Abstract
Background
Community-acquired pneumonia (CAP) is a major driver of hospital antibiotic use. Efficient methods to identify patients treated for CAP in real time using the electronic health record (EHR) are needed. Automated identification of these patients could facilitate systematic tracking, intervention, and feedback on CAP-specific metrics such as appropriate antibiotic choice and duration.
Methods
Using retrospective data, we identified suspected CAP cases by searching for patients who received CAP antibiotics AND had an admitting International Classification of Diseases, Tenth Revision (ICD-10) code for pneumonia OR chest imaging within 24 hours OR bacterial urinary antigen testing within 48 hours of admission (denominator query). We subsequently explored different structured and natural language processing (NLP)–derived data from the EHR to identify CAP cases. We evaluated combinations of these electronic variables through receiver operating characteristic (ROC) curves to assess which best identified CAP cases compared to cases identified by manual chart review. Exclusion criteria were age <18 years, absolute neutrophil count <500 cells/mm3, and admission to an oncology unit.
Results
Compared to the gold standard of chart review, the area under the ROC curve to detect CAP was 0.63 (95% confidence interval [CI], .55–.72; P < .01) using structured data (ie, laboratory and vital signs) and 0.83 (95% CI, .77–.90; P < .01) when NLP-derived data from radiographic reports were included. The sensitivity and specificity of the latter model were 80% and 81%, respectively.
Conclusions
Creating an electronic tool that effectively identifies CAP cases in real time is possible, but its accuracy is dependent on NLP-derived radiographic data.
Keywords: algorithm, antibiotic stewardship, electronic, pneumonia
Key points.
Syndrome-based antibiotic stewardship can be limited by difficulty in finding cases for evaluation. We developed an electronic algorithm to prospectively identify community-acquired pneumonia cases. The use of natural language processing–derived radiographic data was necessary for optimal algorithm performance.
Nationally, there are approximately 600,000 hospital admissions for community-acquired pneumonia (CAP) annually, making CAP one of the most common drivers of antibiotic use in hospitalized patients [1–3]. Furthermore, in a substantial number of cases, antibiotics are not needed because there is no infection (ie, there is an alternative explanation for respiratory symptoms) or CAP therapy is suboptimal due to incorrect therapy choice, dose, route, or duration [4, 5]. Large-scale implementation of successful interventions to optimize CAP treatment has been limited by lack of efficient ways to find cases in real time for intervention.
The Joint Commission recommends that acute-care antimicrobial stewardship programs (ASPs) have procedures to assess appropriateness of antibiotics for CAP [6]. Even though antimicrobial stewardship (AS) dashboards have enhanced AS daily workflow, most of the reports pertain to the drugs (eg, drug-bug mismatch, duplicate therapy, duration >72 hours) rather than syndromes [7–9]. As ASPs may differ in their strategies to optimize antibiotic use (eg, preapproval or postprescription review with feedback) and the antibiotics they choose to target, developing an electronic tool that can efficiently and effectively identify patients with CAP creates a new opportunity for AS intervention regarding need for antibiotics, antibiotic choice, and duration.
Previous approaches to identify CAP relied on International Classification of Diseases (ICD) pneumonia codes, which are known to be imprecise for clinical purposes (eg, they are underutilized, they do not distinguish between different types of pneumonia), or did not identify patients in real time [10–12].
The aim of this study was to determine electronic indicators of CAP that could be incorporated into an automated tool to identify patients started on CAP therapy who likely had CAP.
METHODS
Study Design, Setting, and Population
We conducted a retrospective study of patients ≥18 years of age admitted to The Johns Hopkins Hospital, a 1162-bed tertiary academic hospital in Baltimore, Maryland. To develop a cohort of patients suspected of having CAP, we included admissions between 1 December 2018 and 31 March 2019 as there are usually more CAP cases in the winter months. Individuals who had an absolute neutrophil count ≤500 cells/mm3 or were admitted to an oncology unit were excluded because immunocompromised patients may not have typical CAP findings.
