. rangeli B. The LBT6706 sequence is grouped separately from the T. rangeli A/E branch. The dashes represent lower or absent ultrafast bootstrap values, SH-aLRT values, and posterior probability values.
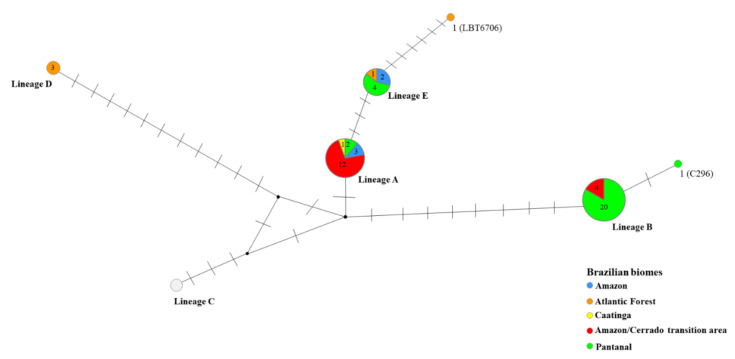
The lineage network (Figure 2) was capable of separating T. rangeli into five groups corresponding to its lineages (A, B, C, D, and E). Two well-defined T. rangeli clusters were observed: one including lineage B and the other including the other lineages (Figure 2). The cluster related to T. rangeli lineage A was found circulating in three biomes and in the transition area between the Amazon and Cerrado biomes and was separated from lineage B with a single mutational step. Additionally, lineages A and E were demonstrated to be closely related since three polymorphic site mutations in 1087, 1113, and 1114 bp of SSU rDNA separated them. The lineage B node was separated from the other lineages by eight polymorphic sites, thus confirming it to be the most distant T. rangeli lineage from the others. The Pantanal biome presented the greatest occurrence of T. rangeli lineage B (Figure 2). Lineages B and C were also separated by four polymorphic sites. Trypanosoma rangeli lineage D was the most distant from lineages A and C, as it was separated by 11 mutation steps (Figure 2).
Figure 2.
SSU rDNA sequence lineage network of Trypanosoma rangeli in different mammalian host species from Brazilian biomes. Networks were constructed with 65 SSU rDNA sequences, and the size of each node is proportional to the lineage frequency. The small black circle represents the median vector, which can be interpreted as an unsampled sequence or an extinct ancestral sequence.
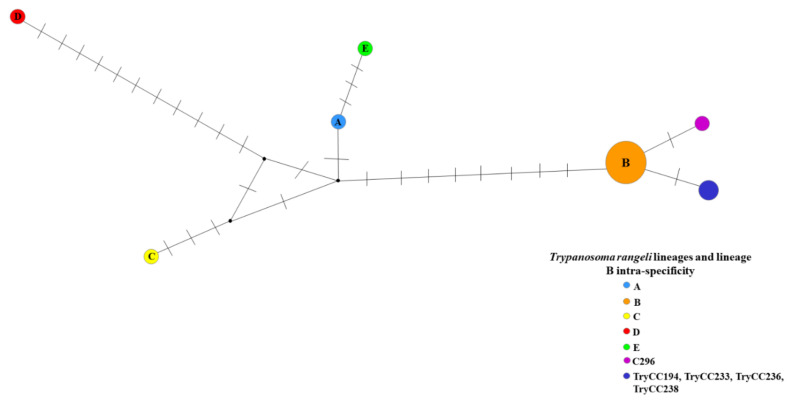
Two new sequences were observed separately from the other recognized lineages: one related to sample C296 (Amazon biome), which was clustered in lineage B in the phylogenetic analysis and separated from this lineage by a single polymorphic site (Figure 2 and Figure 3), and LBT 6706 (Atlantic Forest biome), which was separated from lineage E by six polymorphic sites (Figure 2). In T. rangeli lineage B, which is intraspecific (Figure 3), it is clear that the C296 sequence is different from the other T. rangeli B sequences that have already been described.
Figure 3.
Trypanosoma rangeli lineage B intraspecific network. Networks were constructed with 25 SSU rDNA sequences. The small black circle represents the median vector, which can be interpreted as an unsampled sequence or an extinct ancestral sequence.
The predominant T. rangeli lineages were A and B (Table S1). Lineage A was shown to be the most widely distributed lineage, being detected in five Brazilian states in 7 different mammal species of five orders dispersed in the Caatinga, Pantanal, and Amazon biomes and in the Amazon-Cerrado transition area: Didelphimorphia (n = 2), Carnivora (n = 2), Chiroptera (n = 1), Primates (n = 1), and Rodentia (n = 1) (Figure 1).
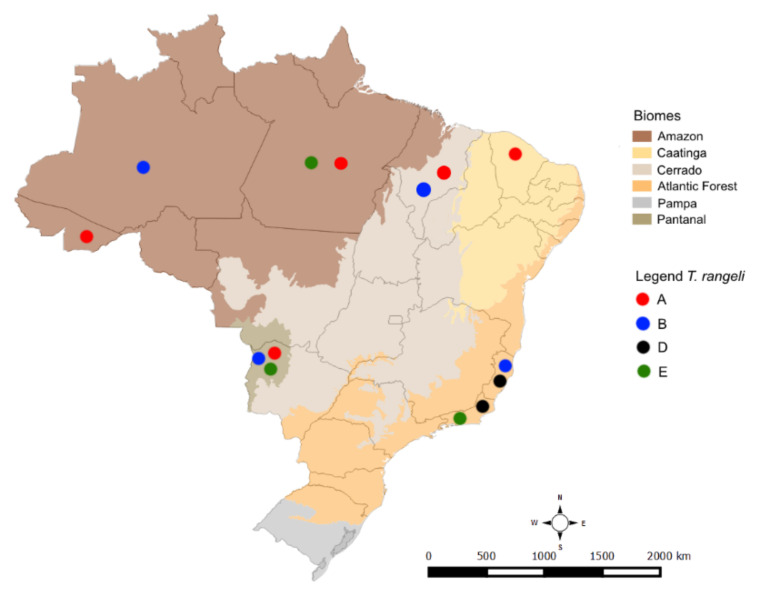
Figure 4 depicts the distribution of T. rangeli lineages in Brazilian biomes: lineages A and B were observed in four biomes, and lineage E was reported in three biomes (Amazon, Atlantic Forest, and Pantanal). Lineage A was the only T. rangeli lineage reported in the Caatinga biome (Figure 2 and Figure 4). The occurrence of lineage D was limited to the Atlantic Forest (Figure 2 and Figure 4). Four mammalian species presented infection by more than one lineage. The bat species (Carollia perspicilata) presented infections by the greatest diversity of T. rangeli lineages (A, B, and D). Sapajus libidinosus and N. nausa were infected by lineages A and B, and C. familiaris was infected by lineages A and E.
Figure 4.
Trypanosoma rangeli lineage distribution map in mammalian hosts in different Brazilian biomes. The lineages are represented by dots of the following colors: red—lineage A, blue—lineage B, black—lineage D, and green—lineage E.
3. Discussion
In this study, we provide an overview of the T. rangeli enzootic scenario in Brazil. T. rangeli is a generalist trypanosome that is capable of infecting a broad range of mammalian species. Trypanosoma rangeli infection has been observed in six mammalian orders, Didelphimorphia, Chiroptera, Carnivora, Rodentia, Cingulata, and Primates, of five Brazilian biomes, thus confirming its generalist characteristics concerning host range. All these mammalian orders were demonstrated to be involved in parasite transmission in the different Brazilian biomes because animals with positive hemocultures suggest competence to be a source of T. rangeli infection of the vectors. Here, the discovery of T. rangeli infecting free-ranging mammals that use diverse habitats provides evidence that T. rangeli is circulating in different forest strata and that these animals are possibly interacting and dispersing the parasite within these areas. In addition, animals may acquire the infection either through the contaminating vectoral route or through the oral route through triatomine ingestion.
According to our results, the carnivore coati (N. nasua) and capuchin monkey (S. libidinosus) were the key hosts of T. rangeli in the Pantanal biome and in the transition area between the Cerrado and Amazon biomes, respectively. Therefore, in this situation, these mammals could be considered a transmission hub of the parasite. The key host transmission role played by these two mammalian taxa can be explained by their habits. Coatis are known to present diurnal, scansorial, and sociable habits, except for adult males, which are solitary [41,42,43,44]. This mammalian taxon uses the ground and tree canopies, where they build nests to rest and reproduce [45]. Coatis present omnivorous feeding behavior, predominantly ingesting invertebrates and fruit, but it is possible that it feeds by vertebrates and carrion [44,46].
