ABSTRACT
In the marine environment, phosphorus availability significantly affects the lipid composition in many cosmopolitan marine heterotrophic bacteria, including members of the SAR11 clade and the Roseobacter clade. Under phosphorus stress conditions, nonphosphorus sugar-containing glycoglycerolipids are substitutes for phospholipids in these bacteria. Although these glycoglycerolipids play an important role as surrogates for phospholipids under phosphate deprivation, glycoglycerolipid synthases in marine microbes are poorly studied. In the present study, we biochemically characterized a glycolipid glycosyltransferase (GTcp) from the marine bacterium “Candidatus Pelagibacter sp.” strain HTCC7211, a member of the SAR11 clade. Our results showed that GTcp is able to act as a multifunctional enzyme by synthesizing different glycoglycerolipids with UDP-glucose, UDP-galactose, or UDP-glucuronic acid as sugar donors and diacylglycerol (DAG) as the acceptor. Analyses of enzyme kinetic parameters demonstrated that Mg2+ notably changes the enzyme’s affinity for UDP-glucose, which improves its catalytic efficiency. Homology modeling and mutational analyses revealed binding sites for the sugar donor and the diacylglycerol lipid acceptor, which provided insights into the retaining mechanism of GTcp with its GT-B fold. A phylogenetic analysis showed that GTcp and its homologs form a group in the GT4 glycosyltransferase family. These results not only provide new insights into the glycoglycerolipid synthesis mechanism in lipid remodeling but also describe an efficient enzymatic tool for the future synthesis of bioactive molecules.
IMPORTANCE The bilayer formed by membrane lipids serves as the containment unit for living microbial cells. In the marine environment, it has been firmly established that phytoplankton and heterotrophic bacteria can replace phospholipids with nonphosphorus sugar-containing glycoglycerolipids in response to phosphorus limitation. However, little is known about how these glycoglycerolipids are synthesized. Here, we determined the biochemical characteristics of a glycolipid glycosyltransferase (GTcp) from the marine bacterium “Candidatus Pelagibacter sp.” strain HTCC7211. GTcp and its homologs form a group in the GT4 glycosyltransferase family and can synthesize neutral glycolipids (monoglucosyl-1,2-diacyl-sn-glycerol [MGlc-DAG] and monogalactosyl [MGal]-DAG) and monoglucuronic acid diacylglycerol (MGlcA-DAG). We also uncovered the key residues for DAG binding through molecular docking, site-direct mutagenesis, and subsequent enzyme activity assays. Our data provide new insights into the glycoglycerolipid synthesis mechanism in lipid remodeling.
KEYWORDS: lipid remodeling, SAR11, marine bacteria
INTRODUCTION
Phospholipids form the structural basis of all cells, but sugar-containing glycoglycerolipids are mainly restricted to marine microbes, cyanobacteria, and higher plants (1, 2). Glycoglycerolipids are found on the lipid bilayer of cell membranes and play critical roles in cell growth, cellular recognition, adhesion, neuronal repair, and signal transduction. These natural glycoglycerolipids often have unusual and sometimes unexpected biological activities, such as antitumor, antiviral, anti-inflammatory, antimalarial, immunostimulatory, and neuritogenic activities, which make them valuable molecular targets for research (3–5). The basic structure of glycoglycerolipids is characterized by a 1,2-diacyl-sn-glycerol moiety with different numbers and types of sugars (glucose, galactose, mannose, rhamnose, or charged sugars like glucuronic acid or sulfoquinovose) attached at the sn-3 position of the glycerol backbone in diacylglycerol (DAG). These sugar attachments have an α- or β-anomeric configuration and are bound via (1→2), (1→3), (1→4), or (1→6) linkages (2, 6). The common glycoglycerolipid structures in marine heterotrophic microbes and cyanobacteria are 1,2-diacyl-3-O-(β-d-galactopyranosyl)-sn-glycerol (monogalactosyl diacylglycerol [MGal-DAG]), 1,2-diacyl-3-O-(α-d-glucopyranosyl)-sn-glycerol (monoglucosyl DAG [MGlc-DAG]),1,2-diacyl-3-O-(α-d-galactopyranosyl)-(1→6)-O-(β-d-galactopyranosyl)-sn-glycerol (digalactosyl DAG [DGal-DAG]), and 1,2-diacyl-3-O-(6-deoxy-6-sulfo-α-d-galactopyranosyl)-sn-glycerol (sulfoquinovosyl DAG [SQDG]) (2, 7).
Glycoglycerolipids are usually synthesized by glycosyltransferases (GTs), which are highly divergent and polyphyletic. The GTs can be categorized into 110 numbered families according to their sequence similarity and signature motifs and the stereochemistry of the glycoside linkage formed (8). Of the 110 families, the families GT4, GT21, and GT28, which are known as glycoglycerolipid synthases, utilize sugar nucleotides as donors and contain a consensus sugar donor binding domain near the C terminus (9, 10). Despite the wide variety of bacterial glycoglycerolipids, only a few bacterial lipid GTs have been identified and characterized so far. The GTs synthesizing MGlc-DAG and DGal-DAG have been isolated from the cell wall-less bacterium Acholeplasma laidlawii and were found to belong to the GT4 family in the carbohydrate-active enzymes (CAZy) database (11). Other known members of bacterial GT4 include the MGlc-DAG synthases from Deinococcus radiodurans and Thermotoga maritima and the MGal-DAG synthase from Borrelia burgdorferi, which was the first cloned galactosyltransferase forming MGal-DAG with the α-anomeric configuration of the sugar (12, 13). A bifunctional GT (designated Agt) from Agrobacterium tumefaciens was found to synthesize MGlc-DAG or monoglucuronic acid diacylglycerol (MGlcA-DAG) with UDP-glucose (UDP-Glc) or UDP-glucuronic acid (UDP-GlcA) as the sugar donor, respectively (6). This enzyme also belongs to the GT4 family and was the first glucuronosyl DAG synthase to be isolated. The processive GTs (Pgts), however, are members of the GT21 family and show high sequence similarity to GT4 family GTs. The Pgts include enzymes from Mesorhizobium loti and A. tumefaciens that synthesize DGal-DAG, glucosylgalactosyl-DAG (GlcGal-DAG), and triglycosyl DAGs (14, 15). To the best of our knowledge, the structure-function relationship of DAG-dependent GT4 glycosyltransferases has not been studied previously due to the lack of a crystal structure of these glycoglycerolipid-producing enzymes, and as such, the binding pockets for UDP-sugars and DAG remain elusive.
Glycoglycerolipids play important roles in marine phytoplankton and heterotrophic bacteria under phosphate deprivation. Lipid remodeling reduces the cellular requirement for phosphorus (P), and the glycoglycerolipids MGlc-DAG/MGlcA-DAG and SQDG replace phospholipids in marine heterotrophic bacteria and marine phytoplankton and cyanobacteria, respectively (16–19). However, little is known about how these glycoglycerolipids are synthesized. Previous studies have shown that a manganese-dependent metallophosphoesterase, PlcP, is essential for lipid remodeling in marine heterotrophs; that the plcP gene is organized in an operon-like structure; and that a putative glycosyltransferase was found down stream of plcP in numerous marine heterotrophic bacteria, such as members of the SAR11 clade (17, 20). Our previous work has shown that the GT from the marine bacterium SAR11 (“Candidatus Pelagibacter sp.” strains HTCC7211 and HTCC1062) is homologous to the Agt GT in A. tumefaciens (17). However, the activity of a SAR11 GT in the synthesis of glycoglycerolipids has not been characterized so far.
In this study, we report the detailed biochemical characterization of a glycoglycerolipid GT (GTcp) from the marine bacterium “Candidatus Pelagibacter sp.” HTCC7211. Our results showed that GTcp has a broad substrate specificity and can synthesize neutral glycolipids (MGlc-DAG and MGal-DAG) and an acidic glycolipid (MGlcA-DAG). GTcp represents the first member of the GT4 family of lipid GTs from marine bacteria. In addition, homology modeling and site-directed mutagenesis analyses revealed details of its substrate recognition mechanism and identified key residues involved in the coordination of DAG in a GT4 family glycosyltransferase for lipid biosynthesis.
RESULTS AND DISCUSSION
GTcp and its homologs form a new group in the GT4 family with a GT-B fold structure.
The gene encoding a putative GTcp (GenBank accession no. WP_008545403.1) consists of 1,005 bp encoding a peptide of 334 amino acids, containing a GT4-like domain (Fig. 1A). This gene was previously hypothesized to be involved in lipid remodeling in “Candidatus Pelagibacter sp.” HTCC7211 for the synthesis of MGlc-DAG and MGlcA-DAG (16, 17). Sequence alignment (Fig. 1B) analyses showed that the GTcp amino acid sequence has between 46% and 58% sequence identity with the putative GT from Labrenzia aggregata (GTla; WP_040439323.1), Thalassospira lucentensis (GTtl; WP_062950653.1), Methylophaga nitratireducenticrescens (GTmn; WP_014706011.1), Desulfobulbus mediterraneus (GTdm; WP_028584068.1), Citromicrobium bathyomarinum JL354 (GTcb; WP_010239457.1), and Kordiimonas gwangyangensis (GTkg; WP_051078133.1) and the characterized Agt (locus tag, atu2297) from A. tumefaciens (6). In the phylogenetic analysis, the GTcp, together with its close homologs (GTla, GTtl, GTmn, GTdm, GTcb, GTkg, and Agt), formed a clade in the GT4 family (Fig. 1A). Sequences from this clade showed low sequence identity (<25%) to other members of the GT4 family, which includes more than 150,000 proteins with at least 22 different enzymatic activities at the time of writing. Purified Agt from A. tumefaciens has been found to synthesize MGlc-DAG or MGlcA-DAG with UDP-glucose or UDP-glucuronic acid as the sugar donor, respectively, and the expression of agt is known to be induced under phosphate deficiency (17). Neither GTcp nor any of its homologs from marine bacteria have been purified or characterized to date.
