ABSTRACT
Many bacteria and other organisms carry out fermentations forming acetate. These fermentations have broad importance for foods, agriculture, and industry. They also are important for bacteria themselves because they often generate ATP. Here, we found a biochemical pathway for forming acetate and synthesizing ATP that was unknown in fermentative bacteria. We found that the bacterium Cutibacterium granulosum formed acetate during fermentation of glucose. It did not use phosphotransacetylase or acetate kinase, enzymes found in nearly all acetate-forming bacteria. Instead, it used a pathway involving two different enzymes. The first enzyme, succinyl coenzyme A (succinyl-CoA):acetate CoA-transferase (SCACT), forms acetate from acetyl-CoA. The second enzyme, succinyl-CoA synthetase (SCS), synthesizes ATP. We identified the genes encoding these enzymes, and they were homologs of SCACT and SCS genes found in other bacteria. The pathway resembles one described in eukaryotes, but it uses bacterial, not eukaryotic, gene homologs. To find other instances of the pathway, we analyzed sequences of all biochemically characterized homologs of SCACT and SCS (103 enzymes from 64 publications). Homologs with similar enzymatic activity had similar sequences, enabling a large-scale search for them in genomes. We searched nearly 600 genomes of bacteria known to form acetate, and we found that 6% encoded homologs with SCACT and SCS activity. This included >30 species belonging to 5 different phyla, showing that a diverse range of bacteria encode the SCACT/SCS pathway. This work suggests the SCACT/SCS pathway is important for acetate formation in many branches of the tree of life.
IMPORTANCE Pathways for forming acetate during fermentation have been studied for over 80 years. In that time, several pathways in a range of organisms, from bacteria to animals, have been described. However, one pathway (involving succinyl-CoA:acetate CoA-transferase and succinyl-CoA synthetase) has not been reported in prokaryotes. Here, we discovered enzymes for this pathway in the fermentative bacterium Cutibacterium granulosum. We also found >30 other fermentative bacteria that encode this pathway, demonstrating that it could be common. This pathway represents a new way for bacteria to form acetate from acetyl-CoA and synthesize ATP via substrate-level phosphorylation. It could be a target for controlling yield of acetate during fermentation, with relevance for foods, agriculture, and industry.
KEYWORDS: acetate, ATP, eubacteria, fermentation
INTRODUCTION
Many bacteria and other organisms carry out anaerobic fermentations forming acetate (1–4). Such acetate-yielding fermentations have importance for foods, agriculture, and industry. Acetate is found in many fermented foods (e.g., Swiss cheese), and it is an energy source for cattle harboring fermentative microbes (5). Further, acetate is an often-unwanted by-product in industrial fermentations (e.g., those producing succinate or propionate) (6). These fermentations also have importance for the energy metabolism of fermentative organisms themselves. If the organism forms acetate from acetyl coenzyme A (acetyl-CoA) or acetyl phosphate, it can generate ATP by substrate-level phosphorylation (7). This is crucial because fermentative metabolism otherwise yields little ATP.
Reflecting this broad importance, the biochemical pathways for forming acetate and ATP during fermentation have been studied for over 80 years (8). Over the course of their study, a total of five pathways have been reported (Fig. 1B and C). For four of the pathways, the high-energy precursor is acetyl-CoA. For one pathway, the precursor is either acetyl-CoA or acetyl phosphate.
FIG 1.
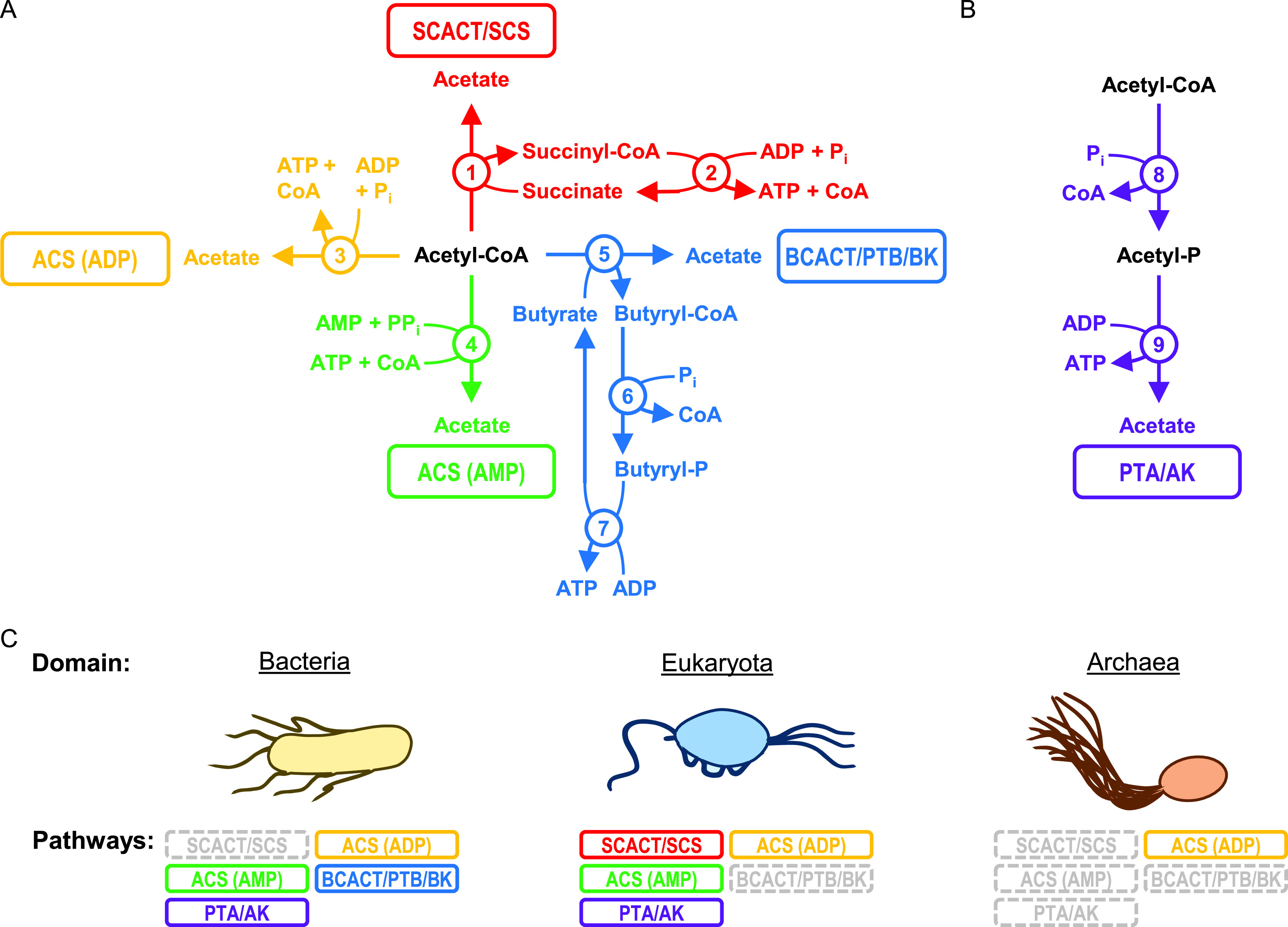
Bacteria use many pathways to form acetate and ATP during fermentation, but none are reported to use the SCACT/SCS pathway. Pathways for forming acetate and ATP from (A) acetyl-CoA and (B) acetyl-CoA or acetyl phosphate. (C) Reported use of pathways by domain. Pathways that form acetate without ATP (e.g., acetyl-CoA hydrolase; EC 3.1.2.1) are not included. The PTA/AK pathway can use both PTA and AK, or it can use AK alone. Enzymes: 1, succinyl-CoA:acetate CoA-transferase (SCACT; EC 2.8.3.18); 2, succinyl-CoA synthetase (ADP forming) (SCS; EC 6.2.1.5); 3, acetyl-CoA synthetase (ADP forming) (ACS [ADP]; EC 6.2.1.13); 4, acetyl-CoA synthetase (ACS [AMP]; EC 6.2.1.1); 5, butyryl-CoA:acetate CoA-transferase (BCACT; EC 2.8.3.8); 6, phosphotransbutyrylase (PTB; EC 2.3.1.19); 7, butyrate kinase (BK; EC 2.7.2.7); 8, phosphotransacetylase (PTA; EC 2.3.1.8); and 9, acetate kinase (AK; EC 2.7.2.1). For bacteria, see reference 52 for the ACS (ADP) pathway, reference 53 for the ACS (AMP) pathway, references 54 and 55 for the BCACT/PTB/BK pathway, and reference 7 for the PTA/AK pathway. For eukaryotes, see references 9 and 56 for the SCACT/SCS pathway, references 57 and 58 for the ACS (ADP) pathway, reference 59 for the ACS (AMP) pathway, and references 60 and 61 for the PTA/AK pathway. For archaea, see references 62 and 63 for the ACS (ADP) pathway. Abbreviations: ACS (ADP), acetyl-CoA synthetase (ADP forming); ACS (AMP), acetyl-CoA synthetase; AK, acetate kinase; BCACT, butyryl-CoA:acetate CoA-transferase; BK, butyrate kinase; CoA, coenzyme A; P, phosphate; Pi, inorganic phosphate; PPi, pyrophosphate; PTA, phosphotransacetylase; PTB, phosphotransbutyrylase; SCACT, succinyl-CoA:acetate CoA-transferase; and SCS, succinyl-CoA synthetase (ADP forming).
Curiously, one of the five pathways has been found to be missing in bacteria (Fig. 1C). The pathway missing in bacteria involves the enzymes succinyl-CoA:acetate CoA-transferase (SCACT; EC 2.8.3.18) and succinyl-CoA synthetase (SCS; EC 6.2.1.5). It was first reported in a flagellate protozoan (9) and then in other eukaryotes (1, 10). No report of this SCACT/SCS pathway in bacteria or archaea has been made. In bacteria, the pathway commonly used instead involves phosphotransacetylase (PTA; EC 2.3.1.8) and acetate kinase (AK; EC 2.7.2.1) (7).
Recently, we found evidence of the SCACT/SCS pathway in some bacteria of the rumen (11). These bacteria form acetate during fermentation yet do not have genes for the PTA/AK pathway. They do have genes of the SCACT/SCS pathway, suggesting they may use this pathway instead. We have since tried to find bacteria with biochemical, not just genomic, evidence of the pathway (12).
Here, we found biochemical evidence of the SCACT/SCS pathway in the bacterium Cutibacterium granulosum. We confirmed that the bacterium has the appropriate genes, the gene products have enzymatic activity, and they form a functional pathway. Strikingly, the genes are bacterial homologs of the SCACT and SCS genes, suggesting that the pathway in this bacterium did not originate from eukaryotes. Moreover, this pathway appears to be common and is encoded by 36 type strains of bacteria that form acetate. Our work shows that the SCACT/SCS pathway is important for acetate formation in many branches of the tree of life.
RESULTS
C. granulosum forms acetate using the SCACT/SCS pathway.
We investigated if C. granulosum uses the SCACT/SCS pathway to form acetate anaerobically, because its genome encodes this pathway but not others (Fig. 1). We included other propionibacteria and bacteria for comparison.
To confirm that C. granulosum forms acetate during fermentation, we measured the concentration of acetate in the culture supernatant when it grew on glucose. We found that C. granulosum and other propionibacteria all formed acetate as a major end product of fermentation (Fig. 2A). The only product produced in larger quantities was propionate (see Table S1 in the supplemental material).
FIG 2.