The capuchin monkey displays arboreal habits, although the young monkeys go to the ground to play and live in small groups [47,48,49,50,51]. Moreover, they can also use the ground in the Pantanal biome [52]. Capuchin monkeys are considered a generalist opportunistic species and present a variable diet that includes primarily fruits; however, they also feed on seeds, nuts, flowers, gums, nectar, fungi, sap, eggs, insects, small vertebrates, and even some oysters and crab species in mangrove regions [48,53]. Here, we also increased the number of monkey species known to be infected with T. rangeli. This clarifies why T. rangeli is so often found in monkeys, as these species consume insects as a part of their diet, and we can speculate that these primates might be infected by T. rangeli through the oral route.
Trypanosoma rangeli was found in four different marsupial species in our study. Infection in marsupials was previously observed in the Amazon and Cerrado biomes in D. albiventris and D. marsupialis [15,54,55] and in Ecuador in D. marsupialis [56]. It is interesting to note that only a few individuals of Didelphis sp. were found with positive hemocultures for T. rangeli. These low positive hemoculture rates may be due to very low parasitemia and may underestimate the real rate of infection, as this non-sensitive parasitological method does not detect cryptic infections. Although we report other marsupial species (Philander opossum and Didelphis aurita) infected with T. rangeli, we expected a greater number of positive samples because marsupials are usually found to be infected with a diversity of trypanosomatid species, which probably would include T. rangeli. In relation to lineage occurrence, we observed marsupials infected with lineage A, as has already been observed [15,28], but here, we also observed marsupials infected with lineage D in the Atlantic Forest. Marsupials are described as bio-accumulators of Trypanosoma species because these animals are able to harbor high Trypanosoma species diversity [57,58]. The occupation of generalist habitats by marsupials may explain why these animals are infected with trypanosomatids, including T. rangeli.
Trypanosoma rangeli has already been shown to infect several rodent species: Phyllomys dasythrix in Brazil [39] and Rhipidomys spp., Sciurus stramineus [56], and Oryzomys xanthaeolus in Ecuador [59]. We added two more rodent species as hosts of T. rangeli in Brazil: Coendou prehensilis and Trinomys dimidiatus. Genera of the orders Trinomys and Coendou have terrestrial and arboreal habits, respectively [60]; C. prehensilis was considered exclusively herbivorous (consuming fruits and seeds) [61,62], but fruits and seeds may easily be contaminated by infected triatomine. The Trinomys diet can be frugivorous, granivorous, herbivorous, or insectivorous [43,63]. It is important to highlight that the Trinomys genus uses hollow trees and burrows in the ground for shelter while resting [63], and these areas can harbor Trypanosoma-infected triatomines.
Additionally, for the first time, we demonstrated T. rangeli infection in a giant armadillo (Priodontes maximus) from the Pantanal biome. This is the first report of a giant armadillo infected with lineage E. These animals dig and live in burrows that can be used by a variety of vertebrates and insects [40,64]. The authors also surmised that the micro-environment inside these burrows can provide favorable conditions for parasite and vector survival and proliferation. P. maximus ingests insects in its diet [65,66], which can also lead to T. rangeli infection, as has already been suggested in the case of T. cruzi [40,67]. One of these scenarios may have led to T. rangeli infection in the giant armadillo. This shows the different manners by which an animal can become infected and how complex the transmission cycle of T. rangeli can be. In addition, the same armadillo demonstrated a stable T. rangeli infection since, in the present study, three positive samples were obtained in different time periods. We do not consider this situation a reinfection since the DNA sequences of the three samples were identical, and other T. rangeli lineages circulate in this biome.
In the case of the giant armadillo infection, our data support the maintenance of the same parasite population and a state of high parasitemia over an extended period. Although there are studies showing that infection can be maintained for a long time [10], it is still unknown how T. rangeli multiplies in or invades vertebrate host cells [2,10,68,69,70]. Additionally, as an extracellular parasite, how it escapes the host’s antibody-based immune response are among questions that need answering to obtain a better understanding of T. rangeli ecology.
We have reported only three fruit-eating bats (C. perpicillata) infected with T. rangeli. Bats are known to be hosts of several trypanosomatid species and are probably the original hosts of the T. cruzi clade [36,37,38,71,72,73,74]. Other authors have also detected low T. rangeli infection rates in bats [6,38,56,75]. It is intriguing to find that T. rangeli infection is rare in bats. Bats usually feed on insects and small vertebrates, and they groom and regurgitate food [76,77,78], so they are frequently in contact with Trypanosoma spp.; due to these habits, they were supposed to have a high rate of exposure to T. rangeli. Experimentally, it was demonstrated that bats acquired T. rangeli infection after triatomine blood feeding [79], so transmission occurred through metacyclic forms of the parasite present during salivary inoculation. We can speculate about some explanations for this finding: (i) bats can rapidly control T. rangeli infection; (ii) they present some sort of unknown mechanism that prevents the infection from being established; or (iii) they present cryptic infections that are not detectable by hemocultural exams.
Trypanosoma rangeli infection in dogs has been reported in some Latin American countries [80,81]. In Brazil, dog infection by T. rangeli was observed for the first time in the Amazon [82], and we are reporting it for the first time in the Atlantic Forest. Lineages A and C were reported in dogs from Venezuela and Colombia, respectively [28]. Here, we added another lineage encountered in dogs; T. rangeli lineage E infection in dogs may be associated with the fact that these animals prey on insects or even small mammals that may be infected. Although these dogs were examined in the domicile environment, the dogs in rural areas on Amazon and Atlantic Forest live in houses that are located at the border and, in several cases, inside the forest, and they are circulating in the sylvatic environment most of the time. That is why we correlate the dogs with the sylvatic area.
Due to its wide distribution and broad range of mammalian host species, T. rangeli probably infects triatomine vectors other than Rhodnius spp., as has been suggested previously. This parasite transmission is correlated to Rhodnius species [7,83] because they present T. rangeli infective forms in salivary glands [8,33,84]. Rhodnius species occur in all the Brazilian biomes, with some species more common and well distributed in the Amazon biome, whereas other species occur in the Pantanal, Cerrado, Caatinga, and Atlantic Forest biomes [85,86]. Although the important role played by Rhodnius triatomine species in T. rangeli transmission is clear, other triatomine species have been reported: Panstrongylus megistus in Minas Gerais and Santa Catarina states [15,21] and Triatoma brasiliensis in the Brazilian semi-arid region [87]. Although DNA from T. rangeli has been reported in the digestive tract of T. brasiliensis, the possibility of an animal ingesting and becoming infected from parasitic forms of triatomine cannot be ruled out. Despite the occurrence of R. domesticus in the Atlantic Forest [85], it is not common to find this species in the sylvatic environment. Dario and coworkers [37] suggested that Triatoma species might be responsible for T. rangeli transmission in Espírito Santo state (Atlantic Forest). In the Atlantic Forest of Santa Catarina, Panstrongylus megistus has already been found to be infected by T. rangeli [21], showing that other species can be involved in transmission. In Rio de Janeiro, the situation might be similar to that observed in Espírito Santo state, where R. domesticus is reported [85]. Therefore, it is likely that triatomine species other than Rhodnius sp. are also able to transmit this trypanosomatid. In areas where Rhodnius presence is confirmed, we suggest that infection might be occurring by the inoculative vectorial route, with contaminative T. rangeli forms present in the salivary gland. In the Atlantic Forest, where Rhodnius species are not found frequently and another genus of triatomine has been found to cause infection [15,21], another T. rangeli transmission route may occur.
Here, we present T. rangeli occurrence in mammals from a new biome—the Caatinga—increasing its distribution beyond the Amazon, Atlantic Forest, Cerrado, and Pantanal biomes. Therefore, T. rangeli is distributed in almost all Brazilian biomes. The Pantanal biome, in this study, was the one with the highest total number of infected animals: N. nasua, P. cancrivorus, and P. maximus. This contradicts the statement of other authors that most cases of T. rangeli infection occur in the Amazon biome [32], where four mammal species were found infected: D. marsupialis, S. b. bicolor, C. familiaris, and C. perspicillata. Although the Pantanal presented more mammals infected with T. rangeli, most of them were observed in coatis; in the Amazon, four different species were found infected, showing the broad range of infected species in this biome. Lineage A was found most often, lineage B was found only in the Amazon region [27,28], and lineage E was observed for the first time in the Pantanal biome [6]. We have demonstrated broad-spectrum circulation since these lineages were observed in different biomes than those in which they were first described. Lineage D was not observed outside the Atlantic Forest, but its occurrence was observed in distant forest fragments, leading to the conclusion that this T. rangeli lineage has an extensive distribution and probably can be observed in other biomes. In light of these findings, previous suggestions that T. rangeli lineages are linked to certain biomes or areas are not supported, but further research is required.