FIG 1.
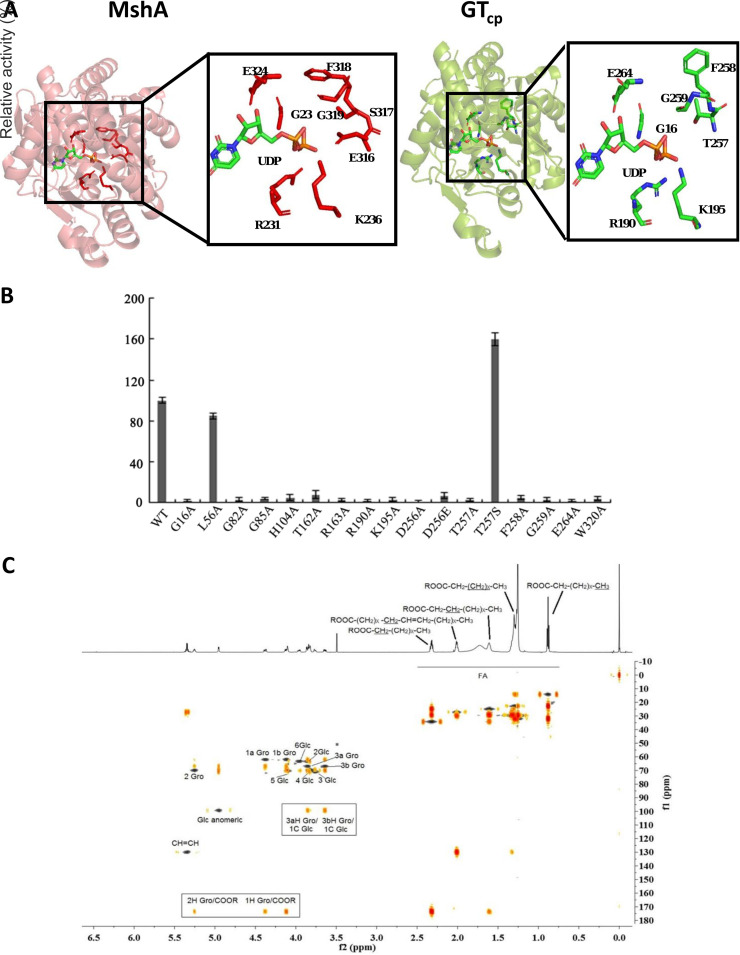
Multiple sequence alignment and functional domain analyses of GTcp protein. (A) A phylogenetic tree of GTcp and its homologs with known 3-dimensional X-ray structures of GTs. Sequences and structures of GTs were obtained from the NCBI database and the PDB database, including GT1 (calG3, SnogD, CalG1, SpnG, UrdGT2, OleD, CalG2, CalG4, GtfD, GtfA, Vinc, UGT78G1, UGT78K6, UGT85H2, UGT71G1, and UGT72B1), GT3 (CeGs), GT5 (Gbss1 and SSI), GT9 (WaaC, WaaF, and Vpar 0760), GT28 (MurG), GT30 (WaaA and KdtA), GT35 (AtPHS2, MalP, and GlgP), GT68 (POFUT2), GT80 (Pdst and Bst), GT4 (PimA, PimB, WsaF, GtfA, TarM, PglH, MshA, BshA, and WaaG), GTcp, and its homologs (GTla, GTtl, GTmn, GTdm, GTcb, GTkg, GTal, GTbb, GTdr, GTpa, and Agt). GTpa is the GT4 glycosyltransferase of Pseudomonas sp. PA14 (Fig. S1). (B) Multiple sequence alignment for GTcp and its homologs using Clustal W program with manual adjusting, including GTcp from “Candidatus Pelagibacter sp.” HTCC7211 (WP_008545403.1), GTla from Labrenzia aggregata (WP_040439323.1), GTtl from Thalassospira lucentensis (WP_062950653.1), GTmn from Methylophaga nitratireducenticrescens (WP_014706011.1), GTdm from Desulfobulbus mediterraneus (WP_028584068.1), GTcb from Citromicrobium bathyomarinum JL354 (WP_010239457.1), GTkg from Kordiimonas gwangyangensis (WP_051078133.1), the characterized Agt from A. tumefaciens (atu2297), and MshA from C. glutamicum (WP_143854623.1). Red bars represent α-helical regions, and blue arrows represent β-sheet from MshA (Protein Data Bank [PDB] entry 3C4Q). Residues interacting with UDP-sugar and DAG are indicated by the closed black circles and squares, respectively. Catalytic dyad composed of His104-Asp256 is shaded in gray. Black boxed regions and red boxed regions in the alignment indicated two conserved UDP-sugar binding motifs of GRVAXEKN and FPSXTDTFG and a conserved Gly-rich motif, respectively. (C) Homology modeling showing the predicted structure of GTcp and the catalytic dyad composed of His104-Asp256. The signature glutamate residue in MshA (Glu316) is substituted to an aspartate residue (Asp256) in GTcp.
Due to the lack of a three-dimensional structure of DAG-dependent GT4 glycosyltransferases to date, a model of GTcp was generated by homology modeling using the X-ray structure of the GT MshA cocrystalized with UDP (Protein Data Bank [PDB] entry 3C4Q; 17% sequence identity) from Corynebacterium glutamicum as the template (Fig. 1C) (21). MshA, also a member of the GT4 family, catalyzes the first step of the biosynthesis of mycothiol in actinobacteria using UDP-N-acetylglucosamine (UDP-GlcNAc) as the sugar donor (21). Using the PDBeFold server, we compared the crystal structure of MshA with the modeled structure of GTcp. The predicted structure of GTcp includes a GT-B fold consisting of two Rossmann-like β-α-β domains, with the N-terminal domain (residues 1 to 160 and residues 320 to 332) and C-terminal domain (residues 169 to 315), separated by a large cleft that includes the catalytic center. The same GT-B fold is also found in several members of the GT4 family enzymes, including MshA and PimA (Fig. 1A) (21, 22).The substrate binding site of the sugar donor is located mainly in the C-terminal domain, where the sugar donor forms a number of hydrogen bonds with the protein. Given that the two enzymes (i.e., MshA and GTcp) share only 17% overall sequence identity, the alignment was manually corrected by incorporating information such as predicted secondary structures and conserved functional residues. Multiple sequence alignment of GTcp and its homologs revealed that GTcp contains a catalytic dyad composed of His104-Asp256, two conserved UDP-sugar binding motifs (GRVAXEKN and FPSXTDTFG), and a conserved Gly-rich motif, of which all are commonly found in the GT4 family (Fig. 1B) (23).
Cloning of putative gene encoding GTcp and functional verification.
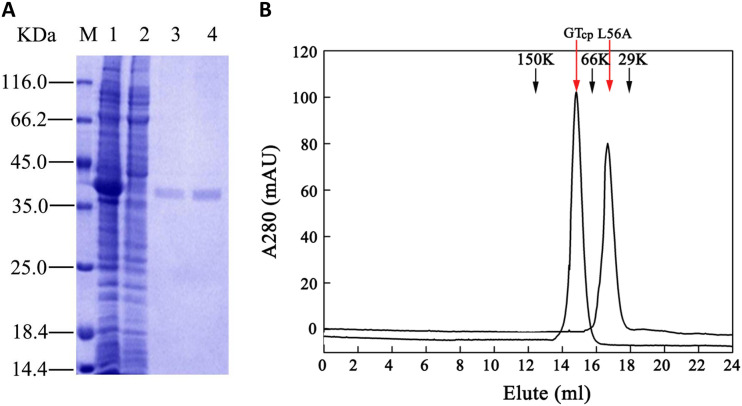
The recombinant plasmid pET22b-GTcp was constructed to purify the enzyme for determination of its catalytic properties. Soluble expression of His-tagged GTcp was achieved in Escherichia coli BL21(DE3) by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The recombinant GTcp was purified to homogeneity by Ni-nitrilotriacetic acid (NTA) affinity followed by gel filtration chromatography using the Superdex 200 column. In SDS-PAGE analyses, the purified recombinant His-tagged GTcp was visible as a major band with a calculated mass of 38 kDa (Fig. 2A). The process used to purify GTcp is summarized in Table 1. The enzyme was purified 3.5-fold with a yield of 17.1% and a specific activity of 47.1 U/mg. The predicted molecular mass of the monomeric form of GTcp is 38 kDa, but GTcp eluted as a peak corresponding to a molecular mass of approximately 70 kDa in the gel filtration chromatography experiments (Fig. 2B). These results suggested that GTcp forms a dimer in solution, consistent with the proposed mechanism that oligomerization is a major factor contributing to the biochemical function and enzymatic activity of GTs (23).