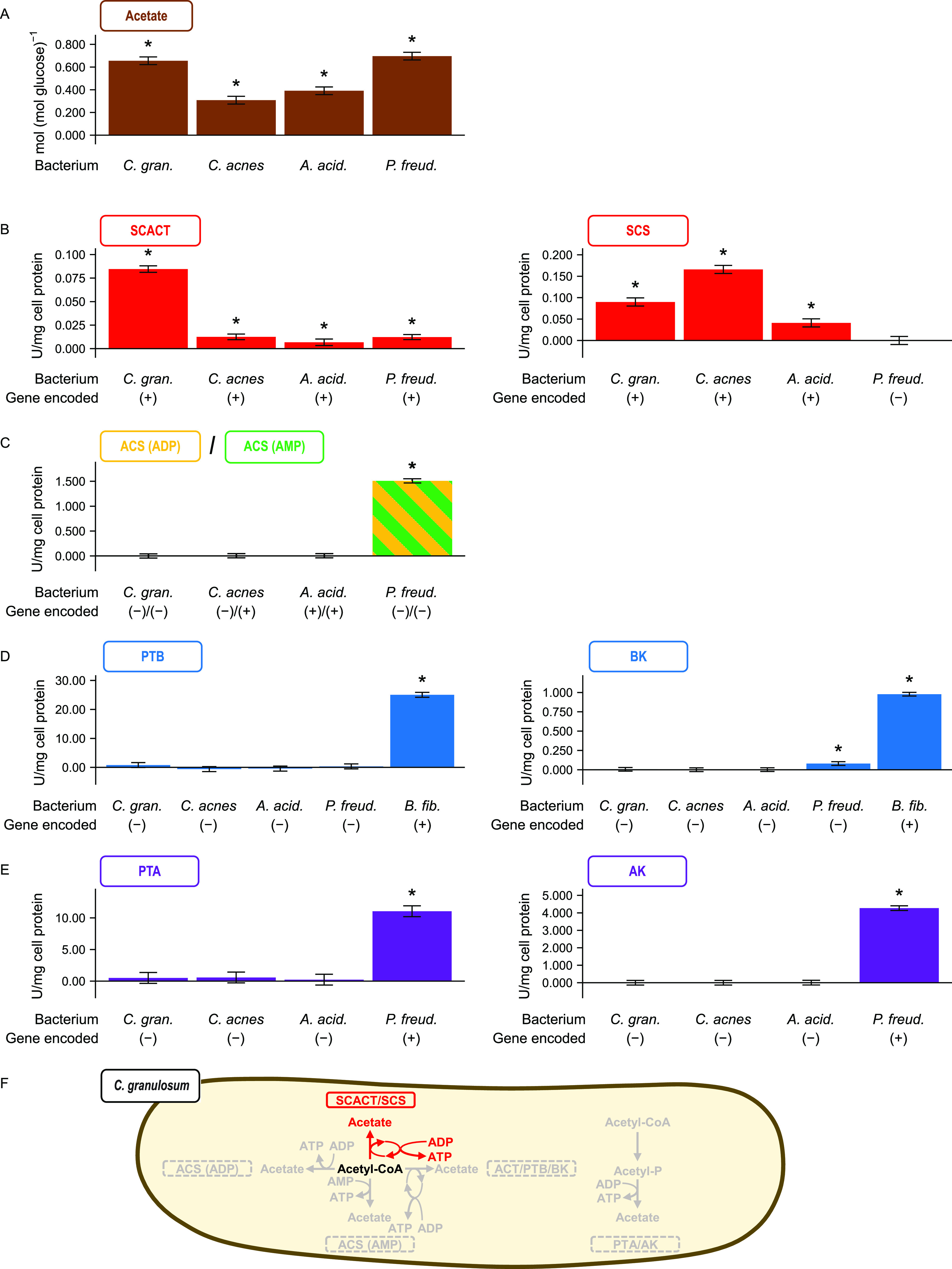
The bacterium Cutibacterium granulosum forms acetate using the SCACT/SCS pathway. Other propionibacteria are included for comparison. (A) Acetate formed during glucose fermentation. (B to E) Activity of enzymes in different pathways for forming acetate. (F) Summary of pathways in C. granulosum. In panel C, the assay cannot distinguish between the enzymes of the ACS (ADP) and ACS (AMP) pathways. In panel D, a nonpropionibacterium (Butyrivibrio fibrisolvens) was included as a control. No attempt was made to measure the activity of BCACT in the BCACT/PTB/BK pathway. Results are means ± standard errors for at least 3 biological replicates (batches of cell extract prepared from independent cultures). Means different from 0 (P < 0.05) are marked with an asterisk. Abbreviations: ACS (ADP), acetyl-CoA synthetase (ADP forming); ACS (AMP), acetyl-CoA synthetase; AK, acetate kinase; BCACT, butyryl-CoA:acetate CoA-transferase; BK, butyrate kinase; PTA, phosphotransacetylase; PTB, phosphotransbutyrylase; SCACT, succinyl-CoA:acetate CoA-transferase; SCS, succinyl-CoA synthetase (ADP-forming); A. acid., Acidipropionibacterium acidipropionici; C. acnes, Cutibacterium acnes; C. gran., Cutibacterium granulosum; P. freud, Propionibacterium freudenreichii.
To determine if the SCACT/SCS pathway was responsible for forming this acetate, we measured activities of the appropriate enzymes (Fig. 2B to E). We found high activity of both SCACT and SCS in cell extracts of C. granulosum. We did not find activity of enzymes from any other pathway to form acetate. These measurements suggest that C. granulosum forms acetate exclusively by the SCACT/SCS pathway (Fig. 2F). Similar measurements suggest that two other propionibacteria (Cutibacterium acnes and Acidipropionibacterium acidipropionici) use this pathway, whereas a third (Propionibacterium freudenreichii) does not.
To support the accuracy of these measurements, we included multiple controls. When measuring the activity of each enzyme, we included one or more bacteria that displayed activity. This supports the idea that our experimental conditions were appropriate to detect activity—i.e., we would have detected activity were any present. Additionally, we spiked cell extracts with purified enzyme at the end of experiments (see Materials and Methods). We observed activity after spiking (data not shown). This again confirms that our conditions were appropriate to detect activity.
We further supported our enzymatic measurements by analyzing which enzymes each bacterium encoded (Fig. 2B to E; also, see Data Set S1). In general, bacteria displayed high activity only when they encoded the appropriate enzymes. For example, C. granulosum encoded only SCACT and SCS and displayed activity of these enzymes only. One exception was for acetyl-CoA synthetase (ACS; EC 6.2.1.13 and EC 6.2.1.1). One bacterium (P. freudenreichii) did not encode ACS, but it still displayed high activity. This is due to interference from AK and PTA activity; our assay cannot distinguish it from ACS activity. Further, two bacteria (A. acidipropionici and P. freudenreichii) encoded ACS, but they did not display high activity. The absence of activity appears genuine, given that we included appropriate controls (see above).
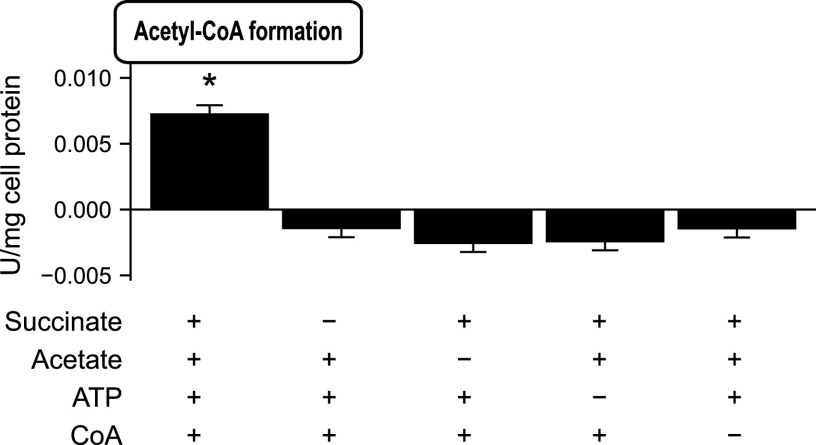
We performed one last experiment to determine if C. granulosum forms acetate through the SCACT/SCS pathway. If C. granulosum uses this pathway, it should be able to form acetyl-CoA from acetate, succinate, ATP, and CoA. Indeed, we found that cell extracts formed acetyl-CoA from these four substrates (Fig. 3). No acetyl-CoA was formed if any of the four substrates was missing, as expected. In sum, enzymatic and genomic evidence supports the idea that C. granulosum forms acetate through the SCACT/SCS pathway.
FIG 3.
The bacterium Cutibacterium granulosum forms acetyl-CoA only when provided with all four substrates of the SCACT/SCS pathway. In different treatments, substrates were provided (+) or withheld (−). Results are means ± standard errors for 3 biological replicates. Means different from 0 (P < 0.05) are marked with an asterisk. Abbreviations: CoA, coenzyme A; SCACT, succinyl-CoA:acetate CoA-transferase; SCS, succinyl-CoA synthetase (ADP forming).
C. granulosum encodes the SCACT/SCS pathway with bacterial gene homologs.
We identified the genes that encode the SCACT/SCS pathway in C. granulosum. We did this not only to confirm that C. granulosum uses this pathway but also to determine if its pathway is bacterial in origin.
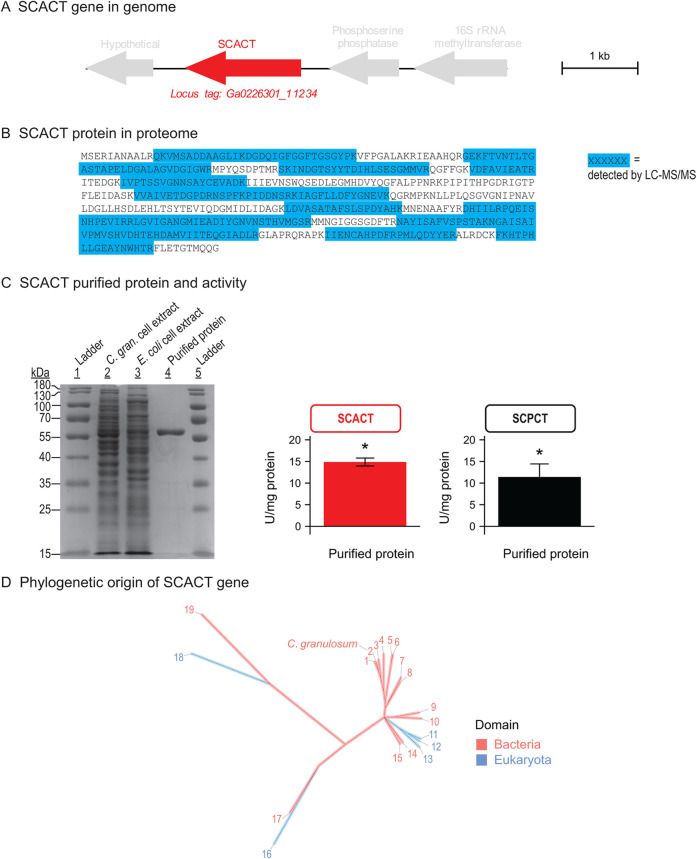
We identified the genes for SCACT and SCS using genomics, proteomics, and enzymatic assays. We searched the genome sequence of C. granulosum, and we found one candidate gene for SCACT (Fig. 4A). By performing untargeted proteomics, we confirmed that this gene was expressed (part of the bacterium’s proteome) (Fig. 4B). We expressed the protein recombinantly in Escherichia coli, and it had SCACT activity (Fig. 4C). This demonstrates that this gene encodes SCACT in C. granulosum. Following a similar process, we found the genes that encode SCS (Fig. 5A to C).
FIG 4.
