ABSTRACT
Within animal-associated microbiomes, the functional roles of specific microbial taxa are often uncharacterized. Here, we use the fungus-growing ant system, a model for microbial symbiosis, to determine the potential defensive roles of key bacterial taxa present in the ants’ fungus gardens. Fungus gardens serve as an external digestive system for the ants, with mutualistic fungi in the genus Leucoagaricus converting the plant substrate into energy for the ants. The fungus garden is host to specialized parasitic fungi in the genus Escovopsis. Here, we examine the potential role of Burkholderia spp. that occur within ant fungus gardens in inhibiting Escovopsis. We isolated members of the bacterial genera Burkholderia and Paraburkholderia from 50% of the 52 colonies sampled, indicating that members of the family Burkholderiaceae are common inhabitants in the fungus gardens of a diverse range of fungus-growing ant genera. Using antimicrobial inhibition bioassays, we found that 28 out of 32 isolates inhibited at least one Escovopsis strain with a zone of inhibition greater than 1 cm. Genomic assessment of fungus garden-associated Burkholderiaceae indicated that isolates with strong inhibition all belonged to the genus Burkholderia and contained biosynthetic gene clusters that encoded the production of two antifungals: burkholdine1213 and pyrrolnitrin. Organic extracts of cultured isolates confirmed that these compounds are responsible for antifungal activities that inhibit Escovopsis but, at equivalent concentrations, not Leucoagaricus spp. Overall, these new findings, combined with previous evidence, suggest that members of the fungus garden microbiome play an important role in maintaining the health and function of fungus-growing ant colonies.
IMPORTANCE Many organisms partner with microbes to defend themselves against parasites and pathogens. Fungus-growing ants must protect Leucoagaricus spp., the fungal mutualist that provides sustenance for the ants, from a specialized fungal parasite, Escovopsis. The ants take multiple approaches, including weeding their fungus gardens to remove Escovopsis spores, as well as harboring Pseudonocardia spp., bacteria that produce antifungals that inhibit Escovopsis. In addition, a genus of bacteria commonly found in fungus gardens, Burkholderia, is known to produce secondary metabolites that inhibit Escovopsis spp. In this study, we isolated Burkholderia spp. from fungus-growing ants, assessed the isolates’ ability to inhibit Escovopsis spp., and identified two compounds responsible for inhibition. Our findings suggest that Burkholderia spp. are often found in fungus gardens, adding another possible mechanism within the fungus-growing ant system to suppress the growth of the specialized parasite Escovopsis.
KEYWORDS: antifungal, attine, burkholderia, burkholdine, defensive symbiosis, escovopsis, fungus-growing ant, pyrrolnitrin
INTRODUCTION
Symbiotic associations are ubiquitous. Organisms do not live in isolation; rather, they exist in complex communities consisting of variable macro- and microorganisms. These symbiotic interactions are known to play a fundamental role in shaping life on earth, and associations can range from transient to obligate and parasitic to beneficial (1). Microbial symbioses span this range and fill a variety of important roles within hosts. Substantial work has been done specifically in association with insect hosts. Research on beneficial microbes in insects has typically focused on the role of symbionts in providing nutrients to their host (2, 3). However, in recent years, it has become clear that microbes often play a critical role in mediating interactions between insects and pathogens or parasites (4, 5). Microbes can provide defense through competitively excluding pathogenic microbes, priming the host immune system, and producing compounds that protect the host (6–8). In this study, we used the fungus-growing ant system to explore a bacterially mediated antifungal defensive symbiosis.
Fungus-growing ants are a well-studied example of a multipartite symbiosis (Fig. 1). Fungus-growing ants (Hymenoptera, Formicidae, Attini, Attina) thrive in the Neotropics and consist of 20 genera and approximately 250 species (9, 10) that have formed ancient and highly evolved symbioses with both fungi and bacteria (11). The ants cultivate fungi in the genus Leucoagaricus (Basidiomycota, Agaricales, Agaricaceae) as a food source. Fungus-growing ants bring an organic substrate to structures known as fungus gardens (Fig. 1B), where Leucoagaricus spp. principally degrade the substrate and produce usable energy for the ants (12, 13). In addition, a consistent bacterial community composed primarily of Proteobacteria exists within fungus gardens (14–18). In leaf-cutter ants, the bacterial community has been shown to help degrade plant secondary compounds and aid in nitrogen acquisition for the ant through biological nitrogen fixation (19, 20). Though the functions of some of the bacterial taxa have been described, the roles of many bacterial fungus garden members are unknown.
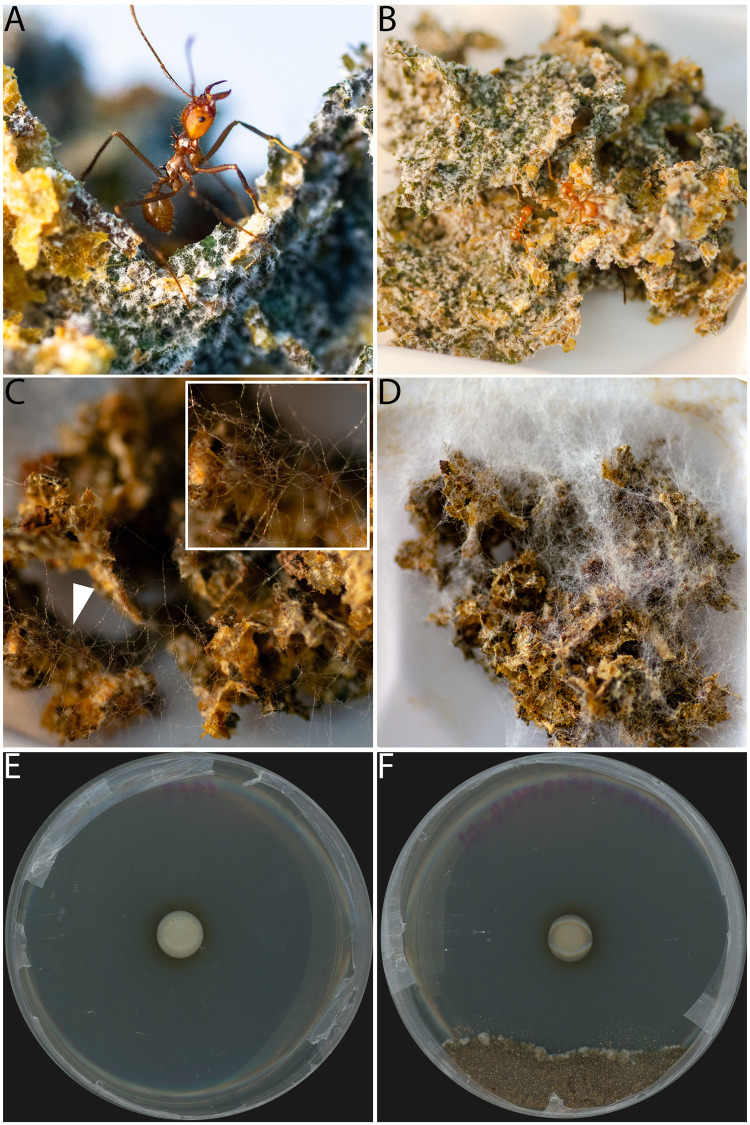
FIG 1.