FIG 2.
(A) Overexpression and purification of GTcp proteins from “Candidatus Pelagibacter sp.” HTCC7211. M, protein molecular weight marker. Lane 1, cell-free supernatant induced with isopropyl β-d-1-thiogalactopyranoside (IPTG); lane 2, cell-free supernatant without IPTG induction; and lanes 3 to 4, purified GTcp (molecular weight estimated to be ∼38 kDa). (B) Gel filtration analysis of the wild-type GTcp and the mutant L56A. The red arrows indicate the eluted position of GTcp and the L56A mutant, and the black arrows indicate protein markers (from left to right: alcohol dehydrogenase [150 kDa, 12.35 ml], albumin [66 kDa, 15.83 ml], and carbonic anhydrase [29 kDa, 17.95 ml]).
TABLE 1.
Purification of the recombinant GTcp from “Candidatus Pelagibacter sp.” HTCC7211
| Step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Purification fold | Yield (%) |
|---|---|---|---|---|---|
| Crude cell extract | 34.8 | 469 | 13.4 | 1 | 100 |
| Ni-NTA affinity | 1.9 | 85 | 44.7 | 3.3 | 18.1 |
| Superdex 200 gel filtration | 1.7 | 80 | 47.1 | 3.5 | 17.1 |
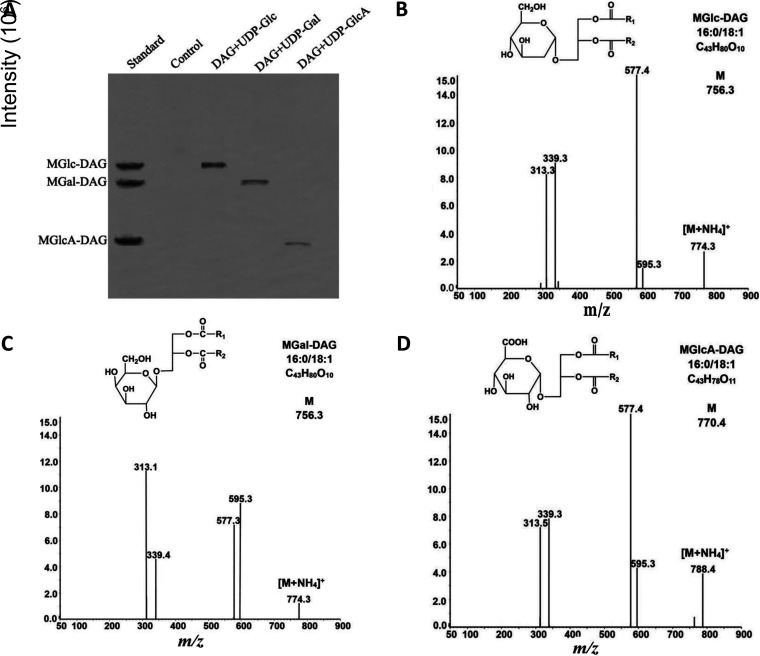
We used two methods to determine the activity of purified GTcp. Previous studies have shown that thin-layer chromatography (TLC) can resolve these selected glycoglycerolipid standards (14, 15). Indeed, as shown in Fig. 3A, the products of the enzymatic reaction with MGlc-DAG, MGal-DAG, and MGlcA-DAG were observed by staining with sulfuric acid-methanol-water and corresponded to the standard markers. These results showed that GTcp is able to transfer galactose, glucose, and hexuronic acid to the DAG acceptor using the respective UDP-sugars.
FIG 3.
Functional characterization of recombinant GTcp. (A) TLC of the enzymatic reaction products with different UDP-sugar donors and diacylglycerol (DAG) as the acceptor by staining with sulfuric acid-methanol-water (45:45:10). (B to D) LC-MS of fragmentation spectra for monohexuronosyl DAGs (MGlc-DAG and MGal-DAG), and MGlcA-DAG were obtained from the products of the GTcp-catalyzed reaction.
For structural identification, the glycoglycerolipids were analyzed by liquid chromatography-mass spectrometry (LC-MS). LC-MS analyses (in the positive ion mode) detected an ammonium adduct (Fig. 3B to D), and fragmentation spectra for monohexuronosyl DAGs (MGlc-DAG and MGal-DAG) and MGlcA-DAG were obtained from the products of the GTcp-catalyzed reactions. The calculated m/z of the parental ion of MGlc-DAG, MGal-DAG, and MGlcA-DAG was 756.3, 756.3, and 770.4, respectively. The two species differed in the neutral loss corresponding to the polar head group (179.0 and 193.1 m/z for the loss of the hexosyl group and the hexuronic acid group, respectively). In each case, this loss yielded a DAG-16:0/18:1 (m/z, 577.3 or 577.4). A further two peaks corresponded to monoacylglycerol with glyceryl-16:0 (m/z, 313.3 or 313.5) and -18:1 (m/z, 339.3 or 339.4) fatty acids, respectively. These results demonstrated that GTcp shows high enzymatic activity toward the synthesis of MGlc-DAG, MGal-DAG, and MGlcA-DAG from DAG and UDP-sugars. Several bacterial lipid GTs from M. loti, A. tumefaciens, Mycoplasma pneumonia, and Mycoplasma genitalium have been found to synthesize different glycoglycerolipids (DGal-DAG, GlcGal-DAG, and triglycosyl DAGs) using UDP-Glu and UDP-galactose (UDP-Gal) as sugar donors (14, 15, 24, 25). The A. tumefaciens GT Agt, which synthesizes neutral glycoglycerolipid (MGlc-DAG) and acidic glycoglycerolipid (MGlcA-DAG) with UDP-Glc or UDP-GlcA as the sugar donor, respectively, has been isolated and characterized (17). To the best of our knowledge, GTcp is the first bacterial lipid GT acting as a multifunctional enzyme to synthesize MGlc-DAG, MGal-DAG, and MGlcA-DAG with different UDP-sugars as donors. It remains to be seen whether bifunctional/multifunctional GT4 enzymes involved in glycoglycerolipid synthesis are a common trait in this group. At least one member of this family, the GT4 homologue in Pseudomonas sp., appears to be specific for UDP-glucose and does not accept UDP-galactose nor UDP-glucuronic acid as the substrate (see Fig. S1 in the supplemental material).
Catalytic properties of GTcp.
The catalytic activity of GTcp was tested with UDP-Glc as the sugar donor substrate and DAG as the potential acceptor substrate. The effect of temperature on GTcp activity was determined in the range of 10 to 50°C (Fig. 4A). GTcp showed maximum activity at around 35°C and more than half of maximum activity at 20 to 45°C. Calculation of the activation energy (Ea) using the Arrhenius plot in a semilogarithmic form [ln(k)→1/T] was shown in Fig. 4B, and the slope of the plot was used to calculate the activation energy which was Ea = 44.9 kJ mol−1. The thermostability of GTcp was evaluated at three different temperatures (30°C, 40°C, and 50°C) with increasing incubation times up to 120 min. Most of the enzyme activity was maintained after incubation at 40°C for at least 120 min, whereas incubation at 50°C for 30 min reduced activity by approximately 50% (Fig. 4C). To investigate the effect of pH on the enzymatic activity of GTcp, the enzymatic reaction was evaluated in different buffers (pH 7.0 to 11.0). The maximum activity of GTcp was at pH 8.5, and it retained more than 50% of maximum activity between pH 7.5 and 9.0 (Fig. 4D). Considering the significance of NaCl for marine enzymes, enzyme activity was determined in the presence of NaCl at different concentrations. The recombinant GTcp maintained 54% of its maximum activity in the presence of 1.5 M NaCl and 30% of its maximum activity when the NaCl concentration was increased to 4 M (Fig. 4E).
FIG 4.
Biochemical characterization of GTcp. (A) Effect of temperature on the activities of GTcp. (B) Arrhenius plot of GTcp. The activation energy of the reaction Ea = 44.9 kJ mol−1 could be determined from the slope of the regression curve with data from 15°C to 30°C (R2 = 0.97). At higher temperatures, enzyme activities decreased, likely due to denaturation of the enzyme. (C) Thermostability of GTcp. The residual enzyme activity was measured after incubation of the purified enzyme at 30°C (diamonds), 40°C (triangles), and 50°C (boxes), respectively. (D) Effect of pH on the activities of GTcp. (E) Effect of NaCl on the activities of GTcp. The enzyme was incubated in buffers containing different concentrations of NaCl (0 to 4 M) at 4°C for 1 h. Residual activity was measured under optimal conditions. (F) Effect of metal ions (5 mM) on the activities of GTcp. The values are means of three independent experiments.
To investigate the substrate specificity of GTcp for DAGs, different species of various chain length of DAGs were tested (Table 2). The Km and kcat values of GTcp were calculated from Hanes-Woolf plots and the Michaelis-Menten equation. For the DAGs with saturated fatty acid chains (di8:0, di10:0, di12:0, di14:0, di16:0, and di18:0), the Km, kcat, and kcat/Km values increased with increasing acyl chain length. GTcp showed the higher activities for the unsaturated DAGs than for the saturated DAGs. The most preferred DAG substrate for GTcp was C16:0/C18:1 DAG, consistent with the fact that C16:0 and C18:1 fatty acids are common in marine bacteria (16, 17).
