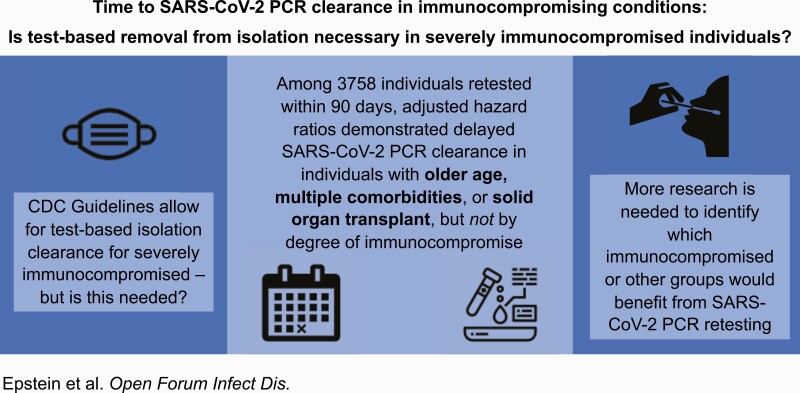
Abstract
To determine the association between immunosuppression and time to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) polymerase chain reaction (PCR) clearance, we studied 3758 adults retested following initial SARS-CoV-2 infection. Cox proportional hazards models demonstrated delayed PCR clearance with older age, multiple comorbidities, and solid organ transplant but not by degree of immunocompromise. These findings challenge current retesting practices.
Keywords: SARS-CoV-2, coronavirus disease 2019 (COVID-19), virus shedding, polymerase chain reaction, immunocompromised host, immunosuppression, disease severity
Few studies support SARS-CoV-2 retesting practices to end isolation. Among 3758 adults retested after SARS-CoV-2 infection, we observed delayed PCR clearance with older age, multiple comorbidities and solid organ transplant, but not other immunocompromising conditions. This challenges current retesting guidance.
Graphical Abstract
Graphical Abstract.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral shedding duration and the length of time individuals remain infectious have important implications for transmission and control of coronavirus disease 2019 (COVID-19). Immunocompromised individuals shed viruses such as influenza, cytomegalovirus, and norovirus longer than immunocompetent persons [1, 2]; however, little is known about the effect of immunosuppression on duration of SARS-CoV-2 culture or polymerase chain reaction (PCR) positivity. At least 1 study demonstrated prolonged SARS-CoV-2 PCR positivity in kidney transplant recipients, albeit without a comparison group [3], and rare cases highlight potential for prolonged, intermittent shedding with immunosuppressive medications [4, 5].
Individuals with SARS-CoV-2 infection may remain PCR-positive for many weeks [6–9], but with RNA concentrations and cultivability of infectious virus generally decreasing over time [8, 10–13]. Although studies are limited, very few have successfully cultured infectious virus past 20 days of illness [4, 5, 8, 10–13]. One high-risk household contact study found that all transmission events occurred within 5 days of symptom onset in the index patient [14]. Similarly, among 285 individuals with COVID-19 symptom and PCR positivity recurrence following initial symptom resolution, no secondary transmission occurred among 790 household contacts during relapse [15]. Collectively, these data suggest that PCR positivity alone after day 10–20 is not associated with transmission likelihood. The US Centers for Disease Control and Prevention (CDC) guidelines recommend symptom-based clearance from transmission-based precautions in most circumstances, but recommend considering test-based clearance in severely immunocompromised individuals [16]. Test-based clearance, however, can increase hospital cost and length of stay by preventing timely discharge or delaying necessary care [17, 18]. The limited studies that compare factors associated with SARS-CoV-2 PCR positivity duration include few immunocompromised patients or lack a comparison group [3, 7, 9, 19]. We therefore sought to compare time to SARS-CoV-2 PCR clearance by degree of immunocompromise, COVID-19 severity, age, and comorbidities in a large inpatient and outpatient sample.
