Abstract
Purpose
To develop a deep transfer learning method that incorporates four-dimensional (4D) information in dynamic contrast-enhanced (DCE) MRI to classify benign and malignant breast lesions.
Materials and Methods
The retrospective dataset is composed of 1990 distinct lesions (1494 malignant and 496 benign) from 1979 women (mean age, 47 years ± 10). Lesions were split into a training and validation set of 1455 lesions (acquired in 2015–2016) and an independent test set of 535 lesions (acquired in 2017). Features were extracted from a convolutional neural network (CNN), and lesions were classified as benign or malignant using support vector machines. Volumetric information was collapsed into two dimensions by taking the maximum intensity projection (MIP) at the image level or feature level within the CNN architecture. Performances were evaluated using the area under the receiver operating characteristic curve (AUC) as the figure of merit and were compared using the DeLong test.
Results
The image MIP and feature MIP methods yielded AUCs of 0.91 (95% CI: 0.87, 0.94) and 0.93 (95% CI: 0.91, 0.96), respectively, for the independent test set. The feature MIP method achieved higher performance than the image MIP method (∆AUC 95% CI: 0.003, 0.051; P = .03).
Conclusion
Incorporating 4D information in DCE MRI by MIP of features in deep transfer learning demonstrated superior classification performance compared with using MIP images as input in the task of distinguishing between benign and malignant breast lesions.
Keywords: Breast, Computer Aided Diagnosis (CAD), Convolutional Neural Network (CNN), MR-Dynamic Contrast Enhanced, Supervised learning, Support vector machines (SVM), Transfer learning, Volume Analysis
© RSNA, 2021
Summary
A deep transfer learning method was developed to use four-dimensional information in dynamic contrast-enhanced MRI by taking the maximum intensity projection of features obtained from convolutional neural networks in the task of distinguishing between benign and malignant breast lesions.
Key Points
■ Reducing volumetric information in dynamic contrast-enhanced MRI to two dimensions by taking the maximum intensity projections (MIPs) of images (image MIP) or of features within the deep neural network architecture (feature MIP) achieved high performance in distinguishing between benign and malignant breast lesions.
■ The feature MIP method demonstrated higher classification performance than the image MIP method (areas under the receiver operating characteristic curve, 0.93 vs 0.91; P = .03).
Introduction
MRI has been established for use in the diagnosis of breast cancer, screening patients at high risk, cancer staging, and monitoring the cancer response to therapies (1,2). In comparison with other commonly used clinical modalities for breast cancer assessment, such as mammography and US, MRI offers much higher sensitivity (3,4). Dynamic contrast-enhanced (DCE) MRI provides high-spatial-resolution volumetric lesion visualization as well as morphologic and functional information by using temporal contrast-enhancement patterns, information that carries substantial clinical value for breast cancer management.
Computer-aided diagnostic systems continue to be developed to assist radiologists in the interpretation of diagnostic images and potentially improve the accuracy and efficiency of breast cancer diagnosis (5). Deep learning methods have demonstrated success in computer-aided detection and diagnostic and prognostic performance based on medical scans (6–10). Training deep neural networks from scratch typically relies on massive datasets for training and is thus often intractable for medical research because of data scarcity. It has been shown that standard transfer learning techniques such as fine-tuning or feature extraction based on ImageNet-trained convolutional neural networks (CNNs) can be used for computer-aided diagnosis (11–13). However, these pretrained CNNs require two-dimensional (2D) inputs, as shown in prior works on breast lesion classification with MRI (14–16), limiting the ability to use three-dimensional (3D; volumetric) or four-dimensional (4D; volumetric and temporal) image information that can contribute to lesion classification.
To take advantage of the rich 4D information inherent in DCE MRI without sacrificing the efficiency provided by transfer learning, a previously proposed transfer learning method, which was shown to outperform methods using only 2D or 3D information, used the maximum intensity projection (MIP) of second postcontrast subtraction images to classify breast lesions as benign or malignant (17). This method has since been adopted by others (18,19). In this study, we propose a transfer learning method that makes use of both volumetric and temporal information in DCE MRI more effectively than image MIP. Instead of collapsing the volumetric information at the image level to form MIP images, we do so at the feature level by taking the maximum of CNN features along the axial dimension for a given lesion directly within the deep neural network architecture, referred to here as feature MIP. Additionally, instead of one postcontrast subtraction image, we incorporate richer temporal information by using four dynamic time points in a DCE sequence in the form of subtraction images in the three channels of the CNN input. Because feature MIP may more effectively leverage the volumetric information in DCE MRI, we hypothesized that the deep learning with feature MIP would achieve higher classification performance than deep learning with image MIP in the task of distinguishing between benign and malignant breast lesions.
