ABSTRACT
Nitrogen limitation has been widely reported to affect the growth and development of fungi, and the transcription factor GCN4 (general control nonderepressible 4) is involved in nitrogen restriction. Here, we found that nitrogen limitation highly induced the expression of GCN4 and promoted the synthesis of ganoderic acid (GA), an important secondary metabolite in Ganoderma lucidum. The activated GCN4 is involved in regulating GA biosynthesis. In addition, the accumulation of reactive oxygen species (ROS) also affects the synthesis of GA under nitrogen restrictions. The silencing of the gcn4 gene led to further accumulation of ROS and increased the content of GA. Further studies found that GCN4 activated the transcription of antioxidant enzyme biosynthesis genes gr, gst2, and cat3 (encoding glutathione reductase, glutathione S-transferase, and catalase, respectively) through direct binding to the promoter of these genes to reduce the ROS accumulation. In conclusion, our study found that GCN4 directly interacts with the ROS signaling pathway to negatively regulate GA biosynthesis under nitrogen-limiting conditions. This provides an essential insight into the understanding of GCN4 transcriptional regulation of the ROS signaling pathway and enriches the knowledge of nitrogen regulation mechanisms in fungal secondary metabolism of G. lucidum.
IMPORTANCE Nitrogen has been widely reported to regulate secondary metabolism in fungi. Our study assessed the specific nitrogen regulatory mechanisms in Ganoderma lucidum. We found that GCN4 directly interacts with the ROS signaling pathway to negatively regulate GA biosynthesis under nitrogen-limiting conditions. Our research highlights a novel insight that GCN4, the nitrogen utilization regulator, participates in secondary metabolism through ROS signal regulation. In addition, this also provides a theoretical foundation for exploring the regulation of other physiological processes by GCN4 through ROS in fungi.
KEYWORDS: nitrogen limitation, GCN4, ROS, ganoderic acids
INTRODUCTION
As an essential nutrient for proteins, nucleic acids, and lipid synthesis, nitrogen plays a critical role in the basal metabolism, growth, and development of fungi. The availability of nitrogen affects fungi extensively. Previous studies indicated that nitrogen limitation triggers morphological transitions in Saccharomyces cerevisiae (1, 2), Aspergillus nidulans (3), and Neurospora crassa (4). Nitrogen limitation also has different effects on secondary metabolism in fungi, such as promoting orsellinic acid and spiroanthrones biosynthesis in A. nidulans (5) and aflatoxin biosynthesis in Aspergillus flavus (6). Numerous studies in microalgae suggest that nitrogen starvation is an effective and widely explored strategy used to induce carbohydrate and lipid accumulation (7, 8). Although there are reports that nitrogen limitation affects the secondary metabolism of fungi, the mechanism is still unclear.
The mechanism of filamentous fungus response to nitrogen source is mainly through the regulation of transcriptional levels. Two transcriptional regulate pathways have been studied well. The first pathway is nitrogen catabolite repression (NCR), which is activated in response to the utilization of nonpreferential nitrogen sources (9). The second pathway is the general amino acid control (GAAC), which stimulates amino acid biosynthesis and nitrogen utilization through the essential transcriptional activator GCN4 (10, 11). GCN4 is a basic region-leucine zipper (bZIP)-type transcription factor containing a C-terminal basic leucine zipper region, which can bind to the UASGCRE (GA[C/G]TCA) motifs in the promoter of its target genes (12). As a global regulator, GCN4 transcriptionally regulates the expression of a wide range of stress response genes to endure stresses. In general, most genes are involved in nutritional stress (13, 14). GCN4 transcriptionally regulates the expression of amino acid-biosynthetic, amino acid transporter, and aminoacyl-tRNA synthetase-encoding genes and genes encoding mitochondrial carrier proteins that modulate nutrition homeostasis (15, 16). Furthermore, the transcription factor CpcA (homologous to GCN4 in yeast) in the ascomycete Leptosphaeria maculans regulates the production of the secondary metabolite toxin sirodesmin PL in response to nitrogen availability (17). In addition, GCN4 also induces the expression of peroxisome biogenesis- and glutathione biosynthesis-related genes as part of the antioxidant response to protect cells against oxidative stress (15, 18) and endoplasmic reticulum (ER) stress (19). Besides its role in regulating gene expression, GCN4 also interacts with other pathways. GCN4 can form a complex with GLN3 (the core transcription factor in NCR) to transcriptionally activate the expression of genes involved in nitrogen utilization (20). Recent studies have also indicated that GCN4 inhibits AMP-activated protein kinase (AMPK) activity by stimulating the expression of hepatic tribbles homolog 3 (TRB3) (21), and ATF4 induces Sesn2 expression to negatively regulate the mechanistic target of rapamycin complex 1 (mTORC1) pathway (22). The function of transcriptional regulation in GCN4 has been extensively studied, but the interaction between GCN4 and reactive oxygen species (ROS) signaling in fungi needs to be further explored.
As a white-rot fungus, Ganoderma lucidum efficiently decomposes lignin and cellulose through a series of lignocellulolytic enzymes to satisfy the need for its growth and development. The study of genome sequencing on G. lucidum revealed that it contains one of the largest sets of enzymes for wood decomposition in the Basidiomycota (23). Beyond that, G. lucidum is also a traditional medicinal fungus in China. It is a valuable herbal medicine that has a history of use for more than 2,000 years. G. lucidum exhibited curative effects based on its antitumor, liver protection, detoxification, anti-aging, stroke, blood lipid-lowering, antidiabetes, and anti-arthritis activities (24, 25). Ganoderic acid (GA) is a major category of pharmacologically active compounds in G. lucidum. A previous study by Zhao et al. showed that GA content increased under conditions of nitrogen limitation, and they hypothesized that the nitrogen regulator AreA regulated the transcription of GA biosynthesis genes (26). Moreover, the addition of calcium ions under conditions of nitrogen limitation could further promote GA biosynthesis (27). However, the molecular mechanism for the increase in GA content under nitrogen limitation in G. lucidum remains unclear. The identification of regulators involved in GA biosynthesis is an important issue that deserves attention. Therefore, it is necessary to thoroughly evaluate the specific regulatory mechanism of the biosynthesis of GA in G. lucidum to provide a theoretical foundation for obtaining the optimal GA contents.