The gene encoding SCACT in C. granulosum is a bacterial homolog, pointing to a bacterial origin of the SCACT/SCS pathway. (A) Location of the SCACT gene in the genome of C. granulosum. (B) Detection of SCACT protein in the proteome of C. granulosum. Specific peptides detected by LC-MS/MS are highlighted in the full protein sequence. Peptides shown were detected in each of two biological replicates (batches of protein prepared from independent cultures). (C) Production and enzymatic activity of recombinant SCACT protein in E. coli. Purity shown by SDS-PAGE was 92%. The amounts of protein loaded were 7.7, 5.8, and 0.8 μg for lanes 2, 3, and 4. Enzymatic activity is mean ± standard error of 3 biological replicates. Means different from 0 (P < 0.05) are marked with an asterisk. (D) Phylogenetic tree of SCACT protein sequences, showing that SCACT in C. granulosum is a bacterial homolog. Proteins included are those with experimental evidence of enzymatic activity. Protein sequences are as follows: 1, Acidipropionibacterium acidipropionici; 2, Cutibacterium granulosum; 3, Propionibacterium freudenreichii; 4, Corynebacterium diphtheriae; 5, Acetobacter cerevisiae; 6, Acetobacter aceti; 7, Snodgrassella alvi; 8, Moraxella catarrhalis; 9, Bacteroides fragilis; 10, Escherichia coli (homolog 1); 11, Aspergillus nidulans; 12, Saccharomyces cerevisiae; 13, Trichomonas vaginalis; 14, Acinetobacter baumannii; 15, Clostridium kluyveri; 16, Fasciola hepatica; 17, Yersinia pestis; 18, Trypanosoma brucei (homolog 1); and 19, Ralstonia eutropha. Full information on proteins is in Data Set S2. Abbreviations: C. gran., Cutibacterium granulosum; SCACT, succinyl-CoA:acetate CoA-transferase; SCPCT, succinyl-CoA:propionate CoA-transferase; SCS, succinyl-CoA synthetase (ADP forming).
FIG 5.
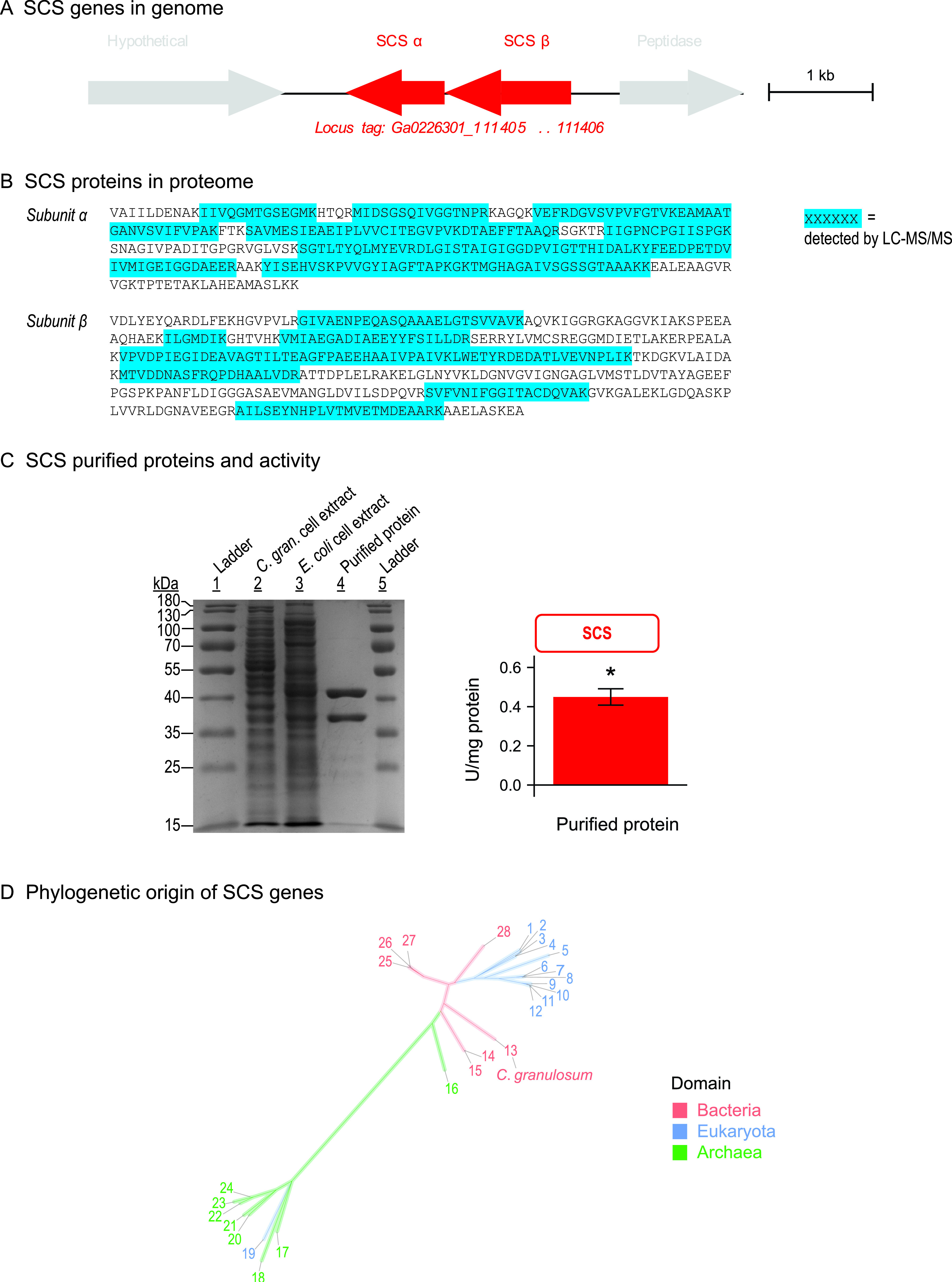
The gene encoding SCS in C. granulosum is a bacterial homolog, pointing to a bacterial origin of the SCACT/SCS pathway. The data were obtained as described for Fig. 4, except that genes are for SCS instead of SCACT. Purity shown by SDS-PAGE was 96%. The amounts of protein loaded were 7.7, 7.2, and 1.4 μg for lanes 2, 3, and 4. Protein sequences are as follows: 1, Toxoplasma gondii; 2, Blastocystis sp,; 3, Solanum lycopersicum (homolog 2); 4, Solanum lycopersicum (homolog 1); 5, Saccharomyces cerevisiae; 6, Columba livia (homolog 2); 7, Mus musculus; 8, Homo sapiens (homolog 1); 9, Columba livia (homolog 1); 10, Homo sapiens (homolog 2); 11, Sus scrofa (homolog 1); 12, Sus scrofa (homolog 2); 13, Cutibacterium granulosum; 14, Thermus thermophilus; 15, Thermus aquaticus; 16, Candidatus “Korarchaeum cryptofilum” (homolog 4); 17, Archaeoglobus fulgidus (homolog 1); 18, Archaeoglobus fulgidus (homolog 2); 19, Giardia lamblia; 20, Thermococcus kodakarensis (homolog 3); 21, Thermococcus kodakarensis (homolog 4); 22, Thermococcus kodakarensis (homolog 1); 23, Pyrococcus furiosus (homolog 5); 24, Pyrococcus furiosus (homolog 6); 25, Escherichia coli; 26, Alcanivorax borkumensis; 27, Pseudomonas aeruginosa; and 28, Advenella mimigardefordensis. Full information on proteins is in Data Set S3. Abbreviations: C. gran., Cutibacterium granulosum; SCACT, succinyl-CoA:acetate CoA-transferase; SCS, succinyl-CoA synthetase (ADP forming).
We compared SCACT and SCS in C. granulosum to all known proteins with SCACT and SCS activity. For SCACT, this included 19 proteins described in 15 publications (Data Set S2). For SCS, it included 28 proteins described in 22 publications (Data Set S3). We found that the sequences of SCACT and SCS in C. granulosum were closely related to bacterial, not eukaryotic, homologs (Fig. 4D and 5D). This suggests that the SCACT/SCS pathway in C. granulosum is bacterial, not eukaryotic, in origin.
Purified SCACT and SCS form a functional pathway.
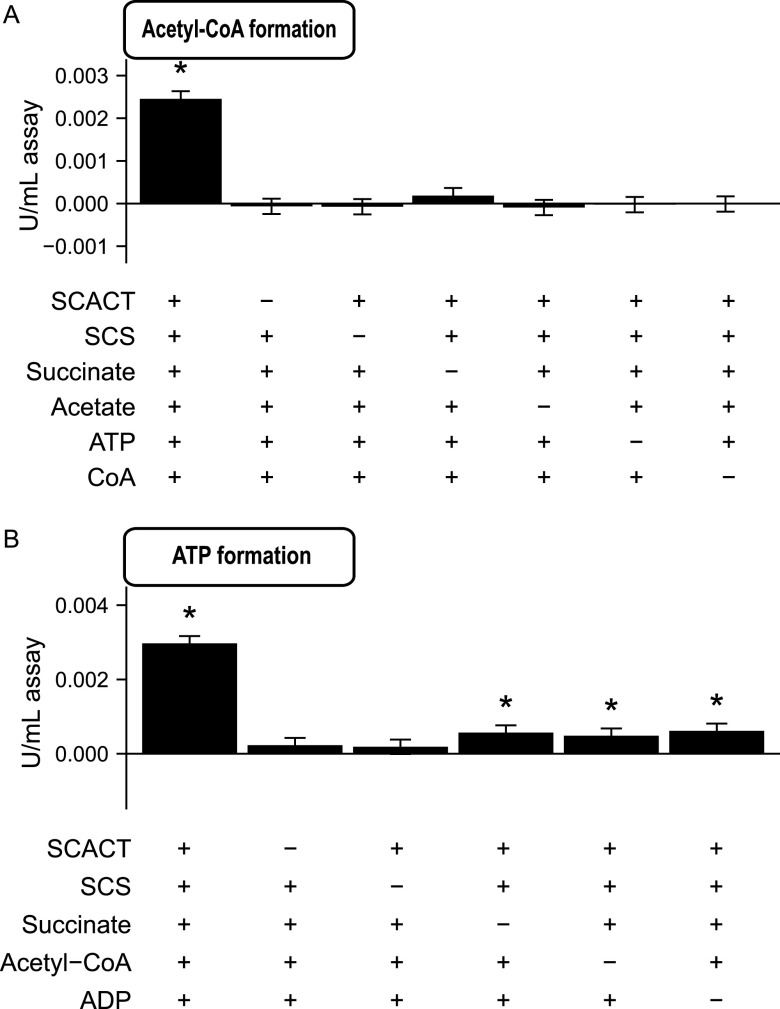
Based on their individual activities, SCACT and SCS together should form the pathway illustrated in Fig. 1. To determine if they indeed form this pathway, we determined if they form acetyl-CoA from acetate, succinate, ATP, and CoA. This experiment was similar to that performed in cell extracts, except that it used purified SCACT and SCS. We found that the enzymes together formed acetyl-CoA from these four substrates (Fig. 6A). When any enzyme or substrate was missing, no acetyl-CoA was formed (Fig. 6A). These results are consistent with the pathway in Fig. 1.
FIG 6.
Purified SCACT and SCS form a functional pathway. (A) Acetyl-CoA formation by SCACT and SCS from succinate, acetate, ATP, and CoA. (B) ATP formation by SCACT and SCS from succinate, acetyl-CoA, and ADP. In different treatments, enzymes or substrates were provided (+) or withheld (−). Each milliliter of assay mix contained 4.5 μg for SCACT and 4.5 μg for SCS. Results are mean ± standard error of 3 biological replicates (batches of SCACT and SCS prepared from independent cultures). Means different from 0 (P < 0.05) are marked with an asterisk. Abbreviations: CoA, coenzyme A; SCACT, succinyl-CoA:acetate CoA-transferase; SCS, succinyl-CoA synthetase (ADP forming).