TABLE 2.
Kinetic parameters of GTcp using UDP-Glc, UDP-Gal, and UDP-GlcA as sugar donors and different DAGs as acceptorsa
| Substrate | kcat (min−1) | Km (μM) | kcat/Km (min−1 mM−1) |
|---|---|---|---|
| UDP-Glc/DAG (C16:0/C18:1) | 5.9 ± 0.4 | 82.0 ± 0.3 | 71.9 ± 2.7 |
| UDP-Gal/DAG (C16:0/C18:1) | 1.0 ± 0.3 | 32.0 ± 0.4 | 31.3 ± 2.9 |
| UDP-GlcA/DAG (C16:0/C18:1) | 4.1 ± 0.1 | 68.0 ± 0.5 | 60.3 ± 3.6 |
| UDP-Glc/DAG (C18:0/C20:4) | 4.8 ± 0.2 | 77.1 ± 0.4 | 62.2 ± 0.9 |
| UDP-Glc/DAG (C18:0/C18:2) | 6.2 ± 1.5 | 102.5 ± 0.8 | 60.5 ± 1.8 |
| UDP-Glc/DAG (di18:1) | 5.0 ± 0.8 | 90.5 ± 1.6 | 55.2 ± 2.2 |
| UDP-Glc/DAG (di18:0) | 3.6 ± 0.7 | 75.3 ± 1.2 | 47.8 ± 1.3 |
| UDP-Glc/DAG (di16:0) | 1.7 ± 1.4 | 55.2 ± 0.9 | 30.8 ± 2.5 |
| UDP-Glc/DAG (di14:0) | 1.0 ± 2.2 | 45.9 ± 1.1 | 21.7 ± 2.8 |
| UDP-Glc/DAG (di12:0) | 0.5 ± 0.4 | 21.6 ± 0.8 | 13.9 ± 1.7 |
| UDP-Glc/DAG (di10:0) | ND | ND | ND |
| UDP-Glc/DAG (di8:0) | ND | ND | ND |
| UDP-Glc/DAG (C16:0/C18:1)b | 8.3 ± 0.3 | 58.0 ± 0.6 | 143.1 ± 3.9 |
The values are means of three independent experiments ± standard deviations. ND, not detectable.
The kinetic parameters were determined using purified GTcp in the presence of Mg2+ (5 mM).
The Michaelis-Menten kinetic parameters for GTcp were determined using UDP-Glc, UDP-Gal, and UDP-GlcA as the sugar donors (Table 3). The Km value for UDP-Glc (82 μM) was higher than those for UDP-Gal and UDP-GlcA, consistent with the fact that UDP-Glc is the preferred substrate under physiological conditions (26). The kcat/Km value for GTcp toward different sugar donors followed the order UDP-Glc (71.4 ± 2.7) > UDP-GlcA (59.6 ± 3.6) > UDP-Gal (32.2 ± 2.9). UDP-xylose, UDP-rhamnose, UDP-mannose, and UDP-fructose were also tested with C16:0/C18:1 DAG, which showed no activity. Thus, GTcp exhibited the highest enzymatic activity for UDP-Glc among the sugar donors tested. In contrast with GTcp, the Pgts from M. loti and A. tumefaciens favor uridine UDP-Gal over UDP-Glc (14, 15). A comparison of GTcp kinetics with other biochemically characterized GT4 glycosyltransferases is listed in Table 4 (21, 27–32).
TABLE 3.
Kinetic parameters of GTcp using UDP-Glc, UDP-Gal, and UDP-GlcA as sugar donors and C16:0/C18:1 DAG as acceptorsa
| Substrate | kcat (min−1) | Km (μM) | kcat/Km (min−1 mM−1) |
|---|---|---|---|
| UDP-Glc | 5.85 ± 0.4 | 82 ± 0.3 | 71.4 ± 2.7 |
| UDP-Gal | 1.03 ± 0.3 | 32 ± 0.4 | 32.2 ± 2.9 |
| UDP-GlcA | 4.05 ± 0.1 | 68 ± 0.5 | 59.6 ± 3.6 |
| UDP-Glc (with 5 mM Mg2+) | 8.25 ± 0.3 | 58 ± 0.6 | 142.2 ± 3.9 |
The values are means ± standard deviations of three independent experiments.
TABLE 4.
Kinetic parameters of GTcp in comparison with GT4 glycosyltransferases using a different sugar donora
| Parameter | Kinetics of GT type with donor substrate |
|||||||
|---|---|---|---|---|---|---|---|---|
| UDP-Glc |
GDP-Man |
UDP-GlcNAC |
||||||
| GTcp | PimA | PimB | MshA | TarM | BshA | GtfA | PglH | |
| Km (μM) | 82 ± 0.3 | 18 ± 2 | 19.0 ± 4.6 | 0.208 ± 0.017 | 65 ± 10 | 180 ± 50 | 11.8 ± 1.5 | 2.6 ± 0.3 |
| kcat (min−1) | 5.8 ± 0.4 | 126 ± 10 | 78.6 | 7.35 ± 0.42 | 4.4 ± 0.2 | |||
| kcat/Km (min−1 mM−1) | 71.4 ± 2.7 | 6.8 | 0.63 | 1.7 ± 0.2 | ||||
aThe values are means ± standard deviations of three independent experiments. UDP-GlcNAC, UDP-N-acetylglucosamine; GDP-Man, GDP-mannose.
Metal ions improve enzyme activity of GTcp.
The effect of various metal ions on the enzyme activity of GTcp is shown in Fig. 4F. Among the tested metal ions (5 mM), Mg2+, Ca2+, and Mn2+ significantly stimulated GTcp activity by up to 223%, 125%, and 138%, respectively, although the as-isolated enzyme is already active. Furthermore, EDTA did not significantly affect the enzymatic activity of GTcp after incubation for 60 min at room temperature. The activity of GTcp was decreased by Ba2+, Cu2+, and Fe2+ to 52%, 23%, and 35%, respectively. Moreover, HgII+, Cd2+, Ni2+, and Co2+ completely abolished enzyme activity. This may have resulted from the binding of these metal ions to the -SH, -CO, and -NH moieties of the amino acids of GTcp, leading to structural changes and inactivation (33).
Given that Mg2+ markedly improved the activity of GTcp, we determined the metal content of the purified enzyme by inductively coupled plasma-mass spectrometry (ICP-MS) and found that the Mg2+: protein molar ratio was <0.03 (see Table S1 in the supplemental material). Similarly, no substantial amount of Ca, Mn, or Zn was found in GTcp, suggesting that this enzyme is unlikely a metalloprotein. The kinetic parameters were subsequently determined using purified GTcp in the presence and absence of Mg2+ (Table 3). In the presence of excess DAG, the Km value of GTcp decreased from 82 μM to 58 μM with the addition of Mg2+, indicating that GTcp has a higher affinity for UDP-Glu in the presence of Mg2+. The catalytic efficiency was nearly 2.0-fold higher than that in the absence of Mg2+. In the presence of excess UDP-Glc, the Km was almost unchanged with/without Mg2+. Metal ions are important regulators of physiological functions and contribute to the preservation of the structural integrity of some proteins (34). In contrast to GT-A fold GTs, GT-B fold GTs, including GTcp, are metal ion independent (23). However, some studies have found that metal ions also change GT-B fold activity, such as GGT58A1 from Absidia coerulea (35), UGT59A1 from Rhizopus japonicas, Bs-PUGT from Bacillus subtilis P118 (36), and human POFUT2 (37). In some cases, the metal ion simultaneously interacts with both the enzyme and the sugar donor in the active site, which causes the glucosyl donor to realign in the active site, and thus affects the activity of the enzyme (38). Interestingly, although human POFUT2 is not a metalloprotein, Mg2+, Mn2+, and Ca2+ are also known to enhance the enzyme activity by facilitating the release of product from the enzyme (37). Therefore, it is tempting to speculate that the addition of Mg2+ may also help enhance GTcp catalysis in a similar manner.
Insights into the structure and glycosylation mechanism of GTcp.