METHODS
Setting
Boston Medical Center (BMC) is an urban academic medical center and the largest safety-net hospital in New England. Per CDC and local department of health guidance, in April 2020, BMC implemented a test-based strategy to end isolation precautions for immunocompromised individuals. We used the CDC’s “severely immunocompromised” definition for removal of transmission-based precautions: individuals receiving active chemotherapy or chronic high-dose steroids (>20 mg/d prednisone equivalent for >14 days), who received a transplant in the past year, and those with HIV with a CD4 count <200 cells/μL or a primary immunodeficiency [16]. Before August 2020, we also retested individuals who received acute or chronic biologic therapy and all transplant recipients. Many immunocompetent patients were retested for precaution removal during prolonged inpatient stays, subsequent admissions or emergency department visits, for clearance for work, or preceding discharge to dialysis centers and/or congregate living facilities, or for visiting a hospitalized newborn. Before retesting, both inpatients and outpatients required symptom-based clearance per CDC recommendations (initially 14 days after symptom onset and ≥3 days after improvement began; later 10 days after symptom onset or positive test and ≥24 hours after improvement [16]). If the initial retest was positive, guidance recommended retesting every 3 days until achieving 2 negative tests ≥24 hours apart.
Retesting occurred inpatient for still-admitted individuals, through a drive-through preprocedure testing site for asymptomatic patients, at home through an ambulance-based hospital partnership for immunocompromised patients and those unable to present for testing, or at an outpatient testing clinic. The SARS-CoV-2 PCR tests utilized varied by supply availability. All BMC samples were nucleic acid amplification tests, which received US Food and Drug Administration Emergency Use Authorization (Supplementary Table 1). Ninety-three percent were nasopharyngeal (NP) specimens performed in-house at BMC; a minority were anterior nares (5.6%) or lower respiratory specimens (1.3%).
Data Collection
We included all adults aged 18–99 years with PCR-confirmed SARS-CoV-2 infection who received ≥1 SARS-CoV-2 PCR retest at BMC within 90 days of initial positive test (as data suggest re-infection is unlikely within the first 90 days [10]). We included all test results recorded in the electronic health record (EHR), including tests performed outside BMC. We extracted data on demographics, medications, anthropomorphic measures, SARS-CoV-2 PCR testing, and underlying medical conditions (classified using International Classification of Diseases, 10th Edition, codes from individuals’ active condition lists or encounter diagnoses) from BMC’s EHR through the COVID Data Repository. Using prospectively designed EHR templates, we captured COVID-19 symptom onset date and medical history. Individuals were assigned 1 of 5 mutually exclusive COVID-19 disease severity categories: exclusive outpatient management, non–intensive care unit (ICU) inpatient hospitalization, ICU hospitalization, deceased, or unknown severity (individuals with initial external testing results imported into BMC’s EHR). For nondeceased individuals, the severity category was based on the highest level of care received within 14 days of the initial positive test; all individuals who died before the study end date were classified as deceased. We obtained cycle thresholds (Cts) or cycle numbers (CNs) for the subset of positive tests with available data (those performed on a platform with recorded quantitative cycle values). The Boston University Medical Campus Institutional Review Board approved this study as exempt human subject research.
Primary Outcome and Predictor
The primary outcome was time to SARS-CoV-2 PCR clearance, defined as the number of days between first positive and first negative PCR without a subsequent repeat positive PCR. Those without a negative PCR or for whom >1 day elapsed between last positive and first negative PCR were right- or interval-censored, respectively. For those with date of symptom onset recorded, we additionally examined time between symptom onset and first negative PCR. Following CDC guidelines, we classified individuals as severely immunocompromised as described above. Moderate immunocompromise included those receiving other immunosuppressive medications, HIV with CD4 ≥200 cells/μL, and transplant >1 year prior (Table 1).
Table 1.
