ABSTRACT
Nitrogen requirements for modern agriculture far exceed the levels of bioavailable nitrogen in most arable soils. As a result, the addition of nitrogen fertilizer is necessary to sustain productivity and yields, especially for cereal crops, the planet’s major calorie suppliers. Given the unsustainability of industrial fertilizer production and application, engineering biological nitrogen fixation directly at the roots of plants has been a grand challenge for biotechnology. Here, we designed and tested a potentially broadly applicable metabolic engineering strategy for the overproduction of ammonia in the diazotrophic symbiont Azospirillum brasilense. Our approach is based on an engineered unidirectional adenylyltransferase (uAT) that posttranslationally modifies and deactivates glutamine synthetase (GS), a key regulator of nitrogen metabolism in the cell. We show that this circuit can be controlled inducibly, and we leveraged the inherent self-contained nature of our posttranslational approach to demonstrate that multicopy redundancy can improve strain evolutionary stability. uAT-engineered Azospirillum is capable of producing ammonia at rates of up to 500 μM h−1 unit of OD600 (optical density at 600 nm)−1. We demonstrated that when grown in coculture with the model monocot Setaria viridis, these strains increase the biomass and chlorophyll content of plants up to 54% and 71%, respectively, relative to the wild type (WT). Furthermore, we rigorously demonstrated direct transfer of atmospheric nitrogen to extracellular ammonia and then plant biomass using isotopic labeling: after 14 days of cocultivation with engineered uAT strains, 9% of chlorophyll nitrogen in Setaria seedlings was derived from diazotrophically fixed dinitrogen, whereas no nitrogen was incorporated in plants cocultivated with WT controls. This rational design for tunable ammonia overproduction is modular and flexible, and we envision that it could be deployable in a consortium of nitrogen-fixing symbiotic diazotrophs for plant fertilization.
IMPORTANCE Nitrogen is the most limiting nutrient in modern agriculture. Free-living diazotrophs, such as Azospirillum, are common colonizers of cereal grasses and have the ability to fix nitrogen but natively do not release excess ammonia. Here, we used a rational engineering approach to generate ammonia-excreting strains of Azospirillum. Our design features posttranslational control of highly conserved central metabolism, enabling tunability and flexibility of circuit placement. We found that our strains promote the growth and health of the model grass S. viridis and rigorously demonstrated that in comparison to WT controls, our engineered strains can transfer nitrogen from 15N2 gas to plant biomass. Unlike previously reported ammonia-producing mutants, our rationally designed approach easily lends itself to further engineering opportunities and has the potential to be broadly deployable.
KEYWORDS: diazotrophs, glutamine synthetase, ammonia, cereal crop, synthetic biology, isotope labeling
INTRODUCTION
Dinitrogen is the most abundant molecule in our atmosphere, yet its inertness has made bioavailable forms of nitrogen the most limiting nutrient for agriculture. Over the past decades, we have depleted organic soil nitrogen in fields used for intensive agriculture faster than it can be biologically replenished (1). As a result, over 100 million tons of synthetic nitrogen fertilizer are produced annually and applied to fields globally, leading to a host of problems, including ecosystem leaching, ground water poisoning, carbon and nitrogen oxide atmospheric pollution, and intense centralized energy consumption (2, 3).
A highly sought-after alternative to chemical fertilization is the biological provision of fixed nitrogen directly at the root of plants (4–8). Across several phyla, about 15% of prokaryotes have evolved diazotrophy, the capability to convert dinitrogen gas to bioavailable ammonia through an enzymatic nitrogen fixation process (9–11). Some plants have evolved intimate symbioses with a subset of these diazotrophs to obtain bioavailable nitrogen, a process primarily limited to nodulating species of the Fabaceae family of plants and diazotrophic rhizobia (12, 13). However, these plants do not include most domesticated crops: 50% of the world’s calories come from the cereal grasses, including corn, rice, and the grains, which are not known to participate in nitrogen transfer symbioses through nodulation (4, 5, 7, 8, 14, 15).
Outside the rhizobia, many bacterial diazotrophs colonize plant roots through looser, nonnodulating associations in the rhizosphere or endophytic compartments. These associative diazotrophs comprise several genera, including Azospirillum, Herbaspirillum, Gluconacetobacter, and Burkholderia, and have been thought to promote plant growth by enhancing plant nitrogen supply. However, despite their potential contribution of nitrogen to plants, associative diazotrophs are unable to keep pace with the nutrient demands required by modern agriculture (16–24). For example, nodulating symbioses are estimated to provide up to 50 to 465 kg N ha−1 year−1, whereas associative diazotrophs provide at best 2 to 170 kg N ha−1 year−1 (4). The direct contributions of diazotrophs to plant nitrogen have been difficult to characterize using biomass measurements alone, in part due to confounding effects such as microbial phytohormone and siderophore production. Isotope dilution and natural abundance approaches are more insightful; however, they are limited by low signal-to-noise readouts and temporal and spatial variability in isotope enrichment of soils, and they require plants with consistent phenotypes (such as root morphology and biomass). These methods also do not directly distinguish between fixed-nitrogen incorporation in plant-tissue colonizing diazotrophs versus the plant itself (4, 25–28). As an alternative to these indirect, dilution-based strategies, direct incorporation of isotopically labeled nitrogen gas into plant biomass can provide more conclusive evidence for diazotrophic contributions to plant nitrogen. For this, isotopes can be tracked from gas into plant-specific molecules, such as chlorophyll, and their signal compared against that in uninoculated plants to control for artifacts and contaminants (22, 29).
There have been major engineering efforts focused on associative diazotrophs to address the shortcoming in biological nitrogen provision for nonnodulating crops (5, 7, 30–34). One target has been the nitrogen fixation step directly: the hetero-octamer nitrogenase NifHDK complex and its many constituents required for its activation and efficient reduction of dinitrogen to ammonia. Optimizing this machinery for higher nitrogen fixation rates, for fixed nitrogen release, or for nitrogen fixation in heterologous organisms, including plants, remains a challenge due to complex multilayered regulation at the transcriptional, translational, and posttranslational levels (see Fig. S1 in the supplemental material) (7, 31, 33, 35–37). Generating an ammonia-releasing diazotroph by engineering the nif genes has been mostly limited to Azotobacter vinelandii knockouts of nifL, encoding an ammonia-responsive repressor of the nitrogen fixation genes present in a subset of diazotrophs (38–40).
Another engineering target has been the nitrogen assimilation pathway rather than the enzymes directly involved in nitrogen fixation. This approach is centered around glutamine synthetase (GS; encoded by glnA), a ubiquitous enzyme that combines ammonia and glutamate to synthesize glutamine. Its activity is required for cellular nitrogen cycling, since it is the primary route by which ammonia initially enters or re-enters the highly branched cellular metabolism of nucleic acid, amino acid, and secondary metabolite biosynthesis (41, 42). Constitutive mutants in the ammonia assimilation pathway of some diazotrophs, mainly Azospirillum brasilense, have previously been stochastically generated by evolution on the toxic ammonia analog ethylenediamine and shown to release ammonia under diazotrophic growth (43–45). These studies lead to the hypothesis that inhibiting GS in a diazotroph would cause ammonia accumulation and release through transmembrane diffusion. In support of this hypothesis, targeted mutation of the GS active site at D49S and substituting the promoter of glnA for an exogenously controlled promoter have also been shown to lead to transient ammonia release upon glnA suppression in A. vinelandii (34, 39). Deregulating GS is attractive because the ratio of glutamine to 2-oxoglutarate is the central intracellular nitrogen status indicator. As the control point for glutamine production and thus nitrogen status, decreased glutamine biosynthesis through GS inhibition natively maximizes the nitrogen fixation machinery without direct interference in the complex regulatory network surrounding NifHDK (46, 47).
Drawing on this prior characterization of diazotrophic nitrogen metabolism, our goal was to design and build a generic system that could cause ammonia overproduction in associative diazotrophs. With the objective of downregulating GS activity, we initially considered synthetic systems to rewire its transcriptional control. However, transcription of the GS-encoding glnA gene is natively coordinated by multiple, intricately balanced promoters, and glnA is also frequently polycistronic with other critical regulatory genes, such as glnB (encoding PII) in Azospirillum brasilense and Gluconacetobacter diazotrophicus, glnL (encoding NtrB), and glnG (encoding NtrC) in Herbaspirillum seropedicae and several Burkholderia strains (see Fig. S1 for regulatory interactions) (48, 49). To avoid interfering with such finely tuned transcriptional regulation and to build a more versatile system, we instead envisioned a posttranslational approach to regulate GS. Additional potential benefits of a posttranslational GS deactivation system include fast, catalytic GS deactivation that does not depend on the half-life of system proteins (e.g., GS) and flexible control over circuit placement and regulation.
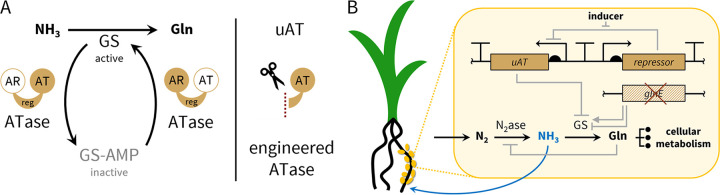
Natively, GS is posttranslationally modified by GS adenylyltransferase (ATase; encoded by glnE), a conserved enzyme that deactivates GS through adenylylation. This process is reversible by hydrolysis of the adenylyl group by the same enzyme (Fig. 1A) and the directionality is determined by a complex system of interregulated proteins (Fig. S1) (41). Previous work has shown that in Escherichia coli, the adenylyl-transferring and the adenylyl-removing activities reside in separate, homologous protein domains and that these can function independently to modulate GS activity in vitro (47, 50).
FIG 1.
