ABSTRACT
The families of copper-containing membrane-bound monooxygenases (CuMMOs) and soluble di-iron monooxygenases (SDIMOs) are involved not only in methane oxidation but also in short-chain alkane oxidation. Here, we describe Rhodococcus sp. strain ZPP, a bacterium able to grow with ethane or propane as the sole carbon and energy source, and report on the horizontal gene transfer (HGT) of actinobacterial hydrocarbon monooxygenases (HMOs) of the CuMMO family and the sMMO (soluble methane monooxygenase)-like SDIMO in the genus Rhodococcus. The key function of HMO in strain ZPP for propane oxidation was verified by allylthiourea inhibition. The HMO genes (designated hmoCAB) and those encoding sMMO-like SDIMO (designated smoXYB1C1Z) are located on a linear megaplasmid (pRZP1) of strain ZPP. Comparative genomic analysis of similar plasmids indicated the mobility of these plasmids within the genus Rhodococcus. The plasmid pRZP1 in strain ZPP could be conjugatively transferred to a recipient Rhodococcus erythropolis strain in a mating experiment and showed similar ethane- and propane-consuming activities. Finally, our findings demonstrate that the horizontal transfer of plasmid-based CuMMO and SDIMO genes confers the ability to use ethane and propane on the recipient.
IMPORTANCE CuMMOs and SDIMOs initiate the aerobic oxidation of alkanes in bacteria. Here, the supposition that horizontally transferred plasmid-based CuMMO and SDIMO genes confer on the recipient similar abilities to use ethane and propane was proposed and confirmed in Rhodococcus. This study is a living example of HGT of CuMMOs and SDIMOs and outlines the plasmid-borne properties responsible for gaseous alkane degradation. Our results indicate that plasmids can support the rapid evolution of enzyme-mediated biogeochemical processes.
KEYWORDS: CuMMO, SDIMO, Rhodococcus, horizontal gene transfer, ethane oxidation, propane oxidation, plasmid
INTRODUCTION
The superfamily of copper-containing membrane-bound monooxygenases (CuMMOs), including ammonia monooxygenases (AMOs), particulate methane monooxygenases (pMMOs), and hydrocarbon monooxygenases (HMOs), plays a key role in biogeochemical cycles (1). Aerobic ammonia oxidizers possess AMOs, and aerobic methanotrophs possess pMMOs; these enzymes catalyze the first steps in the oxidation of ammonia and methane, respectively (1–6). HMOs, novel members of the CuMMOs, provide bacteria with the capacity to grow on short-chain hydrocarbons such as ethane, propane, and ethene as their carbon and energy source (7, 8). HMOs have been found in Proteobacteria (9, 10) and Actinobacteria (7, 8, 11–13). For example, in marine ecosystems, novel short-chain alkane-oxidizing proteobacterial Cycloclasticus symbionts and ethylene-assimilating proteobacterial Haliea species have been described (9, 14). Within the phylum Actinobacteria, a few strains from the order Corynebacteriales have been isolated. Mycolicibacterium (originally classified as Mycobacterium) chubuense NBB4 was isolated by enrichment on ethene as the sole carbon source and could use a broad spectrum of hydrocarbons (C2 to C16 alkanes and C2 to C4 alkenes but not methane) as the substrates (7, 8). Moreover, short-chain alkane oxidation by HMOs in M. chubuense NBB4, Nocardioides sp. strain CF8, and Mycolicibacterium sp. strain JOB5 was inhibited by acetylene and allylthiourea (ATU), also described as typical inhibitors of AMO and pMMO enzymes (15, 16). In addition, soluble di-iron monooxygenases (SDIMOs) are also involved in the oxidation of methane, short-chain alkanes, alkenes, and cyclic, aromatic, and chlorinated hydrocarbons in Actinobacteria and aromatic hydrocarbons and methane in Proteobacteria (12, 17–20). Phylogenies of SDIMO alpha subunit genes have revealed a high diversity of SDIMOs, and these can be classified into six subgroups. The soluble methane monooxygenase (sMMO) (group 3 SDIMO), which is considered an alternative methane monooxygenase (the heterologous pMMO), is apparently an important enzyme for several methanotroph genera within the Alpha- and Gammaproteobacteria (21). Proteobacterial methanotrophic Methylocella species were shown to possess SDIMOs for propane oxidation (19, 22). M. chubuense NBB4 and M. rhodesiae NBB3 also have several types of SDIMOs (21) and sMMO-like SDIMOs, encoded by smoXYB1C1Z, that are known to participate in the oxidation of short-chain alkanes (8, 12, 19, 23). Few studies exist on CuMMOs and SDIMOs in Actinobacteria compared to those on pMMOs/AMOs and sMMOs (9).
Aerobic methane- or ammonia-oxidizing bacteria are limited to a few clusters of microbes, and phylogenies of PmoA/AmoA match closely with 16S rRNA gene- and genome-based phylogenies of the corresponding microbes (1, 3, 24–29). Therefore, targeted primers for pmoA-amoA genes and operational taxonomic unit-based sequence analysis methods have been used to analyze communities of aerobic methanotrophs and ammonia oxidizers, including the complete ammonia-oxidizing (comammox) Nitrospira species (30–32). However, the evolutionary history of CuMMOs shows horizontal gene transfer (HGT) events (1, 28). Phylogenetic and compositional analyses revealed that the adaptation of this enzyme to preferentially oxidize either ammonia or methane has occurred more than once (1). Methane-inactive CuMMOs and bacterial multicomponent monooxygenase genes were also found to be associated with ancestral Actinomycetales. Subsequently, the development of methane oxidation capability in both the particulate and soluble enzyme systems was transferred out of these clades (28). HGT produces extremely dynamic genomes and changes the ecological character of bacterial species (33, 34). It contributes to the spread of genes involved in pathogenesis, heavy metal tolerance, and antibiotic resistance (35, 36). Genomic information has been widely used to identify cases of HGT. The genes in one genome often share the same base compositions, patterns of codon usage, and frequencies of di- and trinucleotides, but these characteristics vary between different microbes. Therefore, HGT events retain the sequence characteristics of the donor genome, deviating from the recipient genome (33). Conflicting branching patterns occur between the gene derived from HGT and the reference phylogenetic trees of conserved core genes (34). DNA segments gained through HGT often display a restricted phylogenetic distribution among related strains or species (33). Most cases of actual HGT events are rarely observed. Therefore, artificial recombinant expression is widely used to obtain the target function using HGT, and CuMMO and SDIMO genes have been used to obtain recombinant expression in Escherichia coli and M. smegmatis (8, 12, 37, 38).
In this study, we report the discovery of HMO and sMMO-like SDIMO genes in Rhodococcus sp. strain ZPP. These genes are located on a megaplasmid. To provide evidence supporting the occurrence of HGT, we performed a mating experiment in which the megaplasmid was transferred to the recipient bacterium Rhodococcus erythropolis.
RESULTS
Physiology of Rhodococcus sp. ZPP on ethane and propane.