Demographic Characteristics, Comorbidities, and Time to SARS-CoV-2 PCR Clearance in Patients Followed at Boston Medical Center for SARS-CoV-2 Infection and Retested Within 90 Days, March 2020–February 2021
| Characteristic | No. (%) (n = 3758) | Median (IQR) Time to First Negative PCR,a d | Unadjusted Hazard Ratio (95% CI)b | Adjusted Hazard Ratio (95% CI)b | Adjusted Hazard Ratio (95% CI)b |
|---|---|---|---|---|---|
| Age, mean (SD, range), y | 48.8 (16.4, 18–99) | — | 0.995 (0.992–0.997) | 0.997 (0.994–1.000) | 0.996 (0.993–0.999) |
| Age quartiles, y | |||||
| 18–35 | 947 (25.2) | 13 (4–28) | Ref. | — | — |
| 36–48 | 937 (24.9) | 16 (5–26) | 0.94 (0.83–1.05) | — | — |
| 49–60 | 933 (24.8) | 17 (8–30) | 0.87 (0.78–0.98) | — | — |
| >60 | 941 (25.0) | 21 (6–33) | 0.81 (0.72–0.90) | — | — |
| Sex | |||||
| Male | 1677 (44.6) | 18 (6–31) | 0.97 (0.89–1.05) | — | — |
| Female | 2081 (55.5) | 14 (7–29) | Ref. | — | — |
| Race/ethnicity | |||||
| Black, non-Hispanic | 1123 (29.9) | 14 (6–27) | 1.10 (1.00–1.21) | — | — |
| Hispanic or Latino | 1739 (46.3) | 18 (6–31) | Ref. | — | — |
| White, non-Hispanic | 509 (13.5) | 18 (6–33) | 0.95 (0.84–1.08) | — | — |
| Other race (non-Hispanic) | 128 (3.4) | 17 (3–29) | 1.06 (0.84–1.34) | — | — |
| Unknown/declined | 259 (6.9) | 15 (7–27) | 1.08 (0.91–1.28) | — | — |
| COVID-19 severity | |||||
| Deceased | 91 (2.4) | 26 (11–33) | 0.67 (0.50–0.91) | 0.77 (0.56–1.04) | 0.76 (0.56–1.03) |
| ICU | 180 (4.8) | 21 (2–25) | 0.93 (0.77–1.12) | 1.01 (0.83–1.22) | 1.04 (0.86–1.27) |
| Hospitalized, no ICU | 664 (17.7) | 21 (7–32) | 0.84 (0.74–0.96) | 0.91 (0.80–1.04) | 0.92 (0.80–1.05) |
| Not hospitalized | 1012 (26.9) | 13 (7–27) | Ref. | Ref. | Ref. |
| Unknown | 1811 (48.2) | 16 (7–29) | 0.88 (0.79–0.97) | 0.86 (0.78–0.95) | 0.87 (0.79–0.96) |
| Immunocompromise categoryc | |||||
| Severe | 277 (7.4) | 22 (4–34) | 0.93 (0.80–1.08) | 0.98 (0.84–1.15) | — |
| Active chemotherapy | 212 (5.6) | 19 (1–34) | 0.98 (0.82–1.16) | — | — |
| High-dose steroids | 55 (1.5) | 23 (6–33) | 0.81 (0.59–1.12) | — | — |
| HIV with CD4 <200 | 10 (0.3) | 27 (8–27) | 0.72 (0.35–1.46) | — | — |
| Moderate | 159 (4.2) | 20 (8–33) | 0.82 (0.67–1.01) | 0.86 (0.70–1.05) | — |
| Solid organ transplantd | 43 (1.1) | 31 (14–47) | 0.60 (0.40–0.90) | — | 0.64 (0.42–0.97) |
| Chronic biologic | 23 (0.6) | 30 (10–33) | 0.78 (0.48–1.26) | — | — |
| HIV with CD4 >200 | 33 (0.9) | 15 (6–29) | 0.85 (0.56–1.29) | — | — |
| Othere | 60 (1.6) | 20 (8–27) | 1.08 (0.78–1.50) | — | — |
| Immunocompetent | 3322 (88.4) | 16 (6–29) | Ref. | Ref. | — |
| Comorbidities | |||||
| Diabetes | 642 (17.1) | 22 (9–36) | 0.77 (0.69–0.86) | — | 0.82 (0.73–0.93) |
| Coronary artery disease | 150 (4.0) | 21 (6–40) | 0.81 (0.66–1.01) | — | 0.94 (0.75–1.17) |
| Hypertension | 1120 (29.8) | 21 (9–33) | 0.90 (0.82–0.98) | — | 1.07 (0.96–1.20) |
| ESRD | 98 (2.6) | 21 (13–33) | 0.76 (0.59–1.00) | — | 0.97 (0.72–1.30) |
| Cirrhosis | 43 (1.1) | 24 (1–30) | 0.83 (0.57–1.20) | — | — |
| Obesity | 1758 (46.8) | 19 (7–31) | 0.89 (0.82–0.96) | — | 0.90 (0.83–0.98) |
| Chronic lung disease | 413 (11.0) | 21 (9–34) | 0.91 (0.80–1.04) | — | 0.94 (0.82–1.07) |
| Rheumatologic disease | 85 (2.3) | 24 (1–36) | 0.72 (0.55–0.94) | — | 0.75 (0.57–0.98) |