Next, we determined if the enzymes could form ATP from acetyl-CoA, succinate, and ADP. This experiment was not possible in cell extracts because of high ATPase activity (∼100-fold higher than the expected rate of ATP formation). We found that the purified enzymes together formed ATP when all three substrates were present (Fig. 6B). Little or no ATP was formed when an enzyme or substrate was missing (Fig. 6B). In sum, SCACT and SCS in C. granulosum together form a functional pathway as illustrated in Fig. 1. This SCACT/SCS pathway forms ATP, representing a new way for bacteria to conserve energy during fermentation.
SCACT also plays a role in producing propionate.
In many organisms, SCACT forms not only acetate from acetyl-CoA but also propionate from propionyl-CoA (13–18). Specifically, it can act as a succinyl-CoA:propionate CoA-transferase (SCPCT; EC 2.8.3.−). This activity is important because it can form propionate during fermentation (17, 18). Indeed, in some eukaryotes, SCACT is thought to be responsible for forming both acetate and propionate (1, 17). We determined if SCACT played a similar role in C. granulosum.
We measured SCPCT activity in cell extracts and purified protein. In cell extracts of C. granulosum, SCPCT activity was 0.0917 (standard error of the mean [SEM], 0.012) U/mg. This was nearly equal to SCACT activity (0.0846 [SEM, 0.0043] U/mg), and the activities did not differ statistically (P = 0.562). We continued measurements with purified protein and again found that the activities of SCPCT and SCACT were nearly equal (Fig. 4C) (P = 0.246). The magnitude of SCPCT and SCACT activities is similar to that of purified enzyme of P. freudenreichii (13). The enzyme of P. freudenreichii has been shown to be responsible for forming propionate during fermentation (13, 18, 19). These findings suggest that SCACT in C. granulosum is important in forming both acetate and propionate, just as in some eukaryotes.
Several bacteria encode the SCACT/SCS pathway in their genomes.
We found that C. granulosum encodes the SCACT/SCS pathway by performing a preliminary search of bacterial genomes. To determine how many other bacteria encode this pathway, we searched for SCACT and SCS genes in 2,733 genomes of bacteria (Data Set S1). These genomes represent all type strains in Bergey's Manual of Systematics of Archaea and Bacteria (20) with an available genome sequence (21). We focused in particular on 585 genomes from bacteria experimentally observed to form acetate during fermentation (21). We searched for these genes using profile hidden Markov models (pHMMs) developed in this work (see below).
With this search, we found 36 type strains that encode the SCACT/SCS pathway and form acetate during fermentation (Fig. 7; Data Set S1). These strains belong to 5 different phyla (Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria). This suggests that many different bacteria could use the SCACT/SCS pathway to form acetate.
FIG 7.
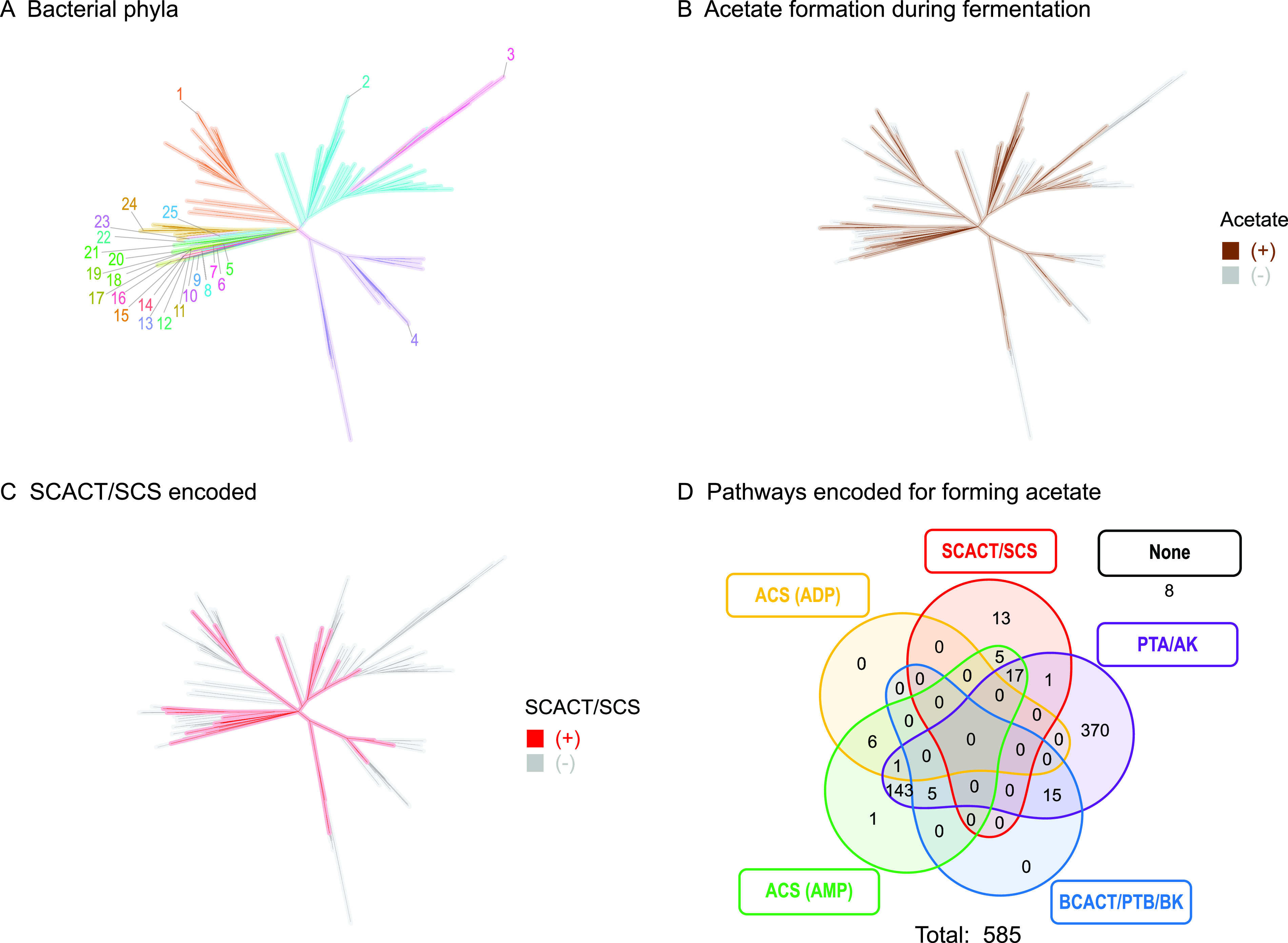
Many bacteria encode enzymes of the SCACT/SCS pathway. Phylogenetic tree of type strains of bacteria in Bergey's Manual of Systematics of Archaea and Bacteria (20), highlighting (A) phyla, (B) strains reported to form acetate during fermentation, and (C) strains that encode the SCACT/SCS pathway in their genome. (D) Venn diagram for strains that encode the SCACT/SCS versus other pathways for forming acetate. In panels C and D, only strains reported to form acetate during fermentation are included. Phyla are as follows: 1, Actinobacteria; 2, Firmicutes; 3, Tenericutes; 4, Proteobacteria; 5, Dictyoglomi; 6, Synergistetes; 7, Thermotogae; 8, Fusobacteria; 9, Lentisphaerae; 10, Spirochaetes; 11, Elusimicrobia; 12, Planctomycetes; 13, Acidobacteria; 14, Verrucomicrobia; 15, Chlamydiae; 16, Caldiserica; 17, Aquificae; 18, Coprothermobacterota; 19, Deinococcus-Thermus; 20, Chloroflexi; 21, Desulfobacterota; 22, Fibrobacteres; 23, Rhodothermaeota; 24, Ignavibacteriae; 25, Bacteroidetes. Abbreviations: ACS (ADP), acetyl-CoA synthetase (ADP forming); ACS (AMP), acetyl-CoA synthetase; AK, acetate kinase; BCACT, butyryl-CoA:acetate CoA-transferase; BK, butyrate kinase; PTA, phosphotransacetylase; PTB, phosphotransbutyrylase; SCACT, succinyl-CoA:acetate CoA-transferase; SCS, succinyl-CoA synthetase (ADP forming).
We did not find any archaea that encoded the SCACT/SCS pathway (Data Set S1). Instead, all archaea encoded the ACS (ADP-forming) pathway. With the exception of archaea, the SCACT/SCS pathway is widely distributed across the tree of life.
SCACT and SCS genes can be accurately identified.
When we first searched bacterial genomes, we failed to find many known SCACT and SCS genes. Databases had annotated many genes with the wrong enzymatic activity, or no activity at all (see Data Sets S2 and S3). Accordingly, we developed our own method, based on pHMMs, to identify genes for these enzymes.
We built and tested pHMMs using 103 biochemically characterized enzymes from 64 publications. To build a pHMM for SCACT, we used sequences of 19 proteins with SCACT activity (Data Set S2). To test it, we used these proteins, plus 32 others that are close homologs but with no SCACT activity (Data Set S2). We followed a similar process for SCS, using 28 proteins with SCS activity and 24 close homologs (Data Set S3). The proteins, phylogenetic trees, and pHMMs are in Fig. S1 and S2.
We tested the pHMM for SCACT, and it accurately predicted which proteins had SCACT activity (Fig. 8A; Data Set S2). Its accuracy also exceeded that of existing databases (Fig. 8A), including TIGRFAM (22), KEGG (23), and COG (24). We found similarly high accuracy for SCS (Fig. 8A; Data Set S3).
FIG 8.
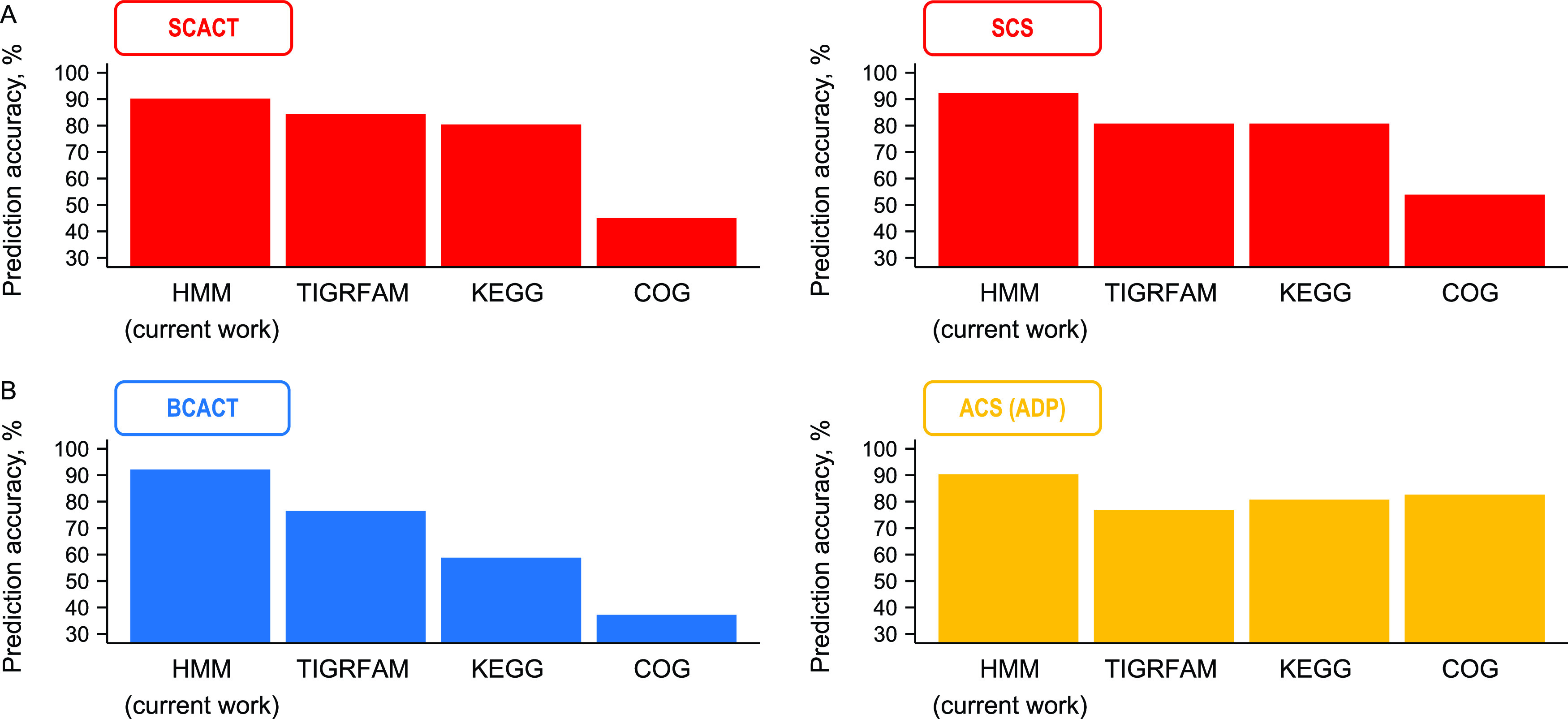
Proteins with SCACT or SCS activity can be accurately predicted using profile hidden Markov models (pHMMs) from this work. The same is observed for proteins with BCACT and ACS (ADP) activity. Accuracy of other databases (TIGRFAM, KEGG, and COG) is shown for comparison. Abbreviations: ACS (ADP), acetyl-CoA synthetase (ADP forming); BCACT, butyryl-CoA:acetate CoA-transferase; SCACT, succinyl-CoA:acetate CoA-transferase; SCS, succinyl-CoA synthetase (ADP forming). See Data Sets S2 and S3 for details of the prediction.