Infection of an Atta cephalotes colony by Escovopsis weberi (CF180408-01) and in vitro petri plate antimicrobial inhibition bioassay with ICBG1719. (A, B) Healthy fungus garden with no Escovopsis infection; (C) day 3 postinfection of fungus garden with Escovopsis; (D) day 7 postinfection of fungus garden with Escovopsis; (E and F) petri plate with only Burkholderia sp. ICBG1719 growing (E) and petri plate inhibition bioassay of ICBG1719 against Escovopsis weberi (CF180408-01), with a clear zone of inhibition (F). Pictures in panels A to D were taken by Caitlin Carlson.
Fungus gardens are threatened by a specialized pathogen, the parasitic fungus Escovopsis (Ascomycota, Hypocreales, Hypocreaceae) (21). In many lineages of fungus-growing ants, the growth of Escovopsis spp. is inhibited by Actinobacteria in the genus Pseudonocardia, defensive symbionts found on the exoskeleton or occurring within specialized structures on the ants (22–24). However, in the two fungus-growing ant genera Atta and Sericomyrmex, this defensive symbiosis with Pseudonocardia has been secondarily lost (23, 24). Despite the lack of the symbiont Pseudonocardia, the fungus gardens are not overrun with Escovopsis spp., suggesting other ways of controlling Escovopsis spp. The ant behaviors of weeding and grooming are known to be crucial for suppressing Escovopsis spp. (25, 26). In addition, there is evidence that antifungal-producing bacteria may colonize the fungus garden and provide some level of inhibition against Escovopsis spp. (27). Santos and colleagues (27) isolated Burkholderia spp. consistently from Atta sexdens fungus gardens (32/57 colonies) and identified isolates that could inhibit the growth of Escovopsis weberi, suggesting a potential role for garden bacteria in the defense of fungus gardens.
The fungus gardens’ Proteobacteria-dominant bacterial community is known to include members of the family Burkholderiaceae (14–18, 27). Burkholderiaceae have diverse metabolic capabilities and inhabit a broad range of ecological niches (28). Members of two closely related genera within the Burkholderiaceae, Burkholderia and Paraburkholderia, have been found and characterized in the context of mammalian and plant pathogenesis, nitrogen fixation, bioremediation, and plant growth stimulation and/or in close association with fungi and insects. These symbiotic Burkholderia and Paraburkholderia spp. can also produce secondary metabolites that are important in ecological interactions. For example, Paraburkholderia rhizoxinica resides in the hyphae of the fungal plant pathogen Rhizopus microsporus. P. rhizoxinica produces an antimitotic macrolide that is converted into rhizoxin, which is the causative agent of rice seedling blight (29, 30). Lagriinae beetles depend on Burkholderia symbionts for the protection of their eggs. Specifically, Burkholderia gladioli produces a blend of antibiotics, including toxoflavin, caryoynencin, lagriene, and sinapigladioside, which protect the egg stage of the beetles against pathogenic microbes (31, 32). Additionally, Burkholderia isolates that produce pyrrolnitrin, a characterized antifungal, have been used as biocontrol agents for plant fungal pathogens (33). Finally, as mentioned previously, the study by Santos and colleagues (27) suggests that antifungal-producing Burkholderia spp. may play a role in suppressing the fungus garden parasite Escovopsis weberi in Atta sexdens.
Here, we conduct a comprehensive investigation of the functional role of fungus garden-associated Burkholderiaceae isolates obtained from colonies of fungus-growing ants that span major clades in the basal and derived lineages to test the hypothesis that fungus garden-associated bacteria can provide protection against the system’s specialized parasites, Escovopsis spp. First, we sampled 52 fungus-growing ant fungus gardens that span eight different ant genera to isolate Burkholderiaceae. We then tested the ability of a subset of Burkholderiaceae isolates to inhibit a panel of 11 fungi, including eight strains of the specialized parasite Escovopsis that were isolated from five different genera of fungus-growing ants. To identify potential biosynthetic gene clusters (BGCs) involved in Escovopsis inhibition, we sequenced the genomes of 30 Burkholderiaceae isolates and performed antiSMASH and BiG-SCAPE analyses. Then, to confirm production of antifungals, organic extracts were prepared from a subset of isolates and analytical chemistry techniques were used to identify antifungals. Finally, the organic extracts were also used to assess the inhibition of Escovopsis spp. and six strains of Leucoagaricus spp.
RESULTS
Burkholderiaceae are consistently present in the fungal gardens of different lineages of attine ants.
Burkholderiaceae were frequently isolated from fungus gardens of fungus-growing ants, indicating that they are common residents of fungus gardens. Nutrient-rich, nonselective medium was used for the Brazilian fungus garden bacterial isolations, and 35% of colonies contained at least one Burkholderiaceae isolate (Table 1). Burkholderiaceae selective medium was used for Costa Rican fungus garden bacterial isolations, and 71% of colonies contained at least one Burkholderiaceae isolate (Table 1).
TABLE 1.
Summary of ant colony collections by geographic location
| Ant genus | No. of colony collections ina: |
|
|---|---|---|
| Brazil | Costa Rica | |
| Attab | 10 (2) | 11 (7) |
| Acromyrmex | 3 (0) | 1 (0) |
| Paratrachymyrmex | 5 (3) | 2 (2) |
| Mycetomoellerius | 0 | 2 (1) |
| Sericomyrmexb | 0 | 5 (5) |
| Mycetophylax | 2 (2) | 0 |
| Cyphomyrmex | 1 (0) | 0 |
| Myrmicocrypta | 1 (0) | 0 |
| Apterostigma | 5 (3) | 0 |
| Unidentified Attini | 4 (1) | 0 |
| Total colonies | 31 (11) | 21 (15) |
The number of colonies that had a Burkholderiaceae isolate is indicated in parentheses.
Two genera, Atta and Sericomyrmex, have secondarily lost Pseudonocardia.
In total, 86 isolates were obtained from these 52 ant colonies, and whole 16S rRNA gene sequences aligned with Burkholderia spp. as the top BLAST hit (see Data Set S1 in the supplemental material) for all isolates. We selected a subset of 30 isolates for whole-genome sequencing that represented unique species-level 16S rRNA gene BLAST hits, such that if two isolates from the same fungus garden sample matched the same species, one was randomly selected for sequencing (Data Set S2). Phylogenetic and average nucleotide identity (ANI) analyses indicated that isolates grouped with both Burkholderia species and Paraburkholderia species (Fig. S1; Data Set S3), in contrast with the original BLAST search using the 16S rRNA gene. Both 16S rRNA gene sequences and whole-genome phylogenies (Fig. S1) indicate that the bacterial isolates from fungus gardens fall among multiple clades, including different lineages within Burkholderia spp., such as plant pathogens (B. gladioli), the B. cepacia complex, and nitrogen-fixing and plant-associated (B. mimosarum, B. nodosa, B. xenovorans, B. phytofirmans), as well as plant-, rhizosphere-, and soil-associated, Paraburkholderia species. ANI analysis confirmed the variety of Burkholderiaceae in fungus gardens. Eighteen out of 30 sequenced isolates shared ≥95% ANI with B. gladioli (5/18), Burkholderia lata (9/18), Burkholderia ambifaria (1/18), B. cepacia (1/18), Burkholderia seminalis (1/18), or Paraburkholderia tropica (1/18), indicating a range of different characterized Burkholderiaceae species. The other 12 isolates shared between 89% and 94% ANI with other Burkholderiaceae isolates, such as Burkholderia ubonensis, Burkholderia pyrrocinia, Paraburkholderia eburnea, and Paraburkholderia caribensis (Data Set S3).