| None of the above | 2166 (57.6) | 15 (6–28) | Ref. | Ref. | — |
| ≥1 of the above | 876 (23.3) | 16 (6–29) | 0.92 (0.83–1.02) | 0.96 (0.86–1.06) | — |
| ≥2 of the above | 507 (13.5) | 20 (9–30) | 0.84 (0.74–0.95) | 0.89 (0.77–1.02) | — |
| ≥3 of the above | 209 (5.6) | 24 (10–36) | 0.68 (0.57–0.82) | 0.73 (0.60–0.88) | — |
| Other conditions (in the absence of other immunocompromising conditions) | |||||
| Acute IL-6 or IL-1 inhibitorf | 154 (4.1) | 24 (11–30) | 0.87 (0.72–1.06) | — | — |
| Acute corticosteroidf | 328 (8.7) | 17 (7–32.5) | 0.95 (0.82–1.10) | — | — |
| B-cell function deficiencyg | 5 (0.1) | 32.5 (18–47.5) | 0.60 (0.22–1.60) | — | — |
| Pregnant | 203 (5.4) | 14 (8–32) | 0.90 (0.74–1.08) | — | — |
Boldface indicates P value <.05.
Abbreviations: COVID-19, coronavirus disease 2019; ESRD, end-stage renal disease; ICD-10, International Classification of Diseases, 10th edition; ICU, intensive care unit; IL, interleukin; IQR, interquartile range; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
aNonparametric survival analysis accounting for interval-censored data utilizing the Expectation–Maximization-Iterative Convex Minorant algorithm to impute median survival estimates and standard errors.
bHazard ratios for time to SARS-CoV-2 PCR clearance obtained using Cox proportional hazards regression accounting for interval-censored data, unadjusted and adjusted for (1) age, disease severity, immunocompromise category, and variables with P < .2 in the univariate analysis and (2) age, disease severity, and individual variables with P < .2 in the univariate analysis.
cImmunocompromise categories are mutually exclusive—if individuals met multiple categories, they were categorized as the most severe (first listed) category.
dSolid organ transplant is considered moderate rather than severe, as all chart-reviewed patients in this category had their transplant >1 year before COVID-19 diagnosis.
eOther immunocompromise includes: rheumatologic conditions, myelofibrosis, asplenia, chronic neutropenia, and other blood and immune disorders (identified by ICD-10 codes on the problem list); immunocompromising medications not included in active chemotherapy category: hydroxyurea, methotrexate, azathioprine.
fFor COVID-19 treatment; only includes individuals not otherwise categorized as immunocompromised.
gOn chronic medication that suppresses B-cell function directly or indirectly (rituximab, belimumab, natalizumab, or daratumumab) in the absence of concurrent cytotoxic chemotherapy.
Analysis
We estimated median time to first negative SARS-CoV-2 PCR using Kaplan-Meier survival estimators and Cox proportional hazards models accounting for interval censoring. We adjusted multivariable Cox regression models for age, immunocompromise category, and COVID-19 severity, as each of these variables has been previously associated with prolonged PCR positivity [3–7], and for all variables with P < .2 in unadjusted analyses. In a second model, we used the same criteria but incorporated only individual immunocompromising conditions or comorbidities with P < .2 rather than the larger categories. All analyses were done in SAS, version 9.4 (Cary, NC, USA).