General strategy for inducible ammonia production from dinitrogen gas built on the conserved central nitrogen metabolism in diazotrophs. (A) Glutamine synthetase (GS) bidirectional regulation by the native adenylyltransferase (ATase) and engineered unidirectional ATase (uAT) schematic. AT and AR denote the adenylyl-transferring and the adenylyl-removing domains, respectively. (B) Genetic engineering strategy showing deletion of the native chromosomal ATase (glnE) and introduction of a uAT on an integrated genetic circuit for inducible heterologous regulation of GS. Gln, glutamine; N2ase, nitrogenase; GS-AMP, adenylylated glutamine synthetase; reg, regulatory domain.
In this work, we present a rationally designed, potentially broadly applicable strategy centered around a unidirectional ATase (uAT) that deactivates GS to yield ammonia production (Fig. 1). We tested our proof of concept in A. brasilense, a previously studied, microaerobic nitrogen-fixing symbiont known to colonize the rhizosphere of grasses (51, 52). We show that our uAT strains release ammonia into culture media, and we verify that its origin is atmospheric nitrogen. We show that our circuit can be controlled inducibly and installed in multiple copies to improve its output and evolutionary stability. In cocultivation with the model monocot Setaria viridis, our uAT strains increased plant biomass and chlorophyll content compared to WT-inoculated controls. To rigorously verify the contribution of biologically fixed nitrogen to plants, we measured direct incorporation of labeled nitrogen from 15N2 gas into the plant-specific chlorophyll derivative pheophytin in S. viridis cocultivated with our engineered uAT strains compared to WT and uninoculated controls (29, 53).
RESULTS
GS activity can be modulated with uATs.
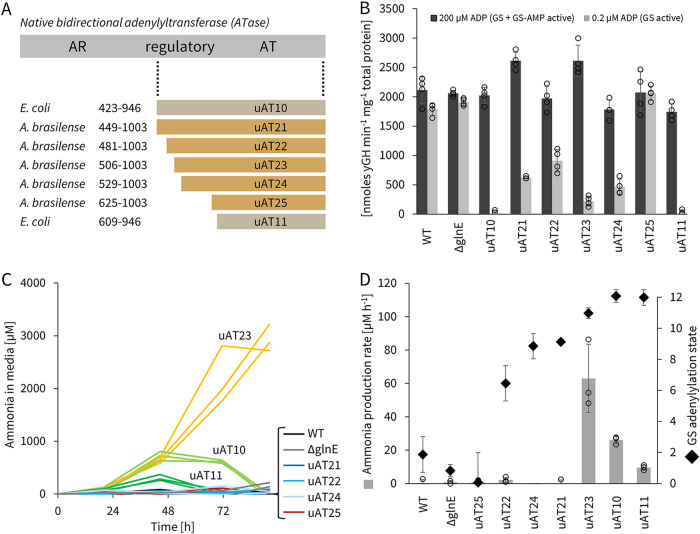
We first sought to generate a set of standard genetic tools for predictable engineering of A. brasilense. Using previously characterized parts, we constructed both replicative (pTS7) and chromosomally integrative (pTS9) plasmids (Fig. S21). Next, we designed several unidirectional ATases (uATs) derived from A. brasilense glnE, alongside two similar E. coli glnE truncations that had been previously characterized in vitro (47, 50). We aligned E. coli and A. brasilense glnE genes to identify interspecies truncation sites homologous to those reported. Since the adenylyl-transferring and adenylyl-removing domains within ATase are also homologous to each other, we predicted their locations through alignments of the A. brasilense glnE N- and C-terminal halves to each other and designed further uATs based on this intraprotein homology (Fig. 2A). We scarlessly deleted the native glnE gene in A. brasilense by double homologous recombination and expressed candidate uATs from a constitutive genetic circuit, R2, on replicative plasmid pTS7 in this ΔglnE strain (Fig. S2A and B and S21).
FIG 2.
GS activity can be modulated by uATs, and GS shutdown leads to ammonia release. (A) Alignment of 7 truncations of ATase from either E. coli and A. brasilense native ATase genes (glnE) that were tested for uAT activity. AT and AR denote the adenylyl-transferring and the adenylyl-removing domains, respectively. Average glutamine synthetase (GS) activity (B) and ammonia concentration in medium (C) of constitutively uAT-expressing A. brasilense ΔglnE strains inoculated at an OD600 of 0.1 in semisolid NFbHP medium (circuit R2 from pTS7 [Fig. S21]). (B) GS activity was measured by γ-glutamyl hydroxamate (γGH) production rates at high (200 μM) and low (0.2 μM) ADP concentrations (see Fig. S3A and B and S4 for the assay); rates are normalized to total protein content determined by the Bradford assay. (C) Ammonia was quantified by the indophenol method (see Fig. S6 for the assay); error bars are for triplicate biological replicates. (D) Comparison of maximal ammonia production rates observed between 20 and 70 h from data in panel C to computed GS adenylylation states (see Fig. S3C for calculations); error bars are standard deviations of biological replicates shown in panels B and C.
Glutamine synthetase activity was determined using the γ-glutamyl hydroxamate (γGH) assay on permeabilized A. brasilense whole cells. The γGH assay measures GS activity by running the enzyme in a quasireverse direction (Fig. S3A) and was chosen for two reasons: one, GS-driven γGH production is easily quantifiable by spectrophotometry, and two, it is sensitive to the adenylylation state of GS depending on the ADP concentration used in the assay. Specifically, prior work establishing this assay has revealed that while all forms of GS are active at a high ADP concentration (200 μM), only unadenylated GS is active at low ADP concentration (0.2 μM) (Fig. 2B; Fig. S3 and S4) (54–56). This behavior likely arises because the affinity of GS for ADP correlates inversely with its adenylylation state.
Expression of all but one of the uAT candidates under nitrogen fixing conditions resulted in significant reduction of GS activity, as measured by the γGH assay at low ADP concentrations, suggesting that GS is mostly in the adenylylated state. Biosynthetically, this means that the enzyme is inactive and thus does not convert ammonia to glutamine. These results are striking, considering the multiple allosteric regulatory networks that control the native ATase (47, 50, 57, 58). For example, normally when cells are glutamine deficient, PII (glnB) and PZ (glnZ) become uridylylated and interact with ATase to deadenylylate and activate GS (Fig. S1); however, with our uAT system, we actuated the opposite response and adenylylate GS in the glutamine-deficient state to keep it inactive. Also notable is that both E. coli uATs are functional in A. brasilense, underscoring the high degree of conservation of this central metabolic pathway and the likely broad applicability of this approach. Finally, we studied the effect of uAT expression on glnA transcription by reverse transcription quantitative PCR (RT-qPCR) and found it to be slightly decreased, at levels of 30 to 40% compared to the wild type (WT) (Fig. S5).
GS shutdown leads to ammonia production.
We next asked whether any of the uAT strains (Fig. 2A) released ammonia. Samples of diazotrophic cultures were taken at time points over several days, clarified, and characterized by the indophenol colorimetric assay to measure ammonia concentrations in the medium (Fig. S6) (59). We did not detect ammonia in the medium of the WT strain, as expected, but several of the uAT-expressing strains did result in 0.3 to 3 mM levels of ammonia accumulation over 2 to 4 days in cultures inoculated at an optical density at 600 nm (OD600) of 0.1 (Fig. 2C). To see if ammonia accumulation correlates with GS adenylylation in the cells, we computed an average adenylylation state using the results shown in Fig. 2B (Fig. S3C) (54, 55). GS is a homododecamer and has 12 total adenylylation sites; thus, the maximal adenylylation (shutdown) state is 12. Based on this analysis and our experimental results, we found that ammonia accumulates in the medium when average adenylylation levels exceed 11 (see results for strains uAT10, uAT11, and uAT23 in Fig. 2D). uAT21 and uAT24 conferred a slightly lower average GS adenylylation level of 9 and did not lead to detectable levels of ammonia in the medium. These data confirm that the reduced ability to assimilate ammonia through GS deactivation results in its extracellular release and suggest that an average adenylylation threshold between 9 and 11 is required to sufficiently deactivate GS for ammonia accumulation.
uAT-driven ammonia production can be inducibly controlled and circuit stability further improved through multicopy redundancy.
Following this initial demonstration that uAT-driven GS deactivation causes ammonia production, we focused further engineering on some of the challenges created by disrupting a central metabolic pathway such as GS (60). Not surprisingly, we observed that diverting nitrogen influx to extracellular ammonia resulted in a growth penalty (Fig. S7 and S8) and strong evolutionary selection for mutants with a disrupted uAT cassette to overtake inoculated cell populations. PCR and sequence analysis of clones isolated from evolved cultures showed several mechanisms of uAT escape, including transposon insertion, point mutation, and recombination (Fig. S9). With the enrichment of these mutants, cultures lost the ability to keep glutamine synthetase fully adenylylated, thus regaining GS activity and depleting their previously accumulated ammonia (Fig. S10). We found that adding exogenous glutamine (1 mM) to cultures does result in an initial boost in ammonia production, but it does not significantly extend their ammonia production lifetime (Fig. S11). These data suggest that glutamine addition at 1 mM does not completely relieve the selective pressure caused by expression of uAT, and mutants (likely already present in the initial culture) still have an advantage. It is possible that glutamine synthetase or ATase has other, yet-undefined, critical roles in the cell.
To improve engineered strain evolutionary stability, several strategies have been proposed (60–62), notably, tight genetic control and multicopy circuit redundancy. As a rationally designed and posttranslational system, our uAT approach is ideally situated to leverage these strategies.