With the same set of 103 enzymes, we discovered that we could build pHMMs for butyryl-CoA:acetate CoA-transferase (BCACT; EC 2.8.3.8) and ACS (ADP) (EC 6.2.1.13) as well. These pHMMs gave more accurate predictions than did existing databases (Fig. 8B; Data Sets S2 and S3).
We built pHMMs using all protein sequences available, and some of the same sequences were later used to test their accuracy. Because building and testing were not independent, accuracy might be overstated. To see if this was a problem, we performed 10-fold cross validation, in which building and testing are independent. To do this, we built pHMMs with 90% of the available sequences. We then tested accuracy with the 10% of sequences we left out. We repeated this process 10 times. Accuracy was still high (90.2% for SCACT, 90.3% for SCS, 92.2% for BCACT, and 88.5% for ACS [ADP]). This supports the idea that our pHMMs would be accurate when tested with new sequences (not used for building).
In sum, we developed a method that accurately identifies proteins with SCACT or SCS activity. With this method, we could confidently search for SCACT and SCS genes in bacterial genomes (Fig. 7). This method has uses beyond the current work and can improve annotation of genes in databases.
DISCUSSION
We have found a pathway for forming acetate and synthesizing ATP that was previously unrecognized in bacteria. Pathways for forming acetate during fermentation have been studied for over 80 years (8), and so our discovery of the SCACT/SCS pathway is surprising. Nonetheless, it appears to be a common pathway, given that it is encoded by 6% of all bacteria that form acetate during fermentation.
The SCACT/SCS pathway has been thought to be exclusively eukaryotic (1). Our study with C. granulosum shows that bacteria can have the pathway, but with bacterial gene homologs. One gene homolog (SCS) originates from the tricarboxylic acid (TCA) cycle (25). The other (SCACT) is used to form propionate during fermentation (13, 18, 19), or it is part of a modified TCA cycle (25–27). In this way, the pathway may have been easy to evolve, and it is unsurprising that bacteria would have it.
We focused on C. granulosum in this study because it is one bacterium that encodes only the SCACT/SCS pathway. Earlier, we had performed experiments with Selenomonas ruminantium HD4, a rumen bacterium that encodes only this pathway (11). In those experiments, we could detect SCS but not SCACT (12). The reason is unknown, but it may be that SCACT was degraded to an acetyl-CoA hydrolase, an enzyme that we did detect. The present experiments confirm that the SCACT/SCS pathway is used in bacteria, even as it leaves the identity of the pathway in S. ruminantium unknown.
Our finding of the SCACT/SCS pathway in bacteria is important for two practical reasons. First, it is important for manipulating yield of fermentation products through genetic engineering. One study tried to engineer A. acidipropionici to produce less acetate (more propionate), but it focused on deleting AK (28). Our study suggests that enzymes of the SCACT/SCS pathway would be more appropriate targets. Indeed, other studies have decreased acetate yield by deleting SCACT, albeit in bacteria not growing by fermentation (29, 30).
Second, our finding is important for predicting metabolic pathways from genome sequences. These predictions are used to determine the metabolic capabilities of bacteria, especially ones not cultured in the lab. When studies predict metabolic pathways for fermentation, most search for enzymes of the PTA/AK pathway only (31). Some also search for enzymes of the ACS (ADP) pathway (32). The current study shows the importance of looking for enzymes of the SCACT/SCS pathway. Otherwise, studies will overlook pathways (or bacteria) that can form acetate.
In sum, our work shows that the SCACT/SCS pathway is important to many fermentative organisms. It suggests there is still much to learn about fermentation, despite its being one of the best-studied types of metabolism.
MATERIALS AND METHODS
Organisms.
The DSMZ was the source for all bacterial strains. The strains used were Acidipropionibacterium acidipropionici 4, Butyrivibrio fibrisolvens D1, Cutibacterium acnes DSM 1897, Cutibacterium granulosum VPI 0507, and Propionibacterium freudenreichii E11.1.
Media and growth.
Strains were grown anaerobically under O2-free CO2 and with Balch tubes with butyl rubber stoppers, using previously described techniques (33, 34). We used PYG medium (DSMZ medium 104). Per liter, PYG medium contained 5 g glucose, 5 g Trypticase peptone (product 211921; BD), 5 g peptone (product 211677; BD), 10 g Bacto yeast extract (product 212750; BD), 5 g beef extract (product LP0029; Oxoid), 2.04 g K2HPO4, 40 mg KH2PO4, 80 mg NaCl, 20 mg MgSO4·7H2O, 10 mg CaCl2·2H2O, 1 ml Tween 80, 5 mg hemin, 1 μl vitamin K1, 4 g NaHCO3, and 449 mg cysteine-HCl. Resazurin was added as a redox indicator. The hemin was added as a 0.5-g/liter solution containing 10 mM NaOH as the diluent. The vitamin K1 was added as a 5-ml/liter solution containing 95% ethanol as the diluent.
B. fibrisolvens D1 was grown on DSMZ medium 712. Per liter, DSMZ medium 712 contained 9 g glucose, 8.3 g Bacto yeast extract (product 212750, BD), 383 mg K2HPO4, 384 mg KH2PO4, 751 mg (NH4)2SO4, 760 mg NaCl, 73 mg MgSO4, 100 mg CaCl2·2H2O, 10.669 g NaHCO3, 16.7 ml vitamin solution, and 1.616 g cysteine hydrochloride monohydrate. Per liter, the vitamin solution contained 2 mg biotin, 2 mg folic acid, 10 mg pyridoxine-HCl, 5 mg thiamine-HCl, 5 mg riboflavin, 5 mg nicotinic acid, 5 mg d-Ca-pantothenate, 0.1 mg vitamin B12, 5 mg p-aminobenzoic acid, and 5 mg lipoic acid. Resazurin was added as a redox indicator.
The temperature for A. acidipropionici and P. freudenreichii was 30°C. The temperature for B. fibrisolvens, C. acnes, and C. granulosum was 37°C.
Cell extracts.
Eight 9-ml cultures were grown to mid-exponential phase and then pooled. Cells were harvested by centrifugation (12,000 × g for 10 min at 4°C; F15-8x50cy rotor and Sorvall Legend XTR centrifuge), washed twice in buffer (50 mM Tris [pH 7.2] and 10 mM MgCl2), and resuspended to 3.8 ml in the same buffer. All steps after growing the cultures were performed aerobically.
The resuspended cells were lysed with a French press (Glen Mills). The resuspended cells were transferred to a mini-cell pressure cell and lysed at 110 MPa. Cell debris was removed by centrifugation (12,000 × g for 15 min at 4°C).
Cell extract was prepared three or more different times for each bacterial strain. It was stored at −80°C until use.
Supernatant.
One 9-ml culture was grown to mid-exponential phase. Cells were removed from supernatant by centrifugation (12,000 × g for 10 min at 4°C).
Supernatants were prepared three or more different times for each bacterial strain. They were stored at −20°C until use.
Enzymatic assays.
We measured enzymatic activity of cell extracts as well as purified SCACT and SCS (see below). Assays were performed at room temperature under the conditions in Table 1. Assay products were monitored by measuring absorbance with a Molecular Devices M3 spectrophotometer. The path length of the assay mix in 96-well plates (0.288 cm) was determined using a solution of known absorbance (NADH). Activity was calculated over the time that absorbance increased (or decreased) linearly. Values were corrected by subtracting the activity of controls (where water replaced cell extract, substrate, or cofactor). One unit of activity is defined as 1 μmol of product formed per min.
TABLE 1.
Conditions used to measure enzymatic activity
| Enzyme or reaction | Reference | Assay mixture components | Product measured (wavelength) | Control | |
|---|---|---|---|---|---|
| Succinyl-CoA:acetate CoA-transferase of C. granulosum (SCACT; EC 2.8.3.18) | Modified from 64 | 50 mM KPO4 buffer (pH 8.0), 100 mM KCl, 1.5 mM 5,5′-dithiobis-(2-nitrobenzoic acid), 30 U/ml phosphotransacetylase,a 300 mM potassium acetate (pH 7.0), 1.5 mM succinyl-CoA | 5-Thio-2-nitrobenzoic acid (412 nm)b | Cell extract replaced with water | |
| Succinyl-CoA:acetate CoA-transferase of other strains (SCACT; EC 2.8.3.18) | 13 | 100 mM Tris (pH 8.0), 0.6 mM succinyl-CoA, 60 mM potassium acetate (pH 7.0), 0.4 mM l-malate, 1 mM NAD, 2.84 mM KPO4 (pH 6.8), 8.8 U/ml malate dehydrogenase,c 1.4 U/ml citrate synthased | Reduced NAD (340 nm)e | Cell extract replaced with water | |
| Succinyl-CoA:propionate CoA-transferase (SCPCT; EC 2.8.3.-) | Modified from 64 | 50 mM KPO4 buffer (pH 8.0), 100 mM KCl, 1.5 mM 5,5′-dithiobis-(2-nitrobenzoic acid), 30 U/ml phosphotransacetylase,a 300 mM potassium propionate (pH 7.0), 1.5 mM succinyl-CoA | 5-Thio-2-nitrobenzoic acid (412 nm)b | Cell extract replaced with water | |
| Succinyl-CoA synthetase (SCS; EC 6.2.1.5) | 65 | 50 mM Tris (pH 7.2), 10 mM MgCl2, 10 mM succinate (pH 7.0), 1.2 mM ATP, 0.4 mM CoA, 200 mM hydroxylamine (pH 7.0) | Succinohydroxamate (540 nm)f | Succinate replaced with water | |
| Acetyl-CoA synthetase (ACS) (EC 6.2.1.13, EC 6.2.1.1) | 65 | 50 mM Tris (pH 7.2), 10 mM MgCl2, 40 mM potassium acetate (pH 7.0), 1.2 mM ATP, 0.4 mM CoA, 200 mM hydroxylamine (pH 7.0) | Aceto-hydroxamate (540 nm)g | Potassium acetate replaced with water | |
| Phosphotransbutyrylase (PTB; EC 2.3.1.19) | Modified from 66 | 76 mM Tris (pH 7.6), 10 mM NH4Cl, 2.5 mM butyryl phosphate, 0.25 mM CoA | Butyryl-CoA (233 nm)h | Butyryl phosphate replaced with water | |
| Butyrate kinase (BK; EC 2.7.2.7) | Modified from 65 | 50 mM Tris (pH 7.2), 10 mM MgCl2, 200 mM butyrate (pH 7.0), 5 mM ATP, 200 mM hydroxylamine (pH 7.0) | Butyryl-hydroxamate (540 nm)g |
Butyrate replaced with water | |
| Phosphotransacetylase (PTA; EC 2.3.1.8) | 66 | 76 mM Tris (pH 7.6), 10 mM NH4Cl, 5 mM acetyl phosphate, 0.25 mM CoA | Acetyl-CoA (233 nm)h | Acetyl phosphate replaced with water | |
| Acetate kinase (AK; EC 2.7.2.1) | Modified from 65 | 50 mM Tris (pH 7.2), 10 mM MgCl2, 100 mM potassium acetate (pH 7.0), 5 mM ATP, 200 mM hydroxylamine (pH 7.0) | Aceto-hydroxamate (540 nm)g | Potassium acetate replaced with water | |
| Acetyl-CoA formation of cell extract | Modified from 13 | 100 mM Tris (pH 8.0), 10 mM MgCl2, 2.84 mM KPO4 (pH 6.8), 1 mM NAD, 4 mM l-malate, 8.8 U/ml malate dehydrogenase, 1.4 U/ml citrate synthase, 30 mM succinate (pH 7.0), 60 mM potassium acetate (pH 7.0), 5 mM ATP, and 2 mM CoA | Reduced NAD (340 nm)e | None | |
| Acetyl-CoA formation of purified SCACT and SCS | Modified from 13 | 100 mM Tris (pH 8.0), 2.84 mM KPO4 (pH 6.8), 10 mM MgCl2, 4 mM l-malate, 1 mM NAD, 8.8 U/ml malate dehydrogenase, 1.4 U/ml citrate synthase, 5 mM succinate (pH 7.0), 10 mM potassium acetate (pH 7.0), 5 mM ATP, and 0.4 mM CoA | Reduced NAD (340 nm)e | None | |
| ATP formation | Modified from 67 | 38 mM triethanolamine (pH 7.4), 3 mM KPO4 (pH 6.8), 6.6 mM MgCl2, 2 mM NADP, 1 mM acetyl-CoA, 25 mM succinate (pH 7.0), 5 mM ADP, 50 mM glucose, 2 U/ml glucose-6-phosphate dehydrogenase,i and 5.4 U/ml hexokinasej | Reduced NADP (340 nm)e | None | |
From Bacillus subtilis (Megazyme E-PTABS).