Development of the Electronic Algorithm to Identify Cap Cases
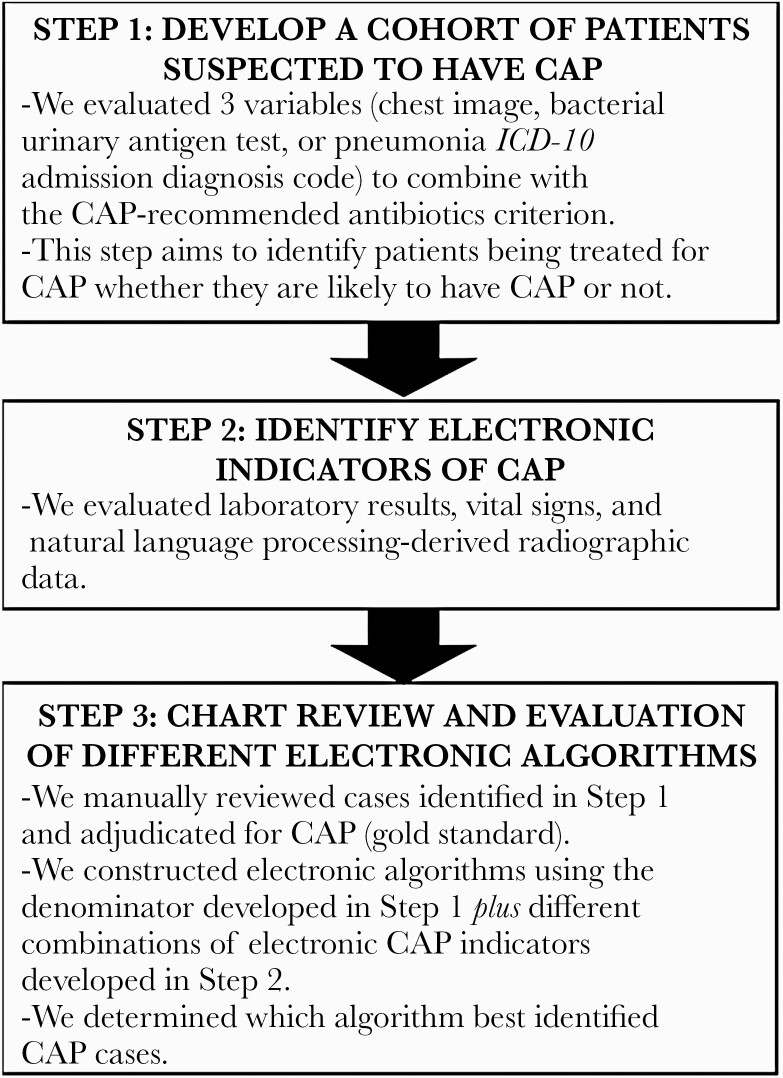
We used an iterative development process to build and refine the algorithm to identify patients likely to have CAP (Figure 1). Details of each step in this process are described below.
Figure 1.
Flow diagram of steps taken to develop the community-acquired pneumonia electronic algorithm. Abbreviations: CAP, community-acquired pneumonia; ICD-10, International Classification of Diseases, Tenth Revision.
Develop a Cohort of Patients Suspected to Have CAP (Denominator Query)
The first step in developing the electronic algorithm was to develop a cohort of patients being treated for CAP (whether they likely have it or not). Because the focus of this electronic tool is to identify opportunities to stop unnecessary antibiotic use when patients lack evidence of infection, and to optimize the antibiotic choice and duration for patients with CAP, we only included patients receiving select antibiotics for ≥48 hours from start in the emergency department (ED). We included the following antibiotics that are commonly prescribed for mild, moderate, and severe CAP at our institution: azithromycin in combination with ampicillin/sulbactam, ceftriaxone, or cefepime; vancomycin with aztreonam (used for patients with severe penicillin allergy); and moxifloxacin. Additional criteria used to identify potential CAP cases included the presence of 1 or more of the following: a chest image (chest radiograph [CXR] or chest computed tomography [CT]) within 24 hours of arrival to the ED, a bacterial urinary antigen test within 48 hours of ED arrival, and an ICD, Tenth Revision (ICD-10) admission diagnosis code for pneumonia documented in the ED or within 6 hours after inpatient admission (Supplementary Table 1). We did not include discharge pneumonia diagnosis codes because our aim was to develop an algorithm that could identify patients in real time. Natural language processing (NLP) was used to exclude chest imaging with an indication for endotracheal tube (ETT) or central line (CL) placement evaluation.