GTcp along with the previously characterized Agt form a group in the GT4 family and are GT-B fold GTs (Fig. 1, Fig. 5A). The GT-B fold GTs are thought to employ the so-called retaining glycosylation for adding the sugar unit to the substrate, although some researchers have proposed an alternative internal return (SNi-like) mechanism (23, 39). In the latter model, the nucleophilic attack and the departure of the leaving group occur on the same face of the sugar and involve the formation of a short-lived oxocarbenium-like transition state with asynchronous acceptor glycoside bond formation and phosphate bond breakdown (40–42). According to the latter model, two conserved amino acid residues are important for catalysis (e.g., Glu316 and His109 in MshA). In MshA, Glu316 and His109 act as catalytic nucleophiles in the SNi-like mechanism, and form hydrogen bonds with the OH-3 and OH-6 of the glucosyl moiety, respectively (21). Indeed, we also found the corresponding residues (Asp256 and His104) in GTcp (Fig. 1B). Asp256 and His104 fulfil a critical role in GTcp catalysis because their substitution to Ala completely abolished catalytic activity (Fig. 5B). Interestingly, mutation of the conserved Asp residue at position 256 to Glu (D256E) reduced activity by more than 90% even though both Asp and Glu residues have a carboxylic acid moiety. Asp256 is essential for the enzymatic activity of GTcp and cannot be replaced by Glu, indicating that the length of the side chain of this residue is important for the activity of GTcp. These results suggested that Asp256 and His104 play important functions in glycosyl transfer while maintaining the α-configuration of the anomeric carbon. To confirm the glycosylated position and the anomeric stereoselectivity, MGlc-DAG was purified and analyzed by 1H-nuclear magnetic resonance (NMR) (Fig. 5C). The glucosyl moiety of the glycolipid was suggested by the characteristic signals for Glc-H1 (δH 5.17, 1H) and Glc-H2-6 (δH 3.20 to 3.70, 6×H) by 1H NMR. The location of the glucosyl moiety was indicated by the correlation between Glc-H1 and C-3 (δH 136.0) in the heteronuclear multiple-bond correlation (HMBC) spectrum. The large coupling constant (J = 3.7 Hz) of Glc-H1 revealed the α-d-configuration of the glucosidic linkage.
FIG 5.
Homology modeling prediction of the UDP-sugar donor binding pocket in the GTcp. (A) UDP-sugar binding site of MshA (21) and predicted UDP-sugar binding site of GTcp. (B) Mutational analysis of the key amino acids involved in the catalytic glycosylation reactions of GTcp. (C) Overlay of 1H, heteronuclear single quantum correlation (HSQC) and heteronuclear multiple-bond correlation (HMBC) spectra of MGlc-DAG.
In terms of the donor binding pocket, GT4 family GTs have two highly conserved sequence motifs in their C-terminal domain that are involved in the binding of the sugar nucleotide with the UDP moiety (22, 28, 43). The sequence alignment analyses indicated that GTcp and its homologs have these two conserved motifs in the donor binding pocket (Fig. 1B), consistent with other GT4 proteins. The highly conserved sites were further investigated to illuminate the mechanism of sugar donor binding (Fig. 5A). Each of the potential active sites in the donor binding pocket (Gly16, Gly82, Gly85, Arg190, Lys195, Thr257, Phe258, Gly259, and Glu264) was mutated to Ala. Enzymatic activity of GTcp in all the mutants was almost completely abolished, supporting our assumption from the homology model that these residues are integral to the sugar donor binding site (Fig. 5B). Thr257 in the structure of GTcp corresponds to MshA residue Ser317, which interacts with the 4-OH of the sugar moiety and forms hydrogen bonds (21). When the residue at position 257 (Thr) in GTcp was substituted with Ser (T257S), the T257S mutant exhibited approximately 1.6-times higher activity than that of wild-type GTcp. Compared with the parental residue Thr, Ser has one fewer methyl group; therefore, there is more free space between the Ser residue and the substrate (44). The generated space might allow more room for the appropriate interaction between the Ser residue and the sugar donor. These findings indicate that the mechanism of the sugar donor recognition of GTcp is similar to that of known GT-B fold GTs, such as MshA. That is, the conserved key residues would form hydrogen bonds with the UDP part and interact with the sugar moiety of the sugar donor.
In order to identify potential residues involved in binding of DAG, we docked C16:1/C18:0 DAG into the homology model of GTcp. DAG is predicted to lay in the groove of the open structure in the model (Fig. 6A and B). Docked DAG interacts with several key resides primarily through hydrophobic interactions but also some polar interactions, e.g., His104, Thr162, Arg163, and Trp320 (Fig. 6C), of which His104, Arg163, and Trp320 are strictly conserved in GTcp in a range of bacteria (Fig. 1B). Indeed, the His104, Arg163, and Trp320 mutants are inactive, supporting a key role in GTcp catalysis (Fig. 5B). Subsequent independent docking of the three UDP-sugars predicted the identical binding pocket for all three (Fig. 6D) in the presence of already docked DAG, and with UDP-galactose as an example, we observe the DAG and UDP-sugars positioned parallel to one another in the “open state” (Fig. 6E). To the best of our knowledge, our study provides the first insight of the binding pocket of DAG in a GT4 glycosyltransferase involved in glycoglycerolipid biosynthesis.
FIG 6.
Identification of key residues for diacylglycerol (DAG) binding in the GTcp. (A) Homology modeling prediction of the open model based on template 2R60. The arrow indicates the wide cleft. (B) C16:1/C18:0-DAG (green) docked in GTcp in the groove (shown as a transparent surface). (C) A detailed depiction of key coordinating hydrophobic (black) and polar (light blue) residues for DAG. His104, Thr162, Arg163, and Trp320 are crucial for enzyme activity (Fig. 5B). (D) An overlay of UDP-glucose (blue), UDP-galactose (cyan), and UDP-glucuronic acid (green) showing all three ligands can occupy the same binding site in similar poses in the DAG-docked GTcp. (E) A top-down view of DAG and UDP-galactose shown docked parallel to each other in their respective binding pockets/grooves.
In MshA, the hydrophobic residue Leu76 helps to stabilize the dimer (21). This residue corresponds to Leu56 in GTcp (Fig. 1B). To investigate the role of Leu56 in GTcp, a site-directed mutant to Ala was made. The mutant L56A protein was loaded onto a Superdex 200 column to analyze its oligomeric state. The eluted peaks of a L56A mutant corresponded to the monomer of GTcp (38 kDa) according to their elution volumes (Fig. 2B). No difference was observed between the wild-type profile and the other mutants during protein purification (data not shown). The mutant L56A showed approximately 85% of wild-type GTcp activity. Structural studies revealed that most of the residues involved in oligomerization are conserved with related GTs, and they appear to be primarily hydrophobic and aromatic residues that form an extensive hydrophobic interface between the monomers (45). These results demonstrated that residue Leu56 is essential for the stable dimerization of the protein but does not play a direct role in the catalytic reaction of GTcp.
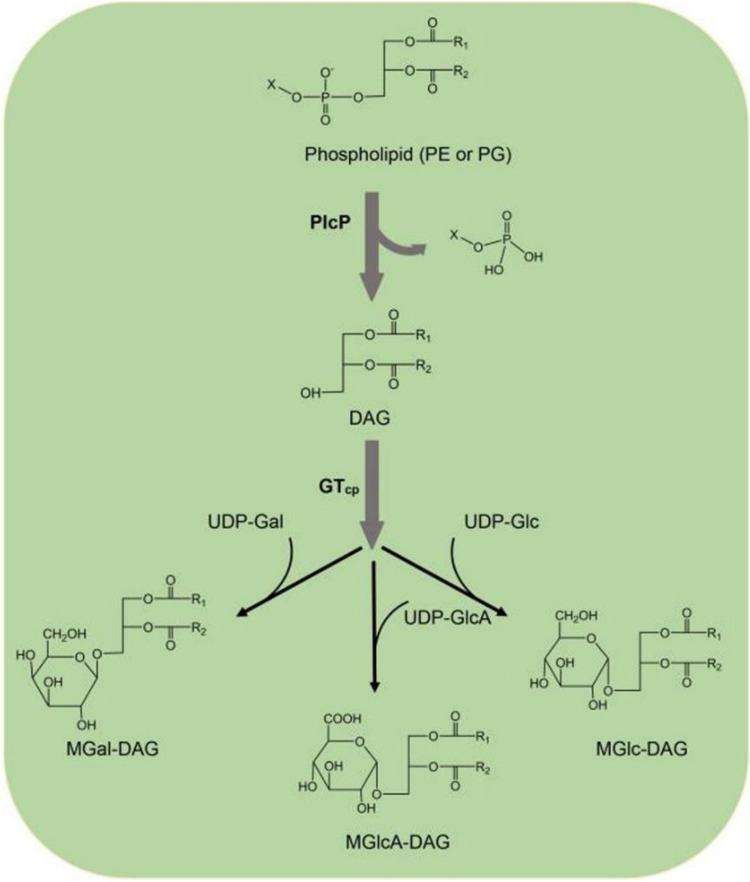
To conclude, our data show that the activity of purified GTcp from the marine bacterium “Candidatus Pelagibacter sp.” HTCC7211 is sufficient for the synthesis of several glycoglycerolipids, including MGlc-DAG, MGal-DAG, and MGlcA-DAG. The ability to synthesize MGlc-DAG and MGlcA-DAG suggests that GTcp may play an important role in lipid remodeling in natural marine systems. GTcp and PlcP, a manganese-dependent metallophosphoesterase, are organized in an operon-like structure in numerous marine heterotrophic bacteria (16, 17). Upon phosphorus (P) deficiency, PlcP selectively degrades phospholipids, such as phosphatidylglycerol (PG) and phosphatidylethanolamine (PE), to DAG, which then serves as the substrate for the biosynthesis of these glycolipids by GTcp using UDP-Glc, UDP-Gal, or UDP-GlcA as sugar donors (Fig. 7). Both the phospholipid PG and the glycoglycerolipid MGlc-DAG are anionic under physiological conditions, and it is likely that they could be interchangeable while maintaining the desirable biophysical properties of the membrane. Indeed, this has also been documented in the SAR11 strain HTCC7211 (16, 17). Similarly, substitution of PG by the anionic sulfur-containing glycolipid SQDG has also been shown for marine cyanobacteria and phytoplankton (46). Together, our work thus points to the important role of glycosyltransferases as key enzymes in the synthesis of glycoglycerolipids in marine bacteria.