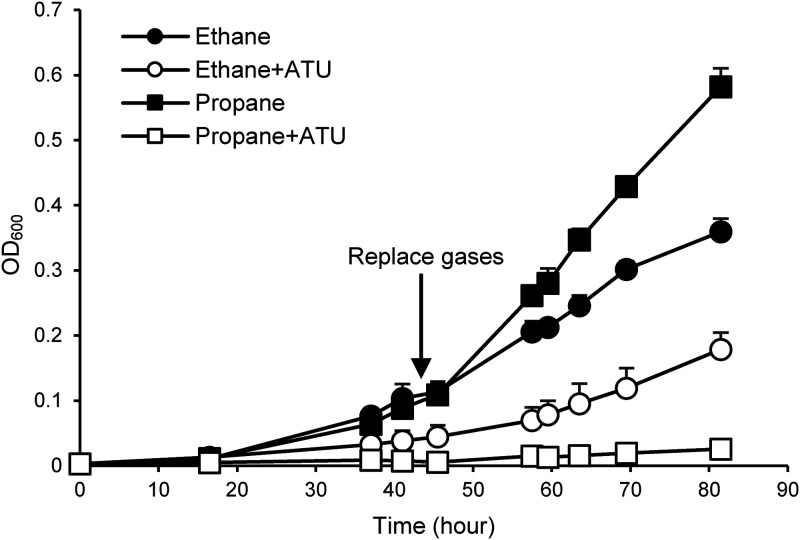
Strain ZPP was isolated after enrichment with ethane as the sole carbon and energy source, using the floating-filter method to prevent the organic compounds in the agar, rather than the ethane, from being used as the carbon and energy source, and purified on 2-fold-diluted R2A agar plates. Next, 10-fold-diluted washed cells of strain ZPP (optical density at 600 nm [OD600] of ∼0.1) were cultured in the ∼1% (vol/vol) gas phase of ethane or propane, and the consumption of these substrates was confirmed (Fig. 1). To observe the growth rate, 1,000-fold-diluted washed cells (OD600 of ∼0.001) were cultured, and the doubling times of strain ZPP were determined to be 16.6 ± 2.0 h for ethane and 11.5 ± 0.3 h for propane by shaking (Fig. 2). Methane was not consumed by strain ZPP (see Table S1 in the supplemental material). When the inhibitor ATU was applied, ethane oxidation was partially inhibited, and propane oxidation was nearly completely inhibited (Fig. 2).
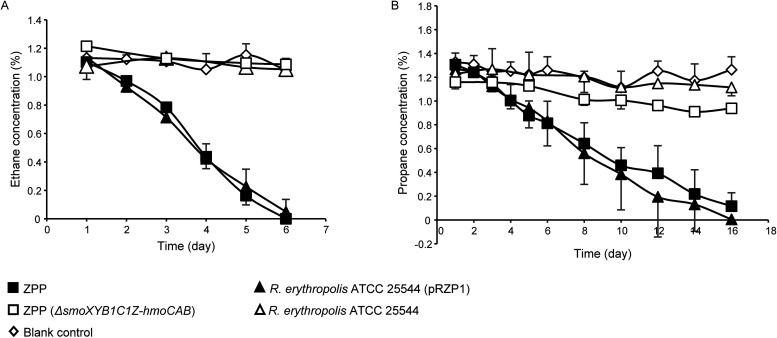
FIG 1.
Ethane (A) and propane (B) consumption over time by the donor strain Rhodococcus sp. ZPP, the transconjugant strain R. erythropolis ATCC 25544(pRZP1), the defective mutant ZPP(ΔsmoXYB1C1Z-hmoCAB), the recipient strain R. erythropolis ATCC 25544, and the blank control that had no cells added to the medium. For all substrates, data are the averages from three independent experiments, and error bars in either direction indicate the standard deviations. For this experiment, 10-fold-diluted washed cells (OD600 of ∼0.1) were used.
FIG 2.
Allylthiourea (ATU) sensitivity of Rhodococcus sp. ZPP for growth on ethane and propane (OD600). For all substrates, data are the averages from three independent experiments, and upper error bars indicate the standard deviations. Lower error bars are not shown in order to avoid overlapping error bars. For these experiments, 1,000-fold-diluted washed cells were used. Gases were replaced after approximately 2 days to maintain sufficient gas concentrations for exponential growth.
Genome anatomy of strain ZPP and detection of linear plasmids.
Combining Illumina short reads with PacBio RSII and MinION long reads, we completed the entire genome of strain ZPP, which is composed of 8.47 Mbp arranged in six linear or circular replicons: one circular chromosome (6.78 Mbp); three linear plasmids, pRZP1 (0.25 Mbp), pRZP2 (0.93 Mbp), and pRZP3 (0.26 Mbp); and two circular plasmids, pRZP4 (0.14 Mbp) and pRZP5 (0.11 Mbp) (Fig. S1).
Genomic BLAST showed that the chromosome of strain ZPP was highly similar to that of Rhodococcus sp. strain S2-17 (GenBank accession no. CP021354) and Rhodococcus sp. strain WAY2 (GenBank accession no. CP046572), and the ANIb (BLAST-based average nucleotide identity) among these three strains was up to 97%. The phylogenetic tree showed that they belong to Rhodococcus clade C (Fig. S2). Interestingly, homologous regions (about 100 kb) of the plasmid pRZP1 were also found in plasmids of seven other strains, and they belonged to Rhodococcus clades C and D as indicated by plasmid BLAST (39) (Fig. S2 and Fig. S3A). These 100-kb homologous regions were located on the edges of the plasmids (Fig. S3A), but the genes encoding HmoCAB and SmoXYB1C1Z were not present in this 100-kb homologous region. Although this region has not been well investigated and annotated, a transfer clostridial plasmid locus, including tcpC (HYG77_35655), was found in this region (40–42) (Fig. S3A), and we designed a primer pair in tcpC to investigate this plasmid.
On the basis of previous research (43, 44), the first 300 bp of the telomeres from these plasmids, as shown in Fig. S3A, and other similar telomeres were added to construct a phylogenetic tree of telomeres to verify the linear plasmids and their clades (43) (Fig. S4). The telomeres of these linear replicons are typical actinobacterial invertrons, similar to those in Rhodococcus jostii RHA1 (43), and the identity between pRZP1L (300 bp on the left edge of pRZP1) and pRHL2L (R. jostii RHA1) was up to 92%. Generally, these telomeres can be divided into two clades, pRZP1L-like and others, which suggests different origins of the pRZP1L-like clade (Fig. S4). All seven homologous plasmids in Fig. S3A belong to the pRZP1L-like clade (although the plasmid pRB29 in Rhodococcus sp. S2-17 was not added as the structure of this plasmid has not been elucidated; it should still be considered a pRZP1L-like plasmid for homologous regions). The linearity of the megaplasmids pRZP2 and pRZP3 was also confirmed by the presence of telomeres (Fig. S4); however, they grouped separately from pRZP1L. PacBio and Nanopore data were also assembled individually to complete the circular replicons of strain ZPP.