Extinction coefficient of 14,150 M−1 cm−1 (68).
From porcine heart (Sigma M1567-5KU).
From porcine heart (Sigma C3260-200UN).
Extinction coefficient of 6,220 M−1 cm−1.
Succinic anhydride was used as a standard and prepared in 50 mM Tris (pH 7.2), 10 mM MgCl2, and 200 mM hydroxylamine (69).
Acetyl phosphate was used as a standard and prepared in 50 mM Tris (pH 7.2), 10 mM MgCl2, and 200 mM hydroxylamine (pH 7.0).
Extinction coefficient of 4,440 M−1 cm−1 (value for S-acetyl-N-succinyl cysteamine) (70).
From Saccharomyces cerevisiae (Sigma G7877-250UN).
From Saccharomyces cerevisiae (Sigma H4502-1KU).
Several assays (for SCS, ACS, butyrate kinase [BK], and AK) measured formation of hydroxamic acids. For these assays, we stopped the reaction at intervals by the addition of 0.286 volume of development solution (0.25 M FeCl3, 2.5 M HCl, and 15% [wt/vol] trichloroacetic acid) (35). Afterwards, we held the assay mix for 5 to 60 min at room temperature, centrifuged it at 18,000 × g for 2 min, and then measured absorbance of the supernatant.
One assay measured formation of acetyl-CoA. The amount of acetyl-CoA formed was calculated from reduced NAD and the equation of Decker (71). The assay mix sometimes formed a precipitate when cell extract was added, and this interfered with measurements. The cause could not be determined, and we discarded results from any affected experiments.
One assay measured formation of ATP from ADP and other substrates. We found that our source of ADP was contaminated with trace amounts of ATP, which interfered with measurements. To overcome this interference, the assay mixture was preincubated for 80 min to allow all ATP to be consumed. Only after this preincubation was the sample (purified SCACT and SCS) added.
When available, purified enzymes were used as additional controls. Specifically, assay mixtures were spiked with purified enzyme at the end of the incubation. The enzymes were obtained from Megazyme. They were SCS from an unspecified prokaryotic source (product code E-SCOAS), ACS (AMP-forming) from Bacillus subtilis (product code E-ACSBS), PTA from B. subtilis (product code E-PTABS), and AK from an unspecified source (product code K-ACETRM).
We determined if activity was different from 0 using an analysis of variance (ANOVA). The statistical model was as follows: Yij = μ + Ti + εij, where Yij is the observation, μ is the overall mean, Ti is the fixed effect of treatment (bacterial strain), and εij is the residual error. For assays measuring acetyl-CoA or ATP formation, Ti refers to treatments with different combinations or substrates or enzymes. All analyses were conducted in R. The model was fit using the aov function. Least-squares means, SEM, and degrees of freedom were taken from the emmeans package. P values were calculated using a one-sided t test.
For purified SCACT and SCS, we determined if activity was different from 0 using a one-sided t test. We determined if SCACT and SCPCT activities were different with a paired, two-sided t test.
Cloning of genes.
Genes encoding SCACT and SCS in C. granulosum were cloned into a plasmid vector. First, genes were PCR amplified from cell extracts of C. granulosum. For SCACT, forward and reverse primers were AAGAAGGAGATATACATATGTCAGAGCGGATTGCCAATGCAG and CAGTGGTGGTGGTGGTGGTGGCCCTGCTGCATGGTGCC. For SCS, they were AAGAAGGAGATATACATATGGACCTGTATGAATACCAAGCC and CAGTGGTGGTGGTGGTGGTGCTTCTTGAGCGACGCCATC. The primers for SCS amplified both α and β subunits as part of the same fragment. Amplification was done with Q5 Hot Start high-fidelity DNA polymerase (New England BioLabs M0493S).
The amplified fragment had overlap sequences with pET-30a-Novagen plasmid (Sigma-Aldrich 69909) and was inserted into the plasmid. The plasmid fragment was PCR-amplified using forward primer CACCACCACCACCACCAC and reverse primer CATATGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGAGG. The insertion was done with NEBuilder HiFi DNA assembly master mix (New England BioLabs E2621S). To enable later purification, the fragment was inserted upstream of a hexahistidine tag sequence.
The assembled plasmids were chemically transformed into E. coli 5α competent cells (New England BioLabs C2987I). Sanger sequencing showed that the transformed plasmid had the correct insert.
Production and purification of proteins.
We produced SCACT and SCS proteins from C. granulosum by using E. coli as the expression system. Plasmids with SCACT or SCS genes were chemically transformed into E. coli BL21(DE3)pLysS (Promega L1191). After transformation, E. coli cells were grown overnight in Terrific Broth (TB) medium with 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. Next, 16 ml of this overnight culture was used to inoculate 0.4 liter of TB medium with kanamycin and chloramphenicol. Cells from the 0.4-liter culture were grown at 37°C to an optical density at 600 nm (OD600) of 0.6 to 0.7, and then the culture bottle was placed in ice water for 4 min with gentle shaking to decrease the culture temperature. After that, expression was induced with 0.1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) for 10 h at 23 to 25°C. Cells were harvested by centrifugation (12,000 × g for 5 min at 4°C), washed with buffer (50 mM Tris [pH 7.2] and 10 mM MgCl2), and then stored at −80°C.
Protein was purified from harvested cells using affinity chromatography. The frozen cells were lysed by thawing and vortex mixing in 12 ml of binding buffer (see below). About 100 U of Pierce Universal nuclease (product PI88700; Fisher) was added to the mixture and incubated on ice for 30 min with frequent vortexing. Then, the mixture was centrifuged at 12,000 × g for 30 min at 4°C. The supernatant (12 ml) was filtered through a 0.2-μm membrane, mixed with 1 ml 50% (vol/vol) slurry of Profinity IMAC resin (Bio-Rad 1560131) for 1.5 h at 4°C with gentle shaking, and transferred to a chromatography column (Bio-Rad 7311550). The column was washed with 13 ml of binding buffer, 2 ml of washing buffer, and 8 ml of elution buffer. After purification, protein was concentrated with an Amicon Ultra-15 centrifugal filter (3-kDa cutoff). It was used immediately or stored at −80°C.
All buffers for affinity chromatography contained 50 mM Tris (pH 7.2) and 100 mM NaCl. Binding buffer additionally contained 10 mM imidazole (pH 8.0). Washing buffer contained either 50 mM imidazole (SCACT) or 10 mM imidazole (SCS). Elution buffer contained either 100 mM imidazole (SCACT) or 20 mM imidazole (SCS).
Purity was checked using SDS-PAGE (12% acrylamide, 29:1 acrylamide–bis-acrylamide) and staining with Coomassie brilliant blue G-250. The sequence of proteins was correct when determined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) (see below).
Untargeted proteomics.
We used LC-MS/MS to determine whether SCACT and SCS proteins were expressed in cell extracts of C. granulosum. We also used it to verify the amino acid sequence of recombinant SCACT and SCS.
Proteins in the sample (e.g., cell extract) were prepared for LC-MS/MS. Proteins were precipitated by adding 1,000 μl of chilled 15% (wt/vol) trichloroacetic acid and 0.2% (wt/vol) dithiothreitol in acetone to 250 μl of sample and incubating at −80°C for 20 min. Incubation was continued at −20°C overnight. Proteins were harvested by centrifugation (21,100 × g for 20 min at 4°C), air dried to remove acetone, and then resuspended in 100 μl of 6 M urea in 50 mM ammonium bicarbonate (AMBIC). They were reduced with 2.5 μl of 200 mM dithiothreitol in AMBIC at 37°C for 30 min and then alkylated with 7.7 μl of 194.6 mM iodoacetamide in AMBIC for 30 min at room temperature in the dark. Alkylation was stopped by adding 20 μl of 200 mM dithiothreitol in AMBIC and incubating at room temperature for 10 min. Proteins were digested to peptides with trypsin/Lys-C mix (Promega V5073) in a 1:25 enzyme-to-protein (wt/wt) ratio at 37°C for 4 h. The trypsin in the mixture was activated by adding 550 μl of AMBIC and then continued for 16 h at 37°C. Digestion was terminated by adding trifluoroacetic acid (TFA) to a final concentration of 1% (vol/vol). Peptides were desalted with Millipore C18 ZipTips (ZTC18S096). The tip was wet by slowly aspiring 10 μl of 100% (vol/vol) acetonitrile (ACN), discarding the solvent, and repeating once. The tip was then equilibrated by aspiring 10 μl of 0.1% TFA, discarding solvent, and repeating once. The digested sample containing 1% TFA was slowly aspirated in and out of the tip for 10 cycles to maximize binding. The tip was rinsed with 20 μl of 0.1% TFA–5% ACN and eluted with 100 μl of 0.1% (vol/vol) acetic acid–60% ACN. The eluted peptides were dried by vacuum centrifugation and resuspended in 40 μl of 0.1% TFA.
The resulting peptides were analyzed using LC-MS. The LC was a Dionex UltiMate 3000 RSLC system (Thermo Fisher) equipped with a PepMap C18 column (75 μm by 25 cm with a 2-μm pore size; Thermo Scientific). The amount of peptide injected was 1 μg, the flow rate of the mobile phase was 200 μl/min, and the column temperature was 40°C. The mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B), and they were used in a gradient elution. The concentration of B was decreased from 10% to 8% over 3 min, increased to 46% over 66 min, increased to 99% over 3 min, held at 99% for 2 min, decreased to 2% over 0.5 min, and held at 2% for 15 min.