We performed 4 sensitivity analyses. First, to mitigate possible bias in PCR clearance time estimates introduced by infrequent testing, we assessed only individuals with ≥2 negative tests ≤7 days apart, as this group was likely retested more frequently. Second, we assessed only individuals with known symptom onset date with a repeat test occurring within 45 days of the initial positive test. We also separately analyzed individuals hospitalized and those not hospitalized for COVID-19. Finally, for the subset of tests with data available, we plotted Cts by days from initial positive SARS-CoV-2 PCR test, stratified by testing location (inpatient/outpatient). We displayed Abbott tests separately as CNs are not comparable with Cts reported by other platforms [20]. We describe patient characteristics of test values below published thresholds of negligible likelihood of infectious virus (24 for E gene, 32 for N gene) [21, 22].
RESULTS
Among 3758 adults meeting inclusion criteria, the mean age (SD) was 48.8 (16.4) years, 1677 (44.6%) were male, 1739 (46.3%) identified as Hispanic or Latinx, 1123 (29.9%) as Black non-Hispanic, and 509 (13.5%) as White non-Hispanic. In total, 277 (7.4%) were severely immunocompromised, 159 (4.2%) were moderately immunocompromised, and 3322 (88.4%) had no documented immunosuppression (Table 1). Approximately half (42.6%) had at least 1 comorbidity, and 935 (24.9%) had a hospital admission at BMC during their COVID-19 episode. Individuals had a median of 3 PCR tests; 1938 received ≥3 tests, 655 received ≥4 tests, and 340 received ≥5 tests. The median time to PCR clearance (interquartile range) was 22 (4–34), 20 (8–33), and 16 days (6–29), respectively, for those severely, moderately, and not immunocompromised (Table 1; Supplementary Figure 1).
Adjusted analyses revealed delayed time to SARS-CoV-2 PCR clearance with solid organ transplant (adjusted hazard ratio [aHR], 0.64; 95% CI, 0.42–0.97), diabetes (aHR, 0.82; 95% CI, 0.73–0.93), obesity (aHR, 0.90; 95% CI, 0.83–0.98), rheumatologic disease (aHR, 0.75; 95% CI, 0.57–0.98), ≥3 comorbidities (aHR, 0.73; 95% CI, 0.60–0.88), and older age (aHR, 0.996, 95% CI, 0.993–0.999), but not immunocompromise severity (aHR, 0.98, 95% CI, 0.84–1.15, and aHR, 0.86, 95% CI, 0.70–1.05, for severely and moderately immunocompromised individuals, respectively, compared with immunocompetent individuals).
Sensitivity analyses demonstrated similar results, though not all associations in the main analysis met the threshold for statistical significance (Supplementary Tables 2–5). Receipt of an interleukin (IL)-1 or IL-6 inhibitor for COVID-19 treatment was associated with delayed PCR clearance in those with known symptom onset date (aHR, 0.68; 95% CI, 0.52–0.89).