Multiple fungus garden Burkholderiaceae isolates inhibit Escovopsis spp.
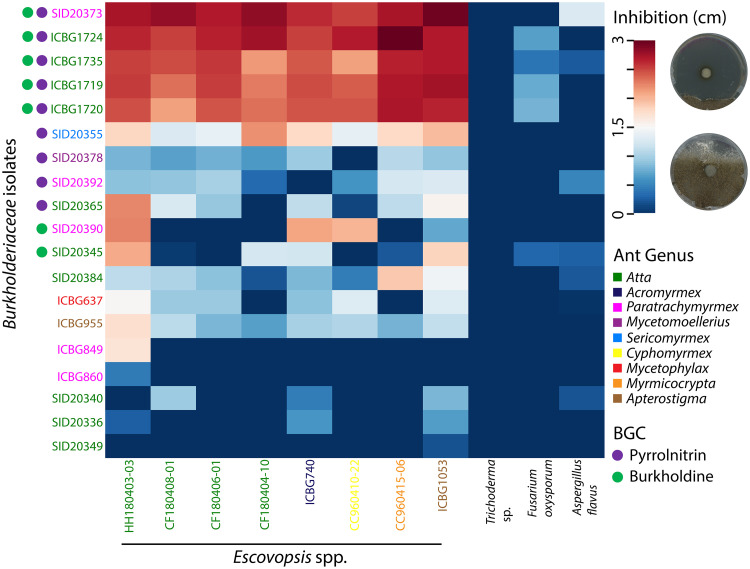
In order to assess the potential of fungus garden-associated Burkholderiaceae to inhibit Escovopsis spp., an in vitro petri plate antimicrobial inhibition bioassay with 19 genome-sequenced Burkholderiaceae isolates from Brazil and Costa Rica was conducted against a panel of 11 fungi: 8 Escovopsis isolates, Aspergillus flavus (Ascomycota, Eurotiales, Trichocomaceae), Fusarium oxysporum (Ascomycota, Hypocreales, Nectriaceae), and Trichoderma sp. (Ascomycota, Hypocreales, Hypocreaceae). After 13 days of coincubation, all 19 Burkholderiaceae isolates had at least one zone of inhibition (ZOI) greater than 0 cm against at least one Escovopsis strain (Fig. 2; Fig. S2 and S3). Another set of in vitro antimicrobial inhibition bioassays with 13 additional isolates from Costa Rican Sericomyrmex colonies were conducted against a panel of six Escovopsis strains and the same three non-Escovopsis fungi noted above. Similar results were found with the 13 additional isolates; all 13 isolates had at least one ZOI greater than 0 cm against at least one Escovopsis strain (Fig. S3). Burkholderia isolates SID20373, ICBG1719, ICBG1720, ICBG1724, and ICBG1735 inhibited all Escovopsis strains significantly more (P < 0.001; average zone of inhibition, ≥ 2.45 cm) (Fig. 2) than the other 27 Burkholderiaceae isolates surveyed (Fig. 2; Fig. S3; Table S1). For clarity, the five isolates listed above that inhibited Escovopsis spp. strongly will be referred to as strong inhibitory isolates. Aspergillus flavus, Trichoderma sp., and F. oxysporum, were not inhibited by any of the Burkholderiaceae isolates.
FIG 2.
Burkholderiaceae-Escovopsis petri plate antimicrobial inhibition bioassays indicated varied levels of inhibition of Escovopsis spp. Zones of inhibition (ZOIs) were measured 13 days after inoculation with Escovopsis spp. Burkholderiaceae and Escovopsis isolates are color coded by the ant colony from which they were isolated. Circles to the left of the Burkholderiaceae isolates indicate the presence of pyrrolnitrin (purple) or burkholdine (green) biosynthetic gene clusters.
Strong inhibitory Burkholderia isolates are predicted to have two antifungal BGCs.
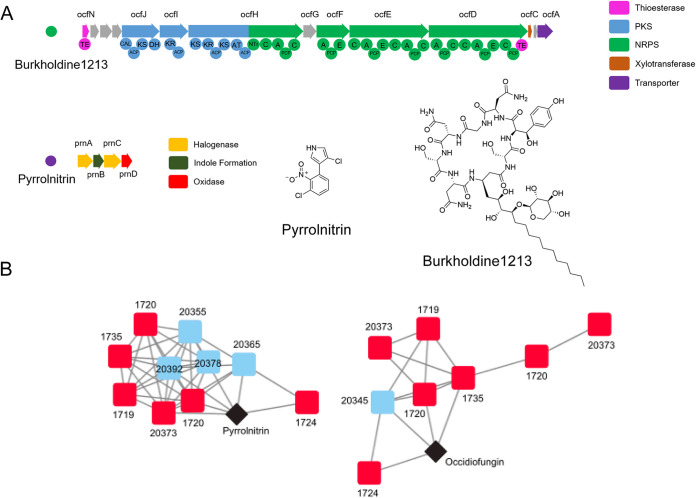
To identify secondary metabolites that might be responsible for the inhibitory activity, the sequenced genome of each Burkholderiaceae isolate was submitted to antiSMASH v4.0 for the detection of biosynthetic gene clusters (BGCs), groupings of genes that encode the production of a secondary metabolite. BiG-SCAPE was used to compare the presence and absence of BGCs across isolates and to search for correlations between the presence of BGCs and Escovopsis inhibition (Fig. S4). Two BGCs were identified in the genomes of all five of the strong inhibitory Burkholderia isolates. These two BGCs had high similarity by BLASTP (>85% identity) to BGCs in the MIBiG database: the BGC for pyrrolnitrin and the BGC for occidiofungin (Fig. S4). Pyrrolnitrin is an antifungal alkaloid biosynthesized from tryptophan and initially isolated from Pseudomonas spp. (34). Occidiofungin is a hybrid nonribosomal peptide/polyketide antifungal glycopeptide and an analog of the burkholdines (35, 36) (Fig. 3). Burkholderia species isolates that did not strongly inhibit Escovopsis contained BGCs for only pyrrolnitrin (SID20355, SID20378, SID20392, SID20365), only antifungal glycopeptides (SID20390, SID20345), or neither of these BGCs. None of the Paraburkholderia species isolates (SID20336 and others) demonstrated inhibition against Escovopsis spp. and were not predicted to contain either antifungal BGC. The distribution of the two antifungal-encoding biosynthetic gene clusters across Burkholderia isolates is demonstrated in Fig. 2.
FIG 3.