The MS was an Orbitrap Fusion Lumos (Thermo Scientific). The instrument was operated in a data-dependent acquisition mode. Peptides were ionized and transferred to the mass analyzer with a spray voltage of 1.8 kV, radio frequency lens level of 46%, and ion transfer tube temperature of 275°C. Survey full-scan MS spectra were then acquired with an m/z range of 375 to 1,575, resolution of 60,000, automatic gain control (AGC) target of 4 × 105, and ion filling time of 50 ms. After the survey scan, MS/MS was performed on the most abundant precursors with a charge state between 2 and 3. Precursors were isolated using a 3-s cycle with window width of 1.2 m/z. Fragmentation of precursors was done by collisionally induced dissociation (CID) with normalized collision energy of 30%, and the resulting fragments were detected using the high rate in the ion trap. MS/MS spectra were acquired with AGC target of 5 × 103, ion filling time of 35 ms, and dynamic exclusion time of 50 s with a 10-ppm mass window.
Peptides and proteins were identified from LC-MS/MS data using X!TandemPipeline (36). The version of X! Tandem (37) used was Alanine. The sequence database contained all proteins in C. granulosum predicted by IMG/M (38). Parameters were set according to Data Set S4.
Analysis of supernatant and media.
Glucose concentration was measured with the glucose oxidase-peroxidase method (39). To overcome interference from cysteine, 2 mol N-ethylmaleimide was added per mol cysteine in samples (40, 41).
Acetate was measured enzymatically. Before analysis, supernatant was heated at 100°C for 10 min to inactivate enzymes. The assay mix contained 150 mM triethanolamine (pH 8.4), 3 mM MgCl2, 10 mM l-malate, 1 mM NAD, 5 mM ATP, 0.4 mM CoA, 1.8 U/ml malate dehydrogenase (product code M1567-5KU; Sigma), 1.4 U/ml citrate synthase (product code C3260-200UN; Sigma), and 0.5 U/ml ACS (AMP forming) (product code E-ACSBS; Megazyme). Supernatant was replaced with water as a control. Reduced NAD was measured, and acetyl-CoA formed was calculated as above.
Formate was measured using an assay mix containing 50 mM KPO4 buffer (pH 7.6), 4 mM NAD, and 0.5 U/ml formate dehydrogenase (product code E-FDHCB; Megazyme). Reduced NAD was measured. Whereas most assays were performed in 96-well plates, this assay was performed in microcuvettes (product code 13-878-122; Fisher). Absorbance was measured with a Thermo Scientific Genesys 20 spectrophotometer.
Succinate, l-lactate, and d-lactate were measured using commercial kits (product code 10176281035 and 11112821035; R-Biopharm).
Other fermentation products (propionate, butyrate, isobutyrate, and isovalerate) were measured using gas chromatography. Samples were prepared by combining supernatant (350 μl), 10 mM 2-ethylbutyric acid (50 μl), formic acid (100 μl), and methanol (500 μl). The gas chromatograph was a Trace 1300 equipped with AI 1310 autosampler, split/splitless injector, and flame ionization detector (FID) (Thermo Scientific). The column was a TraceGOLD-WaxMS (30 m by 0.32 mm [inside diameter] coated with a 1-μm film thickness; Thermo Scientific). N2 (2.5 ml/min) was the carrier gas. The injection was performed in splitless mode (1 min splitless time). The front inlet had a temperature of 230°C. The oven temperature was initially 45°C, maintained for 0.5 min, raised to 235°C at 25°C/min, and finally held at 235°C for 2 min. The FID had a temperature of 240°C and flow rates for air, hydrogen, and nitrogen of 350, 35, and 40 ml/min. The injected sample volume was 1 μl, and the total run time for each analysis was 10.1 min. Data handling was carried out with Chromeleon Chromatography Data System software (Thermo Scientific).
The amount of glucose consumed was calculated from concentrations measured in the medium and supernatant. The amount of fermentation products formed was calculated similarly. Least-squares means and SEM were calculated as for enzymatic assays.
Other analyses.
Protein concentration in the cell extract and purified proteins was measured using a Pierce bicinchoninic acid (BCA) protein assay kit (product 23227; Thermo Scientific).
Annotation of SCACT genes and homologs.
We annotated SCACT genes and homologs with KEGG Orthology (KO) (23), TIGRFAM (22), Pfam (42), or COG (24) IDs. For TIGRFAM and Pfam, annotation was done using HMMER v. 3.3 and hmmscan on the HMMER webserver (43). For KEGG, annotation was done with KofamKOALA (44). For COG, annotation was done with NCBI conserved domain search (45) with COG v.1.0.
Genes were annotated with IDs producing significant bit scores (as defined by the database). For Pfam, outcompeted hits were not kept. For COG, only specific hits were kept.
We also annotated these genes with pHMMs built in this work. Genes were annotated by using pHMMs along with HMMER and hmmsearch (46). For SCACT, a significant bit score was ≥700. For BCACT, it was ≥550.
The pHMMs were built using HMMER and hmmbuild. Protein sequences included are indicated in Fig. S1 and S2, and they were aligned with Clustal Omega (47, 48) beforehand.
From annotations, we predicted if genes had SCACT or other enzymatic activities. Predicted activity was compared against that observed experimentally. The accuracy of prediction was calculated as (TP + TN)/(TP + TN + FP + FN), where TP is true positive, TN is true negative, FP is false positive, and FN is false negative.
Annotation of SCS genes and homologs.
We annotated SCS genes and homologs following the process for SCACT. For SCS, a significant bit score was ≥340 for the α subunit and ≥190 for the β subunit. For ACS, it was ≥550 and ≥270 for the α and β subunits.
Some SCS homologs had α and β subunits that were fused (part of the same protein). To include these subunits in pHMMs, we had to identify the boundaries of the subunits. We did this by using a separate pHMM built with sequences of nonfused subunits.
Gene searches in bacterial genome sequences.
We searched genomes of bacteria and archaea for genes encoding pathways of acetate formation. We used a set of genomes assembled in reference 21. This set includes genomes for 2,925 type strains of bacteria and archaea, including 590 strains that form acetate during fermentation. See Data Set S1 and reference 21 for details.
We performed searches using the IMG/M database (38), the IMG/M genome ID for each genome (Data Set S1), and the KEGG Orthology (KO) ID for each gene (23) (Data Set S5). The locus tags found by these searches are in Data Set S1.
We searched for acetyl-CoA synthetase genes (SCACT and BCACT) using a different method. We searched genomes for these genes first using IMG/M and pfam IDs (42) (Data Set S5). The pfam ID was used because it was the most sensitive; all known acetate-CoA transferase genes were annotated with pfam IDs. We downloaded all sequences and then annotated them as BCACT or SCACT with pHMMs described above. A more direct approach would have been to search IMG/M using the pHMMs, but IMG/M does not support this type of search.
We searched for acyl-CoA synthetase genes (SCS and ACS) similarly to acetyl-CoA transferase genes. Instead of pfam IDs, we used COG IDs to search IMG/M (Data Set S5). We annotated them as SCS or ACS with pHMMs.
Following our previous work (11), we considered that a pathway was encoded if genes for all enzymes was found. If an enzyme had multiple genes (subunits), all genes for that enzyme had to be found. If a gene had multiple domains, database IDs for all domains had to be found also. If a pathway could be catalyzed by multiple isozymes, genes for only one isozyme had to be found.
Construction of phylogenetic trees.
We constructed a phylogenetic tree of the genomes described above. The construction used sequences of 14 ribosomal proteins and was done as in reference 21. Briefly, we downloaded amino acid sequences of the ribosomal proteins from IMG/M (38). We aligned sequences and concatenated these sequences in R. We then used aligned and concatenated sequences to create a phylogenetic tree with RAxML (49). Branch lengths of the consensus tree were calculated using phytools (50). The consensus tree was visualized using ggtree (51). Because no archaea encoded the SCACT/SCS pathway, they were not included in the final visualization of the tree. A total of 2,464 genomes were included in this visualization.
We constructed phylogenetic trees of SCACT and SCS homologs using the same approach as for genome sequences. For SCS, we concatenated α and β subunits (after alignment).
Data availability.
The LC-MS/MS data presented in this paper have been deposited in the Proteomics Identification (PRIDE) Archive with the project accession PXD021231. The pHMMs presented in this paper have been deposited at figshare (https://figshare.com/articles/dataset/Profile_hidden_Markov_models_for_succinyl-CoA_acetate_CoA-transferase_and_other_enzymes/14442749).
ACKNOWLEDGMENTS
We thank Gabriela Grigorean of the UC Davis Proteomics Core for performing LC-MS/MS analysis.
This work was supported by an Agriculture and Food Research Initiative Competitive Grant (grant no. 2018-67015-27495) and Hatch Project (accession no. 1019985) from the United States Department of Agriculture National Institute of Food and Agriculture.
Footnotes
Supplemental material is available online only.
Contributor Information
Timothy J. Hackmann, Email: tjhackmann@ucdavis.edu.