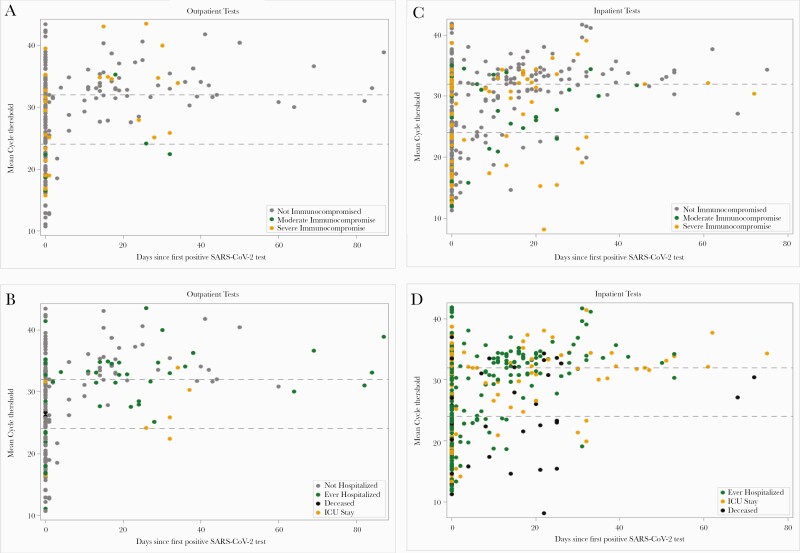
For the subset with Cts or CNs available (n = 754 tests from 601 individuals; 97.5% NP samples), plots stratified by outpatient and inpatient location (Figure 1) demonstrate overall increasing Ct values (decreasing likelihood of infectiousness) over time since infection onset. All tests with Cts <24 after day 20 (the lower threshold cited in the literature above which no tests were culture-positive [22]) occurred during inpatient stays: 8/9 individuals were admitted to the ICU or died, and 6/9 were immunocompromised. Only 5 outpatients had Cts <32 after day 20 (all immunocompetent). Abbott CNs were lower overall (Supplementary Figure 2), but only 3 had CNs <24 after day 40: 1 patient with solid organ transplant and HIV, and 2 nonhospitalized healthy adults under age 30.
Figure 1.
Cycle thresholds for SARS-CoV-2 PCR tests performed in the outpatient (A, B) and inpatient settings (C, D) with available data. Gray dashed lines represent literature-reported thresholds above which there is negligible likelihood of infectious virus (24 for E gene, 32 for N gene) [21, 22]. Each circle represents a single test (there may be multiple tests per individual), and color corresponds to immunocompromise category (A, C) or disease severity (B, D). Mean cycle thresholds refer to mean target 1 and target 2 values within an individual test on a single assay. All assays are displayed together here, except for the Abbott RealTime SARS-CoV-2 assay, which reports cycle number values that incorporate 2 targets into a single cycle number, which are generally lower than and not equivalent to cycle thresholds, and are therefore displayed separately in the supplementary figure. Abbreviations: ICU, intensive care unit; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome 2.
DISCUSSION
We found that solid organ transplant, multiple comorbidities (diabetes, obesity, and rheumatologic disease in particular), and older age were associated with delayed SARS-CoV-2 PCR clearance, but observed no significant differences by degree of immunocompromise. Our study presents the largest sample thus far of SARS-CoV-2 PCR retesting in immunocompromised and immunocompetent patients together, allowing comparison by immunocompromised status. Older age and having ≥3 medical comorbidities were most consistently associated with increased time to PCR clearance in sensitivity analyses, which also revealed delayed clearance in individuals who received an IL-1 or IL-6 inhibitor to treat severe COVID-19. These data align with previous median positive PCR duration in the general population (16 days from first positive test and 21 days from symptom onset in our study vs 17–33 days reported in the literature) [6, 7, 19] and in kidney transplant recipients (50% of solid organ transplant recipients in our study vs >30% in the literature were PCR-positive for >30 days) [3]. We observed prolonged PCR positivity in individuals hospitalized and deceased from COVID-19, but this was not statistically significant; this may have been limited by the 48% without known disease severity (individuals initially tested outside BMC). We did not see associations with sex, general immunosuppression, or corticosteroids reported in smaller studies [7, 9]. Our data suggest that certain groups of immunocompromised, older, and medically complex individuals appear to have delayed PCR clearance compared with otherwise healthy, young individuals. However, even with a robust sample size, sufficient to show prolonged PCR positivity in solid organ transplant recipients, other immunocompromised groups, including 212 on active chemotherapy, did not have delayed PCR clearance, making a prolonged period of transmission from these individuals similarly less likely.