Identification of pyrrolnitrin and burkholdine1213 biosynthetic gene clusters in inhibitory isolates. (A) Genetic architecture of biosynthetic gene clusters encoding the production of burkholdine1213 and pyrrolnitrin identified by AntiSMASH 4.0. Domains of the PKS/NRPS hybrid biosynthetic gene cluster are shown beneath the gene representations for burkholdine1213. PKS, polyketide synthase; NRPS, nonribosomal peptide synthetase. (B) BiG-SCAPE network analysis of the two biosynthetic gene clusters present in Burkholderia isolates. Red squares are strong inhibitory Burkholderia isolates, blue squares are less inhibitory isolates, and black diamonds are database matches to the MIBiG biosynthetic gene cluster database.
Burkholderia extracts containing pyrrolnitrin and burkholdine1213 replicate the results of the petri plate inhibition assay.
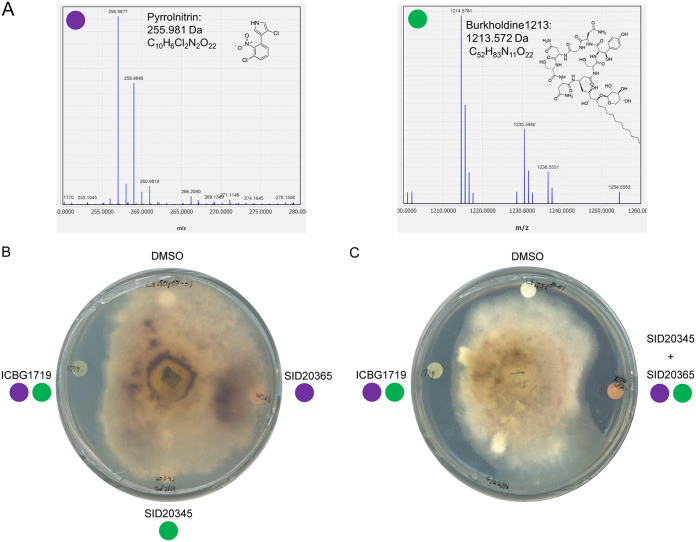
To determine if the antifungal compounds were being produced by the strong inhibitory isolates, we made organic extracts from Burkholderia isolates predicted to have both of the BGCs that encode the production of these compounds (ICBG1719, ICBG1720, ICBG1724, ICBG1735, SID20373) or one or the other compound (SID20365, pyrrolnitrin; SID20345, antifungal glycopeptides). As a negative control, we also included the Paraburkholderia isolate ICBG849, as its genome does not contain either of the BGCs. The extracts were analyzed by ultrahigh-performance liquid chromatography high-resolution electrospray ionization mass spectrometry (UPLC-HRESIMS) for the detection of pyrrolnitrin and antifungal glycopeptides. Pyrrolnitrin was identified by its characteristic two-chlorine isotope pattern and by comparison with a pyrrolnitrin standard purchased from Millipore-Sigma. Burkholdine1213, an antifungal glycopeptide closely related to occidiofungin, was identified by comparison to the published molecular weight and molecular formula of C52H83N11O22 (36). Pyrrolnitrin was identified in ICBG1719, ICBG1720, ICBG1724, ICGB1735, SID20373, and SID20365. Burkholdine1213 was identified in ICBG1719, ICBG1720, ICBG1724, ICBG1735, SID20373, and SID20345.
After the compounds were identified in the extracts, we tested the extracts against one Escovopsis weberi isolate (CF180408-01), A. flavus, F. oxysporum, and Trichoderma sp. in a disc diffusion assay to assess activity (Fig. 4B; Fig. S5). The extracts from an isolate containing both compounds (ICBG1719) demonstrated inhibition against Escovopsis weberi, while extracts containing only one compound, either burkholdine or pyrrolnitrin (SID20365, pyrrolnitrin; SID20345, burkholdine1213), and the dimethyl sulfoxide (DMSO) control did not inhibit Escovopsis weberi. This reflected the results of the previous antimicrobial inhibition bioassays using Burkholderia isolates. Additionally, when the extracts from SID20365 (only pyrrolnitrin) and SID20345 (only burkholdine1213) were combined, creating an extract that artificially contained both compounds, inhibition was observed (Fig. 4C). This suggests that pyrrolnitrin and burkholdine1213 may act additively or synergistically to inhibit Escovopsis spp.
FIG 4.
Detection of pyrrolnitrin and burkholdine1213 in organic extracts of inhibitory isolates and demonstration of extract activity. (A) Extracted ion chromatograms of m/z values that match those of pyrrolnitrin and burkholdine1213 from the organic extract of inhibitory Burkholderia isolate ICBG1719. (B) Disc diffusion assay of extracts from ICBG1719 (both pyrrolnitrin and burkholdine1213), SID20345 (burkholdine1213), and SID20365 (pyrrolnitrin) against Escovopsis weberi (CF180408-01). (C) Disc diffusion assay of extracts from ICBG1719 (both pyrrolnitrin and burkholdine1213) and a combined extract of SID20345 and SID20365 (artificially containing both pyrrolnitrin and burkholdine1213) against Escovopsis weberi (CF180408-01), demonstrating that both compounds must be present for inhibition. Additional disc diffusion assays were conducted on Trichoderma, Aspergillus, and Fusarium (Fig. S5).
Burkholderia extracts inhibit Escovopsis weberi growth at lower concentrations than those used to inhibit Leucoagaricus sp.