FIG 7.
Proposed pathway of the synthesis of nonphosphorus glycoglycerolipids through PlcP and GTcp in “Candidatus Pelagibacter sp.” HTCC7211. PlcP converts phosphatidylglycerol (PG) or phosphatidylethanolamine (PE) to generate diacylglycerol (DAG), and GTcp can synthesize different glycoglycerolipids MGlc-DAG, MGal-DAG, and MGlcA-DAG with UDP-Glc, UDP-Gal, or UDP-GlcA as sugar donors and DAG as the acceptor.
MATERIALS AND METHODS
General materials and microorganisms.
We purchased UDP-glucose (UDP-Glc), UDP-galactose (UDP-Gal), UDP-glucuronic acid (UDP-GlcA), UDP-xylose, UDP-rhamnose, UDP-mannose, UDP-fructose, and diacylglycerols (DAGs) from Sigma-Aldrich Co. (St. Louis, MO, USA). Nickel column and Superdex 200 gel filtration columns were from GE Healthcare (Buckinghamshire, UK). All other chemicals were of the highest reagent grade and were obtained from Sangon (Shanghai, China). The E. coli strains JM109 for DNA manipulation and BL21-CodonPlus (DE3)-RIL for protein expression were obtained from TaKaRa Bio, Inc. (Dalian, China) and Stratagene (La Jolla, CA, USA), respectively.
Cloning, expression, and purification of GTcp.
The gene GTcp from “Candidatus Pelagibacter sp.” HTCC7211 was codon optimized and chemically synthesized by Sangon (Shanghai, China). Several site-directed GTcp mutants (encoding mutations G16A, L56A, G82A, G85A, H104A, T162A, R163A, R190A, K195A, D256A, D256E, T257A, T257S, F258A, G259A, E264A, and W320A) were constructed using the overlapping PCR method with the common primers as for the wild-type GTcp and two site-specific primers for each mutant (Table 5). The genes were then cloned into the pET22b expression vector using NcoI and SalI restriction sites and transformed into the host E. coli BL21-CodonPlus (DE3)-RIL for gene expression. The transformed cells were grown in LB medium containing 100 mg/liter ampicillin and 34 mg/liter chloramphenicol at 37°C with shaking at 180 rpm. When the cultures reached an optical density at 600 nm (OD600) of 0.6, IPTG was added to a final concentration of 0.5 mM. After a further 4 h of growth at 37°C, the cells were harvested by centrifugation and lyophilized by vacuum freezing. The harvested cells were resuspended in buffer A (50 mM Tris-HCl [pH 7.9] and 50 mM NaCl), with 1% (wt/vol) of Triton X-100, and then disrupted by sonication. The cell mixture was then centrifuged at 12,000 × g for 30 min, and the soluble fraction was loaded onto a nickel column (GE Healthcare) pre-equilibrated with buffer A. The recombinant enzymes were eluted with elution buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, and 300 mM imidazole) and dialyzed overnight in buffer A to remove imidazole. For further purification, the enzymes were loaded on a Superdex 200 (16/60) gel filtration column (GE Healthcare), which was pre-equilibrated with buffer B (50 mM Tris-HCl [pH 7.9] and 200 mM NaCl). The fraction size was 0.5 ml and the flow rate was 0.5 ml/min. The peak fractions were collected, concentrated, and analyzed by SDS-PAGE (12% polyacrylamide). The protein concentration was determined using the Bradford method. The purified protein was stored in buffer A containing 25% glycerol at −80°C. SDS-PAGE gels and circular dichroism (CD) spectroscopy analyses of the purified protein and the mutants are shown in Fig. S2 in the supplemental material.
TABLE 5.
Primers used for PCR amplification in this study
| Vector | Primer sequence (5′–3′)a |
|
|---|---|---|
| Upper | Lower | |
| GTcp | GCCGCATATGAAAATTTTAATCGTAAC (NcoI) | GGCGTCGACATTAGGTGATATTAAG (SalI) |
| G16A | CCACTTGTGAATGCTGTAGTTCGAAC | GTTCGAACTACAGCATTCACAAGTGG |
| L56A | GAAATTAGAGCATCATTAAATGTTT | CAAACATTTAATGATGCTCTAATTTC |
| G82A | CATATTGCAACAGAGGCACCTCTTGG | CCAAGAGGTGCCTCTGTTGCAATATG |
| G85A | GGGACCTCTTGCTTTTATGGCAAG | CTTGCCATAAAAGCAAGAGGTCCC |
| H104A | CAACAAGTTTTGCTACAAGATTTG | CAAATCTTGTAGCAAAACTTGTTG |
| T162A | GGTTACATGGGCTAGGGGAGGTAATC | CCATGATTACCTCCCCTAGCCCATGTAAC |
| R163A | GGTTACATGGACTAGCGGAGGTAATCATGG | CACCATGATTACCTCCGCTAGTCCATGTAAC |
| R190A | GGATATACGTCGGTGCAGTTGCAGTTG | CAACTGCAACTGCACCGACGTATATCC |
| K195A | GCAGTTGAAGCAAATATTAAAGC | GCTTTAATATTTGCTTCAACTGC |
| D256A | CCCTAGCAAAACGAATACTTTTGG | CCAAAAGTATGCGTTTTGCTAGGG |
| D256E | CCCTAGCAAAACGAATACTTTTGG | CCAAAAGTATTCGTTTTGCTAGGG |
| T257A | CCCTAGCAAAACCGATGCTTTTGGTATTG | CAATACCAAAAGCATCGGTTTTGCTAGGG |
| T257S | CCCTAGCAAAACCGATAGTTTTGGTATTG | CAATACCAAAACTATCGGTTTTGCTAGGG |
| F258A | CCGATACTGCTGGTATTGTGGTTTTGG | CCAAAACCACAATACCAGCAGTATCGG |
| G259A | CCGATACTTTTGCTATTGTGGTTTTGGAG | CTCCAAAACCACAATAGCAAAAGTATCGG |
| E264A | GTGGTTTTGGAGTCTTTAAGTTGTGG | CCACAACTTAAAGACTCCAAAACCAC |
| W320A | GCTAAAAAATATAGTTCGGAAGAAAC | CCTTGCTGTTTCTTCCGAACTATTTTTTAG |
aThe restriction sites are underlined, and mutations are shown in boldface.
Bioinformatics and homology modeling.
A putative GT gene encoding GTcp was identified in the genome of “Candidatus Pelagibacter sp.” HTCC7211 (GenBank accession no. WP_008545403.1). ClustalW2 software (http://www.ebi.ac.uk/Tools/clustalw2/index.html) was used for multiple sequence alignment analysis of GTcp (47). For the phylogenetic analysis, we used the neighbor-joining method and Molecular Evolutionary Genetic Analysis (MEGA) software (v7.1) (48). The three-dimensional model structure of GTcp was generated using tools at the Phyre 2 protein modeling server (49) and the crystal structure of MshA (Protein Data Bank [PDB] entry 3C4Q) from C. glutamicum as the template. Docking of UDP-Gla/UDP-Glc/UDP-GlcA with GTcp was predicted using the Flexible Docking module in Accelrys Discovery studio (50, 51), and the protein model was imported into Flare (v3.0, Cresset) for docking the DAG substrate and firstly energy minimized with 2,000 iterations with a cut off of 0.200 kcal/mol/A. The DAG lipid was imported as a ligand and energy minimized in Flare before being docked into the active site and the best scoring pose selected. Thereafter we used the DAG-docked ligand with the model as the basis for the follow-up in silico docking of UDP-sugars in the presence of the DAG.
Enzyme activity assay.
The enzymatic activity of GTcp was measured using 0.1 mM UDP-Glc (or UDP-Gal or UDP-GlcA) and 0.1 mM DAG as the substrate and 2.0 μM purified enzyme in 10 mM Tricine-KOH [pH 8.5] and 2 mM dithiothreitol (DTT). The resulting mixture (500 μl) was incubated at 35°C for 60 min with constant shaking at 200 rpm. The products (glycoglycerolipids and DAGs) were extracted using the Floch method with methanol-chloroform-water at a ratio of 1:2:0.6 (vol/vol/vol). The lipid extract was dried under nitrogen conditions at room temperature. The dried lipids were resuspended in acetonitrile and ammonium acetate (10 mM; pH 9.2) at a ratio of 95:5 (vol/vol) and analyzed by LC-MS. One unit of enzymatic activity was defined as the amount of enzyme required to catalyze the conversion of 1 μmol DAG per min under the standard conditions. The measurements were corrected for background hydrolysis in the absence of the enzyme. The Km and Vmax values were calculated using Hanes-Wolff plots with various concentrations of substrate (0.02 to 1.0 mM) and three replicates.
Lipid analysis by TLC and LC-MS.