Intraspecific studies have demonstrated the diversity of the Trypanosoma genus, and the species of this genus were eventually subdivided into genotypes, as already noted, that include T. cruzi [88], T. rangeli [5,6,7,23,27,28], and some Trypanosoma species from bats [57,73]. In the case of T. rangeli, intra-specificity was observed across a variety of molecular markers, such as spliced leaders [89], random amplified polymorphic DNA (RAPD) [5,21], kDNA [23,24,25], and histone H2A genes [90], from which it was possible to differentiate this species into two or three groups. Maia da Silva et al. [5], using the RAPD assay for 22 T. rangeli samples, were able to separate this species into four lineages: A, B, C, and D. This same division was observed when 34 T. rangeli samples were subjected to molecular characterization using two different regions of the 18S rDNA: SSU and ITS1 [27]. These four lineages were corroborated by spliced leader RNA gene sequences [28], which confirmed these sequences as the most valuable and sufficiently defined reliable T. rangeli lineages [25,28,89]. Lineage E was described later [6] using spliced leader sequences and corroborated using other sequences [30,31,32]. In addition, Espinosa-Álvarez et al. [32] used 800 bp sequences from the V7-V8 region of SSU rDNA concatenated with gGAPDH and were able to cluster T. rangeli into the same five lineages. This subdivision is also possible using cathepsin L-like proteases and proline-racemase pseudogenes [30,31]. In our study, we were able to separate T. rangeli into four groups (A/E, B, C, and D) using a smaller SSU rDNA fragment (554 bp), but we were not able to separate lineages A from E in the phylogenetic tree reconstruction using only SSU rDNA. However, using the haplotype network, it was possible to separate all the lineages.
We observed two different sequences of T. rangeli, demonstrating the occurrence of new genotypes that may correspond to new lineages. The two samples showing different SSU rDNA sequences were from hosts collected in the Atlantic Forest and Pantanal biomes, which have high biodiversity [91,92]. We can affirm that this biodiversity extends to parasites, not only for the T. rangeli lineages but also for other trypanosomatid species and lineages, as previously reported [36,37,93]. These results reinforce that the Atlantic Forest is a biodiversity hotspot at all levels, from macro- to microorganisms, and that we are probably dealing with a new parasite hotspot.
We are not proposing any new genotype classification because SSU rDNA sequences alone should not be employed for reliable descriptions of new lineages. The subdivision of T. rangeli into two main groups as initially proposed [94] or into five or more lineages must be supported by multiple genetic or phylogenetic analyses. All the groups contain members with different host species (human, domestic or sylvatic mammals) without any vertebrate host species associations. Grouping was independent of mammalian host species and geographical origin, indicating that other factors determine this segregation. Some associations (not strict) with the Rhodnius species complex have been suggested. However, both the taxonomy and evolutionary history of Rhodnini are far from being clearly understood, and any vector-parasite associations must be interpreted with caution [25,28,32,94]. Most of the studies, including ours, involve subsampling, and a final division/classification of T. rangeli genotypes needs more representative sampling.
To conclude, this study shows how much is still unknown about T. rangeli. Knowledge of mammalian hosts and vector ranges and T. rangeli infection stability in these mammalian hosts and lineage classification is still far from complete and will likely expand in the future as wider geographic areas are explored.
4. Material and Methods
4.1. Trypanosoma Rangeli Study Areas
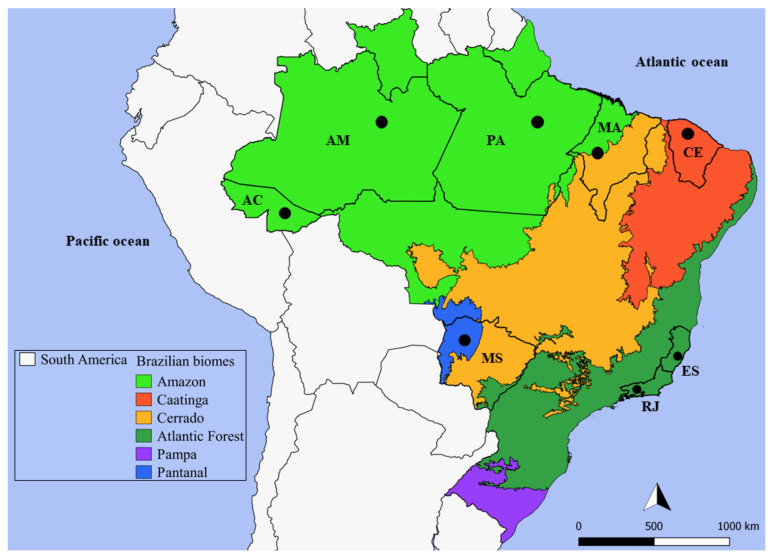
The fieldwork, the aim of which was to study mammalian trypanosomatids, was conducted by our group from 2005 to 2017 [37,38,57,95,96,97] (Dario MA (data not shown) and Lisboa CV (data not shown)). The data are summarized in Figure 5 and Table 2.
Figure 5.
Brazilian biomes investigated for Trypanosoma spp. infection in mammals. Six biomes are represented: Amazon, Cerrado, Caatinga, Atlantic Forest, Pantanal, and Pampa. The acronyms represent the Brazilian states, and the black dots represent the places where fieldwork was performed: AC—Acre, AM—Amazonas, CE—Ceará, ES—Espírito Santo, MA—Maranhão, MS—Mato Grosso do Sul, PA—Pará, and RJ—Rio de Janeiro.
Table 2.
Mammal species captured during fieldwork performed in different Brazilian biomes while surveying for Trypanosoma rangeli between 2005 and 2017.
| Order | Species | Number of Samples |
|---|---|---|
| Carnivora | Nasua nasua | 189 |
| Canis familiaris | 119 | |
| Procyon cancrivorus | 17 | |
| Cingulata | Priodontes maximus | 10 |
| Chiroptera | Carollia perspicillata | 279 |
| Didelphimorphia | Didelphis albiventris | 303 |
| Didelphis aurita | 271 | |
| Didelphis marsupialis | 58 | |
| Philander opossum | 59 | |
| Primates | Alouatta belzebul | 6 |
| Alouatta caraya | 5 | |
| Sapajus libidinosus | 46 | |
| Saguinus bicolor bicolor | 24 | |
| Rodentia | Coendou prehensilis | 5 |
| Trinomys dimidiatus | 1 | |
| Total | 15 | 1392 |
4.2. Trypanosoma Rangeli DNA Samples Origin
DNA samples of the trypanosomatids isolated from hemocultures (n = 55) and blood clots of two animals were deposited in the DNA library of the Trypanosoma from wild and domestic mammals and vectors collection (COLTYP/Fiocruz). The hemocultures were washed with phosphate-buffered saline solution, 100 µg/mL proteinase K (Invitrogen, Carlsbad, CA, USA), and 0.5% sodium dodecyl sulfate was added, and the samples were incubated at 56 °C for 2 h [44]. After this step, the DNA was extracted by the phenol-chloroform method [98]. For the two blood clots, the absolute ethanol was removed, and 50 µL of each blood clot was transferred to a new 1.5 mL tube. DNA extraction was performed according to [58]. The DNA concentration and purity of the DNA samples were quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
4.3. Trypanosoma Rangeli Molecular Characterization
The DNA samples were subjected to two PCR methodologies for T. rangeli identification: (i) 15 samples underwent conventional PCR for amplification of the V7-V8 region of SSU rDNA [99], and (ii) 42 samples underwent nested PCR to amplify the SSU rDNA gene [100,101]. For amplification of the V7-V8 region, 8.5 µL of GoTaq MasterMix (Promega, Madison, WI, USA), 100 ng of DNA template, 10 pmol of the 609F (5′CACCCGCGGTAATTCCAGC3′), and 706R (5′CTGAGACTGTAACCTCAA3′) primers and sterile deionized water were used in a 25 µL final mix solution. Amplification was performed under the following conditions: an initial denaturation cycle at 94 °C for 3 min; 30 cycles at 94 °C for 1 min, 48 °C for 2 min and 72 °C for 2 min; and final elongation at 72 °C for 10 min.