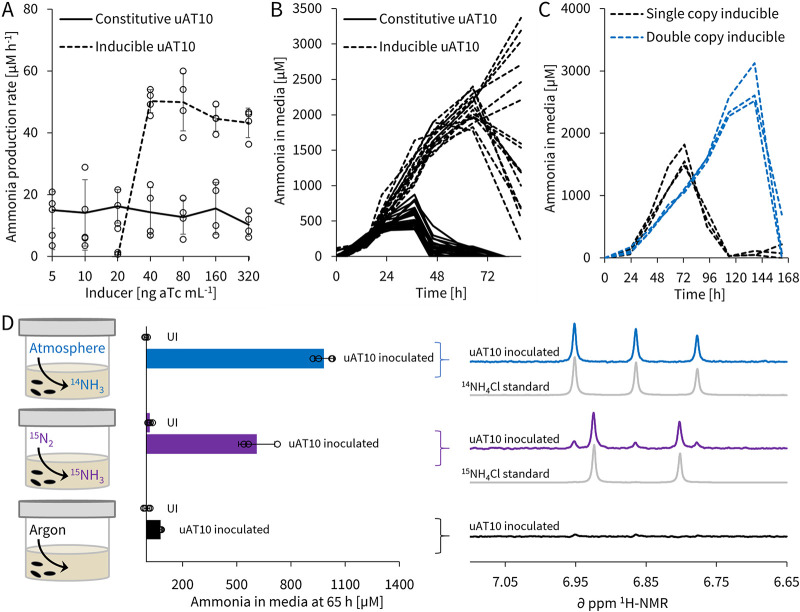
We first sought to build an inducible expression system for A. brasilense in which we could separate growth from production to delay evolutionary pressure until system activation (60, 63, 64). For this, we designed and tested several inducible circuit architectures in A. brasilense using parts from the BioBrick Registry of Standard Biological Parts, until we reached tightly controlled tetracycline repression systems J3 and J4 (Fig. S2A and B and S21B). Following validation with red fluorescent protein (RFP) (Fig. S2A and B), we expressed uAT10 on our inducible circuit J3 from plasmid pTS7: under the same nitrogen-fixing conditions as before, we detected no ammonia production (off-state) until the inducer concentration reached 20 to 40 ng ml−1 anhydrotetracycline, above which ammonia production was constant with respect to inducer concentration at average rates of 50 μM h−1 for 70 h (on-state) (Fig. 3A and B). This inducible on-state ammonia production rate is twice as high and lasts twice as long as the equivalent constitutive strain performance (Fig. 3B and Fig. S12). We also tested our inducible J3 circuit with uAT23 and did not detect ammonia in the media. We suspect that no ammonia was accumulating, because the inducible system on-state protein expression is weaker than the constitutive protein expression (Fig. S2A and B), and that this lower level of uAT expression falls short of the threshold required for GS deactivation (Fig. 2D).
FIG 3.
uAT-driven ammonia production is from de novo nitrogen fixation and can be inducibly controlled, and circuit stability is further improved through multicopy redundancy. (A) Average ammonia production rates of A. brasilense ΔglnE between 15 and 35 h with constitutive and inducible expression of uAT10 at different anhydrotetracycline (aTc) inducer concentrations. (B) Individual time course data, with inducible off-state (below 40 ng ml−1 aTc) lines omitted for clarity (see Fig. S12 for full data). (C) Time course data comparing on-state ammonia production of single- and double-copy uAT10 in A. brasilense ΔglnE at 200 ng aTc ml−1. (D) Controlled atmosphere labeling study of the inducible uAT10 circuit in A. brasilense ΔglnE: medium ammonia concentrations were first determined by the indophenol colorimetric assay 65 h postinduction (200 ng ml−1 aTc) and then analyzed by proton nuclear magnetic resonance (1H-NMR) using an ammonia-selective pulse sequence. All cells were inoculated in semisolid NFbHP medium at OD600 of 0.1 and ammonia concentrations were determined by the indophenol method (see Fig. S4 for assay). UI, uninoculated controls. Error bars show standard deviations for biological quadruplicates (A) and triplicates (D). See Fig. S21 for genetic circuit diagrams and parts; the constitutive circuit used was R2, the single copy inducible circuit was J3, and the double-copy inducible circuit was V2, all on pTS7.
Second, we tested whether multicopy uAT redundancy could further improve ammonia production lifetime. In theory, multicopy integration of genetic material has the potential to increase the effective lifetime of genetic circuits, because all copies must be mutated in the same cell for successful escape and mutant selection. Our results confirm that installing a second inducible uAT10 copy increases ammonia production lifetime, doubling it from 70 h to 140 h (Fig. 3C). We want to highlight that inducibility, such that all copies can be turned off, is a prerequisite for the success of this approach, because cells do not experience additional evolutionary pressure in the off-state. An additional prerequisite is that the circuit affords placement flexibility, such as a posttranslational mechanism does, so that it may be installed in multiple copies; constitutive glnA mutants or synthetic regulation around glnA, for example, could not be augmented by a multicopy strategy. With this initial demonstration of inducibility and multicopy redundancy in hand, we hypothesize that evolutionary stability and circuit performance can be further improved by additional uAT copies and control-circuit architectures.
Based on our initial inducible ammonia production rates for an inoculating cell density at an OD600 of 0.1, or 108 CFU ml−1, and assuming no growth during production (Fig. S7), we computed a specific ammonia production rate of 83,600 molecules of ammonia cell−1 s−1. With estimated nitrogenase dinitrogen reduction rates of 6 s−1, achieving this specific ammonia production rate would require an active nitrogenase complex number of approximately 7,000 per cell (65). Based on native proteomic data of the similarly sized prokaryote E. coli, 7,000 complexes per cell is in the 95th percentile of most abundant proteins, and we thus suspect that the ammonia production rates we measured are close to maximal capacity (66). To test this limit, we attempted to increase ammonia production through further metabolic engineering, by knocking out negative regulators such as draT (encoding nitrogenase ADP-ribosyltransferase; deactivates NifHDK) or mutating nifA (encoding the global nitrogen fixation transcriptional activator/regulator) or by overexpressing the nitrogen fixation-related electron transport genes fdxN, fdxB, and fixX. The nitrogen transporter amtB was also tested in both the overexpressed and knocked-out configurations (Fig. S13) (67–74). None of these approaches proved to increase ammonia production, which is in line with the hypothesis that native fixation capacity would be maximized if the central nitrogen status indicator glutamine is decoupled from ammonia as a result of uAT-induced GS shutdown. We used Western blotting to examine the posttranslational state of nitrogenase in strains expressing uAT. These data show that uAT expression results in the highest ratio of deribosylated (active) to ribosylated (deactive) forms of nitrogenase, compared to WT and ΔglnE strains (Fig. S14). Finally, in trying to maximize ammonia production rates, we also tested the effect of higher and lower inoculating ODs and noted that ammonia production rates scale robustly with OD but require sufficient surface area for gas exchange with the atmosphere at higher OD (Fig. S15).
Isotope labeling confirms de novo nitrogen fixation.
We next performed a labeling experiment to determine whether the ammonia that accumulates in the medium originates from nitrogen fixation that occurs during the course of the assay or whether it is a result of spurious nitrogen sources, such as inducer and antibiotic degradation or intracellular turnover. We tested A. brasilense ΔglnE carrying the J3 inducible circuit with uAT10 in nitrogen-free medium induced with 200 ng ml−1 anhydrotetracycline (on-state) under controlled atmosphere conditions of 20% O2 and 80% either 14N2, 15N2, or argon and analyzed media after 65 h. Since commercial 15N2 gas often contains small amounts of 15NH3, we first verified with the quantitative indophenol assay that our uninoculated controls had no appreciable amounts of ammonia in the media above the assay detection threshold of 10 μM (Fig. 3D). Next, we analyzed the media of inoculated vials using proton nuclear magnetic resonance (1H-NMR) with a previously reported pulse sequence optimized for the detection of ammonia (75). 1H-NMR is ideally suited for this kind of analysis, as isotopically unique differences in nuclear spin manifest as distinctive coupling constants inherently observed by NMR techniques. Thus, 14NH4+ and 15NH4+ are readily distinguishable owing to the different nuclear spins of each isotope. 14NH4+, with a spin of 1, appears as a 1:1:1 triplet with a 1JH-N of 51 Hz, while 15NH4+, with a spin of 1/2, appears as a 1:1 doublet with a 1JH-N of 72 Hz (75). We found that the 15N2 atmosphere samples did indeed show a predominant doublet with a 1JH-N of 73 Hz, from which we conclude that ammonia in the medium is de novo fixed from gaseous dinitrogen (Fig. 3D).
Ammonia-producing strains promote plant growth.
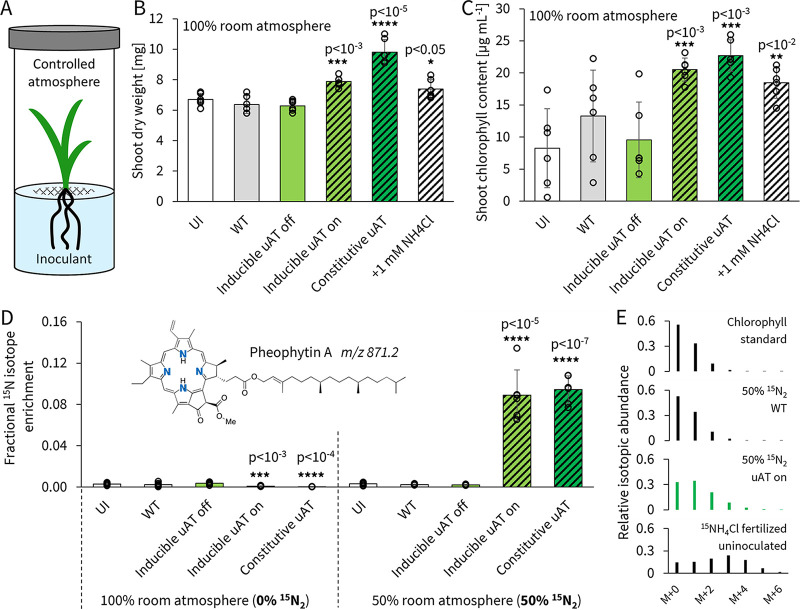
Following NMR validation of our ammonia-producing strains, we next investigated their ability to promote the growth of plants. We chose S. viridis as a previously characterized model monocot, well colonized by A. brasilense and conveniently small for scalable experiments (17, 53). To allow antibiotic-free coculturing of our ammonia-producing strains with plants, we first needed to chromosomally integrate our uAT circuits in the A. brasilense ΔglnE strain (Fig. S21D). Using double homologous recombination, we inserted both constitutive and single-copy-inducible uAT10 circuits and confirmed that the resultant stable strains produce ammonia at rates and evolutionary limit similar to those of plasmid strains (Fig. S16). For the coculture, S. viridis seeds were surface sterilized and germinated on polytetrafluoroethylene (PTFE) rafts in modified 1/5-strength semisolid nitrogen-limiting NFbHP (nitrogen-free basal high-phosphate) medium (Fig. 4A). A. brasilense strains were introduced on day 14 at an OD600 of 0.1, and after an additional 14 days, total shoot dry weight was collected. We observed significant growth promotion with both our constitutively producing and inducible on-state strains at 54% and 24% over WT-inoculated plants, respectively. The wild-type- and inducible off-state-inoculated samples showed no growth promotion, suggesting no native nitrogen transfer (Fig. 4B). Total chlorophyll content was also determined as a metric for plant health (76): the results mirror dry-weight conclusions, with an increase of 71% and 55% over WT for the constitutive and inducible on-state strains, respectively (Fig. 4C). Since our regular coculture setup contained carbon in the medium (9 mM sodium lactate), we separately tested what effect omitting sodium lactate would have on plant growth promotion as an initial step toward soil conditions. The results show slightly reduced but still strong growth promotion of 42% and 18% over WT levels for the constitutive and inducible on-state strains, respectively (Fig. S17).