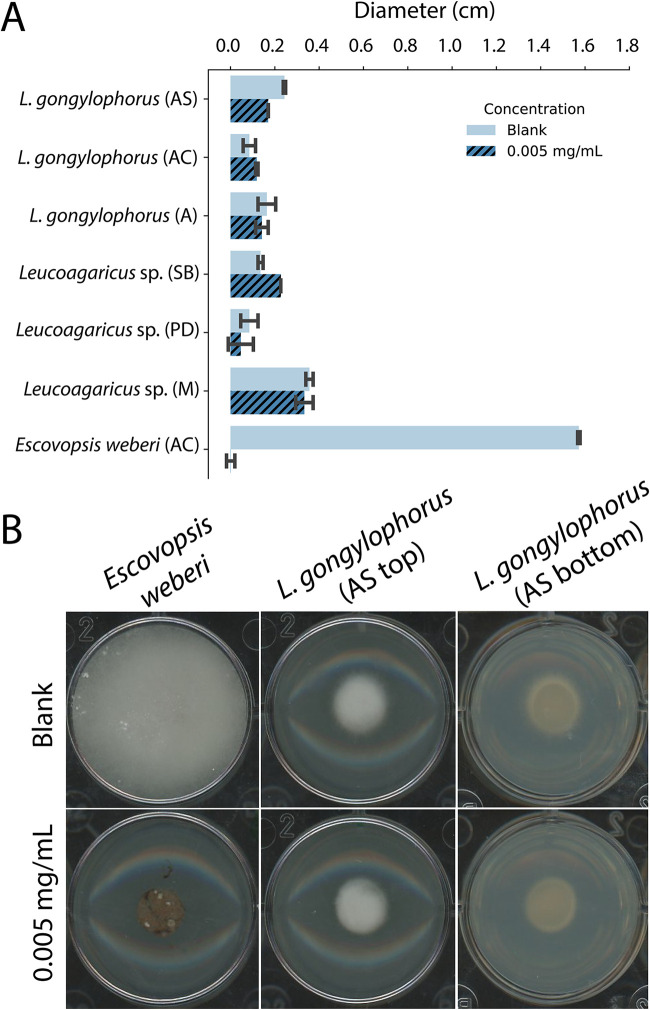
The lack of inhibition of other ecologically relevant fungi suggested that the antifungals produced by inhibitory Burkholderia isolates may be able to inhibit Escovopsis while not harming Leucoagaricus. To test this, six Leucoagaricus species strains and an Escovopsis weberi strain (CF180408-01) were grown individually on plates containing 0.005 mg/ml, 0.05 mg/ml, 0.5 mg/ml, 1 mg/ml, and 2.5 mg/ml Burkholderia extract from an isolate with both pyrrolnitrin and burkholdine1213 (ICBG1719). All plates with extract concentrations above and including 0.5 mg/ml completely inhibited all growth of Leucoagaricus spp. and Escovopsis weberi. After 6 days, at 0.05 mg/ml, the growth rates (i.e., smaller diameters) observed for four out of the six Leucoagaricus spp. were lower than those of the control, while two Leucoagaricus spp. and Escovopsis weberi demonstrated no growth (Fig. S6) (Wilcoxon signed-rank test, Z = –1.51, P = 0.1289). Finally, at 0.005 mg/ml, the diameters of all Leucoagaricus strains grew comparably to the diameter of the control, while the growth of Escovopsis weberi was inhibited (Fig. 5) (Wilcoxon signed-rank test, Z = –2.52, P = 0.0115).
FIG 5.
Leucoagaricus spp. grown on agar containing 0.005 mg/ml ICBG1719 extract (both BGCs) grow comparably to agar containing no extract, while Escovopsis weberi (CF180408-01) from an Atta cephalotes colony is inhibited. (A) The bar graph indicates the growth of Leucoagaricus spp. (n = 2 for each strain) and Escovopsis weberi (n = 3) after 6 days of growth on agar containing no extract (blank) or 0.005 mg/ml. For visual representation, the diameter of the fungal plug (0.6 mm) was subtracted from the overall diameter measurement. (B) Photographs represent typical fungal growth of Escovopsis weberi and Leucoagaricus spp. (photo is from Leucoagaricus gongylophorus from an Atta sexdens colony) on agar with no extract (blank) and 0.005 mg/ml extract. Photos include the top of the plate (AS top) and bottom of the plate (AS bottom). AS, Atta sexdens; AC, Atta cephalotes; A, Acromyrmex sp.; SB, Sericomyrmex bondari; PD, Paratrachymyrmex diversus; M, Myrmicocrypta sp.
DISCUSSION
Here, we demonstrate that Burkholderia fungus garden isolates inhibit the parasitic fungus Escovopsis in vitro. The inhibition of Escovopsis spp. corresponds with the presence of two BGCs that encode the production of two known antifungal compounds, pyrrolnitrin and burkholdine1213. We identified both antifungals in the extracts of inhibitory isolates, confirming the expression of these clusters when cultured. A combination of the extracts of isolates that contained only one or the other antifungal replicated the inhibitory activity, suggesting that both compounds must be present for anti-Escovopsis activity. Additionally, extracts of the inhibitory Burkholderia isolates were capable of inhibiting Escovopsis weberi, but not Leucoagaricus spp., at concentrations as low as 0.05 mg/ml in vitro. These findings suggest an important role for Burkholderia in the defense of fungus gardens from the parasitic fungus Escovopsis but not other ecologically relevant transient fungal invaders (e.g., Trichoderma).
Two lineages of fungus-growing ants, Atta and Sericomyrmex, have secondarily lost the ability to harbor the defensive symbiont Pseudonocardia (23). Four of the five strong inhibitory isolates were isolated from Atta colonies, and one isolate whose genome contained the BGC for pyrrolnitrin, SID20355, was isolated from a Sericomyrmex colony and had an average zone of inhibition of 1.7 cm in inhibition bioassays against the eight Escovopsis species strains (Fig. 2; Fig. S2). We sampled five colonies of Sericomyrmex and were able to obtain at least one Burkholderia isolate from all five colonies, with a range of inhibition zones from 0.29 cm to 1.7 cm. Though we equally sampled between colonies in which Pseudonocardia was present and absent (26 fungus gardens from each group), there was uneven geographic sampling, and different bacterial isolation techniques and specific ant genus sampling may have influenced our results (Table 1). Our results suggest Burkholderia species as potential defensive symbionts; however, additional work must be done to explore whether Burkholderia spp. are a potential replacement (37, 38) for Pseudonocardia in colonies without the symbiont or, more generally, if Burkholderia plays a larger role in defending the garden when Pseudonocardia is absent.
The presence of inhibitory Burkholderia and other known measures of sanitation within the fungus-growing ant system (25, 26) suggest that fungus-growing ants use multiple strategies to promote healthy fungus gardens. Of note, while we focused on strong inhibitory Burkholderia isolates containing two BGCs, some Burkholderia isolates from fungus gardens containing either one BGC or neither were still able to less severely inhibit Escovopsis spp. (Fig. 2; Fig. S2). This suggests that Burkholderia or other garden bacteria might play a role in inhibiting Escovopsis spp. in other fungus-growing ant genera. It appears unlikely that they play a role in the inhibition of other alien fungi, as we saw that Burkholderia isolates barely, if at all, inhibited Trichoderma, Aspergillus, and Fusarium. However, previous work with different fungus garden isolates has indicated the inhibition of other fungi, including Trichoderma (27, 39).
As with internal animal gut microbiomes, the fungus garden microbiome is composed of a complex and diverse microbial community where the roles of resident bacteria are still being established. In recent years, there has been an accumulation of evidence that members of the fungus garden microbiome potentially function as beneficial symbionts, with some taxa aiding in nitrogen fixation, degradation of plant matter, and/or detoxification of plant secondary compounds. In this study, we have described a fungus garden-associated bacterial genus, Burkholderia, as a possible mechanism for certain fungus-growing ant lineages to defend their fungus gardens from parasites. Overall, these new findings combined with past studies suggest that members of the fungus garden microbiome play a key role in facilitating the response of fungus-growing ants to environmental changes and pressures.
MATERIALS AND METHODS
Sampling and bacterial isolations.