The GTcp-synthesized glycoglycerolipids were analyzed by TLC using a Camag Automatic TLC Sampler III (Camag, Muttenz, Switzerland) for spotting. Glycoglycerolipids were separated on silica gel 60 (Merck, Darmstadt, Germany) with chloroform-methanol-water (65:35:4 vol/vol) and stained with sulfuric acid-methanol-water (45:45:10 vol/vol) for visualization. The resulting solutions were further analyzed by high-performance liquid chromatography using a 1290 Infinity II ultraperformance liquid chromatography (UPLC) instrument (Agilent Corp., Santa Clara, CA, USA) coupled with an Triple Quad 5500 (AB Sciex, Framingham, MA, USA) equipped with an electrospray-ion (ESI) detector. A BEH Amide XP column (2.5-μm inner diameter; 3 mm by 150 mm; Waters, Milford, MA, USA) was used for chromatographic separation. The mobile phase consisted of acetonitrile (solvent A) and 10 mM ammonium acetate (pH 9.2) (solvent B). The column was equilibrated for 10 min with 95% A:5% B prior to sample injection. The separation was conducted using a stepwise gradient starting from 95% A:5% B to 70% A:30% B after 15 min with a constant flow rate of 150 μl min−1. Mass spectrometric analysis was performed in the ESI positive ion mode with the ion spray voltage at 3,500 V and temperature at 350°C. The nebulizer gas and heater gas were set at 40 lb/in2. The analytical data were processed by Analyst software (v1.6.3).
Inductively coupled plasma-mass spectrometry.
The metal content of GTcp was measured by using an ICP-MS (Agilent Technologies 7900 ICP-MS). The standards for calibration were freshly prepared by diluting Ca, Mg, Mn, Zn, and S stock solution (at 1,000 mg · liter−1; Sigma-Aldrich, Saint Louis, MO, USA) with 1% (vol/vol) nitric acid with concentrations from 0.1 to 2.0 mg·liter−1 for Ca, Mg, Mn, and Zn and from 1 to 25 mg · liter−1 for S. About 3.0 mg protein was digested in 1% (vol/vol) nitric acid matrix for metal analyses. The content of S was quantified in order to determine the protein concentration. The contents of Ca, Mg, Mn, Zn, and S were measured using the emission lines of 396.847 nm (Ca), 280.270 nm (Mg), 259.373 nm (Mn), 213.856 nm (Zn), and 180.669 nm (S), respectively.
Nuclear magnetic resonance spectroscopy (NMR) spectroscopy.
NMR spectroscopy experiments were carried out in CDCl3 with tetramethylsilane as an internal standard. 1H, 13C, distortionless enhancement by polarization transfer (DEPT), correlation spectroscopy (COSY), heteronuclear single quantum coherence (HSQC), HMBC, and rotating-frame nuclear Overhauser effect spectroscopy (ROESY) experiments were recorded at 298 K with a 600-MHz spectrometer (Bruker Avance 600; Bruker). Bruker standard software Topspin 3.2 was applied to acquire and process all the spectra data. COSY and ROESY experiments were recorded using data sets (t1 by t2) of 2,048 by 256 points, COSY with 4, and ROESY with 16 scans.
Characterization of recombinant GTcp.
The optimum temperature of GTcp was measured in Tricine-KOH buffer (pH 8.5) in the range of 10 to 50°C. The buffer was adjusted to pH 8.5 for each of the assayed temperatures. The activation energy of the cleavage reaction was calculated using the logarithmic form of the Arrhenius equation, as follows: lnk = lnk0 − Ea/R·T. The effect of pH on enzymatic activity was tested at 35°C for pH values in the range of 7.0 to 11.0. The following buffers were used: sodium phosphate (pH 6.0 to 7.5), Tricine-KOH (pH 7.5 to 9.5), and N-cyclohexyl-3-aminopropanesulfonic acid (CAPS; pH 9.5 to 11.0). The thermostability of the purified GTcp was examined by incubating the enzyme in 50 mM Tricine-KOH buffer (pH 8.5) at three different temperatures (30, 40, and 50°C). Samples (80 μl) of the enzyme were collected after incubation periods of 30, 60, 90, and 120 min at each temperature, and the residual activity of each sample was assayed under standard conditions. The enzymatic activity of GTcp was also measured in the presence of various metal salts (MnCl2, ZnCl2, MgCl2, CaCl2, BaCl2, CdCl2, HgCl2, CuCl2, FeSO4, NiSO4, and CoCl2) at 5 mM or 5 mM EDTA. To determine the salt stability of the enzyme, 0 to 4 M NaCl (final concentration) was added to the reaction mixture, and enzyme activity was determined using optimum conditions.
Data availability.
The authors confirm that the data supporting the findings of this study are available within the manuscript and its supplemental materials.
ACKNOWLEDGMENTS
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 726116), Royal Society International Exchanges 2017 Cost Share (China) (IEC\NSFC\170213; grant agreement no. 170213), Program for Science &Technology Innovation Talents in the Universities of Henan Province (18HASTIT040), and projects of science and technology activities for overseas students from department of human resources and social security of Henan Province (2019-3).
Footnotes
Supplemental material is available online only.
Contributor Information
Tao Wei, Email: weit8008@zzuli.edu.cn.
Yin Chen, Email: Y.chen.25@warwick.ac.uk.
Haruyuki Atomi, Kyoto University.
REFERENCES
- 1.Cheng-Sánchez I, Sarabia F. 2018. Chemistry and biology of bioactive glycolipids of marine origin. Mar Drugs 16:294. 10.3390/md16090294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hölzl G, Dörmann P. 2007. Structure and function of glycoglycerolipids in plants and bacteria. Prog Lipid Res 46:225–243. 10.1016/j.plipres.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Kolter TA. 2011. View on sphingolipids and disease. Chem Phys Lipids 164:590–606. 10.1016/j.chemphyslip.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Chirasuwan N, Chaiklahan R, Kittakoop P, Chanasattru W, Ruengjitchatchawalya M, Tanticharoen M, Bunnag B. 2009. Anti HSV-1 activity of sulphoquinovosyl diacylglycerol isolated from Spirulina platensis. Sci Asia 35:137–141. 10.2306/scienceasia1513-1874.2009.35.137. [DOI] [Google Scholar]
- 5.Bergé JP, Debiton E, Dumay J, Durand P, Barthomeuf C. 2002. In vitro anti-inflammatory and anti-proliferative activity of sulfolipids from the red alga Porphyridium cruentum. J Agric Food Chem 50:6227–6232. 10.1021/jf020290y. [DOI] [PubMed] [Google Scholar]
- 6.Semeniuk A, Sohlenkamp C, Duda K, Hölzl G. 2014. A bifunctional glycosyltransferase from Agrobacterium tumefaciens synthesizes monoglucosyl and glucuronosyl diacylglycerol under phosphate deprivation. J Biol Chem 289:10104–10114. 10.1074/jbc.M113.519298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Li CX, Yu GL, Guan HS. 2014. Total synthesis and structure-activity relationship of glycoglycerolipids from marine organisms. Mar Drugs 12:3634–3659. 10.3390/md12063634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lairson LL, Henrissat B, Davies GJ, Withers SG. 2008. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77:521–555. 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 9.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutinho PM, Deleury E, Davies GJ, Henrissat B. 2003. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328:307–317. 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 11.Berg S, Edman M, Li L, Wikström M, Wieslander A. 2001. Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J Biol Chem 276:22056–22063. 10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- 12.Hölzl G, Zähringer U, Warnecke D, Heinz E. 2005. Glycoengineering of cyanobacterial thylakoid membranes for future studies on the role of glycolipids in photosynthesis. Plant Cell Physiol 46:1766–1778. 10.1093/pcp/pci189. [DOI] [PubMed] [Google Scholar]
- 13.Ostberg Y, Berg S, Comstedt P, Wieslander A, Bergström S. 2007. Functional analysis of a lipid galactosyltransferase synthesizing the major envelope lipid in the Lyme disease spirochete Borrelia burgdorferi. FEMS Microbiol Lett 272:22–29. 10.1111/j.1574-6968.2007.00728.x. [DOI] [PubMed] [Google Scholar]
- 14.Hölzl G, Leipelt M, Ott C, Zähringer U, Lindner B, Warnecke D, Heinz E. 2005. Processive lipid galactosyl/glucosyltransferases from Agrobacterium tumefaciens and Mesorhizobium loti display multiple specificities. Glycobiology 15:874–886. 10.1093/glycob/cwi066. [DOI] [PubMed] [Google Scholar]