Materials and Methods
Database
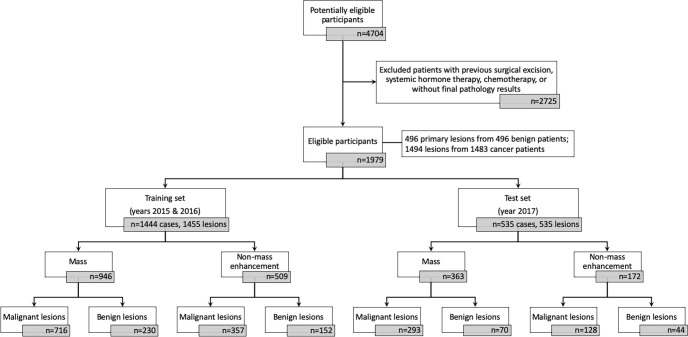
The breast DCE MRI dataset used in this study was retrospectively collected and de-identified prior to analysis, and the study was thus deemed exempt by the institutional review board–approved protocol. The patient population involved in this database has been reported in two previous publications (16,20). The database was acquired consecutively between 2015 and 2017, which initially involved images from 4704 patients who presented for breast DCE MRI examinations. Exclusion criteria included patients with previous surgical excision, systemic hormone therapy, or chemotherapy; examination results that did not exhibit a visible lesion; and lesions without final pathologic results. A total of 1990 distinct lesions from 1979 patients were ultimately included in our study. There were 1494 (75%) malignant lesions from 1483 patients with cancer, including eight bilateral and three bifocal cancers and 496 (25%) benign lesions from 496 patients. The ground truth for each lesion was based on histopathologic findings from surgical specimens. Figure 1 is the flowchart of patients included.
Figure 1:
Flowchart of study participant enrollment.
To minimize the bias in case selection for the computerized image analysis and to mimic a development-then-clinical-use scenario, the dataset was divided into a training and validation dataset as well as into an independent test set solely on the basis of the date of the MRI examinations. The training and validation dataset included 1455 lesions from the years 2015 and 2016, and the test set included 535 lesions from the year 2017. No patient studies were included in both the training and validation set and the test set, and there was one lesion per patient study in the test set.
Image Acquisition
MR images were acquired with 3-T GE scanners using a dedicated eight-channel phased-array breast coil (Discovery 750, GE Medical Systems) with a T1-weighted gradient-spoiled sequence. Sagittal DCE MR images were obtained with the volume imaging for breast assessment (VIBRANT) bilateral breast imaging technique, using typical parameters: repetition time, 3.9 msec; echo time, 1.7 msec; flip angle, 9°; matrix size, 320 × 192; field of view, 22–34 cm; and section thickness, 1.8 mm. The temporal resolution for each dynamic acquisition was 90 seconds. The contrast agent, gadolinium-diethylenetriamine penta-acetic acid (0.1 mmol/kg of body weight, flow rate of 2.0 mL/sec) was injected after the serial mask images were obtained, which was followed by flushing with the same total dose of saline solution.
CNN Input
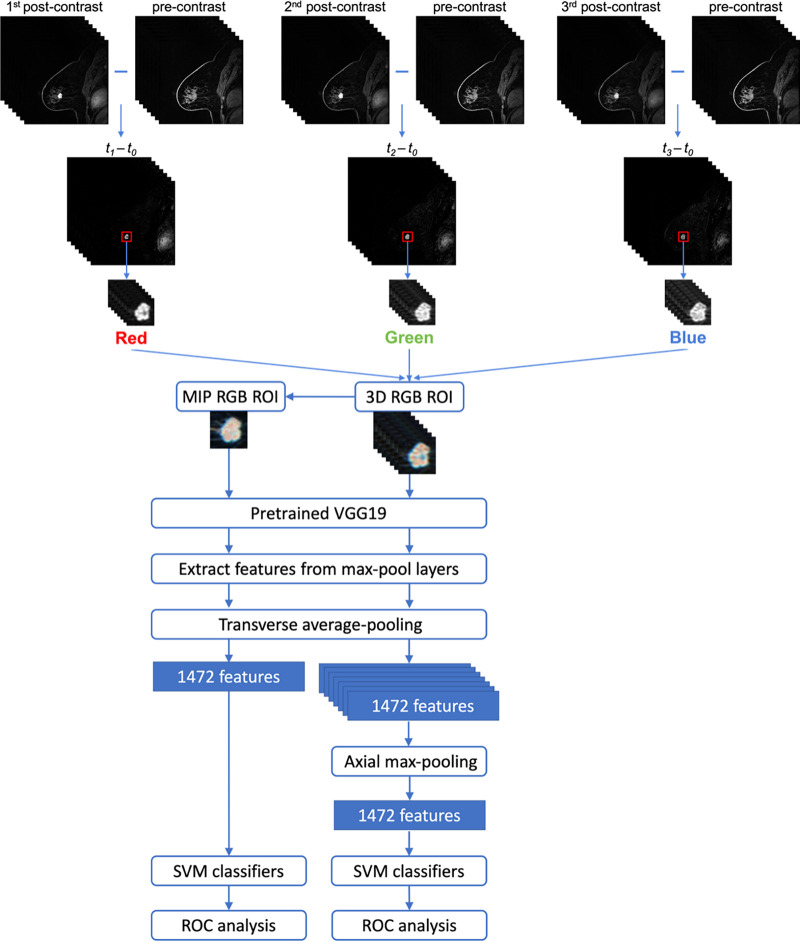
The procedure for creating the input for the CNN architecture from the 4D DCE MRI sequence is illustrated in Figure 2. Subtraction images were created by subtracting the precontrast (t0) images from their corresponding first, second, and third postcontrast images (t1, t2, and t3, respectively) to emphasize the contrast-enhancement pattern within the lesion and suppress the constant background.