Identify Electronic CAP Indicators
The second step was to evaluate potential electronic indicators of CAP. Both structured variables (eg, laboratory test results, vital signs) and NLP-derived radiology results in our electronic health record system (Epic) were identified for inclusion in combination with the aforementioned denominator query (CAP-recommended antibiotic plus chest imaging or bacterial urinary antigen testing). NLP was used to identify radiograph and CT results consistent with an infectious process. Specifically, a structured NLP algorithm electronically assessed radiology reports stored in free-text fields in the database for the presence of “consolidation,” “consolidative opacities,” or “infiltrate” and excluded records containing negation of these terms. NLP-derived radiographic data are summarized in Supplementary Table 2.
Chart Review and Evaluation of Different Electronic Algorithms
Three authors (G. J., H. L., V. F.) reviewed the charts of patients identified as meeting the antibiotic criterion and at least 1 of the 3 other criteria (chest image, bacterial urinary antigen test, and admitting ICD-10 code for pneumonia) to determine which approach was optimally inclusive and parsimonious and to see if there were any additional modifications needed to the criteria (any additional inclusion or exclusion criteria). These cases were manually adjudicated for CAP using a standard CAP definition [13, 14] that included a new pulmonary infiltrate on chest imaging plus clinical evidence of infection (fever/hypothermia, new or increased cough, purulent sputum, pleuritic chest pain, abnormal leukocyte count).
To determine the effectiveness of different algorithms to predict CAP patients, we evaluated different combinations of CAP indicators using a multivariate logistic regression. Each combination of structured indicators and NLP outcomes was evaluated for the sensitivity and specificity of detecting true CAP patients (as determined by chart review), and the most parsimonious model was selected as the most efficacious.
The study was approved by the Johns Hopkins Medicine Institutional Review Board as a quality improvement project.
Statistical Analysis
Categorical and continuous variables were evaluated with χ 2 and Wilcoxon rank-sum tests, respectively. Predictive models of CAP were estimated using multivariate logistic regression for individual-level data including vital signs and laboratory data, radiographic reports, and combinations of these variables. Model fit was assessed by calculating the Akaike information criterion (AIC), the concordance statistic (c-statistic), which estimates the area under the curve, and the Hosmer-Lemeshow goodness-of-fit test against the gold standard of chart review. The value of c-statistics ranges from 0.5 to 1.0. A model with a c-statistic value closer to 1.0 means it has better discrimination (ability to separate patients with CAP from those without CAP). Hosmer-Lemeshow goodness of fit test helps identify models with good calibration (agreement between observed and predicted outcomes), and a P value > .05 indicates adequate calibration. The AIC combines both model fit and parsimony. AIC penalizes models with more parameters, making it a preferred technique to choose the best model. A lower AIC score indicates that the model provides a better fit. If, for example, 2 models had similar c-statistic values, it is best to choose the model with the lower AIC. Our sample size was calculated using the 10 events per variable rule of predictive models to minimize overfitting [15].