This study sought to investigate the regulatory mechanism of GCN4 on secondary metabolism under nitrogen limitation. We report that nitrogen limitation activated GCN4 expression and promoted ROS accumulation. The increased ROS level stimulated the synthesis of GA. Furthermore, GCN4 directly activated the expression of relevant antioxidant genes to attenuate increased ROS levels. In conclusion, GCN4 reduced GA contents by negatively regulating ROS under nitrogen limitation. This finding revealed the innovative view that GCN4 negatively regulates the ROS signaling pathway by directly activating the expression of relevant antioxidant genes. In addition, as an essential regulator in nitrogen utilization, GCN4 participated in the regulation of secondary metabolism through the ROS signaling pathway. Our findings provide unique insights into the interaction between nitrogen utilization and signaling pathways to regulate secondary metabolism.
RESULTS
Increased GA production under nitrogen limitation conditions.
To better understand the effect of nitrogen on the growth and development of Ganoderma lucidum, we first detected mycelial growth on solid medium with limited nitrogen (3 mM asparagine) and rich nitrogen (60 mM Asn) conditions. Mycelia cultured in 3 mM Asn grew slowly and sparsely compared with those in 60 mM Asn (Fig. 1A and B). However, ganoderic acid (GA) contents in mycelia treated with 3 mM Asn were 1.50-fold increased compared with those in mycelia treated with 60 mM Asn (Fig. 1C). The expression of three key genes, hmgr, sqs, and osc (encoding 3-hydroxy-3-methyglutaryl-coenzyme A reductase [HMGR], squalene synthase [SQS], and lanosterol synthase [OSC], respectively), involved in GA biosynthesis were increased 2.25-, 1.45-, and 1.95-fold, respectively, in 3 mM Asn compared with 60 mM Asn (see Fig. S1 in the supplemental material). These results provide preliminary evidence that restriction of nitrogen inhibited mycelial growth but promoted GA biosynthesis.
FIG 1.

GA production increased under nitrogen limitation. (A) Mycelial growth in the WT strain cultivated under nitrogen-limiting conditions. (B) Analysis of the mycelial diameters in the WT strain using a histogram. (C) GA content in WT strain under nitrogen-limiting conditions. The WT strain was cultured with modified CYM with 3 mM and 60 mM asparagine as the sole nitrogen source at 28°C for 7 days for mycelial growth assays. Strains were cultured by using a fermentation strategy in CYM with shaking for 4 days at 28°C. Then, cultures were shifted to modified CYM that contained 3 mM and 60 mM Asn as the sole nitrogen source, cultured with shaking for 5 days at 28°C, and collected for measurement of GA levels. The data are means and SD (n = 3). Statistical significance (*, P < 0.05; **, P < 0.01) was determined by Student's t test.
GCN4 was induced at transcriptional and translational levels under nitrogen-limiting conditions.
To assess whether GCN4 responds to nitrogen-limiting conditions, the gcn4 gene homolog (GL28195) in G. lucidum was identified by alignment with GCN4 protein in Saccharomyces cerevisiae (GenBank accession number NP_010907.3). The gcn4 gene contains an open reading frame with a 903-bp sequence and encodes a protein containing 300 amino acids. A further prediction of the protein domain from the SMART website indicated that the protein contained only one conserved characteristic basic region-leucine zipper (bZIP) domain. This indicated that GCN4 in G. lucidum belongs to the bZIP family. We investigated GCN4 mRNA and protein levels at 3 mM and 60 mM Asn. As shown in Fig. 2, compared with 60 mM Asn, the expression of gcn4 mRNA and protein was upregulated 3.91-fold and 1.49-fold, respectively, at 3 mM Asn (Fig. 2A to C). These results indicate that GCN4 was induced at both transcriptional and translational levels in response to nitrogen limitation.
FIG 2.
GCN4 was induced at transcriptional and translational levels under nitrogen-limiting conditions. (A) Relative transcript levels of gcn4 in the WT strain grown with 3 mM and 60 mM Asn. (B and C) Western blot analysis of GCN4 protein levels in WT strains grown with 3 mM and 60 mM Asn and histogram showing the GCN4/β-tubulin ratio. (D) relative transcript levels of gcn4 in different strains grown with 3 mM and 60 mM Asn. (E and F) Western blot analysis of GCN4 protein levels in different strains grown with 3 mM and 60 mM Asn and histogram showing the GCN4/β-tubulin ratio. The transcript levels of gcn4 and the GCN4/β-tubulin ratio in the WT strain grown with 60 mM Asn were defined as 1. Data are means and SD (n = 3). Statistical significance was determined by Student's t test. **, P < 0.01; ns, not significant. In panel D, bars with different letters are significantly different (P < 0.05) based on Tukey's test.
To better understand the function of GCN4 in nitrogen limitation, we constructed a gcn4-silencing vector and transformed it into G. lucidum (Fig. S2A). Transformants were assessed using quantitative real-time PCR (qRT-PCR). Two transformants, the gcn4i-1 and gcn4i-22 strains (exhibiting 79% and 81% silencing efficacy, respectively), were selected for subsequent studies (Fig. S2B). We also detected GCN4 protein levels in gcn4i-1 and gcn4i-22 strains. Compared with the WT strain, GCN4 protein levels in gcn4-silenced strains were decreased to 59% and 68%, respectively (Fig. S2C and D).
We further detected GCN4 mRNA and protein levels in WT and gcn4-silenced strains that were exposed to different nitrogen concentrations. Silencing in gcn4 decreased the expression levels of gcn4 under both 3 mM and 60 mM Asn conditions (Fig. 2D). In addition, GCN4 protein levels in gcn4i-1 and gcn4i-22 strains grown with 3 mM Asn were decreased approximately 48% and 43% compared with that in the wild type (WT), respectively (Fig. 2E and F). Taken together, the above results indicate that GCN4 is induced in response to nitrogen restriction.
GCN4 regulates growth and GA biosynthesis under nitrogen limitation.
To determine the effect of GCN4 in G. lucidum, we first observed the mycelial growth of WT and gcn4-silenced strains when cultured in 3 mM and 60 mM Asn. Silencing gcn4 inhibited the growth of mycelium and exhibited a stronger inhibition in the presence of 3 mM Asn (Fig. 3A and B). The above results showed that GCN4 plays an important role in the mycelial growth in both nitrogen-sufficient and -limiting conditions.
FIG 3.

GCN4 affected growth and GA biosynthesis under nitrogen-limiting conditions. (A) Mycelial growth in WT, SiControl, and gcn4-silenced strains under nitrogen-limiting conditions. (B) Analysis of mycelial diameters in different strains presented as a histogram. (C) GA content in WT, SiControl, and gcn4-silenced strains. Data are means and SD (n = 3). Statistical significance is represented by different letters corresponding to a P value of <0.05 based on Tukey’s test.