For the nested PCR, 16 pmol of each external primer (TRY927F (5′GAAACAAGAAACACGGGAG3′) and TRY927R (5′CTACTGGGCAGCTTGGA3′)), 8.5 µL of GoTaq MasterMix (Promega, Madison, WI, USA), 50 ng of DNA template, and sterile deionized water (added to reach a final volume of 25 µL) were used. Amplification was performed under the following conditions: initial denaturation at 94 °C for 3 min; 30 cycles at 94 °C for 30 s, 55 °C for 60 s, and 72 °C for 90 s; and final elongation at 72 °C for 10 min. The first-round PCR products were diluted (1:10) in sterile deionized water. Two microlitres of this dilution was used as a template for the second round of PCRs using the internal primers SSU561F (5′TGGGATAACAAAGGAGCA3′) and SSU561R (5′CTGAGACTGTAACCTCAAAGC3′) under the same conditions applied in the first round. For the DNA samples from the blood clots, 12.5 5 µL of GoTaq MasterMix and 20 pmol of each primer were used. All the SSU rDNA amplifications were performed with a Swift MaxPro Thermal Cycle (Esco Scientific, Singapore).
Electrophoresis of the PCR products (~850 bp for the V7-V8 region and ~600 bp for the nested PCR) was performed in 2% agarose gels (Figure S1), which were stained with ethidium bromide solution and visualized under ultraviolet light. The amplified products were purified using the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK). The SSU rDNA amplicons underwent Sanger DNA sequencing reactions for both DNA strands (V7-V8: 609F—5′CACCCGCGGTAATTCCAGC3′ and 706R—5′CTGAGACTGTAACCTCAA3′; SSU rDNA: SSU561F—5′TGGGATAACAAAGGAGCA3′ and SSU561R—5′CTGAGACTGTAACCTCAAAGC3′) with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an ABI 3730 DNA sequencer available at the PDTIS/Fiocruz sequencing facilities.
4.4. Phylogenetic Analysis of Trypanosoma rangeli Lineages
To obtain the consensus SSU rDNA sequences, each forward and reverse sequence was assembled and edited using SeqMan (DNASTAR Lasergene, Gatc, Konstanz, Germany). Trypanosoma rangeli sequences obtained in this study were aligned to an SSU rDNA homologous region in T. rangeli lineages and outgroup sequences deposited in GenBank (Table S2) using the L-INS-i algorithm in MAFFT v.7.0 [102]. The obtained alignment was visualized and manually edited on MegaX software [103]. To determine the T. rangeli lineages, maximum likelihood (ML) estimation and Bayesian inference (BI) were performed. The equal-frequency transition model with gamma-distributed rate variation among sites (TIM2ef+G) was the best model for the dataset, as indicated by the corrected Akaike information criterion (AICc) and Bayesian information criteria (BIC) obtained in jModelTest v.2 [104]. The ML tree reconstruction was performed in the IQ-Tree program [105,106] available on PhyloSuite v.1.2.2. For branch support, ultrafast bootstrapping [107] was performed with 5000 replicates with 1000 maximum interactions and 0.99 minimum correlation coefficients. To validate the ultrafast bootstrap results, the SH-aLRT branch test with 5000 replicates was also applied.
Bayesian tree reconstruction was performed in Bayesian Evolutionary Analysis Sampling Trees (BEAST) v2.6.2 [108]. The Bayesian Markov chain Monte Carlo (MCMC) method was used to assign T. rangeli lineages prior to information. The birth-death model specification was used in tree reconstruction. Four independent runs were performed for 20 M with sampling every 2000 generations. The runs converged, and the effective sample size (ESS) was calculated after 10% of each run was excluded (burn-in) from each run in TRACER v.1.6 [109]. The parameters selected led to ESSs higher than 500 and were considered appropriate. The final tree was generated with maximum clade credibility (MCC) based on 32,404 trees (burn-in = 3600) and a 0.6 posterior probability limit (PP) in Tree Annotator. Both the ML and BI reconstructions were visualized in Figtree v.1.4.3.
Two haplotype networks based on only T. rangeli samples were generated in Network software version 5.0.1.1 (fluxus-engineering.com) to define evolutionary relationships among the T. rangeli lineages and to observe the intra-specificity of lineage B. The SSU rDNA lineage and lineage B intra-specificity networks were built using median-joining [110] and maximum parsimony [111] post-processed clean-up procedure. The SSU rDNA sequences used in the phylogenetic reconstruction and in the haplotype networks are listed in Table S4.
4.5. Statistical Analysis
The overall infection rate and the infection rate for each mammalian species were calculated using the number of species positive for T. rangeli infection divided by the total number of analyzed mammals. In addition, a two-tailed t-test was performed to verify the significance level of infection in mammals. T-tests of paired sample averages and two samples assuming different variances were performed with a significance level of p < 0.05. All the statistical analyses were performed in Microsoft Excel® 365.
Acknowledgments
We would like to express our gratitude to the Laboratório de Biologia de Tripanosomatídeos, to the Research Group InsanaHuna (https://www.insanahuna.com) (accessed on 31 March 2021), and the giant armadillo project members, supporters (https://www.giantarmadillo.org/partners) (accessed on 31 March 2021), and staff for fieldwork support. We thank the Collection of Trypanosoma from Wild and Domestic Mammals and Vectors (Coltryp/Fiocruz) for providing the DNA samples. We would also like to thank the Technological Development and Inputs for Health program from Instituto Oswaldo Cruz Foundation (PDTIS/FIOCRUZ) sequencing platform for sequencing our samples.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10060736/s1. Table S1: Trypanosoma rangeli infection in different mammalian species in Brazilian biomes. Table S2: Two-tailed t-tests of paired average samples of Trypanosoma rangeli infection in mammals. Table S3: Two-tailed t-test of two samples of Trypanosoma rangeli infection assuming different variances. Table S4: Trypanosomatid SSU rDNA sequences retrieved from GenBank used for phylogenetic analysis. Figure S1: Representative 2% agarose gel electrophoresis of 18S rDNA molecular markers for Trypanosoma rangeli molecular identification.
Author Contributions
Conceptualization, A.M.J.; Methodology, M.A.D., M.S.R., M.G.P., C.V.L. and L.L.; Formal Analysis, M.A.D., A.M.J. and M.G.P.; Data Curation, A.M.J., M.S.R. and C.V.L.; Writing—Original Draft Preparation, M.A.D. and A.M.J.; Writing—Review & Editing, M.S.R., C.V.L., L.L., M.G.P., D.K., A.L.J.D., H.M.H., A.L.R.R. and M.M.G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Fundação Oswaldo Cruz (Fiocruz), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and the giant armadillo project (https://www.giantarmadillo.org/partners) (accessed on 31 March 2021). FAPERJ provides a post-doctoral #10 fellowship (2019–current) to M.A.D. H.M.H. received a productivity grant from CNPq (process# 308768/2017-5). ALRR da “Joven Cientista do Nosso Estado” fellowship from FAPERJ (E-26/202.794/2019) and a project grant from CNPq (CNPq/Universal 425293/2018-1). A.M.J. received a “Cientista do Nosso Estado” fellowship from FAPERJ and a “Bolsa de produtividade” fellowship from CNPq.
Institutional Review Board Statement
The sampling procedures reported herein were authorized by the Brazilian Institute of the Environment and Renewable Natural Resources (IBAMA) under license nos. 3365-1 and 27587-12. All procedures followed protocols approved by the Fiocruz Ethics Committee for Animal Research (P0179-03; L0015-07; LW-81/12).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) (accessed on 31 March 2021). The SSU rDNA accession numbers are provided in Table S1.