RESULTS
Cohort of Patients Suspected to Have CAP
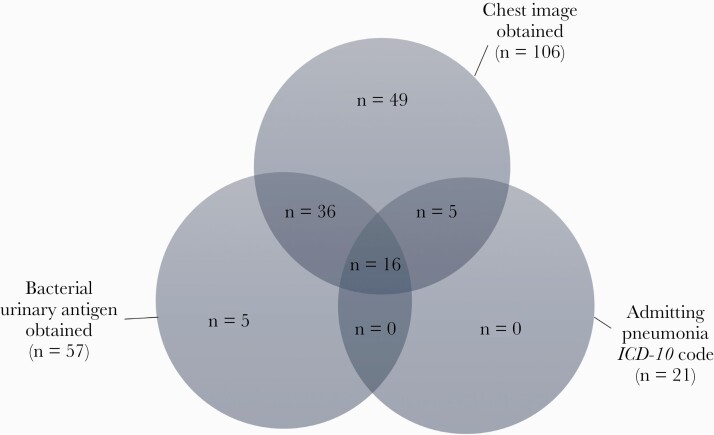
To define the cohort of patients being treated for suspected CAP (whether patients were likely to have CAP or not), we evaluated 3 criteria that could be combined with the receipt of antibiotics criterion. For a month-long period, the antibiotic criterion plus chest imaging yielded 106 cases, the antibiotic criterion plus bacterial urinary antigen testing as the companion criterion yielded 57 cases, and the antibiotic criterion plus admitting ICD-10 diagnosis codes for pneumonia yielded 21 cases (Figure 2). All 21 patients with a pneumonia-related ICD-10 admission diagnosis code were detected by either the chest imaging (5/21) or the chest imaging plus bacterial urinary antigen (16/21) group; thus, ICD-10 codes were not retained as a criterion to define the population of interest. For the chest imaging criterion, we added an exclusion criterion for patients undergoing imaging for ETT or CL placement using NLP. The final denominator query included patients ≥18 years with absolute neutrophil count >500/mm3, admitted to nononcology units who received CAP-recommended antibiotics for ≥48 hours from initiation in the ED AND had at least 1 of the following: bacterial urinary antigen test within 48 hours of ED arrival or chest imaging (CXR or CT) within 24 hours of ED arrival without an indication for ETT or CL placement evaluation.
Figure 2.
Results of the 3 criteria evaluated to select the population of interest (patients treated for suspected pneumonia). Abbreviation: ICD-10, International Classification of Diseases, Tenth Revision.
CAP Indicators and Final Electronic CAP Algorithm
The denominator query described above (CAP-recommended antibiotics plus a chest image or a bacterial urinary antigen test) was applied over a 3-month period, retrieving 173 patients, 41% (71/173) of whom were found to have CAP per the gold standard chart review. Alternative diagnoses among the 102 patients without CAP are summarized in Supplementary Table 3. Characteristics of the 173 patients are shown in Table 1. Age and sex were similar between patients with and without CAP. Variables that were significantly different between the CAP and no CAP groups included fever (45% vs 28%, P = .02), tachypnea (66% vs 47%, P = .01), and consolidation on chest imaging (80% vs 10%, P < .01).
Table 1.
Characteristics of Patients With and Without Community-Acquired Pneumonia per Gold Standard Manual Chart Review
| Characteristic | All Patients (n = 173) |
CAP (n = 71 [41%]) |
No CAP (n = 102 [59%]) |
P Value |
|---|---|---|---|---|
| Male sex | 96 (55.5) | 42 (59.2) | 54 (52.9) | .41 |
| Age, y, median (IQR) | 58 (46–67) | 58 (48–67) | 58 (41–67) | .38 |
| Fever (≥38°C) | 61 (35.3) | 32 (45.1) | 29 (28.4) | .02 |
| Hypothermia (≤36°C) | 84 (48.6) | 36 (50.7) | 48 (47.1) | .63 |
| Subjective fever or chills | 8 (4.6) | 4 (5.6) | 4 (3.9) | .59 |
| Tachypnea (≥24 breaths/min) | 95 (54.9) | 47 (66.2) | 48 (47.1) | .01 |
| Hypoxemia (SpO2 <92% or supplemental O2 received) | 121 (69.9) | 54 (76.1) | 67 (65.7) | .14 |
| WBC count >12 000 cells/mm3 | 84 (48.6) | 39 (54.9) | 45 (44.1) | .16 |
| WBC count <4000 cells/mm3 | 19 (11.0) | 9 (12.7) | 10 (9.8) | .55 |
| Abnormal pro-BNP (>125 pg/mL) | 66 (38.2) | 34 (47.9) | 32 (31.4) | .09 |
| Respiratory viruses | 33 (19.1) | 9 (12.7) | 24 (23.5) | .07 |