Then, we measured the content of GA and the transcript levels of three key genes involved in GA biosynthesis. Silencing of gcn4 significantly increased GA contents by 1.51-fold and 2.20-fold at 3 mM Asn. The expression of hmgr, sqs, and osc was also dramatically increased in the gcn4i-1 and gcn4i-22 strains cultured with 3 mM Asn, compared with WT strains. However, no significant difference was noted between gcn4-silenced and WT strains cultured under nitrogen-sufficient conditions (Fig. S3). In conclusion, gcn4 silencing promotes GA biosynthesis under nitrogen-limiting conditions.
Nitrogen limitation induces the accumulation of intracellular ROS.
ROS serve as signaling molecules involved in many cellular processes, including nitrogen limitation (28). It is reported that ROS affects GA biosynthesis in G. lucidum (29). Therefore, we investigated ROS levels in the WT strain cultivated with different nitrogen concentrations. The fluorescence intensity of the WT strain cultured with 3 mM Asn was about 1.76 times higher than that obtained with 60 mM Asn (Fig. 4A and B). In addition, the H2O2 level in the WT strain cultured with 3 mM Asn increased by 1.88-fold compared with that in 60 mM Asn (Fig. 4C).
FIG 4.

Nitrogen limitation induced the accumulation of intracellular ROS. (A) Images of the mycelial ROS detected by DCFH-DA staining in WT, SiControl, and gcn4-silenced strains grown with 3 mM and 60 mM Asn. The intensity of green fluorescence represents the ROS level. (B) Quantitative analysis of the changes in the ROS fluorescence ratio of the strains grown under nitrogen limitation shown in panel A. (C) Relative H2O2 content in WT, SiControl, and gcn4-silenced strains grown with 3 mM or 60 mM Asn. The ROS level and H2O2 content in the WT strain cultivated under 60 mM Asn were defined as 1. Data are means and SD (n = 3). Statistical significance is represented by different letters corresponding to a P value of <0.05 based on Tukey’s test.
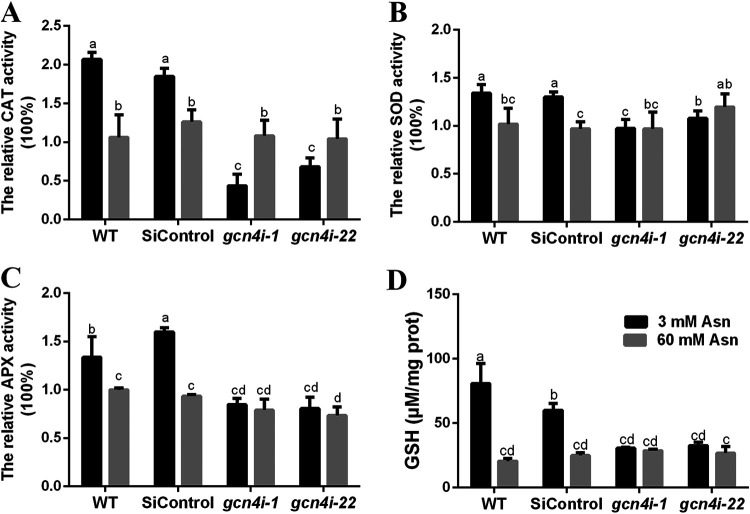
We further measured the activities of ROS-producing enzymes (NADPH oxidase) and antioxidants (including the enzymatic antioxidants catalase [CAT], superoxide dismutase [SOD], ascorbate peroxidase [APX], glutathione peroxidase [GPX], and nonenzymatic antioxidant glutathione [GSH]). As shown in Fig. 5, antioxidant levels in the WT strain were slightly increased in mycelia cultured with 3 mM Asn. The activity of APX, CAT, and SOD in the WT strain cultured at 3 mM Asn increased approximately 1.34-, 2.07-, and 1.31-fold relative to that in 60 mM Asn. Similarly, the GSH levels in the WT strain were also increased 2.93-fold when the strain was treated with 3 mM Asn. However, NOX and GPX activities in the WT strain did not markedly differ between 3 mM and 60 mM Asn (Fig. S4A). Collectively, these data suggest that nitrogen limitation leads to the accumulation of ROS and increased antioxidant activity.
FIG 5.
GCN4 regulated antioxidant activities in response to the ROS accumulation under nitrogen limitation. (A to D) Relative activities of CAT, SOD, APX, and the content of GSH in different strains grown with 3 mM and 60 mM Asn. The antioxidant activity in the WT strain cultivated with 60 mM Asn was defined as 1. The data are means and SD (n = 3). Statistical significance is represented by different letters corresponding to a P value of <0.05 based on Tukey’s test.
GCN4 regulates antioxidant activities in response to the ROS accumulation under nitrogen limitation.
Next, we measured ROS levels in WT and gcn4-silenced strains. At 3 mM Asn, silencing of gcn4 increased the levels of ROS by 1.33- and 1.52-fold and was upregulated by 1.88- and 2.15-fold over that in 60 mM Asn (Fig. 4A and B). This indicates that gcn4 silencing under nitrogen limitation promotes ROS accumulation. We further detected the H2O2 content of these strains under these conditions. H2O2 levels in gcn4-silenced strains cultured under 3 mM Asn increased to 1.52- and 1.22-fold compared with the WT strain, respectively. H2O2 levels in gcn4-silenced strains cultured with 3 mM Asn increased to 2.14- and 1.97-fold that noted for 60 mM Asn (Fig. 4C). These results demonstrated that GCN4 is essential for negatively regulating intracellular ROS levels under the nitrogen-limiting condition.
We further explored the effect of GCN4 on the activities of antioxidants. Compared with that in WT strains cultured with 3 mM Asn, CAT, SOD, and APX activities and GSH contents were dramatically downregulated in gcn4i-1 and gcn4i-22 strains. In particular, CAT activity was significantly reduced by 78.88% and 67.03%, respectively (Fig. 5). The GSH levels in gcn4-silenced strains were also significantly reduced to 38% and 40%, respectively (Fig. 5D). To further explore the mechanism of GCN4 on these antioxidants, the expression of CAT, SOD, APX, and GSH synthesis genes in gcn4-silenced strains was detected. The expression of these genes in gcn4i-1 and gcn4i-22 strains was generally decreased compared with that in the wild-type strain under nitrogen limitation conditions. Among them, the most dramatic changes were observed in cat3, gr, and gst2 genes (Fig. 6). Taken together, these results indicate that GCN4 positively regulates the antioxidant activity and the expression of related genes to alleviate the accumulation of ROS under nitrogen-limiting conditions.
FIG 6.