FIG 4.
(A) Schematic of controlled atmosphere plant-microbe co-culture labeling study. A. brasilense wild-type (WT) and ΔglnE strains with chromosomally integrated constitutive and inducible uAT10 circuits (Fig. S16 and S21) were introduced at an OD600 of 0.1 to 14-day-old S. viridis plants growing gnotobiotically in modified 1/5-strength NFbHP. At the time of inoculation, the on-state replicates were induced with 200 ng ml−1 anhydrotetracycline while off-state replicates had no anhydrotetracycline added. The atmosphere of replicates in the label group was modified by replacing 50% of the volume with 15N2 gas immediately after inoculation (resulting in a final mixture of 90/10 N2/O2). UI, uninoculated controls. After an additional 14 days, samples for determination of shoot dry weight (B) and total chlorophyll content (C) were collected. Chlorophyll was converted to pheophytin for isotopic analysis (D) to determine fractional enrichment of the 15N isotope (see Fig. S18 for calculation and S19 for fertilized controls). (E) Representative pheophytin isotopic envelopes showing mass shifts caused by 15N incorporation. All error bars are standard deviations for 6 biological replicates; stars denote P values determined to be of statistical significance compared to WT using a two-sided homoscedastic t test.
Fixed nitrogen is integrated into plants.
To quantitatively measure the transfer of fixed nitrogen to plant biomass, we looked at direct 15N integration into the plant-specific chlorophyll derivative pheophytin. This gold-standard metric is used to show nitrogen transfer in nodulating symbioses, and we applied it to our engineered monocot system (4). Chlorophyll was extracted from whole shoots and converted to pheophytin by acidification as reported previously (29, 77). Samples were directly analyzed by quantitative time-of-flight (qTOF) mass spectrometry. As a positive control, we confirmed 15N isotope integration in plants fertilized with 15NH4Cl over 14NH4Cl (Fig. S18 and S19). When we displaced 50% of the atmosphere in our coculture setup with 15N2, we observed no isotope enrichment above the 15N natural abundance (0.4%) for uninoculated, WT-inoculated, or inducible off-strain-inoculated plants. For the constitutive-strain- and inducible on-strain-inoculated plants, we observed 9.4% ± 1.3% and 8.9% ± 2.5% integration of 15N atoms in the total N of pheophytin at high statistical significance (P < 10−5) (Fig. 4D and E). Our method was also sensitive enough to detect a statistically significant dilution of natural 15N isotopes in plants grown under regular atmospheric conditions and inoculated with ammonia-producing strains (P < 10−3) (Fig. 4D and E). This isotope dilution occurs under significant de novo gaseous nitrogen influx because natural abundances of 15N in biomass are higher than in the atmosphere. Taken together, our data clearly demonstrate direct transfer of dinitrogen to plants cultivated in the presence of engineered A. brasilense.
DISCUSSION
In summary, we present a modular and controllable posttranslational strategy that causes diazotrophs to release fixed nitrogen. Not only do we show that our strains promote plant growth, but we also present conclusive evidence, obtained with isotope labeling, that atmospheric dinitrogen is fixed by the strain, released as ammonia, and transferred to plant biomass.
Since uAT-induced ammonia production targets highly conserved metabolism, we can now envision its deployment in multiple symbiotic diazotrophs. As we begin onboarding additional strains, we have already successfully obtained equally high and tightly regulated ammonia production in another strain of A. brasilense, Sp7, through one-step disruption of glnE with a uAT circuit (Fig. S20). Bacteria natively adapt to and occupy different niches at the plant root (78); thus, moving our uAT system into a diverse consortium of symbiotic diazotrophs can maximize the amount of fixed nitrogen supplied to the plant. If the specific ammonia production rates achieved by our strains can be upheld in a consortium that colonizes 10% of the root community of 1010 cells g (dry weight) root−1, biological nitrogen fixation could produce 0.2 mg NH3 g (dry weight) root−1 day−1. For an estimated maize root dry weight of 20 g plant−1 over 150 days of the crop cycle (79), our strains could produce 0.6 g fixed ammonia per maize plant. Current U.S. fertilizer nitrogen applications (80) of 224 kg NH3 ha−1 crop cycle−1 at a planting density of 70,000 plants ha−1 convert to 3.2 g NH3 plant−1. Based on these estimates, and also considering that nitrogen supply from diazotrophs at the root will likely have a higher nitrogen transfer efficiency to plants than bulk fertilizer application, we see the potential to significantly contribute to plant nitrogen supply through biological nitrogen fixation in the field.
Strain stability is a general challenge for integrated biological systems that divert energy and resources away from central metabolism and will be of growing relevance as we engineer more complex biological systems (81, 82). This includes the long-term evolutionary stability of engineered diazotrophs. In this work, we have demonstrated proof of concept that inducibility and multicopy redundancy of a self-contained posttranslational circuit can delay the emergence of escape mutants. This strategy could be further augmented or tailored with other control mechanisms, for example, circuits that respond not only to synthetic small chemicals but also to internal nitrogen stress, quorum-sensing molecules, oscillators, native root exudates, or synthetic plant-microbe tethers (7, 83, 84). It is possible that there is an optimal GS shutdown level that maximizes ammonia production rates and strain lifetime as a balance between excreting all fixed nitrogen as ammonia and allowing some “leak” of glutamine synthetase to avoid complete choking of cell metabolism. Additionally, we note that while our current ammonia production lifetime is still about an order of magnitude shorter than what might be desirable on the time scale of agricultural plant growth, a finite circuit lifetime is an advantage for biocontainment.
Finally, this work provides a tool for regulatory understanding of cellular nitrogen metabolism. Several other components have been associated with central nitrogen metabolism in the cell and can now be studied without the confounding effect of immediate ammonia-to-glutamine conversion by GS. For instance, DraT deactivates NifHDK through posttranslational ribosylation under excess nitrogen conditions (67, 68, 85–87). If DraT is responsive to ammonia directly, one would expect that this would interfere with our ammonia-accumulating strains and that knocking out DraT would increase uAT-driven ammonia production. We generated this ΔglnE ΔdraT double knockout uAT strain and found no increase in ammonia levels (Fig. S13). These results suggest that ammonia does not regulate DraT directly and that glutamine is the master regulator. Similarly, and despite previous reports of the ammonia transporter AmtB knockout leading to increased ammonia excretion in A. vinelandii ΔnifL mutants (74), we found no additive effect of the double knockout ΔglnE ΔamtB, raising questions about its role in sensing or transporting ammonia (Fig. S13) (86, 88). Is there anything that senses ammonia directly? Proteomic and blotting analysis of these and additional knockouts in our uAT strains combined with ammonia and glutamine shocks can provide insight into fundamental biology of cellular nitrogen metabolism.
Using engineered bacteria for temporary at-source provision of nitrogen could be an inexpensive, effective, and more efficient strategy of providing nonnodulating plants with the nitrogen they need. Understanding how uAT strains compete in the context of microbial communities and designing additional mechanisms for ensuring maximum and selective strain growth at the roots of crops could further enhance biological nitrogen supply. We anticipate that the rational design of the uAT strategy reported here will readily enable improvements to stability and performance, tailoring to diverse control regimens, and deployment in multiple diazotrophs.
MATERIALS AND METHODS
Study design.
(i) Sample size. Sample size is indicated separately for each experiment. In general, triplicate or quadruplicate biological replicates were used for experiments pertaining to bacteria only. Experiments involving plants used 6 or more replicates due to increased variance inherent to plant experiments.
(ii) Rules for stopping data collection. Ammonia production experiments were run until cultures stopped producing ammonia (due to mutant outgrowth); plant experiments were run until plants started yellowing and visibly stopped growing.
(iii) Data inclusion/exclusion criteria. Only data from contaminated samples or samples lost during assay processing were excluded (e.g., due to pipette error).
(iv) Outliers. No outliers were excluded.
(v) Replicates. Every experiment was performed at least twice, except for the pheophytin isotope labeling study.
(vi) Randomization. Plants were randomly assigned to experimental groups 14 days postgermination and then randomly positioned in the growth chamber and shuffled every 7 days.
(vii) Blinding. No blinding was undertaken.
Experimental design.