To investigate the evolution of ZPP plasmids, phylogenetic analysis of ParA (partition replicons during cell division) proteins was performed (43). As shown in Fig. S5, ParAs of the same genomes were not in one clade. However, eight plasmids in Fig. S3A were clustered into one set of ParAs, with their telomeric sequences being highly similar (Fig. S4).
ISfinder (45) (http://www-is.biotoul.fr) was used to annotate insertion sequence (IS) elements (Table S2 and Fig. S3B). In the homologous regions of plasmids, different types of IS elements were identified. For example, ISRjo4 (family IS256, positions 65651 to 67093), ISRjo4 (family IS256, positions 68606 to 70048), and ISRjo1 (family IS1380, positions 98851 to 100534) were identified in pRZP1. Correspondingly, pRB29 of Rhodococcus sp. S2-17 was found to contain ISRop2 (family ISAzo13) and IS1164 (family IS256).
The clusters of hydrocarbon oxidation genes are located on a plasmid.
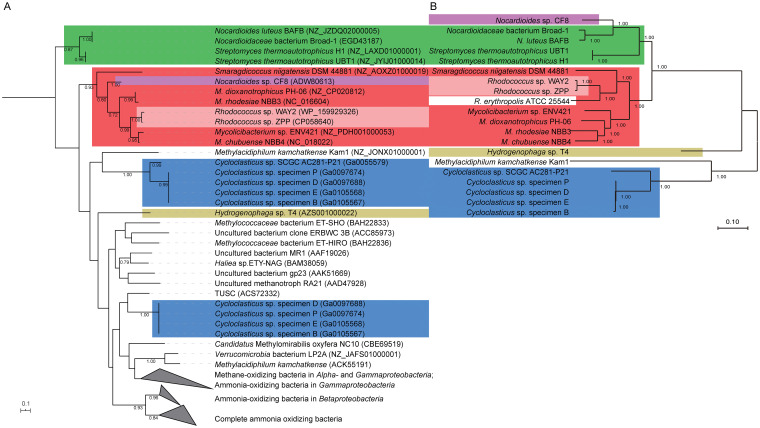
The newly discovered hmoCAB (HYG77_36340, HYG77_36345, and HYG77_36350) and smoXYB1C1Z (HYG77_36175, HYG77_36180, HYG77_36185, HYG77_36190, and HYG77_36195) genes (12) were found on plasmid pRZP1. A phylogenetic tree of CuMMO subunit A was constructed. The HmoA proteins of Rhodococcus species were in a deep-branching position, together with the actinobacterial CuMMO homologs from Mycolicibacterium and Nocardioides species (Fig. 3). The HmoA protein of Rhodococcus sp. WAY2 was identified to be the most similar to the strain ZPP protein. Interestingly, the gene encoding this protein is located on pRWAY02 rather than on pRWAY03. The latter has a homologous region and telomeres in common with pRZP1 (Fig. S3A).
FIG 3.
Phylogenetic tree of copper-containing membrane-bound monooxygenase (CuMMO) subunit A (A) and 92 core metabolic genes (B). CuMMO subunit A proteins were aligned using Muscle with default parameters, 300 bootstraps, 95% partial deletion, and archaeal AmoA (ammonia monooxygenase subunit A) as an outgroup. The nucleotide sequences of single-copy core genes were identified and aligned using the UBCG pipeline (70) and 100 bootstraps with 90% partial deletion. The maximum likelihood (ML) tree was constructed using MEGA X. Nodes whose bootstraps are ≥60 are marked. Two clades of Cycloclasticus sp. specimen D, Cycloclasticus sp. specimen P, Cycloclasticus sp. specimen E, and Cycloclasticus sp. specimen B in panel A are based on data from a previous study (9).
Both Rhodococcus and Mycolicibacterium species have several types of SDIMOs (Fig. S6) (21). Group 3 (sMMO)-like, group 6, and group 5 SDIMOs were found in ZPP and were distributed over plasmids pRZP1 and pRZP3 and the chromosome, respectively. The distances between the positions of hmoCAB and smoXYB1C1Z genes are short on pRZP1 of ZPP, pRWAY02 of Rhodococcus sp. WAY2, pMYCCH.01 of M. chubuense NBB4, and the chromosome of M. rhodesiae NBB3 (Fig. S3B) (8, 12, 23).
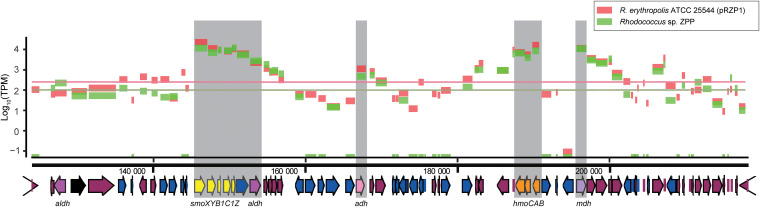
The plasmid pRZP1 contains genes encoding the full pathway for the oxidation of ethane or propane to carbon dioxide, and these genes are actively transcribed in cells growing on propane. The genes encode an alcohol dehydrogenase (adh), an N,N′-dimethyl-4-nitrosoaniline (NDMA)-dependent methanol dehydrogenase (mdh), aldehyde dehydrogenases (aldh), and an acetate-CoA ligase (Fig. 4). Although the NDMA-dependent methanol dehydrogenase is encoded close to the hmoCAB cluster, no methanol-related function was identified (Fig. 4; Table S1). Phylogenetically, all these genes are most similar to those in other Rhodococcus species, which is different for hmoCAB and smoXYB1C1Z.
FIG 4.
Cluster of alkane oxidation genes on the plasmid pRZP1 and its transcriptome using propane as the substrate. Log10-transformed TPM values of the transcriptomes of ZPP and ATCC 25544(pRZP1) are plotted, and the expression levels of housekeeping genes (means of gyrB and recA) are marked as the baselines. The smoXYB1C1Z genes are marked in yellow, and the hmoCAB genes are marked in orange. The alcohol dehydrogenase genes (adh) are marked in light pink, the NDMA-dependent methanol dehydrogenase genes (mdh) are marked in purple, the aldehyde dehydrogenase (aldh) genes are marked in pink, the acetate-CoA ligase gene is marked in black, and the mobile elements (including IS elements, transposases, and phages) are marked in blue.
Mating experiment.
The conjugative transfer of pRZP1 from the wild-type donor strain ZPP to different bacterial strains was investigated, and the transconjugant R. erythropolis ATCC 25544(pRZP1) was obtained from an ampicillin resistance nitrate mineral salts (NMS) plate. The genomic comparison of pRZP1, R. erythropolis ATCC 25544, and R. erythropolis ATCC 25544(pRZP1) directly confirmed the transfer of pRZP1 (Fig. S7). The mating experiments with Rhodococcus rhodochrous ATCC 13808, R. koreensis DSM 44498, Pseudomonas aeruginosa CGMCC 1.1129, R. opacus DSM 43250, Rhodococcus sp. S2-17, and M. smegmatis MC2-155 were not successful.