GCN4 regulated the expression of antioxidant-related genes under nitrogen limitation. Relative expression levels of several antioxidant-related genes (including cat1, cat3, sod2, sod4, gr, and gst2) in the gcn4 transformants under 3 mM and 60 mM Asn conditions. The transcript level in the WT strain cultivated with 60 mM Asn was defined as 1. Data are means and SD (n = 3). Statistical significance is represented by different letters corresponding to a P value of <0.05 based on Tukey’s test.
GCN4 directly binds and activates antioxidant gene expression.
GCN4 induces the expression of genes by interacting with the UASGCRE (GA[C/G]TCA) motif in their promoter regions (30, 31). We further explored the transcriptional activation mechanism of GCN4 on antioxidant-related genes. JASPAR software was used to predict the binding profiles of these antioxidant-related genes. The prediction results show that the promoter regions of gr, gst1, gst2, cat2, cat3, and sod1 genes (involved in the biosynthesis of GSH, CAT, and SOD, respectively [Fig. S5A]) harbor UASGCRE motifs. According to these results, we conducted yeast one-hybrid assays to detect a binding effect. Yeast cell growth on plates demonstrated that GCN4 binds to UASGCRE motifs in the promoter of gr, cat3, and gst2. However, the binding effect disappeared when the UASGCRE motifs were mutated in these genes (Fig. 7A). In addition, GCN4 did not bind to the promoters of gst1, cat2, and sod1 genes (Fig. S5B).
FIG 7.
GCN4 directly binds and activates the expression of antioxidant genes. (A) Detection of the binding effect of GCN4 and the promoter region of antioxidant genes (gr, cat3, and gst2) with the yeast one-hybrid assay. The vector contained GCN4 in Y1HGold yeast strain was mixed with the vector containing the promoter region of antioxidant genes or mutant promoter regions, subjected to gradient dilution, and dotted on plates with SD/−Leu and SD/−Leu containing 300 ng ml−1 AbA (SD/−Leu/AbA300). The consensus binding motif is shown in Fig. S5. (B) EMSA verified the binding effect of GCN4 to the promoter region of antioxidant genes (gr, cat3, and gst2). The amounts of purified recombined GCN4 used were approximately 0 to 0.5 μM, and about 10 ng of the biotin-labeled probe (promoter fragments of gr, cat3, and gst2) was added to each reaction mixture. The 100-fold dilution of the unlabeled specific probe(s) was used as competitor DNA.
To further confirm the direct binding effect of GCN4 in these genes, the electrophoretic mobility shift assay (EMSA) was used to verify the ability of GCN4 to bind to the promoter regions of gr, cat3, and gst2 genes. The full-length sequence of the gcn4 gene was fused to the pET28a expression vector containing a His tag. DNA fragments containing predicted UASGCRE motif regions of gr, cat3, and gst2 labeled with 5′ biotin were used as probes. EMSA results demonstrated that the GCN4 fusion protein bound to the promoter regions of gr, cat3, and gst2 genes, and the binding effects were concentration dependent (Fig. 7B). In addition, the binding effects were eliminated in competitive experiments using a 100-fold-diluted mixture of unlabeled corresponding probes. In conclusion, yeast one-hybrid assay and EMSA results demonstrate that GCN4 directly binds to the UASGCRE motifs in gr, cat3, and gst2 genes to transcriptionally activate the expression of these genes.
GCN4 regulates GA through antioxidant responses under nitrogen-limiting conditions.
We further explored whether the regulation of GA by GCN4 under nitrogen limitation is mediated by ROS. Exogenous H2O2 and GSH (ROS scavenger) were added to determine GA contents. As shown in Fig. 8, exposure to H2O2 increased GA contents in all strains. In particular, the GA contents in gcn4-silenced strains increased to 1.44- and 1.45-fold, respectively, compared to that in the WT. This suggested that GCN4 hindered the promotion of GA synthesis by intracellular ROS. Furthermore, when treated with GSH, the contents of GA in all these strains were decreased, and no significant difference in GA contents was noted between WT and gcn4-silenced strains. The expression of related genes was consistent with the trend noted for GA content. The addition of H2O2 increased the expression levels of genes, and GSH treatment alleviated the trend of gene upregulation (Fig. S6). In summary, we conclude that GCN4 reduces GA content by inhibiting intracellular ROS levels.
FIG 8.

GCN4 regulates GA biosynthesis through antioxidant responses under nitrogen-limiting conditions. GA content in different strains grown with 3 mM Asn is shown. When the strains were shifted to nitrogen-limiting medium, 8 mM H2O2 and 2 mM GSH were added. Data are means and SD (n = 3). Statistical significance is represented by different letters corresponding to a P value of <0.05 based on Tukey’s test.
DISCUSSION
Nitrogen limitation is important nutritional stress that affects invasive growth and differentiation and influences the virulence, pathogenicity, and morphogenetic switches in fungi (32, 33), shoot-root allocation, root architecture, senescence, and flowering in plants (34, 35), and autophagy, growth arrest, and apoptosis in mammalian cells (36, 37). In our study, limited nitrogen inhibited mycelial growth but greatly increased the synthesis of GA. This finding is consistent with studies on secondary metabolism. In Fusarium fujikuroi, a low-nitrogen medium (lower concentration of NH4NO3) produced a higher content of carotenoids, while the synthesis of gibberellins and bikaverins occurred only in the low-nitrogen medium or upon removal the nitrogen (38). Furthermore, low nitrogen concentrations increased gibberellin biosynthesis 3-fold in Gibberella fujikuroi (39). Low concentrations of urea and NaNO3 as the nitrogen-limiting condition is a strategy to increase starch accumulation in algae (40, 41). In conclusion, no matter what nitrogen sources are used, low nitrogen concentration can affect the secondary metabolism. So far, there are few studies on the specific mechanism of secondary metabolism and their involvement by other physiological signaling networks. In this regard, our research found that ROS serves as a signaling molecule in this regulatory process.
ROS plays an important physiological role in the regulation of different cellular functions. A low level of ROS acts as a signaling molecule within cells, while excessive production of ROS can oxidize cell constituents, such as DNA, proteins, and lipids, ultimately leading to cell death (42, 43). Here, we found that nitrogen limitation increased the production of GA. Our previous studies suggested that heat stress or methyl jasmonate (MeJA) treatment induced the accumulation of intracellular ROS to positively regulate GA biosynthesis in G. lucidum (44, 45). In addition, NOX-generated ROS regulates GA biosynthesis (46). In our study, we found that nitrogen limitation leads to the accumulation of ROS. Consistently, studies in Arabidopsis thaliana reported that ROS levels were increased in specific regions of the root in response to nitrogen deprivation (47). A study in pathogenic Colletotrichum acutatum indicated that ROS was minimally detected when mycelia were grown in a complete medium but that ROS levels were significantly enhanced under nitrogen-limiting conditions (48). However, there are relatively few studies on the regulation of ROS through nitrogen limitation in filamentous fungi, and the specific mechanism remains unclear. Research on the interaction mechanism between GCN4 and signaling pathways is limited. In our study, we found that GCN4 activated the transcription of ROS scavenging-related enzyme genes through its binding effect, thereby affecting the ROS signaling pathway. This study identified a new mechanism by which GCN4 affects ROS. In addition to the effects of ROS on secondary metabolism, ROS may exhibit extensive and complex effects on other physiological functions of microorganisms.