(i) DNA assembly. All plasmids were assembled following a standard protocol using Phusion polymerase and Gibson assembly. All DNA synthesis of primers and genes was completed by Integrated DNA Technologies (IDT). Standard synthetic parts from the BioBrick library were used in all genetic circuits. New England Biolabs (NEB) E. coli DH5α cells were used for plasmid propagation. pTS7 was used as the backbone for replicative plasmids, and pTS9 was used as the backbone for integrative plasmids (Fig. S21A). These plasmid backbones were assembled from the following commonly used plasmids: pEAQ-HT (oriV and trfA), pBBR1 (kanR and oriT), pUC19 (bla and colE1), and pVO155 (oriT2). All plasmids were acquired commercially, except that pVO155 was a gift from the Sharon Long laboratory, and pheS* was synthesized based on previously reported literature (89). A PCR extension time of 24 s kb−1 was found to work best for high-GC-content assemblies.
(ii) Azospirillum growth. Unless specified otherwise, Azospirillum strains were prepared for glutamine synthetase assays, ammonia production assays, and plant growth assays as follows: strains were plated on LB agar plates from glycerol stocks and grown at 30°C for 2 days. Single colonies were then inoculated into 5 ml NFbHP medium with 5 mM glutamine and grown for overnight at 30°C and 300 rpm. Cultures were pelleted, triple washed with 1% potassium chloride, and inoculated at an OD600 of 0.1 (unless noted otherwise) at a volume of 3 ml in semisolid NFbHP (without added nitrogen) in 12 ml polypropylene culture tubes. NFbHP medium contained 5 g liter−1 sodium lactate, 1.7 mM sodium chloride, 811 μM magnesium sulfate, 180 μM calcium chloride, and 72 μM iron sulfate. Phosphates were stored separately and added to 34.4 mM dipotassium phosphate and 29.4 mM monopotassium phosphate. Trace elements were added to 1× from the following 50× stock solution: 4.5 mM boric acid, 1.4 mM manganese sulfate, 826 μM sodium molybdate, 86 μM zinc sulfate, and 32 μM copper sulfate. NFbHP was turned semisolid by addition of 0.175% agar prior to inoculation was incubated statically at 30°C postinoculation for experiments characterizing glutamine synthetase activity or ammonia production. For plant experiments, strains were added to plants immediately following the triple wash with 1% potassium chloride.
Plasmids were retained with 50 μg ml−1 of kanamycin at all steps.
The inducible on-state refers to 200 ng ml−1 anhydrotetracycline added immediately following inoculation, unless specified otherwise, and the inducible off-state refers to no anhydrotetracycline added.
(iii) Strain engineering. The A. brasilense Sp245 WT strain was a gift from the Adam Deutschbauer laboratory. Replicative plasmids were transformed into A. brasilense Sp245 by electroporation of 1,000 ng of DNA at 1.25 kV cm−1 and 200 Ω and 25 μF. A brasilense electrocompetent cells were prepared as follows: cells were grown to an OD of 0.5 in LB medium at 30°C and 300 rpm, then rapidly cooled on ice, and pelleted; cells were triple washed with ice-cold 10% glycerol and flash frozen at high concentration in liquid nitrogen. Chromosomally integrative plasmids could be introduced only by conjugation and were moved into A. brasilense by triparental mating using E. coli DH5α as the donor strain and E. coli RK600 as the helper strain. The helper strain was a gift from the Mary Beth Mudgett laboratory. For conjugation, all strains were grown on LB agar plates at 37°C overnight. During growth, the donor plasmids were retained with 50 μg ml−1 kanamycin and the helper plasmid with 100 μg ml−1 chloramphenicol. Cells were scraped from plates at a ratio of 5:1:1 of recipient to helper to donor, resuspended in 1 ml LB, pipette mixed, pelleted, dripped at high concentration onto antibiotic-free LB agar, and incubated at 37°C overnight. Transformants were selected on LB agar with 50 μg ml−1 kanamycin and 10 μg ml−1 streptomycin and restreaked 3 times to ensure proper isolation. For double homologous recombination, negative selection was performed with 5 mM 4-chlorophenylalanine in minimal NFbHP medium with 5 mM glutamine as the nitrogen source. Colonies postconjugation were inoculated until growth was observed (2 to 5 days) at 37°C and 300 rpm, plated on antibiotic-free LB, and screened by PCR for successful double recombination. All strains were stored at −80°C in 1:1 LB-glycerol.
(iv) Fluorescence measurements. RFP strength to characterize genetic circuits in Azospirillum was measured with the Synergy HTX plate reader using a 575/15 excitation filter and 615/16 emission filter. Strains were plated on LB agar plates from glycerol stocks and grown at 30°C for 2 days. Single colonies were then inoculated into 5 ml LB medium and grown for overnight at 30°C and 300 rpm. Cultures were pelleted, washed twice with LB, and inoculated in LB at an OD600 of 0.5 in 200-μl volumes in a 96-well polystyrene plate. Strains were induced with anhydrotetracycline immediately following inoculation, and OD and fluorescence measurements were taken at various time points for both static and shaking plates at 30°C.
(v) Glutamine synthetase activity assay. Strains were cultured as described for Azospirillum growth and cultured statically in semisolid NFbHP medium for 24 h at 30°C to establish nitrogen-fixing conditions prior to glutamine synthetase activity quantification. Glutamine synthetase γ-glutamyl hydroxamate activity was determined based on previously reported procedures (55). All buffers were prepared fresh. Cells were permeabilized by addition of 0.1 mg ml−1 cetyltrimethylammonium bromide (CTAB) and 0.25 mM manganese chloride at room temperature for 5 min and then pelleted. Pellets were washed with ice-cold 1% potassium chloride. Per strain, 6 tubes of each 3-ml culture were combined, concentrated to 4 ml, and stored at 4°C until analysis. A 5× concentrated assay mixture was prepared as follows: 675 mM imidazole hydrochloride buffer, 125 mM potassium arsenate, 100 mM hydroxylamine hydrochloride, 1.25 mM manganese chloride, 0.5 mg ml−1 CTAB. The pH was adjusted to 7.50 at 30°C, the isoactivity point of A. brasilense GS and GS-AMP in this assay, as reported previously (90). A 4× concentrated start mixture was made to 300 mM glutamine and the desired ADP concertation. Then, in a 96-well format, 60 μl of 5× assay mixture was added to 150 μl of cell concentrate and equilibrated at 30°C for 5 min. Reactions were initiated by addition of 75 μl of 4× start mix and took place under static conditions at 30°C. At each time point (0, 5, 10, and 15 min), 66 μl of the reaction mixtures was pipetted into a plate prepared with 133 μl stop mix containing 55 g liter−1 iron chloride, 20 g liter−1 trichloroacetic acid, and 21 ml liter−1 concentrated hydrochloric acid. Absorbance at 540 nm was measured using the Synergy HTX plate reader. All reactions were prepared in 4 technical replicates. Standard curves were determined by adding known concentrations of γ-glutamyl hydroxamate to the stop mix. Rates of γ-glutamyl hydroxamate formation were normalized to total protein content in the reaction mixtures, which was determined using the standard Bradford assay on the cell concentrate.
(vi) Ammonia production assay. Strains were cultured as described for Azospirillum growth. From static semisolid NFbHP cultures, samples of 200 μl were taken at time points and pelleted. A 120-μl portion of supernatant was stored at −20°C until analysis. A 20-μl portion of each sample was analyzed in 96-well PCR plate format using a modified version of the previously described indophenol procedure (59): 120 μl of 150 mM sodium acetate pH 3.7 was added, followed by 80 μl of saturated chlorine water and 30 μl of freshly prepared 8% phenol in water. Plates were immediately incubated at 95°C for 5 min in a thermocycler and then rapidly cooled in cold water. Color was developed by addition of 100 μl of 500 mM borate buffer (pH 12.5). A 200-μl portion of each reaction mixture was transferred to a clear-bottom 96-well polystyrene plate and analyzed for absorbance at 625 nm using the Synergy HTX plate reader. On-plate standards were run for every plate using uninoculated reaction buffer and ammonium chloride as the standard.
Mutants were monitored by plating ammonia-producing cultures onto LB again and analyzing individual colonies by PCR at loci of interest.
(vii) RNA extraction, cDNA synthesis, and qPCR. Strains were cultured as described for Azospirillum growth, with the exception that the diazotrophic inoculation density was increased to an OD600 of 0.5 to increase the material for RNA extraction. Biological replicate tubes were set up such that one 3-ml tube would be harvested at each time point. For this, medium was removed from the tube, leaving only the cellular pellicle (∼200 μl). Total RNA was extracted following Qiagen RNAprotect Bacteria Reagent Handbook protocol 1 (Enzymatic Lysis of Bacteria) and protocol 7 (Purification of Total RNA from Bacterial Lysate using the RNeasy minikit). Appendix B of the handbook (Optional On-Column DNase Digestion using the RNase-Free DNase Set) was followed as part of protocol 7. The kit catalog numbers are as follows: RNAprotect bacterial reagent, 76506; RNeasy Protect bacterial kit, 74524. RNA was eluted in 30 μl RNase-free water with yields between 1 and 8 ng/ml, and cDNA synthesis was immediately performed.
cDNA was synthesized using Invitrogen Superscript IV first-strand synthesis system (catalog number 18091050) following supplier protocol steps 1 to 4. Random hexamers were used as primers (supplied with the kit). cDNA was stored at −20°C.
Quantitative PCR (qPCR) was performed on a QuantStudio 3 real-time PCR system and with the Bioline SensiMix SYBR Hi-ROX kit (catalog number C755J03). The supplier protocol was followed. Primers used for glnA (5′-3′) were GTACACCTCGGACAAGGAC and GTGGTGGTGCTTCTCGA. glyA was used as a previously reported housekeeping gene, with the primers (5′-3′) GGAGATCGCCAAGAAGATCA and GCTCTTGGCGTAGGTCTTGA (91). cDNA template was added to final concentrations of ∼0.25 ng μl−1 in 20-μl reaction mixtures. The thermocycling protocol was 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C 15 s.
Data were analyzed using the Pfaffl method (92).