- 15.Devers EA, Wewer V, Dombrink I, Dörmann P, Hölzl G. 2011. A processive glycosyltransferase involved in glycolipid synthesis during phosphate deprivation in Mesorhizobium loti. J Bacteriol 193:1377–1384. 10.1128/JB.00768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carini P, Van Mooy BA, Thrash JC, White A, Zhao Y, Campbell EO, Fredricks HF, Giovannoni SJ. 2015. SAR11 lipid renovation in response to phosphate starvation. Proc Natl Acad Sci U S A 112:7767–7772. 10.1073/pnas.1505034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebastián M, Smith AF, González JM, Fredricks HF, Van Mooy B, Koblížek M, Brandsma J, Koster G, Mestre M, Mostajir B, Pitta P, Postle AD, Sánchez P, Gasol JM, Scanlan DJ, Chen Y. 2016. Lipid remodelling is a widespread strategy in marine heterotrophic bacteria upon phosphorus deficiency. ISME J 10:968–978. 10.1038/ismej.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Mooy BA, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblízek M, Lomas MW, Mincer TJ, Moore LR, Moutin T, Rappé MS, Webb EA. 2009. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458:69–72. 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- 19.Smith AF, Rihtman B, Stirrup R, Silvano E, Mausz MA, Scanlan DJ, Chen Y. 2019. Elucidation of glutamine lipid biosynthesis in marine bacteria reveals its importance under phosphorus deplete growth in Rhodobacteraceae. ISME J 13:39–49. 10.1038/s41396-018-0249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei T, Quareshy M, Zhang YZ, Scanlan DJ, Chen Y. 2018. Manganese is essential for PlcP metallophosphoesterase activity involved in lipid remodelling in abundant marine heterotrophic bacteria. Appl Environ Microbiol 84:01109-18. 10.1128/AEM.01109-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vetting MW, Frantom PA, Blanchard JS. 2008. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate-assisted catalysis. J Biol Chem 283:15834–15844. 10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerin ME, Kordulakova J, Schaeffer F, Svetlikova Z, Buschiazzo A, Giganti D, Gicquel B, Mikusova K, Jackson M, Alzari PM. 2007. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J Biol Chem 282:20705–20714. 10.1074/jbc.M702087200. [DOI] [PubMed] [Google Scholar]
- 23.Liang D-M, Liu J-H, Wu H, Wang B-B, Zhu H-J, Qiao J-J. 2015. Glycosyltransferases: mechanisms and applications in natural product development. Chem Soc Rev 44:8350–8374. 10.1039/c5cs00600g. [DOI] [PubMed] [Google Scholar]
- 24.Andrés E, Martínez N, Planas A. 2011. Expression and characterization of a Mycoplasma genitalium glycosyltransferase in membrane glycolipid biosynthesis: potential target against Mycoplasma infections. J Biol Chem 286:35367–35379. 10.1074/jbc.M110.214148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klement ML, Ojemyr L, Tagscherer KE, Widmalm G, Wieslander A. 2007. A processive lipid glycosyltransferase in the small human pathogen Mycoplasma pneumoniae: involvement in host immune response. Mol Microbiol 65:1444–1457. 10.1111/j.1365-2958.2007.05865.x. [DOI] [PubMed] [Google Scholar]
- 26.Forget SM, Shepard SB, Soleimani E, Jakeman DL. 2019. On the catalytic activity of a GT1 family glycosyltransferase from Streptomyces venezuelae ISP5230. J Org Chem 84:11482–11492. 10.1021/acs.joc.9b01130. [DOI] [PubMed] [Google Scholar]
- 27.Gu XL, Chen M, Wang QZ, Zhang M, Wang BL, Wang HH. 2005. Expression and purification of a functionally active recombinant GDP-mannosyltransferase (PimA) from Mycobacterium tuberculosis H37Rv. Protein Expr Purif 42:47–53. 10.1016/j.pep.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Batt SM, Jabeen T, Mishra AK, Veerapen N, Krumbach K, Eggeling L, Besra GS, Fütterer k. 2010. Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'. J Biol Chem 285:37741–37752. 10.1074/jbc.M110.165407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobhanifar S, Worrall LJ, Gruninger RJ, Wasney GA, Blaukopf M, Baumann L, Lameignere E, Solomonson M, Brown ED, Withers SG, Strynadka NCJ. 2015. Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid α-glycosyltransferase. Proc Natl Acad Sci U S A 112:E576–E585. 10.1073/pnas.1418084112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Royer CJ, Cook PD. 2019. A structural and functional analysis of the glycosyltransferase BshA from Staphylococcus aureus: insights into the reaction mechanism and regulation of bacillithiol production. Protein Sci 28:1083–1094. 10.1002/pro.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W-W, Jiang Y-L, Zhu F, Yang Y-H, Shao Q-Y, Yang H-B, Ren Y-M, Wu H, Chen YX, Zhou C-Z. 2014. Structure of a novel O-Linked N-aetyl-D-glucosamine (O-GlcNAc) tansferase, GtfA, reveals Insights into the gycosylation of pneumococcal serine-rich repeat adhesions. J Biol Chem 289:20898–20907. 10.1074/jbc.M114.581934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Troutman JM, Imperiali B. 2009. Campylobacter jejuni PglH is a single active site processive polymerase that utilizes product inhibition to limit sequential glycosyl transfer reactions. Biochemistry 48:2807–2816. 10.1021/bi802284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Li W, Zhang G, Lü X, Hwang H, Aker WG, Guan H, Wang P. 2016. Purification and characterization of polysaccharides degradases produced by Alteromonas sp. A321. Int J Biol Macromol 86:96–104. 10.1016/j.ijbiomac.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S, Zhou Z, Yao Z, He BF. 2013. Efficient production of skimmin and 6'-succinylskimmin from umbelliferone by organic solvent-tolerant Bacillus licheniformis ZSP01 using nitrogen sources regulation strategy. Biochem Eng J 71:105–110. 10.1016/j.bej.2012.12.004. [DOI] [Google Scholar]
- 35.Xie K, Dou X, Chen R, Chen D, Fang C, Xiao Z, Dai J. 2017. Two novel fungal phenolic UDP glycosyltransferases from Absidia coerulea and Rhizopus japonicus. Appl Environ Microbiol 83:e03103-16. 10.1128/AEM.03103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitayska T, Petrova P, Ivanova V, Tonkova AI. 2011. Purification and properties of a new thermostable cyclodextrin glucanotransferase from Bacillus pseudalcaliphilus 8SB. Appl Biochem Biotechnol 165:1285–1295. 10.1007/s12010-011-9346-4. [DOI] [PubMed] [Google Scholar]
- 37.Chen CI, Keusch JJ, Klein D, Hess D, Hofsteenge J, Gut H. 2012. Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J 31:3183–3197. 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruitt RN, Chumbler NM, Rutherford SA, Farrow MA, Friedman DB, Spiller B, Lacy DB. 2012. Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J Biol Chem 287:8013–8020. 10.1074/jbc.M111.298414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albesa-Jové D, Guerin ME. 2016. The conformational plasticity of glycosyltransferases. Curr Opin Struct Biol 40:23–32. 10.1016/j.sbi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Errey JC, Lee SS, Gibson RP, Martinez Fleites C, Barry CS, Jung PMJ, O'Sullivan AC, Davis BG, Davies GJ. 2010. Mechanistic insight into enzymatic glycosyl transfer with retention of configuration through analysis of glycomimetic inhibitors. Angew Chem Int Ed Engl 49:1234–1237. 10.1002/anie.200905096. [DOI] [PubMed] [Google Scholar]
- 41.Gómez H, Polyak I, Thiel W, Lluch JM, Masgrau L. 2012. Retaining glycosyltransferase mechanism studied by QM/MM methods: lipopolysaccharyl-a-1, 4-galactosyltransferase C transfers agalactose via an oxocarbenium ion-like transition state. J Am Chem Soc 134:4743–4752. 10.1021/ja210490f. [DOI] [PubMed] [Google Scholar]
- 42.Lee SS, Hong SY, Errey JC, Izumi A, Davies GJ, Davis BG. 2011. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat Chem Biol 7:631–638. 10.1038/nchembio.628. [DOI] [PubMed] [Google Scholar]
- 43.Guerin ME, Kaur D, Somashekar BS, Gibbs S, Gest P, Chatterjee D, Brennan PJ, Jackson M. 2009. New insights into the early steps of phosphatidylinositol mannoside biosynthesis in mycobacteria: PimB′ is an essential enzyme of Mycobacterium smegmatis. J Biol Chem 284:25687–25696. 10.1074/jbc.M109.030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honda Y, Nakano S, Ito S, Dadashipour M, Zhang ZL, Kawarabayasi Y. 2018. Improvement of ST0452 N-Acetylglucosamine-1-phosphate uridyltransferase activity by the cooperative effect of two single mutations identified through structure-based protein engineering. Appl Environ Microbiol 84:e02213-18. 10.1128/AEM.02213-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu CP, Lairson LL, Gilbert M, Wakarchuk WW, Wither SG, Strynadka NC. 2007. Structural analysis of the α-2, 3-sialyltransferase Cst-I from Campylobacter jejuni in apo and substrate-analogue bound forms’. Biochemistry 46:7196–7204. 10.1021/bi602543d. [DOI] [PubMed] [Google Scholar]
- 46.Van Mooy BAS, Rocap G, Fredricks HF, Evans CT, Devol AH. 2006. Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci U S A 103:8607–8612. 10.1073/pnas.0600540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modelling, prediction and analyses. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodis E, Sussman JL. 2009. An encyclopedic effort to make 3D structures easier to understand. Trends Biochem Sci 34:100–101. 10.1016/j.tibs.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Diller DJ, Merz KM, Jr.. 2001. High throughput docking for library design and library prioritization. Proteins 43:113–124. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1 and S2. Download AEM00326-21_Supp_1_seq9.pdf, PDF file, 0.4 MB (398.9KB, pdf)
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the manuscript and its supplemental materials.