The secondary metabolism of fungi is controlled by a complicated regulatory network in response to various environmental stimuli. As a basal nutrient, nitrogen also participates in the regulation of secondary metabolism. A previous study by Zhao et al. demonstrated that the contents of GA were increased under conditions of ammonium sulfate, glutamine, and asparagine limitation (26). Our study confirmed that low concentrations of Asn promoted the increase of GA content and induced the expression of GCN4. The global nitrogen regulator AreA has been extensively proven to participate in regulating secondary metabolism. It can regulate secondary metabolism in either positive or negative ways (49). Our previous study in G. lucidum found that AreA exerts dual regulation on the biosynthesis of GA under different nitrogen sources (50), which indicates that the regulation of secondary metabolism by nitrogen is a complicated process. The regulatory role of GCN4 in the process of nitrogen limitation has rarely been studied in previous studies of filamentous fungi. In this study, we found that the silencing of the gcn4 gene facilitated GA biosynthesis by enhancing the transcript levels of three GA biosynthesis-related genes under nitrogen-limiting conditions. However, our results are insufficient to demonstrate the direct regulation by GCN4 of GA synthesis genes. We provide a novel insight into the specific mechanism by which GCN4 negatively regulates GA biosynthesis. However, it is not clear whether AreA has a similar regulatory effect on GA biosynthesis under nitrogen limitation. In addition, whether AreA and GCN4 cooperate in the regulation of secondary metabolism requires further study.
In our previous studies, we found a close connection between ROS signal and GA biosynthesis. Our present study establishes a framework showing that nitrogen limitation increased the production of GA through the accumulation of ROS. The essential regulator GCN4 was also induced to directly bind to the promoter regions of the antioxidant genes gr, gst2, and cat3 to increase their transcript levels. This helps to alleviate the accumulation of ROS to maintain the redox balance and ultimately relieve the increase in GA content (Fig. 9). Overall, our study strengthens the view that under nitrogen limitation conditions, GCN4 is involved in the negative regulation of secondary metabolism by scavenging intracellular ROS in G. lucidum. These results deepen our understanding of the relationship between GCN4, ROS signaling pathway, and secondary metabolism.
FIG 9.

Model demonstrating that GCN4 negatively regulates the ROS signal pathway to affect GA biosynthesis in G. lucidum under nitrogen-limiting conditions. The nitrogen-limiting condition results in the accumulation of intracellular ROS. Increased ROS levels stimulate GA biosynthesis. Moreover, as signaling molecules, ROS also activate the expression of the transcription factor GCN4. GCN4 carries out transcriptional activation of relevant antioxidant genes by directly binding to the promoters of these genes to relieve ROS damage. Eventually, the content of GA decreases due to the reduction in ROS.
MATERIALS AND METHODS
Strains and culture conditions.
The dikaryotic Ganoderma lucidum strain used in this study, ACCC53264, was obtained from the Agricultural Culture Collection of China (23). The strains were cultured on mushroom complete yeast medium (CYM; 2% glucose, 1% maltose, 0.46% KH2PO4, 0.2% yeast extract, 0.2% tryptone, and 0.05% MgSO4·7H2O) at 28°C. The G. lucidum strains were maintained on potato dextrose agar (PDA) slants. A 0.5-cm3 inoculation block was obtained and inoculated into the CYM plates. After culture at 28°C for 7 days, the mycelial growth was measured. For the nitrogen-limiting experiments, 3 mM or 60 mM asparagine (Asn) was used as the sole nitrogen source and replaced yeast extract and tryptone in CYM. Mycelia were cultured in plates with modified CYM solid medium containing 3 mM or 60 mM Asn as the sole nitrogen source for 7 days at 28°C. For the fermentation experiment, the strains in the CYM plate were punched out in 8 to 10 inoculated blocks of the same size with a hole punch. Then the blocks were transferred and inoculated in the liquid medium. For the nitrogen-limiting condition, we proposed a two-stage cultivation strategy. First, mycelia were grown in the CYM in a flask with shaking for 4 days at 28°C to high biomass levels and then shifted to modified CYM with 3 mM or 60 mM asparagine as the sole nitrogen source in a flask with shaking for 5 days at 28°C. H2O2 (8 mM) and glutathione (GSH, 2 mM) were added when cultures were changed from CYM into a nitrogen-limiting medium. Escherichia coli [DH5α and Rosetta(DE3)] was cultured in LB medium containing ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1).
Transformant construction.
The sequence of the gcn4 gene in G. lucidum was obtained by aligning the amino acid sequence of S. cerevisiae GCN4 with the database of the G. lucidum genome, and the coding sequence of the gcn4 was cloned with the cDNA of G. lucidum with the primers listed in Table S1. The pAN7-dual vector (51) was used to construct gcn4-silencing plasmid and an empty vector control using RNA interference (RNAi) methods. The gcn4-silencing plasmid was transformed into the dikaryotic G. lucidum using the method of Mu et al. (51). For transformant screening, we first selected on CYM with 100 μg ml−1 hygromycin B; then, further screening was performed in CYM with 600 mg ml−1 5-fluoroorotic acid (5-FOA). Two independent silenced strains, the gcn4i-1 and gcn4i-22 strains, with high silencing efficiency were selected for further study. The empty vector control was also selected according to the above-described method and named SiControl.
qRT-PCR analysis.
The strains were cultured in the appropriate media, and mycelia were collected for RNA extraction. Total RNA was extracted using RNAiso Plus reagent (TaKaRa, Dalian, China), and cDNA synthesis was performed using the 5× All-In-One RT MasterMix kit (with an AccuRT genomic DNA removal kit; ABM, Canada). Specific gene expression levels in the WT and transformant strains were detected with quantitative real-time PCR; the primers used are listed in Table S1. The expression of individual genes was normalized against 18S mRNA levels. The relative expression level of genes was evaluated with the 2−ΔΔCT method (52). The transcript levels of all genes were expressed as relative expression levels.
ROS detection.
