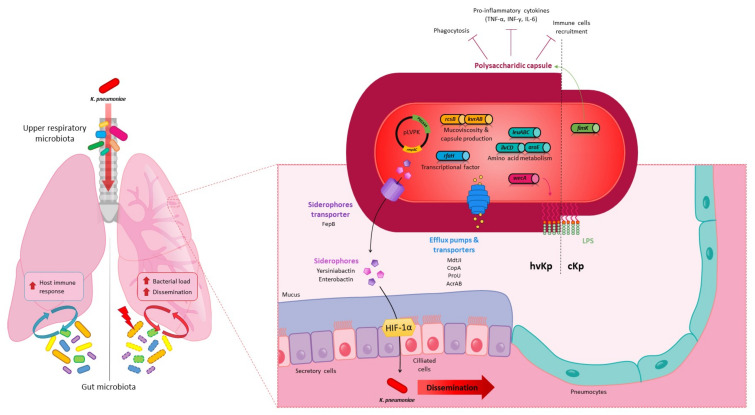
Figure 2.
K. pneumoniae in the lungs in an in vivo murine model Local (respiratory) and distal (gut) microbiota (yellow, blue, green, and purple ovals) can modulate the host immune response to K. pneumoniae (red oval) lung infection and K. pneumoniae pathogenicity unlike dysbiotic gut microbiota (yellow, blue, green, and purple ovals, dotted line). Virulence factors involved in lung colonization by classic (cKp, right side) or hypervirulent (hvKp, left side) K. pneumoniae strains: Capsule, overproduced in hvKp and finely regulated by many pathways (FimK, RcsB, KvrA, KvrB, RmpA, RmpC); Lipopolysaccharide (LPS), which depends on the wecA gene for the initiation of biosynthesis; Membrane proteins such as transporters and efflux pumps (MdtJI, CopA, ProU and AcrAB); Siderophores enterobactin (Ent) and yersiniabactin (Ybt) and FepB siderophore transporters are present in cKp, but their role in virulence and dissemination through the lung epithelial barrier via hypoxia inducible factor-1α (HIF-1 α) stabilization was only characterized for the hvKp strains; Critical fitness genes involved in the synthesis of essential branched chain and aromatic amino acids (leuABC, ilvCD and aroE) and RfaH transcriptional factor.