Figure 2:
Lesion classification pipelines for image maximum intensity projection (MIP) and feature MIP. The top portion illustrates the construction of the region of interest (ROI) that incorporates volumetric and temporal information from the four-dimensional dynamic contrast-enhanced MRI sequence. The same ROI was cropped from the first, second, and third postcontrast subtraction images and combined in the red, green, and blue (RGB) channels to form a three-dimensional (3D) RGB ROI. For image MIP (left branch of the bottom portion), the MIP RGB ROI was generated from the 3D RGB ROI, collapsing volumetric lesion information at the image level. For feature MIP (right branch of the bottom portion), volumetric lesion information was integrated at the feature level by max-pooling the features extracted from all sections. SVM = support vector machine, VGG = Visual Geometry Group model.
To avoid confounding contributions from distant voxels, a region of interest (ROI) around each lesion was automatically cropped from all of its subtraction images with a seed point manually indicated by a breast radiologist (Y.J.) with 5 years of experience in breast DCE MRI. The ROI size was chosen on the basis of the maximum dimension of each lesion, adding a margin of 3 pixels around the lesion to include the parenchyma. The minimum ROI size in the transverse plane was set to 32 × 32 pixels as required by the pretrained CNN architecture. Note that there was only one ROI per lesion. ROIs were not rescaled because our feature extraction method allows for variation in the input size.
For the feature MIP method (described in the next section), the 3D ROIs from the three subtraction image volumes (ie, the first, second, and third postcontrast subtraction 3D ROIs) were input into the network through the red, green, and blue (RGB) channels, respectively, forming a 3D RGB ROI for each lesion. The pixel intensity in each ROI was normalized over the 3D ROI volume. For the image MIP method, the 3D RGB ROI volume for each lesion was collapsed into a 2D MIP ROI by selecting the voxel with the maximum intensity along the axial dimension (ie, perpendicular to the transverse sections).
Classification
Figure 2 also shows a schematic of the transfer learning classification and evaluation process for the two methods. For each lesion, CNN features were extracted from the inputted MIP RGB ROIs and the 3D RGB ROIs separately using a VGG19 (Visual Geometry Group; Oxford University) model pretrained on ImageNet (21,22). Note that these RGB ROIs comply with the desired three-channel input format of VGG19. Feature vectors were extracted at various network depths from the five max pooling layers of the VGGNet. These features were then average pooled along the spatial dimensions, and each resulting feature vector was individually normalized with the Euclidean distance. The pooled features were then concatenated to form a final CNN feature vector of 1472 features for a given lesion and normalized again across all features (13,14).
For our proposed feature MIP method using the 3D RGB ROIs, the 2D feature vectors extracted by VGGNet from each section were further concatenated to form a 3D feature vector, which was subsequently collapsed into a 2D feature vector by selecting the maximum feature value along the axial dimension (ie, taking the MIP of the feature vector along the direction in which sections were stacked; hence the name feature MIP). Max pooling was chosen over average pooling along the axial dimension because it was desirable to select the most prominent occurrence of each feature among all transverse sections of a lesion. Average pooling would have smoothed out the feature map and obscured the predictive features.
Linear support vector machine classifiers were trained on the CNN features extracted from the MIP RGB ROIs and the 3D RGB ROIs separately to differentiate between benign and malignant lesions (Python version 3.7.3, Python Software Foundation). The support vector machine method was chosen over other classification methods because of its ability to handle sparse, high-dimensional data, which is an attribute of the CNN features (23). Compared with support vector machines with nonlinear kernels, linear support vector machines require optimization of one hyperparameter, controlling the trade-offs between misclassification errors and model complexity (24).
Cases from 2015 to 2016 (1455 lesions) were randomly split into 80% for training and 20% for validation under the constraint that lesions from the same patient were kept together in the same set to eliminate the impact of bias from data leakage. In addition, the class prevalence was held constant across the training and validation subsets. Cases from the year 2017 (535 lesions) were used for independent testing in the task of distinguishing malignant from benign lesions. The training set was standardized to zero mean and unit variance, and the validation set and test set were standardized using the statistics of the corresponding training set. Principal component analysis was fit on the training set and subsequently applied to both the validation set and the test set to reduce feature dimensionality (25). When training the classifiers, the class weight was set to be inversely proportional to the class prevalence in the training data to address the problem of class imbalance (ie, 75% cancer prevalence). The support vector machine regularization hyperparameter, C, was optimized using a grid search (24).