The plasmid stability of the wild-type donor and the transconjugant without gases was determined with the content change of plasmid tcpC and hmoA genes compared with the 16S rRNA gene during serial daily subcultures with R2A liquid medium. The results showed that pRZP1 in strain ZPP is much more stable than that in the transconjugant without gases (Fig. S8). After two parallel cultivations with daily subcultures, dilution plate spreading was performed, and 32 single colonies from each culture were selected to verify plasmid loss using the same primers. After 12-day subcultures of strain ZPP, only 1 of 64 colonies lost the plasmid pRZP1. On the contrary, after 6-day subcultures, only 2 colonies of 64 colonies of the transconjugant still had plasmid pRZP1, and 1 of the 2 colonies lost the gene encoding HMO. During the subcultivation of strain ZPP in R2A medium without selection pressure, we also isolated a strain that lost ethane and propane oxidation activity, and the genome sequence of this strain shows that this strain has plasmid pRZP1 but lost its functional region (including hmoCAB and smoXYB1C1Z), so we named this strain ZPP(ΔsmoXYB1C1Z-hmoCAB) (Fig. S9).
The consumption of ethane or propane was verified with strain ZPP, strain ZPP(ΔsmoXYB1C1Z-hmoCAB), R. erythropolis ATCC 25544(pRZP1), and R. erythropolis ATCC 25544 using 10-fold-diluted washed cells (OD600 of ∼0.1) (Fig. 1). Both the wild-type donor and the transconjugant completely consumed ethane more quickly (6 days) than propane (16 days), and the activities of both strains were equal (t test) for ethane (0.23% ± 0.03% and 0.23% ± 0.03% [vol/vol] gas phase per day) or propane (0.09% ± 0.02% and 0.12% ± 0.03% [vol/vol] gas phase per day) (Fig. 1). R. erythropolis ATCC 25544 had no plasmid pRZP1 (Fig. S7), and strain ZPP(ΔsmoXYB1C1Z-hmoCAB) had a partial plasmid of pRZP1 (Fig. S9). The gas consumption of ethane and propane gases by these two negative strains remained undetectable (Fig. 1). Only ZPP and ZPP(ΔsmoXYB1C1Z-hmoCAB) exhibited 2-propanol utilization activity (Table S1).
TPM (transcripts per kilobase per million) values from transcriptome analyses were used to compare the expression levels of the plasmid-borne genes between the two strains. We demonstrated that the trends between the two strains were similar (Fig. 4), with high TPM values for the alkane oxidation pathway-related genes and low TPM values for IS element-related genes.
Strain ZPP also encodes several other types of monooxygenases such as SDIMOs (Fig. S6), AlkB-like monooxygenases, and cytochrome P450s to oxidize alkanes to alcohols, following a series of oxidation steps to acids (46). Previous researchers have identified that these monooxygenases tend to oxidize ≥4-carbon-atom-containing long-chain alkanes and alkenes. R. erythropolis ATCC 25544 also has AlkB-like monooxygenases, but without pRZP1 plasmids, cells could not oxidize ethane and propane. In addition, no high expression levels of these genes were observed in the transcriptome of propane-grown cells (Table S3), indicating that these genes may be not involved in the oxidation of short-chain alkanes.
DISCUSSION
In this study, we described the isolation and genomic characterization of a Rhodococcus strain, ZPP. This bacterium is able to utilize ethane and propane as the sole carbon and energy source. Genome sequencing and assembly revealed the presence of a 6.78-Mbp circular genome together with three linear and two circular megaplasmids. Most genomes of Rhodococcus species are large (5 to 10 Mbp), with various sizes and numbers of large catabolic plasmids (39). These plasmids are frequently transferred by conjugation in the genus Rhodococcus (47–49). Due to repeat regions and the presence of linear plasmids, it is challenging to discriminate plasmids unambiguously from contigs after assembly. To address this, we used PacBio and Nanopore sequencing to solve the problem of assembly and Illumina sequencing for error correction to polish the assembly.
Among the plasmids in strain ZPP, pRZP1 was shown to contain operons encoding both copper-dependent (CuMMO) and iron-dependent (SDIMO) monooxygenases in combination with genes encoding alcohol and aldehyde dehydrogenases. The plasmid stability experiment showed that the pRZP1 plasmid was stable in strain ZPP (see Fig. S8 in the supplemental material). Although this plasmid was unstable in the transconjugant without a gas supply, the transconjugant can grow on ethane and propane, demonstrating the activity of plasmid pRZP1 and its response to gases. The gas consumption also indicated that pRZP1 had similar activities in both strains (Fig. 1). Transcriptome analyses of strain ZPP and the transconjugant growing on propane showed that genes in both operons on the plasmid pRZP1 were highly expressed, supporting their metabolic function in propane oxidation (Fig. 4). The deletion of the CuMMO- and SDIMO-encoding parts of the plasmid pRZP1 (ΔsmoXYB1C1Z-hmoCAB) resulted in a loss of the capacity to grow on ethane and propane (Fig. 1 and 2).
The roles of hmoCAB and smoXYB1C1Z also differed, as became clear from growth experiments. Strain ZPP with propane as the substrate was almost completely inhibited by ATU, a typical inhibitor of CuMMO (15, 16, 50, 51), indicating that this HMO is the key enzyme in propane oxidation. The growth of strain ZPP with ethane oxidation was partially inhibited by ATU (Fig. 1 and 2), which may indicate a role for the sMMO-like proteins (smoXYB1C1Z) encoded on plasmid pRZP1. Heterologous expression experiments on M. chubuense NBB4 indicated that the activities of group 3-like SDIMOs on ethene and ethane were higher than those on propane and butane (8, 12). The proteobacterial methanotroph Methylocella silvestris uses methane or propane as a carbon and energy source and contains methane- and propane-specific SDIMOs (19). In future experiments, mutagenesis or recombination of hmoCAB and smoXYB1C1Z would allow the identification of the relative contributions of each enzyme to the metabolism of ethane and propane. The mating experiment showed that the plasmid used in this study has greater potential than the previously used vector pMycoFos (8). The products of ethane and propane oxidation are ethanol and 1-propanol or 2-propanol. Since strains ZPP and ZPP(ΔsmoXYB1C1Z-hmoCAB) exhibited 2-propanol utilization activity, and the transconjugant is capable of using ethanol and 1-propanol but not 2-propanol as a carbon and energy source, alcohol and aldehyde dehydrogenases harbored by the plasmid pRZP1 may not catalyze 2-propanol oxidation.
Phylogenetic analysis of CuMMO subunit A (Fig. 3) showed that the CuMMO of strain ZPP belongs to a clade of HMO encoded by the genes hmoCAB and related to those of short-chain alkane-degrading Mycolicibacterium (8) and Nocardioides (16) species. These enzymes are thought to have a high affinity for short-chain alkanes (9, 52) and are genetically and phylogenetically distinct from the pMMO genes of methane-oxidizing microbes (7). The group 3-like SDIMO encoded on plasmid pRZP1 differs from the typical group 3 SDIMO (sMMO), mainly existing on chromosomes (Fig. S6) of several methanotroph genera within the Alpha- and Gammaproteobacteria. The phylogeny of sMMOs is coherent with that of the 16S rRNA genes (43, 53); however, other studies indicated that the sMMOs may also have been obtained through several HGT events (17, 18, 28, 54).