Conflicts of Interest
The sponsors had no role in the design, execution, interpretation, or writing of the study.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tereja E. Um nouveau flagela de Rhodnius prolixus. Trypanosoma (ou Crithidia) rangeli n.sp. Bull. Soc. Path. Exot. 1920;13:527–530. [Google Scholar]
- 2.D’Alessandro A., Saravia N.G. Parasitic Protozoa. 2nd ed. Volume 2. Academic Press; San Diego, CA, USA: 1992. Trypanosoma rangeli; pp. 1–54. [Google Scholar]
- 3.Coura J.R., Fernandes O., Arboleda M., Barret T.V., Carrara N., Degrave W., Campbell D.A. Human infection by Trypanosoma rangeli in the Brazilian Amazon. Trans. R. Soc. Trop. Med. Hyg. 1996;90:278–279. doi: 10.1016/S0035-9203(96)90247-3. [DOI] [PubMed] [Google Scholar]
- 4.Grisard E.C., Steindel M., Guarneri A.A., Eger-Mangrich I., Campbell D.A., Romanha A.J. Characterization of Trypanosoma rangeli strains isolated in Central and South America: An Overview. Mem. Inst. Oswaldo Cruz. 1999;94:203–209. doi: 10.1590/S0074-02761999000200015. [DOI] [PubMed] [Google Scholar]
- 5.Maia da Silva F., Rodrigues A.C., Campaner M., Takata C.S.A., Brigido M.C., Junqueira A.C.V., Coura J.R., Takeda G.F., Shaw J.J., Teixeira M.M.G. Randomly amplified polymorphic DNA analysis of Trypanosoma rangeli and allied species from human, monkeys and other sylvatic mammals of the Brazilian Amazon disclosed a new group and a species-specific marker. Parasitology. 2004;128:283–294. doi: 10.1017/S0031182003004554. [DOI] [PubMed] [Google Scholar]
- 6.Maia da Silva F., Marcili A., Lima L., Cavazzana M., Jr., Ortiz P.A., Campaner M., Takeda G.F., Paiva F., Nunes V.L.B., Camargo E.P., et al. Trypanosoma rangeli isolates of bats from Central Brazil: Genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Trop. 2009;109:199–207. doi: 10.1016/j.actatropica.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Vallejo G.A., Guhl F., Schaub G.A. Triatominae-Trypanosoma cruzi/T. rangeli: Vector-parasite interactions. Acta Trop. 2009;110:137–147. doi: 10.1016/j.actatropica.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Guhl F., Vallejo G.A. Trypanosoma (Herpetosoma) rangeli Tejera, 1920: An updated review. Mem. Inst. Oswaldo Cruz. 2003;98:435–442. doi: 10.1590/S0074-02762003000400001. [DOI] [PubMed] [Google Scholar]
- 9.Sousa M.A. Morphobiological characterization of Trypanosoma cruzi Chagas, 1909 and its distinction from other trypanosomes. Mem. Inst. Oswaldo Cruz. 1999;94:205–210. doi: 10.1590/S0074-02761999000700031. [DOI] [PubMed] [Google Scholar]
- 10.Eger-Mangrich I., de Oliveira M.A., Grisard E.C., de Souza W., Steindel M. Interaction of Trypanosoma rangeli Tejera, 1920 with different cell lines in vitro. Parasitol. Res. 2001;87:505–509. doi: 10.1007/s004360000356. [DOI] [PubMed] [Google Scholar]
- 11.Grewal M.S. Pathogenicity of Trypanosoma rangeli Tejera, in the invertebrate host. Exp. Parasitol. 1957;6:123–130. doi: 10.1016/0014-4894(57)90010-3. [DOI] [PubMed] [Google Scholar]
- 12.Grisard E.C. Salivaria or stercoraria? The Trypanosoma rangeli dilemma. Kinetoplast. Biol. Dis. 2002;1:5. doi: 10.1186/1475-9292-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guhl F., Hudson L., Marinkelle C.J., Jaramillo C.A., Bridge D. Clinical Trypanosoma rangeli infection as a complication of Chagas’ disease. Parasitology. 1987;94:475–484. doi: 10.1017/S0031182000055827. [DOI] [PubMed] [Google Scholar]
- 14.Machado P.E., Eger-Mangrich I., Rosa G., Koerich L.B., Grisard E.C., Steindel M. Differential susceptibility of triatominae of the genus Rhodnius to Trypanosoma rangeli strains from different geographical origins. Int. J. Parasitol. 2001;31:631–633. doi: 10.1016/S0020-7519(01)00150-3. [DOI] [PubMed] [Google Scholar]
- 15.Ramirez L.E., Lages-Silva E., Alvarenga-Franco F., Matos A., Vargas N., Fernandes O., Zingales B. High prevalence of Trypanosoma rangeli and Trypanosoma cruzi in opossums and triatomids in a formerly-endemic area of Chagas disease in Southeast Brazil. Acta Trop. 2002;84:189–198. doi: 10.1016/S0001-706X(02)00185-7. [DOI] [PubMed] [Google Scholar]
- 16.Peterson J.K., Graham A.L., Elliott R.J., Dobson A.P., Chávez O.T. Trypanosoma cruzi–Trypanosoma rangeli co-infection ameliorates negative effects of single trypanosome infections in experimentally infected Rhodnius prolixus. Parasitology. 2016;143:1157–1167. doi: 10.1017/S0031182016000615. [DOI] [PubMed] [Google Scholar]
- 17.Hoare C.A. A Zoological Monograph. Blackweel Scientific Publications; Oxford, UK: 1972. The Trypanosomes of mammals; pp. 327–400. [Google Scholar]
- 18.Hecker H., Schwarzenbach M., Rudin W. Development and interactions of Trypanosoma rangeli in and with the reduviid bug Rhodnius prolixus. Parasitol. Res. 1990;76:311–318. doi: 10.1007/BF00928185. [DOI] [PubMed] [Google Scholar]
- 19.Azambuja P., Garcia E.S. Trypanosoma rangeli interactions within the vector Rhodnius prolixus—A mini review. Mem. Inst. Oswaldo Cruz. 2005;100:567–572. doi: 10.1590/S0074-02762005000500019. [DOI] [PubMed] [Google Scholar]
- 20.Macedo A.M., Vallejo G.A., Chiari E., Pena S.D.J. DNA fingerprinting reveals relationships between strains of Trypanosoma rangeli and Trypanosoma cruzi. In: Pena S.D.J., Chakraborty R., Epplen J.T., Jeffreys A.J., editors. DNA Fingerprinting: State of the Science. Birkhauser Verlag; Basel, Switzerland: 1993. pp. 321–329. [DOI] [PubMed] [Google Scholar]
- 21.Steindel M., Dias Neto E., Pinto C.J., Grisard E.C., Menezes C.L., Murta S.M., Simpson A.J., Romanha A.J. Randomly amplified polymorphic DNA (RAPD) and isoenzyme analysis of Trypanosoma rangeli strains. J. Eukaryot. Microbiol. 1994;41:261–267. doi: 10.1111/j.1550-7408.1994.tb01506.x. [DOI] [PubMed] [Google Scholar]
- 22.Henríksson J., Solarj A., Rydaker M., Sousa O.E., Pettersson U. Karyotype variability in Trypanosoma rangeli. Parasitology. 1996;112:385–391. doi: 10.1017/S0031182000066610. [DOI] [PubMed] [Google Scholar]
- 23.Vallejo G.A., Guhl F., Carranza J.C., Lozano L.E., Sánchez J.L., Jaramillo J.C., Gualtero D., Castañeda N., Silva J.C., Steindel M. kDNA markers define two major Trypanosoma rangeli lineages in Latin America. Acta Trop. 2002;81:77–82. doi: 10.1016/S0001-706X(01)00186-3. [DOI] [PubMed] [Google Scholar]
- 24.Vallejo G.A., Guhl F., Carranza J.C., Moreno J., Triana O., Grisard C.E. Parity between kinetoplast: DNA and miniexon gene sequences supports either clonal evolution or speciation in Trypanosoma rangeli strains isolated from Rhodnius colombiensis, R. pallescens and R. prolixus in Colombia. Infect. Genet. Evol. 2003;3:39–45. doi: 10.1016/S1567-1348(02)00150-8. [DOI] [PubMed] [Google Scholar]
- 25.Urrea D.A., Carranza J.C., Cuba Cuba C.A., Gurgel-Gonçalves R., Guhl F., Schofield C.J., Triana O., Vallejo G.A. Molecular characterization of Trypanosoma rangeli strains isolated from Rhodnius ecuadoriensis in Peru, R. colombiensis in Colombia and R. pallescens in Panama, supports a co-evolutionary association between parasites and vectors. Infect. Genet. Evol. 2005;5:123–129. doi: 10.1016/j.meegid.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Cabrine-Santos M., Ferreira K.A.M., Tosi L.R.O., Lages-Silva E., Ramírez L.E., Pedrosa A.L. Karyotype variability in KP1(+) and KP1(−) strains of Trypanosoma rangeli isolated in Brazil and Colombia. Acta Trop. 2009;110:57–64. doi: 10.1016/j.actatropica.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Maia da Silva F., Noyes H., Campaner M., Junqueira A.C.V., Coura J.R., Añez N., Shaw J.J., Stevens J.R., Teixeira M.M.G. Phylogeny, taxonomy and grouping of Trypanosoma rangeli isolates from man, triatomines and sylvatic mammals from widespread geographical origin based on SSU and ITS ribosomal sequences. Parasitology. 2004;129:549–561. doi: 10.1017/S0031182004005931. [DOI] [PubMed] [Google Scholar]