We collected fungus garden samples in January 2017 and April 2018 in the following locations: Anavilhanas, Amazonas, Brazil; Ducke Reserve, Amazonas, Brazil; Itatiaia, Rio de Janeiro, Brazil; Sao Paulo State, Brazil; and La Selva Biological Station, Costa Rica (see Data Set S1 in the supplemental material). Collections of biological samples and research on genetic resources were authorized in Brazil by SISBIO number 46555-5 and CNPq number 010936/2014-9. In Costa Rica, permits were granted by the Comisión Institucional de Biodiversidad of the University of Costa Rica (resolution number 009) and collections were authorized by the Organization of Tropical Studies (OTS), under UCR project 801-B9-515. For the fungus garden samples collected in Brazil, for each colony, a small fungus garden piece collected from the inner region of the garden was put into phosphate-buffered saline (PBS) and then plated onto yeast malt extract agar (YMEA). Plates with bacterial colonies were brought back to the University of Wisconsin—Madison, and pure bacterial isolates were obtained after several rounds of subculturing. We obtained a total of 317 bacterial isolates. We identified 117 isolates to the genus level by whole 16S rRNA gene sequencing. Of those, 33 corresponded to the genus Burkholderia. For the fungus garden samples collected in Costa Rica, ∼0.2 g of fungus garden was collected from the inner region of the garden, put into PBS, vortexed, and homogenized; then the PBS-fungus garden homogenate was serially diluted, and all dilutions were plated on both YMEA and modified APCA (40) [per liter: 0.79 g (NH4)2SO4, 1 g KH2PO4, 0.5 g MgSO4-H2O, 0.2 g KCl, 2 g l-arabinose, 5 mg crystal violet, 50 mg polymyxin B sulfate, 50 mg ampicillin sodium, 10 mg chloramphenicol, 15 g agar].
DNA extraction and assembly.
We extracted DNA from 30 bacterial isolates using the Wizard genomic DNA purification kit (Promega, USA) using the Gram-negative protocol. Genomic DNA libraries for Illumina MiSeq 2× 300-bp paired-end sequencing were prepared by the University of Wisconsin—Madison Biotechnology Center. Reads were corrected with MUSKET v1.1 (41), and paired-ends were merged with FLASH v.1.2.7 (42) and assembled with SPAdes 3.11.0 (43). We taxonomically classified the Burkholderiaceae isolates to the species level by performing a tetra correlation search at JSpeciesWS (44). In addition, ANI was calculated using the anvi’o (45) anvi-compute-genome-similarity program, with the default pyani (46) settings.
Phylogenetic tree.
We generated a genome-based, multilocus Burkholderiaceae phylogeny based on previous methods (47). Briefly, the phylogeny was generated using 93 full TIGRFAM proteins in the “core bacterial protein” set (GenProp0799) as the molecular data set. The protein sequences with the top HMMER bit score for each protein family were aligned using MAFFT (48) and were then converted to codon alignments and concatenated. RAxML-7.2.6 (49) was used to generate the phylogeny using the GTRgamma substitution model and 100 rapid bootstraps on the final, recombination-free alignment. The gene tree-based phylogeny was generated using ASTRAL-II (50). Phylogenies were visualized and edited in FigTree v1.4.3.A. We constructed the 16S rRNA gene phylogenetic tree using BEAST2 v2.5.1 (51) with a TN93 substitution model. The analysis was run for 150,000,000 generations, with every 1,000 generations sampled, and a burn-in of 10% was applied. The code used for this analysis can be accessed at https://github.com/chevrm/core_species_tree.
Escovopsis-Burkholderiaceae bioassays.
In order to assess inhibition profiles of Burkholderiaceae against Escovopsis and other fungi, we performed in vitro plate assays. For each isolate, we grew an overnight culture in yeast malt extract (YME) for 16 to 24 h. We spotted 10 μl of Burkholderiaceae onto the middle of a 100-mm potato-dextrose agar (PDA) plate, placed Parafilm on the plates, and incubated them at room temperature for 7 days. After 7 days, 6-mm fungal plugs of Escovopsis, Trichoderma, Aspergillus, and Fusarium were put on the edge of the plate. Control plates containing only a Burkholderiaceae isolate or only a fungus were also made for each isolate. All plates were covered with two pieces of Parafilm. Over the course of a month, both sides of the plate were scanned every 6 to 7 days, and the zone of inhibition was measured using Fiji (52). The eight Escovopsis strains were chosen because they were isolated from different lineages of fungus-growing ants (Acromyrmex, Apterostigma, Atta, Cyphomyrmex, Myrmicocrypta) (Data Set S1). Escovopsis species IDs are given in Data Set S1. Trichoderma sp., Aspergillus flavus, and Fusarium oxysporum are ecologically relevant fungi and represent different degrees of relatedness to Escovopsis spp.
Biosynthetic gene cluster annotation.
We used antiSMASH v4.0 (53) to predict BGCs and BiG-SCAPE (54) to construct sequence similarity networks of BGCs. In addition to including our predicted BGCs for the BiG-SCAPE analysis, we included the reference BGCs from the MIBiG repository. We used cytoscape (55) to visualize the BGC networks. We color coded the clusters into two categories: strong inhibitory Burkholderia isolates (average ZOI ≥ 2.45 cm against Escovopsis) and noninhibitory Burkholderia isolates (average ZOI ≤ 2.45 cm against Escovopsis) and looked for clusters that contained all the inhibitory isolates. We subsequently used Clinker (56) and BLASTP to determine the similarity of the two antifungal-encoding BGCs from the Burkholderia isolates to the characterized BGCs from the MIBiG repository.
Burkholderia extracts.
Burkholderia isolates ICBG1719, ICBG1720, ICBG1724, ICGB1735, SID20373, SID20345, SID20365, and ICBG849 were inoculated from YMEA plates into 100 ml of YME broth in 500-ml baffled flasks and shaken for 36 h. The 100-ml cultures were used to inoculate two 500-ml cultures of YME broth in 2-liter flasks for each isolate. The isolates were shaken for 36 h with Diaion HP20 resin. The cultures were then vacuum filtered through a Whatman 1-sized filter paper, and the Diaion resin and cell mass was extracted overnight with ethyl acetate. Excess anhydrous sodium sulfate was added to the extraction to remove residual water. The ethyl acetate was filtered and dried in vacuo to yield a Burkholderia extract.
HRESIMS of Burkholderia extracts.
Burkholderia extracts were resuspended in methanol and analyzed for the presence of pyrrolnitrin and burkholdine1213 by UPLC-HRESIMS on a Q Exactive Orbitrap mass spectrometer. A liquid chromatography gradient was run from 5% acetonitrile with 0.1% formic acid to 100% acetonitrile with 0.1% formic acid over 15 min on a Phenomenex XB C18, 2.1-mm by 100-mm, 2.6-μm-particle-size column. The scan range was from 200 m/z to 2,000 m/z in positive mode.
Escovopsis-Burkholderia extract assays.
Escovopsis clavatus (ICBG1053, Apterostigma) and Escovopsis weberi (CF180408-01, Atta cephalotes) plugs were plated onto PDA and grown for 3 days until white mycelia could be seen. We chose these two Escovopsis strains because they were representative of the Escovopsis-Burkholderia inhibition profiles. Additionally, Trichoderma, Aspergillus, and Fusarium were plated on PDA and grown until slight mycelial growth was visible. Burkholderia extracts from ICBG1719, SID20345, and SID20365 were dissolved in dimethyl sulfoxide (DMSO). The extracts were pipetted onto sterile filter paper discs in 10 μl of DMSO at 0.5 mg/disc, 1 mg/disc, and 2 mg/disc and placed onto the PDA plates containing the Escovopsis or other fungi along with a DMSO control disc. After 2 weeks of growth, the ability of each extract to inhibit the growth of the fungi was assessed and pictures were taken of the plates.