Previous compositional analyses of CuMMO operons indicated recent lateral transfer of these operons (1, 33). The evolutionary history of CuMMOs demonstrated that HGT events of these genes, especially AMO/pMMO-encoding genes, do not happen frequently (1, 18, 28). Different from this observation, the phylogeny of HmoAs showed a higher frequency of HGT, not following the phylogeny of the housekeeping genes (Fig. 3).
The comparative genomic analysis of plasmids indicates the wide distribution of pRZP1L-like plasmids in Rhodococcus, but neither the hmoCAB nor smoXYB1C1Z genes were found in these plasmids (Fig. S3A), indicating the mobility of the pRZP1L-like plasmids. Therefore, representative strains of Rhodococcus clade B (R. rhodochrous ATCC 13808), clade C (R. koreensis DSM 44498 and Rhodococcus sp. S2-17), and clade D (R. erythropolis ATCC 25544) were collected for a mating experiment (39). Our conjugation experiments with plasmid pRZP1 resulted in a transconjugant of R. erythropolis ATCC 25544(pRZP1). The transconjugant became capable of utilizing ethane and propane. Conjugative transfer of pRZP1 to several other strains of the genus Rhodococcus failed, possibly due to plasmid incompatibility (55) (R. koreensis DSM 44498 and Rhodococcus sp. S2-17 containing the pRZP1L-like plasmid) or the presence of capsules (for R. rhodochrous ATCC 13808) (56). We used Pseudomonas aeruginosa CGMCC 1.1129 for the mating experiment because it has been successfully used as a recipient in a mating experiment with Rhodococcus sp. strain p52 (8, 12, 48, 49). However, the result showed that conjugative transfer between different genera remained difficult. Our conjugation experiments of strain ZPP with M. smegmatis MC2-155 failed, but the heterologous expression of hmoCAB genes in M. smegmatis MC2-155 was previously reported (8, 12, 48, 49).
It becomes clear that the megaplasmid pRZP1 is a key mobile element of ethane and propane oxidation. Seven pRZP1L-like plasmids could be identified from the complete genomes of Rhodococcus species, and these plasmids grouped in the phylogenetic tree of the typical plasmid protein ParA (Fig. S5), which is involved in the partitioning of replicons during cell division (43). The plasmid-encoded ParA proteins do not always obey organismal phylogeny, indicating the horizontal mobility of plasmids (43). Usually, linear plasmids have more flexibility than circular plasmids. Telomeres are important structures of these linear replicons, and frequent recombination aids in their adaptation to different environments (57). Telomeres of these eight plasmids except the one of Rhodococcus sp. S2-17 are typical actinobacterial invertrons, indicating their linearity and common origin (43). The IS elements and transposase genes were found to be widely distributed on pRZP1L-like plasmids (Fig. 4; Table S2), indicating their active exchanges together with the loss and gain of genes. The specific distance between hmoCAB and smoXYB1C1Z on pRZP1 indicates their relationship in function, but they are more likely not in a single operon as there are IS elements that had low expression levels (58–60) (Fig. 4).
Although strain ZPP, Rhodococcus sp. S2-17, and Rhodococcus sp. WAY2 belong to the same species according to the ANIb values, the differences in their plasmids can lead to a high degree of niche partitioning on carbon sources (61). The cluster of alkane oxidation in pRZP1 makes strain ZPP different from Rhodococcus sp. S2-17, even though these two plasmids possess homologous genes in other regions (Fig. S3A). Interestingly, a similar alkane oxidation cluster was also detected in Rhodococcus sp. WAY2 (Fig. S3B). In addition, the actinobacterial hmoCAB genes have already been found in other types of plasmids in M. chubuense NBB4 and M. dioxanotrophicus PH-06 (7, 8, 11). These results indicate that HGT events of HMO genes may occur widely in Actinobacteria through plasmid-based recombination. Actinobacterial strains often have many types of catabolic genes, which can be lost in the absence of selection pressure (46). However, short-chain alkanes became the sole carbon and energy source in an HMO-based Cycloclasticus (Gammaproteobacteria) symbiosis (9), indicating the potential stable inheritance of HMO genes.
In conclusion, we have demonstrated the growth of the newly isolated Rhodococcus sp. ZPP on ethane and propane and demonstrated this to be dependent on the presence of HMO genes and sMMO-like genes on a plasmid. We showed the potential of HGT in Rhodococcus species. The phenomena observed in this study are living examples of HGT of CuMMOs and SDIMOs. Although HGT often displays a restricted phylogenetic distribution among related strains or species, more clades of CuMMOs and SDIMOs and HGT events of related genes may be discovered in the future, as these important substrates are globally distributed, and the diversity of CuMMOs and SDIMOs in different strains will keep increasing (10). Hydrocarbons are more widely used as carbon and energy sources in the environment than they were previously estimated to be, and HGT may play an important role in the diversity and richness of short-chain alkane degradation-positive microorganisms.
MATERIALS AND METHODS
Bacterial strains, medium, and culture conditions.
R. erythropolis ATCC 25544 (American Type Culture Collection) (NBRC 15567 [Biological Resource Center, NITE] [CGMCC 1.2362]), R. rhodochrous ATCC 13808 (CGMCC 4.1147), R. koreensis DSM 44498 (Deutsche Sammlung von Mikroorganismen [German Collection of Microorganisms]) (CGMCC 4.1724), and Pseudomonas aeruginosa CGMCC 1.1129 were purchased from the China General Microbiological Culture Collection Center (CGMCC). R. opacus DSM 43250 (ACCC 41021) was purchased from the Agricultural Culture Collection of China (ACCC). Rhodococcus sp. S2-17 was donated by CheOk Jeon from Chung-Ang University, South Korea, and M. smegmatis MC2-155 was stored in our laboratory. Rhodococcus sp. ZPP was isolated using the floating-filter method (62) from soils that were enriched with 5% ethane. The supernatant of the enriched soil was transferred to and filtered on a sterile polycarbonate membrane filter (0.2 μm; Millipore, MA), and the filter was floated on NMS liquid medium (63) in a plate that was carefully placed in a 2.5-liter sealed jar filled with a 1% (vol/vol) gas phase of ethane. After 3 weeks of incubation, the lawn on the filter was streaked onto a new filter and floated on NMS liquid medium as described above. After 3 weeks, single colonies from the surface of the filter were transported in NMS liquid medium in a sealed serum bottle and incubated with a 1% (vol/vol) gas phase of ethane for 3 weeks to confirm ethane-oxidizing activity. Pure cultures were isolated from 2-fold-diluted R2A agar (Hopebio Technology, Qingdao, China) with a dilution method. All cultures were grown aerobically at 28°C. Twofold-diluted R2A medium was used to scale up the culture. Cells were harvested and washed with normal saline, and washed cells (OD600 of ∼1.0) were used as the stock. NMS medium was used to verify the growth of strains and activity on different substrates (ethane, propane, methane, methanol, ethanol, 1-propanol, and 2-propanol). An ∼1 to 2% (vol/vol) gas phase was added to the headspace of 125-ml sealed serum bottles with 25 ml NMS medium for oxidation and growth assays. As the plasmid pRZP1 in the transconjugant of R. erythropolis ATCC 25544 was not stable in R2A medium, both the donor and the transconjugant were cultured by adding an ∼5% (vol/vol) gas phase to the headspace of 125-ml sealed serum bottles with 25 ml NMS medium to grow cells for the transconjugant-related experiments. Cells were also harvested and washed with normal saline, and washed cells (OD600 of ∼1.0) were also used as the stock.