Statistical Analysis
Classifier performances were evaluated on the independent test set using receiver operating characteristic (ROC) analysis, with the area under the ROC curve (AUC) serving as the figure of merit (26,27) in the task of distinguishing between benign and malignant lesions. Standard errors and 95% CIs of the AUC values were calculated by bootstrapping (2000 bootstrap samples) (28). The AUC values of the image MIP and the feature MIP schemes were compared using the DeLong test (29). Sensitivity and specificity were also reported for each classifier. The optimal operating point for each classifier was determined, using the ROC curve of the training data, by finding the sensitivity and specificity pair that maximized the function sensitivity – m(1 − specificity), where m is the slope of the ROC curve at the optimal operating point given by
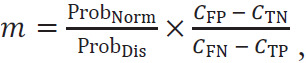 |
with ProbNorm and ProbDis being the probability that a case from the population studied is negative and positive for cancer, respectively, and CFP, CTN, CFN, and CTP being the cost of a false-positive, true-negative, false-negative, and true-positive result, respectively (30,31). We assumed an equal cost for false-positive and false-negative predictions and no cost for the correct predictions. The predicted posterior probabilities of malignancy of the test set were converted to match the cancer prevalence in the training set (32), and the sensitivity and specificity of the test set were reported using the optimal thresholds predetermined on the training data. The two classifiers’ sensitivities and specificities were each compared at the optimal point using the McNemar test (33,34). A P value less than .5 was considered to indicate a statistically significant difference in each performance metric. Statistical analyses were performed in MATLAB (MATLAB R2019b, The MathWorks).
Results
Patient Characteristics
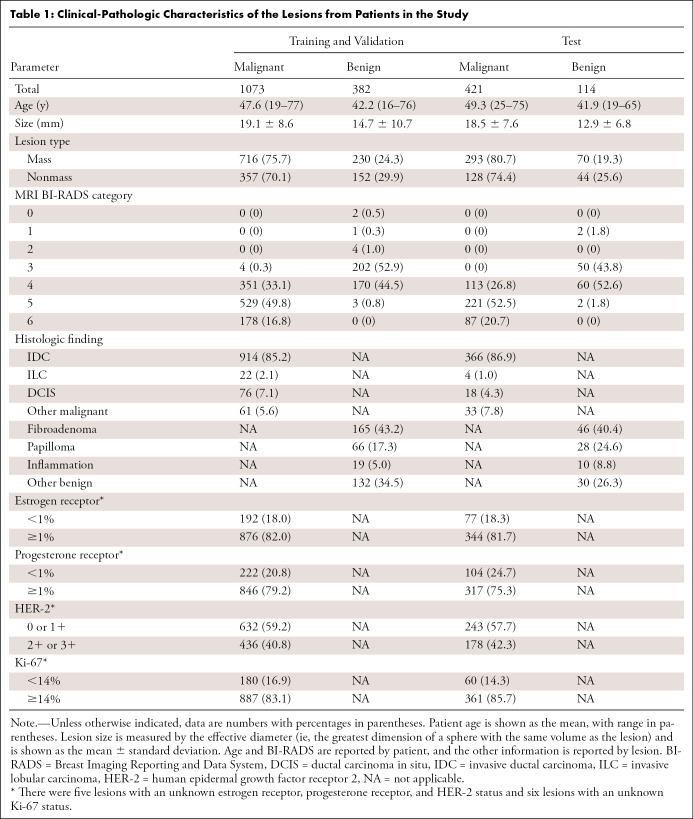
The clinical characteristics of the study population are listed in Table 1. Lesion characteristics were found to be similar in the training and validation data compared with the test data for lesion size for benign (P = .29) and malignant (P = .09) lesions. Similar distributions were noted in other subcategories as well.
Table 1:
Clinical-Pathologic Characteristics of the Lesions from Patients in the Study
Classification Performance
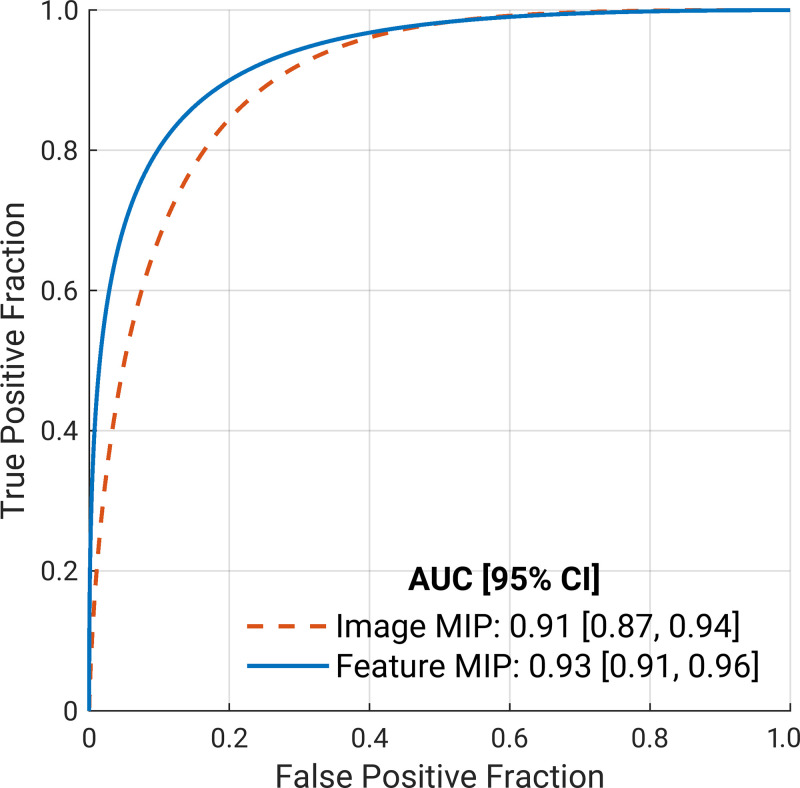
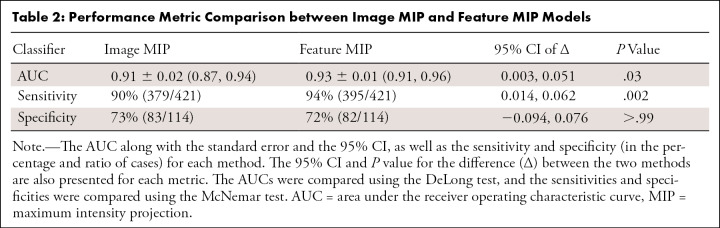