(viii) Western blotting. Strains were cultured as described for Azospirillum growth. At 24 h after inoculation and induction, cells were harvested by 1:1 addition of 2× Laemmli sample buffer buffer, frozen in liquid nitrogen, and stored at −80°C until analysis. Nitrogen shock samples were treated with 20 mM ammonium chloride for 30 min prior to harvesting. Also prior to harvesting, OD600 of cultures were determined.
For blotting analysis, samples were thawed and their concentrations normalized to an OD600 of 0.1 based on OD600 measurements prior to harvesting. Subsamples of 50 μl were taken, to which 1.25 μl β-mercaptoethanol was added, followed by denaturation at 100°C for 1 min. A 10-μl portion of each sample was loaded on a Mini-Protean TGX 4 to 15% precast gel. Gels were run at 100 V for 80 min. Following this, the Trans-Blot Turbo transfer system was used at 25 V for 7 min to transfer samples to a polyvinylidene difluoride (PVDF) membrane.
All subsequent washing, blocking, and staining steps were performed at room temperature on a rocker. Blocker A, blocker B, antibody wash solutions, and chromogenic substrate were from the Invitrogen WesternBreeze chemiluminescent kit (catalog number WB7106). Membranes were first washed twice for 5 min with 10 ml Tris-buffered saline (TBS) and then blocked for 30 min with 10 ml blocking buffer composed of 2 ml blocker A, 3 ml blocker B, and 5 ml water. This was followed by another washing step in 20 ml water for 5 min.
Primary antibody (chicken anti-NifH; Agrisera AS01 021A) was added at a dilution of 1:2,000 for 1 h in 2 ml blocker A, 1 ml blocker B, and 7 ml water. This was followed by 3 washes with 20 ml 1× antibody wash solution. Secondary antibody (goat anti-chicken IgY, alkaline phosphatase [ALP] conjugated; Agrisera AS09 606) was added at a dilution of 1:10,000 for 30 min in 2 ml blocker A, 1 ml blocker B, and 7 ml water. This was followed by 2 washes, 2 min each, with 20 ml water.
Gels were stained in the dark by the addition of 5 ml 1× chromogenic substrate until bands became visible, rinsed in water twice, and photographed.
(ix) NMR analysis. Strains were cultured as described in the Azospirillum growth section with the following modifications: culture volumes were 5 ml in 25 ml scintillation vials capped with rubber stoppers. The headspace was replaced with 20 ml 15N2 gas, 14N2 gas, or argon. Then, 5 ml was displaced with O2, and vials were incubated at 30°C for 65 h. Following incubation, cultures were uncapped and pelleted, and the supernatant was frozen at −20°C until analysis. Samples were quantified for ammonia using the indophenol method as described for the ammonia production assay and prepared for 1H-NMR analysis by addition of 50 μl deuterated dimethyl sulfoxide (DMSO) and 25 μl concentrated HCl to 425-μl samples. Standards were prepared equivalently, using 10 mM 14NH4Cl and 10 mM 15NH4Cl in uninoculated culture buffer. Spectra were collected using an INOVA 600 Oxford NMR with a 64-scan selective pulse sequence for ammonia as reported previously (75).
(x) Setaria plant growth. S. viridis A10 seeds were a gift from the Jose Dinneny laboratory. For all plant experiments, seeds were first surface sterilized in 10% bleach and 0.01% Tween 20 for 10 min, followed by a triple water wash, and then imbibed overnight at 30°C. NFbHP at 1/5 strength with the addition of 1 mM NH4Cl, and 0.5 mM calcium chloride was prepared to a semisolid state (0.175% agar). After imbibation, seeds were planted on 1.2-cm2 PTFE mesh floating on 10 ml medium in 50-ml capped glass tubes. Growth chamber conditions were as follows: 16-h light cycles with 30°C light and 24°C dark temperatures, at a relative humidity of 50%. Strains were prepared as described for Azospirillum growth and inoculated 14 days postplanting to an OD600 of 0.1. Anhydrotetracycline was added to the relevant experimental groups at 200 ng ml−1 following inoculation. At 14 days postinoculation, whole shoots were harvested and lyophilized overnight prior to dry weight determination.
(xi) Pheophytin analysis. Plants were grown as described in the plant growth section. At 14 days postplanting (coincident with microbial inoculation), 50% of the headspace in the label group was displaced with 15N2 gas. After lyophilization and dry weight collection, chlorophyll of shoots was extracted, similarly to previously reported methods (29, 77). Whole shoots were treated as individual samples. Each sample was ball-mill homogenized at 25 Hz for 2 min and extracted in 1.4 ml methanol for 2 h at room temperature. Leaf fragments were precipitated by centrifugation, and 1 ml of supernatant was transferred to a new tube. Then, 145 μl dioxane and 180 μl water were added, and samples were chilled at −80°C for 2 h. Chlorophyll was pelleted at a relative centrifugal force (RCF) of 21,000 for 15 min at 4°C, resuspended in 200 μl 1:1 methanol-acetone, and stored in this state for several days at −80°C until analysis. Chlorophyll was then converted to pheophytin by addition of 1 μl concentrated hydrochloric acid and clarified for 2 min at an RCF of 21,000. Standards were prepared from a chlorophyll authentic standard following the same pheophytin conversion method. Pheophytin isotope abundances were analyzed by quantitative time-of-flight (qTOF) mass spectrometry on an Agilent 6545 liquid chromatograph-mass spectrometer (LC-MS). For this, 5 μl of sample was directly injected into the mass spectrometer in electrospray ion positive mode with a fragmentor voltage of 175 V and capillary voltage of 3,500 V. The solvent was 10% water in acetonitrile and 0.1% formic acid, with a flow rate of 0.6 ml min−1, a sheath gas flow of 12 L min−1 at 300°C, a drying gas flow of 12 liters min−1 at 250°C, and a nebulizer pressure of 10 lb/in2.
Mass-isotopomer distributions (MIDs) were computationally extracted from raw data as previously reported (93). Isotope enrichment was computed as shown in Fig. S18.
(xii) Statistical analysis. Statistical analysis was performed in Excel: P values of statistical significance were computed by two-sided homoscedastic t test. All other data processing is described in the supplemental material, including relative expression unit calculations (Fig. S2B), glutamine synthetase adenylylation state calculations (Fig. S3C), and mass spectrometry data processing (Fig. S18). Sample sizes and units are specified in figure legends for all experiments.
Data availability.
All data generated or analyzed during this study are included in the paper. Numerical values of biological or technical replicates from which graphs were generated may be requested from the corresponding author. All DNA sequences have been deposited in GenBank with accession numbers MW835297 to MW835314. See Fig. S21 for details of sequences.
ACKNOWLEDGMENTS
We thank Kevin Smith, Curt Fisher, and Jay Schwalbe for help with isotope labeling experiments and Katarina Guzman for help with constructing and designing knockout and overexpression lines. We also thank Sharon Long, Drew Endy, and K. C. Huang for helpful comments for this project. We thank the following laboratories for providing reagents for this work: Adam Deutschbauer, Jose Dinneny, Sharon Long, and Mary Beth Mudgett.
We thank the Stanford University departments of Bioengineering and Chemical Engineering, Stanford Bio-X, Ric Weiland Fellowships, and HHMI for funding of this work.
T.S. and E.S. designed experiments and analyzed data; T.S. performed experiments; T.S. wrote the paper with edits from E.S.
T.S., E.S., and Stanford University have filed patent application PCT/US2020/016856 for this work.
Footnotes
Supplemental material is available online only.