M. Julia Pettinari, University of Buenos Aires.
REFERENCES
- 1.Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin WF. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76:444–495. 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottschalk G. 1986. Bacterial metabolism. Springer Verlag, New York, NY, USA. [Google Scholar]
- 3.White D, Drummond J, Fuqua C. 2012. The physiology and biochemistry of prokaryotes. Oxford University Press, New York, NY. [Google Scholar]
- 4.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD. 2016. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44:D471–D480. 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergman EN. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 70:567–590. 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Lin M, Xu M, Yang S-T. 2016. Anaerobic fermentation for production of carboxylic acids as bulk chemicals from renewable biomass, p 323–361. In Hatti-Kaul R, Gashaw M, Mattiasson B (ed), Anaerobes in biotechnology. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 7.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41:100–180. 10.1128/BR.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipmann F. 1939. An analysis of the pyruvic acid oxidation system. Cold Spring Harbor Symp Quant Biol 7:248–259. 10.1101/SQB.1939.007.01.021. [DOI] [Google Scholar]
- 9.Lindmark DG. 1976. Acetate production by Tritrichomonas foetus, p 15–21. In Van den Bossche H (ed), Biochemistry of parasites and host-parasite relationships. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 10.Tielens AG, van Grinsven KW, Henze K, van Hellemond JJ, Martin W. 2010. Acetate formation in the energy metabolism of parasitic helminths and protists. Int J Parasitol 40:387–397. 10.1016/j.ijpara.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Hackmann TJ, Ngugi DK, Firkins JL, Tao J. 2017. Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ Microbiol 19:4670–4683. 10.1111/1462-2920.13929. [DOI] [PubMed] [Google Scholar]
- 12.McCourt C. 2019. Pathways for fermenting lactate to acetate in a rumen bacterium. M.S. thesis. University of Florida, Gainesville, FL. [Google Scholar]
- 13.Schulman M, Wood HG. 1975. Succinyl-CoA: propionate CoA-transferase from Propionibacterium shermanii. EC 2.8.3.6 succinyl-CoA:propionate CoA-transferase. Methods Enzymol 35:235–242. 10.1016/0076-6879(75)35159-8. [DOI] [PubMed] [Google Scholar]
- 14.Fleck CB, Brock M. 2008. Characterization of an acyl-CoA:carboxylate CoA-transferase from Aspergillus nidulans involved in propionyl-CoA detoxification. Mol Microbiol 68:642–656. 10.1111/j.1365-2958.2008.06180.x. [DOI] [PubMed] [Google Scholar]
- 15.Fleck CB, Brock M. 2009. Re-characterisation of Saccharomyces cerevisiae Ach1p: fungal CoA-transferases are involved in acetic acid detoxification. Fungal Genet Biol 46:473–485. 10.1016/j.fgb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA. 2014. Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10:371–377. 10.1038/nchembio.1482. [DOI] [PubMed] [Google Scholar]
- 17.van Grinsven KW, van Hellemond JJ, Tielens AG. 2009. Acetate:succinate CoA-transferase in the anaerobic mitochondria of Fasciola hepatica. Mol Biochem Parasitol 164:74–79. 10.1016/j.molbiopara.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Allen SH, Kellermeyer RW, Stjernholm RL, Wood HG. 1964. Purification and properties of enzymes involved in the propionic acid fermentation. J Bacteriol 87:171–187. 10.1128/JB.87.1.171-187.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deborde C, Rolin DB, Boyaval P. 1999. In vivo 13C NMR study of the bidirectional reactions of the Wood-Werkman cycle and around the pyruvate node in Propionibacterium freudenreichii subsp. shermanii and Propionibacterium acidipropionici. Metab Eng 1:309–319. 10.1006/mben.1999.0124. [DOI] [PubMed] [Google Scholar]
- 20.Whitman WB (ed). 2020. Bergey's manual of systematics of archaea and bacteria. Wiley, Oxford, United Kingdom. https://onlinelibrary.wiley.com/doi/book/10.1002/9781118960608. Accessed 15 April 2020. [Google Scholar]
- 21.Hackmann TJ, Zhang B. 2021. Using neural networks to mine text and predict metabolic traits for thousands of microbes. PLoS Comput Biol 17:e1008757. 10.1371/journal.pcbi.1008757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2012. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D395. 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thauer RK. 1988. Citric-acid cycle, 50 years on. Modifications and an alternative pathway in anaerobic bacteria. Eur J Biochem 176:497–508. 10.1111/j.1432-1033.1988.tb14307.x. [DOI] [PubMed] [Google Scholar]
- 26.Kwong WK, Zheng H, Moran NA. 2017. Convergent evolution of a modified, acetate-driven TCA cycle in bacteria. Nat Microbiol 2:17067. 10.1038/nmicrobiol.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullins EA, Francois JA, Kappock TJ. 2008. A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J Bacteriol 190:4933–4940. 10.1128/JB.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suwannakham S, Huang Y, Yang ST. 2006. Construction and characterization of ack knock-out mutants of Propionibacterium acidipropionici for enhanced propionic acid fermentation. Biotechnol Bioeng 94:383–395. 10.1002/bit.20866. [DOI] [PubMed] [Google Scholar]
- 29.Litsanov B, Brocker M, Bott M. 2012. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337. 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuda K, Jojima T, Suda M, Okino S, Inui M, Yukawa H. 2007. Analyses of the acetate-producing pathways in Corynebacterium glutamicum under oxygen-deprived conditions. Appl Microbiol Biotechnol 77:853–860. 10.1007/s00253-007-1199-y. [DOI] [PubMed] [Google Scholar]
- 31.Magnusdottir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, Greenhalgh K, Jager C, Baginska J, Wilmes P, Fleming RM, Thiele I. 2017. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol 35:81–89. 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 32.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JF. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665. 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 33.Tao J, McCourt C, Sultana H, Nelson C, Driver J, Hackmann TJ. 2019. Use of a fluorescent analog of glucose (2-NBDG) to identify uncultured rumen bacteria that take up glucose. Appl Environ Microbiol 85:e03018-18. 10.1128/AEM.03018-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao JY, Diaz RK, Teixeira CRV, Hackmann TJ. 2016. Transport of a fluorescent analogue of glucose (2-NBDG) versus radiolabeled sugars by rumen bacteria and Escherichia coli. Biochemistry 55:2578–2589. 10.1021/acs.biochem.5b01286. [DOI] [PubMed] [Google Scholar]
- 35.Fowler ML, Ingram-Smith CJ, Smith KS. 2011. Direct detection of the acetate-forming activity of the enzyme acetate kinase. J Vis Exp 2011:3474. 10.3791/3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langella O, Valot B, Balliau T, Blein-Nicolas M, Bonhomme L, Zivy M. 2017. X!TandemPipeline: a tool to manage sequence redundancy for protein inference and phosphosite identification. J Proteome Res 16:494–503. 10.1021/acs.jproteome.6b00632. [DOI] [PubMed] [Google Scholar]
- 37.Craig R, Beavis RC. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466–1467. 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 38.Chen IA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, Huntemann M, Varghese N, White JR, Seshadri R, Smirnova T, Kirton E, Jungbluth SP, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2019. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47:D666–D677. 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karkalas J. 1985. An improved enzymic method for the determination of native and modified starch. J Sci Food Agric 36:1019–1027. 10.1002/jsfa.2740361018. [DOI] [Google Scholar]
- 40.Hackmann TJ, Diese LE, Firkins JL. 2013. Quantifying the responses of mixed rumen microbes to excess carbohydrate. Appl Environ Microbiol 79:3786–3795. 10.1128/AEM.00482-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haugaard N, Cutler J, Ruggieri MR. 1981. Use of N-ethylmaleimide to prevent interference by sulfhydryl reagents with the glucose oxidase assay for glucose. Anal Biochem 116:341–343. 10.1016/0003-2697(81)90368-7. [DOI] [PubMed] [Google Scholar]
- 42.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. 2018. HMMER web server: 2018 update. Nucleic Acids Res 46:W200–W204. 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bodenhofer U, Bonatesta E, Horejš-Kainrath C, Hochreiter S. 2015. msa: an R package for multiple sequence alignment. Bioinformatics 31:3997–3999. [DOI] [PubMed] [Google Scholar]
- 49.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Revell L. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 51.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 52.Schmidt M, Schonheit P. 2013. Acetate formation in the photoheterotrophic bacterium Chloroflexus aurantiacus involves an archaeal type ADP-forming acetyl-CoA synthetase isoenzyme I. FEMS Microbiol Lett 349:171–179. 10.1111/1574-6968.12312. [DOI] [PubMed] [Google Scholar]
- 53.James KL, Ríos-Hernández LA, Wofford NQ, Mouttaki H, Sieber JR, Sheik CS, Nguyen HH, Yang Y, Xie Y, Erde J, Rohlin L, Karr EA, Loo JA, Ogorzalek Loo RR, Hurst GB, Gunsalus RP, Szweda LI, McInerney MJ. 2016. Pyrophosphate-dependent ATP formation from acetyl coenzyme A in Syntrophus aciditrophicus, a new twist on ATP formation. mBio 7:e01208-16. 10.1128/mBio.01208-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buckel W. 2001. Unusual enzymes involved in five pathways of glutamate fermentation. Appl Microbiol Biotechnol 57:263–273. 10.1007/s002530100773. [DOI] [PubMed] [Google Scholar]
- 55.Buckel W, Barker HA. 1974. Two pathways of glutamate fermentation by anaerobic bacteria. J Bacteriol 117:1248–1260. 10.1128/JB.117.3.1248-1260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riviere L, van Weelden SW, Glass P, Vegh P, Coustou V, Biran M, van Hellemond JJ, Bringaud F, Tielens AG, Boshart M. 2004. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J Biol Chem 279:45337–45346. 10.1074/jbc.M407513200. [DOI] [PubMed] [Google Scholar]
- 57.Reeves RE, Warren LG, Susskind B, Lo HS. 1977. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J Biol Chem 252:726–731. 10.1016/S0021-9258(17)32778-3. [DOI] [PubMed] [Google Scholar]
- 58.Sanchez LB, Galperin MY, Muller M. 2000. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming). J Biol Chem 275:5794–5803. 10.1074/jbc.275.8.5794. [DOI] [PubMed] [Google Scholar]
- 59.Takasaki K, Shoun H, Yamaguchi M, Takeo K, Nakamura A, Hoshino T, Takaya N. 2004. Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. Role of acetyl CoA synthetase in anaerobic ATP synthesis. J Biol Chem 279:12414–12420. 10.1074/jbc.M313761200. [DOI] [PubMed] [Google Scholar]
- 60.Atteia A, van Lis R, Gelius-Dietrich G, Adrait A, Garin J, Joyard J, Rolland N, Martin W. 2006. Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J Biol Chem 281:9909–9918. 10.1074/jbc.M507862200. [DOI] [PubMed] [Google Scholar]
- 61.Kreuzberg K, Klock G, Grobheiser DJPP. 1987. Subcellular distribution of pyruvate‐degrading enzymes in Chlamydomonas reinhardtii studied by an improved protoplast fractionation procedure. Physiol Plant 69:481–488. 10.1111/j.1399-3054.1987.tb09229.x. [DOI] [Google Scholar]
- 62.Schäfer T, Schönheit P. 1991. Pyruvate metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus. Arch Microbiol 155:366–377. [Google Scholar]
- 63.Musfeldt M, Selig M, Schonheit P. 1999. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J Bacteriol 181:5885–5888. 10.1128/JB.181.18.5885-5888.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hilpert W, Schink B, Dimroth P. 1984. Life by a new decarboxylation-dependent energy conservation mechanism with Na+ as coupling ion. EMBO J 3:1665–1670. 10.1002/j.1460-2075.1984.tb02030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dijkhuizen L, Van der Werf B, Harder W. 1980. Metabolic regulation in Pseudomonas oxalaticus OX1. Diauxic growth on mixtures of oxalate and formate or acetate. Arch Microbiol 124–124:261–268. 10.1007/BF00427736. [DOI] [Google Scholar]
- 66.Rado TA, Hoch JA. 1973. Phosphotransacetylase from Bacillus subtilis: purification and physiological studies. Biochim Biophys Acta 321:114–125. 10.1016/0005-2744(73)90065-x. [DOI] [PubMed] [Google Scholar]
- 67.Lamprecht W, Trauschold I. 1974. Determination with hexokinase and glucose-6-phosphate dehydrogenase, p 2101–2111. In Bergmeyer HU (ed), Methods of enzymatic analysis, 2nd ed, vol 4. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 68.Eyer P, Worek F, Kiderlen D, Sinko G, Stuglin A, Simeon-Rudolf V, Reiner E. 2003. Molar absorption coefficients for the reduced Ellman reagent: reassessment. Anal Biochem 312:224–227. 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]
- 69.Lipmann F, Tuttle LC. 1945. A specific micromethod for the determination of acyl phosphates. J Biol Chem 159:21–28. 10.1016/S0021-9258(19)51298-4. [DOI] [Google Scholar]
- 70.Michal G, Bergmeyer HU. 1983. Coenzyme A, end-point method with phosphate acetyltransferase, p 165–169. In Bergmeyer HU, Bergmeyer J, Graßl M (ed), Methods of enzymatic analysis, vol 3. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 71.Decker K. 1974. Acetyl-coenzyme A: UV-spectrophotometric assay, p 1988–1993. In Bergmeyer HU (ed), Methods of enzymatic analysis, 2nd ed, vol 4. Verlag Chemie, Weinheim, Germany. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download AEM02959-20_Supp_1_seq12.xls, XLS file, 1.7 MB (1.7MB, xls)
Data Set S2. Download AEM02959-20_Supp_2_seq13.xls, XLS file, 0.08 MB (77.5KB, xls)
Data Set S3. Download AEM02959-20_Supp_3_seq14.xls, XLS file, 0.09 MB (91KB, xls)
Data Set S4. Download AEM02959-20_Supp_4_seq15.xls, XLS file, 0.03 MB (31.5KB, xls)
Data Set S5. Download AEM02959-20_Supp_5_seq16.xls, XLS file, 0.03 MB (33KB, xls)
Table S1, Fig. S1 and S2. Download AEM02959-20_Supp_6_seq17.pdf, PDF file, 1.2 MB (1.2MB, pdf)
Data Availability Statement
The LC-MS/MS data presented in this paper have been deposited in the Proteomics Identification (PRIDE) Archive with the project accession PXD021231. The pHMMs presented in this paper have been deposited at figshare (https://figshare.com/articles/dataset/Profile_hidden_Markov_models_for_succinyl-CoA_acetate_CoA-transferase_and_other_enzymes/14442749).