Figure 3 presents the ROC curves of the image MIP (AUC, 0.91; 95% CI: 0.87, 0.94) and the feature MIP (AUC, 0.93; 95% CI: 0.91, 0.96) approaches, and Table 2 summarizes the classifiers’ performance metrics in the task of distinguishing between benign and malignant breast lesions. A DeLong test comparing the feature MIP method with the image MIP method demonstrated that the feature MIP method achieved a higher classification performance (∆AUC 95% CI: 0.003, 0.051; P = .03). These results suggest that collapsing 3D volumetric information by taking the MIP at the feature level retained higher predictive power than collapsing at the image level. McNemar test results showed that, at the operating point determined using the training set, the sensitivity of using the feature MIP method on the test set was significantly higher than that of the image MIP method, and the specificities failed to demonstrate a significant difference.
Figure 3:
Fitted binormal receiver operating characteristic (ROC) curves for two classifiers that use the four-dimensional volumetric and temporal information from dynamic contrast-enhanced MRI. The dashed orange line represents the image maximum intensity projection (MIP) method, in which the volumetric information is collapsed into two dimensions at the image level. The solid blue line represents the feature MIP method, in which the volumetric information is collapsed at the feature level within the network architecture. The legend gives the area under the ROC curve (AUC) with the 95% CI for each classifier.
Table 2:
Performance Metric Comparison between Image MIP and Feature MIP Models
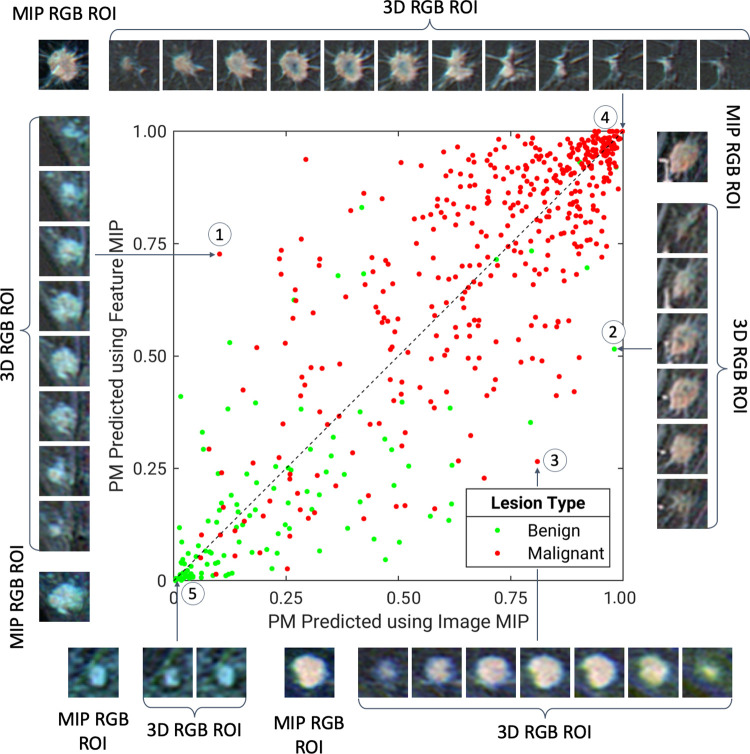
Figures 4 and 5 illustrate the comparison between the probabilities of malignancy predicted using the image MIP method and the feature MIP method. Although the majority of benign and malignant lesions were separated from the other class by both image MIP and feature MIP, these two methods exhibit moderate disagreement between these figures. Overall, the feature MIP method assigned malignant cases with higher probabilities of malignancy and benign cases with lower probabilities of malignancy as compared with image MIP, indicating that the feature MIP classifier has higher discriminatory power than image MIP in distinguishing between benign and malignant lesions. Figure 4 also shows several example lesions for which one method generated more accurate predictions than the other or for which the two methods agreed. The lesions for which feature MIP prediction was more accurate than image MIP prediction, the MIP images either failed to retain important features of the lesions or captured misleading features that did not accurately represent the lesion volumes in the projection process.