This study has limitations. First, only 1 individual received a past-year stem cell transplant, and none received chimeric antigen receptor (CAR) T cell therapy, so generalizability to the stem cell transplant and other extremely immunosuppressed populations is limited. Second, immunocompromised individuals comprised only 11.6% of the population analyzed, and it is possible that we were underpowered to detect significant differences. However, it is also possible that only certain subpopulations of immunocompromised individuals have prolonged PCR positivity, as suggested by differing estimates among subgroups.
Importantly, we did not utilize viral cultures or other means to study transmissibility directly. We therefore cannot conclude anything about infectivity; however, the objective of our study was to determine whether singling out immunocompromised individuals for SARS-CoV-2 PCR retesting for isolation clearance is useful. Time to SARS-CoV-2 PCR clearance did not differ by immunocompromised status, except in the subpopulation of solid organ transplant recipients, and Ct values increased over time, which generally corresponds with minimal ability to culture infectious virus, particularly after day 20 of illness. Interpreted together, we believe it is unlikely that qualitatively PCR testing all immunocompromised individuals after symptom improvement will provide meaningful information to infer infectivity; however, further studies supported by viral culture are necessary to further inform this and help determine which subgroups may benefit from quantitative PCR or other (eg, antibody titer or antigen) testing [12].
As a retrospective EHR record review, systematic retesting at prespecified intervals did not occur, and the exact day of PCR clearance is unknown. To account for the uncertainty of time between last positive and first negative test, we utilized interval censoring techniques. Sensitivity analyses restricted to only individuals frequently tested also failed to show differences in time to PCR clearance by immunocompromised status. The multiple different assays used, with variable sensitivity and cutoffs, also limit findings, though overall Ct trends across assays were similar.
Finally, use of first positive SARS-CoV-2 PCR test, not symptom onset, likely underestimated total time to PCR clearance. Our sensitivity analysis limited to those with known symptom onset date yielded similar estimates but failed to show any significant differences, albeit with a small sample size.
Although initial reports on SARS-CoV-2 and data from other viral infections suggest concern for prolonged SARS-CoV-2 PCR positivity in immunocompromised patients overall, our data do not support this. Furthermore, though sample size prevented multivariable analysis of the relative risk of low Cts of positive PCR tests performed after illness day 20, severe disease was more common than immunosuppression in individuals with prolonged low Ct values in this cohort, and other studies demonstrate that PCR positivity does not imply infectivity. Lower viral loads seen after day 10/11 of illness are associated with lower likelihood of replication-competent virus [12], and most individuals who have recrudescence of positive SARS-CoV-2 PCR after initial symptom resolution do not appear to still be infectious [13]. Even in rare reported cases of infectious virus cultured weeks after initial infection, some had interim negative retesting, and individuals were symptomatic at later positive culture and PCR testing [4, 5]. Without clear evidence of which groups of immunocompromised or other individuals clear PCR later than the general population, or that prolonged asymptomatic PCR positivity corresponds to ongoing transmission risk, the usefulness of qualitative, PCR test–based clearance criteria for severely immunocompromised individuals is unclear. While broad testing strategies are critical to identify new cases, retesting to end isolation may not be warranted, particularly in this resource-constrained global pandemic setting.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Jenny Wang, MS, Boston University School of Public Health, for technical assistance, Elizabeth Duffy, Assistant Professor, Department of Pathology and Laboratory Medicine at Boston University School of Medicine, for assistance in obtaining additional laboratory data, the Boston Medical Center Virtual COVID Data Repository/Clinical Data Warehouse, and most importantly, all the patients and staff involved in BMC COVID-19 efforts, including the COVID results reporting, follow-up monitoring, and retesting teams managed by Elizabeth Keohane, Rory Silvia, and Dominque Hall-Fowler.