Contributor Information
Elizabeth Sattely, Email: sattely@stanford.edu.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Allison FE (ed). 1973. Developments in soil science, vol 3, p 120–138. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 2.Davidson EA, David MB, Galloway JN, Goulds CL, Haeuber R, Harrison JA, Howarth RW, Jaynes DB, Lowrance RR, Nolan BT, Peel JL, Pinder RW, Porter EP, Snyder CS, Townsend AR, Ward MH. 2012. Excess nitrogen in the U.S. environment: trends, risks and solutions. Issues Ecol 2012(15):1–16. [Google Scholar]
- 3.Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320:889–892. 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 4.Pankievicz VCS, Irving TB, Maia LGS, Ané J-M. 2019. Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol 17:99. 10.1186/s12915-019-0710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mus F, Crook MB, Garcia K, Garcia Costas A, Geddes BA, Kouri ED, Paramasivan P, Ryu M-H, Oldroyd GED, Poole PS, Udvardi MK, Voigt CA, Ané J-M, Peters JW. 2016. Symbiotic nitrogen fixation and challenges to extending it to non-legumes. Appl Environ Microbiol 82:3698–3710. 10.1128/AEM.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wurtzel ET, Vickers CE, Hanson AD, Millar AH, Cooper M, Voss-Fels KP, Nikel PI, Erb TJ. 2019. Revolutionizing agriculture with synthetic biology. Nat Plants 5:1207–1210. 10.1038/s41477-019-0539-0. [DOI] [PubMed] [Google Scholar]
- 7.Ryu M-H, Zhang J, Toth T, Khokhani D, Geddes BA, Mus F, Garcia-Costas A, Peters JW, Poole PS, Ané J-M, Voigt CA. 2020. Control of nitrogen fixation in bacteria that associate with cereals. Nat Microbiol 5:314–330. 10.1038/s41564-019-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Triplett EW. 1996. Diazotrophic endophytes: progress and prospects for nitrogen fixation in monocots. Plant Soil 186:29–38. 10.1007/BF00035052. [DOI] [Google Scholar]
- 9.Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R. 2012. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. 10.1186/1471-2164-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd ES, Peters JW. 2013. New insights into the evolutionary history of biological nitrogen fixation. Front Microbiol 4:201. 10.3389/fmicb.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smercina DN, Evans SE, Friesen ML, Tiemann LK. 2019. To fix or not to fix: controls on free-living nitrogen fixation in the rhizosphere. Appl Environ Microbiol 85:e02546-18. 10.1128/AEM.02546-18. (Erratum, 85:e02103-19, 2019, .) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long SR. 2001. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol 125:69–72. 10.1104/pp.125.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scupham AJ, et al. 1996. Inoculation with Sinorhizobium meliloti RMBPC-2 increases alfalfa yield compared with inoculation with a nonengineered wild-type strain. Appl Environ Microbiol 62:4260–4262. 10.1128/AEM.62.11.4260-4262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awika JM. 2011. Major cereal grains production and use around the world. ACS Symp Ser Am Chem Soc 1089:1–13. 10.1021/bk-2011-1089.ch001. [DOI] [Google Scholar]
- 15.Rogers C, Oldroyd GED. 2014. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J Exp Bot 65:1939–1946. 10.1093/jxb/eru098. [DOI] [PubMed] [Google Scholar]
- 16.Rosenblueth M, Ormeno-Orrillo E, Lopez-Lopez A, Rogel MA, Reyes-Hernandez BJ, Martinez-Romero JC, Reddy PM, Martinez-Romero E. 2018. Nitrogen fixation in cereals. Front Microbiol 9:1794. 10.3389/fmicb.2018.01794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankievicz VCS, do Amaral FP, Santos KFDN, Agtuca B, Xu Y, Schueller MJ, Arisi ACM, Steffens MBR, de Souza EM, Pedrosa FO, Stacey G, Ferrieri RA. 2015. Robust biological nitrogen fixation in a model grass–bacterial association. Plant J 81:907–919. 10.1111/tpj.12777. [DOI] [PubMed] [Google Scholar]
- 18.Santos KFDN, Moure VR, Hauer V, Santos ARS, Donatti L, Galvão CW, Pedrosa FO, Souza EM, Wassem R, Steffens MBR. 2017. Wheat colonization by an Azospirillum brasilense ammonium-excreting strain reveals upregulation of nitrogenase and superior plant growth promotion. Plant Soil 415:245–255. 10.1007/s11104-016-3140-6. [DOI] [Google Scholar]
- 19.Christiansen-Weniger C, van Veen JA. 1991. Nitrogen fixation by Azospirillium brasilense in soil and the rhizosphere under controlled environmental conditions. Biol Fertil Soils 12:100–106. 10.1007/BF00341483. [DOI] [Google Scholar]
- 20.Boddey RM, Urquiaga S, Alves BJR, Reis V. 2003. Endophytic nitrogen fixation in sugarcane: present knowledge and future applications. Plant Soil 252:139–149. 10.1023/A:1024152126541. [DOI] [Google Scholar]
- 21.Divan Baldani VL, Baldani JI, Döbereiner J. 2000. Inoculation of rice plants with the endophytic diazotrophs Herbaspirillum seropedicae and Burkholderia spp. Biol Fertil Soils 30:485–491. 10.1007/s003740050027. [DOI] [Google Scholar]
- 22.James EK. 2000. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Res 65:197–209. 10.1016/S0378-4290(99)00087-8. [DOI] [Google Scholar]
- 23.Bremer E, Janzen HH, Gilbertson C. 1995. Evidence against associative N2 fixation as a significant N source in long-term wheat plots. Plant Soil 175:13–19. 10.1007/BF02413006. [DOI] [Google Scholar]
- 24.Chalk PM. 1991. The contribution of associative and symbiotic nitrogen fixation to the nitrogen nutrition of non-legumes. Plant Soil 132:29–39. 10.1007/BF00011009. [DOI] [Google Scholar]
- 25.Bueno Batista M, Dixon R. 2019. Manipulating nitrogen regulation in diazotrophic bacteria for agronomic benefit. Biochem Soc Trans 47:603–614. 10.1042/BST20180342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santi C, Bogusz D, Franche C. 2013. Biological nitrogen fixation in non-legume plants. Ann Bot 111:743–767. 10.1093/aob/mct048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho TLG, Balsemão-Pires E, Saraiva RM, Ferreira PCG, Hemerly AS. 2014. Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria. J Exp Bot 65:5631–5642. 10.1093/jxb/eru319. [DOI] [PubMed] [Google Scholar]
- 28.Lucy M, Reed E, Glick BR. 2004. Applications of free living plant growth-promoting rhizobacteria. Antonie Van Leeuwenhoek 86:1–25. 10.1023/B:ANTO.0000024903.10757.6e. [DOI] [PubMed] [Google Scholar]
- 29.Kahn M, Parra-Colmenares A, Ford CL, Kaser F, McCaskill D, Ketchum RE. 2002. A mass spectrometry method for measuring 15N incorporation into pheophytin. Anal Biochem 307:219–225. 10.1016/s0003-2697(02)00046-5. [DOI] [PubMed] [Google Scholar]
- 30.Temme K, Hill R, Segall-Shapiro TH, Moser F, Voigt CA. 2012. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res 40:8773–8781. 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Temme K, Zhao D, Voigt CA. 2012. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc Natl Acad Sci U S A 109:7085–7090. 10.1073/pnas.1120788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geddes BA, Ryu M-H, Mus F, Costas AG, Peters JW, Voigt CA, Poole P. 2015. Use of plant colonizing bacteria as chassis for transfer of N2-fixation to cereals. Curr Opin Biotechnol 32:216–222. 10.1016/j.copbio.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Curatti L, Rubio LM. 2014. Challenges to develop nitrogen-fixing cereals by direct nif-gene transfer. Plant Sci 225:130–137. 10.1016/j.plantsci.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Ambrosio R, Ortiz-Marquez JCF, Curatti L. 2017. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metab Eng 40:59–68. 10.1016/j.ymben.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2:621–631. 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 36.Li XX, Liu Q, Liu XM, Shi HW, Chen SF. 2016. Using synthetic biology to increase nitrogenase activity. Microb Cell Fact 15:43. 10.1186/s12934-016-0442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smanski MJ, Bhatia S, Zhao D, Park Y, Woodruff LBA, Giannoukos G, Ciulla D, Busby M, Calderon J, Nicol R, Gordon DB, Densmore D, Voigt CA. 2014. Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol 32:1241–1249. 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- 38.Barney BM, Eberhart LJ, Ohlert JM, Knutson CM, Plunkett MH. 2015. Gene deletions resulting in increased nitrogen release by Azotobacter vinelandii: application of a novel nitrogen biosensor. Appl Environ Microbiol 81:4316–4328. 10.1128/AEM.00554-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz-Marquez JC, Do Nascimento M, Curatti L. 2014. Metabolic engineering of ammonium release for nitrogen-fixing multispecies microbial cell-factories. Metab Eng 23:154–164. 10.1016/j.ymben.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Bali A, Blanco G, Hill S, Kennedy C. 1992. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol 58:1711–1718. 10.1128/AEM.58.5.1711-1718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadtman ER. 2004. Regulation of glutamine synthetase activity. EcoSal Plus 1 10.1128/ecosalplus.3.6.1.6. [DOI] [PubMed] [Google Scholar]
- 42.Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol Rev 59:604–622. 10.1128/MMBR.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machado HB, Funayama S, Rigo LU, Pedrosa FO. 1991. Excretion of ammonium by Azospirillum brasilense mutants resistant to ethylenediamine. Can J Microbiol 37:549–553. 10.1139/m91-092. [DOI] [Google Scholar]
- 44.Colnaghi R, Green A, He L, Rudnick P, Kennedy C. 1997. Strategies for increased ammonium production in free-living or plant associated nitrogen fixing bacteria. Plant Soil 194:145–154. 10.1023/A:1004268526162. [DOI] [Google Scholar]
- 45.Van Dommelen A, Keijers V, Wollebrants A, Vanderleyden J. 2003. Phenotypic changes resulting from distinct point mutations in the Azospirillum brasilense glnA gene, encoding glutamine synthetase. Appl Environ Microbiol 69:5699–5701. 10.1128/aem.69.9.5699-5701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu N, Yang M, Gaur U, Xu H, Yao Y, Li D. 2016. Alpha-ketoglutarate: physiological functions and applications. Biomol Ther (Seoul) 24:1–8. 10.4062/biomolther.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang P, Pioszak AA, Ninfa AJ. 2007. Structure-function analysis of glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49) of Escherichia coli. Biochemistry 46:4117–4132. 10.1021/bi0620508. [DOI] [PubMed] [Google Scholar]
- 48.de Zamaroczy M, Paquelin A, Elmerich C. 1993. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J Bacteriol 175:2507–2515. 10.1128/JB.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwab S, Souza EM, Yates MG, Persuhn DC, Stefferns MBR, Chubatsu LS, Pedrosa FO, Rigo LU. 2007. The glnAntrBC operon of Herbaspirillum seropedicae is transcribed by two oppositely regulated promoters upstream of glnA. Can J Microbiol 53:100–105. 10.1139/w06-113. [DOI] [PubMed] [Google Scholar]