- 28.Maia da Silva F., Junqueira A.C., Campaner M., Rodrigues A.C., Crisante G., Ramírez L.E., Caballero Z.C.E., Monteiro F.A., Coura J.R., Añez N., et al. Comparative phylogeography of Trypanosoma rangeli and Rhodnius (Hemiptera: Reduviidae) supports a long coexistence of parasite lineages and their sympatric vectors. Mol. Ecol. 2007;16:3361–3373. doi: 10.1111/j.1365-294X.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- 29.Maia da Silva F., Naiff R.D., Marcili A., Gordo M., D’Affonseca Neto J.A., Naiff M.F., Franco A.M., Campaner M., Valente V., Valente S.A., et al. Infection rates and genotypes of Trypanosoma rangeli and T. cruzi infecting free-ranging Saguinus bicolor (Callitrichidae), a critically endangered primate of the Amazon Rainforest. Acta Trop. 2008;107:168–173. doi: 10.1016/j.actatropica.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Ortiz P.A., Maia da Silva F., Cortez A.P., Lima L., Campaner M., Pral E.M., Alfieri S.C., Teixeira M.M.G. Genes of cathepsin L-like proteases in Trypanosoma rangeli isolates: Markers for diagnosis, genotyping and phylogenetic relationships. Acta Trop. 2009;112:249–259. doi: 10.1016/j.actatropica.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 31.Caballero Z.C., Costa-Martins A.G., Ferreira R.C., Alves J.M.P., Serrano M.G., Camargo E.P., Buck G.A., Minoprio P., Teixeira M.M.G. Phylogenetic and syntenic data support a single horizontal transference to a Trypanosoma ancestor of a prokaryotic proline racemase implicated in parasite evasion from host defences. Parasites Vectors. 2015;8:222. doi: 10.1186/s13071-015-0829-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espinosa-Álvarez O., Ortiz P.A., Lima L., Costa-Martins A.G., Serrano M.G., Herder S., Buck G.A., Camargo E.P., Hamilton P.B., Stevens J.R., et al. Trypanosoma rangeli is phylogenetically closer to Old World trypanosomes than to Trypanosoma cruzi. Int. J. Parasitol. 2018;48:569–584. doi: 10.1016/j.ijpara.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Miles M.A., Arias J.R., Valente S.A., Naiff R.D., de Souza A.A., Povoa M.M., Lima J.A., Cedillos R.A. Vertebrate hosts and vectors of Trypanosoma rangeli in the Amazon Basin of Brazil. Am. J. Trop. Med. Hyg. 1983;32:1251–1259. doi: 10.4269/ajtmh.1983.32.1251. [DOI] [PubMed] [Google Scholar]
- 34.De Araújo V.A., Boité M.C., Cupolillo E., Jansen A.M., Roque A.L. Mixed infection in the anteater Tamandua tetradactyla (Mammalia: Pilosa) from Para State, Brazil: Trypanosoma cruzi, T. rangeli and Leishmania infantum. Parasitology. 2013;140:455–460. doi: 10.1017/S0031182012001886. [DOI] [PubMed] [Google Scholar]
- 35.Alves F.M., Saab de Lima J., Rocha F.L., Herrera H.M., Mourão G.D.M., Jansen A.M. Complexity and multi-factorial of Trypanosoma cruzi sylvatic cycle in coatis, Nasua Nasua (Procynidae), and triatomine bugs in Brazilian Pantanal. Parasites Vectors. 2016;9:378. doi: 10.1186/s13071-016-1649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dario M.A., Moratelli R., Schwabl P., Jansen A.M., Llewellyn M.S. Small subunit ribosomal metabarcoding reveals extraordinary trypanosomatid diversity in Brazilian bats. PLoS Negl. Trop. Dis. 2017;11:e0005790. doi: 10.1371/journal.pntd.0005790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dario M.A., Lisboa C.V., Costa L.M., Moratelli R., Nascimento M.P., Costa L.P., Leite Y.L.R., Llewellyn M.S., Xavier S.C.D.C., Roque A.L.R., et al. High Trypanosoma spp. diversity is maintained by bats and triatomines in Espírito Santo state, Brazil. PLoS ONE. 2017;12:e0188412. doi: 10.1371/journal.pone.0188412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dos Santos F.C.B., Lisboa C.V., Xavier S.C.C., Dario M.A., Verde R.S., Calouro A.M., Roque A.L.R., Jansen A.M. Trypanosoma sp. diversity in Amazonian bats (Chiroptera; Mammalia) from Acre State, Brazil. Parasitology. 2018;145:828–837. doi: 10.1017/S0031182017001834. [DOI] [PubMed] [Google Scholar]
- 39.Steindel M., Carvalho Pinto J.C., Toma H.K., Mangia R.H.R., Ribeiro-Rodrigues R., Romanha A.J. Trypanosoma rangeli (Tejera, 1920) isolated from a sylvatic rodent (Echimys dasythrix) in Santa Catarina Island, Santa Catarina State: First report of this trypanosome in southern Brazil. Mem. Inst. Oswaldo Cruz. 1991;86:73–79. doi: 10.1590/S0074-02761991000100012. [DOI] [PubMed] [Google Scholar]
- 40.Kluyber D., Desbiez A., Attias N., Massocato G., Gennari S., Soares H., Bagagli E., Bosco S.M.G., Garcés H.G., Ferreira J.D.S., et al. Zoonotic parasites infecting free-living armadillos from Brazil. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13839. [DOI] [PubMed] [Google Scholar]
- 41.Crespo J.A. Ecologia de la comunidad de mamiferos del Parque Nacional Iguazu, Misiones. Rev. MACN Ecol. 1982;3:45–162. [Google Scholar]
- 42.Schaller G.B. Mammals and their biomass on a Brazilian ranch. Arq. Zool. São Paulo. 1983;31:1–36. doi: 10.11606/issn.2176-7793.v31i1p1-36. [DOI] [Google Scholar]
- 43.Emmons L.H. Neotropical Rainforest Mammals: A Field Guide. University of Chicago Press; Chicago, IL, USA: 1990. p. 281. [Google Scholar]
- 44.Gompper M.E., Decker D.M. Nasua nasua. Mamm. Species. 1998;580:1–9. doi: 10.2307/3504444. [DOI] [Google Scholar]
- 45.McClearn D. Locomotion, posture, and feeding behavior of kinkajous, coatis, and raccoons. J. Mammal. 1992;73:245–261. doi: 10.2307/1382055. [DOI] [Google Scholar]
- 46.Bianchi R.C., Campos R.C., Xavier-Filho N.L., Olifiers N., Gompper M.E., Mourão G.M. Intraspecific, interspecific, and seasonal differences in the diet of three mid-sized carnivores in a large neotropical wetland. Acta Theriol. 2014;59:13–23. doi: 10.1007/s13364-013-0137-x. [DOI] [Google Scholar]
- 47.Sussman R.W., Garber P.A. A new interpretation of the social organization and mating system of the Callitrichidae. Int. J. Primatol. 1987;8:73–92. doi: 10.1007/BF02737114. [DOI] [Google Scholar]
- 48.Snowdon C.T., Soini P. The tamarins, genus Saguinus. In: MitterMeier A., Rylands A.B., Coimbra-Filho A.F., da Fonseca G.A., editors. Ecology and Behavior of Neotropical Primates. Volume 2. World Wildlife Fund; Washington, DC, USA: 1988. pp. 223–298. [Google Scholar]
- 49.Tardif S.D., Harrison M.L., Simek M.A. Communal infant care in marmosets and tamarins: Relation to energetics, ecology, and social organization. In: Rylands A.B., editor. Marmosets and Tamarins: Systematics, Behaviour and Ecology. Oxford University Press; Oxford, UK: 1993. pp. 220–234. [Google Scholar]
- 50.Auricchio P. Primatas do Brasil. Terra Brasilis Editora; São Paulo, Brazil: 1995. [Google Scholar]
- 51.Garber P.A., Paciulli I.M. Experimental field study of spatial memory and learning in wild capuchin monkeys (Cebus capucinus) Folia Primatol. 1997;68:236–253. doi: 10.1159/000157250. [DOI] [PubMed] [Google Scholar]
- 52.Porfirio G., Santos F.M., Foster V., Nascimento L.F., Macedo G.C., Barreto W.T.G., Fonseca C., Herrera H.M. Terrestriality of wild Sapajus cay (Illiger, 1815) as revealed by camera traps. Folia Primatol. 2017;88:1–8. doi: 10.1159/000464148. [DOI] [PubMed] [Google Scholar]