Figure 4:
A diagonal classifier agreement plot between the image maximum intensity projection (MIP) and feature MIP methods. The x-axis and y-axis denote the probability of malignancy (PM) scores predicted by the image MIP classifier and feature MIP classifier, respectively. Each point represents a lesion for which predictions were made. Points along or near the diagonal from bottom left to top right indicate high classifier agreement; points far from the diagonal indicate low agreement. The insets are the MIP regions of interest (ROIs) and three-dimensional (3D) ROIs, which served as convolutional neural network inputs for the image MIP and feature MIP methods, respectively, of extreme examples for which using feature MIP resulted in more accurate predictions than using image MIP (lesions 1–2), for which using image MIP resulted in more accurate predictions than using feature MIP (lesion 3), and for which the two methods both predict accurately (lesions 4–5). Lesion 1 is an invasive micropapillary carcinoma, lesion 2 is fibromatosis, lesion 3 is a grade II invasive ductal carcinoma, lesion 4 is a grade II invasive ductal carcinoma, and lesion 5 is a nonmass enhancement fibroadenoma. RGB = red, green, and blue.
Figure 5:
Bland-Altman plot for the image maximum intensity projection (MIP) and feature MIP classifiers. The x-axis and y-axis show the mean and difference between the support vector machine output scores (ie, predicted posterior probabilities of malignancy [PMs]) of the two classifiers, respectively. SD = standard deviation.
Discussion
Our study proposed a method to effectively incorporate the 4D volumetric and temporal information inherent in DCE MRI using deep transfer learning. Our method, which uses four DCE time points in the RGB channels of a CNN in the form of subtraction images and takes the MIP of features extracted by the CNN, achieved high performance in the task of distinguishing between benign and malignant breast lesions. Compared with the method of using the MIP of subtraction images, which has been shown in a prior study to outperform the method using only 3D or 2D information (17), our feature MIP method yielded significantly better performance in the breast lesion classification task.
High dimensionality and data scarcity are distinctive challenges in deep learning applications to medical imaging. To exploit the rich clinical information inherent in medical images without sacrificing computational efficiency or model performance, it is important to devise approaches to use transfer learning in creative ways so that volumetric and temporal data can be incorporated even when networks pretrained on 2D images are used. It is worth noting that our finding for the preferable usage of volumetric information in deep learning is relatable to that of human readers. Given the anatomic complexity in the breast parenchyma, the anatomic clutter caused by projecting a 3D volume onto a 2D image is a limiting factor for human readers’ assessment (35–37). Similarly, although conventional clinical MIP images are a convenient way of reducing the dimensionality of DCE MRI when interpreted by radiologists or artificial intelligence algorithms, conventional MIP images are not optimal because of the loss of information and the enhanced anatomic noise in projection images.
In a preliminary study, the feature MIP method showed performance superior to that of image MIP for another dataset (38). However, the work presented here is substantially different in the following ways. First of all, the dataset used in this preliminary study was acquired at an institution in a different country, resulting in major differences in both the image acquisition protocol and the patient population from the dataset in the current study. Thus, findings from the preliminary study cannot be naively generalized to the dataset involved in this study. Moreover, the larger size of the dataset in this study allowed us to perform a more rigorous evaluation through independent training, validation, and testing, rather than through cross-validation as in the previous work. This work also improved the deep learning input by incorporating four DCE time points, instead of only two or three as in the preliminary study.
Another prior study from our group was based on the same dataset and training, validation, and test split as our current study but used a single representative section for each lesion and input the precontrast, first postcontrast, and second postcontrast images’ ROIs into the RGB channels (16). The study reported an AUC of 0.85 using the same VGG19 feature extraction and support vector machine classification approach. Our study using the currently proposed feature MIP method outperformed the previous method by 10% (∆AUC 95% CI: 0.035, 0.120; P < .001).
Training 3D CNNs from scratch is another common approach for taking advantage of high-dimensional information provided by medical images. However, it is computationally expensive and is usually not suited to moderately sized medical datasets. A recent study by Dalmiş et al (18) trained a 3D CNN from scratch on 4D ultrafast DCE MRI data after reducing the dimensionality using MIPs and achieved an AUC of 0.81 (95% CI: 0.77, 0.85). Another study by Li et al (39) trained a 3D CNN on the volume of DCE MRI and incorporated the temporal information in the classification by calculating the enhancement ratio; they reported an AUC of 0.84. Compared with training 3D CNNs from scratch, our method of using transfer learning on 4D medical imaging data trains much fewer free parameters and is therefore computationally more efficient and has demonstrated high performance when used on our moderately sized datasets.
There were a few limitations of this study. First, the database was collected in a single country, and external data would need to be collected to assess the generalizability of our method. Moreover, although this study is focused on the computational aspect of improving the stand-alone performance of a computer-aided diagnostic algorithm, in the future, we plan to perform reader studies to assess the clinical significance of our system when used as a secondary or concurrent reader for radiologists. We also note that without sufficient knowledge about the specific clinical use case, the operating points at which sensitivity and specificity are reported may not be clinically optimal. A different threshold might be chosen if the relative cost of false-positive and false-negative diagnoses were known. In addition, there exist several variations of transfer learning strategies, including optionally adding layers on top of the pretrained network and a final fully connected layer for classification and then fine-tuning the network end to end. In our preliminary investigation, the specific transfer learning strategy employed in this study (namely, extracting features from multiple levels of a pretrained VGG19 network and performing classification using a support vector machine classifier) demonstrated better performance than end-to-end fine-tuning in the task of distinguishing benign and malignant breast lesions in our dataset.