Analytical techniques.
To analyze the ethane- or propane-oxidizing activities, NMS medium (25 ml) with 10-fold-diluted washed cells (final OD600 of ∼0.1) was added to a 125-ml sealed serum bottle. Ethane or propane was added to the headspace (∼1% [vol/vol] gas phase). The contents of ethane and propane in the headspace of serum bottles were analyzed by gas chromatography (GC2020; Ximo, Shanghai, China) with nitrogen as the carrier gas and flame ionization detection. Quantification was performed using a five-point standard curve with different concentrations of calibration gases (Air Liquide, Shanghai, China). To analyze the doubling time and the inhibitory effect of ATU on ethane or propane oxidation, ethane or propane was added to the headspace (∼2% [vol/vol] gas phase), 1,000-fold-diluted washed cells as described above were incubated with or without ATU (0.1 mM) in the cultivation medium with shaking at 150 rpm, the change in the OD600 was monitored, and all gases were replaced after 2 days. Growth on 0.1% (vol/vol) methanol, ethanol, 1-propanol, and 2-propanol was measured by using 1,000-fold-diluted washed cells as described above and monitoring the change in the OD600. Growth on methane was examined by using 1,000-fold-diluted washed cells as described above and monitoring the change in the methane concentration.
Genome sequencing and analysis of plasmids.
DNA was extracted using a DNeasy PowerSoil kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The genome of strain ZPP was sequenced using the Illumina HiSeq PE150 platform (Illumina, San Diego, CA) combined with PacBio RSII sequencing (Pacific Biosciences, Menlo Park, CA) and the MinION sequencing platforms (Oxford Nanopore Technologies, Oxford, UK). De novo assembly was performed using HGAP 7.0.1.66975 and Canu 1.8 with default parameters (53, 64). The genome sequence was then polished using Pilon 1.23 with default parameters (65). The NCBI (National Center for Biotechnology Information) Prokaryotic Genome Annotation Pipeline was used for annotation (66). Other genomes were sequenced using the Illumina HiSeq PE150 platform, and de novo assembly was conducted using SPAdes 3.12.0 with default parameters (53). Homologs of the plasmid pRZP1 were investigated using genomic BLASTn with genome sequences of plasmids downloaded from the NCBI database. Plasmids with homologs were then compared and visualized using Mauve (67).
Phylogenetic constructions and compositional analysis of HGT.
Maximum likelihood (ML) phylogenies were constructed based on related derived gene or protein sequences. The derived nucleotides were aligned using ClustalW, and the derived amino acids were aligned using Muscle in MEGA X (68). MEGA X or RAxML version 8 was used for ML phylogenetic tree construction (69). The nucleotide sequences of single-copy core genes were identified and aligned using the UBCG pipeline (70). ISfinder (45) (http://www-is.biotoul.fr) was used to annotate IS (insertion sequence) elements.
Mating experiment.
Before the mating experiment, the spread plate method with antibiotic gradient plates (up to 20 to 50 μg/ml) was used to detect natural resistance to antibiotics (kanamycin, streptomycin, erythromycin, tetracycline, and ampicillin). The determined concentration of selected antibiotics was confirmed using the corresponding liquid medium. Ampicillin (final concentration, 10 μg/ml) was used to select the transconjugant, as the donor strain ZPP was the nonresistant strain. In contrast, recipient cells were resistant to ampicillin.
In order to achieve conjugation, the donor strain ZPP and each recipient strain were cultivated at 28°C with shaking at 150 rpm for 48 h. Next, donor cells (1 ml) and an equivalent volume of recipient cells were added to 40 ml of 2-fold-diluted R2A liquid medium in Erlenmeyer flasks and mixed briefly. The Erlenmeyer flasks were allowed to stand for 24 h at 28°C. Next, 0.01 ml of the mixed cell cultures was spread on ampicillin (final concentration, 10 μg/ml)-selective NMS plates and cultured in a sealed jar with a 5% (vol/vol) gas phase of propane in the headspace. Seven days later, single colonies were transferred into sealed 125-ml bottles with 25 ml ampicillin (final concentration, 10 μg/ml)-selective NMS liquid medium with a 5% (vol/vol) gas phase of propane. After several rounds of selection, single colonies that possessed the plasmid but were not strain ZPP were transferred to an NMS liquid scale-up culture with propane. Specific primers designed for the detection of hmoA (forward primer e280F [5′-ACCCTATGTGCAGTCGTGTT-3′] and reverse primer e578R [5′-CCGATGTGGAAGGACATTGTG-3′]) and tcpC (forward primer tcpC256f [5′-GACAACGCCACCGAAGACA-3′] and reverse primer tcpC481r [5′-CGTATGTCCGTGACTTCTCCT-3′]) genes in the plasmid and universal primers for the 16S rRNA gene (forward primer 341F [5′-CCTACGGGNGGCWGCAG-3′] and reverse primer 785R [5′-GACTACHVGGGTATCTAATCC-3′]) (71) were used to verify plasmids and strains using real-time quantitative PCR (qPCR) and conventional PCR. Thermal cycling conditions included a 5-min initial denaturation step at 95°C followed by 40 cycles (for qPCR) or 35 cycles (for conventional PCR) of denaturation at 95°C for 30 s, an annealing step at 55°C for 30 s, elongation at 72°C for 30 s, and a final elongation step at 72°C for 10 min (for conventional PCR). Reactions without DNA templates were included in all cases as a negative control.
Measurement of plasmid maintenance.
Strain ZPP and the newly obtained transconjugant colonies were cultivated with NMS medium in sealed bottles with a 5% (vol/vol) gas phase of propane to maintain functional plasmids. The bottles were incubated at 28°C for 4 days with shaking at 150 rpm. Subsequently, the cultures (OD600 of ∼0.1) were transferred to fresh 2-fold-diluted R2A liquid medium every day to retain the cells in logarithmic phase. For each transfer, cells were collected to extract DNA and perform qPCR using primers for hmoA, tcpC, and 16S rRNA genes. During this experiment, the negative mutant of strain ZPP was selected from a 2-fold-diluted R2A plate and verified using genome sequencing.
Transcriptomes of the donor and the transconjugant.