The fluorescent dye DCFH-DA (2′,7′-dichlorodihydrofluorescein diacetate) was used to detect intracellular ROS levels in WT and gcn4-silenced strains according to the previously described method (46). Detection was performed using a Zeiss Axio Imager A1 fluorescence microscope, and images were analyzed using ZEN lite software (Zeiss, Germany). H2O2 levels in WT and gcn4-silenced strains were detected with the hydrogen peroxide assay kit (Jiancheng Bioengineering Institute, Nanjing, China) (53).
Enzymatic activity assays.
The sample of WT and transformant strains were ground with liquid nitrogen and homogenized in 0.5 ml potassium phosphate buffer (phosphate-buffered saline [PBS]; 50 mM, pH 7.0). Then, the supernatant was obtained for enzymatic activity detection. Total NADPH oxidase (NOX), superoxide dismutase (SOD), and catalase (CAT) activity in the WT, SiControl, and gcn4-silenced strains were detected according to the previous methods (53, 54). Ascorbate peroxidase (APX) and glutathione peroxidase (GPX) activities were measured using corresponding assay kits (Solarbio, Beijing, China), and GSH content was measured using GSH and GSSG assay kits (Beyotime Institute of Biotechnology, Shanghai, China). The Bradford method was used to quantify protein concentrations, and the enzyme activity of the control sample, i.e., the WT cultured in 60 mM Asn, was defined as 1. The enzyme activities of other samples are displayed as the fold change relative to the control sample.
Detection of GA content.
The extraction and determination of total ganoderic acid (GA) were performed by a previously described method (55). Mycelia were cultivated with a two-stage cultivation strategy, and the mycelia with different treatments were collected at the same time for GA extraction. Briefly, 200-mg samples were added to 10 ml 95% (wt/vol) ethanol for 2 h ultrasonic extraction. Samples were incubated at 37°C overnight and then centrifuged. The supernatants were dried on a 60°C rotary evaporator to remove the solvent. Finally, 1 ml methanol was added to dissolve the residue. An ultraperformance liquid chromatography system (UPLC; Agilent Technologies, Santa Clara, CA, USA) equipped with a 2.1- by 100-mm Agilent Zorbax Eclipse Plus C18 Rapid Resolution HD 18-μm column was used to detect and analyze the processed samples.
GCN4 polyclonal antibody preparation.
To acquire recombinant GCN4 protein, the coding sequence of gcn4 was inserted into the pET28a(+) expression vector, which was used to transform E. coli Rosetta(DE3) strains. Positive clones were cultivated in LB medium containing kanamycin (50 μg ml−1) and grown at 37°C in a rotary shaker incubator to enrich biomass to a culture density with an optical density at 600 nm (OD600) of approximately 0.4 to 0.6. Then, IPTG (isopropyl-β-d-thiogalactoside) was added, and samples were cultured for 6 h in a rotary shaker incubator at 25°C to induce protein expression. Cells were collected and sonicated using an ultrasonic cell crusher (JY92-IIDN; Scientz, Ningbo, China). Then, His-GCN4 protein was purified using a nickel nitrilotriacetic acid (Ni-NTA) agarose column (Sangon Biotech, Shanghai, China) and detected by SDS-PAGE, and purified His-GCN4 proteins were then sent for antibody preparation (Chemgen Biotech, Shanghai, China).
Western blotting.
The samples were ground in liquid nitrogen, and protein extraction and Western blotting were performed according to methods described previously (56). The total proteins (25 μg) from samples were separated on a 12% (wt/vol) SDS-PAGE gel, transferred to polyvinylidene difluoride membranes (Bio-Rad, USA), incubated with polyclonal antibody to GCN4 (1:1,000), and then incubated with a secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody. A β-tubulin-specific antibody (1:2,000; AT0003; CMCTAG) served as the internal control. Finally, blots were developed using the ECL Western blotting detection system (Amersham Bioscience, Sweden). Images were acquired with the Bio-Rad ChemiDoc Touch imaging system (Bio-Rad, USA), and densitometry was performed using ImageJ v1.8.0 software.
Yeast one-hybrid assay.
The JASPAR database (http://jaspar.genereg.net/) was used to analyze the putative GCN4 binding sites. The yeast one-hybrid assay was performed according to the protocol of the Matchmaker one-hybrid system (Clontech/Biosciences, Palo Alto, CA, USA). DNA fragments of gr, gst1, gst2, cat2, cat3, and sod1 promoter regions that contain the GCN4 binding sites (TGA[G/C]TCA) and promoter regions containing the mutated motif GCGCGCGCGC were cloned into the pAbAi vector (Invitrogen, Carlsbad, CA, USA), and the coding sequence of the gcn4 gene was cloned into the pGADT7 vector. Plasmids were linearized by BstBI (New England Biolabs) digestion and transformed into Y1HGold yeast strains to generate reporter strains (57). Strains were cultured in synthetically defined medium lacking uracil (SD/−Ura) at 30°C for 3 days to screen transformants, and colony PCR was used to verify transformants. Then, the resistance concentration of aureobasidin A (AbA) was determined in transformants. Then, pGADT7-GCN4 and empty negative-control vector were transformed into the above-described Y1HGold yeast transformants and plated on SD/−Leu plates. Plates were incubated at 30°C for 3 days. Transformants were plated onto SD/−Leu and SD/−Leu/AbA plates separately and cultured for 2 to 3 days at 30°C to detect the transcriptional activation effect. All primers used here are listed in Table S1.
Electrophoretic mobility shift assay.
For electrophoretic mobility shift assays (EMSAs), the DNA fragments of gr, gst2, and cat3 were amplified by PCR with primers labeled with or without 5′ biotin probes (Table S1). PCR products and recombinant protein GCN4 were purified, and the concentration was detected using Nanodrop2000 (Thermo Fisher Scientific, USA). Recombinant GCN4 protein (0 to 0.5 μM) was incubated with labeled DNA probes (∼10 ng) individually in binding buffer (10 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol [DTT], 0.1 mM EDTA, 50 mM KCl, and 5% glycerol) at 25°C for 30 min. For competition experiments, 100-fold diluted unlabeled DNA fragment probes are added to the reaction. The mixture without protein served as a negative control. Electrophoresis was performed using nondenaturing 6% polyacrylamide gels with 0.5× Tris-borate-EDTA (TBE) running buffer at 4°C to separate the complex and free probe. The following steps were completed using the chemiluminescent nucleic acid detection module kit (Thermo Fisher Scientific, USA). Images were acquired using the Bio-Rad ChemiDoc Touch imaging system.
Statistical analysis.
Statistical analysis in this study was performed using GraphPad Prism 6 (GraphPad Software, Inc., San Diego, CA, USA). Data from at least three independent sample measurements were averaged, and values shown are the means and standard deviations (SD). Data analyses using Student’s t test were used for two-way comparisons, and one-way analysis of variance (ANOVA) with Duncan’s posttest was used for multiple comparisons.