In conclusion, this study proposes a deep transfer learning approach, referred to as feature MIP, that uses the 4D information in breast DCE MRI and demonstrates its superiority to other approaches through comprehensive statistical comparison. Future work will expand the analysis to include other valuable sequences, such as T2-weighted and diffusion-weighted MRI sequences in multiparametric MRI, rather than including DCE MRI alone. We will also expand the database to include images from different medical centers and populations to evaluate the robustness of our method across imaging manufacturers, facility protocols, and patient populations.
Acknowledgments
Acknowledgments
The authors are grateful for the contributions of Alexandra Edwards and John Papaioannou to this work.
Supported by the National Institutes of Health Quantitative Imaging Network grant U01CA195564, National Institutes of Health National Cancer Institute grant R15 CA227948, and National Institutes of Health grant S10 OD025081 (Shared Instrument Grant), and Radiological Society of North America/American Association of Physicists in Medicine Graduate Fellowship.
Disclosures of Conflicts of Interest: Q.H. Activities related to the present article: institution received grant from National Institutes of Health (NIH NCI U01 CA195564 NIH S10 OD025081 Shared Instrument Grant NIH NCI R15 CA227948). Activities not related to the present article: institution provides board membership for American Association of Physicists in Medicine (AAPM graduate fellowship). Other relationships: disclosed no relevant relationships. H.M.W. Activities related to the present article: institution received NIH NCI R15 grant. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. H.L. disclosed no relevant relationships. Y.J. disclosed no relevant relationships. P.L. disclosed no relevant relationships. M.L.G. Activities related to the present article: institution received grant from National Cancer Institute (NCI Quantitative Imaging Network grant). Activities not related to the present article: author sits on advisory boards for UIUC Biomedical Engineering, Texas A&M BME, and Benedictine University College of Science; scientific consultant for Qlarity Imaging; institution has various existing and pending grants/contracts with NIH that fund author’s lab research at University of Chicago; various patents over the years that are licensed by the University of Chicago; royalties from various patents over the years that are licensed by the University of Chicago; author has stock/stock options from licenses, scientific consultant to Qlarity, and various investments; author sits on NIH NIBIB Advisory Council. Other relationships: institution issued, licensed, received royalties for patents over past 30 years (licensed by the University of Chicago).
Abbreviations:
- AUC
- area under the ROC curve
- CNN
- convolutional neural network
- DCE
- dynamic contrast enhanced
- 4D
- four dimensional
- MIP
- maximum intensity projection
- RGB
- red
- ROC
- receiver operating characteristic
- ROI
- region of interest
- 3D
- three dimensional
- 2D
- two dimensional
References
- 1.Pickles MD, Lowry M, Manton DJ, Turnbull LW. Prognostic value of DCE-MRI in breast cancer patients undergoing neoadjuvant chemotherapy: a comparison with traditional survival indicators. Eur Radiol 2015;25(4):1097–1106. [DOI] [PubMed] [Google Scholar]
- 2.Turnbull LW. Dynamic contrast-enhanced MRI in the diagnosis and management of breast cancer. NMR Biomed 2009;22(1):28–39. [DOI] [PubMed] [Google Scholar]
- 3.Morrow M, Waters J, Morris E. MRI for breast cancer screening, diagnosis, and treatment. Lancet 2011;378(9805):1804–1811. [DOI] [PubMed] [Google Scholar]
- 4.Kuhl CK, Schrading S, Leutner CC, et al. Mammography, breast ultrasound, and magnetic resonance imaging for surveillance of women at high familial risk for breast cancer. J Clin Oncol 2005;23(33):8469–8476. [DOI] [PubMed] [Google Scholar]
- 5.Giger ML, Karssemeijer N, Schnabel JA. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annu Rev Biomed Eng 2013;15(1):327–357. [DOI] [PubMed] [Google Scholar]
- 6.Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional neural networks for medical image analysis: full training or fine tuning?. IEEE Trans Med Imaging 2016;35(5):1299–1312. [DOI] [PubMed] [Google Scholar]
- 7.Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35(5):1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenspan H, Van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans Med Imaging 2016;35(5):1153–1159. [Google Scholar]
- 9.Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin 2019;69(2):127–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheth D, Giger ML. Artificial intelligence in the interpretation of breast cancer on MRI. J Magn Reson Imaging 2020;51(5):1310–1324. [DOI] [PubMed] [Google Scholar]
- 11.Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? In: Ghahramani Z, Welling M, Cortes C, Lawrence N, Weinberger KG, eds.Proceedings of Advances in Neural Information Processing Systems 27.San Diego, Calif:Neural Information Processing Systems,2014;3320–3328. [Google Scholar]
- 12.Donahue J, Jia Y, Vinyals O, et al. DeCAF: a deep convolutional activation feature for generic visual recognition. In: Proceedings of the 31st International Conference on Machine Learning.Cambridge, Mass:ML Research Press,2014x,647–655. [Google Scholar]