The donor and the transconjugant were also cultured in sealed bottles with 25 ml NMS medium and a 5% (vol/vol) gas phase of propane at 28°C. The cells of the donor and the transconjugant were harvested in the logarithmic phase after approximately 10 days by centrifugation at 7,000 × g. The supernatant was then filtered through a filter membrane. The total RNA was extracted from this mixture of precipitated cells and the filter membrane residue using a mirVana microRNA (miRNA) isolation kit (Thermo Fisher Scientific, Waltham, MA). The genomic DNA was digested using a recombinant DNase I kit (TaKaRa, Dalian, China). RNA quality was assessed with a Qubit 2.0 instrument and the Qubit RNA HS assay kit. Next, a QIAseq FastSelect 5S/16S/23S kit was used to remove 5S/16S/23S rRNA, according to the manufacturer’s instructions. A QIAseq stranded total RNA library kit was used to build the library according to the manufacturer’s instructions, and cDNA was sequenced on the Illumina PE150 platform. The quality-trimmed reads were mapped to the annotated genome, and TPM values were calculated using RSEM (72) with Bowtie 2 and default parameters (73). The expression levels of housekeeping genes with high general expression stability, specifically those encoding DNA gyrase subunit B and RecA, were marked as the baselines.
Data availability.
Genomes were submitted to GenBank under accession no. CP058638 to CP058643 (ZPP) and JACJUL000000000 [R. erythropolis ATCC 25544(pRZP1)]. Genomic and transcriptomic reads were also submitted to NODE (The National Omics Data Encyclopedia) (https://www.biosino.org/node/) under project identifier OEP001070 and GenBank under BioProject accession no. PRJNA643244.
ACKNOWLEDGMENTS
This research was financially supported by the National Key R&D Program of China (grant no. 2018YFC0310600), the National Natural Science Foundation of China (NSFC) (grant no. 91751107, 31870109, 31811540398, and 31470222), and the Science and Technology Research Program of Shanghai (grant no. 19DZ2282100).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Zhe-Xue Quan, Email: quanzx@fudan.edu.cn.
Alfons J. M. Stams, Wageningen University
REFERENCES
- 1.Khadka R, Clothier L, Wang L, Lim CK, Klotz MG, Dunfield PF. 2018. Evolutionary history of copper membrane monooxygenases. Front Microbiol 9:2493. 10.3389/fmicb.2018.02493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. 1995. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett 132:203–208. 10.1111/j.1574-6968.1995.tb07834.x. [DOI] [PubMed] [Google Scholar]
- 3.Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712. 10.1128/AEM.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688. 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lucker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman NV, Yau S, Wilson NL, Nolan LM, Migocki MD, Ly MA, Crossett B, Holmes AJ. 2011. Untangling the multiple monooxygenases of Mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environ Microbiol Rep 3:297–307. 10.1111/j.1758-2229.2010.00225.x. [DOI] [PubMed] [Google Scholar]
- 8.Coleman NV, Le NB, Ly MA, Ogawa HE, McCarl V, Wilson NL, Holmes AJ. 2012. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J 6:171–182. 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin-Blum M, Antony CP, Borowski C, Sayavedra L, Pape T, Sahling H, Bohrmann G, Kleiner M, Redmond MC, Valentine DL, Dubilier N. 2017. Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat Microbiol 2:17093. 10.1038/nmicrobiol.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rochman FF, Kwon M, Khadka R, Tamas I, Lopez-Jauregui AA, Sheremet A, Smirnova AV, Malmstrom RR, Yoon S, Woyke T, Dunfield PF, Verbeke TJ. 2020. Novel copper-containing membrane monooxygenases (CuMMOs) encoded by alkane-utilizing Betaproteobacteria. ISME J 14:714–726. 10.1038/s41396-019-0561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y, Wei K, Si K, Mathieu J, Li M, Alvarez PJJ. 2017. Whole-genome sequence of the 1,4-dioxane-degrading bacterium Mycobacterium dioxanotrophicus PH-06. Genome Announc 5:e00625-17. 10.1128/genomeA.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin KE, Ozsvar J, Coleman NV. 2014. SmoXYB1C1Z of Mycobacterium sp. strain NBB4: a soluble methane monooxygenase (sMMO)-like enzyme, active on C2 to C4 alkanes and alkenes. Appl Environ Microbiol 80:5801–5806. 10.1128/AEM.01338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda H, McClay K, Steffan RJ, Zylstra GJ. 2012. Characterization of three propane-inducible oxygenases in Mycobacterium sp. strain ENV421. Lett Appl Microbiol 55:175–181. 10.1111/j.1472-765X.2012.03290.x. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T, Nakamura T, Fuse H. 2012. Isolation of two novel marine ethylene-assimilating bacteria, Haliea species ETY-M and ETY-NAG, containing particulate methane monooxygenase-like genes. Microbes Environ 27:54–60. 10.1264/jsme2.me11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith CA, Reilly KT, Hyman MR. 2003. Characterization of the initial reactions during the cometabolic oxidation of methyl-butyl ether by propane-grown Mycobacterium vaccae JOB5. Appl Environ Microbiol 69:796–804. 10.1128/aem.69.2.796-804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamamura N, Arp DJ. 2000. Isolation and characterization of alkane-utilizing Nocardioides sp. strain CF8. FEMS Microbiol Lett 186:21–26. 10.1111/j.1574-6968.2000.tb09076.x. [DOI] [PubMed] [Google Scholar]
- 17.Osborne CD, Haritos VS. 2019. Beneath the surface: evolution of methane activity in the bacterial multicomponent monooxygenases. Mol Phylogenet Evol 139:106527. 10.1016/j.ympev.2019.106527. [DOI] [PubMed] [Google Scholar]
- 18.Kang CS, Dunfield PF, Semrau JD. 2019. The origin of aerobic methanotrophy within the Proteobacteria. FEMS Microbiol Lett 366:fnz096. 10.1093/femsle/fnz096. [DOI] [PubMed] [Google Scholar]
- 19.Crombie AT, Murrell JC. 2014. Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris. Nature 510:148–151. 10.1038/nature13192. [DOI] [PubMed] [Google Scholar]
- 20.Horz H-P, Yimga MT, Liesack W. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl Environ Microbiol 67:4177–4185. 10.1128/aem.67.9.4177-4185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes AJ. 2009. The diversity of soluble di-iron monooxygenases with bioremediation applications, p 91–102. In Singh A, Kuhad RC, Ward OP (ed), Advances in applied bioremediation. Springer, Berlin, Germany. [Google Scholar]
- 22.Farhan Ul Haque M, Crombie AT, Murrell JC. 2019. Novel facultative Methylocella strains are active methane consumers at terrestrial natural gas seeps. Microbiome 7:134. 10.1186/s40168-019-0741-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirajuddin S, Rosenzweig AC. 2015. Enzymatic oxidation of methane. Biochemistry 54:2283–2294. 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382. 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumont MG, Lüke C, Deng Y, Frenzel P. 2014. Classification of pmoA amplicon pyrosequences using BLAST and the lowest common ancestor method in MEGAN. Front Microbiol 5:34. 10.3389/fmicb.2014.00034. [DOI] [Google Scholar]