Leucoagaricus-Burkholderia extract assays.
We plated six Leucoagaricus strains isolated from Atta sexdens, Atta cephalotes, Acromyrmex octospinosus, Sericomyrmex bondari, Paratrachymyrmex diversus, Myrmicocrypta sp., and an Escovopsis weberi isolate (CF180408-01) from Atta cephalotes on PDA and let them grow for a month at room temperature. We prepared PDA plates containing 0 mg/ml, 0.005 mg/ml, 0.05 mg/ml, 0.5 mg/ml, 1 mg/ml, and 2.5 mg/ml Burkholderia extract in DMSO. Media were vigorously mixed for homogenous distribution of the extract, and then 3 ml was pipetted into each well of a 12-well plate (catalog no. 82050-926; Greiner Bio-One) and left to dry overnight in the dark. Then, 0.6-mm plugs taken from the outer edges of Leucoagaricus species or Escovopsis fungal plates were deposited into the center of each well (n = 2 for each fungal strain at 6 concentrations). Pictures were taken 6 days postexposure, and the diameter of fungal growth was measured in Fiji. To test if Leucoagaricus species growth was significantly greater than Escovopsis weberi growth with 0.05 mg/ml and 0.005 mg/ml Burkholderia extract, we performed a Wilcoxon signed-rank test in JMP Pro 14 using the average growth of all Leucoagaricus species strains (n = 2 replicates × 6 strains = 12) and the average growth of Escovopsis weberi (n = 3 replicates × 1 strain = 3) for each concentration.
Data availability.
All sequencing data have been uploaded into the NCBI databases under BioProject numbers PRJNA564151 and PRJNA603049. Whole-genome and SRA accession numbers for each isolate can be found in Data Set S2. Whole 16S rRNA gene sequences are under the GenBank accession numbers MW756842 to MW756915 and MW772240 to MW772251. Data Set S1 includes individual accession numbers.
ACKNOWLEDGMENTS
We are grateful to Ted Schultz, Jeffrey Sosa-Calvo, and Eugenia Okonski for identification of our fungus-growing ant specimens. We thank Camila Carlos-Shanley for her guidance and support at the conception of this project. We thank the staff and scientists at the La Selva Biological Station for allowing field collections and the scientists Allan Artavia and Miguel Pacheco in the Pinto-Tomás lab at the University of Costa Rica for their assistance with field collections.
This material is based upon work supported in part by National Institutes of Health grant U19 TW009872, National Institutes of Health grant U19 AI142720, National Institute of Allergy and Infectious Diseases, National Institutes of Health, grant T32 AI055397, National Science Foundation grant DEB-1927155, and São Paulo Research Foundation (FAPESP) grant 2013/50954-0.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material is available online only.
Contributor Information
Cameron R. Currie, Email: currie@bact.wisc.edu.
Knut Rudi, Norwegian University of Life Sciences.
REFERENCES
- 1.Skelton J, Doak S, Leonard M, Creed RP, Brown BL. 2016. The rules for symbiont community assembly change along a mutualism-parasitism continuum. J Anim Ecol 85:843–853. 10.1111/1365-2656.12498. [DOI] [PubMed] [Google Scholar]
- 2.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37. 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 3.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 4.Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M. 2015. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep 32:904–936. 10.1039/c5np00010f. [DOI] [PubMed] [Google Scholar]
- 5.Van Arnam EB, Currie CR, Clardy J. 2018. Defense contracts: molecular protection in insect-microbe symbioses. Chem Soc Rev 47:1638–1651. 10.1039/c7cs00340d. [DOI] [PubMed] [Google Scholar]
- 6.Dillon RJ, Dillon VM. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49:71–92. 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 7.Piel J. 2009. Metabolites from symbiotic bacteria. Nat Prod Rep 26:338–362. 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 8.Pan X, Zhou G, Wu J, Bian G, Lu P, Raikhel AS, Xi Z. 2012. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A 109:E23–E31. 10.1073/pnas.1116932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solomon SE, Rabeling C, Sosa-Calvo J, Lopes CT, Rodrigues A, Vasconcelos HL, Bacci M, Mueller UG, Schultz TR. 2019. The molecular phylogenetics of Trachymyrmex Forel ants and their fungal cultivars provide insights into the origin and coevolutionary history of ‘higher-attine’ ant agriculture. Syst Entomol 44:939–956. 10.1111/syen.12370. [DOI] [Google Scholar]
- 10.Branstetter MG, Ješovnik A, Sosa-Calvo J, Lloyd MW, Faircloth BC, Brady SG, Schultz TR. 2017. Dry habitats were crucibles of domestication in the evolution of agriculture in ants. Proc Biol Sci 284:20170095. 10.1098/rspb.2017.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Currie CR. 2001. A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu Rev Microbiol 55:357–380. 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 12.Aylward FO, Currie CR, Suen G. 2012. The evolutionary innovation of nutritional symbioses in leaf-cutter ants. Insects 3:41–61. 10.3390/insects3010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khadempour L, Burnum-Johnson KE, Baker ES, Nicora CD, Webb-Robertson B-JM, White RA, Monroe ME, Huang EL, Smith RD, Currie CR. 2016. The fungal cultivar of leaf-cutter ants produces specific enzymes in response to 2 different plant substrates. Mol Ecol 25:5795–5805. 10.1111/mec.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aylward FO, Burnum KE, Scott JJ, Suen G, Tringe SG, Adams SM, Barry KW, Nicora CD, Piehowski PD, Purvine SO, Starrett GJ, Goodwin LA, Smith RD, Lipton MS, Currie CR. 2012. Metagenomic and metaproteomic insights into bacterial communities in leaf-cutter ant fungus gardens. ISME J 6:1688–1701. 10.1038/ismej.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG, Pinto-Tomás AA, Foster CE, Pauly M, Weimer PJ, Barry KW, Goodwin LA, Bouffard P, Li L, Osterberger J, Harkins TT, Slater SC, Donohue TJ, Currie CR. 2010. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet 6:e1001129. 10.1371/journal.pgen.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khadempour L, Fan H, Keefover-Ring K, Carlos-Shanley C, Nagamoto NS, Dam MA, Pupo MT, Currie CR. 2020. Metagenomics reveals diet-specific specialization of bacterial communities in fungus gardens of grass- and dicot-cutter ants. Front Microbiol 11:570770. 10.3389/fmicb.2020.570770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barcoto MO, Carlos-Shanley C, Fan H, Ferro M, Nagamoto NS, Bacci M, Currie CR, Rodrigues A. 2020. Fungus-growing insects host a distinctive microbiota apparently adapted to the fungiculture environment. Sci Rep 10:12384. 10.1038/s41598-020-68448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronque MUV, Lyra ML, Migliorini GH, Bacci M, Oliveira PS. 2020. Symbiotic bacterial communities in rainforest fungus-farming ants: evidence for species and colony specificity. Sci Rep 10:10172. 10.1038/s41598-020-66772-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francoeur C, Khadempour L, Moreira-Soto R, Gotting K, Book A, Pinto-Tomás A, Keefover-Ring K, Currie C. 2020. Bacteria contribute to plant secondary compound degradation in a generalist herbivore system. mBio 11:e02146-20. 10.1128/mBio.02146-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinto-Tomás AA, Anderson MA, Suen G, Stevenson DM, Chu FST, Cleland WW, Weimer PJ, Currie CR. 2009. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326:1120–1123. 10.1126/science.1173036. [DOI] [PubMed] [Google Scholar]