| Influenza viruses | 17 (9.8) | 7 (9.9) | 10 (9.8) | |
| Respiratory syncytial virus | 8 (4.6) | 1 (1.4) | 7 (6.9) | |
| Rhinoviruses/enteroviruses | 6 (3.5) | 1 (1.4) | 5 (4.9) | |
| Adenoviruses | 1 (0.6) | … | 1 (1.0) | |
| Metapneumovirus | 1 (0.6) | … | 1 (1.0) | |
| Blood culture for CAP pathogen | 4 (2.3) | 2 (2.8) | 2 (2.0) | |
| Staphylococcus aureus | 2 (1.2) | … | 2 (2.0) | |
| Streptococcus pneumoniae | 1 (0.6) | 1 (1.4) | … | |
| Haemophilus influenzae | 1 (0.6) | 1 (1.4) | … | |
| Bacterial urinary antigen | 4 (2.3) | 3 (4.2) | 1 (1.0) | .16 |
| Streptococcus pneumoniae | 4 (2.3) | 3 (4.2) | 1 (1.0) | |
| Sputum culture | 7 (4.0) | 7 (9.9) | … | |
| Staphylococcus aureus | 3 (1.7) | 3 (4.2) | … | |
| Haemophilus influenzae | 3 (1.7) | 3 (4.2) | … | |
| Moraxella catarrhalis | 1 (0.6) | 1 (1.4) | … | |
| Consolidation on either CXR or CTa | 69 (40) | 52 (73.3) | 17 (16.7) | <.01 |
| Consolidation on CXR | 30 (17) | 23 (32.4) | 7 (6.9) | <.01 |
| Consolidation on CT | 52 (30) | 41 (58) | 11 (11) | <.01 |
| Infiltrate on either CXR or CTa | 10 (5.8) | 6 (8.5) | 4 (3.9) | .20 |
Data are presented as No. (%) unless otherwise indicated. Bold P values indicate statistical significance.
Abbreviations: BNP, brain natriuretic peptide; CAP, community-acquired pneumonia; CXR, chest radiograph; CT, computed tomography; IQR, interquartile range; O2, oxygen; SpO2, oxygen saturation; WBC, white blood cell.
aPatients who had consolidation on both CXR and chest CT were counted only once.
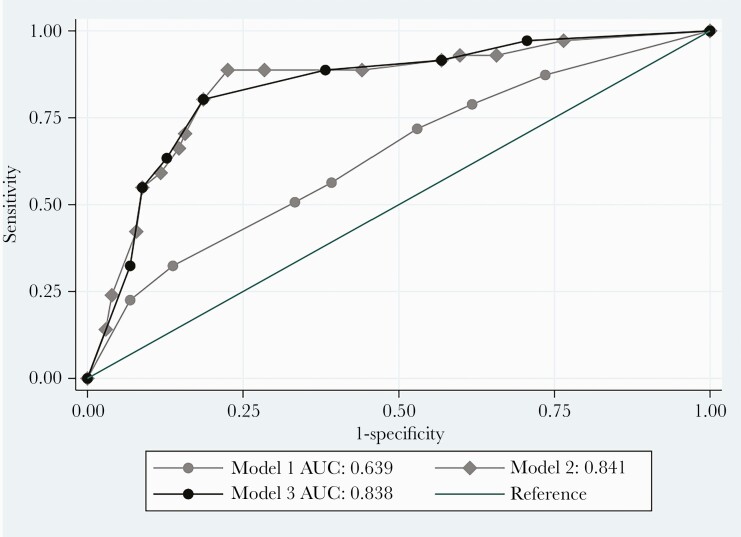
A full list of the structured variables derived from laboratory data and vital signs and NLP-derived data from radiographic reports that were investigated for their sensitivity and specificity of detecting CAP indicators are summarized in Supplementary Table 4. Models that included both laboratory and vital signs data plus NLP-derived radiographic data demonstrated the best performance (Figure 3, Supplementary Table 5). Compared to the gold standard, the area under the ROC curve (AUC) was 0.63 (95% confidence interval [CI], .55–.72; P = .04) for an algorithm that included fever, tachypnea, and leukocytosis (Figure 3, model 1), 0.84 (95% CI, .77–.90; P < .01) when NLP-derived data from radiographic reports were added to fever, tachypnea, and leukocytosis (Figure 3, model 2), and 0.83 (95% CI, .77–.90) for tachypnea, leukocytosis, and NLP-derived radiographic data (Figure 3, model 3). The sensitivity and specificity were 77% and 84%, respectively, for model 2, and 80% and 81%, respectively, for model 3. Adding positive sputum/blood culture or positive bacterial urinary antigen did not improve performance of the algorithm (Supplementary Table 5). In evaluating NLP-derived radiographic data, we found that inclusion of both CXR and chest CT performed better than either image alone (Supplementary Table 5).
Figure 3.
Receiver operating characteristic curves for composite community-acquired pneumonia indicators. Model 1 (no natural language processing): temperature ≥38°C plus tachypnea plus leukocytosis. Model 2: model 1 plus “consolidation” or “infiltrate” on chest radiograph (CXR) or chest computed tomography (CT). Model 3: tachypnea plus leukocytosis plus “consolidation” or “infiltrate” on CXR or CT. Abbreviation: AUC, area under the curve.
DISCUSSION