Data availability.
The nucleotide sequences of gcn4 in the G. lucidum strain ACCC53264 have been deposited in GenBank under the accession number MN380309.
ACKNOWLEDGMENTS
This work was supported by China Agriculture Research System of MOF and MARA (grant CARS20), the National Natural Science Foundation of China (grants 31972059 and 31900065), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant KYCX19_0613).
Footnotes
Supplemental material is available online only.
Contributor Information
Mingwen Zhao, Email: mwzhao@njau.edu.cn.
Emma R. Master, University of Toronto
REFERENCES
- 1.Mitchell AP. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev 58:56–70. 10.1128/MR.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090. 10.1016/0092-8674(92)90079-R. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann B, Wanke C, LaPaglia SK, Braus GH. 2000. c-Jun and RACK1 homologues regulate a control point for sexual development in Aspergillus nidulans. Mol Microbiol 37:28–41. 10.1046/j.1365-2958.2000.01954.x. [DOI] [PubMed] [Google Scholar]
- 4.Müller B, Russo V. 1989. Nitrogen starvation or glucose limitation induces conidiation in constantly shaken liquid cultures of Neurospora crassa. Fungal Genet Newsl 36:58–60. 10.4148/1941-4765.1508. [DOI] [Google Scholar]
- 5.Scherlach K, Sarkar A, Schroeckh V, Dahse H-M, Roth M, Brakhage AA, Horn U, Hertweck C. 2011. Two induced fungal polyketide pathways converge into antiproliferative spiroanthrones. Chembiochem 12:1836–1839. 10.1002/cbic.201100132. [DOI] [PubMed] [Google Scholar]
- 6.Ehrlich KC, Cotty PJ. 2002. Variability in nitrogen regulation of aflatoxin production by Aspergillus flavus strains. Appl Microbiol Biotechnol 60:174–178. 10.1007/s00253-002-1094-5. [DOI] [PubMed] [Google Scholar]
- 7.Burch AR, Franz AK. 2016. Combined nitrogen limitation and hydrogen peroxide treatment enhances neutral lipid accumulation in the marine diatom Phaeodactylum tricornutum. Bioresour Technol 219:559–565. 10.1016/j.biortech.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Longworth J, Wu D, Huete-Ortega M, Wright PC, Vaidyanathan S. 2016. Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res 18:213–224. 10.1016/j.algal.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, van Helden J, André B. 2007. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol 27:3065–3086. 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch AG, Fink GR. 1986. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisia [sic]. CRC Crit Rev Biochem 21:277–317. 10.3109/10409238609113614. [DOI] [PubMed] [Google Scholar]
- 11.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21:4347–4368. 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. 2014. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 38:254–299. 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolfes RJ, Hinnebusch AG. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol Cell Biol 13:5099–5111. 10.1128/mcb.13.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valenzuela L, Aranda C, González A. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J Bacteriol 183:2331–2334. 10.1128/JB.183.7.2331-2334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinnebusch AG, Natarajan K. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell 1:22–32. 10.1128/ec.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ljungdahl PO, Daignan-Fornier B. 2012. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190:885–929. 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott CE, Fox EM, Jarvis RS, Howlett BJ. 2011. The cross-pathway control system regulates production of the secondary metabolite toxin, sirodesmin PL, in the ascomycete, Leptosphaeria maculans. BMC Microbiol 11:169. 10.1186/1471-2180-11-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11:619–633. 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 19.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108. 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 20.Hernández H, Aranda C, Riego L, González A. 2011. Gln3–Gcn4 hybrid transcriptional activator determines catabolic and biosynthetic gene expression in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 404:859–864. 10.1016/j.bbrc.2010.12.075. [DOI] [PubMed] [Google Scholar]
- 21.Li K, Xiao Y, Yu J, Xia T, Liu B, Guo Y, Deng J, Chen S, Wang C, Guo F. 2016. Liver-specific gene inactivation of the transcription factor ATF4 alleviates alcoholic liver steatosis in mice. J Biol Chem 291:18536–18546. 10.1074/jbc.M116.726836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin HR, Du CH, Wang C-Z, Yuan C-S, Du W. 2019. Ginseng metabolite protopanaxadiol induces Sestrin2 expression and AMPK activation through GCN2 and PERK. Cell Death Dis 10:311. 10.1038/s41419-019-1548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S, Li C, Wang L, Guo X, Sun Y, Luo H, Li Y, Song J, Henrissat B, Levasseur A, Qian J, Li J, Luo X, Shi L, He L, Xiang L, Xu X, Niu Y, Li Q, Han MV, Yan H, Zhang J, Chen H, Lv A, Wang Z, Liu M, Schwartz DC, Sun C. 2012. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun 3:913. 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jong S, Birmingham J. 1992. Medicinal benefits of the mushroom Ganoderma. Adv Appl Microbiol 37:101–134. 10.1016/s0065-2164(08)70253-3. [DOI] [PubMed] [Google Scholar]
- 25.Xu Z, Chen X, Zhong Z, Chen L, Wang Y. 2011. Ganoderma lucidum polysaccharides: immunomodulation and potential anti-tumor activities. Am J Chin Med 39:15–27. 10.1142/S0192415X11008610. [DOI] [PubMed] [Google Scholar]
- 26.Zhao W, Xu JW, Zhong JJ. 2011. Enhanced production of ganoderic acids in static liquid culture of Ganoderma lucidum under nitrogen-limiting conditions. Bioresour Technol 102:8185–8190. 10.1016/j.biortech.2011.06.043. [DOI] [PubMed] [Google Scholar]
- 27.Li H-J, Zhang D-H, Han L-L, Yu X, Zhao P, Li T, Zhong J-J, Xu J-W. 2016. Further improvement in ganoderic acid production in static liquid culture of Ganoderma lucidum by integrating nitrogen limitation and calcium ion addition. Bioprocess Biosyst Eng 39:75–80. 10.1007/s00449-015-1491-7. [DOI] [PubMed] [Google Scholar]