- 53.Visalberghi E. Responsiveness to objects in two social groups of tufted capuchin monkeys (Cebus apella) Am. J. Primatol. 1988;15:349–360. doi: 10.1002/ajp.1350150408. [DOI] [PubMed] [Google Scholar]
- 54.Deane L. Finding of Trypanosoma of the rangeli type in opossums of the species Didelphis marsupialis marsupialis in the state of Pará. Rev. Bras. Malariol. Doencas. Trop. 1958;10:451–458. [PubMed] [Google Scholar]
- 55.Brandão E.M.V., Xavier S.C.C., Carvalhaes J.G., D’Andrea P.S., Lemos F.G., Azevedo F.C., Cássia-Pires R., Jansen A.M., Roque A.L.R. Trypanosomatids in small mammals of an agroecosystem in central Brazil: Another piece in the puzzle of parasite transmission in an anthropogenic landscape. Pathogens. 2019;8:190. doi: 10.3390/pathogens8040190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ocaña-Mayorga S., Aguirre-Villacis F., Pinto C.M., Vallejo G.A., Grijalva M.J. Prevalence, genetic characterization, and 18S small subunit ribosomal RNA diversity of Trypanosoma rangeli in triatomine and mammal hosts in endemic areas for Chagas disease in Ecuador. Vector Borne Zoonotic Dis. 2015;15:732–742. doi: 10.1089/vbz.2015.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen A.M., Xavier S.C.C., Roque A.L.R. Trypanosoma cruzi transmission in the wild and its most important reservoir hosts in Brazil. Parasites Vectors. 2018;11:502. doi: 10.1186/s13071-018-3067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrigues M.S., Lima L., Xavier S.C.D.C., Herrera H.M., Rocha F.L., Roque A.L.R., Teixeira M.M.G., Jansen A.M. Uncovering Trypanosoma spp. diversity of wild mammals by the use of DNA from blood clots. Int. J. Parasitol. Parasites Wildl. 2019;8:171–181. doi: 10.1016/j.ijppaw.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miguel Pinto C., Ocaña-Mayorga S., Lascano M.S., Grijalva M.J. Infection by trypanosomes in marsupials and rodents associated with human dwellings in Ecuador. J. Parasitol. 2006;92:1251–1255. doi: 10.1645/GE-886R.1. [DOI] [PubMed] [Google Scholar]
- 60.Oliveira J.A., Bonvicino C.R. Ordem rodentia. In: Reis N.R.D., Peracchi A.L., Pedro W.A., de Lima I.P.d., editors. Mamíferos do Brasil. Suprema; Londrina, Brazil: 2006. pp. 347–400. [Google Scholar]
- 61.Charles-Dominique P., Atramentowicz M., Charles-Dominique M., Gerard H., Hladik A., Hladik C.M., Prévost M. Les mammiferes frugivores arboricoles nocturnes d’une foret Guyanaise: Inter-relations plants-animaux. Terre Vie. 1981;35:341–435. [Google Scholar]
- 62.Eisenberg J.F. The evolution of arboreal herbivores in the Class Mammalia. In: Montgomery G., editor. The Ecology of Arboreal Folivores. Smithsonian Inst. Press; Washington, DC, USA: 1978. pp. 135–152. [Google Scholar]
- 63.Attias N., Raíces D.S.L., Pessoa F.S., Albuquerque H., Jordão-Nogueira T., Modesto T.C., Bergallo H.D.G. Potential distribution and new records of Trinomys species (Rodentia: Echimyidae) in the state of Rio de Janeiro. Zoologia. 2009;26:305–315. doi: 10.1590/S1984-46702009000200013. [DOI] [Google Scholar]
- 64.Desbiez A.L.J., Kluyber D. The role of giant armadillos (Priodontes maximus) as physical ecosystem engineers. Biotropica. 2013;45:537–540. doi: 10.1111/btp.12052. [DOI] [Google Scholar]
- 65.Anacleto T.C.S., Marinho-Filho J. Hábito alimentar do tatu-canastras (Xenarthra, Dasypodidae) em uma área do cerrado do Brasil central. Rev. Bras. Zool. 2001;18:681–688. doi: 10.1590/S0101-81752001000300003. [DOI] [Google Scholar]
- 66.Vaz V.C., Santori R.T., Jansen A.M., Delciellos A.C., D’Andrea P.S. Notes on food habits of armadillos (Cingulata, Dasypodidae) and anteaters (Pilosa, Myrmecophagidae) at Serra da Capivara National Park (Piauí State, Brazil) Edentata. 2012;13:84–89. doi: 10.5537/020.013.0107. [DOI] [Google Scholar]
- 67.D’Alessandro A., Barreto P., Saravia N., Barreto S. Epidemiology of Trypanosoma cruzi in the oriental plains of Colombia. Am. J. Trop. Med. Hyg. 1984;33:1084–1095. doi: 10.4269/ajtmh.1984.33.1084. [DOI] [PubMed] [Google Scholar]
- 68.Scorza C., Urdaneta-Morales S., Tejero F. Trypanosoma (Herpetosoma) rangeli Tejera, 1920. Preliminary report on histopathology in experimentally infected mice. Rev. Inst. Med. Trop. Sao Paulo. 1986;28:371–378. doi: 10.1590/S0036-46651986000500014. [DOI] [PubMed] [Google Scholar]
- 69.Osório Y., Travi B.L., Palma G., Saravia N.G. Infectivity of Trypanosoma rangeli in a promonocytic mammalian cell line. J. Parasitol. 1995;81:687–693. doi: 10.2307/3283955. [DOI] [PubMed] [Google Scholar]
- 70.Ferreira L.D.L., Pereira M.H., Guarneri A.A. Revisiting Trypanosoma rangeli transmission involving susceptible and non-susceptible Hosts. PLoS ONE. 2015;10:e0140575. doi: 10.1371/journal.pone.0140575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lima L., Espinosa-Alvarez O., Hamilton P.B., Neves L., Takata C.S., Campaner M., Attias M., de Souza W., Camargo E.P., Teixeira M.M.G. Trypanosoma livingstonei: A new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasites Vectors. 2013;6:221. doi: 10.1186/1756-3305-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramirez J.D., Tapia-Calle G., Munoz-Cruz G., Poveda C., Lina M.R., Hincapié E., Guhl F. Trypanosome species in neo-tropical bats: Biological, evolutionary and epidemiological implications. Infect. Genet. Evol. 2014;22:250–256. doi: 10.1016/j.meegid.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lima L., Espinosa-Alvarez O., Pinto C.M., Cavazzana M., Jr., Pavan A.C., Carranza J.C., Lim B., Campaner M., Takata C.S.A., Camargo E.P., et al. New insights into the evolution of the Trypanosoma cruzi clade provided by a new trypanosome species tightly linked to Neotropical Pteronotus bats and related to an Australian lineage of trypanosomes. Parasites Vectors. 2015;8:657. doi: 10.1186/s13071-015-1255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rangel D.A., Lisboa C.V., Novaes R.L.M., Silva B.A., Souza R.D.F., Jansen A.M., Moratelli R., Roque A.L.R. Isolation and characterization of trypanosomatids, including Crithidia mellificae, in bats from the Atlantic Forest of Rio de Janeiro, Brazil. PLoS Negl. Trop. Dis. 2019;13:1–17. doi: 10.1371/journal.pntd.0007527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cavazzana M., Marcili A., Lima L., da Silva F.M., Junqueira A.C.V., Veludo H.H., Viola L.B., Campaner M., Nunes V.L.B., Paiva F., et al. Phylogeographic, ecological and biological patterns shown by nuclear (ssrRNA and gGAPDH) and mitochondrial (Cyt b) genes of trypanosomes of the subgenus Schizotypal parasitic in Brazilian bats. Int. J. Parasitol. 2010;40:345–355. doi: 10.1016/j.ijpara.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 76.Wilkinson G.S. Social grooming in the common 855 vampire bat, Desmodus rotundus. Anim. Behav. 1986;34:1880–1889. doi: 10.1016/S0003-3472(86)80274-3. [DOI] [PubMed] [Google Scholar]
- 77.Findley J.S. Bats: A Community Perspective. 1st ed. Cambridge University Press; New York, NY, USA: 1993. [Google Scholar]
- 78.Kalko E.K.V. Neotropical leaf-nosed bats (Phyllostomidae): “Whispering” bats or candidates for acoustic survey? In: Brigham M., Jones G., Kalko E.K.V., editors. Proceedings of a Workshop on Identification and Acoustic Monitoring of Bats. Bat Conservation International; Austin, TX, USA: 2002. pp. 63–69. [Google Scholar]