Using CAP-recommended antibiotics plus bacterial urinary antigen testing and/or chest imaging in combination with laboratory (leukocytosis), vital signs (tachypnea), and NLP-derived radiographic data, we developed an electronic algorithm that effectively identifies CAP cases among nonneutropenic hospitalized adults.
Lower respiratory infections are a major driver of antibiotic use in hospitalized patients [2], and up to 30% of patients receiving CAP therapy are found to have alternative diagnoses [4]. Approaches to increase ASP efficiency in finding cases for intervention are urgently needed. Having an electronic algorithm that can identify patients receiving antibiotics for suspected CAP in real time and distinguishing which patients likely have CAP from those who may have an alternative explanation for their respiratory symptoms provides an intervention opportunity for ASPs that may not be able to systematically perform postprescription review and feedback on all antibiotics. Furthermore, this algorithm would allow for tracking of CAP-specific antibiotic use metrics and inform syndrome-specific interventions and feedback to clinicians. In this study, we report step-by-step how we developed such an electronic algorithm. The first step was to generate the population of interest (those receiving antibiotics for suspected CAP). Because CAP antibiotics are prescribed in many other circumstances, we narrowed the search by adding additional clinical criteria that can be electronically retrieved. We found that admission ICD-10 codes for pneumonia were underutilized as previously reported in the literature [11, 12] and did not retrieve any unique cases. We explored 2 additional criteria, chest imaging and bacterial urinary antigen tests, because these are commonly included in the workup of CAP. Although we limited the generalizability of the electronic algorithm by including bacterial urinary antigen orders, only 5% of the denominator cases would have been missed without this criterion that were not captured by the chest image order criterion, and the algorithm likely would be valuable for institutions where bacterial urinary antigen testing is not routinely used or available. The second step in developing the electronic algorithm was to identify indicators of CAP that could be combined with the denominator criteria to determine which combinations worked best to detect CAP cases. In ROC curve analysis, inclusion of NLP-derived radiographic data along with vital signs and laboratory data (tachypnea and leukocytosis) significantly improved CAP case identification, and inclusion of both CXR and CT resulted in a better AUC than models with either of these alone. These findings are in agreement with prior studies that showed that inclusion of text analyses of chest imaging reports improves the performance of case detection algorithms [10]. The electronic algorithm can be adjusted further and adapted to local needs. For example, while we included azithromycin in the antibiotic criterion, doxycycline could be added if regularly used for atypical coverage in CAP regimens in combination with other CAP antibiotics. Similarly, the antibiotic criterion of the algorithm would need to be updated if there are local updates to CAP antibiotic recommendations. The electronic algorithm also identifies patients receiving antibiotics for suspected CAP who do not appear to have CAP (eg, patients with heart failure), allowing ASPs to intervene to stop or modify antibiotic therapy.