- 28.Scherz‐Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26:1749–1760. 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu R, Cao P, Ren A, Wang S, Yang T, Zhu T, Shi L, Zhu J, Jiang A-L, Zhao M-W. 2018. SA inhibits complex III activity to generate reactive oxygen species and thereby induces GA overproduction in Ganoderma lucidum. Redox Biol 16:388–400. 10.1016/j.redox.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59:407–450. 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 31.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC. 2010. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem 285:16893–16911. 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talbot NJ, Ebbole DJ, Hamer JE. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575–1590. 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau G, Hamer JE. 1996. Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea. Plant Cell 8:771–781. 10.2307/3870280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stitt M. 1999. Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2:178–186. 10.1016/S1369-5266(99)80033-8. [DOI] [PubMed] [Google Scholar]
- 35.Brouquisse R, Masclaux C, Feller U, Raymond P. 2001. Protein hydrolysis and nitrogen remobilisation in plant life and senescence, p 275–293. In Lea PJ, Morot-Gaudry J-F (ed), Plant nitrogen. Springer, Berlin, Germany. [Google Scholar]
- 36.Pavlova NN, Hui S, Ghergurovich JM, Fan J, Intlekofer AM, White RM, Rabinowitz JD, Thompson CB, Zhang J. 2018. As extracellular glutamine levels decline, asparagine becomes an essential amino acid. Cell Metab 27:428–438.e5. 10.1016/j.cmet.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Jiménez C, Goding CR. 2019. Starvation and pseudo-starvation as drivers of cancer metastasis through translation reprogramming. Cell Metab 29:254–267. 10.1016/j.cmet.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodríguez-Ortiz R, Limón MC, Avalos J. 2009. Regulation of carotenogenesis and secondary metabolism by nitrogen in wild-type Fusarium fujikuroi and carotenoid-overproducing mutants. Appl Environ Microbiol 75:405–413. 10.1128/AEM.01089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brückner B, Blechschmidt D. 1991. Nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi. Appl Microbiol Biotechnol 35:646–650. 10.1007/BF00169631. [DOI] [Google Scholar]
- 40.Dragone G, Fernandes BD, Abreu AP, Vicente AA, Teixeira JA. 2011. Nutrient limitation as a strategy for increasing starch accumulation in microalgae. Appl Energy 88:3331–3335. 10.1016/j.apenergy.2011.03.012. [DOI] [Google Scholar]
- 41.Shang C, Bi G, Yuan Z, Wang Z, Alam MA, Xie J. 2016. Discovery of genes for production of biofuels through transcriptome sequencing of Dunaliella parva. Algal Res 13:318–326. 10.1016/j.algal.2015.12.012. [DOI] [Google Scholar]
- 42.Aguirre J, Ríos-Momberg M, Hewitt D, Hansberg W. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol 13:111–118. 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Scherz-Shouval R, Elazar Z. 2007. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 17:422–427. 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Shi L, Gong L, Zhang X, Ren A, Gao T, Zhao M. 2015. The regulation of methyl jasmonate on hyphal branching and GA biosynthesis in Ganoderma lucidum partly via ROS generated by NADPH oxidase. Fungal Genet Biol 81:201–211. 10.1016/j.fgb.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Liu R, Zhang X, Ren A, Shi D-K, Shi L, Zhu J, Yu H-S, Zhao M-W. 2018. Heat stress-induced reactive oxygen species participate in the regulation of HSP expression, hyphal branching and ganoderic acid biosynthesis in Ganoderma lucidum. Microbiol Res 209:43–54. 10.1016/j.micres.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Mu D, Li C, Zhang X, Li X, Shi L, Ren A, Zhao M. 2014. Functions of the nicotinamide adenine dinucleotide phosphate oxidase family in Ganoderma lucidum: an essential role in ganoderic acid biosynthesis regulation, hyphal branching, fruiting body development, and oxidative-stress resistance. Environ Microbiol 16:1709–1728. 10.1111/1462-2920.12326. [DOI] [PubMed] [Google Scholar]
- 47.Shin R, Berg RH, Schachtman DP. 2005. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46:1350–1357. 10.1093/pcp/pci145. [DOI] [PubMed] [Google Scholar]
- 48.Brown SH, Yarden O, Gollop N, Chen S, Zveibil A, Belausov E, Freeman S. 2008. Differential protein expression in Colletotrichum acutatum: changes associated with reactive oxygen species and nitrogen starvation implicated in pathogenicity on strawberry. Mol Plant Pathol 9:171–190. 10.1111/j.1364-3703.2007.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tudzynski B. 2014. Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol 5:656. 10.3389/fmicb.2014.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Sun Z, Shi D, Song S, Lian L, Shi L, Ren A, Yu H, Zhao M. 2019. Dual functions of AreA, a GATA transcription factor, on influencing ganoderic acid biosynthesis in Ganoderma lucidum. Environ Microbiol 21:4166–4179. 10.1111/1462-2920.14769. [DOI] [PubMed] [Google Scholar]
- 51.Mu D, Shi L, Ren A, Li M, Wu F, Jiang A, Zhao M. 2012. The development and application of a multiple gene co-silencing system using endogenous URA3 as a reporter gene in Ganoderma lucidum. PLoS One 7:e43737. 10.1371/journal.pone.0043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Zhang G, Sun Z, Ren A, Shi L, Shi D, Li X, Zhao M. 2017. The mitogen-activated protein kinase GlSlt2 regulates fungal growth, fruiting body development, cell wall integrity, oxidative stress and ganoderic acid biosynthesis in Ganoderma lucidum. Fungal Genet Biol 104:6–15. 10.1016/j.fgb.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Zhang G, Ren A, Shi L, Zhu J, Jiang A, Shi D, Zhao M. 2018. Functional analysis of an APSES transcription factor (GlSwi6) involved in fungal growth, fruiting body development and ganoderic-acid biosynthesis in Ganoderma lucidum. Microbiol Res 207:280–288. 10.1016/j.micres.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 55.You B-J, Lee H-Z, Chung K-R, Lee M-H, Huang M-J, Tien N, Chan C-W, Kuo Y-H. 2012. Enhanced production of ganoderic acids and cytotoxicity of Ganoderma lucidum using solid-medium culture. Biosci Biotechnol Biochem 76:1529–1534. 10.1271/bbb.120270. [DOI] [PubMed] [Google Scholar]
- 56.Hu Y, Xu W, Hu S, Lian L, Zhu J, Shi L, Ren A, Zhao M. 2020. In Ganoderma lucidum, Glsnf1 regulates cellulose degradation by inhibiting GlCreA during the utilization of cellulose. Environ Microbiol 22:107–121. 10.1111/1462-2920.14826. [DOI] [PubMed] [Google Scholar]
- 57.Duan Z, Chen Y, Huang W, Shang Y, Chen P, Wang C. 2013. Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy 9:538–549. 10.4161/auto.23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6, Table S1. Download AEM00156-21_Supp_1_seq3.pdf, PDF file, 0.7 MB (746KB, pdf)
Data Availability Statement
The nucleotide sequences of gcn4 in the G. lucidum strain ACCC53264 have been deposited in GenBank under the accession number MN380309.