Financial support. This work was supported by the National Institutes of Health (T32DA013911 to T.C.B., R01HD095630 to J.M.); the National Institute of Allergy and Infectious Diseases through the Providence/Boston Center for AIDS Research (P30AI042853 to B.P.L.); the Burroughs Wellcome Fund/American Society for Tropical Medicine and Hygiene Postdoctoral Fellowship in Tropical Infectious Diseases (T.C.B.); the US National Institute of General Medical Sciences (R01GM122876 to L.F.W.); the Boston University Clinical and Translational Science Institute via a Clinical and Translational Science Award from the National Center for Advancing Translational Sciences, National Institutes of Health (UL1TR001430 to H.E.H. and M.H.), and by internal funding from Boston Medical Center for the retesting clinical efforts.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. Epstein RL, Marathe JG, Bouton TC, Barocas JA participated in study design and conception; Epstein RL, Xiao Y, and Marathe J led clinical retesting efforts and designed data templates; Hofman M and Hsu HE extracted and cleaned data from the electronic medical record; Epstein RL and Sperring H analyzed data with the help of Lodi S, White LF, Miller NS. All authors contributed to, critically revised, and approved the final draft for publication.
Patient consent. The Boston University Medical Campus Institutional Review Board approved this study as exempt human subject research. As such, patient consent was not required for this retrospective review of electronic health record data.
Prior presentation. These data have not previously been presented and are not publicly available.
References
- 1. Manuel O, Estabrook M; American Society of Transplantation Infectious Diseases Community of Practice. RNA respiratory viral infections in solid organ transplant recipients: guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 2019; 33:e13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davis A, Cortez V, Grodzki M, et al. Infectious norovirus is chronically shed by immunocompromised pediatric hosts. Viruses 2020; 12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benotmane I, Gautier Vargas G, Wendling M, et al. In-depth virological assessment of kidney transplant recipients with COVID-19. Am J Transplant 2020; 20:3162–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aydillo T, Gonzalez-Reiche AS, Aslam S, et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med 2020; 383:2586–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun J, Xiao J, Sun R, et al. Prolonged persistence of SARS-CoV-2 RNA in body fluids. Emerg Infect Dis 2020; 26:1834–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19). Clin Infect Dis 2020; 71:799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Q, Zheng XS, Shen XR, et al. Prolonged shedding of severe acute respiratory syndrome coronavirus 2 in patients with COVID-19. Emerg Microbes Infect 2020; 9:2571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vena A, Taramasso L, Di Biagio A, et al. Prevalence and clinical significance of persistent viral shedding in hospitalized adult patients with SARS-CoV-2 infection: a prospective observational study. Infect Dis Ther 2021; 10:387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention. Duration of isolation and precautions for adults with COVID-19. 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html. Accessed February 13, 2021.
- 11. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 12. van Kampen JJA, van de Vijver DAMC, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 2021; 12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu J, Peng J, Xiong Q, et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine 2020; 59:102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng HY, Jian SW, Liu DP, et al. ; Taiwan COVID-19 Outbreak Investigation Team. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med 2020; 180:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korea Centers for Disease Control and Prevention. Findings from investigation and analysis of re-positive cases. 2020. Available at: https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030&act=view&list_no=367267&nPage=1. Accessed October 19, 2020.
- 16. Centers for Disease Control and Prevention. Discontinuation of transmission-based precautions and disposition of patients with COVID-19 in healthcare settings (interim guidance). 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html. Accessed March 10, 2021.
- 17. Millard H, Wilson C, Fortunati F, Li L. COVID-19 psychiatric patients: impact of variability in testing on length of hospital stay and disposition back to congregate care settings. Psychiatry Res 2020; 292:113324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C, Glass S, Demars S, Tulloch-Palomino LG, Wander PL. Estimated excess acute-care length of stay and extra cost of testing-based versus symptom-based isolation strategies among veterans hospitalized with coronavirus disease 2019 (COVID-19) discharging to a congregate setting. Infect Control Hosp Epidemiol 2021; 42:356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu W, Liu Y, Xu Z, et al. Clinical characteristics and predictors of the duration of SARS-CoV-2 viral shedding in 140 healthcare workers. J Intern Med 2020; 288:725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marais G, Naidoo M, Hsiao NY, et al. The implementation of a rapid sample preparation method for the detection of SARS-CoV-2 in a diagnostic laboratory in South Africa. PLoS One 2020; 15:e0241029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Basile K, McPhie K, Carter I, et al. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 2020; 71:2663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.