- 50.Jaggi R, van Heeswijk WC, Westerhoff HV, Ollis DL, Vasudevan SG. 1997. The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J 16:5562–5571. 10.1093/emboj/16.18.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okon Y. 1985. Azospirillum as a potential inoculant for agriculture. Trends Biotechnol 3:223–228. 10.1016/0167-7799(85)90012-5. [DOI] [Google Scholar]
- 52.Steenhoudt O, Vanderleyden J. 2000. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev 24:487–506. 10.1111/j.1574-6976.2000.tb00552.x. [DOI] [PubMed] [Google Scholar]
- 53.Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu X-G, Kellogg E, Van Eck J. 2010. Setaria viridis: a model for C4 photosynthesis. Plant Cell 22:2537–2544. 10.1105/tpc.110.075309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shapiro BM, Stadtman ER. 1970. Glutamine synthetase (Escherichia coli). Methods Enzymol 17:910–922. 10.1016/0076-6879(71)17305-3. [DOI] [PubMed] [Google Scholar]
- 55.Goldberg RB, Hanau R. 1979. Relation between the adenylylation state of glutamine synthetase and the expression of other genes involved in nitrogen metabolism. J Bacteriol 137:1282–1289. 10.1128/JB.137.3.1282-1289.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bender RA, Janssen KA, Resnick AD, Blumenberg M, Foor F, Magasanik B. 1977. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol 129:1001–1009. 10.1128/JB.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Dommelen A, Spaepen S, Vanderleyden J. 2009. Identification of the glutamine synthetase adenylyltransferase of Azospirillum brasilense. Res Microbiol 160:205–212. 10.1016/j.resmic.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Stadtman ER, Mura U, Chock PB, Rhee SG. 1980. The interconvertible enzyme cascade that regulates glutamine synthetase activity, p 41–59. In Mora J, Palacios R (ed) Glutamine: metabolism, enzymology, and regulation. Academic Press, New York, NY. [Google Scholar]
- 59.Bolleter WT, Bushman CJ, Tidwell PW. 1961. Spectrophotometric determination of ammonia as indophenol. Anal Chem 33:592–594. 10.1021/ac60172a034. [DOI] [Google Scholar]
- 60.Rugbjerg P, Sommer MOA. 2019. Overcoming genetic heterogeneity in industrial fermentations. Nat Biotechnol 37:869–876. 10.1038/s41587-019-0171-6. [DOI] [PubMed] [Google Scholar]
- 61.Tyo KEJ, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760. 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 62.Bergthorsson U, Andersson DI, Roth JR. 2007. Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A 104:17004–17009. 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holtz WJ, Keasling JD. 2010. Engineering static and dynamic control of synthetic pathways. Cell 140:19–23. 10.1016/j.cell.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 64.Peng B, Plan MR, Carpenter A, Nielsen LK, Vickers CE. 2017. Coupling gene regulatory patterns to bioprocess conditions to optimize synthetic metabolic modules for improved sesquiterpene production in yeast. Biotechnol Biofuels 10:43–43. 10.1186/s13068-017-0728-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffman BM, Lukoyanov D, Yang Z-Y, Dean DR, Seefeldt LC. 2014. Mechanism of nitrogen fixation by nitrogenase: the next stage. Chem Rev 114:4041–4062. 10.1021/cr400641x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. 2008. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9:102–102. 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moure VR, et al. 2013. The nitrogenase regulatory enzyme dinitrogenase reductase ADP-ribosyltransferase (DraT) is activated by direct interaction with the signal transduction protein GlnB. J Bacteriol 195:279–286. 10.1128/JB.01517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moure VR, Costa FF, Cruz LM, Pedrosa FO, Souza EM, Li X-D, Winkler F, Huergo LF. 2015. Regulation of nitrogenase by reversible mono-ADP-ribosylation, p 89–106. In Koch-Nolte F (ed), Endogenous ADP-ribosylation. Springer International Publishing, Cham, Switzerland. [DOI] [PubMed] [Google Scholar]
- 69.Aquino B, Stefanello AA, Oliveira MAS, Pedrosa FO, Souza EM, Monteiro RA, Chubatsu LS. 2015. Effect of point mutations on Herbaspirillum seropedicae NifA activity. Braz J Med Biol Res 48:683–690. 10.1590/1414-431X20154522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, Xie X, Yang M, Dixon R, Wang Y-P. 2017. Modular electron-transport chains from eukaryotic organelles function to support nitrogenase activity. Proc Natl Acad Sci U S A 114:E2460–E2465. 10.1073/pnas.1620058114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgren T, Nordlund S. 2006. Two pathways of electron transport to nitrogenase in Rhodospirillum rubrum: the major pathway is dependent on the fix gene products. FEMS Microbiol Lett 260:30–35. 10.1111/j.1574-6968.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- 72.Poudel S, Colman DR, Fixen KR, Ledbetter RN, Zheng Y, Pence N, Seefeldt LC, Peters JW, Harwood CS, Boyd ES. 2018. Electron transfer to nitrogenase in different genomic and metabolic backgrounds. J Bacteriol 200:e00757-17. 10.1128/JB.00757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ledbetter RN, et al. 2017. The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 56:4177–4190. 10.1021/acs.biochem.7b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plunkett MH, Knutson CM, Barney BM. 2020. Key factors affecting ammonium production by an Azotobacter vinelandii strain deregulated for biological nitrogen fixation. Microb Cell Fact 19:107. 10.1186/s12934-020-01362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nielander AC, McEnaney JM, Schwalbe JA, Baker JG, Blair SJ, Wang L, Pelton JG, Andersen SZ, Enemark-Rasmussen K, Čolić V, Yang S, Bent SF, Cargnello M, Kibsgaard J, Vesborg PCK, Chorkendorff I, Jaramillo TF. 2019. A versatile method for ammonia detection in a range of relevant electrolytes via direct nuclear magnetic resonance techniques. ACS Catal 9:5797–5802. 10.1021/acscatal.9b00358. [DOI] [Google Scholar]
- 76.Liang Y, Urano D, Liao K-L, Hedrick TL, Gao Y, Jones AM. 2017. A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings. Plant Methods 13:26. 10.1186/s13007-017-0174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parra-Colmenares A, Kahn ML. 2005. Determination of nitrogen fixation effectiveness in selected Medicago truncatula isolates by measuring nitrogen isotope incorporation into pheophytin. Plant Soil 270:159–168. 10.1007/s11104-004-1308-y. [DOI] [Google Scholar]
- 78.Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. 2014. Niche and host-associated functional signatures of the root surface microbiome. Nat Commun 5:4950. 10.1038/ncomms5950. [DOI] [PubMed] [Google Scholar]
- 79.Yu P, Li X, White PJ, Li C. 2015. A large and deep root system underlies high nitrogen-use efficiency in maize production. PLoS One 10:e0126293. 10.1371/journal.pone.0126293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Banger K, Nafziger ED, Wang J, Muhammad U, Pittelkow CM. 2018. Simulating nitrogen management impacts on maize production in the U.S. Midwest. PLoS One 13:e0201825. 10.1371/journal.pone.0201825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Calles J, Justice I, Brinkley D, Garcia A, Endy D. 2019. Fail-safe genetic codes designed to intrinsically contain engineered organisms. Nucleic Acids Res 47:10439–10451. 10.1093/nar/gkz745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fernandez-Rodriguez J, Yang L, Gorochowski TE, Gordon DB, Voigt CA. 2015. Memory and combinatorial logic based on DNA inversions: dynamics and evolutionary stability. ACS Synth Biol 4:1361–1372. 10.1021/acssynbio.5b00170. [DOI] [PubMed] [Google Scholar]
- 83.Geddes BA, Paramasivan P, Joffrin A, Thompson AL, Christensen K, Jorrin B, Brett P, Conway SJ, Oldroyd GED, Poole PS. 2019. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat Commun 10:3430. 10.1038/s41467-019-10882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mondy S, Lenglet A, Beury-Cirous A, Libanga C, Ratet P, Faure D, Dessaux Y. 2014. An increasing opine carbon bias in artificial exudation systems and genetically modified plant rhizospheres leads to an increasing reshaping of bacterial populations. Mol Ecol 23:4846–4861. 10.1111/mec.12890. [DOI] [PubMed] [Google Scholar]
- 85.Huergo LF, Souza EM, Steffens MBR, Yates MG, Pedrosa FO, Chubatsu LS. 2005. Effects of over-expression of the regulatory enzymes DraT and DraG on the ammonium-dependent post-translational regulation of nitrogenase reductase in Azospirillum brasilense. Arch Microbiol 183:209–217. 10.1007/s00203-005-0763-z. [DOI] [PubMed] [Google Scholar]
- 86.Huergo LF, Pedrosa FO, Muller-Santos M, Chubatsu LS, Monteiro RA, Merrick M, Souza EM. 2012. PII signal transduction proteins: pivotal players in post-translational control of nitrogenase activity. Microbiology 158:176–190. 10.1099/mic.0.049783-0. [DOI] [PubMed] [Google Scholar]
- 87.Moure VR, Siöberg CLB, Valdameri G, Nji E, Oliveira MAS, Gerdhardt ECM, Pedrosa FO, Mitchell DA, Seefeldt LC, Huergo LF, Högbom M, Nordlund S, Souza EM. 2019. The ammonium transporter AmtB and the PII signal transduction protein GlnZ are required to inhibit DraG in Azospirillum brasilense. FEBS J 286:1214–1229. 10.1111/febs.14745. [DOI] [PubMed] [Google Scholar]
- 88.Lamoureux G, Javelle A, Baday S, Wang S, Berneche S. 2010. Transport mechanisms in the ammonium transporter family. Transfus Clin Biol 17:168–175. 10.1016/j.tracli.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 89.Miyazaki K. 2015. Molecular engineering of a PheS counterselection marker for improved operating efficiency in Escherichia coli. Biotechniques 58:86–88. 10.2144/000114257. [DOI] [PubMed] [Google Scholar]
- 90.Pirola MC, Monopoli R, Aliverti A, Zanetti G. 1992. Isolation and characterization of glutamine synthetase from the diazotroph Azospirillum brasilense. Int J Biochem 24:1749–1754. 10.1016/0020-711X(92)90124-J. [DOI] [Google Scholar]
- 91.McMillan M, Pereg L. 2014. Evaluation of reference genes for gene expression analysis using quantitative RT-PCR in Azospirillum brasilense. PLoS One 9:e98162. 10.1371/journal.pone.0098162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nett RS, Guan X, Smith K, Faust AM, Sattely ES, Fischer C. 2018. D2O labeling to measure active biosynthesis of natural products in medicinal plants. AIChE J 64:4319–4330. 10.1002/aic.16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S21. Download AEM00582-21_Supp_1_seq4.pdf, PDF file, 1.7 MB (1.7MB, pdf)
Data Availability Statement
All data generated or analyzed during this study are included in the paper. Numerical values of biological or technical replicates from which graphs were generated may be requested from the corresponding author. All DNA sequences have been deposited in GenBank with accession numbers MW835297 to MW835314. See Fig. S21 for details of sequences.