- 26.Lüke C, Frenzel P. 2011. Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl Environ Microbiol 77:6305–6309. 10.1128/AEM.05355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knief C. 2015. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 6:1346. 10.3389/fmicb.2015.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne CD, Haritos VS. 2018. Horizontal gene transfer of three co-inherited methane monooxygenase systems gave rise to methanotrophy in the Proteobacteria. Mol Phylogenet Evol 129:171–181. 10.1016/j.ympev.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. 2012. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539. 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JG, Xia F, Zeleke J, Zou B, Rhee SK, Quan ZX. 2017. An improved protocol with a highly degenerate primer targeting copper-containing membrane-bound monooxygenase genes for community analysis of methane- and ammonia-oxidizing bacteria. FEMS Microbiol Ecol 93:fiw244. 10.1093/femsec/fiw244. [DOI] [PubMed] [Google Scholar]
- 31.Wen X, Yang S, Liebner S. 2016. Evaluation and update of cutoff values for methanotrophic pmoA gene sequences. Arch Microbiol 198:629–636. 10.1007/s00203-016-1222-8. [DOI] [PubMed] [Google Scholar]
- 32.Xia F, Wang JG, Zhu T, Zou B, Rhee SK, Quan ZX. 2018. Ubiquity and diversity of complete ammonia oxidizers (comammox). Appl Environ Microbiol 84:e01390-18. 10.1128/AEM.01390-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 34.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet 16:472–482. 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 35.Babic A, Berkmen MB, Lee CA, Grossman AD. 2011. Efficient gene transfer in bacterial cell chains. mBio 2:e00027-11. 10.1128/mBio.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson C, Hall JPJ, Harrison E, Wood A, Brockhurst MA. 2017. Gene mobility promotes the spread of resistance in bacterial populations. ISME J 11:1930–1932. 10.1038/ismej.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. 2010. Oxidation of methane by a biological dicopper centre. Nature 465:115–119. 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liew EF, Tong D, Coleman NV, Holmes AJ. 2014. Mutagenesis of the hydrocarbon monooxygenase indicates a metal centre in subunit-C, and not subunit-B, is essential for copper-containing membrane monooxygenase activity. Microbiology (Reading) 160:1267–1277. 10.1099/mic.0.078584-0. [DOI] [PubMed] [Google Scholar]
- 39.Cappelletti M, Zampolli J, Di Gennaro P, Zannoni D. 2019. Genomics of Rhodococcus, p 23–60. In Alvarez HM (ed), Biology of Rhodococcus, 2nd ed. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 40.Bannam TL, Teng WL, Bulach DM, Lyras D, Rood JI. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J Bacteriol 188:4942–4951. 10.1128/JB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsons JA, Bannam TL, Devenish RJ, Rood JI. 2007. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J Bacteriol 189:7782–7790. 10.1128/JB.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng WL, Bannam TL, Parsons JA, Rood JI. 2008. Functional characterization and localization of the TcpH conjugation protein from Clostridium perfringens. J Bacteriol 190:5075–5086. 10.1128/JB.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLeod MP, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJM, Holt R, Brinkman FSL, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci U S A 103:15582–15587. 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalkus J, Menne R, Reh M, Schlegel HG. 1998. The terminal structures of linear plasmids from Rhodococcus opacus. Microbiology (Reading) 144:1271–1279. 10.1099/00221287-144-5-1271. [DOI] [PubMed] [Google Scholar]
- 45.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cappelletti M, Fedi S, Zannoni D. 2019. Degradation of alkanes in Rhodococcus, p 137–171. In Alvarez HM (ed), Biology of Rhodococcus, 2nd ed. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 47.Valero-Rello A, Hapeshi A, Anastasi E, Alvarez S, Scortti M, Meijer WG, MacArthur I, Vázquez-Boland JA. 2015. An invertron-like linear plasmid mediates intracellular survival and virulence in bovine isolates of Rhodococcus equi. Infect Immun 83:2725–2737. 10.1128/IAI.00376-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J, Qiu Y, Ding P, Peng P, Yang H, Li L. 2017. Conjugative transfer of dioxin-catabolic megaplasmids and bioaugmentation prospects of a Rhodococcus sp. Environ Sci Technol 51:6298–6307. 10.1021/acs.est.7b00188. [DOI] [PubMed] [Google Scholar]
- 49.Ren C, Wang Y, Tian L, Chen M, Sun J, Li L. 2018. Genetic bioaugmentation of activated sludge with dioxin-catabolic plasmids harbored by Rhodococcus sp. strain p52. Environ Sci Technol 52:5339–5348. 10.1021/acs.est.7b04633. [DOI] [PubMed] [Google Scholar]
- 50.McCarty GW. 1999. Modes of action of nitrification inhibitors. Biol Fertil Soils 29:1–9. 10.1007/s003740050518. [DOI] [Google Scholar]
- 51.Bédard C, Knowles R. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev 53:68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redmond MC, Valentine DL, Sessions AL. 2010. Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl Environ Microbiol 76:6412–6422. 10.1128/AEM.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 54.Villada JC, Duran MF, Lee PKH. 2019. Genomic evidence for simultaneous optimization of transcription and translation through codon variants in the pmoCAB operon of type Ia methanotrophs. mSystems 4:e00342-19. 10.1128/mSystems.00342-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drlica K, Gennaro ML. 2001. Plasmids, p 1485–1490. In Brenner S, Miller JH (ed), Encyclopedia of genetics. Academic Press, New York, NY. [Google Scholar]
- 56.Sutcliffe IC, Brown AK, Dover LG. 2010. The rhodococcal cell envelope: composition, organisation and biosynthesis, p 29–71. In Alvarez HM (ed), Biology of Rhodococcus. Springer, Berlin, Germany. [Google Scholar]
- 57.Volff J-N, Altenbuchner J. 2000. A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol Lett 186:143–150. 10.1111/j.1574-6968.2000.tb09095.x. [DOI] [PubMed] [Google Scholar]
- 58.Price MN, Arkin AP, Alm EJ. 2006. The life-cycle of operons. PLoS Genet 2:e96. 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocha EPC. 2008. The organization of the bacterial genome. Annu Rev Genet 42:211–233. 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- 60.Bratlie MS, Johansen J, Drabløs F. 2010. Relationship between operon preference and functional properties of persistent genes in bacterial genomes. BMC Genomics 11:71. 10.1186/1471-2164-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874–878. 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 63.Xing X-H, Wu H, Luo M-F, Wang B-P. 2006. Effects of organic chemicals on growth of Methylosinus trichosporium OB3b. Biochem Eng J 31:113–117. 10.1016/j.bej.2006.06.001. [DOI] [Google Scholar]
- 64.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J. 2018. UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285. 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 71.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2, Fig. S1 to S9. Download AEM00227-21_Supp_1_seq6.pdf, PDF file, 4.6 MB (7.7MB, pdf)
Table S3. Download AEM00227-21_Supp_2_seq7.xlsx, XLSX file, 0.03 MB (26.7KB, xlsx)
Data Availability Statement
Genomes were submitted to GenBank under accession no. CP058638 to CP058643 (ZPP) and JACJUL000000000 [R. erythropolis ATCC 25544(pRZP1)]. Genomic and transcriptomic reads were also submitted to NODE (The National Omics Data Encyclopedia) (https://www.biosino.org/node/) under project identifier OEP001070 and GenBank under BioProject accession no. PRJNA643244.