- 13.Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging (Bellingham) 2016;3(3):034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017;44(10):5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herent P, Schmauch B, Jehanno P, et al. Detection and characterization of MRI breast lesions using deep learning. Diagn Interv Imaging 2019;100(4):219–225. [DOI] [PubMed] [Google Scholar]
- 16.Whitney HM, Li H, Ji Y, Liu P, Giger ML. Comparison of breast MRI tumor classification using human-engineered radiomics, transfer learning from deep convolutional neural networks, and fusion method. Proc IEEE 2020;108(1):163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antropova N, Abe H, Giger ML. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging (Bellingham) 2018;5(1):014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalmiş MU, Gubern-Mérida A, Vreemann S, et al. Artificial intelligence-based classification of breast lesions imaged with a multiparametric breast MRI protocol with ultrafast DCE-MRI, T2, and DWI. Invest Radiol 2019;54(6):325–332. [DOI] [PubMed] [Google Scholar]
- 19.Hu Q, Whitney HM, Giger ML. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci Rep 2020;10(1):10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji Y, Li H, Edwards AV, et al. Independent validation of machine learning in diagnosing breast Cancer on magnetic resonance imaging within a single institution. Cancer Imaging 2019;19(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. ArXiv 1409.1556 [preprint] https://arxiv.org/abs/1409.1556. Posted September 4, 2014. Accessed June 26, 2019. [Google Scholar]
- 22.Deng J, Dong W, Socher R, Li LJ, Li K, Li FF. ImageNet: a large-scale hierarchical image database. In: Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition.Piscataway, NJ:Institute of Electrical and Electronics Engineers,2009;248–255. [Google Scholar]
- 23.Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis 2015;115(3):211–252. [Google Scholar]
- 24.Shawe-Taylor J, Sun S. A review of optimization methodologies in support vector machines. Neurocomputing 2011;74(17):3609–3618. [Google Scholar]
- 25.Jolliffe IT. Principal component analysis. 2nd ed. New York, NY:Springer,2011. [Google Scholar]
- 26.Metz CE, Herman BA, Shen JH. Maximum likelihood estimation of receiver operating characteristic (ROC) curves from continuously-distributed data. Stat Med 1998;17(9):1033–1053. [DOI] [PubMed] [Google Scholar]
- 27.Metz CE, Pan X. "Proper" binormal ROC curves: theory and maximum-likelihood estimation. J Math Psychol 1999;43(1):1–33. [DOI] [PubMed] [Google Scholar]
- 28.Efron B. Better bootstrap confidence intervals. J Am Stat Assoc 1987;82(397):171–185. [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44(3):837–845. [PubMed] [Google Scholar]
- 30.Obuchowski NA. Receiver operating characteristic curves and their use in radiology. Radiology 2003;229(1):3–8. [DOI] [PubMed] [Google Scholar]
- 31.Metz CE. Basic principles of ROC analysis. Semin Nucl Med 1978;8(4):283–298. [DOI] [PubMed] [Google Scholar]
- 32.Horsch K, Giger ML, Metz CE. Prevalence scaling: applications to an intelligent workstation for the diagnosis of breast cancer. Acad Radiol 2008;15(11):1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947;12(2):153–157. [DOI] [PubMed] [Google Scholar]
- 34.Hawass NE. Comparing the sensitivities and specificities of two diagnostic procedures performed on the same group of patients. Br J Radiol 1997;70(832):360–366. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Abbey CK, Boone JM. Association between power law coefficients of the anatomical noise power spectrum and lesion detectability in breast imaging modalities. Phys Med Biol 2013;58(6):1663–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Abbey CK, Nosratieh A, Lindfors KK, Boone JM. Anatomical complexity in breast parenchyma and its implications for optimal breast imaging strategies. Med Phys 2012;39(3):1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrett JW, Li Y, Li K, Chen GH. Reduced anatomical clutter in digital breast tomosynthesis with statistical iterative reconstruction. Med Phys 2018;45(5):2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Q, Whitney HM, Giger ML. Transfer learning in 4D for breast cancer diagnosis using dynamic contrast-enhanced magnetic resonance imaging. ArXiv 1911.03022 [preprint] https://arxiv.org/abs/1911.03022. Posted November 8, 2019. Accessed February 3, 2020. [Google Scholar]
- 39.Li J, Fan M, Zhang J, Li L. Discriminating between benign and malignant breast tumors using 3D convolutional neural network in dynamic contrast enhanced-MR images. In: Cook TS, Zhang J, eds.Proceedings of SPIE: medical imaging 2017—imaging informatics for healthcare, research, and applications.Vol 10138.Bellingham, Wash:International Society for Optics and Photonics,2017;1013808. [Google Scholar]



![Bland-Altman plot for the image maximum intensity projection (MIP) and feature MIP classifiers. The x-axis and y-axis show the mean and difference between the support vector machine output scores (ie, predicted posterior probabilities of malignancy [PMs]) of the two classifiers, respectively. SD = standard deviation.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/adb0/8231792/afa764e3622f/ryai.2021200159.fig5.jpg)