- 79.Thomas M.E., Rasweiler J.J., D’Alessandro A. Experimental transmission of the parasitic flagellates Trypanosoma cruzi and Trypanosoma rangeli between triatomine bugs or mice and captive neotropical bats. Mem. Inst. Oswaldo Cruz. 2007;102:559–565. doi: 10.1590/S0074-02762007005000068. [DOI] [PubMed] [Google Scholar]
- 80.Pífano F., Peñalver L.M., Medina R., Dominguez E. La infección natural del perro por el Trypanosoma rangeli. Primera comprobación de un reservorio vertebrado extra-humano de protozoario de la natureza. Gac. Med. Car. 1948;56:7. [Google Scholar]
- 81.D’alessandro A. Biology of Trypanosoma (Herpetosoma) rangeli, Tejera, 1920. In: Lumsden W.H.R., Evans D.A., editors. The Biology of the Kinetoplastida. Academic Press; New York, NY, USA: 1976. pp. 327–403. [Google Scholar]
- 82.Xavier S.C., Roque A.L., Bilac D., de Araújo V.A., da Costa Neto S.F., Lorosa E.S., da Silva L.F., Jansen A.M. Distantiae transmission of Trypanosoma cruzi: A new epidemiological feature of acute Chagas disease in Brazil. PLoS Negl. Trop. Dis. 2014;8:e2878. doi: 10.1371/journal.pntd.0002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.D’Alessandro A., de Hincapie O. Rhodnius neivai: A new experimental vector of Trypanosoma rangeli. Am. J. Trop. Med. Hyg. 1986;35:512–514. doi: 10.4269/ajtmh.1986.35.512. [DOI] [PubMed] [Google Scholar]
- 84.De Stefani Marquez D., Rodrigues-Ottaiano C., Monica Oliveira R., Pedrosa A.L., Cabrine-Santos M., Lages-Silva E., Ramírez L.E. Susceptibility of different triatomine species to Trypanosoma rangeli experimental infection. Vector Borne Zoonotic Dis. 2006;6:50–56. doi: 10.1089/vbz.2006.6.50. [DOI] [PubMed] [Google Scholar]
- 85.Gurgel-Gonçalves R., Galvão C., Costa J., Peterson A.T. Geographic distribution of Chagas disease vectors in Brazil based on ecological niche modelling. J. Trop. Med. 2012;2012:1–15. doi: 10.1155/2012/705326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Santos F.M., Jansen A.M., Mourão G.M., Jurberg J., Nunes A.P., Herrera H.M. Triatominae (Hemiptera, Reduviidae) in the Pantanal region: Association with Trypanosoma cruzi, different habitats and vertebrate hosts. Rev. Soc. Bras. Med. Trop. 2015;48:532–538. doi: 10.1590/0037-8682-0184-2015. [DOI] [PubMed] [Google Scholar]
- 87.Lima-Oliveira T.M., Fontes F.V.H.M., Lilioso M., Pires-Silva D., Teixeira M.M.G., Meza J.G.V., Harry M., Fileé J., Costa J., Valença-Barbosa C., et al. Molecular eco-epidemiology on the sympatric Chagas disease vectors Triatoma braziliensis and Triatoma petrocchiae: Ectopes, genetic variation, natural infection prevalence by trypanosomatids and parasite genotyping. Acta Trop. 2020;201:105188. doi: 10.1016/j.actatropica.2019.105188. [DOI] [PubMed] [Google Scholar]
- 88.Ingales B., Miles M.A., Campbell D.A., Tibayrenc M., Macedo A.M., Teixeira M.M., Schijman A.G., Llewellyn M.S., Lages-Silva E., Machado C.R., et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012;12:240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 89.Grisard E.C., Campbell D.A., Romanha A.J. Mini-exon gene sequence polymorphism among Trypanosoma rangeli strains isolated from distinct geographical regions. Parasitology. 1999;118:375–382. doi: 10.1017/S0031182098003928. [DOI] [PubMed] [Google Scholar]
- 90.Cuervo C., Lopez M.C., Puerta C. The Trypanosoma rangeli histone H2A gene sequence serves as a differential marker for KP1 strains. Infect. Genet. Evol. 2006;6:401–409. doi: 10.1016/j.meegid.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 91.Mittermeier R.A., Turner W.R., Larsen F.W., Brooks T.M., Gascon C. Global biodiversity conservation: The critical role of hotspots. In: Zachos F.E., Habel J.C., editors. Biodiversity Hotspots. Springer Publishers; London, UK: 2011. pp. 3–22. [Google Scholar]
- 92.Junk W.J., Cunha C.N., Wantzen K.M., Petermann P., Strussmann C., Marques M.I., Adis J. Biodiversity and its conservation in the Pantanal of Mato Grosso, Brazil. Aquat. Sci. 2006;68:278–309. doi: 10.1007/s00027-006-0851-4. [DOI] [Google Scholar]
- 93.Jansen A.M., Xavier S.C.D.C., Roque A.L.R. Landmarks of the Knowledge and Trypanosoma cruzi Biology in the Wild Environment. Front. Cell. Infect. Microbiol. 2020;6:10. doi: 10.3389/fcimb.2020.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sincero T.C., Stoco P.H., Steindel M., Vallejo G.A., Grisard E.C. Trypanosoma rangeli displays a clonal population structure, revealing a subdivision of KP1(−) strains and the ancestry of the Amazonian group. Int. J. Parasitol. 2015;45:225–235. doi: 10.1016/j.ijpara.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 95.Lisboa C.V., Mangia R.H., Luz S.L., Kluczkovski A., Jr., Ferreira L.F., Ribeiro C.T., Fernandes O., Jansen A.M. Stable infection of primates with Trypanosoma cruzi I and II. Parasitology. 2006;133:603–611. doi: 10.1017/S0031182006000722. [DOI] [PubMed] [Google Scholar]
- 96.Xavier S.C., Roque A.L., Lima V.D.S., Monteiro K.J., Otaviano J.C., Ferreira da Silva L.F., Jansen A.M. Lower richness of small wild mammal species and chagas disease risk. PLoS Negl. Trop. Dis. 2012;6:e1647. doi: 10.1371/journal.pntd.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malavazi P.F.N.S., Daudt C., Melchior L.A.K., Meneguetti D.U.O., Xavier S.C.C., Jansen A.M., Souza S.F., Roque A.L.R. Trypanosomes of vectors and domestic dogs in Trypanosoma cruzi transmission areas from Brazilian southwestern amazon: New mammalian host for Trypanosoma janseni. Acta Trop. 2020;210:105504. doi: 10.1016/j.actatropica.2020.105504. [DOI] [PubMed] [Google Scholar]
- 98.Vallejo G.A., Guhl F., Chiari E., Macedo A.M. Species specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammal hosts by polymerase chain reaction amplification of kinetoplast minicircle DNA. Acta Trop. 1999;72:203–212. doi: 10.1016/S0001-706X(98)00085-0. [DOI] [PubMed] [Google Scholar]
- 99.Borghesan T.C., Ferreira R.C., Takata C.S., Campaner M., Borda C.C., Paiva F., Milder R.V., Teixeira M.M., Camargo E.P. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013;164:129–152. doi: 10.1016/j.protis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 100.Noyes H.A., Stevens J.R., Teixeira M., Phelan J., Holz P. A nested PCR for the ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int. J. Parasitol. 1999;29:331–339. doi: 10.1016/S0020-7519(98)00167-2. [DOI] [PubMed] [Google Scholar]
- 101.Smith A., Clark P., Averis S., Lymbery A.J., Wayne A.F., Morris K.D., Thompson R.C.A. Trypanosomes in a declining species of threatened Australian marsupial, the brush-tailed bettong Bettongia penicillata (Marsupialia: Potoroidae) Parasitology. 2008;135:1329–1335. doi: 10.1017/S0031182008004824. [DOI] [PubMed] [Google Scholar]
- 102.Katoh S. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Darriba D., Taboada G., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chernomor O., von Haeseler A., Minh B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systemat. Biol. 2018 doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bandelt H.-J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 111.Polzin T., Daneschmand S.V. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 2003;31:12–20. doi: 10.1016/S0167-6377(02)00185-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are openly available in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) (accessed on 31 March 2021). The SSU rDNA accession numbers are provided in Table S1.