Previous electronic pneumonia algorithms have concentrated on ICD diagnosis codes for case identification; however, because they do not accurately identify all patients with CAP or distinguish CAP from health care–associated pneumonia, their use alone is not helpful to guide stewardship interventions [10, 12, 16]. Our tool is innovative because it includes receipt of CAP-recommended antibiotics for ≥48 hours from ED initiation as a selection criterion to identify patients who were thought to have an infection by the treating providers. Next steps include using this algorithm to build reports into our AS dashboard as an actionable tool to promote appropriate use of antibiotics in CAP patients, including integration with additional clinical data such as allergy history and microbiologic data to flag inappropriate antibiotic choices based on local recommendations (eg, moxifloxacin for a patient without a penicillin allergy).
There are limitations to our study. Antibiotic receipt was one of the inclusion criteria to find cases; hence, patients who may have had CAP but were not treated with antibiotics are not identified with the electronic algorithm. However, this algorithm is intended to address inappropriate antibiotic use rather than missing treatment. Similarly, patients with CAP who received antibiotics not included in the algorithm will not be identified; however, guideline compliance is high at our institution. We evaluated performance of the electronic algorithm against a gold standard of manual chart review using the Infectious Diseases Society of America definition of CAP [14]; however, a true gold standard CAP diagnosis remains elusive [17]. We evaluated ICD-10 pneumonia codes as a possible criterion to select patients suspected of having CAP. Because a diagnosis of pneumonia may not be entered by ED providers due to overlapping symptoms of CAP and noninfectious processes such as heart failure, we included ICD pneumonia codes entered in the ED or up to 6 hours after inpatient admission. The algorithm depends on radiographic data, which may represent a barrier to reproducibility of the tool in institutions with electronic health record systems that do not integrate radiographic data or when most patients are not admitted through the emergency department and have their CXRs performed outside of the hospital. Although the NLP-derived radiographic variables were easily found in electronic data, information technology resources and expertise were required to extract them; it may be challenging for sites without these resources to operationalize such an algorithm. Additionally, as the tool was built as a predictive algorithm, its effectiveness may differ between institutions, and extensions to examine the generalizability of the tool in other institutions are ongoing. We have not yet implemented this tool within our AS dashboard, and further research is needed to determine the impact of the tool on patient management, outcomes, and antibiotic use data. The number of patients evaluated was relatively small, although a parsimonious number of predictor variables were evaluated and methods to avoid overfitting the model were used [15]. We chose winter months to develop the algorithm because CAP cases peak in these months. Additional validation may be needed to ensure that the electronic algorithm maintains accuracy when applied to other hospitals and other seasons and during viral pandemics. Similarly, further research is needed to validate the electronic CAP algorithm in neutropenic and oncology patients who were excluded from this study due to their atypical CAP presentations, and to characterize the impact of other immunocompromising conditions such as human immunodeficiency virus infection on the accuracy of the electronic CAP algorithm.
In summary, we developed an electronic algorithm that can effectively identify CAP in adult hospitalized patients; however, the use of an NLP-derived radiographic criterion appears necessary for optimal performance, limiting its reproducibility in settings without access to such data. Our algorithm is a practical starting point for syndrome-based AS interventions.
Supplementary Material
Notes
Patient consent statement. The design of this work was approved by The Johns Hopkins Medicine Institutional Review Board.
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: IDWeek 2020, Virtual, 21–25 October 2020. Poster 910938.