- 21.Currie CR, Mueller UG, Malloch D. 1999. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci U S A 96:7998–8002. 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. 2006. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311:81–83. 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Sosa-Calvo J, Horn HA, Pupo MT, Clardy J, Rabeling C, Schultz TR, Currie CR. 2018. Convergent evolution of complex structures for ant-bacterial defensive symbiosis in fungus-farming ants. Proc Natl Acad Sci U S A 115:10720–10725. 10.1073/pnas.1809332115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Currie CR, Summerbell RC, Scott JA, Malloch D. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–704. 10.1038/19519. [DOI] [Google Scholar]
- 25.Currie CR, Stuart AE. 2001. Weeding and grooming of pathogens in agriculture by ants. Proc Biol Sci 268:1033–1039. 10.1098/rspb.2001.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Marín H, Nash DR, Higginbotham S, Estrada C, Van Zweden JS, D’Ettorre P, Wcislo WT, Boomsma JJ. 2015. Functional role of phenylacetic acid from metapleural gland secretions in controlling fungal pathogens in evolutionarily derived leaf-cutting ants. Proc Biol Sci 282:20150212. 10.1098/rspb.2015.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos AV, Dillon RJ, Dillon VM, Reynolds SE, Samuels RI. 2004. Occurrence of the antibiotic producing bacterium Burkholderia sp. in colonies of the leaf-cutting ant Atta sexdens rubropilosa. FEMS Microbiol Lett 239:319–323. 10.1016/j.femsle.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Kaltenpoth M, Flórez LV. 2020. Versatile and dynamic symbioses between insects and Burkholderia bacteria. Annu Rev Entomol 65:145–170. 10.1146/annurev-ento-011019-025025. [DOI] [PubMed] [Google Scholar]
- 29.Partida-Martinez LP, Hertweck C. 2005. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884–888. 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 30.Partida-Martinez LP, Groth I, Schmitt I, Richter W, Roth M, Hertweck C. 2007. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporous. Int J Syst Evol Microbiol 57:2583–2590. 10.1099/ijs.0.64660-0. [DOI] [PubMed] [Google Scholar]
- 31.Flórez LV, Scherlach K, Gaube P, Ross C, Sitte E, Hermes C, Rodrigues A, Hertweck C, Kaltenpoth M. 2017. Antibiotic-producing symbionts dynamically transition between plant pathogenicity and insect-defensive mutualism. Nat Commun 8:15172. 10.1038/ncomms15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flórez LV, Scherlach K, Miller IJ, Rodrigues A, Kwan JC, Hertweck C, Kaltenpoth M. 2018. An antifungal polyketide associated with horizontally acquired genes supports symbiont-mediated defense in Lagria villosa beetles. Nat Commun 9:2478. 10.1038/s41467-018-04955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung BK, Hong SJ, Park GS, Kim MC, Shin JH. 2018. Isolation of Burkholderia cepacia JBK9 with plant growth-promoting activity while producing pyrrolnitrin antagonistic to plant fungal diseases. Appl Biol Chem 61:173–180. 10.1007/s13765-018-0345-9. [DOI] [Google Scholar]
- 34.Hammer PE, Hill DS, Lam ST, Van Pée KH, Ligon JM. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl Environ Microbiol 63:2147–2154. 10.1128/AEM.63.6.2147-2154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu G, Smith L, Liu A, Lu SE. 2011. Genetic and biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl Environ Microbiol 77:6189–6198. 10.1128/AEM.00377-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Z, Falkinham JO, Tawfik KA, Jeffs P, Bray B, Dubay G, Cox JE, Schmidt EW. 2012. Burkholdines from Burkholderia ambifaria: antifungal agents and possible virulence factors. J Nat Prod 75:1518–1523. 10.1021/np300108u. [DOI] [PubMed] [Google Scholar]
- 37.Husnik F, McCutcheon JP. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci U S A 113:E5416–E5424. 10.1073/pnas.1603910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudakaran S, Kost C, Kaltenpoth M. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol 25:375–390. 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Schoenian I, Spiteller M, Ghaste M, Wirth R, Herz H, Spiteller D. 2011. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc Natl Acad Sci U S A 108:1955–1960. 10.1073/pnas.1008441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawanishi T, Uematsu S, Nishimura K, Otani T, Tanaka-Miwa C, Hamamoto H, Namba S. 2009. A new selective medium for Burkholderia caryophylli, the causal agent of carnation bacterial wilt. Plant Pathol 58:237–242. 10.1111/j.1365-3059.2008.01980.x. [DOI] [Google Scholar]
- 41.Liu Y, Schröder J, Schmidt B. 2013. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics 29:308–315. 10.1093/bioinformatics/bts690. [DOI] [PubMed] [Google Scholar]
- 42.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platformfor ’omics data. PeerJ 3:e1319–e1329. 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 47.McDonald BR, Currie CR. 2017. Lateral gene transfer dynamics in the ancient bacterial genus Streptomyces. mBio 8:e00644-17. 10.1128/mBio.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 50.Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31:i44–i52. 10.1093/bioinformatics/btv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, Matschiner M, Mendes FK, Müller NF, Ogilvie HA, Du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu CH, Xie D, Zhang C, Stadler T, Drummond AJ. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol 15:e1006650-28. 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, De Los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. 2017. AntiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, De Los Santos ELC, Yeong M, Cruz-Morales P, Abubucker S, Roeters A, Lokhorst W, Fernandez-Guerra A, Cappelini LTD, Goering AW, Thomson RJ, Metcalf WW, Kelleher NL, Barona-Gomez F, Medema MH. 2020. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol 16:60–68. 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models. Genome Res 13:2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilchrist CLM, Chooi Y-H. 2021. clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 2021:btab007 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S3. Download AEM00178-21_Supp_3_seq1.xlsx, XLSX file, 0.2 MB (198.8KB, xlsx)
Data Set S1. Download AEM00178-21_Supp_1_seq2.xlsx, XLSX file, 0.02 MB (19.4KB, xlsx)
Data Set S2. Download AEM00178-21_Supp_2_seq3.xlsx, XLSX file, 0.01 MB (15.9KB, xlsx)
Table S1, Figures S1 to S6. Download AEM00178-21_Supp_4_seq11.pdf, PDF file, 4.2 MB (4.2MB, pdf)
Data Availability Statement
All sequencing data have been uploaded into the NCBI databases under BioProject numbers PRJNA564151 and PRJNA603049. Whole-genome and SRA accession numbers for each isolate can be found in Data Set S2. Whole 16S rRNA gene sequences are under the GenBank accession numbers MW756842 to MW756915 and MW772240 to MW772251. Data Set S1 includes individual accession numbers.