References
- 1. Fridkin S, Baggs J, Fagan R, et al. ; Centers for Disease Control and Prevention. Vital signs: improving antibiotic use among hospitalized patients. MMWR Morb Mortal Wkly Rep 2014; 63:194–200. [PMC free article] [PubMed] [Google Scholar]
- 2. Magill SS, Edwards JR, Beldavs ZG, et al. . Prevalence of antimicrobial use in US acute care hospitals, May-September 2011. JAMA 2014; 312:1438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee JS, Giesler DL, Gellad WF, Fine MJ. Antibiotic therapy for adults hospitalized with community-acquired pneumonia: a systematic review. JAMA 2016; 315:593–602. [DOI] [PubMed] [Google Scholar]
- 4. Avdic E, Cushinotto LA, Hughes AH, et al. . Impact of an antimicrobial stewardship intervention on shortening the duration of therapy for community-acquired pneumonia. Clin Infect Dis 2012; 54:1581–7. [DOI] [PubMed] [Google Scholar]
- 5. Tamma PD, Avdic E, Keenan JF, et al. . What is the more effective antibiotic stewardship intervention: preprescription authorization or postprescription review with feedback? Clin Infect Dis 2017; 64:537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The Joint Commission. New antimicrobial stewardship standard. Available at: https://www.jointcommission.org/-/media/enterprise/tjc/imported-resource-assets/documents/new_antimicrobial_stewardship_standardpdf.pdf?db=web&hash=69307456CCE435B134854392C7FA7D76. Accessed 12 May 2021.
- 7. Forrest GN, Van Schooneveld TC, Kullar R, et al. . Use of electronic health records and clinical decision support systems for antimicrobial stewardship. Clin Infect Dis 2014; 59(Suppl 3):S122–33. [DOI] [PubMed] [Google Scholar]
- 8. Ghamrawi RJ, Kantorovich A, Bauer SR, et al. . Evaluation of antimicrobial stewardship-related alerts using a clinical decision support system. Hosp Pharm 2017; 52:679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dzintars K FV, Avdic E, Smith J, et al. . Development of the antimicrobial stewardship module in an electronic health record: options to enhance daily antimicrobial stewardship activities [manuscript published online ahead of print 27 May 2021]. Am J Health-Syst Pharm 2021. doi:10.1093/ajhp/zxab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeLisle S, Kim B, Deepak J, et al. . Using the electronic medical record to identify community-acquired pneumonia: toward a replicable automated strategy. PLoS One 2013; 8:e70944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Livorsi DJ, Linn CM, Alexander B, et al. . The value of electronically extracted data for auditing outpatient antimicrobial prescribing. Infect Control Hosp Epidemiol 2018; 39:64–70. [DOI] [PubMed] [Google Scholar]
- 12. Higgins TL, Deshpande A, Zilberberg MD, et al. . Assessment of the accuracy of using ICD-9 diagnosis codes to identify pneumonia etiology in patients hospitalized with pneumonia. JAMA Netw Open 2020; 3:e207750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bordón J, Peyrani P, Brock GN, et al. ; CAPO Study Group. The presence of pneumococcal bacteremia does not influence clinical outcomes in patients with community-acquired pneumonia: results from the Community-Acquired Pneumonia Organization (CAPO) international cohort study. Chest 2008; 133:618–24. [DOI] [PubMed] [Google Scholar]
- 14. Mandell LA, Wunderink RG, Anzueto A, et al. ; Infectious Diseases Society of America; American Thoracic Society. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007; 44(Suppl 2):S27–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riley RD, Ensor J, Snell KIE, et al. . Calculating the sample size required for developing a clinical prediction model. BMJ 2020; 368:m441. [DOI] [PubMed] [Google Scholar]
- 16. Aronsky D, Chan KJ, Haug PJ. Evaluation of a computerized diagnostic decision support system for patients with pneumonia: study design considerations. J Am Med Inform Assoc 2001; 8:473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Musher DM, Thorner AR. Community-acquired pneumonia. N Engl J Med 2014; 371:1619–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.